Abstract
Understanding how Hofmeister salt ions and other solutes interact with proteins, nucleic acids, other biopolymers and water and thereby affect protein and nucleic acid processes as well as model processes (e.g solubility of model compounds) in aqueous solution is a longstanding goal of biophysical research. Empirical Hofmeister salt and solute “m-values” (derivatives of the observed standard free energy change for a model or biopolymer process with respect to solute or salt concentration m3) are equal to differences in chemical potential derivatives: m-value = Δ(dμ2/dm3) = Δμ23 which quantify the preferential interactions of the solute or salt with the surface of the biopolymer or model system (component 2) exposed or buried in the process. Using the SPM, we dissect μ23 values for interactions of a solute or Hofmeister salt with a set of model compounds displaying the key functional groups of biopolymers to obtain interaction potentials (called α-values) that quantify the interaction of the solute or salt per unit area of each functional group or type of surface. Interpreted using the SPM, these α-values provide quantitative information about both the hydration of functional groups and the competitive interaction of water and the solute or salt with functional groups. The analysis corroborates and quantifies previous proposals that the Hofmeister anion and cation series for biopolymer processes are determined by ion-specific, mostly unfavorable interactions with hydrocarbon surfaces; the balance between these unfavorable nonpolar interactions and often-favorable interactions of ions with polar functional groups determine the series null points. The placement of urea and glycine betaine (GB) at opposite ends of the corresponding series of nonelectrolytes results from the favorable interactions of urea, and unfavorable interactions of GB, with many (but not all) biopolymer functional groups. Interaction potentials and local-bulk partition coefficients quantifying the distribution of solutes (e.g. urea, glycine betaine) and Hofmeister salt ions in the vicinity of each functional group make good chemical sense when interpreted in terms of competitive noncovalent interactions. These interaction potentials allow solute and Hofmeister (noncoulombic) salt effects on protein and nucleic acid processes to be interpreted or predicted, and allow the use of solutes and salts as probes of interface formation and large-scale conformational changes in the steps of a biopolymer mechanism.
I) Introduction
A) Solute and Salt Ion Series in Aqueous Chemistry and Biochemistry
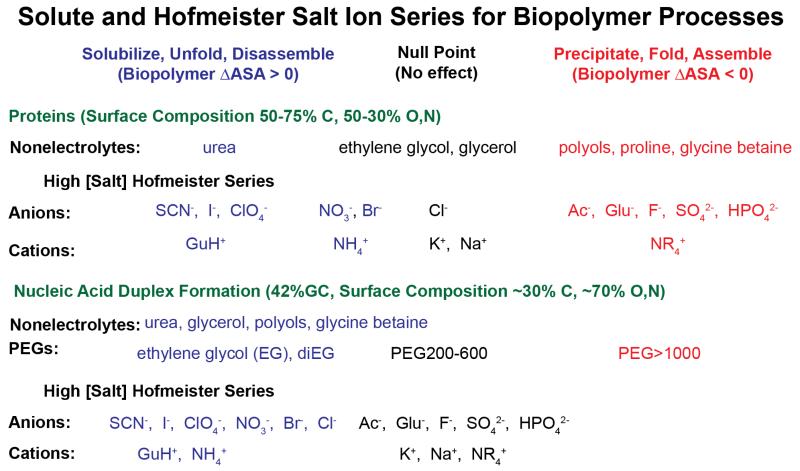
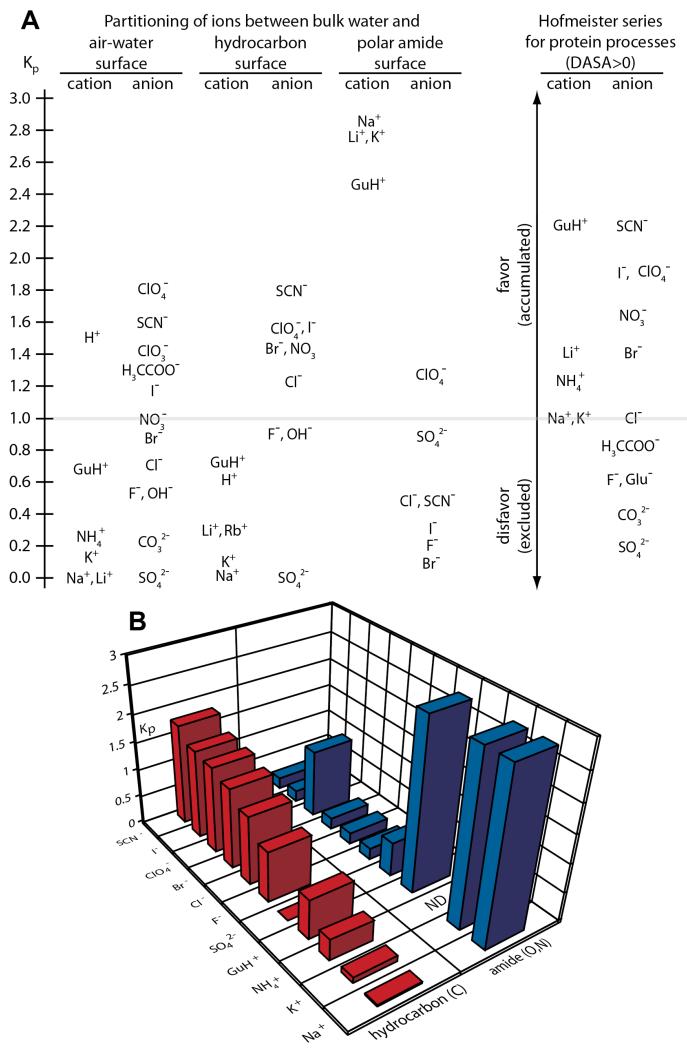
Solutes and salts are universally used in vitro to optimize or shift the thermodynamics and kinetics of biopolymer processes, including protein precipitation/purification, protein crystallization, protein folding, nucleic acid helix formation and RNA folding, and protein-protein, protein-ligand, and protein-nucleic acid interactions1-4. Dating back to Hofmeister’s (1888) studies of the effectiveness of different salts as protein precipitants5, effects of high concentrations of salt anions, cations and nonelectrolyte solutes on many noncovalent protein (also nucleic acid, small molecule) processes in aqueous solution have been observed to fit the regular series listed in Figure 11, 6, 7. Solutes and salt ions at the right end of these series are the most stabilizing; that is, the most effective at driving protein folding, protein assembly and other noncovalent protein self-assembly and binding processes that remove large amounts of protein molecular surface from water. Solutes and salt ions at the left end of these series are most destabilizing, driving unfolding, disassembly, and the exposure of protein surface to the solution. In the assembly direction, all these processes replace protein-water and protein-solute interactions with protein-protein interactions. Generally, the protein surface buried or exposed (i.e. the ΔASA) is primarily hydrocarbon. Approximately two-thirds of the surface buried in folding, and half of native protein surface, is aliphatic or aromatic hydrocarbon; typically about 90% of this hydrocarbon surface is aliphatic. Both folded and unfolded surfaces of proteins are about 15-20% amide oxygen and nitrogen8-10. Most of these amide groups are in the peptide backbone, with a O:N ASA ratio of ~2.4:1.
Figure 1.
Solute and Hofmeister Salt Ion Series for Protein and Nucleic Acid Processes. Solutes and salt ions are arranged from those that promote processes exposing biopolymer surface (blue) to those that promote burial of biopolymer surface (red) for effects on protein processes (top) and nucleic acid duplex formation (bottom). Approximate positions of the null (no effect) points in these series are indicated. Hofmeister salt ion series are those observed for folding or duplex formation at high salt concentration where coulombic effects are minimized.
Addition of small nonelectrolytes such as glycine betaine (GB), proline, and trehalose and other sugars and larger polyols favors processes that remove protein surface from water11, 12. Trimethylamine oxide (TMAO), a close chemical analog of glycine betaine, has a similar effect. Though used as an osmolyte in some eukaryotic cells, TMAO is much less suitable than GB for in vitro biophysical and biochemical studies, because its aqueous solutions are quite alkaline and because it is chemically less stable than GB, decomposing into volatile trimethyl amine. Effects of solutes like glycerol and ethylene glycol on processes that bury or expose protein surface are typically small, neither favoring nor disfavoring them (hence the “null” (i.e. no large effect) designation for these solutes in Figure 1). Urea and urea derivatives, on the other hand, favor processes that expose protein surface to water and hence destabilize protein assemblies and solubilize protein precipitates8, 13.
Studies of effects of these small nonelectrolyte solutes on stability of nucleic acid duplexes reveals a similar rank order7, 14-19 summarized in Fig 1. However, all small nonelectrolytes examined to date destabilize nucleic acid duplexes, favoring melting and exposure of heterocyclic base surface to water. One of the least destabilizing is GB, which has no effect on stability of AT base pairs, but destabilizes GC base pairs16. The surfaces buried in nucleic acid duplex formation and in protein folding or self-assembly processes differ greatly in composition: the nucleic acid base surface buried in helix formation is approximately two-thirds polar O and N and one-third nonpolar C, while that buried in protein folding is approximately two-thirds nonpolar C and one-third polar O and N7, 9, 20. Can these differences in surface composition explain the similar order but shifted null point of the series of nonelectrolyte solutes for protein and nucleic acid processes?
Another series of effects of uncharged solutes is the molecular weight (i.e. degree of polymerization) series from monomer to polymer, investigated most extensively for the series from ethylene glycol to polyethylene glycol21-25 (Figure 1). For nucleic acid duplex formation, ethylene glycol is destabilizing, but high molecular weight polyethylene glycols are stabilizing. Quantitative investigation of the series between EG and PEG provides an excellent method of separating the contributions of small solute effects (chemical preferential interactions) from those of large solute effects (formation of a flexible coil domain; excluded volume effects arising from the inability of the protein or nucleic acid being investigated to access the substantial volume of solution or interact with the chain segments in the interior of flexible coils). As discussed in more detail below, these physical effects arising from solute size are only observed for glycols larger than tri- or tetraEG. For protein folding, ethylene glycol is a relatively nonperturbing solute, neither stabilizing nor destabilizing, while polymeric PEG can be stabilizing or destabilizing22, 23, 25. Pure excluded volume effects of a polymeric solute (which experimentally must be separated from the effect of segregating many segments of that polymeric solute in the interior of the flexible coil) are significantly larger for processes involving association of polymeric strands or protomers into multimers, aggregates or precipitates than for conformational changes without a change in strandedness21.
As in the case of large solutes, effects of salts also include both chemical and physical effects of interactions of salt ions with the biopolymers or model compound involved in the process of interest. These can include short-range chemical preferential interactions of the individual salt ions with the biopolymer surface buried or exposed in the process, chemical site binding of ions to initial and/or final conformations/states of the biopolymers, and long-range physical coulombic interactions of these salt ions with the fixed charges on the initial and final conformations/states of the biopolymers or model compound. Coupling between these various modes of interaction is observed for polyions at low salt concentration, where coulombically-driven accumulation of salt counterions can drive site binding or preferential interactions of the salt counterions with the polyion surface26, 27. The term “Hofmeister effects” of salts usually refers to the weaker chemical (preferential interaction) effects of the individual salt ions manifested at relatively high salt concentration, as in Hofmeister’s original studies of protein precipitation. For some polyion processes, “reverse” series and/or amplifications of Hofmeister effects of salt counterions are observed at low salt concentration where coulombic effects of salts are significant, as discussed in subsequent sections26. The use of high salt concentrations and weakly charged (or uncharged) model compounds, biopolymers and surfaces minimizes coulombic effects, allowing Hofmeister effects of salts to be most unambiguously observed.
Figure 1 shows typical series of Hofmeister anions and univalent cations for protein self-assembly processes and for nucleic acid duplex formation. Anions favoring protein self-assembly include sulfate, phosphate, carboxylates including the physiological anion glutamate, and fluoride; cations favoring protein self-assembly include alkyl ammonium cations (NR4+), K+ and Na+. Most of these are strongly hydrated ions that prefer to interact with water, rather than with biopolymer surfaces. At the other end of these cation and anion series are ions that favor protein disassembly; these ions (e.g. thiocyanate, perchlorate and iodide anions and guanidinium cation) are less strongly hydrated and/or have more tendency to interact with biopolymer surfaces. Nucleic acid duplex formation exhibits the same rank order of Hofmeister anion and cation effects, but a very different null point that likely varies with base composition. No salt exerts a stabilizing Hofmeister effect on a 12 bp (7 AT, 5 GC) nucleic acid duplex, as shown in Fig 5B below7.
Figure 5.
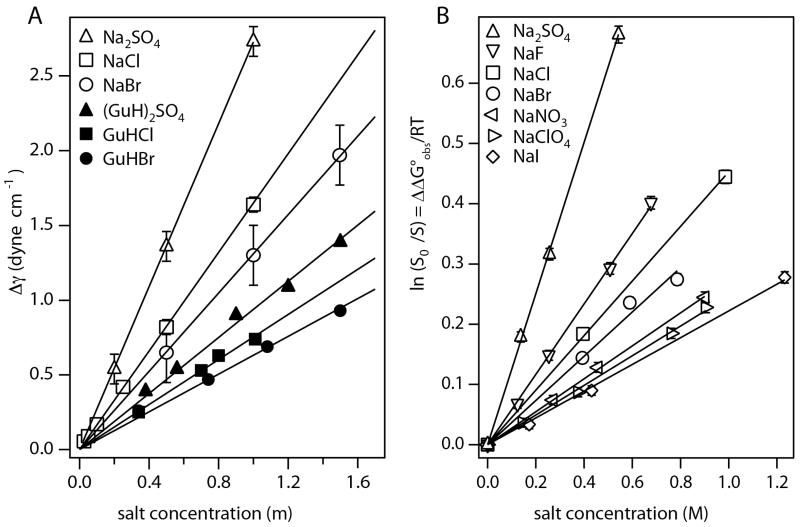
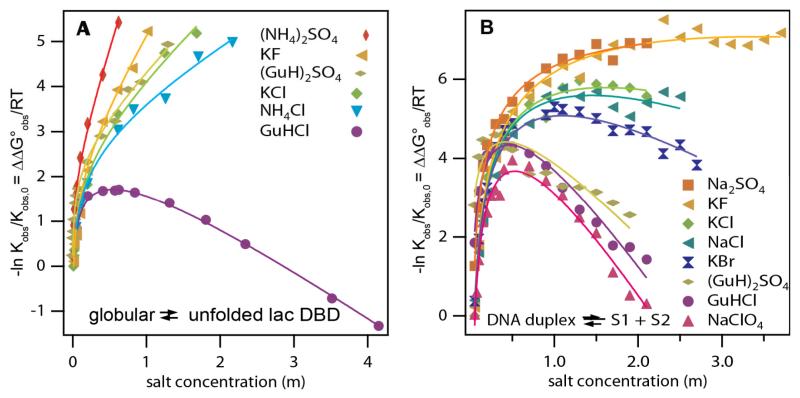
Hofmeister Salt Effects on Model Processes involving Nonpolar Surfaces. Panel A: Increase in surface tension of water (Δγ) as a function of the molality of Na+ and GuH+ salts of SO42−, Cl− and Br − (see Pegram et al34 for literature references; constant temperatures for each series in range 15-30°C). Slopes are surface tension increments (STI) (eq. 13) characterizing effects of these salts on the free energy of transfer of water from bulk to surface. Panel B) Effects of Na+ salts on the logarithm of the relative solubility of benzene (ln(S0/S) = ΔΔG°obs/RT, where S is the solubility in the presence of salt and S0 is the solubility in the absence of salt) at 25°C as a function of salt molarity for the anion series SO42−, F−, Cl−, Br −, NO3−, ClO4−, and I−. Slopes are m-values/RT for the effects of these salts on transfer of benzene from the pure liquid to water. Figure reused with permission from Pegram et al34.
B. Explanations of Chemical Effects of Small Solutes and Salt Ions
What is the molecular origin of these ubiquitous solute and salt ion series? Early interpretations of chemical effects of salts and solutes on aqueous processes were largely structural. Processes exhibiting these effects include the protein and nucleic acid processes discussed above, the model processes of dissolving organic compounds and disassembling micelles, as well as the process of transferring water from bulk water to the air-water interface. Salt and solute effects on these processes were attributed to their ability to affect the hydrogen-bonded structure of water. However, recent spectroscopic studies show that the effect of solutes and salt ions on water structure is confined to the first one or two layers of water of hydration of the solute or salt ion28.
One quantitative explanation of the thermodynamics of solute and Hofmeister salt effects invokes molecular-scale cavity formation1, 29. The process of dissolving a model compound or a protein, or unfolding a protein, is divided into two steps: creating a cavity in water for the model compound or newly exposed protein surface, followed by making the interactions across that interface. To quantify the thermodynamics of cavity formation as a function of solute or salt concentration, surface tension increments are used; these quantify the effect of solute or salt concentration on the free energy of transfer of water from bulk to surface, per unit area of surface created. At a semi-quantitative level, surface tension increments of Hofmeister salts are consistent with effects of Hofmeister salts on processes that expose or bury primarily hydrocarbon surface1. Most fundamentally, this means that Hofmeister salt effects on surface tension (i.e. salt effects on the transfer of water from bulk to the nonpolar interface with air) and on hydrocarbon solubility (i.e. salt effects on the process of exposing hydrocarbon surface to water and hydrating this nonpolar molecular surface) have similar thermodynamic origins. However, this correlation does not necessarily mean that cavity formation and molecular-scale surface tension effects are the fundamental physical phenomena responsible for effects of Hofmeister salts on processes exposing hydrocarbon surface to water.
The solute partitioning model (SPM) is an alternative model for the interpretation of chemical (preferential interaction) effects of nonelectrolyte solutes and Hofmeister salts on all aqueous processes8, 9, 30-34. Recent molecular thermodynamic analysis of both surface tension and hydrocarbon solubility data using the SPM, detailed below, interprets solute and salt effects on surface tension and on solubility of uncharged model compounds quantitatively in terms of a surface hydration b1 (H2O/Å2) and a solute partition coefficient Kp. Kp, the ratio of local to bulk solute concentration, quantifies the competition between the solute or salt ions and water for a molecular or macroscopic surface, and the extent to which the solute or salt ion accumulates at the surface, replacing local water, or is excluded from that surface, leaving it hydrated. For most Hofmeister salt ions, Kp values quantifying accumulation (Kp > 1) or exclusion (Kp < 1) from the air water interface and molecular hydrocarbon surface in water are similar, thereby providing a quantitative explanation of the parallels between salt effects on surface tension and hydrocarbon solubility in terms of ion partitioning between local and bulk water, not cavity formation33, 34.
Timasheff recognized that the interactions of Hofmeister salts with proteins, like those of nonelectrolyte solutes, were preferential (chemical) interactions between the salt (or solute) component (3) and the protein component (2)3. His data are presented as preferential interaction coefficients Γ32 and chemical potential derivatives μ23 = dμ2/dm3 (where Γ32 = − μ23/μ33) and discussed qualitatively using both the cavity model and the local-bulk model (the precursor of the SPM). Tanford35 and Schellman36, 37 developed weak binding models based on exchange between the solute/salt ion and water to discuss preferential interactions of urea and other solutes and salts with proteins and interpret μ23 values. Tanford38 and more recently Bolen and collaborators39, 40 analyzed solubility increments quantifying the effects of urea and other nonelectrolytes on the solubility of amino acids and dipeptides to obtain side chain and backbone transfer free energies, which Bolen and collaborators have used in conjunction with a surface area analysis to predict effects of these solutes on the standard free energy change for protein unfolding and other protein processes39, 40.
The SPM-based analysis reviewed here quantifies the competition between a solute or salt ion and water for any molecular or macroscopic surface, using structural input (water-accessible surface area (ASA) and composition). This analysis yields two levels of useful information. Interaction potentials, obtained from osmometric and solubility data for model compounds and from surface tension increments for the air water interface, quantify the interaction of a solute with a unit area of a particular functional group, atom, or surface. These interaction potentials are very useful for interpreting or predicting interactions of solutes or salts with biopolymer or other surfaces, as well as interpreting or predicting m-values or changes in surface area (ΔASA) in protein or nucleic acid processes7, 8, 12. From these interaction potentials, local-bulk partition coefficients quantifying the accumulation or exclusion of the solute from the vicinity of this group or surface are obtained. These partition coefficients for solutes like urea, GB, and salt ions and functional groups like amide and anionic oxygens, amide and cationic nitrogens, and aliphatic and aromatic carbons, are consistent with expectations based on competitive hydrogen bonding capabilities of the solute and water to interact with polar groups, and also provide evidence for the significance of cation-π and π-π interactions between solute and aromatic groups on model compounds8, 12.
The SPM-based analysis not only predicts and interprets the interactions of solutes and salt ions with the functional groups of biopolymers, but also is applicable to predict and interpret effects of small solutes and salts on the surface tension of water and on all biopolymer and model compound processes in water7, 8, 12, 33, 41. The fundamental premise of this analysis, in common with the backbone and side chain transfer free energy analysis38-40, is that these contributions to solutes and salt effects are independent and additive. For interactions of two larger solutes, each with multiple functional groups, especially when presented on constrained surfaces, deviations from additivity are more likely. Deviations would indicate that favorable (complementary) or unfavorable nearest neighbor effects are significant, as in forming arrays of noncovalent interations in a ligand-biopolymer or biopolymer-biopolymer interface. Interactions of larger or more highly charged solutes with biopolymers may include significant contributions of physical (e.g. excluded volume, coulombic) effects as well as chemical (preferential interaction) effects7, 21.
C. m-values: Experimental Thermodynamic Inputs to Characterize Solute and Hofmeister Salt Effects on Biopolymer and Model Processes
Solute and Hofmeister salt effects on aqueous processes are characterized by m-values, which are derivatives of the standard free energy change ΔGoobs = − RTlnKobs with respect to solute or salt concentration:
| Eq. 1 |
In Eq 1, Kobs is the equilibrium constant for the process expressed in terms of equilibrium concentrations of products and reactants, and ΔGoobs is the corresponding standard free energy change. Interactions of solutes or salt ions (component 3) with biopolymers or model compounds (component 2) in aqueous solution affect the activity coefficient of component 2.
m-values quantify solute and Hofmeister salt effects on standard free energy changes, not only for biopolymer self-assembly or disassembly processes, but also for model processes that change the exposure of nonpolar or polar surface to water, including dissolving a model compound in water (where Kobs is the solubility and the m-value is proportional to the Setchenow coefficient1, 42), micelle formation (where Kobs is the reciprocal of the critical monomer concentration (CMC)), and transferring water from bulk to the air-water interface, where the surface tension is the free energy change per unit area and the surface tension increment (STI) is the m-value.
II. Examples of m-Value Plots Illustrating Effects of Solutes and Hofmeister Salt Ions on Aqueous Biopolymer and Model Processes
A. Effects of Small Nonelectrolyte Solutes on Protein Unfolding and Protein Stability
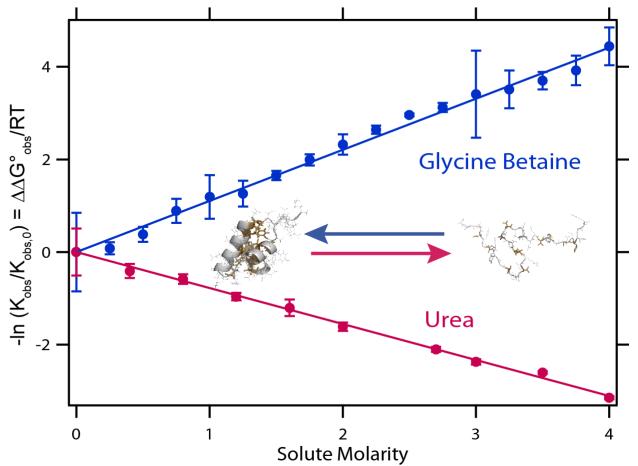
Figure 2 plots the logarithm of the equilibrium concentration quotient (observed equilibrium constant) Kobs for unfolding the DNA binding domain of lac repressor, a small (51 folded residues) single domain globular protein, as a function of urea and GB concentration; lnKobs varies linearly with solute concentration over a wide range. For this very small protein, addition of 2.1 M GB reduces Kobs for unfolding by tenfold; addition of 3.0 M urea increases Kobs by tenfold. Slopes of these plots, when multiplied by −RT, yield urea and GB unfolding m-values. Solute m-values are generally independent of solute concentration over a wide range; typically molar scale plots are more linear than molal scale plots, and both molar and molal scale plots have the same initial slope9, 13. For small nonelectrolyte solutes like urea and GB, these m-values are entirely determined by chemical (preferential) interactions3, 8, 12. Urea favors protein unfolding because it is locally accumulated in the vicinity of the protein surface exposed in unfolding (ΔASA); the composition of this surface is approximately 65-70% hydrocarbon (C) and 15-20% amide (O, N). GB favors folding because it is locally excluded from these surfaces, which prefer to remain hydrated. Urea unfolding m-values are proportional to ΔASA of unfolding (Fig. 3), a key finding of Myers et al13, which is predicted and interpreted using the Solute Partitioning Model (SPM9, 20). From SPM-based analysis of m-values one also obtains partition coefficients Kp which relate the local concentration of the solute (here urea, GB) near the surface of interest to the bulk solute concentration. For the surface exposed in unfolding, the composite Kp value for urea is greater than unity (accumulation) while that for GB is less than unity (exclusion)8, 12.
Figure 2.
Effects of Urea and Glycine Betaine (GB) on Protein Unfolding. Plots of -ln(Kobs/Kobs,0) = ΔΔG°obs/RT vs solute molarity for effects of urea (25°C) and GB (59°C) on unfolding of the lac repressor DNA binding domain (lacDBD)75. Kobs is the unfolding equilibrium constant in the presence of solute and Kobs,0 is the unfolding equilibrium constant in the absence of solute; slopes are m-values/RT. Inset figure shows the folded structure (one monomer of dimer in PDB 1OSL) and one unfolded structure (determined by ProtSA76) of lacDBD with the surface exposed on unfolding highlighted in yellow.
Figure 3.
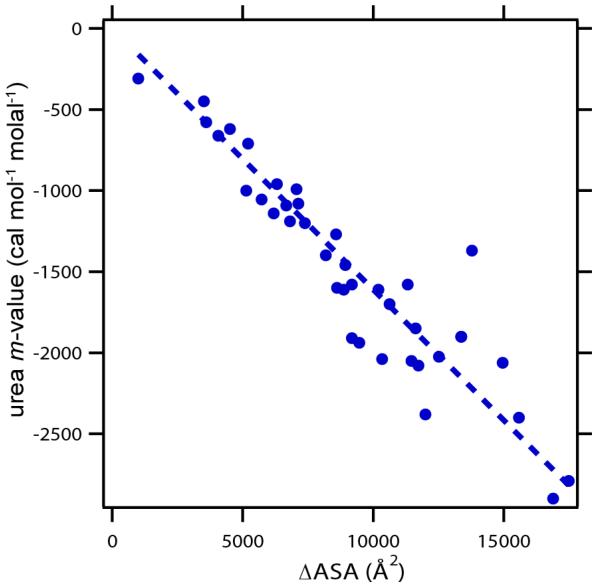
Myers, Pace, and Scholtz13 Plot of Urea m-values for Protein Unfolding as a Function of ΔASA of Unfolding. Protein data set and ΔASA determinations (for a fully extended model of the urea-denatured state) from Guinn et al8. Unfolding m-values determined at temperatures ranging from 0 to 40°C with most at 20-25°C (for conditions see Hong et al61 and refs therein).
B. Effects of varying the degree of polymerization of the solute (ethylene glycol to polyethylene glycol) on nucleic acid helix melting
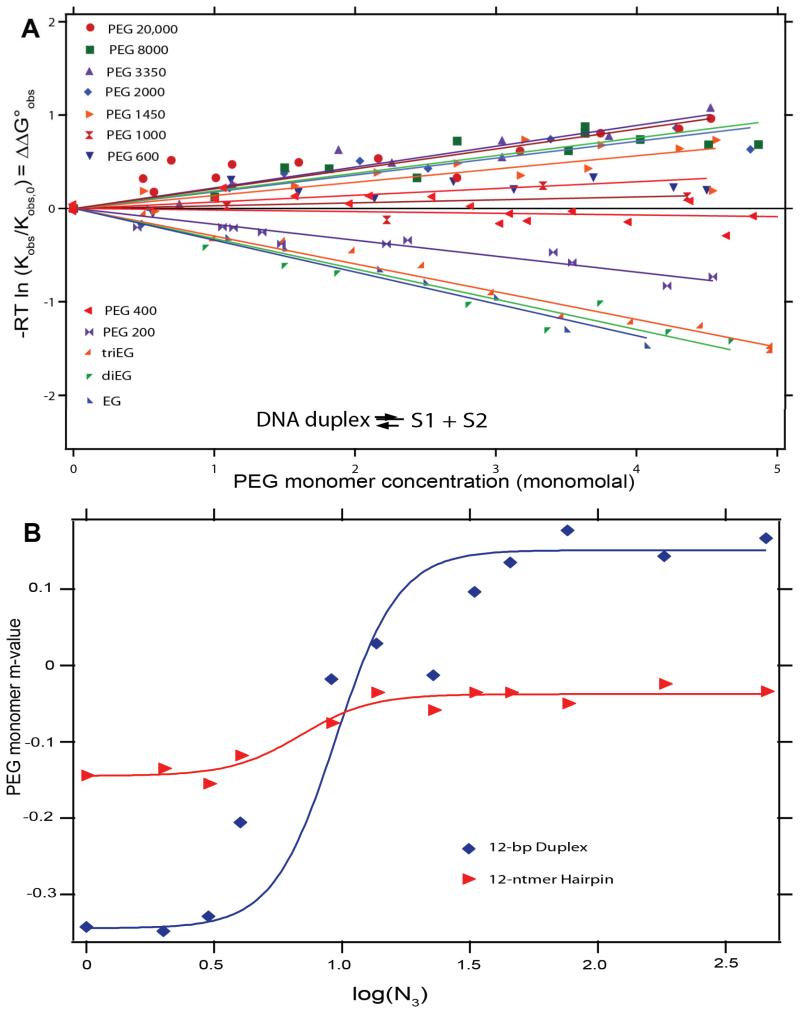
Figure 4A summarizes the effects of the solute series from ethylene glycol (EG) to polyethylene glycol (PEG) on Kobs for melting of a 12 bp DNA duplex. All PEG concentrations are expressed on a monomolal scale; use of this PEG monomer concentration scale not only allows data for the entire series to be represented clearly on one plot but also illustrates the very different effects of low, moderate and high molecular weight PEG on the duplex melting process. Low molecular weight PEGs favor melting; for EG, diEG and triEG, monomer m-values are the same, indicating that the favorable chemical (preferential) interaction of the DNA surfaces exposed in melting with the two –CH2OH end groups of diEG and triEG (and with the two halves of EG) is the same as with the –CH2OCH2- interior groups. As the PEG molecular weight is increased from PEG 200 (tetraethylene glycol) to PEG 1450, the favorable effect on melting per PEG monomer, which is the same for EG, di- and tri-EG, is first reduced and then replaced by a strong unfavorable effect on melting (i. e. duplex stabilization) of higher molecular weight PEG. Above PEG 1450, this stabilization becomes independent of PEG molecular weight. A summary plot of the variation in PEG monomer m-value with the number of residues N3 in the PEG molecule (log scale) is shown in Figure 4B.
Figure 4.
Effects of Polyethylene Glycol (PEG) Degree of Polymerization (N3) on DNA Duplex and Hairpin Melting. (A) Plots of −RTlnKobs/Kobs,0 = ΔΔG°obs vs PEG monomer concentration (monomolal) for the series from ethylene glycol to PEG 20,000 on DNA duplex melting at 40°C, where Kobs is the melting equilibrium constant in the presence of PEG and Kobs,0 is the melting equilibrium constant in the absence of PEG. (B) PEG monomer m-values (slopes from A) for melting a 12bp DNA duplex and 4bp hairpin plotted as a function of log(N3) where N3 is the degree of polymerization of PEG. Figure adapted from Knowles et al21.
For a 4 bp hairpin helix, where melting is not accompanied by strand separation, qualitatively similar but smaller effects of increasing PEG molecular weight on the PEG monomer m-value are observed (see Fig 4 B). Monomer m-values of EG, diEG and triEG for hairpin melting are all the same, but are somewhat less than half as large in magnitude as for the 12 bp duplex, consistent with an interaction with the ASA exposed in melting, which is less than half as large for the 4 bp hairpin as for the 12 bp duplex. For the 4 bp hairpin, the high PEG molecular weight plateau occurs at a m-value near zero; the favorable chemical interaction of functional groups of low molecular weight glycols in this series (EG, di- and triEG) with the nucleic acid base surface exposed in melting is largely eliminated by a physical effect of high molecular weight PEG, but not replaced by a helix-stabilizing effect as in the case of the 12 bp duplex.
Two large-solute effects become significant for PEG oligomers larger than tri- or tetraEG. One is excluded volume; the flexible PEG polymer excludes the DNA strands from the volume of solution occupied by the chain molecule. This effect is much more important for 12 bp duplex melting, with strand separation, than for 4 bp hairpin melting, which only involves a change in shape and not a change in strandedness. The other is sequestration; the large fraction of PEG monomers which transiently are located in the “interior” of the chain molecule cannot interact chemically with the DNA surface exposed in melting, so the chemical effect of EG and PEG is gradually eliminated with increasing PEG molecular weight. Knowles et al21 interpret these behaviors quantitatively. Figure 4 B demonstrates that for these systems only chemical (preferential interaction) effects of the solute are observed up to a solute molecular weight of approximately 200 (tetraEG).
C. Hofmeister Effects of Salts on Model Processes that Expose an Uncharged, Nonpolar Surface to Water
Figures 5 A and B provide classic examples of Hofmeister effects, where coulombic effects are largely absent and the surfaces involved are entirely nonpolar. Surface tension differences for sodium and guanidinium salts, plotted vs salt concentration in Fig 5 A, are free energy differences per unit of surface area for the transfer of water from bulk to the nonpolar air-water surface in a salt solution as compared to pure water. These free energy differences increase linearly with salt concentration and yield salt-specific m-values for this transfer process. Logarithms of solubility ratios for dissolving benzene in water and in salt solutions are plotted vs salt concentration in Fig 5 B. These plots are also linear with slopes that are interpreted as salt m-values/RT for this benzene transfer process. Most (but not all) salts increase surface tension and reduce benzene solubility. The cation and anion series deduced from these surface tension and hydrocarbon solubility data are the same as the Hofmeister series of cations and anions determined for protein solubility and other protein processes (Figure 1), though the null points of many of these series differ as discussed below. Analysis of these and other data for these model transfer processes demonstrates that the cation and anion of the salt make additive, independent contributions to the m -value, and provide values for the amount of local water at the air-water and molecular hydrocarbon surface and for the partition coefficients of individual salt ions between bulk water and this local water33, 34.
D. Salt Effects (Coulombic, Hofmeister) on Protein and Nucleic Acid Melting and Protein Aggregation
1) Melting
Figure 6 summarizes effects of a wide range of Hofmeister salts on the logarithm of the equilibrium constant Kobs for unfolding the lac repressor DNA binding domain (Fig 6A; cf. Fig 2) and for melting a 12 bp DNA duplex (Fig. 6B). At low salt concentration (< 0.5 molal), coulombic effects are large and cause the stability (- RTln Kobs) of the folded and helical forms to increase with increasing salt concentration. Plots of lnKobs vs ln[salt] are linear in this low salt range (data of ref 7; not shown), with slopes which are similar for all salts of the same valence, as expected for a coulombic effect43. This coulombic effect is particularly pronounced for the 12 bp DNA oligoanion; transition from the duplex to two separate strands reduces the number of negatively charged phosphates from 22 (duplex) to 11 for each melted strand and also reduces the axial charge density from 2 phosphate charges to less than 1 per 3.4 Å. The 51-residue DBD has a net charge of +2, with 7 positive and 5 negative charges which presumably become more separated in the unfolded form and cause its stability to increase more modestly with salt concentration in this range.
Figure 6.
Effects of Hofmeister Salts on Protein Unfolding and DNA Melting. Panels show the effects of low and high concentrations of salts spanning the Hofmeister series on -ln(Kobs/Kobs,0) = ΔΔG°obs/RT for A) lacDBD unfolding at 37°C and B) DNA helix melting at 40°C. Kobs is the unfolding or melting equilibrium constant in the presence of salt and Kobs,0 is the reference equilibrium constant in low-salt buffer. Fitted curves allow a separation of Hofmeister and coulombic effects of these salts at high salt concentration, as described in Pegram et al7. Figure adapted with permission from Pegram et al.
Above 0.1 m salt (lacDBD) or 0.5 m salt (12-mer DNA) where coulombic effects are expected to be small, large salt-specific differences in Kobs and in its salt derivative (i.e. m-value/RT) are observed. Salt series for both processes follow the rank order of the Hofmeister series, with very different null points. The stability of lacDBD increases with increasing salt concentration for all salts investigated except GuHCl, but the stability of the DNA duplex is not increased by any salt in this range. These very different null points of the Hofmeister salt series for lacDBD and the 12bp DNA duplex must be a consequence of the very different compositions of the surfaces exposed in melting (ΔASA). The lacDBD surface exposed in unfolding is primarily (~70%) nonpolar C and only (~30%) polar N, O, while the DNA surface exposed in melting is primarily (~70%) polar N and O, and only ~30% nonpolar C.
2) Protein Aggregation and Phase Separation
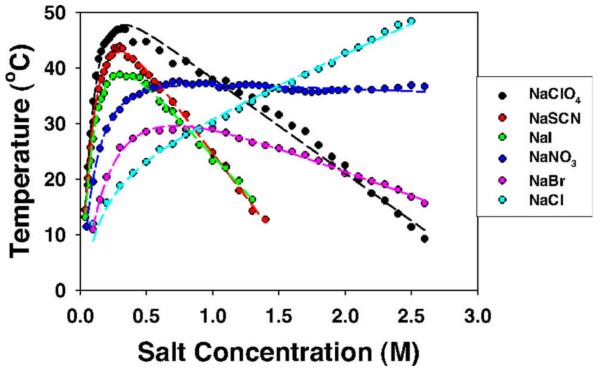
Figure 7 (Zhang & Cremer26) shows the dramatic and complicated effects of different sodium salts with anions from the middle to the solubilizing end of the Hofmeister series on the cloud point transition temperature Tcp for liquid-liquid phase separation and aggregation of lysozyme upon cooling at pH 9.4 where it is positively charged. Derivatives dTcp/d[salt] measure the ratio of the salt m-value to the enthalpy change for aggregation. At high [salt], where coulombic salt effects are minimized and Hofmeister effects of these sodium salts are dominant, Tcp varies linearly with salt concentration. These high-[salt] slopes follow the normal protein Hofmeister anion series, (positive for Cl−, null point near NO3−, and increasingly negative for Br−, ClO4−, and SCN−). At lower salt concentration (below 0.4 M), a reverse order of highly magnified effects of all these sodium salts is observed, where SCN−, I−, and ClO4− favor aggregation much more than Cl− .These magnified, reverse-order Hofmeister effects of salt counterions (here anions) at low [salt] are the result of coulombically driven anion binding (most significant for SCN−, least for Cl−) to positively charged lysozyme at low salt concentration, reducing its charge and favoring aggregation and phase separation26.
Figure 7.
Reverse and Normal Hofmeister Salt Effects. The cloud-point temperature of the cationic protein lysozyme (pH 9.4) as a function of anion type and concentration (Zhang and Cremer26), showing high salt (normal) and low salt (magnified, reversed) Hofmeister salt effects on liquid-liquid phase separation and aggregation. Figure reprinted with permission.
III) Analysis of Solute and Hofmeister Salt Effects
A) Interpretation of m-Values in terms of Chemical Potential Derivatives μ23
1) m-Values for Biopolymer Processes: Relationship to Δμ23
Interactions of solutes or salt ions (component 3) with biopolymers (component 2) in aqueous solution affect the activity coefficient (f2) of the biopolymer, giving rise to significant derivatives dlnf2/dm3. These activity coefficient derivatives are to an excellent approximation the same as reduced chemical potential derivatives (1/RT)dμ2 /dm3 = μ23/RT. Because the observed equilibrium constant Kobs for the biopolymer process is defined in terms of product and reactant concentrations and not activities, Kobs depends on m3 and this dependence (i.e the solute m-value) is determined by the dependence of f2 on m3, or equivalently by μ23:
| Eq. 2 |
Chemical potential derivatives μ23 are closely related to preferential interaction coefficients (dialysis or Donnan coefficients) Γ32 = − μ23/μ33 where μ33 = dμ3/dm3. For an uncharged solute dμ3/dm3 = (RT/m3)(1 + ε3) where ε3 = dlnf3/dlnm3 represents an often-small correction for solute-solute nonideality (i.e. ε3 is small in magnitude in comparison to unity) that can be approximated by its value in the absence of the biopolymer or model compound species 2. Experimentally determined values of Γ32 and μ23 may be compared with theoretical (e.g. Poisson-Boltzmann) or computational (e.g. Monte Carlo, molecular dynamics) predictions of the radial distribution of solute 3 in the vicinity of solute 2 using Kirkwood-Buff theory and integrations over the local excess or deficit in the radial distribution of solute 344, 45.
For biopolymer processes where the solute effect is characterized by a m-value = Δμ23, we assume additivity of free energy contributions from these short-range interactions so that the m-value can be interpreted as the value of μ23 for interaction of solute (3) with the biopolymer surface area exposed (or buried) in the process (i.e. the ΔASA)9, 20. The extensive data base of urea m-values for protein unfolding reveals that these m-values are proportional to ΔASA for this homologous series of surfaces (varying in total area but with very similar compositions (65-70% hydrocarbon (C), 15-20% amide (O,N), the remainder other polar (O, N). This proportionality of urea m-values to ΔASA, first established by Myers et al13 and interpreted using the SPM by Courtenay et al9, is shown for an updated set of small, single domain globular proteins lacking disulfides in Figure 2.
2) m-Values for Model Processes; Relationship to μ23
m-Values also quantify solute and Hofmeister salt effects on standard free energy changes of model processes that expose a molecular or macroscopic surface to water. These processes, their solute or salt m-value, and the relation to μ23, include:
- Transfer of model compounds from the pure liquid or solid phase to water, quantified by the solubility m2ss:
In Eq. 4, the solubility m-value, determined at constant a2, is equal to μ23 (at constant m3) only for sparingly soluble model compounds for which the activity coefficient γ2 is not a function of m2 even at the solubility limit. Osmometry is a good alternative to quantify μ23 for interactions of solutes with highly soluble model compounds, as reviewed below.Eq 4 - Transfer of amphiphiles from a micelle to water, quantified by the critical monomer concentration CMC:
where Δμ23 is interpreted as μ23 for the interaction of solute (3) with that portion of the surface of the amphiphile (2) that is buried in the micelle and exposed to water in micelle dissociation.Eq 5 - Transfer of water from bulk to the air-water surface, quantified by the surface tension γ:
where α2 is a solute-surface interaction potential, quantifying the direction and strength of the solute-surface interaction per unit area of surface (see below). STI m-values, while for a two component system, contain the same quantitative information about the interaction of the solute with the air-water surface (relative to interactions with water) as do values of μ23 for a molecular surface in water.Eq 6
3) Determination of μ23 for Solute-Model Compound Interactions by Osomometry
Values of μ23 quantifying preferential interactions of solutes and Hofmeister salts with soluble, non-volatile model compounds are obtained directly from the difference ΔOsm46, 47 between the three component osmolality (Osm(m2, m3)) and the two component osmolalities (Osm(m2), Osm(m3)), determined by vapor pressure osmometry8, 11, 12, 15, 48 or isopiestic distillation49:
| Eq 7 |
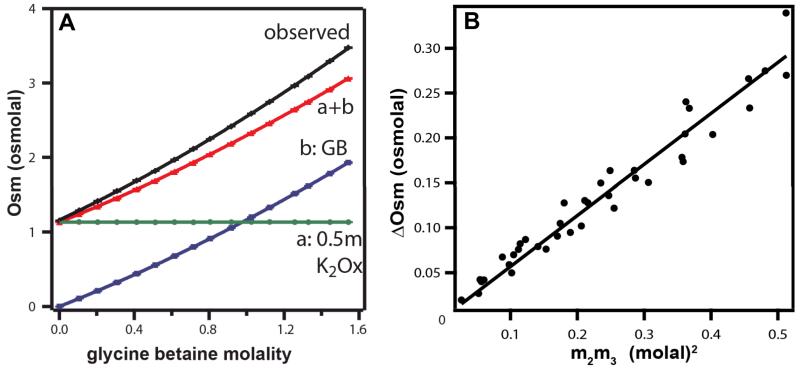
The slope of a plot of ΔOsm vs. the concentration product m2m3 is therefore equivalent to a free energy derivative m-value/RT quantifying the effect of one solute on the chemical potential of the other. As an example, figure 8A shows osmolality measurements on a constant-molality K2Oxalate solution as a function of the molal concentration of GB added. The black curve is the osmolality Osm(m2,m3) of the three-component GB-K2Oxalate aqueous solution. The red curve is the sum of the osmolalities of the individual two-component solutions (variable GB Osm(m3) (blue curve); constant K2Oxalate Osm(m2) (green line)). The difference between the black and red curves is ΔOsm (Eq. 7). Figure 8B shows the plot of ΔOsm vs the product m2m3 for all concentrations of GB and K2Oxalate investigated; the plot is linear and yields (from the slope) a constant value for the chemical potential derivative μ23/RT = 0.55 m−1 for the interaction of GB and K2Oxalate (μ23= μ32).
Figure 8.
VPO Determination of ΔOsm and μ23 for the Interaction of Glycine Betaine (GB) with K2Oxalate. A) Determination of ΔOsm for a series of experiments at 25°C in which the molality of K2Oxalate is fixed at 0.5 m and the molality of GB is increased. Green line (a) is the osmolality of 0.5 m K2Oxalate in the absence of GB. Blue curve (b) is the osmolality of GB solutions as a function of its molal concentration. Red curve (a + b) is the sum of the osmolalities of these two component solutions: 0.5 m K2Oxalate (a) and variable GB molality (b), plotted against GB molality. Top black curve is the observed osmolality of the three component solutions (0.5 m K2Oxalate, variable GB) plotted vs GB concentration. The difference between the red and black curves is the excess osmolality ΔOsm; this is positive and increases with increasing GB concentration. B) A plot of ΔOsm against m2m3, the product of molal concentrations of K2Oxalate and GB, using K2Oxalate data from Capp et al12; slope is μ23/RT (Eq. 7).
Vapor pressure osmometry (VPO) and solubility measurements are complementary methods for quantifying interactions of a solute or Hofmeister salt with model compounds or folded (native) biopolymers. VPO is most useful for relatively soluble, nonvolatile compounds while solubility is most useful for sparingly soluble compounds. Eq 7 for analysis of VPO data is not applicable to situations where both the solute and the model compound or biopolymer are salts.
B. Analysis of Chemical Potential Derivatives (μ23) and m-Values (Δμ23) Using Surface Areas and the Solute Partitioning Model (SPM)
Guggenheim proposed a two-state (local, bulk) model of water near the air-water interface31, 41 and others proposed an analogous (local, bulk) model of water in the vicinity of a molecular surface30, 50. From these precedents, we developed the solute partitioning model (SPM) to describe the competitive short range interactions of solutes or salt ions with water for an interface or surface. In the SPM, the number of water molecules per Å2 of interface or molecular surface is designated b1, and a microscopic partition coefficient Kp quantifies the local accumulation or exclusion of the solute from the local water at the interface or surface.
1) Analysis of μ23 for Interaction of a Solute or Hofmeister Salt with a Surface
From a molecular thermodynamic analysis using the SPM, the chemical potential derivative μ23 quantifying the interaction of a nonelectrolyte solute with a homogeneous molecular surface (e.g aromatic or aliphatic hydrocarbon) is predicted to be proportional to ASA, with a proportionality constant α which is determined by b1 and Kp. If the molecular surface is chemically heterogeneous, then b1 and Kp are the average hydration and solute partition coefficient of the different functional groups tha make up the surface.
| Eq 8 |
where the microscopic partition coefficient Kp = m3loc/m3bulk and the solute(3)-solute(3) self-nonideality correction term is ε3 = dlnf3/dlnm3. The proportionality constant α3,2 defined in Eq. 8 is called an interaction potential because it quantifies the strength and direction of the preferential interaction of the solute with the surface, is independent of ASA, and to a first approximation is independent of solute concentration. At solute concentrations above 1 m, some dependence of the quantities Kp, b1, and ε3 on m3 is expected9, 32. If the solute is completely excluded from the surface, so that Kp,3 = 0, then measurement of α3,2 provides a determination of the hydration of the surface b1.
Where the solute is a salt with ν ions per formula unit (ν+ cations and ν− anions; ν = ν+ + ν−), SPM analysis yields the following expressions to interpret the chemical potential derivative μ23 for the Hofmeister (noncoulombic) interaction of the salt component and the individual salt ions with the surface31, 33:
| Eq 9 |
In Eq. 9, the individual ion interaction potentials are α+,2 = − (Kp,+ − 1)b1(1 + ε±)/55.5 and α−,2 = − (Kp,− − 1)b1(1 + ε±)/55.5 and the individual ion partition coefficients are Kp+, = m+loc/m+bulk, Kp− = m−loc/m−bulk. The term ε± = dlnf±/dlnm3 corrects for self-nonideality of the salt component (3). If both cation and anion of a salt are completely excluded from the surface, so that Kp,+ = Kp,− = 0, then measurement of α3,2 for that salt provides a determination of the hydration of the surface b1 .
2) Analysis of m-Values (μ23) for the Effect of a Solute or Hofmeister Salt on a Process with a Change in Surface Area ΔASA
From the SPM-based Eq. 8, the m-value quantifying the effect of a nonelectrolyte solute on a process is interpreted a
| Eq 10 |
where α3,2is the interaction potential for the surface specified by the ΔASA.
From Eq. 9, the m-value quantifying the noncoulombic (Hofmeister) effect of a salt on a process is interpreted as
| Eq 11 |
The SPM-based analysis therefore predicts that solute and Hofmeister salt m-values are a) independent of solute concentration, in agreement with experiment (see for example Fig 2), and b) proportional to ΔASA for homologous series of biopolymer surfaces with same composition (same Kp and b1), like those exposed in unfolding globular proteins (Fig 3).
3) SPM-based Interpretation of Surface Tension Increments
Completely analogous expressions to Eqs 8 and 9 are obtained to interpret surface tension increments STI of nonelectrolytes51 and salts31, 33, 41. For nonelectrolyte solutes,
| Eq. 12 |
and, for salts,
| Eq. 13 |
Measurements of STI for solutes and salts which are completely excluded from the air-water surface allow determination of b1, the amount of local water per unit area of that surface.
C. Dissection of Solute-Surface Interaction Potentials α For Heterogeneous Molecular Surfaces
For interactions of a solute or salt (component 3) with the chemically heterogeneous surface of a biopolymer or a small model compound, different functional groups are expected to interact differently with the solute and exhibit different values of Kp (and possibly b1). Given the short-range character of these interactions, we proposed that the observed μ23/RT is the sum of contributions αiASAi from interactions of the solute with the different types of biopolymer surface (functional groups)8, 12, 21:
| Eq. 14 |
where each αi for the interaction of solute 3 with a functional group on solute 2 is related to a local partition coefficient for partitioning of solute 3 between bulk water and the local water of hydration of that functional group:
| Eq. 15 |
as in Eq 8. Values of μ23/RT for interactions of a solute with inorganic salts and electrolytes as model compounds include the contribution of the inorganic salt ions:
| Eq. 16 |
where βion is the contribution to μ23/RT from the solute-ion interaction, and νion is the stoichiometric number of that ion per formula unit of the salt.
A “two way” breakdown of μ23 values into interactions of individual functional groups on solute 3 with functional groups on solute 2 is currently being investigated (Knowles et al, in preparation):
| Eq. 17 |
Data is being obtained to test the dissection of μ23 values for model compounds, and the buildup of μ23 values or m-values for biopolymer processes from the interactions of the solute with the individual functional groups on the biopolymer surface involved in the process. We are systematically determining αi values for the interaction of urea, GB, Hofmeister salts and other small solutes of biochemical significance (proline, glycols, polyols, sugars, TFE, etc) with different biopolymer functional groups from model compound data. We are using this database to predict or interpret values of μ23/RT for the interactions of these solutes with native biopolymers, and to predict or interpret m-values for biopolymer processes, as probes of interface formation and large-scale coupled conformational changes in the steps of biopolymer processes including open complex formation and transcription initiation by RNA polymerase at promoter DNA.
IV. Case Studies: Quantifying the Interactions of GB, Urea, Hofmeister Salts and Salt Ions with Functional Groups of Proteins and Nucleic Acids and the Hydration of these Groups; Applications to Protein and Nucleic Acid Processes
A) Introduction
Here we summarize the determination of solute-surface interaction potentials (α values) and partition coefficients Kp for the interactions of GB, urea and Hofmeister salts with the functional groups of proteins, and the hydration (b1) of some of these groups, from the database of model compound interaction data (μ23 values). Quantitative thermodynamic information about these very important noncovalent interactions of biochemistry and aqueous chemistry was not previously available. Interaction potentials for urea and GB with polar and charged groups make good chemical sense in terms of competitive hydrogen bonding. With these α values (or equivalently Kp values) and structural information about the composition and amount of protein surface exposed or buried in a protein process, solute or Hofmeister salt m-values for the process can be predicted. Alternatively, from experimental measurements of solute and/or Hofmeister salt m-values for the steps of a protein process or mechanism, information about the amount and composition of the protein surface exposed or buried in interfaces and coupled conformational changes in those steps can be obtained. In this way solutes and Hofmeister salts can serve as very useful probes of steps involving interface formation or large scale conformational changes in the mechanism of action of protein molecular machines.
B. Determining the Hydration b1 of Different Groups and Surfaces
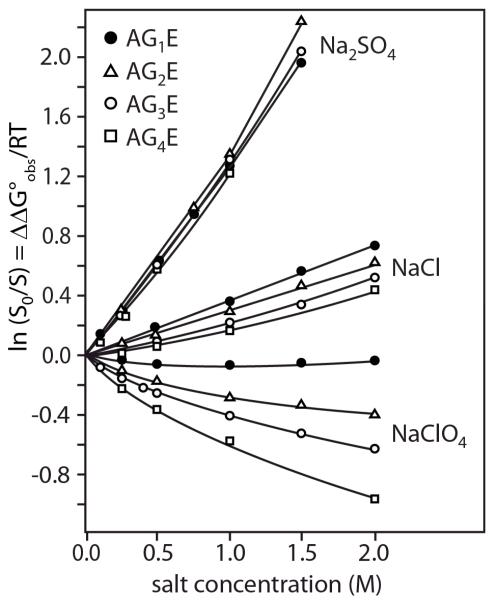
To use solutes as probes or to predict or interpret solute or Hofmeister salt m-values, only α-values (and ASA information for a prediction) are required. The SPM-based analysis provides justification for the use of ASA information to interpret m-values or values of μ23 and obtain α-values, but the SPM is not otherwise needed for this more empirical analysis. To interpret α values and obtain solute partition coefficients (Kp) using the SPM-based analysis, it is necessary to determine the hydration b1 of each functional group and type of surface. Lower bound values of the hydration b1 of some functional groups and surfaces can be determined by assuming complete exclusion (Kp = 0) of the solute or salt exhibiting the largest positive α-value (cf. Eq. 8, 9, 12,13). Na2SO4 and sucrose have the most positive surface tension increments and surface α-values, indicating that Na+, SO42− and sucrose are the most highly excluded cation, anion and nonelectrolyte from the air-water interface. Na2SO4 has the largest unfavorable effect of any salt on solubility of benzene and toluene and the largest aromatic hydrocarbon α-value33, indicating that Na+ and SO42− are the most highly excluded cation and anion from aromatic hydrocarbon surface. α-values for interactions of the excluded solute glycine betaine with anionic and amide O surface are more positive than for interactions of GB with any other functional group12.
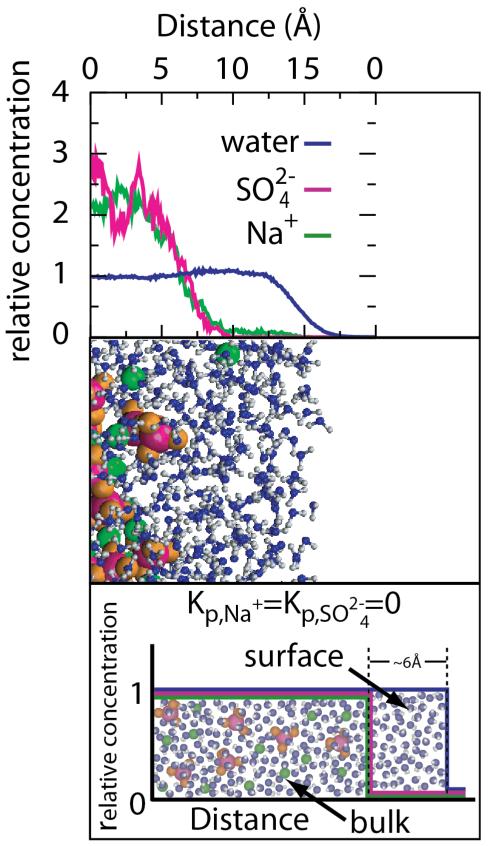
From Na2SO4-aromatic hydrocarbon solubility m-values and Na2SO4 and sucrose STI values, analyzed using Eqs. 9, 12, and 13, lower bound values of b1 of 0.18 H2O/A2 to 0.20 H2O/A2 are obtained for the amount of local water at the molecular surface of aromatic hydrocarbons in water and at the air-water interface. This experimentally-derived lower bound on the amount of local (hydration or surface) water corresponds to two layers of water at bulk density, an eminently reasonable result. MD simulations predict complete exclusion of Na+ and SO42− ions of Na2SO4 from approximately this amount of water at the air-water interface (see Figure 12 below)52, 53. Recent spectroscopic studies show that the effect of solutes and salt ions on water structure is confined to the first one or two layers of water of hydration of the solute or salt ion28. From analysis of the GB data, similar lower bounds on b1 are obtained for anionic carboxylate and amide O groups, though anionic phosphate O may be more hydrated. We therefore use a hydration b1 = 0.18 H2O/A2 to calculate solute partition coefficients Kp for all types of protein surface. Recent MD simulations of water at the surface of the villin headpiece subdomain (HP-36) conclude that the thickness of “biological water” is uniform for different segments of the protein (~5Å)54.
Figure 12.
Ion distributions (panel A) and molecular snapshot (panel B) of the air–water interface from molecular dynamics (MD) simulations of Gopalakrishnan et al52. Relative Concentration means the local concentration at each point of water and ions relative to the average concentration for the entire simulation cell. The complete exclusion of Na+ and SO2−4 from the surface water in panels A and B agrees well with the treatment of Na2SO4 in the SPM as a completely excluded reference salt (panel C). Adapted with permission from Pegram and Record34.
C) Why Glycine Betaine(GB) is a strong osmoprotectant and protein stabilizer while Urea is a protein denaturant: opposite preferential interactions with key protein functional groups
Glycine betaine and urea have opposite effects on protein self-assembly processes (cf Figure 1). Addition of GB favors the assembly direction (e.g. folding (e.g. Figure 3), precipitation 55, which removes protein surface from water, while urea favors disassembly (unfolding (e.g. Figure 3), dissolving2 which exposes protein surface to water. But GB does not drive nucleic acid helix formation, having little or no effect on stability of AT-rich DNA and destabilizing GC-rich DNA14-16, 18, while urea is destabilizing for all base compositions17, 56, 57. GB is an exceptionally good solute to reduce water activity and boost osmolality in vivo 58, in concentrated protein or nucleic acid solutions 14, 15, 59, and more generally in vitro 12, while urea is not15, 20. What are the thermodynamic and molecular origins of these very different interactions of GB and urea with biopolymers, and very different effects of GB and urea on protein and nucleic acid processes? What information is needed to predict or interpret GB and urea m-values (i.e. differences in μ23; Eq 2) for protein processes?
2) Obtaining μ23 Values For Interactions of Urea and GB with Functional Groups of Proteins by Osmometry and Solubility Assays
To determine the interactions of a solute with the functional groups and types of surface of proteins and nucleic acids using the analysis of section III above requires measurement of the preferential interactions of these solutes with model compounds displaying various combinations of these functional groups and surfaces. For proteins, these types of surface include: nonpolar (aliphatic, aromatic) hydrocarbon (C), polar (amide, hydroxyl) and anionic (carboxylate) oxygen (O), and polar (amide) and cationic (ammonium, guanidinium) nitrogen (N).
Preferential interactions of small solutes like GB and urea with these functional groups of proteins are short-range (noncoulombic) and relatively weak, because they are competitive interactions, in which the solute and the functional group can interact with water or with each other. A reasonable starting point, motivated by the data, is to assume that the free energy consequences of these preferential interactions are additive, and that the preferential interaction of GB, urea or any other small solute or salt ion with a protein surface can be built up as the sum of its interactions with individual functional groups on that surface, quantified using model compound data (cf. Eq 16). Large datasets (μ23 values) quantifying the interaction of urea and GB with model compounds displaying one or more protein functional group have been obtained by VPO, solubility, and phase partitioning studies, and deduced (for hydrocarbon surfaces) from micelle formation assays8, 12.
Figure 9 shows VPO data for the interaction of urea and GB with sufficiently soluble model compounds plotted as ΔOsm vs m2m3 as in Fig 8B. (From Eq 7, the slopes of these plots are μ23/RT.) Addition of urea lowers the chemical potential of all nonelectrolyte model compounds studied (i.e. μ23 < 0; Fig 9A panels a and b), indicating that urea interacts favorably with most protein functional groups. Interactions of urea with Na+ and K+ salts of organic ions (Fig 9A panels c and d) are less favorable, indicating most simply that preferential interactions of urea with these cations are unfavorable. On the other hand GB raises the chemical potential of most model compounds studied (both nonelectrolytes and electrolytes; Fig 9B), indicating that preferential interactions of GB with most protein surface types are unfavorable. GB exhibits favorable preferential interactions only with NaBenzoate, NaCl, and ArgHCl, indicating most simply that at least its interactions with aromatic hydrocarbon surface and Cl− are favorable. The VPO data sets for interactions of urea and GB with nonelectrolyte solutes in Fig 9 clearly demonstrate the same trends as those observed for the effects of these solutes on biopolymer processes (see Fig. 2). For both urea and GB, VPO data sets were augmented by solubility determinations of μ23 for sparingly soluble model compounds8, 12.
Figure 9.
Interactions of Urea (panel A) and GB (panel B) with Model Compounds. Excess osmolalities ΔOsm (Eq. 7) determined by VPO at 25°C are plotted vs m2m3, the product of molal concentrations of the model compound and urea or GB. Slopes are μ23/RT. VPO data for nonelectrolytes are grouped in panels (a) and (b) for urea and panel (a) for GB; the other panels show VPO data for salts. Adapted with permission from Guinn et al8, Capp et al12.
3) Analysis of Urea- and GB-Model Compound μ23 Values into Interactions with Individual Functional Groups of Proteins
Values of μ23 for interactions of urea and GB with model compounds were dissected, using Equation 16, into individual contributions from interactions of these solutes with each model compound functional group and inorganic ion. The resulting αi and βion values are shown in Table 1. Since the model compounds studied differ greatly in surface composition, and the number analyzed (45 for urea, 27 for GB) greatly exceeds the number of αi and βion values fitted for, these values are well-determined. When these αi and βion values are used to predict μ23/RT values for interactions of urea and GB with all the model compounds in the training set, very good agreement with the experimental values is obtained8, 12.
Table 1. Interactions of Urea and Glycine Betaine with Functional Groups of Proteins (αi and Kp values)a and Inorganic Ions (βion values)b.
| 103αi (m−1 Å−2) |
SPM Kp |
|||
|---|---|---|---|---|
| Surface Type (i) | Urea | GB | Urea | GB |
| Aromatic C | −0.89 ± 0.05 | −2.3 ± 0.4 | 1.28 ± 0.02 | 1.62 ± 0.11 |
| Amide O | −0.87 ± 0.18 | 2.8 ± 1.0 | 1.28 ± 0.06 | 0.24 ± 0.27 |
| Carboxylate O | −0.40 ± 0.15 | 2.9 ± 0.2 | 1.13 ± 0.05 | 0.22 ± 0.06 |
| Amide N | −0.32 ± 0.23 | −2.0 ± 0.7 | 1.10 ± 0.07 | 1.54 ± 0.19 |
| Hydroxyl O | −0.25 ± 0.06 | 0.1 ± 0.2 | 1.08 ± 0.02 | 0.97 ± 0.06 |
| Aliphatic C | −0.11 ± 0.05 | 0.3 ± 0.3 | 1.03 ± 0.02 | 0.92 ± 0.08 |
| Cationic N | 0.18 ± 0.16 | −1.2 ± 0.4 | 0.94 ± 0.05 | 1.32 ± 0.11 |
| Inorganic Ion | βion (m−1)c | |||
|
|
||||
| Na+ | 0.104 ± 0.013 | 0 | ||
| K+ | 0.146 ± 0.014 | 0.05 ± 0.02 | ||
| Cl− | −0.170 ± 0.021 | −0.04 ± 0.04 | ||
Using the SPM-based molecular thermodynamic interpretation of α values (Eq 15), and b1 = 0.18 H2O/Å2 (as discussed above), local-bulk partition coefficients Kp are obtained for the distribution of urea and GB between each functional group or surface type and bulk water. Comparison of these partition coefficients (Kp) for urea and GB (Table 1) provides molecular insight into the competitive interactions involving water, solute and model compound functional group that determine these Kp and α values. For polar functional groups, hydrogen bonding interactions appear most important; urea and GB differ greatly in their interactions with these O and N moieties because urea has multiple hydrogen bond donors and acceptors while GB has only hydrogen bond acceptors 8, 12.
Table 1 quantifies the extent to which the functional groups of proteins prefer to interact with urea and GB, as compared to interactions with water. The strongest preferential interactions of urea are favorable interactions with amide O and aromatic C; interactions of urea with anionic and hydroxyl O, amide N and aliphatic C are also favorable but significantly weaker, while the urea-cationic N interaction is unfavorable. Urea accumulates to a moderate extent in the vicinity of both amide O and aromatic C (local concentration ~28% greater than bulk) and to a lesser extent at anionic O (~13% above bulk), amide N (~10% greater than bulk) and hydroxyl O (~8% greater than bulk). Very modest accumulation of urea is observed at aliphatic C (only ~3% greater than bulk). Cationic N surface (and Na+ and K+ ions) prefer to interact with water than with urea; as a result the local concentration of urea in the vicinity of cationic N is 6% less than its bulk concentration. Cl−, on the other hand, exhibits a favorable preferential interaction with urea. The favorable or unfavorable character of the urea-ion interaction is consistent with expectation based on the surface composition of urea (~ 2/3 partially-positive amide N, ~ 1/3 partially-negative amide O).
By far the strongest preferential interactions of GB are its highly unfavorable interactions with anionic and amide O. As discussed above, GB is highly excluded from two layers of water at the surface of these oxygens (local concentration less than 25% of the bulk GB concentration). GB is weakly excluded from aliphatic C (local concentration ~8% less than bulk) and hydroxyl O (local concentration ~3% less than bulk). For urea, all four of these interactions are favorable. GB accumulates to a very significant extent in the vicinity of aromatic C and amide and cationic N (local concentrations 62%, 54% and 32% greater than bulk). Comparison of βion values shows that net interactions of GB with inorganic ions (Na+, K+, Cl−) are relatively weak by comparison with urea-ion interactions.
4) Molecular Interpretation of Kp values for Urea and GB Accumulation or Exclusion In the Vicinity of Functional Groups of Proteins
A “two-way” dissection (see Eq 17) of μ23 values for interactions of urea and GB with model compounds into contributions from interactions of individual functional groups on both solutes has not yet been accomplished. Nevertheless a qualitative interpretation of Kp values for interactions of urea and GB with the polar and charged functional groups of proteins is possible, leading to very reasonable deductions about the competitive hydrogen bonding capabilities of these solutes and water. For example, the favorable interactions of urea with amide O and anionic O indicate that the hydrogen bonds formed with amide-NH2 groups of urea as the donor and these oxygens as acceptor are more favorable (in part because there are more ways of forming them) than the corresponding hydrogen bonds with water, resulting in accumulation of urea at these groups. GB, on the other hand, lacks a hydrogen bond donor and so has no way to compete with water to interact with these amide and anionic oxygens, which therefore prefer to remain hydrated, largely excluding GB.
The observed accumulation of GB in the vicinity of amide and cationic N indicates that the anionic carboxyl oxygens of GB are a better acceptor of hydrogen bonds from these nitrogens than is the polar oxygen of water. Urea is much less accumulated at amide N than at amide O and is somewhat excluded from cationic N. These results indicate that while urea–NH….O=C-amide hydrogen bonds are quite favorable, relative to hydrogen bonds to water, urea–NH….N-amide hydrogen bonds are unfavorable relative to interactions with water. Amide hydrogen bonds in α-helices and β-sheets are of course –NH….O=C-. The shift from a modest favorable interaction of urea with amide N to a slightly unfavorable interaction of urea with cationic N indicates that protonating the nitrogen disfavors urea–NH….+NH (cationic N) hydrogen bonds (and eliminates the possibility of hydrogen bonds where urea N is the donor) by more than it favors (cationic N)+NH…O=C(urea) hydrogen bonds.
Both urea and GB are accumulated at aromatic C; for both solutes this interaction is the strongest favorable preferential interaction. For GB, this quite favorable interaction is almost certainly a cation-π interaction of the trimethyl ammonium group of GB with the aromatic ring12. The more modest accumulation of urea at aromatic surface presumably arises from hydrogen bonding or partial-cation π between amide N groups on urea and the aromatic π-system, and/or π-π stacking interactions between the π-system or urea and the ring60. Urea and GB exhibit weak favorable and unfavorable interactions with aliphatic C. The interaction of water with both aromatic and aliphatic C surface is unfavorable, relative to water-water interactions (the hydrophobic effect), and the interactions of urea and GB with these hydrocarbon surfaces need to be interpreted relative to that interaction.
5) Application of Interaction Potentials (α values) or Kp values to Interpret and Predict Effects of Urea and GB on Protein Processes (Folding, DNA Binding)
Interaction potentials (α values) or Kp values in Table 1 allow one to interpret and predict urea and GB m-values for steps of protein processes in terms of the amount and composition of the surface (ΔASA) involved in the process. Values of μ23 for interactions of urea with folded and unfolded protein surfaces can also be predicted from ASA information. Because urea has been widely used to determine protein stability by extrapolation to zero urea concentration, a large collection of urea m-values is available for analysis13, 61.
a) Protein Folding
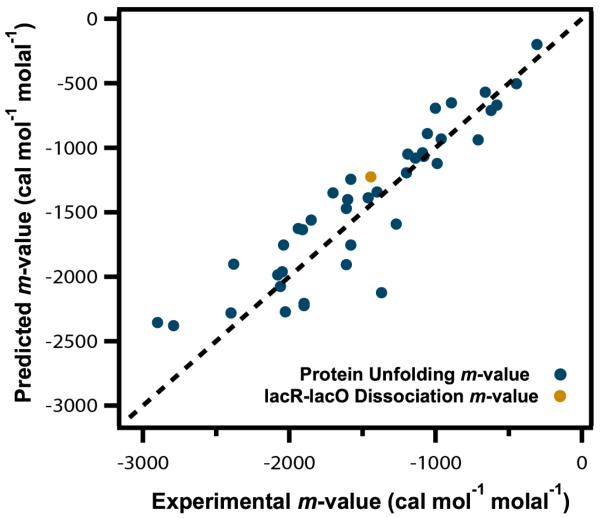
Using the α values in Table 1 and ΔASA values calculated using an extended chain model, we predicted m-values for unfolding a set of well-characterized globular proteins and for dissociation of lac repressor from lac operator DNA and found good agreement between predicted and experimental values (Figure 10). Since these unfolding processes form a homologous series with similar compositions of the protein surface exposed in unfolding and differing primarily in the total ASA exposed, a systematic offset of predicted from experimental m-values would indicate a systematic error in the ASA analysis or an error in one or more of the α values, but no such systematic deviation is observed, and the scatter of the data is about the line is similar to that observed in Figure 3. Only one isothermal GB m-value has been determined; for unfolding of the lac DNA binding domain (Figure 2) the experimental m-value is 790 cal mol−1 molal−1 at 59°C, similar to the predicted m-value of 690 cal mol−1 molal−1 at 25°C. While this 12% difference between predicted and experimental m-values is not outside of the uncertainty in both values, it is interesting to observe that predicted and observed values of the quantity m-value/RT differ by only 3%. If the stabilizing effect of GB were entirely entropic, as would be predicted to be the case for a completely excluded solute, then m-value/RT would be independent of temperature while the m-value itself would be more temperature dependent.
Figure 10.
Predicting Urea m-Values. Comparison of predicted and experimental m-values for effects of urea on protein unfolding and dissociation of lac repressor from operator DNA. Predicted values calculated from Eq 14 using urea α-values (Table 1) and published amounts and composition of the ΔASA8, 12. Dashed line represents equality of predicted and experimental values. Experimental unfolding m-values are from Fig 7 and lacR-lacO dissociation m-value is from Fig 11. Predicted protein unfolding m-values are from Guinn et al8 and predicted lacR-lacO m-value is from Fig 1112, 61.
b) Protein Interface Formation and Coupled Conformational Changes
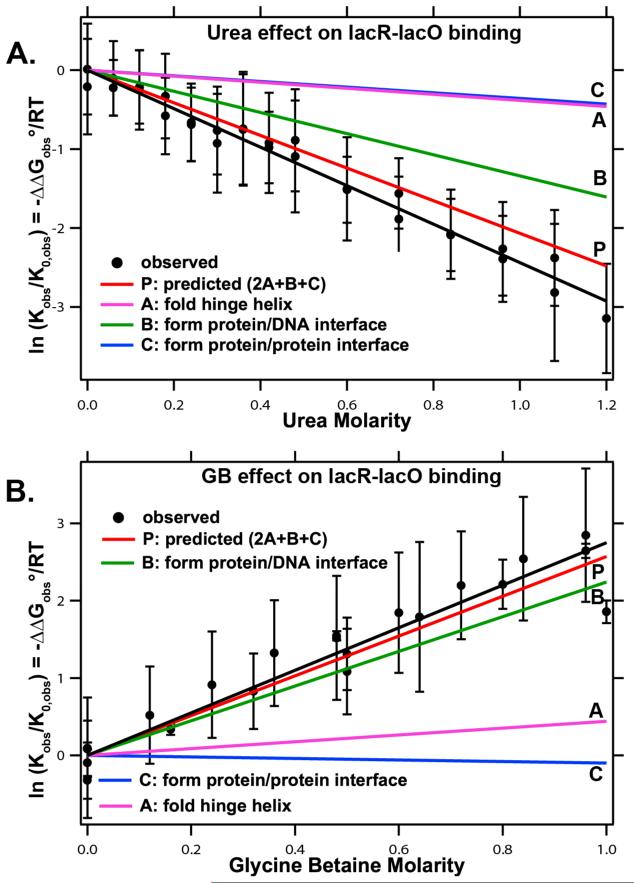
The α-values in Table 1 are very useful to predict and interpret effects of interface formation and coupled conformational changes in protein processes. Effects of urea and GB on binding of lac repressor tetramer to 40 bp lac operator DNA are shown in Figure 11. Logarithms of normalized binding constants (lnKobs/Kobs,0), determined from urea and GB titrations of poised lac repressor-lac operator mixtures, are plotted as functions of urea (A) and GB (B) concentration at 25°C. In this binding interaction, not only is an extensive interface formed between two DBD of lac repressor and 20 bp of operator DNA, but also two protein interfaces form as the two flexible hinge regions that tether the DBD to core repressor fold and the DBD-operator assembly then interacts with the core repressor61-63. Formation of these three interfaces buries 6900 Å2 of protein and DNA surface12. From structural data on the amount and composition of the protein and DNA surfaces buried, we predict an overall urea m-value = 1.24±0.19 kcal mol−1 m−1 and GB m-value = 1.54±0.84 kcal mol−1 m−1, in agreement within the experimental uncertainty with experimental values of 1.42±0.21 kcal mol−1 m−1 and 1.66±0.24 kcal mol−1 m−1 respectively (Figure 11). (The large uncertainty in the GB prediction reflects the uncertainties in the GB α values for aliphatic C and amide O (Table 1). Investigation of interactions of GB with additional model compounds will reduce these uncertainties.)
Figure 11.
Effect of Urea and GB on Binding of lac Repressor Protein to lac Operator DNA. Comparison of observed and predicted effects of urea (panel A) and GB (panel B) on the lac repressor-lac operator binding constant Kobs relative to its value Ko in the absence of solute. (Experiments performed at constant salt molality at 25°C.) Predicted contributions to the dependence of ln (Kobs/Ko) on solute concentration from the three interfaces formed during this assembly process are also shown. Experimental lacR-lacO binding data in Panel A from unpublished data (M. W. Capp); binding data for panel B and ASA calculations are from Capp et al12.
Formation of the repressor-operator interface buries 630Å2 of anionic DNA phosphate O surface, at which urea accumulates strongly and from which GB is strongly excluded12, 56. Formation of this interface is predicted to contribute −1.3 m−1 (62%) to the urea m-value and 2.2 m−1 (85%) to the GB m-value. Folding the hinge helices, which buries 520 Å2 of amide surface, is predicted to contribute −0.4 m−1 (19%) to the urea m-value and 0.4 m−1 (15%) to the GB m-value. Formation of the core repressor-DBD interface is predicted to contribute −0.4 m−1 (19%) to the urea m-value and to make no significant contribution to the GB m-value as a result of relatively small and compensating effects (Figure 11). Therefore, while urea and GB are predicted to detect primarily lacR-lacO interface formation, formation of the two protein-protein lacR interfaces also contributes significantly (38%) to the urea m-value.
c) Predicting the Most Important Interactions Responsible for Urea and GB Effects
Dissection by functional group of the effects of urea and GB on different protein processes shows quantitatively why urea is a denaturant and GB is a stabilizer. Table 2 shows predicted contributions of different surface types to urea and GB m-values for unfolding a globular protein, melting an α-helix (which exposes a higher proportion of amide ASA) and for the interaction with native protein surface. All values in the table are for 1000 Å2 of ASA.
Table 2. Predicted contributions to an m-value or μ23 at 25°C (cal mol−1 molar−1) for exposure of 1000 Å2 of Protein Surfacea.
| Surface Type (i) | Process |
Surface of Native Proteinb | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Globular Protein Unfolding | α helix melting | ||||||||
| ΔASA | Urea | GB | ΔASA | Urea | GB | ASA | Urea | GB | |
| Amide O | 120 | −62 | 199 | 480 | −247 | 796 | 100 | −52 | 166 |
| Carboxylate O | 40 | −9 | 69 | 100 | −24 | 172 | 160 | −38 | 275 |
| Hydroxyl O | 30 | −4 | 2 | 0 | 0 | 0 | 20 | −3 | 1 |
| Amide N | 50 | −9 | −59 | 90 | −17 | −107 | 40 | −8 | −47 |
| Cationic N | 40 | 4 | −28 | 0 | 0 | 0 | 130 | 14 | −92 |
| Aliphatic C | 650 | −42 | 115 | 330 | −21 | 59 | 530 | −35 | 94 |
| Aromatic C | 70 | −37 | −95 | 0 | 0 | 0 | 20 | −11 | −27 |
|
| |||||||||
| Predicted m-value or μ23 per 1000 Å2 of surface: |
−159 | 203 | −309 | 920 | −133 | 370 | |||
| Experimentally derived value: | − 184c |
224d | −598 | ND | −124 | 776 | |||
Guinn et al8
From osmometric data for interaction of solutes with native BSA14, 20. BSA is anionic with a net charge of approximately −17 at the pH of these experiments74, so the BSA component includes at least the corresponding number of Na+ ions. Interactions of solutes with these cations are not included in the predictions. Inclusion of these interactions would not significantly affect the predicted GB μ23 value, but would make the urea μ23 value less favorable (Table 1).
Urea globular protein m-value from average of Fig. 7 dataset for urea scaled to 1000Å2.
GB globular protein m-value from lacDBD data in Fig 2 scaled to 1000Å2. Data taken at 59°C but prediction determined from 25°C αi values.
For protein unfolding, interactions of urea and GB with amide O, aliphatic C and aromatic C surface exposed in unfolding are predicted to make the largest contributions to the m-value (Table 2). Interactions of urea with amide O and aromatic C are similarly favorable and moderately strong: because the ΔASA of amide O significantly exceeds that of aromatic C in unfolding, amide O is predicted to contribute ~40% and aromatic C ~20% of the unfolding m-value. While the α value for urea and aliphatic C is only marginally different from zero, its contribution to the ΔASA is largest and this interaction is predicted to contribute ~25% of the m-value. GB has both favorable and unfavorable interactions with protein functional groups; the net interaction with both folded and unfolded protein surface is predicted to be unfavorable, resulting in exclusion of GB. For unfolding, the unfavorable interaction of GB with amide O is predicted to be approximately as large as the overall GB m-value. The next two largest contributions are predicted to be from interactions of GB with aliphatic C and aromatic C surface; these are about equal in magnitude and opposite in sign.
For melting of a series of α-helices, urea m-values appear to be proportional to ΔASA, but with a very different proportionality constant than that observed for unfolding of globular proteins; the urea m-value/ΔASA ratio for α-helix melting20, 64 is more than three times larger than for unfolding of globular proteins. Using the α values of Table 1, the m-value/ΔASA ratio for α-helix melting is predicted to be twice as large as for unfolding of globular proteins (Table 2), in semi-quantitative agreement with experiment. The predicted GB m-value for α-helix melting is also significantly larger (~4.4X) than the predicted globular protein unfolding m-value; data is not yet available to test this prediction. Table 2 shows that these differences arise primarily from the much larger proportion of amide ASA exposed in α-helix melting, and the strong favorable (urea) and unfavorable (GB) interactions with amide O surface, as previously proposed20.
Native protein surface displays a smaller proportion of hydrocarbon surface (55% of total) than is exposed in protein unfolding (72%) and so displays a larger proportion of polar and charged surface. GB interacts unfavorably with native protein surface due to its significant unfavorable interaction with polar and anionic oxygen surface, making GB a good osmoprotectant because its chemical potential is raised in the presence of protein surface increasing its contribution to the solution’s osmolality. Urea on the other hand interacts favorably with native protein surface due to its favorable interaction with most surface types and so will be a poor osmoprotectant because its osmolality will be lowered in the presence of protein surface. However, we predict urea to be an effective protein solubilizer because its favorable interactions with native protein surface will increase protein solubility in water (GB’s unfavorable interactions will lower protein solubility).
6) Comparison with other Approaches
a) Transfer Free Energy Analysis
In the transfer free energy analysis of Auton and Bolen39, 65, preferential interactions of urea and other solutes with amino acids and small peptides were determined from solubility measurements and dissected to quantify transfer free energies of the 20 amino acid side chains and the peptide backbone unit from water to one molar solute, closely related to μ23 values. From these data they concluded that urea denatures proteins primarily because of its favorable interaction with the peptide backbone. On the other hand, from the SPM-based α value analysis described above, Guinn et al8 find that favorable interactions of urea with amide O (mostly backbone), aliphatic C (mostly side chain) and aromatic C (all side chain) make the most significant contributions. Predictions of the transfer free energy analysis agree well with m-values for urea and other solutes for protein unfolding and other protein processes40. In the transfer free energy analysis of protein unfolding, the average accessibility of backbone and individual side chains for two models (extended, more compact) of the unfolded state is used to calculate the ΔASA; Guinn et al used an extended chain model of the unfolded state. Left to be resolved are some significant differences between values of μ23 for interactions of urea with the subset of five amino acids determined by osmometry and those calculated from the solubility data of ref 39. In addition, values of μ23 predicted for interactions of urea from the best fit set of model compound α values disagree significantly with those calculated from the solubility data of ref 39 for 10 of the 18 amino acids and the cyclic dipeptide investigated8. By contrast predicted values of μ23 for amino acids and the cyclic dipeptide agree in 9 of 11 cases with experimental values of μ23 from osmometry and solubility in the data set of ref 8.
b) Molecular Dynamics Simulations
While preferential interactions represent the strength of solute interactions with biopolymer functional groups relative to water they do not give the molecular origin of these interactions. Insight into the noncovalent interactions between solutes and biopolymers is provided by a statistical thermodynamic analysis (most simply the SPM, more generally Kirkwood Buff (KB) integrals). A Kirkwood-Buff integral relates the radial distribution function of species 2 around species 3 to the chemical potential derivative μ2345. Hence radial distributions of one solute in the vicinity of another predicted by simulation can be compared with values of μ23 from thermodynamic experiments by Kirkwood Buff integrals.
Because solute effects are relatively subtle, MD simulation parameters must be carefully chosen to properly describe them. In fact, the standard Charmm force field actually predicts accumulation of GB at triglycine surface where it should be excluded 66. In recent studies, Smith et al. have successfully applied atomistic simulations to obtain KB integrals for a number of binary and ternary solution systems; they have also used the KB integrals to improve atomistic force fields for small molecules such as urea44.
Using the KB force field, simulations of urea with the trp cage miniprotein67 and polyglycine68 indicate that urea is significantly hydrogen bonded to the protein backbone and that the number of hydrogen bonds to the backbone increases with urea concentration. Horinek et al computed that in 60% of hydrogen bonds with polyglycine urea is acting as a donor, presumably to amide O, consistent with the relatively strong favorable preferential interaction of urea with amide O we observe. Of the computed 40% of hydrogen bonds where urea acts as an acceptor, urea O is the only acceptor seen supporting our interpretation that amide NH-O hydrogen bonds are favorable while NH-N bonds are not8, 68. Urea interactions with hydrocarbon are also found to be significant in simulations since interactions with trp cage side chains, which contain significant aromatic and aliphatic hydrocarbon surface, contribute 60% to the urea m-value67 (compared to 55% predicted from our preferential interaction data). Simulations of urea’s interaction with an RNA hairpin show that urea destabilizes through hydrogen bonds to the bases as well as π-π stacking interactions with the heterocyclic nucleic acid ring indicating urea could be stacking on protein aromatic rings as well60.
c) Spectroscopic Studies of Urea-Amide Interactions
Evidence for hydrogen bonds of urea with amide groups has been obtained by FTIR and NMR spectroscopy. Sagle et al69 used FTIR to study urea’s hydrogen bonding to the amide O of Poly(N-isopropylacrylamide) (PNIPAM). When urea is added, the amide I absorption band shifts to a frequency indicative of CO-HN hydrogen bonding and collapsed state of the polymer is destabilized, presumably because urea hydrogen bonds to PNIPAM better than water does 69. These spectroscopic measurements were used to calculate the fraction θ of PNIPAM amide O hydrogen bonded to urea as a function of urea concentration. This fraction θ can be predicted from the SPM Kp value8:
The fraction of PNIPAM amide O that are hydrogen bonded to urea agrees with the SPM prediction within error below 4 molar urea.
Lim et al studied the effect of urea, GuHCl and polyols on acid and base catalyzed amide hydrogen exchange (HX) using 1D NMR spectroscopy. They observed that urea slows the rate of amide HX of the diamide aAma, presumably by hydrogen bonding with amide N and so inhibiting HX. However, GuHCl had very little effect on the HX rate. Analysis of model compound solubility data (figX) shows that GuH+ accumulates at amide surface. From the discussion above, this interaction appears to involve hydrogen bonding of GuH+ to amide O and not to amide N. The absence of an effect of GuH+ on exchange of the amide N is consistent with this interpretation. The polyols sorbitol and glycerol also do not affect the HX rate, indicating that they are excluded from amide surface70.
D) Molecular Interactions Responsible for the Hofmeister Anion and Cation Series and their Different Null Points for Different Aqueous Processes
What is the molecular basis of the Hofmeister series of cations and anions observed for such diverse processes in aqueous solution as unfolding proteins, dissolving hydrocarbons or proteins, and transferring water from bulk to the air-water interface? Do the Hofmeister ion series of Figure 1 primarily reflect the very different tendencies of different ions to interact with a macroscopic or molecular nonpolar surface in water (classical refs) or are other types of surface or combinations of surfaces (like the methylene and amide groups of the peptide backbone refs) involved? Why are the null points of these series different for model processes like increasing the amount of air-water surface or dissolving hydrocarbons than for protein unfolding and other protein processes, and different yet again for nucleic acid melting? For protein, nucleic acid and other polyampholyte or polyelectrolyte processes, when and to what extent can Coulombic and Hofmeister effects of salts be separated? For processes involving polyions, how can amplified or reverse Hofmeister effects of counterions at low salt be explained?
1) Analysis of μ23 Values and Surface Tension Increments for Interactions of Hofmeister Salts with Molecular and Macroscopic Surfaces to Obtain Single Ion Kp Values
Analysis of the extensive literature data for surface tension increments (STI; i.e. air-water surface free energy m-values per unit area) and hydrocarbon solubility free energy increments (i.e. solubility m-values for hydrocarbons), like those displayed in Fig 5A and 5B, using an SPM-based analysis (Eqs 10 and 11)31, 33, 41, 51 provides answers to the above questions, and provides a quantitative database to predict and interpret Hofmeister salt effects. For these homogenous nonpolar surfaces, interaction potentials (α) quantifying the preferential interaction of the salt component (the composite effect of the salt cation and anion) per unit area of the surface are obtained from (STI)/kBT for air-water surface, where kB is Boltzmann’s constant, and from μ23/ASA for molecular hydrocarbon surface. These are listed in Table 3 for the complete set of Na+ salts where both types of data (STI and solubility increments) are available, and for a few other cases of interest. Table 3 shows that the rank orders of salt interaction potentials for these two nonpolar surfaces are quite similar, but that interaction potentials for the air-water surface are systematically larger (less favorable preferential interaction of the salt) for air-water than for aromatic hydrocarbon-water surface.
Table 3. Hofmeister Salts: Comparison of Interaction Potentials (αi) for Molecular Hydrocarbon Surfaces with Reduced Surface Tension Increments (STI/kT).
| αi×103 (m−1Å−1)a |
||||
|---|---|---|---|---|
| Salt | STI/kT×103 (m−1Å−1) | AIiphatic C | Aromatic C | Amide (O,N) |
| Na2SO4 | 6.7±0.1 | 6.0±0.1 | 5.9±0.2 | −6.8±0.5 |
| (NH4)2SO4 | 5.6±0.3 | -- | 3.8±0.1 | −3.3±0.3 |
| KF | 4.4±0.3 | 3.4±0.1 | 2.4±0.1 | −4.6±0.3 |
| GuH2SO4 | 2.3±0.5 | 2.2±0.3 | 0.3±0.5 | −5.8±1.0 |
| NaCI | 4.2±0.4 | 2.2±0.1 | 2.0±0.1 | −4.2±0.2 |
| KCI | 3.9±0.3 | 2.0±0.1 | 1.7±0.1 | −4.2±0.2 |
| KBr | 3.3±0.1 | 1.7±0.1 | 1.2±0.2 | −4.1±0.4 |
| NaCIO4 | 0.5±0.1 | 0.9±0.1 | 1.1±0.1 | −7.0±0.3 |
| NH4CI | 3.4±0.4 | -- | 0.8±0.3 | −1.2±0.8 |
| GuHCI | 1.8±0.6 | 0.3±0.1 | −0.8±0.2 | −3.7±0.4 |
αi, values determined at 25-30°C
These trends of surface interaction potentials are clarified when interpreted to obtain microscopic local-bulk partition coefficients (Kp values) obtained from a SPM-based analysis. To determine Kp values requires a determination of the amount of local water b1 per unit surface area obtained from the STI or solubility increment for a completely excluded salt. MD simulations indicate that Na2SO4 is completely excluded from the air-water interface, unlike other salts and acids52, 53. Fig. 12 shows distributions of Na+ and SO42− near the air-water interface predicted from molecular dynamics simulations52. Both Na+ and SO42− are completely excluded from a surface region of comparable thickness (5-10 Å) to that deduced from the SPM-based analysis34, 41. This supports the assumption that Na2SO4, the salt with the largest positive STI and largest effect on hydrocarbon solubility, is completely excluded (Kp = 0) from the local water at both these surfaces. As discussed earlier, this assumption yields b1 = 0.18 ± 0.01 H2O/A2, corresponding to approximately two layers of water at bulk density. If Na2SO4 were not completely excluded (Kp > 0), then b1 for these surfaces would of necessity be larger than 0.18 H2O/A2. We assume that b1 = 0.18 H2O/A2 for other molecular surfaces; at present anionic phosphate oxygen appears to be the only surface where more than two layers of local water are present12
Table 4 and Figure 14 summarize partition coefficients Kp for individual salt ions at the nonpolar air-water surface and at the molecular surface of aromatic and aliphatic hydrocarbons in water. To within the experimental uncertainty, these single-ion Kp values for both cations and anions are independent of salt concentration up to approximately 1 molal or higher. Values of Kp for cation and anion are additive, as predicted by Eq. 9, and are independent of the salt investigated. For both the air-water nonpolar surface and the molecular nonpolar surface of aromatic hydrocarbons (and aliphatic hydrocarbons, where data is available), these Kp values define cation and anion series which closely follow the cation and anion Hofmeister series for protein processes listed in Fig 1. However, the null points (Kp = 1) for both cations and anions differ for the different surfaces.
Table 4. SPM partition coefficients (Kp=m3loc/m3bulk)a of Individual Hofmeister cations and anionsb.
| Cation | Kpc | Anion | Kpc | ||||
|---|---|---|---|---|---|---|---|
| Air/Water | Aromatic | Aliphatic | Air/Water | Aromatic | Aliphatic | ||
|
|
|
||||||
| GuH+ | 0.67 | 0.99 | 0.70 | SCN− | 1.64 | 1.80 | -- |
| NH4+ | 0.25 | 0.49 | 0.28 | I− | 1.18 | 1.62 | 1.54 |
| Li+ | 0.08 | 0.28 | 0.23 | ClO4− | 1.77 | 1.54 | -- |
| K+ | 0.12 | 0.11 | 0.08 | NO3− | 0.98 | 1.47 | -- |
| Na+ | 0.00 | 0.03 | -- | Br− | 0.86 | 1.40 | -- |
| Cl− | 0.69 | 1.23 | 1.23 | ||||
| H − | 0.53 | 0.92 | -- | ||||
| SO4− | 0.00 | 0.00 | -- | ||||
| H+ | 1.50 | 0.61 | 0.59 | OH− | 0.58 | 1.03 | -- |
| Acetate- | 1.30 | -- | -- | ||||
Figure 14.
Single Ion Partition Coefficients for Hofmeister Cations and Anions at Nonpolar and Polar Surfaces. (A) Rank orders of cation and anion partition coefficients Kp = mionlocal/mionbulk quantifying local accumulation (Kp > 1) or exclusion (Kp < 1) of these ions in the vicinity of the macroscopic air-water surface and the molecular surface of hydrocarbons and amide groups in water. The null point of each series is at Kp = 1. At right is an approximate alignment of the ion series for protein processes (Fig 1) with these Kp series. (B) Projection bar graph displaying Kp values for interactions of key Hofmeister cations and anions with molecular hydrocarbon (red) and amide (blue) surfaces (from panel A left), arranged according to the Hofmeister series order for effects of these ions on protein processes (Fig 1). Both panels adapted with permission from Pegram et al33, 34.
This dissection of individual Hofmeister ion Kp values indicates that all Hofmeister cations are excluded from the air-water interface. (The proton is accumulated but is not part of the Hofmeister cation series for protein processes.) Cations like GuH+ and NH4+, which are at the accumulated, destabilizing end of the protein Hofmeister series, are moderately excluded from the air-water interface. Concentrations of GuH+ and NH4+ in the two layers of water at the air-water surface are predicted to be approximately 67% and 25% as large as their bulk concentrations, respectively; no Hofmeister cation yet investigated is more accumulated at the air water interface than GuH+. Anions from the favorably-interacting (accumulated) end of the protein Hofmeister series (SCN−, I−, ClO4−) also accumulate at the air water interface. Local concentrations of SCN− and ClO4− are predicted to exceed their bulk concentrations by 1.6 and 1.8 fold, respectively, while I− is predicted to be less dramatically accumulated (1.2 fold greater surface concentration compared to bulk). Acetate, which typically ranks with other carboxylates and with F− near the excluded end of the Hofmeister series for protein processes, accumulates at the air water surface. We hypothesized that accumulation of acetate allows its methyl end to be at the surface while the carboxylate end remains in water41.
Hofmeister anions accumulate to a greater extent at the surface of aromatic hydrocarbons (and also aliphatic hydrocarbons, where data is available) in water than at the air-water surface. For interactions with aromatic hydrocarbon surface, the null point of the Hofmeister anion series is F−. This is a very different null point than those observed for air-water (NO3−) and protein (Cl−) surfaces. F− is highly excluded from the air-water surface (Kp = 0.53) and is net excluded from protein surface. For Hofmeister cations, the differences are smaller and may not be as significant as for anions. For interactions with aromatic hydrocarbon surface, GuH+ is at the null point (Kp = 1) of the cation series. Both GuH+ and NH4+ interact more favorably with aromatic than with aliphatic surface. Extents of exclusion of GuH+ and NH4+ from aliphatic and from air-water surface are similar. Since GuH+ and NH4+ are the cationic functional groups of arginine and lysine side chains of proteins, their interactions with aromatic and aliphatic hydrocarbon surface are relevant in many biological contexts. The finding that GuH+ is much less strongly excluded from hydrocarbon surfaces than are alkali metal cations is the principal explanation for why GuHCl is a protein unfolding agent and NaCl or KCl are not, as discussed below.
2) Interactions of Hofmeister salts with amide-containing model compounds: deducing interactions of cations and anions with amide groups
To interpret or predict Hofmeister salt effects on the thermodynamics of protein processes, individual thermodynamic contributions of interactions of anions and cations with amide and other types of polar biopolymer molecular surface as a function of salt concentration are needed. Literature data exists to accomplish part of this goal; more experiments are needed to complete this analysis. Salt effects on solubility of oligopeptides and amide model compounds follow the classical Hofmeister series, but the null point is shifted from that observed for molecular hydrocarbon or air-water surfaces. Figure 13 shows literature data for a series of end-capped oligoglycine peptides (N-acetyl(glycine)nethyl esters, or AGnE, with surface compositions ranging from 13% to 28% amide O,N, 75% to 66% aliphatic C, and 12% to 7% ester O surface as n increases from 1 to 4). While all Na salts reduce the solubility of benzene (Fig 5), salts at the favorably interacting end of the Hofmeister series (e.g. NaClO4, NaSCN) increase the solubility of some of these peptides. In addition, for all salts, the solubility m-value, or μ23, becomes less positive (or more negative) as the amount of amide surface increases (Fig 13). Robinson et al concluded that the amide is “salted in” while hydrocarbon groups are “salted out”, and that these groups contributed additively to the solubility m-value71.
Figure 13.
Hofmeister Salt Effects on Solubility of End-capped Glycine Peptides (NR ref). Values of the logarithm of the relative solubility (ln(S0/S) = ΔΔG°obs/RT) of a series of glycine peptides (acetyl glycine ethyl esters; AGnE with n = 1 to 4) at 25°C plotted as a function of concentration of Na+ salts of anions spanning the Hofmeister series (SO42−, Cl−, ClO4−). S is the solubility of the peptide in the presence of salt and S0 is the solubility of the peptide in the absence of salt. Initial slopes are m-values/RT for the effect of these salts on the peptide transfer process. Figure redrawn from Nandi and Robinson71 by Pegram et al33 and used with permission.
Pegram and Record dissected values of μ23 obtained for each salt and peptide assuming additive contributions from the end groups, methylene carbons and amide groups33. End group contributions could only be assessed semiquantitatively from solubility data on the highly soluble model compound ethyl acetate; errors introduced from use of these data do not appear large, and the contribution of the end groups of course is constant as n is varied in a series of peptides. Partition coefficients Kp quantifying the distribution of the salt component in the vicinity of the amide group were obtained by application of the SPM-based analysis to these data; these are tabulated for the Na-based series of Hofmeister salts and the AGnE series of peptides in Supplemental33.
Separation of salt-interior amide Kp values into single-ion Kp values for the cation and anion of the salt is more ambiguous than for salt-hydrocarbon Kp values, because no salt component or salt ion is completely excluded from the amide group the way that Na2SO4 is completely excluded from aromatic hydrocarbon or air-water surface. To obtain single ion Kp values for amide interactions, use was made of the classical observations that NaCl and KCl typically exhibit little if any Hofmeister (i.e. non-Coulombic) effect on unfolding transitions of globular proteins and that Cl−, Na+, and K+ ions are thus inferred to be centrally positioned in individual anion and cation series derived from unfolding data7, 33. The surface composition of the AG4E peptide is similar to that of the surface exposed in unfolding a globular protein (approximately 2/3 nonpolar C and 1/3 polar N,O). Indeed, for this peptide, overall partition coefficients Kp for NaCl and KCl, calculated using the same hydration b1 = 0.18 H2O/A2 as determined for hydrocarbon and amide O data, are equal and close to unity (Kp,NaCl = Kp, KCl = 0.97).
For this AG4E peptide, overall single ion partition coefficients of 0.97 were assigned to Na+, K+ and Cl−. From this assignment and single ion partition coefficents for hydrocarbon surface (Table 4), estimates of single ion partition coefficients for amide (O,N) surface are obtained. For both cations and anions, these predicted single ion Kp values make good chemical sense, as discussed below. Fig 14a compares single ion partition coefficients for air-water, aromatic hydrocarbon, and amide surface; Fig 14b is a summary bar graph representation which places single ion partition coefficients for hydrocarbon and amide surfaces in the order predicted by the protein Hofmeister series. Both figures show clearly that the rank order of the Hofmeister anion and cation series for protein processes result from the rank order of preferential interactions of these ions with hydrocarbon, not amide surface.
The chart of Kp values in Fig 14a emphasizes that single ion partition coefficients for amide surface are remarkably different for cations and anions, and partition coefficients for all cations and some anions are remarkably different for amide and hydrocarbon surfaces. The SPM-based analysis was required to determine these striking single ion behaviors, which had not been previously appreciated. In particular, all cations investigated (including Na+, K+, GuH+) are highly accumulated at amide surface. Presumably part of the explanation for this is that ~¾ of the surface of an interior amide group (e. g. amides of the peptide backbone) is partially-negative O. Na+ and K+ are highly excluded from hydrocarbon (and air-water) surface, while GuH+ is randomly distributed between aromatic hydrocarbon surface and bulk water and slightly excluded from aliphatic hydrocarbon surface. Indeed, as discussed below, the difference in denaturing abilities of these chloride salts results from the highly unfavorable (protein stabilizing) interaction of Na+ and K+ but not GuH+ with hydrocarbon surface. If the assumption used to generate single ion Kp values for the AG4E peptide were incorrect, this would shift the cation and anion Kp values but would not affect the relative rankings nor the conclusion that cations are highly accumulated and that most anions are excluded from interior amide groups of the peptide backbone of AGnE peptides and proteins.
3) Prediction of Hofmeister Salt Effects on Biopolymer Processes
From these ion partition coefficients for nonpolar and polar amide surfaces, one can now interpret or predict, for the first time, most of the non-Coulombic effect of Hofmeister salts on amide or peptide solubility, protein unfolding, and other protein processes in terms of structural information (the amount and composition of the ΔASA in the process)7, 33. For example, protein unfolding exposes surface (ΔASA) which is ~ 1/3 polar (divided equally between amide groups and other polar or charged surface) and 2/3 nonpolar; therefore, quantitative analysis of the thermodynamic consequences of accumulation (or exclusion) of salt cations and anions near both nonpolar and polar surface is of key importance for the interpretation of Hofmeister salt effects on protein unfolding. Because most anions are excluded to a similar extent from amide surface (Fig. 14), the Hofmeister anion series for unfolding is determined by whether the anion is accumulated (like iodide, perchlorate) or excluded (like fluoride, sulfate) near nonpolar surface. Most cations are excluded from nonpolar surface but strongly accumulated at polar amide surface and as a result are nearly neutral at driving unfolding. Guanidinium cation is much less excluded from nonpolar surface than other cations, but equally accumulated at amide surface; this behavior provides a novel quantitative explanation of its effect as a protein denaturant. The Hofmeister order of anions and cations arises from the extent of accumulation or exclusion of these ions near nonpolar surface. Although interactions of cations and anions with polar amide surface do not exhibit the Hofmeister order, favorable cation-amide interactions are crucial for determining whether a given Hofmeister salt will favor or disfavor a biopolymer or model process in which both nonpolar and amide surface are exposed. For example, DNA duplex melting exposes a surface which is only ~35% hydrocarbon with the rest being primarily polar N and O. The favorable cation interaction with polar surface, and more favorable anion interaction with polar than with hydrocarbon surface, explains the observation (Fig 6) that no Hofmeister salt stabilizes a 42% GC DNA duplex at high salt concentration, resulting in the shifted null point of the DNA Hofmeister series in Fig 1.
To predict or interpret Hofmeister salt effects on processes involving charged biopolymers in an analogous manner to that done for urea and GB, Hofmeister and coulombic effects must be separated. At high salt concentration such a separation appears feasible, and is described for lac DBD and the 12 base pair DNA duplex from Fig 6 in ref 7. Once this has been accomplished using modeling or a simple functional form suggested by Poisson Boltzmann calculations (as in ref 7), the results of ASA calculations on the native and unfolded states can be used (as described above) to make a quantitative comparison of predicted m-values (using the α values for the functional groups of proteins; Table 1) with experimental “Hofmeister m-values”. Predicted and experimental m-values agree well for lacDBD, and appear consistent for the DNA duplex. Work in progress to obtain α values for Hofmeister salt-DNA base interactions will provide a more complete comparison with the experimental m-values.
4) Comparison to Other Approaches
von Hippel and collaborators found that while Hofmeister salts interact unfavorably with a homogeneous hydrocarbon surface, vicinal methylene groups significantly affect the affinity of salts for amide model compounds72. Recently, Rembert et al73 used thermodynamic and proton NMR experiments along with MD simulations to study the interactions of Hofmeister ions with the polypeptide (VPGVG)120. They concluded that destabilizing anions like SCN− and I− bind to amide N and the adjacent α-carbon (but not the valine side chains), neutral anions like Cl− bind weakly to this site and stabilizing anions like SO42− are excluded from all peptide surface types. The result for SO42− is consistent with the model compound analysis showing strong exclusion from both nonpolar and polar amide surface. As in the case of urea and GB effects, a current point of debate is therefore whether interactions of Hofmeister ions with protein functional groups and Hofmeister salt effects on protein processes are dominated by backbone interactions or whether the most important distinction is in how the salt ion or solute interacts with different types of surface (C, N, O), irrespective of location on backbone or side chains.
Acknowledgment
We thank members of our laboratory, both current (Ben Knowles, Irina Shkel) and previous (Elizabeth Courtenay, Wentao Zhang, Dan Felitsky, Jiang Hong, Jonathan Cannon, Charles Anderson, Ruth Saecker) and a dedicated group of undergraduates (Megan Kratz, Brad Nelms, Robert Erdman, Timothy Wendorff, Dana Bellissimo, Andrew LaCroix, Nickolas Deines, Michelle Pollock, Joanne Tsarouha, Xiaoyi Qu, Roger Diehl, Michael Kerins, Tyler Weid, Hyo Cha, Rachel Wong, Chau Phan, Emily Lingeman, Katie Bieda, Shaenah Maguire, Matt Sternke, Shane Zhou, Kayla Coppens, Audrey Hartzler, Laura Vian, Evan Buechel, August Melcher) for their contributions to the research reviewed here. Research from our laboratory was supported primarily by the NIH (GM47022). We also thank Pavel Jungwirth, Paul Cremer and other members of the committee for organizing the Faraday Discussion at which this paper was presented.
Supported by NIH Grant GM47022 (MTR)
References
- 1.Baldwin RL. Biophysical Journal. 1996;71:2056–2063. doi: 10.1016/S0006-3495(96)79404-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.von Hippel PH, Schleich T. Biological Macromolecules. 1969;2:417–574. [Google Scholar]
- 3.Timasheff SN. Adv. Protein Chem. 1998;51:351–432. doi: 10.1016/s0065-3233(08)60656-7. [DOI] [PubMed] [Google Scholar]
- 4.Record MT, Zhang W, Anderson CF. Advances in Protein Chemistry. 1998;51:281–353. doi: 10.1016/s0065-3233(08)60655-5. [DOI] [PubMed] [Google Scholar]
- 5.Hofmeister F. Arch Exp Pathol Pharmakol. 1888;24:247–260. [Google Scholar]
- 6.von Hippel PH, Schleich T. Accounts Chem Res. 1969;2:257–265. [Google Scholar]
- 7.Pegram LM, Wendorff T, Erdmann R, Shkel I, Bellisimo D, Felitsky DJ, Record MT. PNAS. 2010;107:7716–7721. doi: 10.1073/pnas.0913376107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guinn EJ, Pegram LM, Capp MW, Pollock MN, Record MT. proceedings of the national academy of science. 2011 doi: 10.1073/pnas.1109372108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Courtenay ES, Capp MW, Saecker RM, Record MT. Proteins: Struct., Funct., and Genetics. 2000;41:72–85. doi: 10.1002/1097-0134(2000)41:4+<72::aid-prot70>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 10.Holbrook JA, Capp MW, Saecker RM, Record MT. Biochemistry. 1999;38:8409–8422. doi: 10.1021/bi990043w. [DOI] [PubMed] [Google Scholar]
- 11.Courtenay ES, Capp MW, Anderson CF, Record MT. Biochemistry. 2000;39:4455–4471. doi: 10.1021/bi992887l. [DOI] [PubMed] [Google Scholar]
- 12.Capp MW, Pegram LM, Saecker RM, Kratz M, Riccardi D, Wendorff T, Record J. G. C. a. M. T. Biochemistry. 2009;48:10372–10379. doi: 10.1021/bi901273r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Myers JK, Pace CN, Scholtz JM. Protein Science. 1995;4:2138–2148. doi: 10.1002/pro.5560041020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Felitsky DJ, Cannon JG, Capp MW, Hong J, Wynsberghe AWV, Anderson CF, Record MT. Biochemistry. 2004;43:14732–14743. doi: 10.1021/bi049115w. [DOI] [PubMed] [Google Scholar]
- 15.Hong J, Capp MW, Anderson CF, Saecker RM, Felitsky DJ, Anderson MW, Record MT. Biochemistry. 2004;43:14744–14758. doi: 10.1021/bi049096q. [DOI] [PubMed] [Google Scholar]
- 16.Rees WA, Yager TD, Korte J, von Hippel PH. Biochemistry. 1993;32:137–144. doi: 10.1021/bi00052a019. [DOI] [PubMed] [Google Scholar]
- 17.Shelton VM, Sosnick TR, Pan T. Biochemistry. 1999;38:16831–16839. doi: 10.1021/bi991699s. [DOI] [PubMed] [Google Scholar]
- 18.Lambert D, Draper DE. Journal of Molecular Biology. 2007;370:993–1005. doi: 10.1016/j.jmb.2007.03.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levine L, Gordon JA, Jencks WP. Biochemistry. 1963;2:168–175. doi: 10.1021/bi00901a030. [DOI] [PubMed] [Google Scholar]
- 20.Courtenay ES, Capp MW, Record MT. Prot.Sci. 2001;10:2485–2497. doi: 10.1110/ps.ps.20801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Knowles DB, LaCroix AS, Deines NF, Shkel I, Record MT. PNAS. 2011;108:12699–12704. doi: 10.1073/pnas.1103382108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kramer RM, Shende VR, Motl N, Pace CN, Scholtz JM. Biophysical Journal. 2012;102:1907–1915. doi: 10.1016/j.bpj.2012.01.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee LL-Y, Lee JC. Biochemistry. 1987;26:7813–7819. doi: 10.1021/bi00398a042. [DOI] [PubMed] [Google Scholar]
- 24.H SC, B CJ. Biochemistry. 1999;38:496–508. doi: 10.1021/bi9820154. [DOI] [PubMed] [Google Scholar]
- 25.Bhat R, Timasheff SN. Protein Science. 1992;1:1133–1143. doi: 10.1002/pro.5560010907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Y, Cremer PS. Proc Natl Acad Sci. 2009;106:15249–15253. doi: 10.1073/pnas.0907616106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gross LM, Strauss UP. In: in Chemical Physics of Ionic Solutions. Conway BE, Barradas RG, editors. Wiley; New York: 1966. pp. 361–389. [Google Scholar]
- 28.Omta AW, Kropman MF, Woutersen S, Bakker HJ. Science. 2003;301:347–349. doi: 10.1126/science.1084801. [DOI] [PubMed] [Google Scholar]
- 29.Melander W, Horvath C. Arch Biochem. 1977;183:200–215. doi: 10.1016/0003-9861(77)90434-9. [DOI] [PubMed] [Google Scholar]
- 30.Record MT, Anderson CF. Biophys.J. 1995;68:786–794. doi: 10.1016/S0006-3495(95)80254-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pegram LM, Record MT. PNAS. 2006;103:14278–14281. doi: 10.1073/pnas.0606256103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Felitsky DJ, Record MT. Biochemistry. 2004;43:9276–9288. doi: 10.1021/bi049862t. [DOI] [PubMed] [Google Scholar]
- 33.Pegram LM, Record MT. J.Phys.Chem.B. 2008;112:9428–9436. doi: 10.1021/jp800816a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pegram LM, Record MT. Chem. Phys. Letters. 2008;467:1–8. doi: 10.1016/j.cplett.2008.10.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tanford C. J. Mol. Biol. 1969;39:539–544. doi: 10.1016/0022-2836(69)90143-0. [DOI] [PubMed] [Google Scholar]
- 36.Schellman JA. Biopolymers. 1994;34:1015–1026. doi: 10.1002/bip.360340805. [DOI] [PubMed] [Google Scholar]
- 37.Schellman JA. Biophys.J. 2003;85:108–125. doi: 10.1016/S0006-3495(03)74459-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nozaki Y, Tanford C. J.Biol.Chem. 1963;238:4074–4081. [PubMed] [Google Scholar]
- 39.Auton M, Holthauzen LMF, Bolen DW. PNAS. 2007;104:15317–15322. doi: 10.1073/pnas.0706251104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Auton M, Rosgen J, Mikhail S, Holthauzen LMF, Bolen DW. Biophysical Chemistry. 2011;159:90–99. doi: 10.1016/j.bpc.2011.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pegram LM, Record MT. J.Phys.Chem.B. 2007;111:5411–5417. doi: 10.1021/jp070245z. [DOI] [PubMed] [Google Scholar]
- 42.Setchenow M. Ann. Chim. Phys. 1892;25:226–270. [Google Scholar]
- 43.Shkel IA, Record MT. Soft Matter. 2012;8:9345–9355. doi: 10.1039/C2SM25607J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weerasinghe S, Smith PE. J.Phys.Chem.B. 2003;107:3891–3898. [Google Scholar]
- 45.Pierce V, Kang M, Aburi M, Weerasinghe S, Smith PE. Cell Biochemistry and Biophysics. 2008;50:1–22. doi: 10.1007/s12013-007-9005-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Robinson RA, Stokes RH. J. Phys. Chem. 1961;65:1954–1958. [Google Scholar]
- 47.Anderson CF, Record MT. Biophys.Chem. 2004;112:165–175. doi: 10.1016/j.bpc.2004.07.016. [DOI] [PubMed] [Google Scholar]
- 48.Zhang W, Capp MW, Bond JP, F. AC, Record MT. Biochemistry. 1996;35:10506–10516. doi: 10.1021/bi960795f. [DOI] [PubMed] [Google Scholar]
- 49.Cannon JG, Anderson CF, Record MT. J.Phys.Chem.B. 2007;111:9675–9685. doi: 10.1021/jp072037c. [DOI] [PubMed] [Google Scholar]
- 50.Timasheff SM, Inoue H. Biochemistry. 1968;7:2501–2513. doi: 10.1021/bi00847a009. [DOI] [PubMed] [Google Scholar]
- 51.Pegram LM, Record MT. J. Phys. Chem. C. 2009;113:2171–2174. doi: 10.1021/jp8073305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gopalakrishnan S, Jungwirth P, Tobias DJ, Allen HC. J. Phys. Chem. B. 2005;109:8861–8872. doi: 10.1021/jp0500236. [DOI] [PubMed] [Google Scholar]
- 53.Mucha M, Frigato T, Levering LM, Allen HC, Tobias DJ, Dang LX, Jungwirth P. J. Phys. Chem. B. 2009;105:7617–7623. doi: 10.1021/jp0445730. [DOI] [PubMed] [Google Scholar]
- 54.Sunha SK, Chakraborty S, Bandyopadhyay S. J. Phys. Chem. B. 2008;112:8203–8209. doi: 10.1021/jp8000724. [DOI] [PubMed] [Google Scholar]
- 55.Jeruzalmi D, Steitz TA. J. Mol. Biol. 1997;274:748–756. doi: 10.1006/jmbi.1997.1366. [DOI] [PubMed] [Google Scholar]
- 56.Guinn EJ, Schwinefus J, Cha H, Record MT. J. Am. Chem. Soc. 2012 In preparation. [Google Scholar]
- 57.Lambert D, Draper DE. Biochemistry. 2012;51:9014–9026. doi: 10.1021/bi301103j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cayley S, Record MT. Biochemistry. 2003;42:12596–12609. doi: 10.1021/bi0347297. [DOI] [PubMed] [Google Scholar]
- 59.Arakawa T, Timasheff SN. Biophys. J. 1985;14:411–414. doi: 10.1016/S0006-3495(85)83932-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Priyakumar UD, Hyeon C, Thirumalai D, MacKerell AD. J. Am. Chem. Soc. :17759–17761. doi: 10.1021/ja905795v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hong J, Capp MW, Saecker RM, Record MT. Biochemistry. 2005;44:16896–16911. doi: 10.1021/bi0515218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kalodimos CG, Folkers GE, Boelens R, Kaptein R. Proc. Natl. Acad. Sci. U.S.A. 2001;98:6039–6044. doi: 10.1073/pnas.101129898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kalodimos CG, Bonvin AMJJ, Salinas RK, Wechselberger R, Boelens R, Kaptein R. EMBO J. 2002;21:2866–2876. doi: 10.1093/emboj/cdf318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Scholtz JM, Barrick D, York EJ, Stewart JM, Baldwin RL. PNAS. 1995;92:185–189. doi: 10.1073/pnas.92.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Auton M, Bolen DW. PNAS. 2005;102:15065–15068. doi: 10.1073/pnas.0507053102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ma L, Pegram L, Record MT, Cui Q. Biochemistry. 2010;49:1954–1962. doi: 10.1021/bi9020082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Canchi DR, Garcia AE. Biophysical Journal. 2011;100:1526–1533. doi: 10.1016/j.bpj.2011.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Horinek D, Netz RR. J. Phys. Chem. A. 2011;115:6125–6136. doi: 10.1021/jp1110086. [DOI] [PubMed] [Google Scholar]
- 69.Sagle LB, Zhang Y, Litosh VA, Chen X, Cho Y, Cremer PS. Journal of the American Chemical Society. 2009;131:9304–9310. doi: 10.1021/ja9016057. [DOI] [PubMed] [Google Scholar]
- 70.Lim WK, Rosgen J, Englander SW. PNAS. 2009;106:2595–2600. doi: 10.1073/pnas.0812588106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nandi PK, Robinson DR. J. Am. Chem. Soc. 1972;94:1299–1308. doi: 10.1021/ja00759a042. [DOI] [PubMed] [Google Scholar]
- 72.Hamabata A, von Hippel PH. Biochemistry. 1973;12:1264–1271. doi: 10.1021/bi00731a004. [DOI] [PubMed] [Google Scholar]
- 73.Rembert KB, Paterova J, Heyda J, Hilty C, Jungwirth P, Cremer PS. J. Am. Chem. Soc. 2012;134:10039–10046. doi: 10.1021/ja301297g. [DOI] [PubMed] [Google Scholar]
- 74.Peters T. All about Albumin: Biochemistry, Genetics, and Medical Applications. Academic Press, Inc.; San Diego, CA: 1996. [Google Scholar]
- 75.Felitsky DJ, Record MT. Biochemistry. 2003;42:2202–2217. doi: 10.1021/bi0270992. [DOI] [PubMed] [Google Scholar]
- 76.Estrada J, Bernado P, Blackledge M, Sancho J. BMC Bioinf. 2009;10:104. doi: 10.1186/1471-2105-10-104. [DOI] [PMC free article] [PubMed] [Google Scholar]