Abstract
Our objective was to design and implement a clinical history database capable of linking to our database of quantitative results from 99mTc-mercaptoacetyltriglycine (MAG3) renal scans and export a data summary for physicians or our software decision support system.
Methods
For database development, we used a commercial program. Additional software was developed in Interactive Data Language. MAG3 studies were processed using an in-house enhancement of a commercial program. The relational database has 3 parts: a list of all renal scans (the RENAL database), a set of patients with quantitative processing results (the Q2 database), and a subset of patients from Q2 containing clinical data manually transcribed from the hospital information system (the CLINICAL database). To test interobserver variability, a second physician transcriber reviewed 50 randomly selected patients in the hospital information system and tabulated 2 clinical data items: hydronephrosis and presence of a current stent. The CLINICAL database was developed in stages and contains 342 fields comprising demographic information, clinical history, and findings from up to 11 radiologic procedures. A scripted algorithm is used to reliably match records present in both Q2 and CLINICAL. An Interactive Data Language program then combines data from the 2 databases into an XML (extensible markup language) file for use by the decision support system. A text file is constructed and saved for review by physicians.
Results
RENAL contains 2,222 records, Q2 contains 456 records, and CLINICAL contains 152 records. The interobserver variability testing found a 95% match between the 2 observers for presence or absence of ureteral stent (κ = 0.52), a 75% match for hydronephrosis based on narrative summaries of hospitalizations and clinical visits (κ = 0.41), and a 92% match for hydronephrosis based on the imaging report (κ = 0.84).
Conclusion
We have developed a relational database system to integrate the quantitative results of MAG3 image processing with clinical records obtained from the hospital information system. We also have developed a methodology for formatting clinical history for review by physicians and export to a decision support system. We identified several pitfalls, including the fact that important textual information extracted from the hospital information system by knowledgeable transcribers can show substantial interobserver variation, particularly when record retrieval is based on the narrative clinical records.
Keywords: MAG3 renography, databases, decision support systems
Publication in medical informatics has grown exponentially in the last 20 y (1), concurrent with the development of electronic medical records and general improvement in information technology. An important component of this growth has been the use of database technology.
Several national and multiinstitutional databases have been developed to hold information on patients with kidney disease (2), including databases for the Health Resources and Services Administration (3), the National Multicystic Kidney Registry (4), and the Dialysis Outcomes and Practice Patterns study (5). The largest such database is probably the U.S. Renal Data System, maintained by the National Institute of Diabetes and Digestive and Kidney Diseases of the National Institutes of Health. As of 2008, this database was aware of more than 2 million Medicare patients with chronic kidney disease (6).
In the field of nuclear medicine, it is not uncommon for individual laboratories to maintain databases of patient records as teaching files, for business purposes, or for tracking patients with specific diseases (7–9). Our use of renal databases began with 2 initiatives. The first was to collect quantitative results from the computerized analysis of 99mTc-mercaptoacetyltriglycine (MAG3) renal scans. For this analysis, we used QuantEM-II, an in-house–developed enhanced version (10) of the commercial QuantEM program (GE Healthcare). We wanted to collect the results of many patient studies in a form that would be organized and searchable, convenient for research projects.
The second initiative was to prepare input for RENEX, a software decision support system under development for interpreting MAG3 renal scans (11–14). A specific goal has been to extend RENEX by incorporating knowledge of the patient’s clinical history into the decision support algorithm for scan interpretation. This goal requires clinical data to be provided in a strictly defined format. Moreover, to obtain valid comparisons between scan interpretations provided by RENEX and the interpretations of physicians, the clinical data provided to RENEX must also be summarized in a human-readable format for physicians.
Our objective was to develop a relational database system to organize patient history data, relate these data to the quantitative output of QuantEM-II, and prepare a clinical summary formatted for use by either human readers or the RENEX decision support system. We also wanted to establish the robustness of the relational database by determining the interobserver agreement of pertinent clinical variables.
MATERIALS AND METHODS
Institutional Review Board approval was obtained to retrospectively examine patient records in the hospital information system (HIS). To hold data collected from HIS, we used FileMaker Pro (FileMaker, Inc.), a commercial database management application that is relational, scriptable, and highly customizable. We used the developer version of FileMaker Pro, which allows a database to be deployed as an application on any computer using the same operating system, without requiring the commercial FileMaker program. Software for combining clinical data and MAG3 quantitative results was developed using Interactive Data Language (ITT Visual Information Solutions).
Database Design
To create a database file using the FileMaker Pro application, a table to hold conceptually related data is first defined. Individual data items are defined within the table as fields, and these can be presented to the user in a visual arrangement, or layout. Our database is organized into 3 parts. The first (the RENAL database) is a listing of all retrievable renal scans acquired in our department. Records are created manually and contain a minimal number of fields to identify and categorize each patient according to the clinical indication for MAG3 imaging. The second part (the Q2 database) represents patients from RENAL whose images have been processed using QuantEM-II. Records are created automatically by importing of the XML (extensible markup language) results file saved by QuantEM-II. Fields in this database include patient demographics, calculated functional values, quality control findings determined from QuantEM-II (15), and names of research projects in which the study was included. The third part (the CLINICAL database) represents patients from Q2 for whom a clinical history has been compiled from the HIS. Patient records in CLINICAL are created manually, each with a study date that corresponds to a particular MAG3 scan date. If a patient has more than one MAG3 scan, each scan has its own record and its own relevant clinical history. The 3 databases are profiled in Table 1.
TABLE 1.
Profile of Databases
| Profile | RENAL (1 table) | Q2 (1 table) | CLINICAL (2 tables) | ||
|---|---|---|---|---|---|
| Total | History | Imaging | |||
| Fields | 14 | 288 | 355 | 275 | 80 |
| Layouts | 3 | 22 | 20 | 15 | 5 |
| Scripts | 4 | 11 | 49* | ||
| Value lists | 0 | 5 | 27* | ||
Same scripts and value lists are used for history and imaging tables.
Records in all 3 databases can be browsed with fields shown either in tabular format, similar to a spreadsheet, or in various graphic layouts that organize records visually through use of color and grouping of conceptually related fields (16).
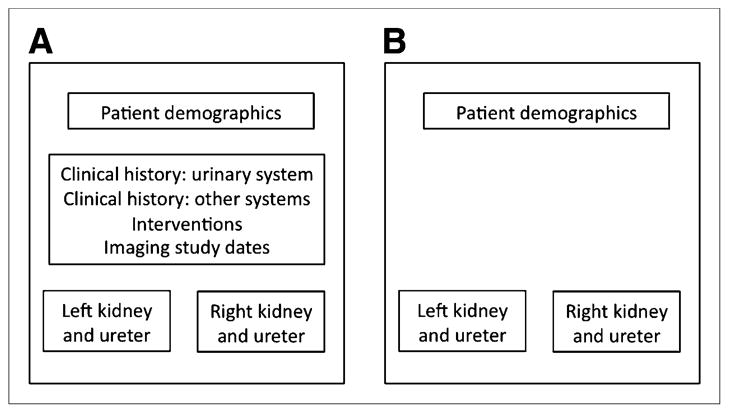
Two independent tables are defined for CLINICAL, with records related between tables by a unique identifier. The conceptual organization is shown in Figure 1.
FIGURE 1.
Organization of CLINICAL. History table (A) includes fields summarizing patient’s history of conditions affecting urinary system and other systems, interventional procedures that have been performed, and left and right kidney and ureter findings. Imaging results table (B) contains same set of left and right kidney and ureter fields, derived from imaging study reports rather than narrative history. Interventions and other history are not included on this table. Demographics are present on both tables so that patient records can be matched to their imaging studies.
The first table in CLINICAL (Fig. 1A) holds demographics, clinical history, a brief summary of findings from prior MAG3 scans, and the dates of other imaging studies. The second Table (Fig. 1B) holds the results of up to 11 additional radiologic reports: CT scans (up to 3), sonograms (up to 2), CT angiograms, MR images, MR angiograms, retrograde contrast images, kidney–ureter–bladder radiographs, and intravenous pyelograms. The CT and ultrasound dates on a patient’s records are automatically sorted chronologically on entry. History and imaging findings up to the day of the MAG3 scan are entered for each patient record. Fifty-six of the field names relating to left and right kidney history are also used on the imaging table.
Clinical Database Development
To protect the privacy of patient information, access to HIS and to computer systems containing our databases is controlled by user name and password.
The design of CLINICAL was developed using the technique of continual refinement with feedback (17,18). The initial list of database fields was developed by the nuclear medicine physician who served as a domain expert for renal imaging. Another physician transcriber began adding patient records to this database and populating those with data from HIS. Meetings were held regularly between the domain expert, the transcriber, and the database implementer, and after each meeting the user interface of CLINICAL was revised as necessary. The number of fields or the structure and behavior of fields were modified to reflect the breadth of content available in HIS, to add new terminology, or to address the formatting needs of RENEX. Whenever new fields were added to CLINICAL, existing patient records were revisited in HIS. Each iteration in the development of CLINICAL was given a unique version number. The final database contains 342 fields.
The database design uses structured data entry, with narrowly defined fields whose contents are restricted by a value list—a list of allowable values from which the user may choose. Value lists are intended to provide the most appropriate descriptors for various clinical conditions. The complete list of fields present in both the left and the right kidney and ureter sections of CLINICAL, along with their complete value lists where applicable, is given in Table 2.
TABLE 2.
Database Fields for Clinical History Table
| Field | Type of data | Value list contents |
|---|---|---|
| History | Value list | Normal, absent, no comment |
| Urinoma | Value list | No data, not present, equivocal, present |
| Renal parenchyma | Value list | No data, normal, equivocal, atrophied |
| Renal scar | Value list | No data, not present, equivocal, present |
| Hydronephrosis | Value list | No data, not present, equivocal, present, mild, moderate, severe |
| Hydroureter | Value list | No data, not present, equivocal, present, mild, moderate, severe |
| Stricture | Value list | No data, not present, equivocal, present |
| Renal calculus | Value list | No data, not present, equivocal, present |
| Largest size (mm) | Literal value | |
| Ureteropelvic junction calculus | Value list | No data, not present, equivocal, present |
| Largest size (mm) | Literal value | |
| Ureteral calculus | Value list | No data, not present, equivocal, present |
| Largest size (mm) | Literal value | |
| Calculus, obstructive | Value list | No data, not obstructive, obstructive |
| Solid renal mass | Value list | No data, not present, equivocal, present |
| Largest size (cm) | Literal value | |
| Cystic renal mass | Value list | No data, not present, equivocal, present |
| Largest size (cm) | Literal value | |
| Mixed renal mass | Value list | No data, not present, equivocal, present |
| Largest size (cm) | Literal value | |
| Mass, obstructive | Value list | No data, not obstructive, obstructive |
| Renal artery stenosis | Value list | No data, not present, equivocal, mild, moderate, severe |
| Surgery | Checkbox options (1 or more) | Prior stent, current stent, prior nephrostomy, current nephrostomy, ureteral reimplantation, pyeloplasty, total nephrectomy, partial nephrectomy, nephrolithotomy |
| Stent removal date | Literal date | |
| Nephrostomy removal date | Literal date | |
| Flank pain on arrival | Value list | No data, not present, equivocal, present |
| Flank pain after diuretic | Value list | No data, not present, equivocal, present |
| Ureterocele | Value list | No data, not present, equivocal, present |
| Duplicated urinary system | Value list | No data, not present, equivocal, present |
When a new patient record is created in CLINICAL, the default value for most fields is “no data,” which means the value is unknown. For many fields, the standard value list consists of “no data,” “not present,” “equivocal,” and “present.” “Not present” means there is evidence in HIS that a clinical condition is not present or that an intervention has not been performed. “Patient category” and “notes” fields are available to capture information that is more general or to indicate the need for new field definitions.
Data Export
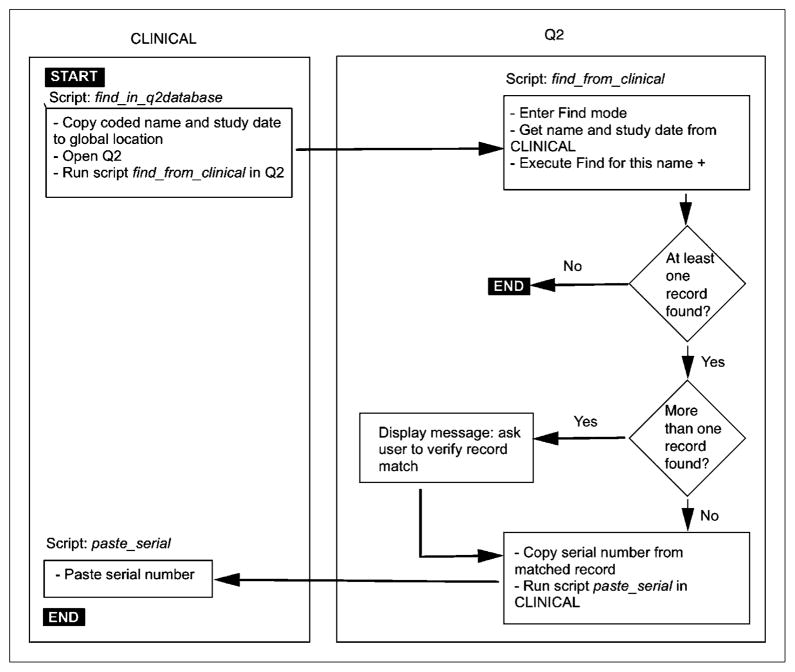
The contents of a patient’s records can be exported from CLINICAL in 1 of 2 formats. The first format is XML, used to create input for the RENEX system. Database scripts build XML format tags for all fields to be exported and concatenate these into a file saved to disk. Q2 and CLINICAL export their results to 2 separate XML files, and these must be matched and combined into a single file for use by RENEX. To match records reliably, relationships were defined between database files. Q2 was designated the master file, with a serial number automatically assigned when a new record is created (19). Serial numbers are never reused, even if the associated record is permanently deleted. The 2 databases communicate via program scripts, and if a matching record is found, the serial number of the record is copied from Q2 to CLINICAL. Under script control, a combined XML file is not created unless a valid serial number is present for that patient. The record-matching algorithm is shown in Figure 2. Once the record match is successful, an Interactive Data Language program is invoked to read the separate XML files from Q2 and CLINICAL and combine these into a single file that can then be used by RENEX.
FIGURE 2.
Algorithm for matching patient records across database tables.
The second method of export from CLINICAL is to create a text file that is readable by a physician interpreting a MAG3 study. A subset of fields in CLINICAL is used, excluding any field whose value is “no data.” Compilation of the patient history makes extensive use of calculated fields, whose content is dynamic and is built from other fields by applying a sequence of text-manipulation functions available in FileMaker Pro. Functions automatically add or change punctuation and add words to form complete sentences within the calculated fields. Next, scripted algorithms poll all the calculated fields, assembling their contents into a single text field that is a structured narrative of patient history (20). Finally, this field is automatically exported to disk as a text file. The process is illustrated in Figure 3.
FIGURE 3.
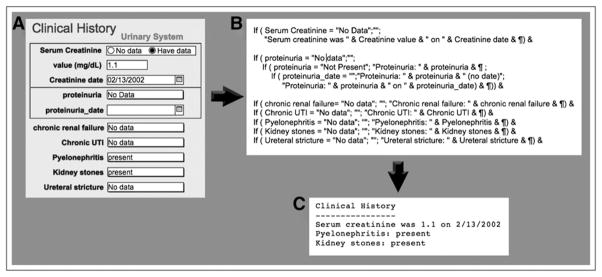
Example of how small subset of fields in CLINICAL would be converted to structured text for physician review. (A) Fields as they appear on graphic layout with which user interacts. (B) Database’s internal manipulation of same fields by text functions, performed in calculated field not seen by user. (C) Text file extracted by database script. Fields with value “no data” are not included, greatly simplifying narrative.
After all patient records were entered in the clinical database, interobserver variability was evaluated by selecting 2 clinical variables (hydronephrosis and presence of a ureteral stent) that may affect MAG3 scan interpretation regarding the presence or absence of obstruction. Fifty patients were selected at random from CLINICAL, using an algorithm that generates random numbers. A second physician transcriber searched HIS for the 2 clinical fields: presence of a ureteral stent was tabulated from history documents, and hydronephrosis was tabulated both from the narrative summaries of hospitalizations/patient visits and separately from the actual imaging reports. The 2 transcribers were considered to agree if both found a stent to be present, if both found a stent to be absent, if both found hydronephrosis to be present, or if both found hydronephrosis to be absent. Agreement was evaluated using the κ-statistic (21). The field value “no data” was considered a match only if the second transcriber also entered that value.
RESULTS
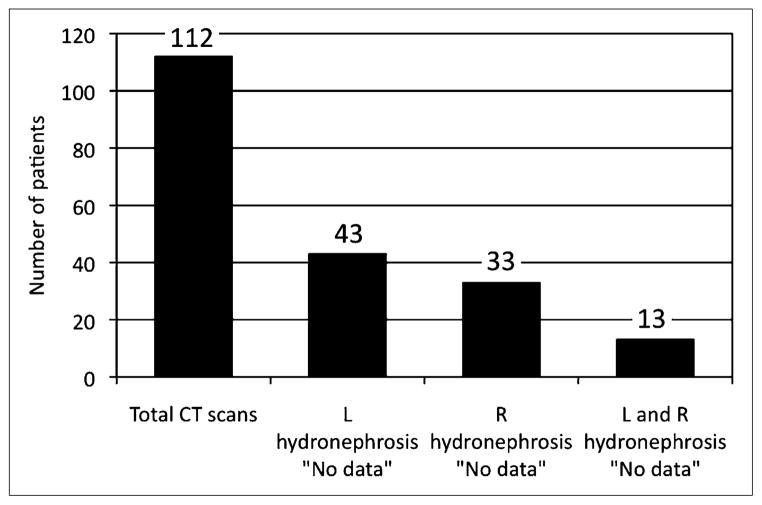
RENAL contains 2,222 records spanning more than 10 y, Q2 contains 456 records, and CLINICAL contains 152 records in the history table and 302 records in the imaging studies table. CLINICAL allows many combinations of values to be searched and tabulated. As an example, because presence or absence of hydronephrosis is an important clinical variable, CLINICAL was queried to determine, first, the number of CT scans performed within 1 y of the MAG3 scan for obstruction and, second, the frequency that an important clinical finding (presence or absence of hydronephrosis) was omitted from the CT report. Figure 4 indicates that 112 CT scans were performed within 1 y before MAG3 imaging for suspected obstruction, yet there was no comment on the presence or absence of hydronephrosis in 38%, 29%, and 12% for the left kidney, right kidney, or both kidneys, respectively.
FIGURE 4.
Reporting of hydronephrosis for 112 CT scans performed within 1 y of MAG3. Hydronephrosis was not reported as either present or absent for 38% (43/112) of left kidneys, 29% (33/112) of right kidneys, and 12% (13/112) of both left and right kidneys.
We encountered several pitfalls in transcribing from HIS. One patient’s MAG3 scan was not entered in HIS, resulting in a study date in CLINICAL that was for a later scan. Once the error had been detected, the CT findings, which were later than the true MAG3 date, were removed from CLINICAL. One patient had several alias names in HIS and had history items associated with some names but not others. We noted that renal ultrasound studies can be performed in the physician’s office, and although these have formal reports, they are not archived in the radiology section of our HIS. Occasionally, an imaging study report used terminology different from our value lists. For example, the value list used mild, moderate, and severe to describe the degree of hydronephrosis, but radiologists were not consistent in applying this terminology to describe hydronephrosis and the data recorder had to use judgment to place terms such as “marked hydronephrosis” or combined terms such as “mild to moderate hydronephrosis” into the mild, moderate, or severe categories of our value list.
The algorithm developed to match records between Q2 and CLINICAL—and produce an input for RENEX—failed to find an exact match in 6 of 152 cases. There were 3 typographic errors in entering the MAG3 study date in CLINICAL. In 2 cases, the date of a different MAG3 study was used, and in 1 case the MAG3 date differed by 1 d between HIS and CLINICAL because HIS used the date the interpretation was finalized, which was the morning after a late-afternoon study. The 6 discrepancies were manually resolved by reviewing the records in Q2 and CLINICAL.
In the 50 patients selected to test interobserver variability for retrieval of significant database fields, the presence of a current ureteral stent from history was matched between the 2 transcribers for 95% of kidneys (κ = 0.52; Table 3). The presence or absence of hydronephrosis based on the narrative history in the summaries of hospitalizations or patient visits was matched for 75% of kidneys (κ = 0.41; Table 4). Of the 50 patients reviewed, 34 had 49 imaging studies found by both transcribers; the presence or absence of hydronephrosis determined from these imaging study reports was matched for 92% of kidneys (κ = 0.84; Table 5).
TABLE 3.
Joint Judgment of 2 Readers Regarding Presence of Current Ureteral Stent Based on Narrative Summaries of Hospitalizations and Patient Visits
| Reader 1 | Reader 2 | ||
|---|---|---|---|
| Stent present | Stent absent | Total | |
| Stent present | 3 | 0 | 3 |
| Stent absent | 5 | 92 | 97 |
| Total | 8 | 92 | 100 |
TABLE 4.
Joint Judgment of 2 Readers Regarding Presence of Obstruction Based on Narrative Summaries of Hospitalizations and Patient Visits
| Reader 1 | Reader 2 | ||
|---|---|---|---|
| Hydronephrosis present | Hydronephrosis absent | Total | |
| Hydronephrosis present | 17 | 19 | 36 |
| Hydronephrosis absent | 6 | 58 | 64 |
| Total | 23 | 77 | 100 |
TABLE 5.
Joint Judgment of 2 Readers Regarding Presence of Obstruction Based on Imaging Reports
| Reader 1 | Reader 2 | ||
|---|---|---|---|
| Hydronephrosis present | Hydronephrosis absent | Total | |
| Hydronephrosis present | 41 | 8 | 49 |
| Hydronephrosis absent | 0 | 49 | 49 |
| Total | 41 | 57 | 98 |
DISCUSSION
We have developed a database system to relate clinical history to findings from quantitative analysis of MAG3 studies. The database consists of 3 parts: RENAL, a list of all renal scans; Q2, patients with quantitative processing; and CLINICAL, patients from Q2 who also have clinical history data. The fact that Q2 includes measures of scan quality is potentially just as important for clinicians as for the decision support system, analogous to the way image quality measures are useful in image databases for teaching (22). For exchanging data between databases, we use XML, a text-based format developed to facilitate data interchange between applications (23).
The design of a database is particularly important if the system is to be clinically useful (24). After the initial design of CLINICAL, we refined it in an iterative fashion (17,18). The use of version numbers allowed tracking of the evolution of the application and reversion of the database to an earlier version if necessary. Information was entered manually because we lack an electronic connection to HIS. Structured entry was used to limit semantic errors in the entering of textual information (25) and to help ensure uniformity in the use of clinical terminology. Moreover, structured entry helps to generate data that can more easily be reused for other purposes (26), such as for export to our decision support system (27). Value lists (Table 2) were developed to correspond as closely as possible to the most commonly encountered terminology in HIS. Our value lists for most clinical conditions consisted of “no data,” “not present,” “equivocal,” and “present.” Because it was not possible to use the same value list for all fields, however, in some fields “present” might be expanded and qualified by such terms as “mild,” “moderate,” or “severe” or other appropriate values. A general-purpose “notes” field holds information that does not fit elsewhere. Decisions made by the transcriber when interpreting textual information in HIS can be described in the “notes” field for possible later clarification (28), potentially reducing the number of times HIS must be accessed. This feature addresses the inflexibility that is a disadvantage of the structured-entry paradigm (29).
Of the 3 databases we developed, CLINICAL has been the most challenging to design and populate with meaningful data. A patient history is assembled from many kinds of text documents in HIS—documents that vary in completeness and may use abbreviations or unfamiliar or nonstandard terminology. HIS, although being our primary data source, can never be assumed to be free of errors and ambiguities. This limitation is a recognized challenge in the use of textual information (28).
Verifying the completeness and logical consistency of the information in HIS, and maintaining consistency after the information is transferred to CLINICAL (which is always a small subset of HIS), depends on the medical knowledge, insight, and judgment of the transcriber. One example is the use of different terminology by different clinical interpreters to describe the same imaging findings.
As an example of disambiguation, CLINICAL has separate fields for recording the presence of a current ureteral stent, the presence of a previous stent, and the date of stent removal. We encountered 2 scenarios for possible uncertainty. In scenario 1, the stated reason for performing a urologic procedure is to remove a stent, but there is no specific mention in the operative notes that this was actually done. This seems inconsistent, but it may follow institutional standard practice (the procedure indication always reflects what was actually done), or else it may simply reflect an individual physician’s dictation style. In scenario 2, a stent is removed and another is placed during the same procedure. In either of these scenarios, should a stent removal be entered in CLINICAL? In scenario 1, we did not count a stent removal; in scenario 2 we did, as well as recording the presence of a current stent.
The meaning of “no data” in imaging studies may be different from that in history. In most radiology reports, presence or absence of specific conditions (such as hydronephrosis) may be noted, depending on the indication for imaging. However, most reports do not list the absence of all conditions that could possibly be seen by that imaging modality. Thus “no data” might reasonably be interpreted to mean “condition not present.” We chose the more conservative use of “no data” as offering no evidence for or against the presence of the condition.
Many fields in the left and right kidney/ureter history are also available on the imaging table in CLINICAL. Presence or absence of a condition could be reported from an ultrasound or other imaging study and would be entered in the imaging table only. Conversely, evidence of a condition could be derived from several nonimaging sources in HIS and would then be entered in the history table only. Having a “history” of a condition such as hydronephrosis (apart from its notation on a specific interventional procedure report) implies that some imaging study may have been done in the past but that the date and detailed findings from that study are not available. In this sense, fields on the history table that derive from sources such as admission interviews, hospitalization discharge summaries, or office visit notes represent less precise data than fields on the imaging table. Consideration must also be given to what is and is not reported after an imaging study. Of the 112 abdominal CT scans obtained during the year before a MAG3 scan for suspected obstruction, 13 CT reports failed to comment on the presence of hydronephrosis (Fig. 4), suggesting that a pertinent negative finding was probably omitted, highlighting the benefit of structured reporting (30).
If the same patient is present in more than one of our databases, it is essential that records can be matched with certainty in all files (31,32). A possible matching method is to use one or a combination of several key fields the files have in common (18). This approach may be sufficient when the user is browsing records, but combining data from different files for external use requires greater security. To identify patients, we use unique serial numbers, controlled by a single database, and software methods for matching and combining XML files.
Correct filling of any database field requires several steps: the document must be appropriately populated and entered into HIS, the relevant document must then be located in HIS and interpreted by the transcriber (perhaps including correlation with other documents), and the findings must be correctly entered into the database. Our review of CT scan reports indicated that relevant information may be omitted and emphasizes the advantages of structured reports (30). Physician interobserver agreement in identifying the presence or absence of hydronephrosis based on the narrative summaries was 75%; expressed differently, the observers failed to obtain the same information regarding hydronephrosis in more than 20% of kidneys (κ = 0.41). κ is a measure of agreement that is corrected for chance. Landis and Koch have suggested that κ-values of 0.00–0.19 indicate poor agreement, 0.20–0.39 indicate fair agreement, 0.40–0.59 indicate moderate agreement, 0.60–0.79 indicate substantial agreement, and 0.80–1.00 indicate almost perfect agreement (33). A limitation of κ is its dependence on the prevalence or number in each of the rating categories; the high κ-values derived for detecting the presence of a stent in Table A, for example, are partially due to the high prevalence of “stent present” results. In our study, the observers were not pressured to complete their chart review in a limited time; interobserver variability would likely be greater in a time-pressured clinical setting. These results highlight the limitations of clinical data, particularly since the presence or absence of hydronephrosis can influence the interpretation of a MAG3 renal scan for obstruction. The results also point out the utility of structured reporting and data mining to develop more reliable and consistent databases.
There are several limitations in this development. Manual creation of records in CLINICAL requires significant time and expertise. Transcribers were not given specific guidelines on how to review HIS records systematically; however, both were physicians and were instructed to review the patient records to determine whether a patient had hydronephrosis or a ureteral stent. This approach is similar to the approach used by a physician to search HIS for relevant information that might assist in clinical image interpretation. Electronic exchange of information between databases may be available in the future, but automatic extraction of facts from narrative text in HIS would remain problematic. The task could be approached using natural language processing algorithms. There is growing database literature that addresses this complex challenge (34).
Control of data consistency within CLINICAL includes the use of value lists and monitoring of the date order of imaging studies. However, there is no methodology in the database to search for errors or to test logical consistency, such as checking to see if 2 imaging reports have contradictory findings.
We did not attempt to build our value lists to match an established formal taxonomy such as SNOMED CT (Systematized Nomenclature of Medicine–Clinical Terms) (35). Rather, we relied on the terminology understood from interaction with referring clinicians, as well as terms commonly used in our HIS.
CLINICAL contains only records for patients with suspected obstruction. The addition of patients in other clinical categories will probably require new fields and value lists. Finally, there are a limited number of records in CLINICAL. User-interface or other design deficiencies may become apparent when a larger variety of patient data is entered and a much larger number of records are browsed, searched, or sorted.
CONCLUSION
A relational database system has been developed that organizes the renal studies performed in our laboratory, compiles the results of quantitative image processing using QuantEM-II software, and holds patient clinical and history information using an iterative development cycle, a combination of free-form text fields, and structured data entry with defaults, flexible field layouts, and XML data interchange. Database functionality was extensively augmented through the use of program scripts and calculated fields, and several potential pitfalls were identified. This new system allows patient records to be reliably matched across files and formatted and exported for use by physicians or by a software decision support system. Finally, this system can also serve as a template for developing similar database systems in other laboratories.
Manual data transcription from HIS is time-intensive and often relies on the transcriber’s familiarity with clinical reports and the structure of HIS. Important textual information extracted from HIS by knowledgeable transcribers can show substantial interobserver variation (>20%), may not be as robust as is commonly assumed, and emphasizes the advantages of structured reporting and the potential of data mining.
Acknowledgments
This work was supported by grant R01-LM007595 from the National Institute of Biomedical Imaging and Bio-engineering and the National Institute of Diabetes and Digestive and Kidney Diseases. Russell Folks, Ernest Garcia, and Andrew Taylor receive royalties from the sale of QuantEM software.
Footnotes
This arrangement has been reviewed and approved by Emory University in accordance with its conflict-of-interest policy. No other potential conflict of interest relevant to this article was reported.
References
- 1.Deshazo JP, Lavallie DL, Wolf FM. Publication trends in the medical informatics literature: 20 years of “Medical Informatics” in MeSH. BMC Med Inform Decis Mak. 2009;9:7. doi: 10.1186/1472-6947-9-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaplan B, Schold J, Meier-Kriesche HU. Overview of large database analysis in renal transplantation. Am J Transplant. 2003;3:1052–1056. doi: 10.1034/j.1600-6143.2003.00193.x. [DOI] [PubMed] [Google Scholar]
- 3.OPTN/SRTR Annual Report. [Accessed September 6, 2012.];Health Resources and Services Administration (HRSA) Web site. Available at: http://optn.transplant.hrsa.gov/ar2009/chapter_iii_AR_cd.htm?cp=4.
- 4.Wacksman J, Phipps L. Report of the multicystic kidney registry: preliminary findings. J Urol. 1993;150:1870–1872. doi: 10.1016/s0022-5347(17)35918-9. [DOI] [PubMed] [Google Scholar]
- 5.Pisoni RL, Gillespie BW, Dickinson DM, Chen K, Kutner MH, Wolfe RA. The dialysis outcomes and practice patterns study (DOPPS): design, data elements, and methodology. Am J Kidney Dis. 2004;44:7–15. doi: 10.1053/j.ajkd.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 6.2010 Annual Report. [Accessed September 19, 2012.];United States Renal Data System Web site. Available at: http://www.usrds.org/2010/slides/indiv/v1index.html.
- 7.Morioka CA, El-Saden S, Pope W, et al. A methodology to integrate clinical data for the efficient assessment of brain-tumor patients. Inform Health Soc Care. 2008;33:55–68. doi: 10.1080/17538150801956762. [DOI] [PubMed] [Google Scholar]
- 8.Ruchin PE, Baron DW, Wilson SH, Boland J, Muller DWM, Roy PR. Long-term follow-up of renal artery stenting in an Australian population. Heart Lung Circ. 2007;16:79–84. doi: 10.1016/j.hlc.2006.12.008. [DOI] [PubMed] [Google Scholar]
- 9.Pouliot F, Lebel MH, Audet J-F, Dujardin T. Determination of success by objective scintigraphic criteria after laparoscopic pyeloplasty. J Endourol. 2010;24:299–304. doi: 10.1089/end.2009.0134. [DOI] [PubMed] [Google Scholar]
- 10.Folks R, Garcia E, Taylor A. Development of a software application for quantitative processing of nuclear renography [abstract] J Nucl Med. 2008;49 (suppl):157P. [Google Scholar]
- 11.Garcia EV, Taylor A, Halkar R, et al. RENEX: an expert system for the interpretation of 99mTc-MAG3 scans to detect renal obstruction. J Nucl Med. 2006;47:320–329. [PubMed] [Google Scholar]
- 12.Manatunga AK, Binongo JN, Taylor AT. Computer-aided diagnosis of renal obstruction: utility of log-linear modeling versus standard ROC and kappa analysis. EJNMMI Res. 2011;1:1–8. doi: 10.1186/2191-219X-1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bao J, Manatunga A, Binongo JNG, Taylor A. Key variables for interpreting MAG3 diuretic scans: development and validation of a predictive model. AJR. 2011;197:325–333. doi: 10.2214/AJR.10.5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taylor A, Garcia E, Binongo J, et al. Diagnostic performance of an expert system for interpretation of Tc-99m MAG3 scans in suspected renal obstruction. J Nucl Med. 2008;49:216–224. doi: 10.2967/jnumed.107.045484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Folks RD, Garcia E, Taylor A. Development and prospective evaluation of an automated software system for quality control of quantitative Tc-99m MAG3 renal studies. J Nucl Med Technol. 2007;35:27–33. [PMC free article] [PubMed] [Google Scholar]
- 16.Gulliksen J, Sandblad B. Domain-specific design of user interfaces. Int J Hum Comput Interact. 1995;7:135–151. [Google Scholar]
- 17.Patel VL, Kushniruk AW. Interface design for health care environments: the role of cognitive science. Proc AMIA Symp. 1998:29–37. [PMC free article] [PubMed] [Google Scholar]
- 18.Stewart M, Thind A, Terry AL, Chevendra V, Marshall JN. Implementing and maintaining a researchable database from electronic medical records: a perspective from an academic family medicine department. Healthc Policy. 2009;5:26–39. [PMC free article] [PubMed] [Google Scholar]
- 19.Lyons RA, Jones KH, John G, et al. The SAIL databank: linking multiple health and social care datasets. BMC Med Inform Decis Mak. 2009;9:3. doi: 10.1186/1472-6947-9-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson SB, Bakken S, Dine D, et al. An electronic health record based on structured narrative. J Am Med Inform Assoc. 2008;15:54–64. doi: 10.1197/jamia.M2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kundel HL, Polansky M. Measurement of observer agreement. Radiology. 2003;228:303–308. doi: 10.1148/radiol.2282011860. [DOI] [PubMed] [Google Scholar]
- 22.Lyman JA, Hersh W, Spackman K. Representing clinical information in an internal medicine teaching image database [abstract] Proc AMIA Symp. 2000:1074. [Google Scholar]
- 23.Mesiti M, Jimenez-Ruiz E, Sanz I, et al. XML-based approaches for the integration of heterogeneous bio-molecular data. BMC Bioinformatics. 2009;10:S7. doi: 10.1186/1471-2105-10-S12-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saleem JJ, Patterson ES, Militello L, Asch SM, Doebbeling BN, Render ML. Using human factors methods to design a new interface for an electronic medical record. Proc AMIA Symp. 2007:640–644. [PMC free article] [PubMed] [Google Scholar]
- 25.Grefen PWPJ, Apers PMG. Integrity control in relational database systems—an overview. Data Knowl Eng. 1993;10:187–223. [Google Scholar]
- 26.Rosenbloom ST, Stead WW, Denny JC, et al. Generating clinical notes for electronic health record systems. Appl Clin Inform. 2010;1:232–243. doi: 10.4338/ACI-2010-03-RA-0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Los RK, Ginneken AMV, Roukema J, Moll HA, Lei JVD. Why are structured data different? Relating differences in data representation to the rationale of OpenSDE. Med Inform Internet. 2005;30:267–276. doi: 10.1080/14639230500367563. [DOI] [PubMed] [Google Scholar]
- 28.Pestian JP, Itert L, Andersen C, Duch W. Preparing clinical text for use in biomedical research. J Database Manage. 2006;17:1–11. [Google Scholar]
- 29.Ash JS, Berg M, Coiera E. Some unintended consequences of information technology in health care: the nature of patient care information system-related errors. J Am Med Inform Assoc. 2004;11:104–112. doi: 10.1197/jamia.M1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taylor AT, Blaufox MD, De Palma D, et al. Guidance document for structured reporting of diuresis renography. Semin Nucl Med. 2012;42:41–48. doi: 10.1053/j.semnuclmed.2010.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Black N. Secondary use of personal data for health and health services research: why identifiable data are essential. J Health Serv Res Policy. 2003;8(suppl 1):36–40. doi: 10.1258/135581903766468873. [DOI] [PubMed] [Google Scholar]
- 32.Durham E, Xue Y, Kantarcioglu M, Malin B. Private medical record linkage with approximate matching. AMIA Annu Symp Proc. 2010;2010:182–186. [PMC free article] [PubMed] [Google Scholar]
- 33.Landis JR, Koch G. The measurement of observer agreement for categorical data. Biometrics. 1977;33:159–174. [PubMed] [Google Scholar]
- 34.Wei C, Sung S, Doong S, Ng P. Integration of structured and unstructured text data in a clinical information system. J Integrated Des Process Sci. 2006;10:61–77. [Google Scholar]
- 35.SNOMED Clinical Terms. [Accessed September 6, 2012.];National Library of Medicine Web site. Available at: http://www.nlm.nih.gov/research/umls/Snomed/snomed_main.html.