Abstract
Cells possess internal ~24-hour or circadian clocks that synchronize physiological processes with daily cycles of light and nutrient availability. In this issue, Asher et al. (2010) find that PARP-1 (Poly(ADP-ribose) polymerase-1) modifies components of the clock machinery in response to feeding, providing a mechanism for how metabolic rhythms coordinate with circadian rhythms.
Many organisms synchronize their behavior, physiology, and metabolism with the 24-hour rotation of the Earth. In mammals, a master pacemaker is located in the hypothalamic region called the suprachiasmatic nucleus (SCN), which contains a cluster of ~10,000 neurons. Each SCN neuron expresses a transcriptional feedback loop that self-generates an oscillation with a period of ~24 hours (Green et al. 2008). Cells in peripheral tissues, such as the liver, contain similar cell-autonomous clocks, and the SCN synchronizes the oscillation of their internal clocks to coordinate rhythms throughout the body. However, in both the SCN and peripheral tissues, the molecular clocks must integrate extracellular cues to maintain synchrony with the environment. Light is the dominant cue for the central clock in the SCN, but for peripheral tissues, metabolic cycles, such as feeding and fasting, can also regulate the internal clocks.
The link between circadian and metabolic rhythms is an area of intense study because disrupting the synchrony is thought to contribute to the etiology of disorders such as diabetes, obesity, and cardiovascular disease (Green et al. 2008). Nonetheless, how circadian and metabolic systems interact remains largely undefined; in particular, how feeding directly modulates the molecular oscillators in peripheral tissues has been a mystery. In this issue of Cell, Asher et al. (2010) find that that the activity of Poly(ADP-ribose) polymerase-1 (PARP-1) in the liver of mice oscillates in synchrony with the feeding-fasting cycle, providing a new link between metabolism and circadian rhythms.
PARP-1 is a highly conserved nuclear protein that adds chains of ADP-ribose molecules to proteins in a process called PARylation (or polyADP-ribosylation). PARP-1 uses nicotinamide adenine dinucleotide (NAD+) to synthesize these polymers, which can include up to 200 ADP-ribose units. Like other post-translational modifications, PARylation alters protein function; chains of ADP-ribose are negatively charged and thus, their addition is believed to disrupt electrostatic interactions, such as those involved in DNA binding. The major substrate of PARP-1 is itself, and auto-PARylation inhibits DNA binding of PARP-1 (D’Amours et al., 1999).
Although PARP-1 was originally characterized as a protein involved in the DNA repair pathway, recent evidence suggests that PARP-1 plays crucial roles in many processes, including chromatin remodeling and transcription (Krishnakumar and Kraus, 2010). Furthermore, the catalytic activity of PARP-1 is regulated by many factors, including NAD+ levels, allosteric interactions with chromatin and transcription factors, and numerous types of post-translational modifications, such as acetylation, phosphorylation, sumolyation and auto-ADP-ribosylation (D’Amours et al., 1999).
Now Asher et al. (2010) show that the PARylation activity of PARP-1 oscillates with a circadian pattern in the liver of mice. Using a combination of in vitro and in vivo approaches, the authors find that PARP-1 PARylates itself in a rhythmic manner, with the peak activity occurring at the beginning of the light phase when mice consume little food. Surprisingly, this rhythmic activity of PARP-1 is still present when the circadian clock in the cell has been suppressed; however, its phase can be shifted by altering the feeding time. Thus, oscillations of PARP-1 activity in the liver appear to be regulated by feeding signals rather than by components of the circadian clock.
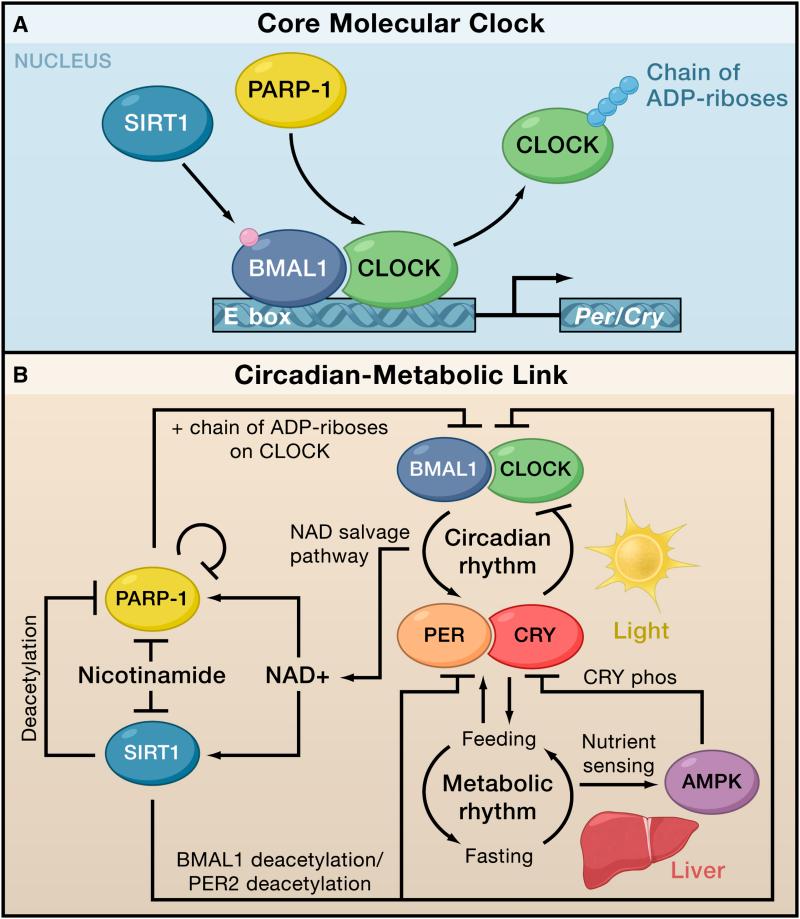
But what about the converse? Could the oscillations in PARP-1 activity help to synchronize the endogenous clock with the feeding-fasting cycle? To address this question, the authors determined if PARP-1 modified two central components of the cellular clock, CLOCK and BMAL1 (brain and muscle aryl hydrocarbon receptor nuclear translocator (ARNT)-like). CLOCK and BMAL1 heterodimerize to activate transcription of their repressors, Period (Per) and Cryptochrome (Cry), whose proteins products form a complex involved in the negative feedback loop of the cell’s internal clock (Figure 1). Asher and colleagues find that PARP-1 not only interacts with these transcriptions factors, but it also PARylates CLOCK with a circadian rhythm that mirrors the oscillations of its own auto-PARylation. Moreover, PARylation of CLOCK reduces its binding to DNA and possibly alters transcription of its target genes (Figure 1). Taken together, these results indicate that PARP-1 is a critical player in a pathway that connects nutrient intake to the molecular clock of liver cells.
Figure 1. PARP-1 links the circadian clock with metabolic rhythms.
(A) The molecular clock in cells is composed of an autoregulatory feedback loop that takes ~24 hours to complete. The transcription factor CLOCK and BMAL1 bind to E-boxes and activate the negative arm of the feedback loop by triggering gene expression of their own repressors, Cryptochrome (Cry) and Period (Per). In response to feeding, PARP-1 poly-ADP-ribosylates (i.e., PARlyates) CLOCK (Asher et al. 2010), which inhibits DNA binding by CLOCK/BMAL1. In contrast, Sirtuin I (SIRT1) modulates BMAL1 by deacetylating it.
(B) Circadian rhythms exert a strong influence over metabolic rhythms by controlling the timing of behaviors, such as feeding and sleep. Metabolic genes, such as AMP-activated protein kinase (AMPK) regulate levels of NAD+ and are themselves under circadian control. The activities of PARP-1 and SIRT1 are directly modulated by NAD+, and these enzymes feedback to regulate circadian genes.
The regulation of PARP-1 by nutrient intake is the latest discovery linking circadian rhythm to metabolism and energy balance (Green et al., 2008). Many metabolic hormones, including insulin, leptin, and corticosterone, exhibit circadian oscillations, and the SCN also regulates metabolism by modulating key transport systems and metabolic enzymes. CLOCK and BMAL1 themselves directly regulate numerous nuclear receptors involved in lipid and glucose metabolism and adipogenesis, including REV-ERBα and PPARα (peroxisome proliferator-activated receptors) (Froy and Miskin, 2010). Disrupting the Clock gene in mice induces many symptoms of metabolic syndrome, including obesity, hyperlipidemia, high circulating glucose, and low circulating insulin (reviewed in Green et al., 2008). Similarly, deleting Bmal1 in the pancreas leads to hypoinsulinaemia and diabetes (Marcheva et al., 2010).
Although metabolism is thought to be primarily downstream of the cellular clock, numerous studies provide evidence that metabolic cycles can operate independently from or even influence circadian rhythms. For example, when animals are given meals at only one particular time each day, their behavior and physiology adjust to the restricted feeding regime; over a period of time, they anticipate the arrival of food. This behavior can be entrained by altering feeding time. This observation lead to the hypothesis that a food-entrainable oscillator exists in the peripheral tissue that is distinct from the circadian oscillator in the SCN (Green et al., 2008).
Asher and colleagues find that disrupting the Parp-1 gene in mice impairs the ability of liver cells to adapt to changes in their feeding schedule. When the authors switch the feeding time from night to day, the oscillations of PARP-1 activity in liver cells of wild-type mice synchronize with the feeding schedule after ~4 days, but this entrainment is significantly delayed in the mice lacking Parp-1. Thus, PARP-1 appears to contribute to the cellular machinery that entrains with the feeding-fasting cycle. Furthermore, mice lacking the Parp-1 gene have a slightly longer circadian period than wild-type mice, suggesting that PARP-1 may feedback and alter the master circadian clock in the SCN
Daily oscillations in PARP-1 activity also have implications for cellular levels of NAD+ and the function of Sirtuin 1 (SIRT1). An NAD+-dependent deacetylase, SIRT1 is involved with energy homeostasis together with the AMP-activated protein kinase (AMPK) (Figure 1). AMPK is another nutrient sensor, and it regulates SIRT1 activity by controlling levels of NAD+. Thus, AMPK may also control PARP-1 activity (Canto and Auwerx, 2009). SIRT1 can regulate the activity of BMAL1 and PER2 by deacetylation (Figure 1) (Asher et al., 2008; Nakahata et al., 2008). Furthermore, PARP-1 and SIRT1 potentially compete for a limited pool of NAD+; SIRT1 can directly regulate PARP-1 activity by deacetylation (Krishnakumar and Kraus, 2010); and, these two enzymes appear to antagonize each other functionally (Kolthur-Seetharam et al., 2006). Therefore, characterizing the interplay among PARP-1, SIRT1, and AMPK in response to nutrients will be important in determining how circadian and metabolic rhythms are linked.
Asher and colleagues describe a new role for PARP-1, and as is the case in most new discoveries, their results raise as many questions as they answer. Foremost, it is not clear what regulates the rhythmic activation of PARP-1. Although an obvious candidate is NAD+ availability, Asher and colleagues find that excess NAD+ does not alter the oscillations in PARP-1 activity. Given the numerous mechanisms through which PARP-1 can be activated, it will be challenging to pinpoint the exact one operating here. Nevertheless, the work presented by Asher and colleagues adds a new dimension to the biology of PARP-1 and identifies an important link between metabolic and circadian rhythms.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Asher G, Gatfield D, Stratmann M, Reinke H, Dibner C, Kreppel F, Mostoslavsky R, Alt FW, Schibler U. Cell. 2008;134:317–328. doi: 10.1016/j.cell.2008.06.050. [DOI] [PubMed] [Google Scholar]
- Asher G, Reinke H, Altmeyer M, Gutierrez-Arcelus M, Hottiger MO, Schibler U. Cell. 2010;(this issue) doi: 10.1016/j.cell.2010.08.016. [DOI] [PubMed] [Google Scholar]
- Canto C, Auwerx J. Curr Opin Lipidol. 2009;20:98–105. doi: 10.1097/MOL.0b013e328328d0a4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Amours D, Desnoyers S, D’Silva I, Poirier GG. Biochem J. 1999;34:249–268. [PMC free article] [PubMed] [Google Scholar]
- Froy O, Miskin R. Aging. 2010;2:7–27. doi: 10.18632/aging.100116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green CB, Takahashi JS, Bass J. Cell. 2008;134:728–742. doi: 10.1016/j.cell.2008.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolthur-Seetharam U, Dantzer F, McBurney MW, de Murcia G, Sassone-Corsi P. Cell Cycle. 2006;5:873–877. doi: 10.4161/cc.5.8.2690. [DOI] [PubMed] [Google Scholar]
- Krishnakumar R, Kraus WL. Mol Cell. 2010;39:8–24. doi: 10.1016/j.molcel.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcheva B, Ramsey KM, Buhr ED, Kobayashi Y, Su H, Ko CH, Ivanova G, Omura C, Mo S, Vitaterna MH, et al. Nature. 2010;466:627–631. doi: 10.1038/nature09253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, Chen D, Guarente LP, Sassone-Corsi P. Cell. 2008;134:329–340. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]