Summary
Telomeres are essential for nuclear organization in yeast and during meiosis in mouse. Exploring telomere dynamics in living human cells by advanced time-lapse confocal microscopy allowed us to evaluate the spatial distribution of telomeres within the nuclear volume. We discovered an unambiguous enrichment of telomeres at the nuclear periphery during post-mitotic nuclear assembly, while telomeres were localized more internally during the rest of the cell cycle. Telomere enrichment at the nuclear rim was mediated by physical tethering of telomeres to the nuclear envelope, likely via specific interactions between the shelterin subunit RAP1 with the nuclear envelope protein Sun1. Genetic interference revealed a critical role in cell cycle progression for Sun1, but no effect on telomere positioning for RAP1. Our results shed light on the dynamic relocalization of human telomeres during the cell cycle and suggest redundant pathways for tethering telomeres to the nuclear envelope.
Introduction
Telomeres protect genetic material from degradation and our chromosomes from end-to-end fusion (O’Sullivan and Karlseder, 2010) and also function in nuclear organization, which has emerged as an essential aspect of gene regulation and genome stability. One of the best examples is the segregation of gene-rich and gene-poor chromatin domains, which tend to localize towards the nuclear interior or the periphery, respectively. In budding yeast, telomeres are organized in clusters enriched at the nuclear periphery (Hediger et al., 2002), critical for telomere silencing, likely by creating a nuclear sub-compartment enriched for silencing factors. Moreover, it was reported that the binding of telomeres to the nuclear envelope protects telomeric repeats from recombination (Schober et al., 2009). Moreover, telomere clustering to the envelope is crucial for proper meiotic pairing and the recombination of homologous chromosomes during meiosis in yeast and mammals (Chikashige et al., 2006; Ding et al., 2007).
The impact of telomere positioning on genome organization in human cells is still unclear. 3D-fluorescent in situ hybridization (FISH) experiments suggested that telomeres were on average nearer to the center of the cell and not enriched at the nuclear periphery in lymphocytes (Amrichova et al., 2003). A similar approach established that telomeres assemble into a telomeric disk in G2 phase, suggesting cell cycle regulation of telomere position (Vermolen et al., 2005). The dynamic behavior of telomeric repeats was also studied in living human U2OS osteosarcoma cells (Jegou et al., 2009), where the majority followed a constrained diffusive movement, and some associated and dissociated to form dynamic clusters. These studies suggest that telomere distribution in human cells is dynamic, but failed to address the spatial organization of telomeres in real time during the whole cell cycle.
Here, we used advanced time-lapse confocal microscopy to explore telomere dynamics in human primary and cancer cells. We followed chromatin and telomeres via live imaging over a period of at least 20 hours, giving us an unprecedented level of insight into the spatial distribution of telomeres within the nuclear volume in real time, enabling the discovery that telomeres associate with the nuclear matrix during nuclear assembly after mitosis.
Results and Discussion
Visualization of human telomeres and chromatin in live cells
To follow the positioning of human telomeres in living cells, fluorescent versions of telomeric proteins were stably expressed in human primary fibroblasts (IMR90) and HeLa cells with long telomeres (HeLa1.2.11). The fusion between TRF1 and EGFP was properly targeted to telomeres as attested by its colocalization with endogenous TRF2 (Supplementary Figure 1A). The combined expression of EGFP-TRF1 and the histone H2B-mCherry allowed the concomitant visualization of telomeres and chromatin in living cells (Supplementary Figure 1B). Overexpression of EGFP-TRF1 and/or H2B-mCherry in IMR90 and HeLa1.2.11 cells did not initiate checkpoint activation, as indicated by the failure to phosphorylate H2AX (γH2AX), indicating that no telomere uncapping occurred (Supplementary Figure 1C).
Human telomeres localize to the nuclear periphery following mitosis
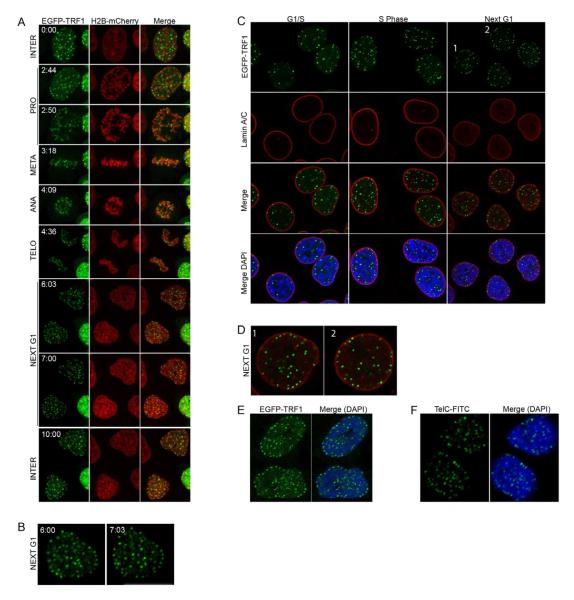
HeLa1.2.11 cells expressing EGFP-TRF1 and H2B-mCherry were monitored by live imaging on a spinning disk confocal microscope every six minutes over at least 20 hours to visualize cell cycle dynamics (Supplementary Movie 1). Time-lapse images show that telomeres were randomly distributed throughout the nucleus during interphase (Figure 1A, panels “Interphase”). Telomere distribution dramatically changed just at the end of mitosis. An enrichment of telomeres at the nuclear rim was observed after the completion of cell division, and lasted during the entire process of nuclear assembly (Figure 1A, B, panels “Next G1”). Live imaging was also performed in primary IMR90 cells expressing EGFP-TRF1 and H2B-mCherry. Following telomere dynamics during one cell cycle again revealed their repositioning to the nuclear periphery during post-mitotic nuclear assembly (Supplementary Movie 2, Supplementary Figure 1D).
Figure 1. Live cell imaging of human telomeres.
(A) Time-lapse images of HeLa1.2.11 cells expressing EGFP-TRF1 and H2B-mCherry. A single focal plane is presented per time-lapse (hr:min). (B) Magnification of the 6:03 and 7:03 images. (C) Immunofluorescence staining of synchronized HeLa1.2.11 cells expressing EGFP-TRF1 in G1/S, S phase, or after mitosis (next G1). (D) Magnification of two nuclei from panel C, Next G1. (E) Staining of synchronized IMR90 cells expressing EGFP-TRF1 after mitosis (next G1). (F) FISH staining of HeLa1.2.11 cells. See also Figures S1, S2 and Movies S1, S2.
Telomere enrichment at the periphery was then investigated in synchronized and fixed cells. EGFP-TRF1-expressing cells were synchronized at the G1-S boundary and cell cycle progression was determined by DNA content. EGFP-TRF1 expression did not affect cell cycle progression (Supplementary Figure 1E). Cells were fixed for immunofluorescence at the G1-S block, 4 hours after release (S Phase), and then after cell division at 10 and 12 hr (Next G1) (Figure 1C). The nuclear periphery was visualized by co-staining of Lamin A/C, a structural component of the lamina that is specifically associated with the nuclear envelope. Similarly to live-imaging, telomeres were found enriched at the nuclear periphery after cell division (Figure 1C, panels “Next G1”, Figure 1D), whereas cells in S phase or at the G1-S boundary displayed a random distribution (Figure 1C, “G1/S” and “S Phase” panels). Comparable results were obtained with primary IMR90 cells (Figure 1E).
To exclude the possibility that overexpression of EGFP-TRF1 itself could affect telomere distribution in the nucleus, positioning of telomeres was assessed by telomere FISH on fixed and synchronized untransfected HeLa1.2.11 cells (Figure 1F), where a specific enrichment of telomeres at the nuclear periphery could be detected after cell division. Together, these observations indicate that telomeres dynamically relocalize to the nuclear periphery during nuclear assembly in primary and in cancer cells.
Quantification of the subnuclear position of telomeres
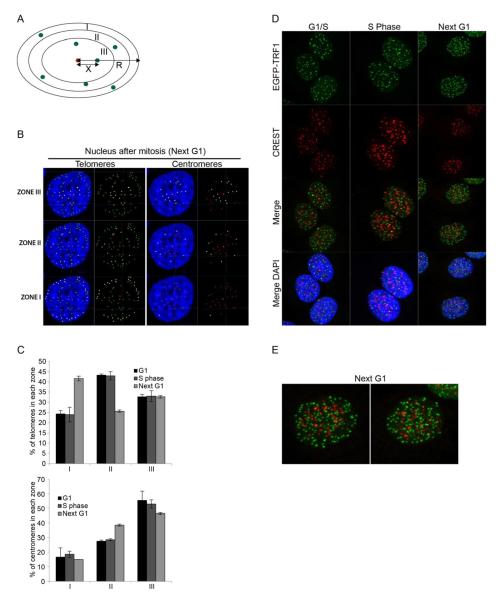
To objectively quantify telomere positioning on the central focal planes of confocal microscopy images, the distances between each fluorescent focus of interest and the center of the ellipsoid-shaped nucleus were scored and divided by the center-to-periphery distance through each specific dot. This allows the evaluation of telomere subnuclear position, independently from the size of the nucleus. The ratios obtained were scored relative to three pre-defined nuclear zones of equal surface (Figure 2A). Signals from telomeres were reconstructed in 2D and the distance ratios for each dot automatically calculated and attributed to one of the three zones (Figure 2B). A random distribution of any telomere throughout the nucleus should lead to positioning in each zone with equal frequency (33%).
Figure 2. Quantification of telomere and centromere position in the nucleus.
(A) Schematic of the quantification method. The ratio between × (distance from the signal of interest to the center) and R (radius of the ellipse through that signal) is calculated. This ratio allows to position the signal in one of the three zones of equal surface I, II and III. (B) Representative example of a 2D reconstruction of telomeres and centromeres and their classification into three zones. Telomeres and centromeres that belong to each specific zone are displayed in yellow. The center of the nucleus is displayed in red. (C) Quantification of telomere and centromere position in the nucleus of HeLa1.2.11 cells. G1, S phase and Next G1 data are represented as a percentage of spots (y axis) per zone (x axis); as mean +/− SD. The number of nuclei, telomeres and centromeres is shown in Supplementary Table 1. (D) Immunofluorescence staining of synchronized HeLa1.2.11 cells stably expressing EGFP-TRF1 (maximum intensity projection). (E) Magnification of two individual nuclei from panel D (next G1). See also Figure S2.
We first analyzed telomere distribution in fixed HeLa1.2.11 cells from three independent synchronizations (Supplementary Table 1A). We noted that the number of telomeres found at the G1/S transition and in S phase was lower compared to that of cells in early G1, suggesting the formation of telomere clusters that assemble as the cells progress in the cell cycle.
At the G1/S transition and during S phase, telomeres were found excluded from the periphery, with only 25% in zone I, 40% in the intermediate zone II, and 32% in the most central zone III (Figure 2B, C). This changed after mitosis when more than 40% of telomeres localized to the nuclear periphery (Figure 2B, C). The redistribution seemed to mostly originate from telomeres in zone II, whose occupation dropped to 25%, while zone III still contained about 30% of telomeres. We performed a similar analysis on synchronized IMR90 fibroblasts (Supplementary Figure 2A). While invagination of the nuclear envelope rendered the automated distribution impossible on many nuclei, the limiting the number of nuclei that could be analyzed (Supplementary Figure 2B) led to comparable results as in HeLa cells (Supplementary Figure 2C).
These data clearly demonstrate that almost half of all telomeres localize to the nuclear periphery after mitosis, during the time of reassembly of the nuclear envelope.
Enrichment at the nuclear periphery is not a common feature of repressed chromatin
Telomere enrichment at the periphery could be a result of the repressive nature of telomeric chromatin and therefore be shared by other heterochromatic regions throughout the genome. To test this hypothesis the position of centromeres was assessed using a CREST antibody (Figure 2D, Supplementary Table 1A). CREST staining in G1-S, S phase or after cell division did not reveal any specific localization or redistribution of centromeres in the nucleus. In fact, during nuclear reassembly, when telomeres are enriched at the periphery, centromeres still positioned close to the nuclear center (Figure 2D, E). Quantification of centromere localization indicated that they were preferentially localized internally within the nucleus, with about 50% present in zone III in G1, S phase, before and after cell division (Figure 2B, C). Centromeres were excluded from the nuclear periphery, with less than 20% of them localized in zone I throughout the cell cycle, suggesting that enrichment of telomeres at the periphery is not a common feature of repressed chromatin.
Telomeres are physically anchored to the nuclear envelope during nuclear reassembly
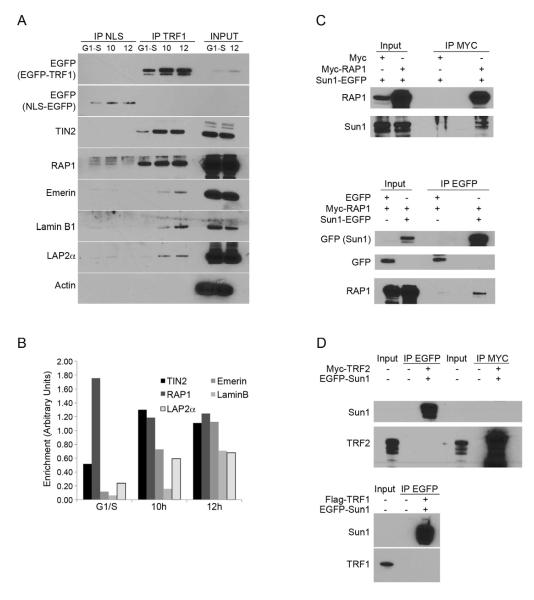
To test for a physical anchoring of chromosome ends to the envelope, chromatin immunoprecipitations (ChIP) were carried out in synchronized HeLa1.2.11 cells in G1-S, during S phase, and after mitosis (next G1). As expected, the shelterin components TRF1 and RAP1 bound telomeric repeats throughout the cell cycle (Supplementary Figure 3A). We then assessed whether telomeres could interact with A-type lamin (Lamin A/C) and B-type Lamin (Lamin B1), or with the inner nuclear membrane protein Sun1, a component of the LINC (linkers of the nucleoskeleton to the cytoskeleton) complex that bridges inner and outer membranes of the nuclear envelope. Binding of these proteins to TTAGGG repeats was minimal and did not vary throughout the cell cycle (Supplementary Figure 3A). We reasoned that at the time telomeres are enriched at the nuclear periphery during nuclear assembly, a significant portion of nuclear envelope proteins is not yet localized at the envelope, but stored in the endoplasmic reticulum with no access to chromatin (Kutay and Hetzer, 2008). Sun1 can be visualized as protein aggregate around the nucleus (Supplementary Figure 3B, panel “Next G1”). As it is not possible to specifically precipitate the proteins that are already readdressed at the membrane or diffuse in the nucleoplasm, we developed a modified version of the ChIP protocol, termed Telomere Association Assay, in which EGFP-TRF1 and its associated partners were precipitated from crosslinked HeLa1.2.11 cell lysates via EGFP bead pull downs. Instead of purifying the DNA from the precipitated material, associated proteins were extracted and the crosslink reversed. The presence of specific co-purified proteins was then tested by western analysis. HeLa1.2.11 cells expressing NLS-EGFP were used as a control. We found that TRF1 was efficiently precipitated in extracts from EGFP-TRF1 cells arrested in G1-S, and in extracts from cells collected 10 or 12 hr after release from the G1-S block (Figure 3A). As expected, other shelterin components were efficiently co-purified in all three extracts (Figure 3A-3B).
Figure 3. Telomeres are physically tethered to the nuclear envelope.
(A) Telomere association assay of HeLa1.2.11 cells expressing NLS-EGFP or EGFP-TRF1. Cells were collected at the G1/S boundary or 10 and 12 hours after release. (B) Quantification of signals from panel A. (C) Co-immunoprecipitation of HeLa1.2.11 cells expressing, Myc alone, Myc-RAP1 and/or EGFP-Sun1. Myc beads or GFP beads were used for immunoprecipitation. (D) Co-immunoprecipitation of HeLa1.2.11 cells expressing Myc-TRF2 or Flag-TRF1 and EGFP-Sun1. Myc beads or GFP beads were used for immunoprecipitation as indicated (IP MYC, IP EGFP). See also Figure S3.
Next, we used the Telomere Association Assay to test for the co-purification of proteins from, or associated with, the nuclear envelope. Lamin B1, the lamin-associated polypeptides LAP2alpha and Emerin, co-precipitated with TRF1, but only in the sample enriched for cells that had just gone through mitosis (Figure 3A, B). These time points precisely corresponded to the period when telomeres were found enriched at the nuclear periphery. No co-precipitation could be detected in extracts from cells blocked at G1-S, or in S phase extracts (Figure 3A, B; Supplementary Figure 3C, D). The specificity of the experiment was carefully addressed: First, EGFP immunoprecipitation from NLS-EGFP extracts did not retrieve TRF1, TIN2, RAP1, or any of the nuclear envelope proteins tested (Figure 3A). Second, the cytoplasmic protein Actin did not co-purify with TRF1 (Figure 3A). Third, immunoprecipitation with beads that specifically recognize Myc-tagged proteins failed to pull down TIN2, Lamin B1 or Emerin (Supplementary Figure 3E), suggesting that the co-purifications are not due to an unspecific pull-down of these proteins in the EGFP-TRF1 extracts. Taken together, these results imply that telomeres form protein-protein or DNA-protein interactions with factors from or associated with the nuclear envelope during the process of nuclear assembly. Two reports previously showed that laminB1 and LAP2alpha bind chromosome ends in late anaphase to serve as a nucleation site for nuclear membrane assembly (Dechat et al., 2004; Martin et al., 2010). Accordingly, our experiments suggest that telomeres are tethered to nuclear envelop factors during the process of membrane assembly, from late anaphase until the nucleus has regained its mature size.
RAP1 can interact with Sun1
Next, we investigated the mechanisms of telomere recruitment to the nuclear periphery. Sun domain proteins have been involved in the tethering of telomeres to the nuclear envelope in both fission and budding yeast, as well as during meiosis in mouse cells (Bupp et al., 2007; Chikashige et al., 2007; Ding et al., 2007). We therefore tested whether telomeres could be tethered to the nuclear envelope through the association of Shelterin with Sun1. We performed co-immunoprecipitation experiments using extracts from HeLa1.2.11 cells expressing a fusion between EGFP and Sun1 (EGFP-Sun1), and beads that specifically recognize the EGFP tag. As expected, immunoblotting of the recovered Sun1 complexes identified Lamin A/C, which was previously shown to interact with Sun1 (Supplementary Figure 3F). Of all the endogenous Shelterin proteins tested, only RAP1 could be recovered in association with Sun1 (Supplementary Figure 3F and data not shown).
In our hands, the RAP1 antibody reveals doublet bands and their respective intensities vary between protein extraction methods. Only the lower band was detected in the Sun1 precipitates. The reciprocal precipitation was unsuccessful due to the small amount of soluble Sun1 protein. To circumvent this we used HeLa1.2.11 cells co-expressing Myc-RAP1 and EGFP-Sun1 and performed immunoprecipitations against EGFP or Myc. This reveled specific associations of EGFP-Sun1 in the anti-Myc precipitations and Myc-RAP1 in the EGFP precipitations (Figure 3C). These interactions were considered as specific, since extracts from cells expressing the tag alone yielded neither Sun1 nor RAP1 (Figure 3C).
Similar experiments were performed with co-expression of Sun1 with TRF1 or TRF2 (Figure 3D). As anticipated, endogenous Lamin A/C and RAP1 were efficiently recovered in association with EGFP-Sun1 and Myc-TRF2, respectively (Supplementary Figure 3G). However, Myc-TRF2 or Flag-TRF1 could not be recovered in association with EGFP-Sun1 (Figure 3D). Reversely, Myc-TRF2 precipitation did not recover EGFP-Sun1 (Figure 3D). Sun1 is potentially bridged to TRF2 via RAP1, but this interaction could not be detected, likely due to the much higher level of overexpressed TRF2, as compared to the endogenous RAP1.
These results demonstrate that Sun1 and RAP1 interact. RAP1 is required for attachment of telomeres to the nuclear envelope and their clustering during the meiotic prophase I in fission yeast (Kanoh and Ishikawa, 2001), and based on the Sun1-RAP1 interaction suggested here, we can postulate that a similar mechanism is conserved in human mitotic cells.
RAP1 independent telomere tethering to the nuclear envelope
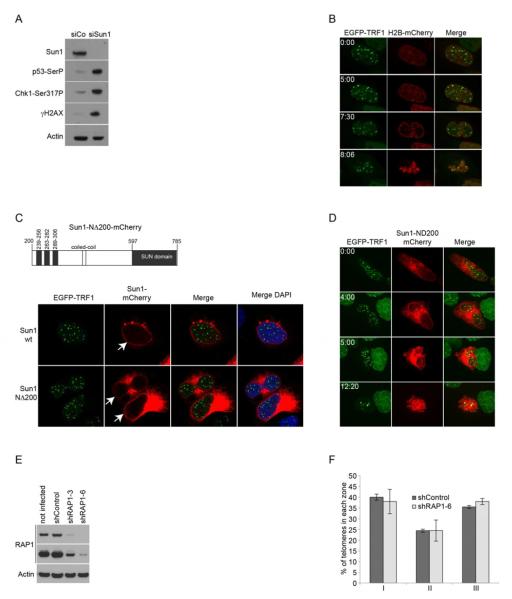
To evaluate if the Sun1/RAP1 interaction is essential to tether telomeres to the nuclear rim, we suppressed Sun1 expression. Transfection of Sun1-specific siRNA oligonucleotides in HeLa1.2.11 cells induced a very effective reduction of Sun1 expression, and simultaneously a strong activation of a checkpoint response (Figure 4A). Sun1-suppressed HeLa1.2.11 cells expressing EGFP-TRF1 and H2B-mCherry were imaged live, using the same parameters as before (Figure 1). Movies from these experiments revealed that Sun1 siRNA had a dramatic effect on the cell cycle with most cells dying from apoptosis before entering mitosis, while cells transfected with scrambled control siRNAs divided normally (Figure 4B and data not shown). In an attempt to circumvent lethality, we overexpressed a truncated form of Sun1 lacking the first N-terminal 200 amino acids (Sun1-NΔ200) defining the region of Sun1 that is located in the nucleoplasm. This mutant conserves its three putative transmembrane domains as well as the conserved C-terminal SUN domain located in the perinuclear space (Figure 4C). Expressed in HeLa1.2.11 cells, Sun1-NΔ200-mCherry was found partially localized to the nuclear envelope, with some of the protein retained in the endoplasmic reticulum (ER) (Figure 4C). Similarly to what was observed after Sun1 knockdown, HeLa1.2.11 cells expressing Sun1-NΔ200-mCherry died of apoptosis, and none of the cells imaged live passed through mitosis (Figure 4D, Supplementary Movie 3). Over time, expression of this Sun1 mutant dramatically altered the structure of the nuclear envelope (Figure 4D, panel 4:00hr), reminiscent of a mouse Sun1 mutant lacking its N terminus (Padmakumar et al., 2005). The dramatic effect associated with impaired Sun1 function clearly argues for a critical role of Sun1 in cell cycle progression, but hinders our ability to specifically test for its role in telomere tethering.
Figure 4. RAP1-independent telomere tethering.
(A) Western blotting of HeLa1.2.11 cells transfected with Sun1 or a control siRNA. (B) Time-lapse images from movies of HeLa1.2.11 cells stably expressing EGFP-TRF1 and H2B-mCherry and transfected with Sun1 siRNA. A single focal plane is presented per time-lapse (indicated in hr:min). (C) Schematic of the Sun1-NΔ200 mutant and immunofluorescence of cells expressing EGFP-TRF1 and Sun1 wt or Sun1-NΔ200-mCherry. (D) Time-lapse images from movies of HeLa1.2.11 cells stably expressing EGFP-TRF1 and transfected with Sun1-NΔ200-mCherry. A maximum intensity projection of all focal planes is presented per time-lapse (hr:min). (E) Western blotting of HeLa1.2.11 cells expressing RAP1 or control shRNA. (F) Quantification of telomere position in the nucleus post mitosis of HeLa1.2.11 cells. Nuclei after mitosis were selected from time-lapse images and used for quantification. Data are represented as a percentage of spots (y axis) per zone (x axis); as mean +/− SD. The number of nuclei and telomeres is shown in Table S1. See also Movie S3.
We next targeted RAP1 expression using two stably expressing shRNAs (Figure 4E). HeLa1.2.11 cells expressing the most effective shRNA construct (RAP1-6) did not display major cell cycle defects, nor trigger a detectable DNA damage response (data not shown). We used these cells for live imaging and compared telomere distribution after mitosis with cells expressing a scrambled shRNA control. Quantification of telomere positioning revealed that about 40% of telomeres still localize to the peripheral zone I after mitosis in RAP1 suppressed cells, similar to what was found in cells expressing a control shRNA (Figure 4F, Supplementary Table 1). This is reminiscent of RAP1-deficient meiotic telomeres in mice that still undergo bouquet formation through telomere tethering to the nuclear envelope-localized Sun1 (Scherthan et al., 2011).
Several hypotheses can be drawn to explain why we did not observe a decrease in the numbers of telomeres at the periphery in RAP1 suppressed cells. First, RAP1 has recently been proposed to have extra-telomeric functions in human cells (Martinez et al., 2010; Teo et al., 2010). It is therefore possible that the pool of RAP1 binding to Sun1 is not localized to telomeres, although this is unlikely, considering that telomeres were found physically bound to the nuclear envelope (Figure 3). Second, we cannot exclude that the remaining RAP1 expression after RAP1 knockdown is sufficient to promote telomere tethering. Third, multiple pathways could be involved in human telomere tethering, as is the case in budding yeast, in which telomere tethering to the nuclear envelope is dependent on two redundant pathways (Hediger et al., 2002).
In this study we demonstrate that telomeres are tethered to the nuclear envelope in human primary and cancer cells during the process of nuclear reassembly. In mitosis, chromosome condensation and nuclear envelope breakdown disrupt nuclear organization, which must be reestablished during nuclear reassembly (Kutay and Hetzer, 2008). Attachment of chromosome ends to the nuclear envelope while the daughter nuclei are shaped suggests that telomeres could serve as a nuclear envelope anchor point to reorganize chromatin domains after each cell division. This process appears to be dependent on the nuclear envelope protein Sun1, which is part of the LINC complex and has been shown to move chromosomes for bouquet formation during meiosis in fission yeast and mice. The transient proximity of telomeres to the nuclear envelope could also have an impact on telomere maintenance, by establishing specific chromatin marks or regulating telomere transcription and replication. This hypothesis is supported by the fact that A-type lamins have a role in the maintenance of telomeres in mouse cells (Gonzalez-Suarez et al., 2009). Telomere tethering to the nuclear envelope in human cells brings a new dimension to telomere regulation, with a potential impact on nuclear organization, regulation of cell division, gene expression and telomere maintenance.
Experimental Procedures
Cell culture and treatments
Human IMR90 fibroblasts and HeLa1.2.11 cells were grown, synchronized and transfected as described (O’Sullivan et al., 2010). Viral infections. Lentivirus stocks were generated by the Salk Institute Gene Transfer Core. HeLa1.2.11 cells were plated in growth media containing 4 μg.mL−1 polybrene (Sigma) and lentivirus at a multiplicity of infection of 10 and cultured for 2 d. The cells were then split into media containing 1 μg.mL−1 puromycin and selected for 7 d before analysis.
Plasmids (Primers used are listed in Supplementary Table 2)
Cloning strategies are described in the supplemental data.
siRNA
Sun1 siRNA suppression was done as described (Talamas and Hetzer, 2011).
shRNA
pLKO.1 TRCN0000010356 shRNA against RAP1 (Open Biosystem, sense sequence 5−-AGAGTTCTTGCATTGGAACT-3′); pLKO.1 control shRNA (sense sequence 5′-CCTAAGGTTAAGTCGCCCTCG-3′).
Antibodies
anti-γ-H2AX (613402 clone 2F3, Biolegend); anti-Sun1 (Ab1: ab74758, Abcam; Ab2: HPA008346, Sigma Prestige Antibodies); anti-phospho-Chk1-Ser317 (2348, Cell Signaling Technology); anti-phospho-p53-Ser15 (9284, Cell Signaling Technology); anti-TRF1/2 (Karlseder lab); anti-RAP1 (A300-306A, Bethyl Laboratories); anti-TIN2 (T. de Lange); anti-Actin (A1978, SIGMA); anti-Emerin (06-1052, Millipore); anti-LaminA/C (L1293, Sigma); anti-LaminB1 (sc-6216, Santa Cruz Biotechnology); anti-BAF (3F10-4G12, Novus Biological); anti-LAP2alpha (04-640, Millipore); anti-GFP-HRP (12-002-105, Miltenyi Biotec). HPR-linked anti-mouse or anti-rabbit (NXA931 or NA934V, GE Healthcare); Alexa-488-conjugated anti-mouse or anti-rabbit (Invitrogen); Alexa-594-conjugated anti-mouse or anti-rabbit (Invitrogen).
Live Cell Imaging
Cells were grown in 35 mm Ibidi dishes. Images were acquired with a Cell Observer SD spinning disk confocal microscope (Axio Observer Z1 platform with a Yokagawa CSU-X1 Nipkow spinning disk head, Zeiss) equipped with an incubation chamber, a 512×512 16bit Evolve EMCCD camera (Photometrics, Tucson, AZ), and Axiovision software (Zeiss). 8 to 9-image focal planes were taken with a 63× Plan-Apochromat objective and a 1 μm step size every 6 min for at least 20 h.
Confocal Microscopy
Fixed cells were imaged on an LSM 780 laser scanning confocal microscope (Zeiss).
Quantitative Analysis of telomere and centromere Position
Imaris 7.4.0 (Andor) image analysis software was used to calculate distances between each telomere of interest and the center of the nucleus, and divided by the center-to-periphery distance through each given telomere. The center and periphery of each nucleus was determined by DAPI staining, and the center of the telomeres by fitting a circle around the diameter. Signals from telomeres were reconstructed in 2D, and the distance ratios for each telomere automatically calculated and attributed by the software to one of nuclear zones of equal surface. The most peripheral zone I has a width of 0.816 × the nuclear radius (calculated for each telomere), zone II = 0.816 × r to 0.577 × r, and zone III = 0.577 × r. The same method was used for centromere position analysis.
Western blotting, immunofluorescence, telomere FISH, chromatin immunoprecipitation
Carried out as described (O’Sullivan et al., 2010).
Telomere Association Assay
Cells were washed with PBS twice before fixation with 1% formaldehyde in PBS during 15 min at RT. Cells were collected by scraping and washed again by centrifugation. The cell pellet was lysed in 1% SDS, 50mM Tris-HCl, pH 8.0, 10mM EDTA. Lysates were sonicated to obtain chromatin fragments < 1 kb, and centrifuged for 10 min at 4°C. 1 mg of proteins lysate, diluted 10 times with 0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl, pH 8.0, and 150 mM NaCl was used per immunoprecipitation and supplemented with 50 μl μMACṢAnt̃iGFP microbeads (Miltenyi Biotec) for 2 h at 4°C. Labeled cell lysates were applied to MACS column placed in a separator (Miltenyi Biotec) and washed with 0.1% SDS, 1% Triton X-100, 2 mM EDTA, pH 8.0, 20 mM Tris-HCl, pH 8.0 containing 150 mM NaCl for the first 3 washes, and 500 mM NaCl for the next 3 washes. After 1 last wash with 20 mM Tris-HCl, pH 7.5, targeted proteins were eluted in 50 μl of Elution Buffer for SDS-PAGE (Miltenyi Biotec) while the microbeads remained in the columns. 10 μl of 4× LDS (Invitrogen) with DTT were added to the eluates before reversing the crosslink at 95°C for 30 min. For Input samples, 10% of the lysa tes was diluted in 4× LDS (Invitrogen) with DTT. Image J software was used to quantify the signals obtained by western blotting relative to the amount of EGFP-TRF1 precipitated for each cell lysate.
Co-Immunoprecipitations
Performed using the μMACS GFP and c-myc Epitope Tag Isolation kits (Miltenyi Biotec) according to the Manufacturer’s instructions.
Supplementary Material
Highlights.
Telomeres are tethered to the nuclear envelope after mitosis
Sun1 interacts with RAP1
Telomeres impact chromosome organization in human cells
Acknowledgements
We thank the Hetzer laboratory for reagents. L.C was supported by the Glenn Center for Aging Research at the Salk Institute and A.J.C. is supported by a NIH training grant (5T32CA009370-29). JK is the Shiley Chair Professor for Aging Research and supported by the Cancer Center Core Grant P30 CA014195-38, the NIH (GM087476), the Szabo Trust and the Highland Street Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amrichova J, Lukasova E, Kozubek S, Kozubek M. Nuclear and territorial topography of chromosome telomeres in human lymphocytes. Exp Cell Res. 2003;289:11–26. doi: 10.1016/s0014-4827(03)00208-8. [DOI] [PubMed] [Google Scholar]
- Bupp JM, Martin AE, Stensrud ES, Jaspersen SL. Telomere anchoring at the nuclear periphery requires the budding yeast Sad1-UNC-84 domain protein Mps3. J Cell Biol. 2007;179:845–854. doi: 10.1083/jcb.200706040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikashige Y, Haraguchi T, Hiraoka Y. Another way to move chromosomes. Chromosoma. 2007;116:497–505. doi: 10.1007/s00412-007-0114-8. [DOI] [PubMed] [Google Scholar]
- Chikashige Y, Tsutsumi C, Yamane M, Okamasa K. Meiotic proteins bqt1 and bqt2 tether telomeres to form the bouquet arrangement of chromosomes. Cell. 2006 doi: 10.1016/j.cell.2006.01.048. [DOI] [PubMed] [Google Scholar]
- Dechat T, Gajewski A, Korbei B, Gerlich D, Daigle N, Haraguchi T, Furukawa K, Ellenberg J, Foisner R. LAP2alpha and BAF transiently localize to telomeres and specific regions on chromatin during nuclear assembly. J Cell Sci. 2004;117:6117–6128. doi: 10.1242/jcs.01529. [DOI] [PubMed] [Google Scholar]
- Ding X, Xu R, Yu J, Xu T, Zhuang Y, Han M. SUN1 is required for telomere attachment to nuclear envelope and gametogenesis in mice. Dev Cell. 2007;12:863–872. doi: 10.1016/j.devcel.2007.03.018. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Suarez I, Redwood A, Perkins S, Vermolen B, Lichtensztejin D, Grotsky D, Morgado-Palacin L, Gapud E, Sleckman B, Sullivan T. Novel roles for A-type lamins in telomere biology and the DNA damage response pathway. The EMBO journal. 2009;28:2414–2427. doi: 10.1038/emboj.2009.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hediger F, Neumann FR, Van Houwe G, Dubrana K, Gasser SM. Live imaging of telomeres: yKu and Sir proteins define redundant telomere-anchoring pathways in yeast. Curr Biol. 2002;12:2076–2089. doi: 10.1016/s0960-9822(02)01338-6. [DOI] [PubMed] [Google Scholar]
- Jegou T, Chung I, Heuvelman G, Wachsmuth M, Gorisch SM, Greulich-Bode KM, Boukamp P, Lichter P, Rippe K. Dynamics of telomeres and promyelocytic leukemia nuclear bodies in a telomerase-negative human cell line. Mol Biol Cell. 2009;20:2070–2082. doi: 10.1091/mbc.E08-02-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanoh J, Ishikawa F. spRap1 and spRif1, recruited to telomeres by Taz1, are essential for telomere function in fission yeast. Curr Biol. 2001;11:1624–1630. doi: 10.1016/s0960-9822(01)00503-6. [DOI] [PubMed] [Google Scholar]
- Kutay U, Hetzer MW. Reorganization of the nuclear envelope during open mitosis. Curr Opin Cell Biol. 2008;20:669–677. doi: 10.1016/j.ceb.2008.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C, Chen S, Jackson DA. Inheriting nuclear organization: can nuclear lamins impart spatial memory during post-mitotic nuclear assembly? Chromosome Res. 2010;18:525–541. doi: 10.1007/s10577-010-9137-8. [DOI] [PubMed] [Google Scholar]
- Martinez P, Thanasoula M, Carlos AR, Gomez G, Tejera AM, Schoeftner S, Dominguez O, Pisano DG, Tarsounas M, Blasco MA. Mammalian RAP1 exerts a dual control of telomere function and gene expression through binding to both telomeric and extra-telomeric sites. Nat Cell Biol. 2010 doi: 10.1038/ncb2081. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Sullivan RJ, Karlseder J. Telomeres: protecting chromosomes against genome instability. Nat Rev Mol Cell Biol. 2010;11:171–181. doi: 10.1038/nrm2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Sullivan RJ, Kubicek S, Schreiber SL, Karlseder J. Reduced histone biosynthesis and chromatin changes arising from a damage signal at telomeres. Nat Struct Mol Biol. 2010;17:1218–1225. doi: 10.1038/nsmb.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmakumar VC, Libotte T, Lu W, Zaim H, Abraham S, Noegel AA, Gotzmann J, Foisner R, Karakesisoglou I. The inner nuclear membrane protein Sun1 mediates the anchorage of Nesprin-2 to the nuclear envelope. J Cell Sci. 2005;118:3419–3430. doi: 10.1242/jcs.02471. [DOI] [PubMed] [Google Scholar]
- Scherthan H, Sfeir A, de Lange T. Rap1-independent telomere attachment and bouquet formation in mammalian meiosis. Chromosoma. 2011;120:151–157. doi: 10.1007/s00412-010-0295-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schober H, Ferreira H, Kalck V, Gehlen LR, Gasser SM. Yeast telomerase and the SUN domain protein Mps3 anchor telomeres and repress subtelomeric recombination. Genes Dev. 2009;23:928–938. doi: 10.1101/gad.1787509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talamas JA, Hetzer MW. POM121 and Sun1 play a role in early steps of interphase NPC assembly. J Cell Biol. 2011;194:27–37. doi: 10.1083/jcb.201012154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo H, Ghosh S, Luesch H, Ghosh A, Wong ET, Malik N, Orth A, de Jesus P, Perry AS, Oliver JD, et al. Telomere independent Rap1 is an IKK-adaptor and regulates NFκB- dependent gene expression. Nat Cell Biol. 2010 doi: 10.1038/ncb2080. in press. [DOI] [PubMed] [Google Scholar]
- Vermolen BJ, Garini Y, Mai S, Mougey V, Fest T, Chuang TC, Chuang AY, Wark L, Young IT. Characterizing the three-dimensional organization of telomeres. Cytometry A. 2005;67:144–150. doi: 10.1002/cyto.a.20159. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.