Abstract
Mammalian cells respond to virus infections by eliciting both innate and adaptive immune responses. One of the most effective innate antiviral responses is the production of alpha/beta interferon and the subsequent induction of interferon-stimulated genes (ISGs), whose products collectively limit virus replication and spread. Following viral infection, interferon is produced in a biphasic fashion that involves a number of transcription factors, including the interferon regulatory factors (IRFs) 1, 3, 7, and 9. In addition, virus infection has been shown to directly induce ISGs in the absence of prior interferon production through the activation of IRF3. This process is believed to require virus replication and results in IRF3 hyperphosphorylation, nuclear localization, and proteasome-mediated degradation. Previously, we and others demonstrated that herpes simplex virus type 1 (HSV-1) induces ISGs and an antiviral response in fibroblasts in the absence of both interferon production and virus replication. In this report, we show that the entry of enveloped virus particles from diverse virus families elicits a similar innate response. This process requires IRF3, but not IRF1, IRF7, or IRF9. Following virus replication, the large DNA viruses HSV-1 and vaccinia virus effectively inhibit ISG mRNA accumulation, whereas the small RNA viruses Newcastle disease virus, Sendai virus, and vesicular stomatitis virus do not. In addition, we found that IRF3 hyperphosphorylation and degradation do not correlate with ISG and antiviral state induction but instead serve as a hallmark of productive virus replication, particularly following a high-multiplicity infection. Collectively, these data suggest that virus entry triggers an innate antiviral response mediated by IRF3 and that subsequent virus replication results in posttranslational modification of IRF3, such as hyperphosphorylation, depending on the nature of the incoming virus.
Viral infections of mammalian cells lead to an innate immune response characterized by the induction and secretion of alpha/beta interferon (IFN-α/β) and the subsequent transcriptional upregulation of IFN-stimulated genes (ISGs) (21, 59). Many of the known ISGs render cells resistant to viral infections, while others mediate antiproliferative or antiapoptotic responses and modulate the immune system. Virus-mediated IFN production occurs in a biphasic manner (52, 61) and involves several proteins from the IFN regulatory factor (IRF) family, a growing collection of transcription factors that play distinct roles in many biological processes (25, 38, 52, 61). In the early phase, constitutively produced IRF3, along with several other transcription factors, including IRF1, NF-κB, and ATF-2/c-Jun (31), weakly activates the IFN-β promoter. Secreted IFN-β binds to the IFN receptor on surrounding cells, initiating the Jak/Stat/IRF9 IFN signal transduction cascade (30, 33, 70) and resulting in the induction of IRF7. In the late phase, IRF3 and IRF7 collectively amplify IFN-α/β gene induction, leading to full ISG stimulation through the IFN signaling cascade.
It has become clear over the last several years that ISGs can also be activated by virus infection or double-stranded RNA (dsRNA), a by-product of virus infection, in the absence of IFN production (1, 63, 66-68, 72). For example, ISG56, an IFN-stimulated protein involved in translational regulation via binding to eukaryotic initiation factor 3 (23), has been shown to be upregulated by IFN, dsRNA, and viruses via independent pathways (24). Both viruses and dsRNA induce IRF3/CBP/p300 complexes which can directly bind to the IFN-stimulated response element (ISRE) in the promoter region of ISGs, implicating IRF3 in direct, IFN-independent induction of ISGs (35, 68, 69, 71).
The current model of virus-induced IRF3 and IRF7 activation consists of phosphorylation by a cellular kinase(s), a conformational change resulting in protein homo- or heterodimerization, nuclear translocation, and an association with the CREB-binding protein (CBP) and/or p300 coactivators (29, 35, 54). Virus infection activates the cellular DNA-dependent protein kinase DNA-PK, leading to the phosphorylation of IRF3 on Thr135 followed by IRF3 nuclear localization (27). However, the role of N-terminal IRF3 phosphorylation is unclear, as other studies have found that virus-mediated IRF3 dimerization, nuclear translocation, and cofactor binding require C-terminal phosphorylation (35, 36, 71). Recently, the cellular kinases TBK-1 and IKKɛ were found to phosphorylate IRF3 and IRF7 following infections with Sendai virus (18, 55). In particular, IKKɛ was found to phosphorylate IRF3 on Ser396, the minimal phosphoacceptor site required for virus-mediated IRF3 activation in vivo (53, 55).
Previously, we and others reported that herpes simplex virus type 1 (HSV-1), a large enveloped DNA virus, triggers a host cellular response that is characterized by the upregulation of a specific set of genes, most of which are also inducible by IFN, resulting in the activation of an antiviral state in an IFN-independent fashion (40, 45). This cellular response is mediated by HSV-1 virion particles and is inhibited by virus replication, suggesting that a newly synthesized viral protein(s) inhibits the response. Recently, the HSV-1 immediate-early protein ICP0 was shown to block ISG induction through a proteasome-dependent mechanism that remains to be determined (16). The related herpesvirus human cytomegalovirus (HCMV) induces expression of ISGs both in the presence and absence of viral gene expression, although significantly more ISGs are induced when viral gene expression is inhibited (10, 72, 73). With HSV-1, virus binding and penetration are essential for antiviral state induction (40, 48), whereas with HCMV, soluble glycoprotein B is sufficient to trigger the response (9, 56). The IFN-independent induction of ISGs by HSV-1 and HCMV can be dissociated from the IFN pathway and appears to involve IRF3 (9, 40, 43, 45, 48).
The focus of this paper is to elucidate the role of IRFs in the IFN-independent activation of ISGs following virus particle entry. While experimental evidence suggested that IRF3 plays a key role in this process, a number of discrepancies needed to be addressed. A recent study demonstrated that overexpression of a constitutively activated form of IRF3 is sufficient to modulate cellular gene expression, including the induction of a limited set of ISGs, in the absence of IFN production (22). However, in previous studies, IRF3-induced antiviral effects were thought to require the activity of IFN genes and a functional IFN pathway (11, 26, 47), whereas overexpression of IRF1 was found to lead to the IFN-independent activation of ISGs and resistance to virus infection (46). In addition, although IRF3 activation by Sendai virus and other related RNA viruses appears to require virus replication (57, 62), IRF3 binding activity following infection with the herpesviruses HSV-1 and HCMV is maximal when virus replication is inhibited (43, 48). Thus, the precise activation and function of IRFs, particularly IRF3, in the IFN-independent induction of ISGs in response to viral infection were unclear. In this study, we show that enveloped particles from both RNA and DNA virus families induce ISGs in the absence of IFN production and virus replication and that this event requires IRF3, but not IRF1, IRF7, or IRF9. Collectively, these data further our understanding of the initial events that occur following virus entry into a susceptible cell.
MATERIALS AND METHODS
Cells and viruses.
Human embryonic lung (HEL) fibroblasts (American Type Culture Collection [ATCC]) were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS). A549 cells (ATCC) and wild-type (IRF+/+), IRF1−/−, IRF3−/−, and IRF3−/− IRF9−/− mouse embryo fibroblasts (39, 51) (obtained from T. Taniguchi) were maintained in alpha minimal essential medium supplemented with 10% FBS. Vero cells (ATCC) were maintained in DMEM supplemented with 5% FBS. 293 cells (ATCC) were maintained in minimal essential medium-F11 supplemented with 10% FBS. Vesicular stomatitis virus (VSV; strain Indiana), vaccinia virus (VV; strain WR) and HSV-1 (strain KOS) were propagated on Vero cells, while adenovirus type 5 (Ad) and all Ad derivatives were propagated on 293 cells. AdE1E3 is an Ad derivative with deletions in early regions 1 and 3 (5). AdIRF3 and AdIRF7 contain wild-type human IRF3 and IRF7, respectively, under the control of a CMV immediate-early promoter in the AdE1E3 background (J. Bramson, McMaster University). Newcastle disease virus (NDV; strain La Sota) and Sendai virus (SV; strain Cantell) were kindly provided by E. Nagy and J. Hiscott, respectively. For viral infections, cells were split and seeded into dishes 24 h prior to infection. Infections occurred in serum-free DMEM for 1 h, followed by replacement with complete medium, with the exception of infection with adenoviruses, which occurred in phosphate-buffered saline (PBS) supplemented with 0.9 mM CaCl2 and 0.5 mM MgCl2 for 30 min. Infections with HSV-1, VV, Ad, and VSV utilized a multiplicity of infection of 10 PFU per cell, whereas infections with SV and NDV utilized 40 hemagglutination units (HAU) per 106 cells, unless otherwise specified. UV inactivation was performed with a UV Stratalinker 2400 instrument (Stratagene) for a period of time sufficient to reduce viral titers by a factor of 105 (data not shown). VSV plaque reduction assays were performed as previously described (40).
RT-PCR analysis.
Total cellular RNAs were harvested by using Trizol (Gibco BRL) according to the manufacturer's instructions. For reverse transcription (RT)-PCR, aliquots (2 μg) were reverse transcribed by use of 200 ng of random hexamer primer and Superscript II reverse transcriptase (Invitrogen). PCRs were subsequently performed per the manufacturer's specifications with the following primers: human ISG56 forward and reverse, 5′-ACGGCTGCCTAATTTACAGC-3′ and 5′-AGTGGCTGATATCTGGGTGC-3′, respectively; human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) forward and reverse, 5′-CGGAGTCAACGGATTTGGTCGTA-3′ and 5′-AGCCTTCTCCATGGTGGTGAAGAC-3′, respectively; murine ISG56 forward and reverse, 5′-ACAGCTACCACCTTTACAGC-3′ and 5′-TTAACGTCACAGAGGTGAGC-3′, respectively; murine IP-10 forward and reverse, 5′-TCATCCTGCTGGGTCTGAGT-3′ and 5′-CTGGGTAAAGGGGAGTGATG-3′, respectively; and murine actin forward and reverse, 5-GATGACGATATCGCTGCGCTG-3′ and 5′-GTACGACCAGAGGCATACAGG-3′, respectively.
Western blot and immunofluorescence analyses.
For Western blot analysis, cells were washed twice and then harvested in cold PBS, followed by centrifugation at 200 × g for 3 min at 4°C. Cell pellets were resuspended in whole-cell-extract buffer (20 mM HEPES [pH 7.4], 100 mM NaCl, 10 mM β-glycerophosphate, 0.2% Triton X-100, 50 mM NaF, 1 mM sodium orthovanadate, 1 mM phenylmethylsulfonyl fluoride, 2 mM dithiothreitol, and 1× protease inhibitor cocktail [Sigma]), lysed on ice for 15 min, and then centrifuged for 10 min at 13,000 × g at 4°C. Extract concentrations were determined by using a Bradford assay kit (Bio-Rad), and the indicated amounts of extracts were run on 9% polyacrylamide gels, transferred to nitrocellulose, and probed with either an anti-IRF3, anti-IRF1, anti-IRF7 (SC-9082, SC-13041, and SC-9083, respectively; Santa Cruz), anti-ISG56 (provided by G. Sen), anti-HSV-1 (DAKO), or anti-VSV-G (Roche) antibody at a dilution of 1:1,000. To ensure equal loading, all Western blots were reprobed with an anti-actin antibody (SC-1616; Santa Cruz). For immunofluorescence analysis, cells grown on coverslips were fixed for 2 min with methanol, followed by blocking with PBS containing 2% goat serum for 1 h at room temperature. Endogenous IRF3 was detected by using a 1:100 dilution of SC-9082 followed by a 1:800 dilution of a goat anti-rabbit secondary antibody conjugated to fluorescein isothiocyanate (Jackson Laboratory).
RESULTS
Virus particles from diverse families induce ISGs and an antiviral state in the absence of gene expression and interferon production.
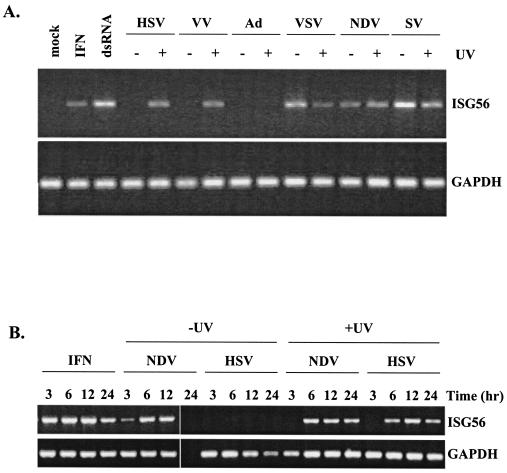
To further study the requirement of viral gene expression for ISG induction, we screened a number of different viruses for the ability to induce ISGs in the presence or absence of viral gene expression. Nonimmortalized, untransformed HEL fibroblasts were infected with HSV-1, VV, Ad, VSV, NDV, and SV (refer to Table 1 for virus characteristics). Viral gene expression was inhibited by UV inactivation. Induction of the ISG56 gene was used as a measure of ISG regulation, as we and others have found that ISG56 is reproducibly one of the most highly expressed genes following stimulation with IFN, dsRNA, or a virus infection (13, 20, 24, 40).
TABLE 1.
Viruses used in this study and their properties
| Virus | Family | Genome | Location of replication | Envelope |
|---|---|---|---|---|
| HSV-1 | Herpesvirus | dsDNA | Nucleus | Yes |
| VV | Poxvirus | dsDNA | Cytoplasm | Yes |
| Ad | Adenovirus | dsDNA | Nucleus | No |
| VSV | Rhabdovirus | ssRNA (−ve)a | Cytoplasm | Yes |
| NDV | Paramyxovirus | ssRNA (−ve) | Cytoplasm | Yes |
| SV | Paramyxovirus | ssRNA (−ve) | Cytoplasm | Yes |
-ve, negative strand.
As expected, ISG56 RNA accumulation in HEL fibroblasts was readily detected by RT-PCR at 6 h posttreatment with IFN-α and poly(I) · poly(C), a synthetic form of dsRNA (Fig. 1A). With the exception of Ad, all viruses efficiently accumulated ISG56 mRNA in the absence of virus replication (+UV). Following virus replication (−UV), however, ISG56 mRNA was only observed in cells infected with VSV, NDV, or SV. Similar results were found when RT-PCR analysis was performed using primers specific for IP-10 (see Fig. 3; also data not shown), another ISG originally identified by microarray analysis to be induced by virus particles in the absence of IFN (40). Where detected, the kinetics of ISG56 mRNA accumulation following viral infection were delayed compared to treatment with IFN (Fig. 1B). As expected, we failed to detect ISG56 mRNA following infection with replicating HSV-1 at any time point, and by 24 h postinfection, replicating NDV showed significantly lower total mRNA levels, as evidenced by the disappearance of both ISG56 and GAPDH mRNA species. Indeed, following 24 h of infection with wild-type NDV, complete cellular cytotoxicity was noted and the amount of total RNA harvested was drastically decreased (data not shown). Western blot analysis on whole-cell extracts prepared at 12 h postinfection (Fig. 1C) demonstrated the presence of ISG56 protein following all treatments for which ISG56 mRNA was detected, with one exception. Infection with replication-competent VSV yielded ISG56 mRNA but not protein. This observation is consistent with reports that the VSV matrix protein mediates the inhibition of mRNA nuclear export (64). To confirm that our UV inactivation was effectively preventing virus gene expression, we subjected the above extracts from HSV- and VSV-infected samples to Western blot analysis, using a polyclonal antibody directed against HSV-1 proteins and a monoclonal antibody specific for the VSV-G protein, respectively (Fig. 1D). In both cases, the level of UV inactivation chosen for these studies was sufficient to completely inhibit viral protein production. In addition, RT-PCR analysis was used to monitor viral transcript levels to ensure that UV inactivation prevented viral transcription (data not shown). Furthermore, following 48 h of infection with all UV-inactivated viruses, no cytopathic effects were observed in fibroblasts, whereas complete cellular cytotoxicity (cell rounding and detachment) was observed by 24 h postinfection with all replication-competent viruses, with the exception of Ad, with which complete toxicity was delayed several days (data not shown).
FIG. 1.
Induction of ISGs and a cellular antiviral state following virus infection does not require virus replication. HEL fibroblasts were left untreated (mock), treated with 1,000 U of human IFN-α per ml (IFN) or 100 μg of poly(I) · poly(C) per ml (dsRNA), or infected with wild-type or UV-inactivated virus. (A) Accumulation of ISG56 and GAPDH transcripts was determined by RT-PCR at 6 h posttreatment or postinfection. (B) Accumulation of ISG56 and GAPDH transcripts was determined by RT-PCR at different times posttreatment or postinfection. (C) Western blot analysis of 40 μg of whole-cell extract was performed at 12 h posttreatment or postinfection by use of a polyclonal ISG56 antibody. (D) Western blot analysis of the HSV and VSV samples from panel C with a pan-HSV-1 polyclonal antibody and a VSV-G monoclonal antibody to confirm the effectiveness of UV inactivation. *, location of a nonspecific band occurring with the pan-HSV-1 polyclonal antibody.
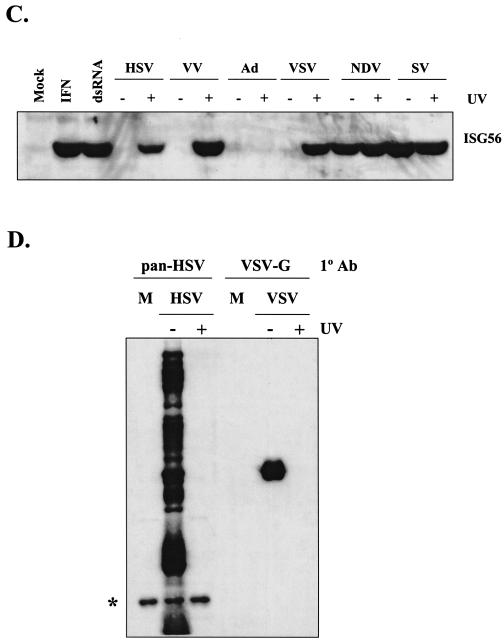
FIG. 3.
IRF3 is necessary to restore ISG induction in the absence of virus replication and IFN production. Wild-type (IRF+/+) MEFs were mock infected and IRF3−/− IRF9−/− MEFs (IRF3&9−/−) were infected with AdE1E3, AdIRF3, or AdIRF7 at a multiplicity of infection of 10. Twelve hours later, monolayers were treated with 100 U of universal IFN-α/β per ml or 100 μg of poly(I) · poly(C) per ml (dsRNA) or were infected with UV-inactivated HSV-1, NDV, or VSV. RT-PCR analysis was performed 6 h later to assess levels of ISG56, IP-10, and actin transcripts.
When UV-inactivated virus particles were tested in a virus plaque reduction assay, all viruses tested, with the exception of Ad, induced a complete antiviral state in fibroblasts that prevented the subsequent replication of wild-type virus (Table 2). To test for the secretion of a biologically active compound such as IFN that would account for the antiviral state induction, supernatants from infected fibroblasts were transferred to naïve Vero cell monolayers that were subsequently tested in a VSV plaque reduction assay (Table 2). Vero cells were chosen because they fail to produce IFN due to a genetic lesion in the IFN locus (15, 17) yet are able to respond to IFN and induce an antiviral state that is capable of decreasing the replication efficiency of VSV by 5 to 6 logs (41, 66). Whereas supernatants from HEL fibroblasts treated with IFN and dsRNA significantly decreased the plaquing efficiency of wild-type VSV on Vero cells, supernatants from HEL fibroblasts treated with UV-inactivated viruses did not. To ensure that the virus inoculum itself did not elicit an antiviral response, either banded virus was used or mock lysates from cells in which the virus was propagated were tested. As previously reported (40), the virus inoculum itself did not play a role in antiviral state induction (data not shown).
TABLE 2.
IFN-independent induction of an antiviral state in response to virus particles
| Treatment | No. of plaques on indicated cells
|
|||
|---|---|---|---|---|
| HEL fibroblastsa
|
Vero cellsb
|
|||
| Expt 1 | Expt 2 | Expt 1 | Expt 2 | |
| Mock | 116 | 80 | 137 | 146 |
| IFN | 0 | 0 | 2 | 12 |
| dsRNA | 0 | 0 | 6 | 11 |
| UV-inactivated HSV | 0 | 0 | 139 | 149 |
| UV-inactivated VV | 0 | 0 | 152 | 129 |
| UV-inactivated VSV | 0 | 0 | 156 | 123 |
| UV-inactivated NDV | 0 | 0 | 155 | 127 |
| UV-inactivated Ad | 125 | 78 | 148 | 138 |
Following the addition of wild-type VSV at 16 h posttreatment.
Following the addition of wild-type VSV 6 h after the transfer of HEL fibroblast supernatants to naïve Vero monolayers.
The innate cellular response to virus particles and dsRNA requires IRF3.
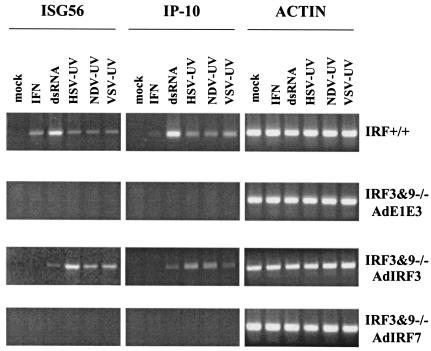
Initial studies of IFN gene regulation identified IRF1 as a positive regulator in response to viral infections (61). Subsequently, IRF3 was shown to play an important role in both IFN gene expression and ISG induction (25). Whereas overexpression of IRF1 leads to the IFN-independent activation of ISGs and resistance to virus infection (46), IRF3-induced antiviral effects were initially thought to require the activation of IFN genes (26). To assess the roles of IRF1 and IRF3 in the induction of the cellular antiviral response to either viral particles or dsRNA, we used primary mouse embryo fibroblasts (MEFs) derived from wild-type (IRF+/+), IRF1 null, and IRF3 null animals (39, 51). Wild type, IRF1−/−, and IRF3−/− MEFs were left untreated (mock), treated with universal IFN-α/β or poly(I) · poly(C) (dsRNA), or infected with UV-inactivated HSV-1 or Ad for 12 h, followed by a standard VSV plaque reduction assay. As shown in Fig. 2, wild-type, IRF1−/−, and IRF3−/− MEFs responded to IFN and induced an antiviral state in cells that was sufficient to prevent VSV plaque production. Wild-type and IRF1−/− MEFs also induced an antiviral state in response to dsRNA and UV-inactivated HSV-1, whereas MEFs lacking IRF3 failed to respond to either stimulus. Similar results were found with UV-inactivated VSV, VV, NDV, and SV (data not shown). When supernatants from treated IRF3−/− monolayers were applied to naïve murine fibroblasts, biologically active IFN was not detected, similar to the results outlined in Table 2 (data not shown). In accordance with the results outlined in Fig. 1, UV-inactivated Ad was unable to induce an antiviral response, regardless of the presence or absence of IRF1 or IRF3. This result was also observed when an Ad mutant with deletions in the E1 and E3 regions (AdE1E3) was used (data not shown).
FIG. 2.
IRF3 is essential for induction of an antiviral state in fibroblasts in response to dsRNA or enveloped virus particles. MEFs derived from wild-type (IRF+/+), IRF1 null (IRF1−/−), or IRF3 null (IRF3−/−) mice were left untreated (mock), treated with 100 U of universal IFN-α/β per ml or 100 μg of poly(I) · poly(C) per ml (dsRNA), or infected with UV-inactivated HSV-1 or Ad. A wild-type VSV virus plaque reduction assay was performed 12 h later.
To confirm that IRF3 is necessary to induce the IFN-independent antiviral state, we utilized a recombinant Ad expressing wild-type IRF3 (AdIRF3) in IRF3 and -9 null MEFs. IRF9 (also known as ISGF3γ and p48) is an essential component of the IFN signal transduction pathway. Thus, IRF3−/− IRF9−/− MEFs fail to respond to exogenous IFN and fail to induce both IFN-α/β and IRF7 following stimulation (51). We also utilized an AdIRF7 construct to assess the role of IRF7 in the cellular antiviral response. Wild-type MEFs and IRF3−/− IRF9−/− MEFs that were preinfected with AdE1E3 (parental control), AdIRF3, or AdIRF7 were mock treated, treated with IFN or dsRNA, or infected with UV-inactivated HSV, NDV, or VSV. Wild-type MEFs responded to all treatments by inducing ISG56 and IP-10 messages (Fig. 3) and an antiviral state (data not shown). Infection of IRF3−/− IRF9−/− MEFs with AdIRF3, but with neither the parental AdE1E3 nor AdIRF7, restored responsiveness to dsRNA and UV-inactivated virus particles. In all cases, the IRF3−/− IRF9−/− MEFs failed to respond to IFN due to the absence of IRF9. Thus, in the absence of virus gene expression, the host response to incoming virus particles in primary fibroblasts is dependent on IRF3 but does not require IRF1, IRF7, IRF9, or IFN production. These experiments also demonstrate that IRF3 is essential for IFN-independent, dsRNA-mediated ISG induction in fibroblasts.
Virus-mediated ISG production does not correlate with IRF3 hyperphosphorylation and subsequent degradation and is virus and cell type dependent.
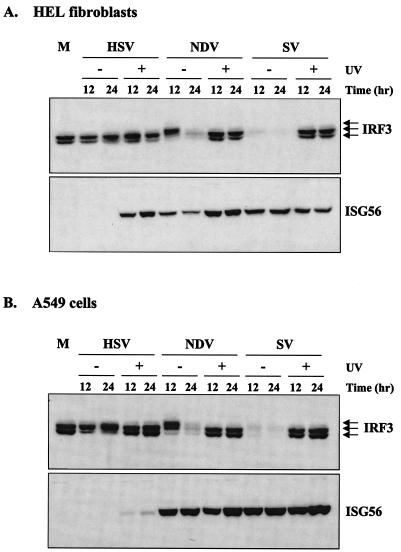
Since IRF3 appears to be essential for the cellular response to virus particles in primary mouse fibroblasts, we looked at IRF3 in nonimmortalized, untransformed human HEL fibroblasts. Wild-type NDV and SV induced the hyperphosphorylation and subsequent degradation of IRF3 (Fig. 4A), consistent with previous reports of proteasome-mediated degradation of IRF3 following its nuclear translocation and transactivation activities (35). By 12 h postinfection, IRF3 was barely detectable in SV-infected cells, although hyperphosphorylation was observable at earlier times (see Fig. 6). In contrast, wild-type HSV-1 failed to induce hyperphosphorylation and degradation of IRF3 and, consistent with the data shown in Fig. 1, also failed to induce ISG56 protein. Wild-type HSV-1 did, however, induce a shift from the smaller to the larger form of IRF3. In response to UV-inactivated HSV, NDV, or SV, IRF3 hyperphosphorylation and subsequent degradation were not observed, yet ISG56 protein was efficiently induced. Similar results were found when cells were treated with dsRNA, in that ISGs were induced in the absence of detectable IRF3 hyperphosphorylation and degradation (data not shown). Since the majority of the previous studies on IRF3 phosphorylation were performed in immortalized cells such as A549 and 293, we repeated the experiment in A549 human lung adenocarcinoma cells to determine whether the differences we observed were cell type specific (Fig. 4B). Similar results were observed in both cell types, with two exceptions. First, in A549 cells, UV-inactivated HSV-1 induced very low levels of ISG56 protein and, unlike UV-inactivated NDV or SV, failed to induce a complete antiviral state in these cells (data not shown). Second, the shift from the smaller to the larger form of IRF3 following infection with replicating HSV-1 occurred with faster kinetics and was more obvious in A549 cells than in HEL fibroblasts. The significance of these observations is currently under investigation.
FIG. 4.
ISG56 protein production does not require virus replication and does not correlate with IRF3 hyperphosphorylation. Untransformed HEL fibroblasts (A) or immortalized A549 cells (B) were left untreated (M) or infected with wild-type or UV-inactivated HSV, NDV, or SV. Whole-cell extracts were harvested at 12 or 24 h postinfection, and 40 μg of sample was run on an sodium dodecyl sulfate-9% polyacrylamide gel electrophoresis (SDS-9% PAGE) gel, followed by Western blot analysis. All blots were reprobed with an antibody against β-actin to ensure equal loading (data not shown).
FIG. 6.
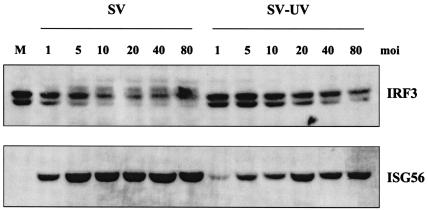
Low-multiplicity SV infection induces ISG56 in the absence of detectable IRF3 hyperphosphorylation. A549 cells were infected with replication-competent and UV-inactivated SV at the indicated multiplicities of infection (HAU per 106 cells). Whole-cell extracts were harvested at 8 h postinfection, and 40 μg of sample was run on an SDS-9% PAGE gel, followed by Western blot analysis using antibodies specific for IRF3 and ISG56.
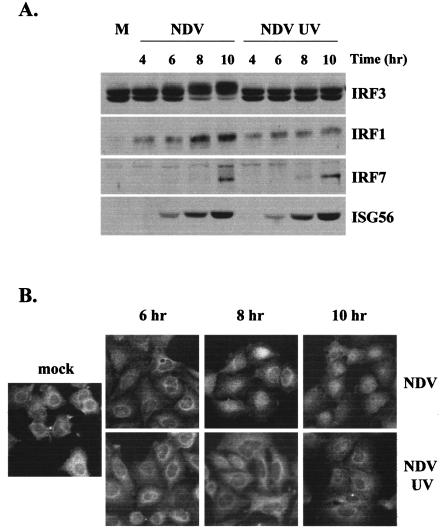
To further analyze the role of IRF3 hyperphosphorylation following virus infection, we performed a time course analysis with NDV in A549 cells. As shown in Fig. 5A, both virus particles and replicating viruses accumulated ISG56 protein with similar kinetics, while IRF3 hyperphosphorylation was only observed at later times after virus replication. Similar results were found with HEL fibroblasts (data not shown). While in MEFs we found that IRF1 and IRF7 were not required for ISG induction in response to virus particles, we reprobed the above blots for IRF1 and IRF7 to assess their possible involvement in human cells. IRF1 was barely detectable in untreated cells but was subsequently induced following replication-competent and, to a lesser extent, UV-inactivated virus infection. IRF7 was undetectable in untreated cells but accumulated following infection with wild-type and UV-inactivated virus at late times postinfection, notably after ISG56 protein accumulation. In a parallel time course study using immunofluorescence, infection with replicating NDV resulted in the nuclear accumulation of endogenous IRF3 in the majority of cells in the culture, whereas IRF3 nuclear translocation was difficult to detect in cultures infected with UV-inactivated NDV (Fig. 5B). Collectively, these data argue that IRF3 hyperphosphorylation and subsequent degradation do not correlate with ISG induction, but may instead serve as a hallmark of efficient virus replication, and that very low levels of IRF3 nuclear translocation are sufficient for ISG and antiviral state induction.
FIG. 5.
ISG induction does not correlate with IRF3 hyperphosphorylation or nuclear translocation. A549 cells were left untreated (M) or infected with wild-type or UV-inactivated NDV. (A) Whole-cell extracts were harvested at the indicated times postinfection, and 40 μg of sample was run on an SDS-9% PAGE gel, followed by Western blot analysis using antibodies specific for IRF3, IRF1, IRF7, or ISG56. (B) Immunofluorescence was performed with methanol-fixed A549 cells with an antibody specific for IRF3.
IRF3 hyperphosphorylation correlates with high levels of virus replication.
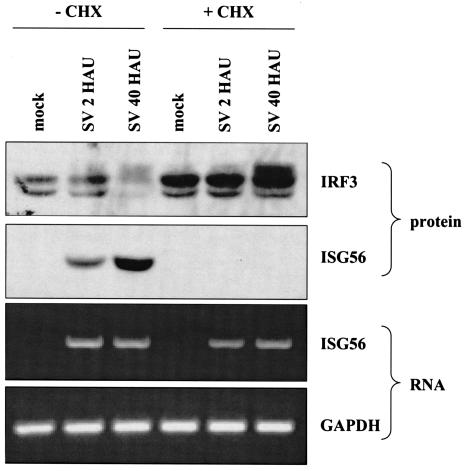
Given the difference in the modification of IRF3 in the presence or absence of virus replication, particularly with the paramyxovirus SV (Fig. 4), we sought to determine whether the multiplicity of infection of the virus played a significant role. In the majority of published studies on IRF3, paramyxoviruses are utilized at 40 to 80 HAU per 106 cells. For this study, A549 cells were infected for 8 h with either replicating or UV-inactivated SV at multiplicities of infection ranging from 1 to 80 HAU per 106 cells. While ISG56 protein accumulated in all samples, detectable IRF3 hyperphosphorylation was consistently observed with wild-type SV at 5 to 10 HAU per 106 cells (Fig. 6). In addition, as the multiplicity of infection increased, the ability to detect IRF3 nuclear translocation with replicating SV increased accordingly (data not shown). To confirm that replication-competent SV could induce ISG56 in an IFN-independent fashion, we repeated the experiment in the presence and absence of cycloheximide, using a high (40 HAU per 106 cells) and low (2 HAU per 106 cells) multiplicity of infection (Fig. 7). In the presence and absence of cycloheximide, hyperphosphorylation of IRF3 was only readily observed with a high multiplicity of infection, yet ISG56 mRNA was detected with high and low multiplicities of infection. Similar results were found with IP-10 (data not shown). Since cycloheximide inhibits protein synthesis, ISG56 protein was only observed in the absence of the drug. Interestingly, in the presence of cycloheximide, the levels of IRF3 were elevated, even following a high-multiplicity infection. This experiment confirms our previous results that the production of IFN, IRF1, and IRF7 is not required for ISG56 induction. Furthermore, ISG mRNA and protein are produced following low-multiplicity infections of replication-competent virus in the absence of detectable IRF3 hyperphosphorylation.
FIG. 7.
IRF3 is differentially modified following low- and high-multiplicity infections in the presence or absence of de novo protein synthesis. A549 cells were either mock infected or infected with SV at a low (2 HAU per 106 cells) or high (40 HAU per 106 cells) multiplicity of infection in the presence or absence of 100 μg of cycloheximide (CHX) per ml. Whole-cell extracts and total RNA were harvested at 8 h postinfection for use in Western blot and RT-PCR analyses, respectively.
DISCUSSION
This report documents a generalized innate response to virus particle entry. Viruses from diverse families, including paramyxoviruses, rhabdoviruses, poxviruses, and herpesviruses, were found to induce an antiviral response in cells in the absence of both IFN production and virus replication. This cellular response is characterized by the direct induction of a limited set of ISGs, including, but not limited to, ISG56, ISG54, ISG15, and IP-10 (40; also data not shown). Although it has been estimated that the human genome contains 600 to 2,000 ISGs (14), induction of a small subset of ISGs appears to be sufficient to induce an antiviral state capable of inhibiting the replication of wild-type virus. ISG56 inhibits protein synthesis by binding to the p48 subunit of eukaryotic initiation factor 3 (23), while the function of the related protein ISG54 remains to be determined. IP-10 is a C-X-C chemokine that is thought to mediate Th1-dominant immune responses (7, 44, 65), and ISG15 is a ubiquitin-like protein that amplifies and directs some of the immunomodulatory properties of IFN (12, 37).
The viruses used for this study are quite diverse, ranging from small, single-stranded RNA viruses to large, double-stranded DNA viruses. The sole common feature among the viruses that are capable of inducing ISGs and a cellular antiviral response in human and mouse fibroblasts is the presence of an envelope. In other cell types, including HeLa, U373 astrocytoma, and renal epithelial cells, adenoviruses lacking the E1 and/or E3 regions were found to induce ISGs (8, 32, 49). In fibroblasts, however, we failed to observe an induction of ISGs or an antiviral response with replicating, nonreplicating, or UV-inactivated adenoviruses. This discrepancy could be due to the nature of the cells used in this study. In general, when screening a variety of cell types with UV-inactivated enveloped viruses, we observed consistent induction of ISGs and antiviral responses in untransformed cells (fibroblast and epithelial) but inconsistent results in transformed or immortalized cells, as demonstrated with UV-inactivated HSV-1 in HEL fibroblasts versus A549 cells (Fig. 4). Since a wide array of enveloped virus particles induce a cellular antiviral response, it is tempting to speculate that fusion between cellular membranes and viral envelopes initiates the response. Indeed, lipid transfection reagents have been found to induce ISGs, although this phenomenon appears to occur via the production of IFN (34). How membrane fusion would initiate the antiviral cascade is unclear, as the different viruses used in this study enter cells through different mechanisms. For example, HSV-1 predominantly enters cells via direct, pH-independent fusion at the plasma membrane, whereas VSV enters via an indirect, pH-dependent endocytic mechanism (6, 58).
Recent studies have convincingly demonstrated that IRF1, IRF3, IRF7, and IRF9 are important cellular mediators of both IFN and ISGs in response to viral infections through binding to the positive regulatory domain and/or ISRE element within the promoter region of IFN and ISGs, respectively (25, 28, 43, 61, 68, 69). Although viral infections predominantly induce ISGs via the production of IFN, it is clear that ISGs can also be induced directly by dsRNA or by replicating virus in the absence of IFN, presumably via direct binding of IRFs to ISRE elements within ISG promoters (1, 24, 66, 67). In a recent study, the induction of a subset of ISGs following infection with replicating NDV was found to rely on the presence of IRF3 (42). In this report, we show that IRF3 is essential for the IFN-independent induction of ISGs and an antiviral response following the entry of enveloped virus particles in the absence of virus replication. Although IRF1 and IRF7 were induced following infection with UV-inactivated NDV, several lines of evidence indicate that these two transcription factors are not critical for the innate antiviral response. First, an antiviral response was observed following infection of UV-inactivated viruses in IRF1 null but not IRF3 null fibroblasts. Since the antiviral response occurred in the absence of IFN, IRF9 activity was not required and IRF7 was not induced. Second, in fibroblasts lacking both IRF3 and IRF9, reconstitution of IRF3 but not IRF7 restored the induction of ISGs and an antiviral state in response to UV-inactivated virus. Since these fibroblasts are deficient for IRF9 in addition to IRF3, they fail to respond to IFN and thus are also unable to produce IRF7 (51). This observation parallels the results of a study published by Nakaya et al. in which IRF7 was found to contribute little, if nothing, to ISG induction following infection with replicating virus (42). Third, in the presence of cycloheximide, a protein synthesis inhibitor that prevents the accumulation of both IRF1 and IRF7, ISG56 mRNA was readily observed following SV infection. Fourth, although IRF7 was produced in HEL fibroblasts infected with wild-type and UV-inactivated NDV, detectable levels were observed at several hours post-ISG induction. Finally, the spectrum of ISGs induced in untransformed human fibroblasts in response to UV-inactivated virus overlaps significantly with that of ISGs induced in Jurkat cells with a constitutively activated form of IRF3 in the absence of virus infection (22, 40). While these data suggest that IRF3, but not IRF1, IRF7, or IRF9, is critical for the initial response to virus particle entry, they do not discount the likelihood that these factors play a role at later times postentry, following virus replication.
Our data also indicate that hyperphosphorylation and subsequent degradation of IRF3, often used as readouts of IRF3 activation, do not correlate with ISG induction but rather serve as markers of robust RNA virus replication. It is likely that the IRF3 pathway has evolved to be very sensitive to low numbers of incoming virus particles, whereby only a small fraction of the endogenous IRF3 protein needs to be activated in order to elicit a biological response. We failed to observe hyperphosphorylation, nuclear localization, and subsequent degradation of endogenous IRF3, yet readily observed induction of ISGs, with either UV-inactivated virus or a low-multiplicity infection. Although some level of IRF3 modification and nuclear localization would presumably occur under these conditions, our methods of detection were insufficient to monitor these events. One possible outcome of our findings is that hyperphosphorylation may not be required for the biological activity of IRF3 but rather may be an anti-immune system defense strategy utilized by replicating RNA viruses to induce the degradation of IRF3. Several lines of evidence support this observation. First, infection with high, but not low, multiplicities of infection of the paramyxoviruses NDV and SV led to the disappearance of IRF3 following hyperphosphorylation and nuclear translocation. Similar results were observed with VSV (data not shown). Interestingly, these events were not observed following infection with the large DNA viruses HSV-1 and VV. However, the HSV-1 immediate-early protein ICP0 has been found to inhibit IRF3- and IRF7-mediated activation of ISGs (36a), while the VV E3L protein inhibits NDV-mediated hyperphosphorylation of both IRF3 and IRF7 (57), indicating that these viruses have alternative methods for counteracting the effects of this cellular pathway. Indeed, many viruses block IRF3, attesting to its importance in protecting cells from virus infections (2-4, 19, 26, 50, 60). Second, IRF3 hyperphosphorylation and detectable nuclear translocation occurred at late times postinfection, approximately 2 h after the accumulation of ISG56 protein was observed. Similar results were reported for a related paramyxovirus, measles virus, in A549 cells, in which IRF3 hyperphosphorylation and DNA binding were observed following virus replication at late times (16 to 20 h postinfection) (62).
The dose-response experiment in the presence or absence of cycloheximide raises an interesting question. This experiment demonstrated that de novo protein synthesis is not required for either IRF3 hyperphosphorylation or ISG induction, although the former is only detected following a high-multiplicity infection. At the same (and higher) multiplicities of infection, however, UV-inactivated virus failed to induce the hyperphosphorylation of IRF3. This observation suggests that a high level of transcription and/or dsRNA production might be important to mediate IRF3 hyperphosphorylation. Previous work has suggested that for small RNA viruses, de novo protein synthesis is required for IRF3 activation. In one study, cycloheximide was found to prevent the hyperphosphorylation of IRF3 and IRF7 in 293T cells (57). However, along with a difference in the cell type used, the previous study assayed transfected IRF3 and IRF7, whereas we monitored endogenous protein. Interestingly, one conclusion from the previous study was that the vaccinia virus E3L dsRNA-binding protein effectively inhibited IRF3 and IRF7 phosphorylation following SV infection, implying that dsRNA production plays a role in the modification of IRF3. In another study using A549 cells and measles virus, UV inactivation was found to prevent virus-mediated IRF3 hyperphosphorylation, thus leading to the conclusion that IRF3 activation requires viral replication (62). However, for this particular experiment, neither downstream ISG induction nor antiviral state induction was measured.
From this study, we are developing a model in which the entry of (enveloped) virus particles elicits a general, innate immune response that relies on the direct induction of a limited set of ISGs through the activation of IRF3. Following virus replication, a number of events may occur, depending on the particular virus, the multiplicity of infection, and the cell type. For example, large DNA viruses such as herpesviruses and poxviruses have the capacity to encode immune response modifiers that may block a specific portion of the IRF3 pathway, thus reducing or eliminating ISG induction. Other viruses, such as the paramyxoviruses, may eliminate endogenous IRF3 by inducing its hyperphosphorylation and subsequent degradation. Still other viruses, such as the rhabdoviruses, may prevent ISG protein production by blocking the export of ISG mRNAs from the nucleus. Within the context of this model, many outstanding questions remain. These include the mechanism(s) by which cellular kinases capable of phosphorylating IRF3 become activated, which step of the virus cycle and/or which viral component(s) induces their activation, what level of IRF3 phosphorylation (or other modification) is required for its biological activity, what percentage of endogenous IRF3 is required to become activated to induce a biologically relevant response, and what modifications of IRF3 occur as a result of high levels of virus replication. Identifying and characterizing the primary interactions between a virus and its host cell are essential to the understanding of virus pathogenesis and will enable the development of effective antiviral therapies.
Acknowledgments
We thank J. Hiscott, E. Nagy, T. Taniguchi, G. Sen, and J. Bramson for reagents, J. Smiley and J. Hummel for critical reading of the manuscript, and K. Laryea for providing technical assistance.
The Canadian Institutes of Health Research sponsored this work.
REFERENCES
- 1.Bandyopadhyay, S. K., G. T. Leonaard, T. Bandyopadhyay, G. R. Stark, and G. C. Sen. 1995. Transcriptional induction by double-stranded RNA is mediated by interferon-stimulated response elements without activation of interferon-stimulated gene factor 3. J. Biol. Chem. 270:19624-19629. [DOI] [PubMed] [Google Scholar]
- 2.Barnes, B., B. Lubyova, and P. M. Pitha. 2002. On the role of IRF in host defense. J. Interferon Cytokine Res. 22:59-71. [DOI] [PubMed] [Google Scholar]
- 3.Basler, C. F., A. Mikulasova, L. Martinez-Sobrido, J. Paragas, E. Muhlberger, M. Bray, H. D. Klenk, P. Palese, and A. Garcia-Sastre. 2003. The Ebola virus VP35 protein inhibits activation of interferon regulatory factor 3. J. Virol. 77:7945-7956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Basler, C. F., X. Wang, E. Muhlberger, V. Volchkov, J. Paragas, H. D. Klenk, A. Garcia-Sastre, and P. Palese. 2000. The Ebola virus VP35 protein functions as a type I IFN antagonist. Proc. Natl. Acad. Sci. USA 97:12289-12294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bett, A., W. Haddara, L. Prevec, and F. Graham. 1994. An efficient and flexible system for construction of adenovirus vectors with insertions or deletions in early regions 1 and 3. Proc. Natl. Acad. Sci. USA 91:8802-8806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bomsel, M., and A. Alfsen. 2003. Entry of viruses through the epithelial barrier: pathogenic trickery. Nat. Rev. Mol. Cell Biol. 4:57-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bonecchi, R., G. Bianchi, P. P. Bordignon, D. D'Ambrosio, R. Lang, A. Borsatti, S. Sozzani, P. Allavena, P. A. Gray, A. Mantovani, and F. Sinigaglia. 1998. Differential expression of chemokine receptors and chemotactic responsiveness of type 1 T helper cells (Th1s) and Th2s. J. Exp. Med. 187:129-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borgland, S. L., G. P. Bowen, N. C. W. Wong, T. A. Libermann, and D. A. Muruve. 2000. Adenovirus vector-induced expression of the C-X-C chemokine IP-10 is mediated through capsid-dependent activation of NF-κB. J. Virol. 74:3941-3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boyle, K. A., R. L. Pietropaolo, and T. Compton. 1999. Engagement of the cellular receptor for glycoprotein B of human cytomegalovirus activates the interferon-responsive pathway. Mol. Cell. Biol. 19:3607-3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Browne, E. P., B. Wing, D. Coleman, and T. Shenk. 2001. Altered cellular mRNA levels in human cytomegalovirus-infected fibroblasts: viral block to the accumulation of antiviral mRNAs. J. Virol. 75:12319-12330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Darnell, J. E., Jr., I. M. Kerr, and G. R. Stark. 1994. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science 264:1415-1421. [DOI] [PubMed] [Google Scholar]
- 12.D'Cunha, J., E. Knight, Jr., A. L. Haas, R. L. Truitt, and E. C. Borden. 1996. Immunoregulatory properties of ISG15, an interferon-induced cytokine. Proc. Natl. Acad. Sci. USA 93:211-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Der, S. D., A. Zhou, B. R. Williams, and R. H. Silverman. 1998. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc. Natl. Acad. Sci. USA 95:15623-15628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.deVeer, M. J., M. Holko, M. Frevel, E. Walker, S. Der, J. M. Paranjape, R. H. Silverman, and B. R. G. Williams. 2001. Functional classification of interferon-stimulated genes identified using microarrays. J. Leukoc. Biol. 69:912-920. [PubMed] [Google Scholar]
- 15.Diaz, M. O., S. Ziemin, M. M. Le Beau, P. Pitha, S. D. Smith, R. R. Chilcote, and J. D. Rowley. 1988. Homozygous deletion of the alpha- and beta 1-interferon genes in human leukemia and derived cell lines. Proc. Natl. Acad. Sci. USA 85:5259-5263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eidson, K. M., W. E. Hobbs, B. J. Manning, P. Carlson, and N. A. DeLuca. 2002. Expression of herpes simplex virus ICP0 inhibits the induction of interferon-stimulated genes by viral infection. J. Virol. 76:2180-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Emeny, J. M., and M. J. Morgan. 1979. Regulation of the interferon system: evidence that Vero cells have a genetic defect in interferon production. J. Gen. Virol. 43:247-252. [DOI] [PubMed] [Google Scholar]
- 18.Fitzgerald, K. A., S. M. McWhirter, K. L. Faia, D. C. Rowe, E. Latz, D. T. Golenbock, A. J. Coyle, S. M. Liao, and T. Maniatis. 2003. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat. Immunol. 4:491-496. [DOI] [PubMed] [Google Scholar]
- 19.Foy, E., K. Li, C. Wang, R. Sumpter, Jr., M. Ikeda, S. M. Lemon, and M. Gale, Jr. 2003. Regulation of interferon regulatory factor-3 by the hepatitis C virus serine protease. Science 300:1145-1148. [DOI] [PubMed] [Google Scholar]
- 20.Geiss, G., G. Jin, J. Guo, R. Bumgarner, M. G. Katze, and G. C. Sen. 2001. A comprehensive view of regulation of gene expression by double-stranded RNA-mediated cell signaling. J. Biol. Chem. 276:30178-30182. [DOI] [PubMed] [Google Scholar]
- 21.Goodbourne, S., L. Didcock, and R. E. Randall. 2000. Interferons: cell signalling, immune modulation, antiviral responses and virus countermeasures. J. Gen. Virol. 81:2341-2364. [DOI] [PubMed] [Google Scholar]
- 22.Grandvaux, N., M. J. Servant, B. tenOever, G. C. Sen, S. Balachandran, G. N. Barber, R. Lin, and J. Hiscott. 2002. Transcriptional profiling of interferon regulatory factor 3 target genes: direct involvement in the regulation of interferon-stimulated genes. J. Virol. 76:5532-5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo, J., D. J. Hui, W. C. Merrick, and G. C. Sen. 2000. A new pathway of translational regulation mediated by eukaryotic initiation factor 3. EMBO J. 19:6891-6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo, J., K. L. Peters, and G. C. Sen. 2000. Induction of the human protein P56 by interferon, double-stranded RNA, or virus infection. Virology 267:209-219. [DOI] [PubMed] [Google Scholar]
- 25.Hiscott, J., P. Pitha, P. Genin, H. Nguyen, C. Heylbroeck, Y. Mamane, M. Algarte, and R. Lin. 1999. Triggering the interferon response: the role of IRF-3 transcription factor. J. Interferon Cytokine Res. 19:1-13. [DOI] [PubMed] [Google Scholar]
- 26.Juang, Y., W. Lowther, M. Kellum, W. C. Au, R. Lin, J. Hiscott, and P. M. Pitha. 1998. Primary activation of interferon A and interferon B gene transcription by interferon regulatory factor 3. Proc. Natl. Acad. Sci. USA 95:9837-9842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karpova, A. Y., M. Trost, J. M. Murray, L. C. Cantley, and P. M. Howley. 2002. Interferon regulatory factor-3 is an in vivo target of DNA-PK. Proc. Natl. Acad. Sci. USA 99:2818-2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kroger, A., M. Koster, K. Schroeder, H. Hauser, and P. P. Mueller. 2002. Activities of IRF-1. J. Interferon Cytokine Res. 22:5-14. [DOI] [PubMed] [Google Scholar]
- 29.Kumar, K. P., K. M. McBride, B. K. Weaver, C. Dingwall, and N. C. Reich. 2000. Regulated nuclear-cytoplasmic localization of interferon regulatory factor 3, a subunit of double-stranded RNA-activated factor 1. Mol. Cell. Biol. 20:4159-4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leonard, W. J. 2001. Role of Jak kinases and STATs in cytokine signal transduction. Int. J. Hematol. 73:271-277. [DOI] [PubMed] [Google Scholar]
- 31.LePage, C., P. Genin, M. G. Baines, and J. Hiscott. 2000. Interferon activation and innate immunity. Rev. Immunogenet. 2:374-386. [PubMed] [Google Scholar]
- 32.Lesokhin, A. M., F. Delgado-Lopez, and M. S. Horwitz. 2002. Inhibition of chemokine expression by adenovirus early region three (E3) genes. J. Virol. 76:8236-8243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Levy, D. E. 1995. Interferon induction of gene expression through the Jak-Stat pathway. Semin. Virol. 6:181-189. [Google Scholar]
- 34.Li, X. L., M. Boyanapalli, X. Weihua, D. V. Kalvakolanu, and B. A. Hassel. 1998. Induction of interferon synthesis and activation of interferon-stimulated genes by liposomal transfection reagents. J. Interferon Cytokine Res. 18:947-952. [DOI] [PubMed] [Google Scholar]
- 35.Lin, R., C. Heylbroeck, P. M. Pitha, and J. Hiscott. 1998. Virus-dependent phosphorylation of the IRF-3 transcription factor regulates nuclear translocation, transactivation potential, and proteasome-mediated degradation. Mol. Cell. Biol. 18:2986-2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin, R., Y. Mamane, and J. Hiscott. 1999. Structural and functional analysis of interferon regulatory factor 3: localization of the transactivation and autoinhibitory domains. Mol. Cell. Biol. 19:2465-2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36a.Lin, R.., R. S. Noyce, S. E. Collins, R. D. Everett, and K. L. Mossman. 2004. The herpes simplex virus ICP0 RING finger domain inhibits IRF3- and IRF7-mediated activation of interferon-stimulated genes. 78:1675-1684. [DOI] [PMC free article] [PubMed]
- 37.Malakhova, O. A., M. Yan, M. P. Malakhov, Y. Yuan, K. J. Ritchie, K. I. Kim, L. F. Peterson, K. Shuai, and D.-E. Zhang. 2003. Protein ISGylation modulates the JAK-STAT signaling pathway. Genes Dev. 17:455-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mamane, Y., C. Heylbroeck, P. Genin, M. Algarte, M. Servant, C. LePage, C. DeLuca, H. Kwon, R. Lin, and J. Hiscott. 1999. Interferon regulatory factors: the next generation. Gene 237:1-14. [DOI] [PubMed] [Google Scholar]
- 39.Matsuyama, T., T. Kimura, M. Kitagawa, K. Pfeffer, T. Kawakami, N. Watanabe, T. M. Kundig, R. Amakawa, K. Kishihara, A. Wakeham, J. Potter, C. L. Furlonger, A. Narendran, H. Suzuki, P. S. Ohashi, C. J. Paige, T. Taniguchi, and T. W. Mak. 1993. Targeted disruption of IRF-1 or IRF-2 results in abnormal type 1 IFN gene induction and aberrant lymphocyte development. Cell 75:83-97. [PubMed] [Google Scholar]
- 40.Mossman, K. L., P. F. Macgregor, J. J. Rozmus, A. B. Goryachev, A. M. Edwards, and J. R. Smiley. 2001. Herpes simplex virus triggers and then disarms a host antiviral response. J. Virol. 75:750-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mossman, K. L., H. A. Saffran, and J. R. Smiley. 2000. Herpes simplex virus ICP0 mutants are hypersensitive to interferon. J. Virol. 74:2052-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nakaya, T., M. Sato, N. Hata, M. Asagiri, H. Suemori, S. Noguchi, N. Tanaka, and T. Taniguchi. 2001. Gene induction pathways mediated by distinct IRFs during viral infection. Biochem. Biophys. Res. Commun. 283:1150-1156. [DOI] [PubMed] [Google Scholar]
- 43.Navarro, L., K. Mowen, S. Rodems, B. Weaver, N. Reich, D. Spector, and M. David. 1998. Cytomegalovirus activates interferon immediate-early response gene expression and an interferon regulatory factor 3-containing interferon-stimulated response element-binding complex. Mol. Cell. Biol. 18:3796-3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Neville, L. F., G. Mathiak, and O. Bagasra. 1997. The immunobiology of interferon-gamma inducible protein 10 kD (IP-10): a novel, pleiotropic member of the C-X-C chemokine superfamily. Cytokine Growth Factor Rev. 8:207-219. [DOI] [PubMed] [Google Scholar]
- 45.Nicholl, M. J., L. H. Robinson, and C. M. Preston. 2000. Activation of cellular interferon-response genes after infection of human cells with herpes simplex virus type 1. J. Gen. Virol. 81:2215-2218. [DOI] [PubMed] [Google Scholar]
- 46.Pine, R. 1992. Constitutive expression of an ISGF2/IRF1 transgene leads to interferon-independent activation of interferon-inducible genes and resistance to virus infection. J. Virol. 66:4470-4478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pitha, P. M., W. C. Au, W. Lowther, Y. T. Juang, S. L. Schafer, L. Burysek, J. Hiscott, and P. A. Moore. 1998. Role of the interferon regulatory factors (IRFs) in virus-mediated signaling and regulation of cell growth. Biochimie 80:651-658. [DOI] [PubMed] [Google Scholar]
- 48.Preston, C. M., A. N. Harman, and M. J. Nicholl. 2001. Activation of interferon response factor-3 in human cells infected with herpes simplex virus type 1 or human cytomegalovirus. J. Virol. 75:8909-8916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reich, N., R. Pine, D. Levy, and J. E. Darnell, Jr. 1988. Transcription of interferon-stimulated genes is induced by adenovirus particles but is suppressed by E1A gene products. J. Virol. 62:114-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ronco, L. V., A. Y. Karpova, M. Vidal, and P. M. Howley. 1998. Human papillomavirus 16 E6 oncoprotein binds to interferon regulatory factor-3 and inhibits its transcriptional activity. Genes Dev. 12:2061-2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sato, M., H. Suemori, N. Hata, M. Asagiri, K. Ogasawara, K. Nakao, T. Nakay, M. Katsuki, S. Noguchi, N. Tanaka, and T. Taniguchi. 2000. Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-α/β gene induction. Immunity 13:539-548. [DOI] [PubMed] [Google Scholar]
- 52.Sato, M., T. Taniguchi, and N. Tanaka. 2001. The interferon system and interferon regulatory factor transcription factors—studies from gene knockout mice. Cytokine Growth Factor Rev. 12:133-142. [DOI] [PubMed] [Google Scholar]
- 53.Servant, M. J., N. Grandvaux, B. R. tenOever, D. Duguay, R. Lin, and J. Hiscott. 2003. Identification of the minimal phosphoacceptor site required for in vivo activation of interferon regulatory factor 3 in response to virus and double-stranded RNA. J. Biol. Chem. 278:9441-9447. [DOI] [PubMed] [Google Scholar]
- 54.Servant, M. J., B. tenOever, C. LePage, L. Conti, S. Gessani, I. Julkunen, R. Lin, and J. Hiscott. 2001. Identification of distinct signaling pathways leading to the phosphorylation of interferon regulatory factor 3. J. Biol. Chem. 276:355-363. [DOI] [PubMed] [Google Scholar]
- 55.Sharma, S., B. R. tenOever, N. Grandvaux, G. P. Zhou, R. Lin, and J. Hiscott. 2003. Triggering the interferon antiviral response through an IKK-related pathway. Science 300:1148-1151. [DOI] [PubMed] [Google Scholar]
- 56.Simmen, K. A., J. Singh, B. G. Luukkonen, M. Lopper, A. Bittner, N. E. Miller, M. R. Jackson, T. Compton, and K. Fruh. 2001. Global modulation of cellular transcription by human cytomegalovirus is initiated by viral glycoprotein B. Proc. Natl. Acad. Sci. USA 98:7140-7145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Smith, E. J., I. Marie, A. Prakash, A. Garcia-Sastre, and D. E. Levy. 2001. IRF3 and IRF7 phosphorylation in virus-infected cells does not require double-stranded RNA-dependent protein kinase R or IκB kinase but is blocked by vaccinia virus E3L protein. J. Biol. Chem. 276:8951-8957. [DOI] [PubMed] [Google Scholar]
- 58.Spear, P. G., and R. Longnecker. 2003. Herpesvirus entry: an update. J. Virol. 77:10179-10185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stark, G. R., I. M. Kerr, B. R. G. Williams, R. H. Silverman, and R. D. Schreiber. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67:227-264. [DOI] [PubMed] [Google Scholar]
- 60.Talon, J., C. M. Horvath, R. Polley, C. F. Basler, T. Muster, P. Palese, and A. Garcia-Sastre. 2000. Activation of interferon regulatory factor 3 is inhibited by the influenza A virus NS1 protein. J. Virol. 74:7989-7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Taniguchi, T., K. Ogasawara, A. Takaoka, and N. Tanaka. 2001. IRF family of transcription factors as regulators of host defense. Annu. Rev. Immunol. 19:623-655. [DOI] [PubMed] [Google Scholar]
- 62.tenOever, B. R., M. J. Servant, N. Grandvaux, R. Lin, and J. Hiscott. 2002. Recognition of the measles virus nucleocapsid as a mechanism of IRF-3 activation. J. Virol. 76:3659-3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tiwari, R. K., J. Kusari, and G. C. Sen. 1987. Functional equivalents of interferon-mediated signals needed for induction of an mRNA can be generated by double-stranded RNA and growth factors. EMBO J. 6:3373-3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.von Kobbe, C., J. M. van Deursen, J. P. Rodrigues, D. Sitterlin, A. Bachi, X. Wu, M. Wilm, M. Carmo-Fonseca, and E. Izaurralde. 2000. Vesicular stomatitis virus matrix protein inhibits host cell gene expression by targeting the nucleoporin Nup98. Mol. Cell 6:1243-1252. [DOI] [PubMed] [Google Scholar]
- 65.Ward, S. G., K. Bacon, and J. Westwick. 1998. Chemokines and T lymphocytes: more than an attraction. Immunity 9:1-11. [DOI] [PubMed] [Google Scholar]
- 66.Wathelet, M. G., P. M. Berr, and G. A. Huez. 1992. Regulation of gene expression by cytokines and virus in human cells lacking the type-I interferon locus. Eur. J. Biochem. 206:901-910. [DOI] [PubMed] [Google Scholar]
- 67.Wathelet, M. G., I. M. Clauss, J. Content, and G. A. Huez. 1988. Regulation of two interferon-inducible human genes by interferon, poly(rI)-poly(rC) and viruses. Eur. J. Biochem. 174:323-329. [DOI] [PubMed] [Google Scholar]
- 68.Wathelet, M. G., C. H. Lin, B. S. Parekh, L. V. Ronco, P. M. Howley, and T. Maniatis. 1998. Virus infection induces the assembly of coordinately activated transcription factors on the IFN-β enhancer in vivo. Mol. Cell 1:507-518. [DOI] [PubMed] [Google Scholar]
- 69.Weaver, B. K., K. P. Kumar, and N. C. Reich. 1998. Interferon regulatory factor 3 and CREB-binding protein/p300 are subunits of double-stranded RNA-activated transcription factor DRAF1. Mol. Cell. Biol. 18:1359-1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Williams, B. R. G., and S. J. Haque. 1997. Interacting pathways of interferon signaling. Semin. Oncol. 24(Suppl. 9):S9-70-S9-77. [PubMed] [Google Scholar]
- 71.Yoneyama, M., W. Suhara, Y. Fukuhara, M. Fukuda, E. Nishida, and T. Fujita. 1998. Direct triggering of the type I interferon system by virus infection: activation of a transcription factor complex containing IRF-3 and CBP/p300. EMBO J. 17:1087-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhu, H., J. Cong, G. Mamtora, T. Gingeras, and T. Shenk. 1998. Cellular gene expression altered by human cytomegalovirus: global monitoring with oligonucleotide arrays. Proc. Natl. Acad. Sci. USA 95:14470-14475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhu, H., J. Cong, and T. Shenk. 1997. Use of differential display analysis to assess the effect of human cytomegalovirus infection on the accumulation of cellular RNAs: induction of interferon-responsive RNAs. Proc. Natl. Acad. Sci. USA 94:13985-13990. [DOI] [PMC free article] [PubMed] [Google Scholar]