Abstract
Two families of E3 ubiquitin ligases are prominent in cell cycle regulation and mediate the timely and precise ubiquitin-proteasome-dependent degradation of key cell cycle proteins: the SCF (Skp1/Cul1/F-box protein) complex and the APC/C (Anaphase Promoting Complex or Cyclosome). While certain SCF ligases drive cell cycle progression throughout the cell cycle, APC/C (in complex with either of two substrate recruiting proteins: Cdc20 and Cdh1) orchestrates exit from mitosis (APC/CCdc20) and establishes a stable G1 phase (APC/CCdh1). Upon DNA damage or perturbation of the normal cell cycle, both ligases are involved in checkpoint activation. Mechanistic insight into these processes has significantly improved over the last ten years, largely due to a better understanding of APC/C and the functional characterization of multiple F-box proteins, the variable substrate recruiting components of SCF ligases. Here, we review the role of SCF- and APC/C-mediated ubiquitylation in the normal and perturbed cell cycle and discuss potential clinical implications of SCF and APC/C functions.
Keywords: Ubiquitin proteasome system, cell cycle, SCF complex, F-box, APC/C, cancer
1. Introduction
The mammalian cell cycle is a strictly regulated process controlled by the oscillating activities of cyclin-dependent kinases (CDKs), which are activated by cyclins and inhibited by CDK inhibitors (CKIs). Many cyclins and CDKs have been described, with each cyclin associating with one or more CDKs, and most CDKs interacting with one or more cyclins [1]. The prototypical cyclins driving cell cycle progression are cyclin A, B, and E, which are expressed in a cell cycle dependent manner and associate with Cdk1 or Cdk2 to mediate downstream events, such as DNA replication and mitosis. Specifically, cyclin E-Cdk2 is activated in late G1 phase to promote S-phase entry and DNA replication, cyclin A-Cdk2 and cyclin A-Cdk1 are involved in S-phase progression and the G2/M transition respectively, while cyclin B-Cdk1 is activated to promote mitotic entry.
This oscillating activity of CDKs is regulated by diverse mechanisms, starting from the transcriptional and translational level, to posttranslational modifications (phosphorylation in particular), and the periodic degradation of cyclins and CKIs by the ubiquitin-proteasome system [2, 3]. Contrary to reversible modifications, such as phosphorylation or association with CKIs, ubiquitin-mediated proteasomal degradation is an irreversible mechanism that assures the strict unidirectionality of the cell cycle, and it plays a key role in cell cycle regulation by mediating the precise spatial and temporal proteolysis of the main components of the cell cycle machinery.
Ubiquitylation, the covalent attachment of the small 76-amino-acid polypeptide ubiquitin to a target protein, occurs through a three-step enzymatic cascade. Ubiquitin is first bound and activated by the E1 ubiquitin activating enzyme in an ATP dependent manner. It is subsequently transferred to the E2 ubiquitin conjugating enzyme, before the E3 ubiquitin ligase enzyme specifically binds the substrate protein to mediate the transfer of ubiquitin to a lysine residue in the target [4]. Several rounds of ubiquitin conjugation produce long ubiquitin chains in which each ubiquitin molecule is covalently bound to a specific lysine of the previous ubiquitin moiety (polyubiquitylation). In the case of polyubiquitylation via lysine 48 or lysine 11, the polyubiquitylated substrate is committed to association with, and subsequent degradation by, the 26S proteasome. However, there are different types of ubiquitylation, which can lead to other molecular consequences, depending on the nature of the E2. Indeed, substrates can be monoubiquitylated or polyubiquitylated through five other internal lysine residues (K6, K27, K29, K33, and K63) or the N-terminus of ubiqutin [5–7]. While monoubiquitylation or K63-linked polyubiquitylation specify non-proteolytic fates for a substrate, the implications of other chain linkages are less understood [5, 7, 8]. Ubiquitylation itself can be reversed by specific deubiquitinating enzymes (DUBs) leading to a model in which ubiquitylation is controlled by the balance between an E3 ligase and a DUB [9].
To allow for high specificity in the ubiquitylation of target proteins, more than 600 different E3 ligases exist in the human genome [10]. Depending on their homology domains, these ligases can be subdivided in two major classes, the HECT (Homologous to E6-AP C-Terminus) family E3 ligases and the RING (Really Interesting New Gene) family E3 ligases [11]. The approximately 30 different HECT ligases form transient covalent linkages with ubiquitin during the ubiquitylation process, while RING ligases only mediate the transfer of ubiquitin from the E2 directly to the substrate. RING E3 ligases can be further subcategorized into those in which one protein contains both the RING and the substrate adaptor domain or multi-subunit complexes, in which these domains are part of distinct proteins within the complex [11]. One of the best described E3 families within the multi-subunit RING ligases is the cullin RING ligase (CRL) superfamily [12], which includes the CRL1 (better known as SCF, standing for Skp1-Cul1-F-box protein complex) and the APC/C (anaphase promoting complex/cyclosome, which is also referred to as a CRL-like ligase) E3 ligases.
The SCF complex consists of Cul1, the scaffold of the complex, Rbx1, the RING protein, which eventually recruits the E2, and Skp1 (S-phase kinase associated protein 1), which serves as an adaptor to bind the F-box protein, the substrate-binding subunit (Fig. 1) [12–14]. F-box proteins are named after the F-box domain, a 40-amino-acid motif initially identified in cyclin F (aka Fbxo1), which binds Skp1 [15]. They contain an additional motif, which constitutes the substrate binding domain. Depending on these domains, they are classified into three groups. F-box proteins containing WD-40 domains are called Fbxws, while those with leucine-rich repeats are called Fbxls. The remaining F-box proteins, with diverse motifs (e.g., tetratrico peptide repeats, kelch repeats, proline-rich motifs) are named Fbxos [13, 16, 17].
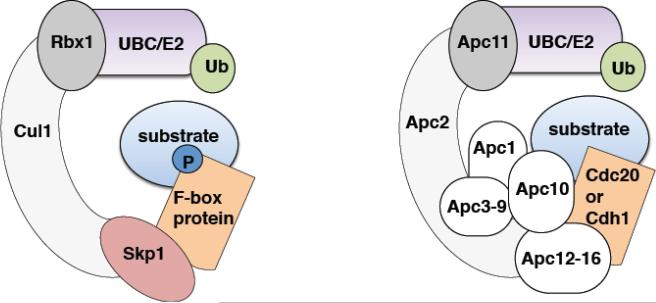
Figure 1. Structure of the SCF and the APC/C ubiquitin ligases.
The SCF and APC/C E3 ligases are both members of the Cullin-RING ligase (CRL) superfamily. Cul1 and Apc2 (light grey) are the scaffold proteins of SCF and APC/C, respectively. On one end, they bind to a RING finger protein, Rbx1 or Apc11 (dark grey), which recruits the E2 ubiquitin conjugating enzyme (UBC). On their other end, they connect to the substrate specific unit via an adaptor molecule (red). In the case of SCF, the F-box proteins are the variable substrate binding components (orange), while Cdh1 and Cdc20 (in somatic cells), together with Apc10, recruit substrates to the APC/C. The scheme illustrates the relationship between APC/C and SCF components and does not represent the topology of APC/C subunits.
Sixty-nine different F-box proteins are encoded by the human genome, and individual F-box proteins recognize different substrates. Therefore, these proteins determine the broad functional bandwidth of SCF complexes. Importantly, specific substrates or biological activities have so far been assigned to only a few F-box proteins, and each of these play a key role in essential cellular processes, such as cell cycle control, apoptosis, DNA damage responses, gene transcription, or translation [3, 18].
The APC/C is a prominent E3 ubiquitin ligase involved in cell cycle regulation. It consists of thirteen different subunits and, in somatic cells, has two co-activators: Cdc20 and Cdh1 (also known as Fzr1), which define substrate specificity and associate with the APC/C core at defined stages of the cell cycle (Fig. 1) [19]. While SCF ligases can be active throughout the cell cycle, APC/C activity is restricted to the time between metaphase and the end of the next G1, and during this time, APC/CCdc20 initiates anaphase and mitotic exit, while APC/CCdh1 contributes to mitotic exit and establishment of a stable G1 state [3, 20–22].
In the following sections, we outline the current understanding of how SCF and APC/C E3 ligases regulate cell cycle progression, their response to environmental cues in each phase of the cell cycle, and potential clinical and therapeutic implications of SCF and APC/C biology.
2. The G0 and G1 phases
2.1 Quiescence and G1
In response to mitogen withdrawal (or after exit from mitosis), a stable G0 or G1 state has to be established. The APC/CCdh1 complex is the central player in this task, and it is activated in late mitosis to complete mitotic exit and reset the cell cycle by targeting a variety of proteins involved in DNA replication, cycle progression, and mitosis. Among the targets that promote DNA replication is Cdc6, which binds to the origin recognition complex (ORC) to form pre-replication complexes (preRCs). Cdc6 is kept at low levels during early G1 through APC/CCdh1-mediated degradation (Fig. 2A) [23]. APC/CCdh1 also negatively regulates pro-proliferative signal transduction by targeting Ets2 (Fig. 2A), a transcription factor involved in Ras-Raf-MAPK signaling that induces the expression of cyclin D1 [24, 25]. APC/CCdh1 activity also precludes early accumulation of positive cell cycle regulators [21, 26]. Specifically, Skp2 (S phase kinase-associated protein, aka Fbl1) is targeted for proteolysis to avert premature formation of the SCFSkp2 complex, which promotes cell cycle progression by mediating the degradation of CKIs (see below and Fig. 2A) [26–28]. Importantly, the Cdc25A phosphatase, which promotes S-phase entry and mitosis by dephosphorylating and thus activating Cdk2 and Cdk1, is also kept at low levels during early G by APC/CCdh1 (Fig. 2A) [29]. Finally, APC/CCdh1 targets mitotic proteins for proteasomal degradation [19–21, 30]. Among the most prominent targets are cyclin A and cyclin B (Fig. 2A), the main drivers of mitotic entry [31, 32]. Furthermore, Aurora A, Aurora B, and polo-like kinase 1 (Plk1), important mitotic kinases involved in centrosome maturation, spindle assembly, and chromosome separation, are additional targets of APC/CCdh1 (Fig. 2A) [33–36].
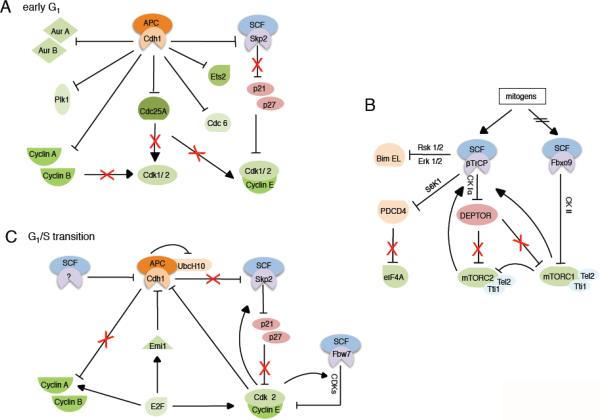
Figure 2. SCF and APC/C mediated degradation processes in G1 phase and the response to mitogen stimulation.
A. APC/CCdh1 maintains a stable G1 phase by targeting mitotic proteins like Aurora A, Aurora B, Plk1, cyclin A, and cyclin B for degradation. In addition, APC/CCdh1 prevents premature accumulation of positive cell cycle regulators, such as Ets2, Cdc6, and Cdc25A. The CDK inhibitors p21 and p27 are stabilized via APC/CCdh1-dependent degradation of Skp2 to keep CDK activity low during G1.
B. Upon mitogen stimulation, SCFβTrCP mediates the degradation of inhibitory or proapoptotic proteins, such as PCDC4, DEPTOR, and BimEL, promoting cell growth. Upon mitogen withdrawal, SCFFbxo9 targets Tel2 and Tti1 for degradation within mTORC1 to attenuate mTORC1 activity and sustain mTORC2 signaling.
C. At the G1/S transition, APC/CCdh1 activity is inhibited through several mechanisms, such as inhibitory phosphorylation by CDKs and binding to Emi1. Skp2-dependent degradation of p21 and p27 leads to activation of cyclin E-Cdk2, which initiates S phase before being degraded via SCFFbw7. See main text for more details.
Hence, by targeting protooncogenic proteins, such as Skp2 and Ets2, APC/CCdh1 exerts an anti-proliferative activity. In fact, APC/CCdh1 acts as a tumor suppressor in mice, where Fzr1 heterozygous mice have elevated rates of spontaneous tumor formation [37].
In response to mitogen withdrawal, cells must also switch to a state of nutritional saving and restrict protein translation and cell growth to sustain survival. SCFFbxo9 contributes to this process via differential regulation of mTORC1 and mTORC2 signaling. Specifically, the Tel2 (telomere maintenance 2) and Tti1 (Tel2 interacting protein 1) proteins, integral components of both the mTORC1 and mTORC2 complexes, are phosphorylated by CKII to target them for degradation by SCFFbxo9 upon growth factor withdrawal. This pathway specifically targets Tel2 and Tti1 in the mTORC1 complex (Fig. 2B) [38]. As a consequence, mTORC1 signaling is attenuated to restrain protein translation and cell growth, but via relief of mTORC1-mediated feedback inhibition, mTORC2 signaling is sustained to promote survival [38].
2.2 The growth factor response
If growth factors and nutrients are available, cells respond with a number of events that lead to growth and, eventually, cell division. Certain SCF ligases are acutely involved in the orchestration of growth factor responses during G1 phase. For example, the tumor suppressor PDCD4 (programmed cell death protein 4), which inhibits the translation initiation factor eIF4A, is degraded in response to mitogens in a two-step mechanism. First, PDCD4 is phosphorylated on serine 67 (and possibly adjacent serine residues) by S6K1, and it is subsequently ubiquitylated by SCFβTrCP (Fig. 2B), resulting in increased protein synthesis and cell growth [39]. In response to mitogens and survival signals, SCFβTrCP furthermore promotes survival by targeting the proapoptotic BH3-only protein BimEL for degradation in an Rsk1/2- and Erk1/2-dependent manner (Fig. 2B) [40]. Another important target of SCFβTrCP in growth factor responses is the mTOR inhibitor DEPTOR (Fig. 2B). After mitogen stimulation, DEPTOR is phosphorylated by mTORC1 or mTORC2 and CK1α, which directs ubiquitylation and degradation, relieving mTOR inhibition [41, 42]. Thus, in cooperation with SCFβTrCP, mTOR signaling promotes further mTOR activation via this auto-amplification loop.
2.3 Post-restriction point G1 and the G1 to S phase transition
With sufficient mitogen stimulation and cell growth, cells commit to cell division, passing a point of G1 known as “restriction point”, after which cells will proceed through a round of cell division even if mitogens are removed. The transition from G1 to S phase results from decreasing APC/CCdh1 activity, decreasing CKIs levels, rising cyclin expression, increasing CDK activity, phosphorylation and inactivation of Rb (retinoblastoma) protein family members, and activation of E2F transcription factor family members.
Inactivation of APC/CCdh1 during progression through the late G1 phase occurs via different mechanisms. First, UbcH10, the APC/C specific E2 ubiquitin conjugating enzyme, is ubiquitylated by APC/CCdh1 (Fig. 2C), thereby providing a negative feedback loop limiting APC/CCdh1 activity [43]. Likewise, Cdh1 initiates autoubiquitylation within APC/C (Fig. 2C), thus limiting its activity [44]. Furthermore, rising levels of CDK activity lead to phosphorylation of Cdh1, disrupting its binding to APC/C (Fig. 2C) [3, 45]. Later, in S-Phase, an unidentified SCF complex contributes to Cdh1 degradation (Fig. 2C), either directly or indirectly, and this process may depend on previous Cdh1-phosphorylation [46]. Finally, at the G1/S transition, the transcription factor E2F induces expression of Emi1 (an F-box protein also known as Fbxo5), which binds to APC/CCdh1 as a pseudosubstrate inhibitor, inhibiting APC/CCdh1 throughout S and G2 (Fig. 2C) [47, 48].
Rising CDK activity during G1 phase occurs through several different mechanisms. Primarily, the expression of cyclins is induced on a transcriptional level, with cyclin E and cyclin A expression being initiated by E2F (Fig. 2C) [49, 50]. However, the majority of cyclin-Cdk complexes are inhibited via association with CDK inhibitors (CKIs), such as p21 or p27. In late G1, Cdk4 and Cdk6 bound to cyclin D1 catalyze the phosphorylation (and consequent inactivation) of Rb family proteins (pRb, p107, and p130), which bind and inhibit E2F transcription factors, preventing their promotion of cell cycle progression. With decreasing APC/CCdh1 activity in late G1, Skp2 accumulates and forms the SCFSkp2 complex (Fig. 2C), which targets cell cycle inhibitors for degradation, promoting cell cycle progression throughout S, G2, and M [22]. Importantly, SCFSkp2 binds p27 and mediates its proteasomal degradation, liberating cyclin E-Cdk2 from p27 inhibition (Fig. 2C) [51–54]. Ubiquitylation of p27 depends on phosphorylation of threonine 187 by cyclin E-Cdk2 and the co-factor Cks1 [55–59]. Other CKIs ubiquitylated by SCFSkp2 include p21 and p57, whose degradation further reinforces cyclin-Cdk1/2 activation [60, 61]. By analogy to p27, phosphorylation by cyclinECdk2 and binding of Cks1 are likely prerequisites for SCFSkp2 dependent ubiquitiylation of p21 and p57 [60, 61]. These functions distinguish Skp2 as an E3 ligase with oncogenic properties, which is further supported by the frequent overexpression of Skp2 in many tumors [62].
3. S and G2 Phases of the cell cycle
3.1 S phase
Cyclin E-Cdk2 and (later) cyclin A-Cdk2 are the two main CDK complexes in S phase and mediate the initiation of DNA and centrosome duplication. At the same time, cyclin A-Cdk2 further inhibits APC/CCdh1 via Cdh1 phosphorylation to ensure cyclin stability (Fig. 3A) [43]. Upon successful S-phase entry, cyclin D1 is phosphorylated by GSK3β, targeting it for degradation by an SCF ligase containing Fbxo4 and the small heat-shock protein α/B-crystallin (the latter being a substrate adaptor) [63]. As cells progress through S and approach G2, the levels of S phase cyclins decrease again. Notably, cyclin E abundance is regulated via a negative feedback loop. Specifically, as Cdk2-activity increases, Cdk2-bound cyclin E is phosphorylated on threonine 380 in a Cdk2-dependent manner, whereupon phosphorylated cyclin E is targeted for degradation by the SCFFbw7 complex (Figs. 2C and 3A) [64, 65].
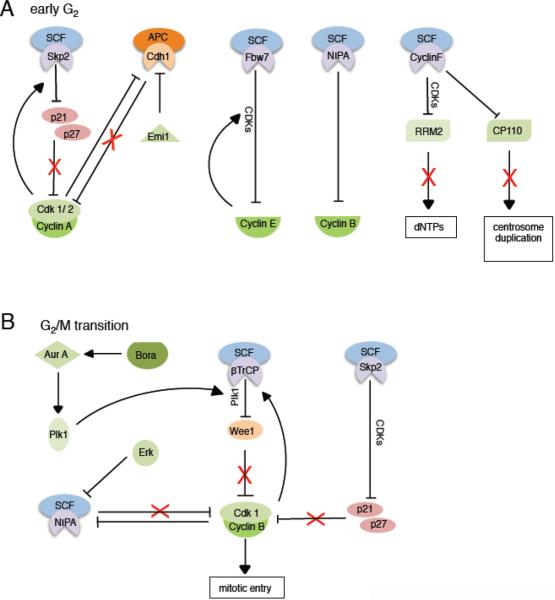
Figure 3. SCF and APC/C mediated degradation processes in S and G2 phases of the cell cycle.
A. In early G2, APC/CCdh1 remains inactivated, SCFSkp2 maintains cyclin A-Cdk1/2 activity via degradation of p21 and p27, SCFFbw7 promotes cyclin E degradation, and SCFNIPA prevents premature accumulation of nuclear cyclin B. Other S phase proteins (e.g., RRM2 and CP110) are also degraded via cyclin F to stop dNTP production and assure centrosomes are only replicated once per cell cycle.
B. At the G2/M transition, SCFSkp2 continues to target p21 and p27, while Wee1 is degraded in a Plk1- and SCFβTrCP-dependent manner to release cyclin B-Cdk1 from inhibition. See main text for more details.
3.2 DNA damage checkpoint
During S and G2 phases, it is crucial for cells to assure accurate and complete DNA replication. Therefore, DNA damage detection mechanisms and checkpoints provide precise surveillance. Upon DNA damage, ATM activates the checkpoint kinase Chk2 and ATR activates Chk1 in a Claspin-dependent process (Fig. 4). Chk1 and Chk2 then mediate inhibitory phosphorylation of Cdc25A on serine 216 [66, 67]. Cdc25A promotes cell cycle activation by virtue of its ability to remove inhibitory phosphorylations from Cdk1 and Cdk2, but Cdc25A phosphorylation by Chk1/2 results in ubiquitylation by SCFβTrCP and degradation, attenuating CDK activity (Fig. 4) [68]. Degradation of Cdc25A leads to a pause in cell cycle progression during S or G2, depending on the timing of the DNA damage.
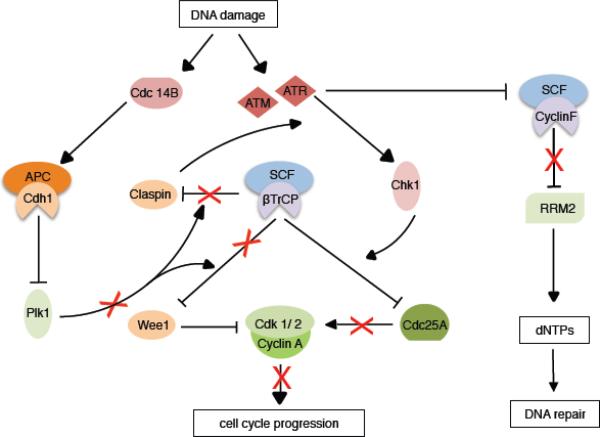
Figure 4. Role of SCF and APC/C ligases in the response to DNA damage.
Upon DNA damage, SCF and APC/C activate S and G2 checkpoints to halt cell cycle progression. Chk1 (which is activated by ATR) promotes the SCFβTrCP-dependent of Cdc25A to inhibit CDK activity, while dephosphorylation of Cdh1 by Cdc14B allows for Plk1 degradation and subsequent stabilization of Claspin and Wee1. See main text for more details.
Another target of the DNA damage checkpoint in G2 phase is Plk1 [69, 70]. In response to DNA damage, the phosphatase Cdc14B is released from the nucleolus to the nucleoplasm, where it dephosphorylates Cdh1 (Fig. 4). As a consequence, APC/CCdh1 becomes reactivated to target Plk1 for degradation (Fig. 4). In this way, Claspin and Wee1 are stabilized to activate the checkpoint and inhibit cell cycle progression (Fig. 4) [70]. Notably, other known targets of APC/CCdh1 are likely protected during this process by DUBs, particularly USP28 [70, 71].
SCFβTrCP also plays a role during recovery from the checkpoint. Claspin, which mediates ATR-induced activation of Chk1 upon DNA damage, is phosphorylated by Plk1 during checkpoint recovery and subsequently degraded in an SCFβTrCP dependent manner [72, 73]. As a consequence, activation of Chk1 declines, and the cell recovers from the checkpoint. Moreover, SCFβTrCP also regulates translation during the silencing of the G2 DNA damage checkpoint. In response to genotoxic stress, AMPK (Adenosine Monophosphate-activated Protein Kinase) phosphorylates and activates eEF2K (eukaryotic Elongation Factor 2 Kinase), which, in turn, inhibits elongation by phosphorylating and inhibiting eEF2, a protein that mediates movement of the ribosome along mRNA [74]. During checkpoint silencing, SCFβTrCP targets eEF2K for degradation to enable rapid resumption of translation elongation [74].
3.3 G2 phase and G2/M transition
After successful and accurate completion of DNA replication, production of deoxyribonucleotides (dNTPs) must be avoided. To this end, RRM2 (Ribonucleotide Reductase family Member 2), which converts ribonucleotides to dNTPs for DNA synthesis, is phosphorylated on threonine 33 by CDKs and ubiquitylated by the SCFCyclinF complex, reducing the availability of dNTPs (Fig. 3A) [75]. In contrast, cyclin F is downregulated in response to DNA damage in an ATR-dependent manner to stabilize RRM2 (Fig. 4). By this means, production of dNTPs is enhanced to allow for efficient DNA repair [75].
Beyond DNA replication, SCFCyclinF also regulates centrosome duplication. Cyclin F and CP110, a protein essential for centrosome duplication, physically associate on the centrioles during G2 [76]. By targeting CP110 for proteasomal degradation (Fig. 3A), SCFCyclinF assures that centrosomes are replicated only once during the cell cycle, preventing centrosome overduplication, which could lead to multipolar or asymmetric mitotic spindles and subsequent chromosome aberrations [76].
With progression through G2 phase, the cell prepares for mitotic entry, which is orchestrated by cyclin B-Cdk1. Therefore, cyclin B-Cdk1 is a strictly regulated kinase complex. In late G2 phase, SCFβ-TrCP promotes activation of Cdk1 by mediating the degradation of Wee1, a Cdk1 inhibitory kinase (Fig. 3B). A prerequisite for SCFβ-TrCP binding to Wee1 is phosphorylation of serine 53 and serine 123 by Plk1 and Cdk1, respectively [77]. Thus, a positive feedback loop between Cdk1 activation and Wee1 degradation assures rapid activation of Cdk1 upon mitotic entry (Fig. 3B). The activity of Plk1 itself is also tightly controlled through the synergistic actions of Bora and Aurora A kinase. Accumulation of Bora during G2 leads to Aurora A-mediated activation of Plk1, which contributes to Wee1 degradation and subsequent activation of Cdk1 (Fig. 3B) [78].
To preclude premature mitotic entry, early activation of cyclin B-Cdk1 must be prevented. To this end, SCFNIPA targets nuclear cyclin B1 during S and G2 (Fig. 3A) [79]. However, in late G2, NIPA is phosphorylated at serine 395 by cyclin B-Cdk1, after initial phosphorylation on serine 354 and serine 359 by Erk2 (Fig. 3B) [80, 81]. Phosphorylation inhibits assembly of a functional SCFNIPA complex, allowing nuclear accumulation of cyclin B1 and subsequent mitotic entry. In this context, cyclin B1 regulates its own abundance through a positive feedback loop (Fig. 3B).
4. Mitosis
4.1 Prometaphase and spindle assembly checkpoint
Cyclin B-Cdk1 orchestrates essential steps of early mitosis, including the assembly of the mitotic spindle, breakdown of the nuclear envelope, cessation of gene transcription, and condensation of chromosomes. These processes drive the cell through prophase until metaphase.
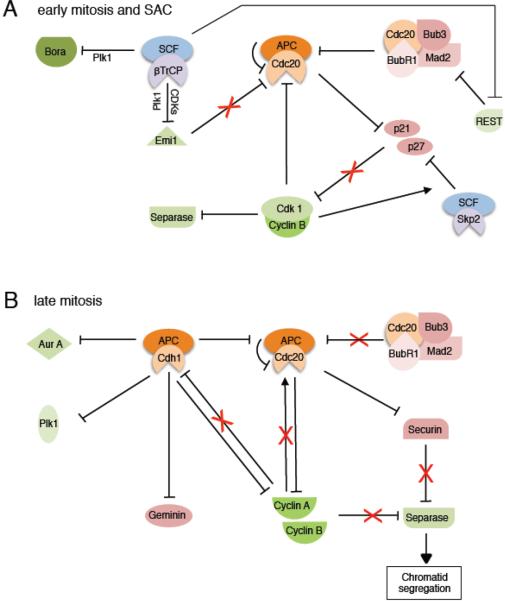
Starting from metaphase, the APC/C becomes a key player in promoting mitotic progression and, eventually, mitotic exit. Thus, inhibitory mechanisms that keep the APC/C in check throughout S- and G2 phases must be removed. In this regard, SCFβTrCP mediates ubiquitylation and degradation of the APC/C inhibitor Emi1 following phosphorylation by cyclin B-Cdk1 and Plk1 in early mitosis (Fig. 5A) [82, 83]. However, until all kinetochores are correctly attached to spindle fibers and the chromosomes are properly aligned on the metaphase plate, the Spindle Assembly Checkpoint (SAC) inhibits APC/C activation, blocking the onset of anaphase [84]. In the presence of unattached kinetochores, the mitotic checkpoint proteins, BubR1, Bub3, and Mad2, bind to Cdc20 to form the Mitotic Checkpoint Complex (MCC), which sequesters Cdc20 and directly inhibits activation of the APC/CCdc20 (Fig. 5A) [84–86]. Another model suggests that Cdc20 is targeted for degradation by APC/C in an MCC-dependent manner during SAC activation [87, 88]. Furthermore, inhibitory phosphorylation of Cdc20 by protein kinases, such as Cdk1 (Fig. 5A), MAPK, or Bub1, contributes to Cdc20 inhibition during SAC activation [89–91]. Interestingly, the transcriptional activation of Mad2 is also regulated by SCFβTrCP. During late G2, SCFβTrCP targets REST (Repressor-Element-1-Silencing Transcription factor) for degradation, allowing transcriptional derepression of Mad2 (Fig. 5A) [92].
Figure 5. SCF and APC/C mediated degradation processes during mitosis.
A. In early mitotis, SCFβTrCP ubiquitylates Emi1 to eliminate the repression of APC/CCdh1 (which remains inactive because of Cdh1 phosphorylation by CDKs) and, possibly, APC/CCdc20. However, during activation of the Spindle Assembly Checkpoint (SAC), APC/CCdc20 activity is repressed, with cyclin B-Cdk1 contributing to its inhibition.
B. After fulfillment of the SAC, APC/CCdc20 inhibition is relieved, allowing it to target mitotic cyclins and Securin for degradation, the latter leading to Separase activation and sister chromatid separation. With mitotic progression, APC/CCdc20 activity decreases via several mechanisms, and APC/CCdh1 assembles to target cell cycle proteins for degradation. See main text for more details.
Despite the many mechanisms keeping APC/CCdc20 in check, its activity is not completely inhibited; a minor fraction remains active even during SAC activation. This subpopulation sustains cyclin B-Cdk1 activity during prometaphase by targeting p21 for degradation (Fig. 5A) [93]. Although p21 is degraded via SCFSkp2 at the G1/S transition, it re-accumulates during G2 and interacts with cyclin B-Cdk1 before it is ubiquitylated by APC/CCdc20. Interestingly, APC/CCdc20 and APC/CCdh1 have opposing effects on p21 levels. While p21 is degraded via APC/CCdc20 in early mitosis, APC/CCdh1 indirectly stabilizes p21 in G1 phase by targeting Skp2 and Cdc20 for degradation [27, 94, 95]. In addition, APC/CCdc20 targets two other substrates for degradation in early mitosis, cyclin A and Nek2A, even when the spindle checkpoint is active. How the checkpoint-resistant activity of the APC/CCdc20 is regulated remains unclear. Potential mechanisms include Cdc20-independent binding of substrates to APC10 or an increase in substrate affinity for Cdc20, which allows substrates to outcompete SAC proteins [96–98].
4.2 Metaphase to anaphase transition and mitotic exit
Upon proper attachment of all spindle microtubules to chromosome kinetochores, the SAC is rapidly inactivated, and APC/CCdc20 is released from inhibition via an uncertain mechanism (Fig. 5B). Checkpoint inactivation depends on Cdc20-dependent ubiquitylation. According to some, this leads to dissociation of the MCC from APC/CCdc20, but others suggest that it causes degradation of Cdc20 [99, 100]. An additional, ATP-dependent step is also required for both dissociation of the MCC from APC/C and disassembly of the free MCC [101, 102]. This step has been shown to involve β–γ bond cleavage of ATP, suggesting the presence of an additional ubiquitin independent process, which may be kinase- or chaperone-related [102]. The understanding of SAC inactivation is further complicated by the fact that Cdc20 proteolysis is also stimulated by Mad2 and BubR1 to keep levels of Cdc20 low in order to avoid premature activation of APC/C and maintain the checkpoint [87, 88]. Eventually, after kinetochore attachment, activated APC/CCdc20 initiates anaphase by targeting Securin for degradation (Fig. 5B) [20, 103, 104]. During metaphase, sister chromatids are held together by Cohesin. Securin is a chaperone that binds and inhibits Separase, an enzyme capable of cleaving Cohesin. With Securin degradation, Separase is released, and centromeric Cohesin is cleaved, which leads to sister chromatid segregation (Fig. 5B) [105]. Moreover, Shugoshin, a protector of centromeric sister chromatid cohesion, is targeted for degradation by APC/CCdh1; however, this step is not indispensable for chromatid segregation and seems more important during meiosis [106, 107].
Both mitotic cyclins, cyclin A and cyclin B, are targeted for degradation by the APC/C (Fig. 5B) [108]. Whereas cyclin A degradation starts in prometaphase, cyclin B ubiquitylation starts at the metaphase/anaphase transition, mediated first by APC/CCdc20 and later by APC/CCdh1 [109–111]. Notably, APC/CCdc20 has different effects on Cdk1 activity depending on the mitotic phase. While it augments cyclin B-Cdk1 activity during prometaphase via degradation of p21 (Fig. 5A), it suppresses cyclin B-Cdk1 via degradation of cyclin B in late mitosis (Fig. 5B) [93]. Mitotic cyclin-CDK activity inhibits Separase, so the degradation of cyclin A and cyclin B by APC/CCdc20 is directly involved in sister chromatid separation (Fig. 5B) [20]. Furthermore, drastically reduced CDK activity in anaphase allows for disassembly of the mitotic spindle, chromosome decondensation, cytokinesis, and reconstitution of the nuclear envelope [20].
Although APC/C is the most prominent E3 ubiquitin ligase orchestrating the metaphase to anaphase transition, SCF ubiquitin ligases are also involved in the regulation of anaphase onset. For instance, Bora, which works together with Plk1 and Aurora A to regulate microtubule polymerization, spindle stability, and tension between kinetochores, is targeted for degradation by SCFβTrCP at the metaphase to anaphase transition (Fig. 5A) [112, 113]. Interestingly, a prerequisite for Bora ubiquitylation is phosphorylation by Plk1, which itself is activated in a Bora-dependent manner at the G2/M transition [78, 112]. Timely activation and degradation of Bora assures regulated cell cycle progression, as inhibition of Bora degradation delays anaphase onset and knockdown of Bora activates the spindle checkpoint [112, 113].
In late mitosis, Aurora A and Plk1 are eventually targeted for degradation via the APC/CCdh1 to promote mitotic exit (Fig. 5B) [33, 34, 36]. Another important substrate of APC/C in late mitosis is Geminin (Fig. 5B), which inhibits DNA re-replication by precluding formation of the pre-replication complex (pre-RC) after S-phase, rendering replication possible during the subsequent cell cycle [114].
Overall, the APC/C is the central E3 ligase that coordinates mitotic progression, mitotic exit, and the subsequent establishment of a G1 state. While anaphase onset is induced by APC/CCdc20, both APC/CCdc20 and APC/CCdh1 orchestrate mitotic exit, and APC/CCdh1 contributes to establishment and maintenance of the G1 phase of the next cell cycle. This distinct activation profile is the result of different regulatory steps. As mentioned earlier, Cdh1 is kept inactivated via phosphorylation by CDKs during S, G2 and part of mitosis [115]. Degradation of cyclin B reduces CDK activity at the metaphase to anaphase transition, so the levels of dephosphorylated Cdh1 rise. Dephosphorylated Cdh1 binds APC/C and negatively regulates APC/CCdc20 by targeting Cdc20 for degradation (Fig. 5B) [94, 95]. Moreover, APC/C dependent autoubiquitylation of Cdc20 in anaphase has been proposed to further contribute to the inactivation of APC/CCdc20 (Fig. 5B) [99, 100]. The oscillation between Cdc20 and Cdh1 expression partly explains how the APC/C times the degradation of its various substrates. Depending on their destruction box and their affinity for Cdc20 or Cdh1, APC/C substrates will be targeted earlier or later during mitosis or G1 for proteasomal degradation [116, 117].
5. Other CRLs involved in cell cycle control
Other CRLs are also implicated in cell cycle control and the DNA damage response. CRL3 complexes appear to control mitosis via proteolytic and non-proteolytic mechanisms. The best understood cell cycle-related CRL3 substrate is Aurora B. K63-type ubiquitylation of Aurora B targets it to mitotic chromosomes to control their alignment [118]. The CRL4 ligases also control features of the cell cycle. In addition to Cul4 and Rbx1, CRL4 ligases contain the adaptor protein DDB1 (DNA Damage Binding protein 1), which recruits members of the DCAF (DDB1–Cul4 Associated Factors) family to dictate the specificity of substrate degradation [119–121]. CRL4Cdt2 is a prominent member of this family implicated in DNA replication and the DNA damage response. Notably, CRL4Cdt2 interacts with different UBCs (UBCH8, UBE2G1, and UBE2G2) to mediate ubiquitylation of its various substrates [122]. An important target of CRL4Cdt2 is Cdt1, a replication licensing factor and pre-replication complex (pre-RC) component that is tightly regulated to ensure that DNA replication only occurs once per cell cycle. After S phase, Cdt1 activity is attenuated both by Geminin binding and by proteasomal degradation. Importantly, CRL4Cdt2-mediated degradation of Cdt1 is initiated shortly after S phase onset [123–126]. This event requires Cdt1 binding to PCNA via a PIP (PCNA Interacting Protein)-box motif present in Cdt1. Upon DNA damage, Cdt1 is degraded in the same way to preclude DNA replication [127]. (Interestingly, Cdt1 is also targeted for degradation by SCFSkp2 in response to CDK-dependent phosphorylation [123, 128].)
An additional target of CRL4Cdt2 is p21, which is degraded both in response to UV-induced DNA damage and during the unperturbed cell cycle. In the later context, CRL4Cdt2 collaborates with SCFSkp2 to mediate timely degradation of p21. Like Cdt1, p21 degradation by CRL4Cdt2 requires interaction with PCNA via a PIP-box [129]. CRL4Cdt2 is further involved in cell cycle control via regulation of histone methylation by ubiquitylation of Set8. Set8 mono-methylates Lys 20 of histone H4 (H4K20me1) during G2 to promote chromatin compaction. During S phase and upon DNA damage, Set8 is degraded by CRL4Cdt2 in a PCNA-dependent manner [130–132]. Perturbation of this mechanism leads to accumulation of H4K20me1, premature chromatin compaction, and activation of the G2/M checkpoint [130, 131].
Interestingly, Cdt2 levels are regulated via SCFFbxo11 during the cell cycle [133, 134] and inhibition of Cdt2 degradation delays cell cycle exit. In contrast to most other SCF substrates, whose phosphorylation promotes binding to the F-box protein, CDK-dependent phosphorylation of the Cdt2 degron inhibits binding to and degradation via Fbxo11 [134]. Thus, crosstalk between an SCF ligase, CDKs, and a CRL4 complex controls the abundance of Cdt2 to regulate the timing of cell cycle exit.
6. Clinical implications
6.1 SCF complexes as targets in the therapy of cancer and other diseases
The SCF complexes and the APC/C are key players in cell cycle control and the DNA damage response. As such, they govern cell proliferation and genome stability, which, when deregulated, contribute to tumorigenesis. Among the SCF ligases controlling cell growth and proliferation (discussed above), this situation has been described in particular for SCFSkp2, SCFTrCP, SCFFbxw7, and SCFFbxo9. The recent therapeutic success of the proteasome inhibitor bortezomib in multiple myeloma and certain types of B-cell Non-Hodgkin's lymphoma has expedited the efforts to develop inhibitors of ubiquitin ligases or its components [135, 136]. While bortezomib exhibits a certain degree of selectivity towards cancer cells and has a noticeable therapeutic index, it non-specifically blocks the ubiquitin-proteasome-dependent degradation of all cellular proteins, resulting in considerable side effects and the development of resistance. Therapies directed against individual E3 ligases or ligase families known to be deregulated in human cancers may thus prove more efficacious. To qualify as a target for therapeutic inhibition, a ligase should be an oncoprotein (based on the activity of its substrates - e.g., tumor suppressors) and deregulated in tumors.
6.1.1 Skp2
Skp2 is an oncoprotein by virtue of its function in degrading negative cell cycle regulators, including p27, p21, p130, and p57 [137, 138]. Skp2 is overexpressed in various malignancies, such as gastric cancer [139, 140], colon cancer [141], and breast cancer [142], with an associated decrease in p27 levels and indicating a poor prognosis. The oncogenic function of Skp2 is further underscored by mouse genetic studies. Targeted expression of Skp2 in the T-lymphoid lineage cooperates with activated N-Ras to induce T-cell lymphomas [143], while tissue specific expression of Skp2 in the prostate induces hyperplasia and low grade carcinoma [144]. In addition, a critical role for Skp2-dependent degradation of p27 in colon adenoma-carcinoma development was demonstrated using a Skp2 knock-in model [145]. Similarly, in Skp2-null cells, aberrant oncogenic signaling or inactivation of tumor suppressor genes trigger a potent, tumor-suppressive senescence response, although Skp2 inactivation alone does not induce senescence [146]. Finally, RNAi-mediated knockdown of Skp2 inhibits the growth of tumor cells, including cell lines derived from glioblastoma [147], melanoma [148], and oral cancer [149]. Specific inhibition of Skp2 would thus be expected to be effective in the treatment of cancers with activated Skp2 signaling.
Interestingly, Skp2 also interacts with a number of proteins produced by pathogenic viruses, such as X-protein (HBV), EBNA3C (EBV), and E7 (HPV16/18) [150], suggesting that Skp2 inhibitors may be also utilized in certain viral infections.
6.1.2 βTrCP
Given the diversity of its substrates (reviewed in [151]), βTrCP might be expected to exert both oncogenic and tumor suppressor activities. However, in certain tissues, βTrCP is clearly an oncoprotein based on its ubiquitylation activity against tumor suppressors, such as IκB, a negative regulator of NFκB [152], PDCD4, an inhibitor of eIF4A-mediated protein translation [39], and BimEL, a potent proapoptotic protein [40]. Indeed, βTrCP is overexpressed in breast cancer [153], and forced overexpression of βTrCP induces transformation in breast epithelium [154]. Likewise, upregulation of βTrCP has been shown in colon cancer, where it correlates with elevated NFκB activity and poor prognosis [155] and in pancreatic cancer cells, where it impacts chemoresistance [156]. Further evidence demonstrating the oncogenic potential of βTrCP stems from transgenic mouse models, in which tumor formation was observed in the mammary gland, liver, and kidney when βTrCP expression was directed to these tissues [154, 157]. Interestingly, no obvious phenotype was observed upon targeted expression of βTrCP in lymphoid organs, suggesting that βTrCP-dependent tumorigenesis is tissue specific [154]. Therefore, disrupting the interaction of βTrCP and its tumor suppressive substrates may be an attractive therapeutic strategy in defined tumor entities.
Similarly to Skp2, βTrCP interacts with a number of proteins produced by pathogenic viruses, such as Tax (HTLV1). Moreover, the HIV-1 protein Vpu targets SCFβTrCP to CD4 [158]. This finding, together with the evidence that other HIV-1 proteins (i.e. Vif and Vpr) bind CRLs to eliminate cellular proteins with antiviral activity (e.g., APOBEC3), indicates that SCFβTrCP and other CRLs are involved in the HIV life cycle and represent potential targets in the fight against this virus.
6.1.3 Fbxo9 and Fbxw7
The rationale for targeting the ubiquitin-proteasome system is particularly evident in multiple myeloma, due to the high efficacy of proteasome inhibitors in this disease. Indeed, Bortezomib, a reversible proteasome inhibitor, has been approved as the first line treatment for multiple myeloma. In addition, Carfilzomib, a second generation irreversible proteasome inhibitor has recently been approved by the Food and Drug Administration (FDA) for the treatment of patients with relapsed and refractory multiple myeloma, and demonstrates increased potency and an improved therapeutic index [159, 160]. Two ubiquitin ligases of the SCF family have recently been reported to contribute to the pathogenesis of multiple myeloma, SCFFbxo9 and SCFFbxw7α.
As outlined above, Fbxo9 targets Tel2/Tti1 proteins in a CK2-dependent manner to adjust mTOR signaling to the availability of growth factors. Significantly, Fbxo9 is overexpressed in multiple myeloma and drives constitutive activation of the PI3K/mTORC2/Akt pathway to promote survival. In multiple myelomas with elevated Fbxo9 expression, inhibition of Fbxo9 represents an attractive approach to inhibit Akt signaling, one of the most important mediators of cell survival in this disease [159]. Inhibition of CK2, the kinase that promotes Fbxo9-mediated ubiquitylation of Tel/Tti1, may also be an interesting therapeutic approach in multiple myeloma, particularly since CK2 inhibitors are readily available [161].
Although Fbxw7 behaves as a tumor suppressor due to its ubiquitylation activity against mitogenic substrates (e.g., Notch, c-Myc, cyclin E), Fbxw7 functions as a pro-survival gene in multiple myeloma by constitutively targeting the NFκB inhibitor p100 in a GSK3-dependent manner [162]. While a number of cancers, including T-ALL, breast cancers, and gastric adenocarcinoma often carry mutations in the Fbxw7 gene, leading to an accumulation of mitogenic substrates [163–165], these mutations do not occur in B-cell malignancies like multiple myeloma [166]. Fbxw7 and GSK3 may thus serve as promising targets for the treatment of multiple myelomas with constitutive activation of the NFκB pathway.
In a short term setting, acute delivery of an Fbxw7 inhibitor could also be effective in tumors where this F-box protein plays a tumor suppressive role by inducing cancer stem cells to proliferate and, therefore, become susceptible to traditional chemotherapies. However, this hypothesis remains to be proved experimentally.
6.1.3 Other F-box proteins
Fbxo11 mutations are present in human cancers, such as diffuse large B cell lymphomas (DLBCLs), colon, lung, ovary, and head and neck tumors [167–171] (and Staudt L., personal communication). At least in DLCBLs, these mutations inhibit SCFFbxo11. The presence of inactivating mutations suggests that Fbxo11 may function as a tumor suppressor, whose loss of function contributes to the pathogenesis of DLBCL (via BCL6 accumulation) and other cancers (through the stabilization of unidentified oncogenic substrates). Furthermore, inactivating mutations have been reported for Fbxo4 in esophageal carcinoma, and these mutations are associated with an increase in cyclin D1 levels. The mutations occur in the N-terminal regulatory regions of Fbxo4 and disrupt dimerization and activation [172]. Moreover, there is evidence for the loss of α/B crystallin in breast cancer and a corresponding increase in cyclin D1 levels. α/B crystallin is the substrate adaptor for cyclin D1 together with Fbxo4 [63, 173]. These data suggest a tumor suppressor role for the SCFFbxo4-α/B crystallin ligase in human tumors based on its ability to destabilize cyclin D1. Finally, cyclin F has been reported to be downregulated in hepatocelular carcinoma (HCC). In this context, low cyclin F expression was an independent poor prognostic marker for overall survival and correlated with tumor size and clinical stage [174]. Given the role of cyclin F in regulating centrosome homeostasis and the balanced abundance of dNTPs, these findings hint at a function for cyclin F as a tumor suppressor in HCC by maintaining genomic stabilty [75, 76].
6.2 Strategies to target CRLs
In theory, targets for inhibiting substrate degradation by the ubiquitin proteasome system can involve any of the three enzymes involved in ubiquitin transfer, including the E1 activating, the E2 conjugating, and the E3 ligase enzymes. These approaches would be expected to inhibit most or certain ubiquitin ligase families, when targeting E1 or E2 enzymes, respectively, and become highly specific when targeting single E3 ligases.
6.2.1 Targeting E1 ubiquitin activating enzymes
E1s (of which there are two in mammals) catalyze the initial step in ubiquitin conjugation, which involves an ATP-dependent covalent attachment of ubiquitin to its active cysteine site [4]. Efforts have been undertaken to target E1. These studies have yielded candidate compounds like PYR-41, an irreversible ubiquitin E1 inhibitor, but further investigations will be necessary to assess if they are clinical candidates [175]. Additional efforts have been directed to the E1s for ubiquitin like proteins (such as NEDD8, SUMO, etc.). Recently, MLN4924, an adenosine sulfamate analog, was reported to inhibit the E1 responsible for NEDDylation, the covalent addition of NEDD8 to substrates [176]. The cullin family of proteins are the most important substrates for neddylation, and the activity of SCF and other CRLs requires cullin neddylation [12, 177, 178]. In contrast to an inhibitor for the ubiquitin E1s, a small molecule inhibitor of the NEDD8 E1 would only inhibit CRLs. Currently, MLN4924 is in phase I/II clinical trials for the treatment of multiple myeloma and non-Hodgkins lymphoma, and the preclinical data suggest high efficacy in AML [179, 180].
6.2.2 Targeting E2 ubiquitin conjugating enzymes
Together with the E3 ligase, E2s mediate the transfer of ubiquitin to the target protein and govern the type and extent of ubiquitin linkage [181]. The human genome encodes at least 38 E2 enzymes, and Cdc34 (aka Ubc3) appears to be the major E2 for SCF ligases in promoting K48-linked polyubiquitylation and proteasomal degradation [12]. Recently, CC0651, a small molecule allosteric inhibitor of Cdc34 was reported [182]. CC0651 inhibits p27 ubiquitylation and degradation, and it demonstrates convincing specificity for human Cdc34, as evidenced by a lack of reactivity against any other E2/E3 pairs tested [182]. While current data on other E2 inhibitors is limited, the development of CC0651 indicates the feasibility of developing highly selective inhibitors of E2s and encourages efforts to target other E2s in a similar manner.
6.2.3 Targeting E3 ubiquitin ligases
E3 ligases determine which target protein becomes ubiquitylated. Inhibition of a single E3 ligase would be the most selective targeting approach, since it would affect a limited number of proteins, potentially translating into a better therapeutic ratio and fewer side effects. Most E3 ligases do not feature a canonical active site, and instead, the active site is a protein-protein interaction with a substrate. Inhibition of these protein-protein interactions is typically considered more difficult than inhibition of a catalytic site. However, this task appears increasingly feasible with advances in the structural understanding of these interactions [183]. MDM2 is an extensively studied ubiquitin E3 ligase with strong clinical relevance by virtue of its ability to regulate the abundance of the tumor suppressor p53. Previously, the Nutlin class of imidazoline chemotypes was identified, and they specifically disrupt the protein-protein binding interface between MDM2 and p53, stabilizing p53 [184]. Nutlin-3a is the most promising candidate and has favorable preclinical characteristics in terms of pharmacological properties and toxicity [185, 186]. Currently, Nutlin-3a is being investigated in early phase clinical trials for several solid and hematological tumor entities, setting a precedent for the specific inhibition of a ligase-substrate pair as a promising clinical approach.
In the case of SCF complexes, inhibition of either the F-box protein-substrate interface or the recruitment of the F-box protein to the SCF core are attractive strategies to selectively inhibit individual ubiquitylation events. In this regard, the small molecule inhibitor CpdA has previously been found to prevent the binding of Skp2 to the SCF ligase complex, inducing G1/S arrest and apoptosis by stabilizing p27, p21, and other Skp2 target proteins. In addition, CpdA sensitized multiple myeloma cells to cytostatic agents and bortezomib, and it was active against both myeloid and lymphoblastoid leukemia blasts [187].
Independent studies have recently demonstrated the feasibility of direct inhibition of the interfaces between the F-box protein and either the substrate or SCF core. SCF-I2 is an allosteric inhibitor of substrate recognition by the yeast F-box protein Cdc4 and inhibits binding and ubiquitylation of substrates. SCF-I2 inserts itself between the β-strands of the WD-40 propeller domain of Cdc4, impairing recognition of Cdc4-specific phosphodegrons [188]. In addition, a recent study screened for inhibitors that selectively target the p27-binding interface formed by Skp2-Cks1. This approach yielded four compounds that stabilize the expression of p27 in different human cancer cell lines and induce cell cycle arrest in G1 [189]. Finally, a chemical genetics screen for enhancers of rapamycin identified SMER3, an inhibitor of the SCFMet30 ligase. SMER3 specifically inhibits binding of the F-box protein subunit Met30 to the SCF core, stabilizing substrates such as Met4 [190].
Another approach to prevent substrate degradation at the E3 ligase level is to inhibit the kinase that phosphorylates a substrate to mark it for degradation. This approach particularly applies to SCF ubiquitin ligases, as it is well established that substrate phosphorylation is frequently a prerequisite for F-box protein binding [13, 151, 191]. Indeed, kinases like Plk1, which control the degradation of tumor suppressors, are potential targets, and inhibitors have already entered early clinical trials [192]. Notably, this approach could quickly impact the clinic, as inhibitors to numerous kinases are readily available, but broader application will require further investigations.
Together, SCF ligases are promising therapeutic target structures, whose pharmacological modulation would offer highly specific therapeutic approaches to a wide variety of malignancies. The success of this undertaking will largely depend on the pharmacological progress in developing specific inhibitors of F-box proteins and a better mechanistic understanding of the many uncharacterized F-box proteins, both with regard to their substrates and the posttranslational modifications that prime them for SCF-dependent ubiquitylation. Finally, it will be critical to further define tumor entities in which distinct SCF ligases are deregulated. This data will help define distinct F-box proteins as biomarkers and allow the selection of patient subpopulations that would profit from a targeted approach against SCF ligases.
Highlights
Role of Cullin RING ligases (CRLs) in the normal cell cycle
Involvement of CRLs in the cellular response to different environmental cues
Particular focus on ligases of the SCF and APC/C families
Clinical implications in targeted cancer therapies
Acknowledgements
We thank Jeffrey Skaar for critically reading the manuscript. We apologize to colleagues whose work could not be mentioned owing to space limitations. Work in the Bassermann laboratory is supported by grants from the German Research Foundation (Emmy Noether Program, BA 2851/3-1) and the German Cancer Aid (#109543), and in the Pagano laboratory by grants from the NIH (R01-GM057587, R37-CA076584, and R21-CA161108). MP is an investigator with the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Murray AW. Recycling the cell cycle: cyclins revisited. Cell. 2004;116:221–234. doi: 10.1016/s0092-8674(03)01080-8. [DOI] [PubMed] [Google Scholar]
- [2].Glotzer M, Murray AW, Kirschner MW. Cyclin is degraded by the ubiquitin pathway. Nature. 1991;349:132–138. doi: 10.1038/349132a0. [DOI] [PubMed] [Google Scholar]
- [3].Skaar JR, Pagano M. Control of cell growth by the SCF and APC/C ubiquitin ligases. Curr Opin Cell Biol. 2009;21:816–824. doi: 10.1016/j.ceb.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- [5].Clague MJ, Urbe S. Ubiquitin: same molecule, different degradation pathways. Cell. 2010;143:682–685. doi: 10.1016/j.cell.2010.11.012. [DOI] [PubMed] [Google Scholar]
- [6].Behrends C, Harper JW. Constructing and decoding unconventional ubiquitin chains. Nat Struct Mol Biol. 2011;18:520–528. doi: 10.1038/nsmb.2066. [DOI] [PubMed] [Google Scholar]
- [7].Xu P, Duong DM, Seyfried NT, Cheng D, Xie Y, Robert J, Rush J, Hochstrasser M, Finley D, Peng J. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell. 2009;137:133–145. doi: 10.1016/j.cell.2009.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Jacobson AD, Zhang NY, Xu P, Han KJ, Noone S, Peng J, Liu CW. The lysine 48 and lysine 63 ubiquitin conjugates are processed differently by the 26 s proteasome. J Biol Chem. 2009;284:35485–35494. doi: 10.1074/jbc.M109.052928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Clague MJ, Coulson JM, Urbe S. Cellular functions of the DUBs. J Cell Sci. 2012;125:277–286. doi: 10.1242/jcs.090985. [DOI] [PubMed] [Google Scholar]
- [10].Li W, Bengtson MH, Ulbrich A, Matsuda A, Reddy VA, Orth A, Chanda SK, Batalov S, Joazeiro CA. Genome-wide and functional annotation of human E3 ubiquitin ligases identifies MULAN, a mitochondrial E3 that regulates the organelle's dynamics and signaling. PLoS One. 2008;3:e1487. doi: 10.1371/journal.pone.0001487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Metzger MB, Hristova VA, Weissman AM. HECT and RING finger families of E3 ubiquitin ligases at a glance. J Cell Sci. 2012;125:531–537. doi: 10.1242/jcs.091777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- [13].Cardozo T, Pagano M. The SCF ubiquitin ligase: insights into a molecular machine. Nat Rev Mol Cell Biol. 2004;5:739–751. doi: 10.1038/nrm1471. [DOI] [PubMed] [Google Scholar]
- [14].Zheng N, Schulman BA, Song L, Miller JJ, Jeffrey PD, Wang P, Chu C, Koepp DM, Elledge SJ, Pagano M, Conaway RC, Conaway JW, Harper JW, Pavletich NP. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature. 2002;416:703–709. doi: 10.1038/416703a. [DOI] [PubMed] [Google Scholar]
- [15].Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper JW, Elledge SJ. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell. 1996;86:263–274. doi: 10.1016/s0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- [16].Cenciarelli C, Chiaur DS, Guardavaccaro D, Parks W, Vidal M, Pagano M. Identification of a family of human F-box proteins. Curr Biol. 1999;9:1177–1179. doi: 10.1016/S0960-9822(00)80020-2. [DOI] [PubMed] [Google Scholar]
- [17].Jin J, Cardozo T, Lovering RC, Elledge SJ, Pagano M, Harper JW. Systematic analysis and nomenclature of mammalian F-box proteins. Genes Dev. 2004;18:2573–2580. doi: 10.1101/gad.1255304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Silverman JS, Skaar JR, Pagano M. SCF ubiquitin ligases in the maintenance of genome stability. Trends Biochem Sci. 2012;37:66–73. doi: 10.1016/j.tibs.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].van Leuken R, Clijsters L, Wolthuis R. To cell cycle, swing the APC/C. Biochim Biophys Acta. 2008;1786:49–59. doi: 10.1016/j.bbcan.2008.05.002. [DOI] [PubMed] [Google Scholar]
- [20].Acquaviva C, Pines J. The anaphase-promoting complex/cyclosome: APC/C. J Cell Sci. 2006;119:2401–2404. doi: 10.1242/jcs.02937. [DOI] [PubMed] [Google Scholar]
- [21].Li M, Zhang P. The function of APC/CCdh1 in cell cycle and beyond. Cell Div. 2009;4:2. doi: 10.1186/1747-1028-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Nakayama KI, Nakayama K. Regulation of the cell cycle by SCF-type ubiquitin ligases. Semin Cell Dev Biol. 2005;16:323–333. doi: 10.1016/j.semcdb.2005.02.010. [DOI] [PubMed] [Google Scholar]
- [23].Mailand N, Diffley JF. CDKs promote DNA replication origin licensing in human cells by protecting Cdc6 from APC/C-dependent proteolysis. Cell. 2005;122:915–926. doi: 10.1016/j.cell.2005.08.013. [DOI] [PubMed] [Google Scholar]
- [24].Li M, Shin YH, Hou L, Huang X, Wei Z, Klann E, Zhang P. The adaptor protein of the anaphase promoting complex Cdh1 is essential in maintaining replicative lifespan and in learning and memory. Nat Cell Biol. 2008;10:1083–1089. doi: 10.1038/ncb1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Albanese C, Johnson J, Watanabe G, Eklund N, Vu D, Arnold A, Pestell RG. Transforming p21ras mutants and c-Ets-2 activate the cyclin D1 promoter through distinguishable regions. J Biol Chem. 1995;270:23589–23597. doi: 10.1074/jbc.270.40.23589. [DOI] [PubMed] [Google Scholar]
- [26].Bashir T, Pagano M. Don't skip the G1 phase: how APC/CCdh1 keeps SCFSKP2 in check. Cell Cycle. 2004;3:850–852. [PubMed] [Google Scholar]
- [27].Bashir T, Dorrello NV, Amador V, Guardavaccaro D, Pagano M. Control of the SCF(Skp2-Cks1) ubiquitin ligase by the APC/C(Cdh1) ubiquitin ligase. Nature. 2004;428:190–193. doi: 10.1038/nature02330. [DOI] [PubMed] [Google Scholar]
- [28].Wei W, Ayad NG, Wan Y, Zhang GJ, Kirschner MW, Kaelin WG., Jr. Degradation of the SCF component Skp2 in cell-cycle phase G1 by the anaphase-promoting complex. Nature. 2004;428:194–198. doi: 10.1038/nature02381. [DOI] [PubMed] [Google Scholar]
- [29].Donzelli M, Squatrito M, Ganoth D, Hershko A, Pagano M, Draetta GF. Dual mode of degradation of Cdc25 A phosphatase. EMBO J. 2002;21:4875–4884. doi: 10.1093/emboj/cdf491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Skaar JR, Pagano M. Cdh1: a master G0/G1 regulator. Nat Cell Biol. 2008;10:755–757. doi: 10.1038/ncb0708-755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].King RW, Peters JM, Tugendreich S, Rolfe M, Hieter P, Kirschner MW. A 20S complex containing CDC27 and CDC16 catalyzes the mitosis-specific conjugation of ubiquitin to cyclin B. Cell. 1995;81:279–288. doi: 10.1016/0092-8674(95)90338-0. [DOI] [PubMed] [Google Scholar]
- [32].Geley S, Kramer E, Gieffers C, Gannon J, Peters JM, Hunt T. Anaphase-promoting complex/cyclosome-dependent proteolysis of human cyclin A starts at the beginning of mitosis and is not subject to the spindle assembly checkpoint. J Cell Biol. 2001;153:137–148. doi: 10.1083/jcb.153.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Honda K, Mihara H, Kato Y, Yamaguchi A, Tanaka H, Yasuda H, Furukawa K, Urano T. Degradation of human Aurora2 protein kinase by the anaphase-promoting complex-ubiquitin-proteasome pathway. Oncogene. 2000;19:2812–2819. doi: 10.1038/sj.onc.1203609. [DOI] [PubMed] [Google Scholar]
- [34].Castro A, Arlot-Bonnemains Y, Vigneron S, Labbe JC, Prigent C, Lorca T. APC/Fizzy-Related targets Aurora-A kinase for proteolysis. EMBO Rep. 2002;3:457–462. doi: 10.1093/embo-reports/kvf095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Stewart S, Fang G. Destruction box-dependent degradation of aurora B is mediated by the anaphase-promoting complex/cyclosome and Cdh1. Cancer Res. 2005;65:8730–8735. doi: 10.1158/0008-5472.CAN-05-1500. [DOI] [PubMed] [Google Scholar]
- [36].Lindon C, Pines J. Ordered proteolysis in anaphase inactivates Plk1 to contribute to proper mitotic exit in human cells. J Cell Biol. 2004;164:233–241. doi: 10.1083/jcb.200309035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Garcia-Higuera I, Manchado E, Dubus P, Canamero M, Mendez J, Moreno S, Malumbres M. Genomic stability and tumour suppression by the APC/C cofactor Cdh1. Nat Cell Biol. 2008;10:802–811. doi: 10.1038/ncb1742. [DOI] [PubMed] [Google Scholar]
- [38].Fernandez-Saiz V, Targosz BS, Lemeer S, Eichner R, Langer C, Bullinger L, Reiter C, Slotta-Huspenina J, Schroeder S, Knorn AM, Kurutz J, Peschel C, Pagano M, Kuster B, Bassermann F. SCF(Fbxo9) and CK2 direct the cellular response to growth factor withdrawal via Tel2/Tti1 degradation and promote survival in multiple myeloma. Nat Cell Biol. 2012 doi: 10.1038/ncb2651. [DOI] [PubMed] [Google Scholar]
- [39].Dorrello NV, Peschiaroli A, Guardavaccaro D, Colburn NH, Sherman NE, Pagano M. S6K1- and betaTRCP-mediated degradation of PDCD4 promotes protein translation and cell growth. Science. 2006;314:467–471. doi: 10.1126/science.1130276. [DOI] [PubMed] [Google Scholar]
- [40].Dehan E, Bassermann F, Guardavaccaro D, Vasiliver-Shamis G, Cohen M, Lowes KN, Dustin M, Huang DC, Taunton J, Pagano M. betaTrCP- and Rsk1/2-mediated degradation of BimEL inhibits apoptosis. Mol Cell. 2009;33:109–116. doi: 10.1016/j.molcel.2008.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Duan S, Skaar JR, Kuchay S, Toschi A, Kanarek N, Ben-Neriah Y, Pagano M. mTOR generates an auto-amplification loop by triggering the betaTrCP- and CK1alpha-dependent degradation of DEPTOR. Mol Cell. 2011;44:317–324. doi: 10.1016/j.molcel.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Gao D, Inuzuka H, Tan MK, Fukushima H, Locasale JW, Liu P, Wan L, Zhai B, Chin YR, Shaik S, Lyssiotis CA, Gygi SP, Toker A, Cantley LC, Asara JM, Harper JW, Wei W. mTOR drives its own activation via SCF(betaTrCP)-dependent degradation of the mTOR inhibitor DEPTOR. Mol Cell. 2011;44:290–303. doi: 10.1016/j.molcel.2011.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Rape M, Kirschner MW. Autonomous regulation of the anaphase-promoting complex couples mitosis to S-phase entry. Nature. 2004;432:588–595. doi: 10.1038/nature03023. [DOI] [PubMed] [Google Scholar]
- [44].Listovsky T, Oren YS, Yudkovsky Y, Mahbubani HM, Weiss AM, Lebendiker M, Brandeis M. Mammalian Cdh1/Fzr mediates its own degradation. EMBO J. 2004;23:1619–1626. doi: 10.1038/sj.emboj.7600149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Lukas C, Sorensen CS, Kramer E, Santoni-Rugiu E, Lindeneg C, Peters JM, Bartek J, Lukas J. Accumulation of cyclin B1 requires E2F and cyclin-A-dependent rearrangement of the anaphase-promoting complex. Nature. 1999;401:815–818. doi: 10.1038/44611. [DOI] [PubMed] [Google Scholar]
- [46].Benmaamar R, Pagano M. Involvement of the SCF complex in the control of Cdh1 degradation in S-phase. Cell Cycle. 2005;4:1230–1232. doi: 10.4161/cc.4.9.2048. [DOI] [PubMed] [Google Scholar]
- [47].Miller JJ, Summers MK, Hansen DV, Nachury MV, Lehman NL, Loktev A, Jackson PK. Emi1 stably binds and inhibits the anaphase-promoting complex/cyclosome as a pseudosubstrate inhibitor. Genes Dev. 2006;20:2410–2420. doi: 10.1101/gad.1454006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Hsu JY, Reimann JD, Sorensen CS, Lukas J, Jackson PK. E2F-dependent accumulation of hEmi1 regulates S phase entry by inhibiting APC(Cdh1) Nat Cell Biol. 2002;4:358–366. doi: 10.1038/ncb785. [DOI] [PubMed] [Google Scholar]
- [49].Geng Y, Eaton EN, Picon M, Roberts JM, Lundberg AS, Gifford A, Sardet C, Weinberg RA. Regulation of cyclin E transcription by E2Fs and retinoblastoma protein. Oncogene. 1996;12:1173–1180. [PubMed] [Google Scholar]
- [50].Schulze A, Zerfass K, Spitkovsky D, Middendorp S, Berges J, Helin K, Jansen-Durr P, Henglein B. Cell cycle regulation of the cyclin A gene promoter is mediated by a variant E2F site. Proc Natl Acad Sci U S A. 1995;92:11264–11268. doi: 10.1073/pnas.92.24.11264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Tsvetkov LM, Yeh KH, Lee SJ, Sun H, Zhang H. p27(Kip1) ubiquitination and degradation is regulated by the SCF(Skp2) complex through phosphorylated Thr187 in p27. Curr Biol. 1999;9:661–664. doi: 10.1016/s0960-9822(99)80290-5. [DOI] [PubMed] [Google Scholar]
- [52].Carrano AC, Eytan E, Hershko A, Pagano M. SKP2 is required for ubiquitin-mediated degradation of the CDK inhibitor p27. Nat Cell Biol. 1999;1:193–199. doi: 10.1038/12013. [DOI] [PubMed] [Google Scholar]
- [53].Pagano M, Tam SW, Theodoras AM, Beer-Romero P, Del Sal G, Chau V, Yew PR, Draetta GF, Rolfe M. Role of the ubiquitin-proteasome pathway in regulating abundance of the cyclin-dependent kinase inhibitor p27. Science. 1995;269:682–685. doi: 10.1126/science.7624798. [DOI] [PubMed] [Google Scholar]
- [54].Sutterluty H, Chatelain E, Marti A, Wirbelauer C, Senften M, Muller U, Krek W. p45SKP2 promotes p27Kip1 degradation and induces S phase in quiescent cells. Nat Cell Biol. 1999;1:207–214. doi: 10.1038/12027. [DOI] [PubMed] [Google Scholar]
- [55].Sheaff RJ, Groudine M, Gordon M, Roberts JM, Clurman BE. Cyclin E-CDK2 is a regulator of p27Kip1. Genes Dev. 1997;11:1464–1478. doi: 10.1101/gad.11.11.1464. [DOI] [PubMed] [Google Scholar]
- [56].Vlach J, Hennecke S, Amati B. Phosphorylation-dependent degradation of the cyclin-dependent kinase inhibitor p27. EMBO J. 1997;16:5334–5344. doi: 10.1093/emboj/16.17.5334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Montagnoli A, Fiore F, Eytan E, Carrano AC, Draetta GF, Hershko A, Pagano M. Ubiquitination of p27 is regulated by Cdk-dependent phosphorylation and trimeric complex formation. Genes Dev. 1999;13:1181–1189. doi: 10.1101/gad.13.9.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Ganoth D, Bornstein G, Ko TK, Larsen B, Tyers M, Pagano M, Hershko A. The cell-cycle regulatory protein Cks1 is required for SCF(Skp2)-mediated ubiquitinylation of p27. Nat Cell Biol. 2001;3:321–324. doi: 10.1038/35060126. [DOI] [PubMed] [Google Scholar]
- [59].Spruck C, Strohmaier H, Watson M, Smith AP, Ryan A, Krek TW, Reed SI. A CDK-independent function of mammalian Cks1: targeting of SCF(Skp2) to the CDK inhibitor p27Kip1. Mol Cell. 2001;7:639–650. doi: 10.1016/s1097-2765(01)00210-6. [DOI] [PubMed] [Google Scholar]
- [60].Bornstein G, Bloom J, Sitry-Shevah D, Nakayama K, Pagano M, Hershko A. Role of the SCFSkp2 ubiquitin ligase in the degradation of p21Cip1 in S phase. J Biol Chem. 2003;278:25752–25757. doi: 10.1074/jbc.M301774200. [DOI] [PubMed] [Google Scholar]
- [61].Kamura T, Hara T, Kotoshiba S, Yada M, Ishida N, Imaki H, Hatakeyama S, Nakayama K, Nakayama KI. Degradation of p57Kip2 mediated by SCFSkp2-dependent ubiquitylation. Proc Natl Acad Sci U S A. 2003;100:10231–10236. doi: 10.1073/pnas.1831009100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Hershko DD. Oncogenic properties and prognostic implications of the ubiquitin ligase Skp2 in cancer. Cancer. 2008;112:1415–1424. doi: 10.1002/cncr.23317. [DOI] [PubMed] [Google Scholar]
- [63].Lin DI, Barbash O, Kumar KG, Weber JD, Harper JW, Klein-Szanto AJ, Rustgi A, Fuchs SY, Diehl JA. Phosphorylation-dependent ubiquitination of cyclin D1 by the SCF(FBX4-alphaB crystallin) complex. Mol Cell. 2006;24:355–366. doi: 10.1016/j.molcel.2006.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Clurman BE, Sheaff RJ, Thress K, Groudine M, Roberts JM. Turnover of cyclin E by the ubiquitin-proteasome pathway is regulated by cdk2 binding and cyclin phosphorylation. Genes Dev. 1996;10:1979–1990. doi: 10.1101/gad.10.16.1979. [DOI] [PubMed] [Google Scholar]
- [65].Koepp DM, Schaefer LK, Ye X, Keyomarsi K, Chu C, Harper JW, Elledge SJ. Phosphorylation-dependent ubiquitination of cyclin E by the SCFFbw7 ubiquitin ligase. Science. 2001;294:173–177. doi: 10.1126/science.1065203. [DOI] [PubMed] [Google Scholar]
- [66].Matsuoka S, Huang M, Elledge SJ. Linkage of ATM to cell cycle regulation by the Chk2 protein kinase. Science. 1998;282:1893–1897. doi: 10.1126/science.282.5395.1893. [DOI] [PubMed] [Google Scholar]
- [67].Zeng Y, Forbes KC, Wu Z, Moreno S, Piwnica-Worms H, Enoch T. Replication checkpoint requires phosphorylation of the phosphatase Cdc25 by Cds1 or Chk1. Nature. 1998;395:507–510. doi: 10.1038/26766. [DOI] [PubMed] [Google Scholar]
- [68].Busino L, Donzelli M, Chiesa M, Guardavaccaro D, Ganoth D, Dorrello NV, Hershko A, Pagano M, Draetta GF. Degradation of Cdc25A by beta-TrCP during S phase and in response to DNA damage. Nature. 2003;426:87–91. doi: 10.1038/nature02082. [DOI] [PubMed] [Google Scholar]
- [69].Smits VA, Klompmaker R, Arnaud L, Rijksen G, Nigg EA, Medema RH. Polo-like kinase-1 is a target of the DNA damage checkpoint. Nat Cell Biol. 2000;2:672–676. doi: 10.1038/35023629. [DOI] [PubMed] [Google Scholar]
- [70].Bassermann F, Frescas D, Guardavaccaro D, Busino L, Peschiaroli A, Pagano M. The Cdc14B-Cdh1-Plk1 axis controls the G2 DNA-damage-response checkpoint. Cell. 2008;134:256–267. doi: 10.1016/j.cell.2008.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Bassermann F, Pagano M. Dissecting the role of ubiquitylation in the DNA damage response checkpoint in G2. Cell Death Differ. 2010;17:78–85. doi: 10.1038/cdd.2009.104. [DOI] [PubMed] [Google Scholar]
- [72].Peschiaroli A, Dorrello NV, Guardavaccaro D, Venere M, Halazonetis T, Sherman NE, Pagano M. SCFbetaTrCP-mediated degradation of Claspin regulates recovery from the DNA replication checkpoint response. Mol Cell. 2006;23:319–329. doi: 10.1016/j.molcel.2006.06.013. [DOI] [PubMed] [Google Scholar]
- [73].Mailand N, Bekker-Jensen S, Bartek J, Lukas J. Destruction of Claspin by SCFbetaTrCP restrains Chk1 activation and facilitates recovery from genotoxic stress. Mol Cell. 2006;23:307–318. doi: 10.1016/j.molcel.2006.06.016. [DOI] [PubMed] [Google Scholar]
- [74].Kruiswijk F, Yuniati L, Magliozzi R, Low TY, Lim R, Bolder R, Mohammed S, Proud CG, Heck AJ, Pagano M, Guardavaccaro D. Coupled activation and degradation of eEF2K regulates protein synthesis in response to genotoxic stress. Sci Signal. 2012;5:ra40. doi: 10.1126/scisignal.2002718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].D'Angiolella V, Donato V, Forrester FM, Jeong YT, Pellacani C, Kudo Y, Saraf A, Florens L, Washburn MP, Pagano M. Cyclin F-mediated degradation of ribonucleotide reductase M2 controls genome integrity and DNA repair. Cell. 2012;149:1023–1034. doi: 10.1016/j.cell.2012.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].D'Angiolella V, Donato V, Vijayakumar S, Saraf A, Florens L, Washburn MP, Dynlacht B, Pagano M. SCF(Cyclin F) controls centrosome homeostasis and mitotic fidelity through CP110 degradation. Nature. 2010;466:138–142. doi: 10.1038/nature09140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Watanabe N, Arai H, Nishihara Y, Taniguchi M, Hunter T, Osada H. M-phase kinases induce phospho-dependent ubiquitination of somatic Wee1 by SCFbeta-TrCP. Proc Natl Acad Sci U S A. 2004;101:4419–4424. doi: 10.1073/pnas.0307700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Seki A, Coppinger JA, Jang CY, Yates JR, Fang G. Bora and the kinase Aurora a cooperatively activate the kinase Plk1 and control mitotic entry. Science. 2008;320:1655–1658. doi: 10.1126/science.1157425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Bassermann F, von Klitzing C, Munch S, Bai RY, Kawaguchi H, Morris SW, Peschel C, Duyster J. NIPA defines an SCF-type mammalian E3 ligase that regulates mitotic entry. Cell. 2005;122:45–57. doi: 10.1016/j.cell.2005.04.034. [DOI] [PubMed] [Google Scholar]
- [80].Bassermann F, von Klitzing C, Illert AL, Munch S, Morris SW, Pagano M, Peschel C, Duyster J. Multisite phosphorylation of nuclear interaction partner of ALK (NIPA) at G2/M involves cyclin B1/Cdk1. J Biol Chem. 2007;282:15965–15972. doi: 10.1074/jbc.M610819200. [DOI] [PubMed] [Google Scholar]
- [81].Illert AL, Zech M, Moll C, Albers C, Kreutmair S, Peschel C, Bassermann F, Duyster J. Extracellular signal-regulated kinase 2 (ERK2) mediates phosphorylation and inactivation of Nuclear interaction partner of Anaplastic lymphoma kinase (NIPA) at G2/M. J Biol Chem. 2012 doi: 10.1074/jbc.M112.373464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Margottin-Goguet F, Hsu JY, Loktev A, Hsieh HM, Reimann JD, Jackson PK. Prophase destruction of Emi1 by the SCF(betaTrCP/Slimb) ubiquitin ligase activates the anaphase promoting complex to allow progression beyond prometaphase. Dev Cell. 2003;4:813–826. doi: 10.1016/s1534-5807(03)00153-9. [DOI] [PubMed] [Google Scholar]
- [83].Guardavaccaro D, Kudo Y, Boulaire J, Barchi M, Busino L, Donzelli M, Margottin-Goguet F, Jackson PK, Yamasaki L, Pagano M. Control of meiotic and mitotic progression by the F box protein beta-Trcp1 in vivo. Dev Cell. 2003;4:799–812. doi: 10.1016/s1534-5807(03)00154-0. [DOI] [PubMed] [Google Scholar]
- [84].Fang G, Yu H, Kirschner MW. The checkpoint protein MAD2 and the mitotic regulator CDC20 form a ternary complex with the anaphase-promoting complex to control anaphase initiation. Genes Dev. 1998;12:1871–1883. doi: 10.1101/gad.12.12.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Sudakin V, Chan GK, Yen TJ. Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20, and MAD2. J Cell Biol. 2001;154:925–936. doi: 10.1083/jcb.200102093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Braunstein I, Miniowitz S, Moshe Y, Hershko A. Inhibitory factors associated with anaphase-promoting complex/cylosome in mitotic checkpoint. Proc Natl Acad Sci U S A. 2007;104:4870–4875. doi: 10.1073/pnas.0700523104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Nilsson J, Yekezare M, Minshull J, Pines J. The APC/C maintains the spindle assembly checkpoint by targeting Cdc20 for destruction. Nat Cell Biol. 2008;10:1411–1420. doi: 10.1038/ncb1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Ge S, Skaar JR, Pagano M. APC/C- and Mad2-mediated degradation of Cdc20 during spindle checkpoint activation. Cell Cycle. 2009;8:167–171. doi: 10.4161/cc.8.1.7606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].D'Angiolella V, Mari C, Nocera D, Rametti L, Grieco D. The spindle checkpoint requires cyclin-dependent kinase activity. Genes Dev. 2003;17:2520–2525. doi: 10.1101/gad.267603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Chung E, Chen RH. Phosphorylation of Cdc20 is required for its inhibition by the spindle checkpoint. Nat Cell Biol. 2003;5:748–753. doi: 10.1038/ncb1022. [DOI] [PubMed] [Google Scholar]
- [91].Tang Z, Shu H, Oncel D, Chen S, Yu H. Phosphorylation of Cdc20 by Bub1 provides a catalytic mechanism for APC/C inhibition by the spindle checkpoint. Mol Cell. 2004;16:387–397. doi: 10.1016/j.molcel.2004.09.031. [DOI] [PubMed] [Google Scholar]
- [92].Guardavaccaro D, Frescas D, Dorrello NV, Peschiaroli A, Multani AS, Cardozo T, Lasorella A, Iavarone A, Chang S, Hernando E, Pagano M. Control of chromosome stability by the beta-TrCP-REST-Mad2 axis. Nature. 2008;452:365–369. doi: 10.1038/nature06641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Amador V, Ge S, Santamaria PG, Guardavaccaro D, Pagano M. APC/C(Cdc20) controls the ubiquitin-mediated degradation of p21 in prometaphase. Mol Cell. 2007;27:462–473. doi: 10.1016/j.molcel.2007.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Pfleger CM, Kirschner MW. The KEN box: an APC recognition signal distinct from the D box targeted by Cdh1. Genes Dev. 2000;14:655–665. [PMC free article] [PubMed] [Google Scholar]
- [95].Reis A, Levasseur M, Chang HY, Elliott DJ, Jones KT. The CRY box: a second APCcdh1-dependent degron in mammalian cdc20. EMBO Rep. 2006;7:1040–1045. doi: 10.1038/sj.embor.7400772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Di Fiore B, Pines J. How cyclin A destruction escapes the spindle assembly checkpoint. J Cell Biol. 2010;190:501–509. doi: 10.1083/jcb.201001083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Hayes MJ, Kimata Y, Wattam SL, Lindon C, Mao G, Yamano H, Fry AM. Early mitotic degradation of Nek2A depends on Cdc20-independent interaction with the APC/C. Nat Cell Biol. 2006;8:607–614. doi: 10.1038/ncb1410. [DOI] [PubMed] [Google Scholar]
- [98].van Zon W, Wolthuis RM. Cyclin A and Nek2A: APC/C-Cdc20 substrates invisible to the mitotic spindle checkpoint. Biochem Soc Trans. 2010;38:72–77. doi: 10.1042/BST0380072. [DOI] [PubMed] [Google Scholar]
- [99].Varetti G, Guida C, Santaguida S, Chiroli E, Musacchio A. Homeostatic control of mitotic arrest. Mol Cell. 2011;44:710–720. doi: 10.1016/j.molcel.2011.11.014. [DOI] [PubMed] [Google Scholar]
- [100].Foe IT, Foster SA, Cheung SK, DeLuca SZ, Morgan DO, Toczyski DP. Ubiquitination of Cdc20 by the APC occurs through an intramolecular mechanism. Curr Biol. 2011;21:1870–1877. doi: 10.1016/j.cub.2011.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Reddy SK, Rape M, Margansky WA, Kirschner MW. Ubiquitination by the anaphase-promoting complex drives spindle checkpoint inactivation. Nature. 2007;446:921–925. doi: 10.1038/nature05734. [DOI] [PubMed] [Google Scholar]
- [102].Miniowitz-Shemtov S, Teichner A, Sitry-Shevah D, Hershko A. ATP is required for the release of the anaphase-promoting complex/cyclosome from inhibition by the mitotic checkpoint. Proc Natl Acad Sci U S A. 2010;107:5351–5356. doi: 10.1073/pnas.1001875107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Zou H, McGarry TJ, Bernal T, Kirschner MW. Identification of a vertebrate sister-chromatid separation inhibitor involved in transformation and tumorigenesis. Science. 1999;285:418–422. doi: 10.1126/science.285.5426.418. [DOI] [PubMed] [Google Scholar]
- [104].Peters JM. The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat Rev Mol Cell Biol. 2006;7:644–656. doi: 10.1038/nrm1988. [DOI] [PubMed] [Google Scholar]
- [105].Hauf S, Waizenegger IC, Peters JM. Cohesin cleavage by separase required for anaphase and cytokinesis in human cells. Science. 2001;293:1320–1323. doi: 10.1126/science.1061376. [DOI] [PubMed] [Google Scholar]
- [106].Karamysheva Z, Diaz-Martinez LA, Crow SE, Li B, Yu H. Multiple anaphase-promoting complex/cyclosome degrons mediate the degradation of human Sgo1. J Biol Chem. 2009;284:1772–1780. doi: 10.1074/jbc.M807083200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Kitajima TS, Kawashima SA, Watanabe Y. The conserved kinetochore protein shugoshin protects centromeric cohesion during meiosis. Nature. 2004;427:510–517. doi: 10.1038/nature02312. [DOI] [PubMed] [Google Scholar]
- [108].Sudakin V, Ganoth D, Dahan A, Heller H, Hershko J, Luca FC, Ruderman JV, Hershko A. The cyclosome, a large complex containing cyclin-selective ubiquitin ligase activity, targets cyclins for destruction at the end of mitosis. Mol Biol Cell. 1995;6:185–197. doi: 10.1091/mbc.6.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Raff JW, Jeffers K, Huang JY. The roles of Fzy/Cdc20 and Fzr/Cdh1 in regulating the destruction of cyclin B in space and time. J Cell Biol. 2002;157:1139–1149. doi: 10.1083/jcb.200203035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].den Elzen N, Pines J. Cyclin A is destroyed in prometaphase and can delay chromosome alignment and anaphase. J Cell Biol. 2001;153:121–136. doi: 10.1083/jcb.153.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Clute P, Pines J. Temporal and spatial control of cyclin B1 destruction in metaphase. Nat Cell Biol. 1999;1:82–87. doi: 10.1038/10049. [DOI] [PubMed] [Google Scholar]
- [112].Seki A, Coppinger JA, Du H, Jang CY, Yates JR, 3rd, Fang G. Plk1- and beta-TrCP-dependent degradation of Bora controls mitotic progression. J Cell Biol. 2008;181:65–78. doi: 10.1083/jcb.200712027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Chan EH, Santamaria A, Sillje HH, Nigg EA. Plk1 regulates mitotic Aurora A function through betaTrCP-dependent degradation of hBora. Chromosoma. 2008;117:457–469. doi: 10.1007/s00412-008-0165-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].McGarry TJ, Kirschner MW. Geminin, an inhibitor of DNA replication, is degraded during mitosis. Cell. 1998;93:1043–1053. doi: 10.1016/s0092-8674(00)81209-x. [DOI] [PubMed] [Google Scholar]
- [115].Kramer ER, Scheuringer N, Podtelejnikov AV, Mann M, Peters JM. Mitotic regulation of the APC activator proteins CDC20 and CDH1. Mol Biol Cell. 2000;11:1555–1569. doi: 10.1091/mbc.11.5.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Zur A, Brandeis M. Timing of APC/C substrate degradation is determined by fzy/fzr specificity of destruction boxes. EMBO J. 2002;21:4500–4510. doi: 10.1093/emboj/cdf452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Rape M, Reddy SK, Kirschner MW. The processivity of multiubiquitination by the APC determines the order of substrate degradation. Cell. 2006;124:89–103. doi: 10.1016/j.cell.2005.10.032. [DOI] [PubMed] [Google Scholar]
- [118].Sumara I, Maerki S, Peter M. E3 ubiquitin ligases and mitosis: embracing the complexity. Trends Cell Biol. 2008;18:84–94. doi: 10.1016/j.tcb.2007.12.001. [DOI] [PubMed] [Google Scholar]
- [119].Angers S, Li T, Yi X, MacCoss MJ, Moon RT, Zheng N. Molecular architecture and assembly of the DDB1-CUL4A ubiquitin ligase machinery. Nature. 2006;443:590–593. doi: 10.1038/nature05175. [DOI] [PubMed] [Google Scholar]
- [120].He YJ, McCall CM, Hu J, Zeng Y, Xiong Y. DDB1 functions as a linker to recruit receptor WD40 proteins to CUL4-ROC1 ubiquitin ligases. Genes Dev. 2006;20:2949–2954. doi: 10.1101/gad.1483206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Lee J, Zhou P. DCAFs, the missing link of the CUL4-DDB1 ubiquitin ligase. Mol Cell. 2007;26:775–780. doi: 10.1016/j.molcel.2007.06.001. [DOI] [PubMed] [Google Scholar]
- [122].Shibata E, Abbas T, Huang X, Wohlschlegel JA, Dutta A. Selective ubiquitylation of p21 and Cdt1 by UBCH8 and UBE2G ubiquitin-conjugating enzymes via the CRL4Cdt2 ubiquitin ligase complex. Mol Cell Biol. 2011;31:3136–3145. doi: 10.1128/MCB.05496-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Nishitani H, Sugimoto N, Roukos V, Nakanishi Y, Saijo M, Obuse C, Tsurimoto T, Nakayama KI, Nakayama K, Fujita M, Lygerou Z, Nishimoto T. Two E3 ubiquitin ligases, SCF-Skp2 and DDB1-Cul4, target human Cdt1 for proteolysis. EMBO J. 2006;25:1126–1136. doi: 10.1038/sj.emboj.7601002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Arias EE, Walter JC. PCNA functions as a molecular platform to trigger Cdt1 destruction and prevent re-replication. Nat Cell Biol. 2006;8:84–90. doi: 10.1038/ncb1346. [DOI] [PubMed] [Google Scholar]
- [125].Jin J, Arias EE, Chen J, Harper JW, Walter JC. A family of diverse Cul4-Ddb1-interacting proteins includes Cdt2, which is required for S phase destruction of the replication factor Cdt1. Mol Cell. 2006;23:709–721. doi: 10.1016/j.molcel.2006.08.010. [DOI] [PubMed] [Google Scholar]
- [126].Zhong W, Feng H, Santiago FE, Kipreos ET. CUL-4 ubiquitin ligase maintains genome stability by restraining DNA-replication licensing. Nature. 2003;423:885–889. doi: 10.1038/nature01747. [DOI] [PubMed] [Google Scholar]
- [127].Hu J, McCall CM, Ohta T, Xiong Y. Targeted ubiquitination of CDT1 by the DDB1-CUL4A-ROC1 ligase in response to DNA damage. Nat Cell Biol. 2004;6:1003–1009. doi: 10.1038/ncb1172. [DOI] [PubMed] [Google Scholar]
- [128].Li X, Zhao Q, Liao R, Sun P, Wu X. The SCF(Skp2) ubiquitin ligase complex interacts with the human replication licensing factor Cdt1 and regulates Cdt1 degradation. J Biol Chem. 2003;278:30854–30858. doi: 10.1074/jbc.C300251200. [DOI] [PubMed] [Google Scholar]
- [129].Abbas T, Sivaprasad U, Terai K, Amador V, Pagano M, Dutta A. PCNA-dependent regulation of p21 ubiquitylation and degradation via the CRL4Cdt2 ubiquitin ligase complex. Genes Dev. 2008;22:2496–2506. doi: 10.1101/gad.1676108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Abbas T, Shibata E, Park J, Jha S, Karnani N, Dutta A. CRL4(Cdt2) regulates cell proliferation and histone gene expression by targeting PR-Set7/Set8 for degradation. Mol Cell. 2010;40:9–21. doi: 10.1016/j.molcel.2010.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Centore RC, Havens CG, Manning AL, Li JM, Flynn RL, Tse A, Jin J, Dyson NJ, Walter JC, Zou L. CRL4(Cdt2)-mediated destruction of the histone methyltransferase Set8 prevents premature chromatin compaction in S phase. Mol Cell. 2010;40:22–33. doi: 10.1016/j.molcel.2010.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Jorgensen S, Eskildsen M, Fugger K, Hansen L, Larsen MS, Kousholt AN, Syljuasen RG, Trelle MB, Jensen ON, Helin K, Sorensen CS. SET8 is degraded via PCNA-coupled CRL4(CDT2) ubiquitylation in S phase and after UV irradiation. J Cell Biol. 2011;192:43–54. doi: 10.1083/jcb.201009076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Abbas T, Mueller A, Shibata E, Keaton M, Rossi M, Dutta A. CRL1-FBXO11 promotes Cdt2 ubiquitylation and degradation and regulates Pr-Set7/Set8-mediated cellular migration. Mol Cell. 2013 doi: 10.1016/j.molcel.2013.02.003. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Rossi M, Duan S, Jeong YT, Horn M, Saraf A, Florens L, Washburn MP, Antebi A, Pagano M. Regulation of the CRL4Cdt2 ubiquitin ligase and cell cycle exit by the SCFFbxo11 ubiquitin ligase. Mol Cell. 2013 doi: 10.1016/j.molcel.2013.02.004. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Rajkumar SV, Richardson PG, Hideshima T, Anderson KC. Proteasome inhibition as a novel therapeutic target in human cancer. J Clin Oncol. 2005;23:630–639. doi: 10.1200/JCO.2005.11.030. [DOI] [PubMed] [Google Scholar]
- [136].Moreau P, Richardson PG, Cavo M, Orlowski RZ, San Miguel JF, Palumbo A, Harousseau JL. Proteasome inhibitors in multiple myeloma: 10 years later. Blood. 2012;120:947–959. doi: 10.1182/blood-2012-04-403733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Nakayama KI, Nakayama K. Ubiquitin ligases: cell-cycle control and cancer. Nat Rev Cancer. 2006;6:369–381. doi: 10.1038/nrc1881. [DOI] [PubMed] [Google Scholar]
- [138].Skaar JR, D'Angiolella V, Pagan JK, Pagano M. SnapShot: F Box Proteins II. Cell. 2009;137:1358, e1351. doi: 10.1016/j.cell.2009.05.040. 1358. [DOI] [PubMed] [Google Scholar]
- [139].Masuda TA, Inoue H, Sonoda H, Mine S, Yoshikawa Y, Nakayama K, Nakayama K, Mori M. Clinical and biological significance of S-phase kinase-associated protein 2 (Skp2) gene expression in gastric carcinoma: modulation of malignant phenotype by Skp2 overexpression, possibly via p27 proteolysis. Cancer Res. 2002;62:3819–3825. [PubMed] [Google Scholar]
- [140].Masuda TA, Inoue H, Nishida K, Sonoda H, Yoshikawa Y, Kakeji Y, Utsunomiya T, Mori M. Cyclin-dependent kinase 1 gene expression is associated with poor prognosis in gastric carcinoma. Clinical cancer research : an official journal of the American Association for Cancer Research. 2003;9:5693–5698. [PubMed] [Google Scholar]
- [141].Shapira M, Ben-Izhak O, Linn S, Futerman B, Minkov I, Hershko DD. The prognostic impact of the ubiquitin ligase subunits Skp2 and Cks1 in colorectal carcinoma. Cancer. 2005;103:1336–1346. doi: 10.1002/cncr.20917. [DOI] [PubMed] [Google Scholar]
- [142].Signoretti S, Di Marcotullio L, Richardson A, Ramaswamy S, Isaac B, Rue M, Monti F, Loda M, Pagano M. Oncogenic role of the ubiquitin ligase subunit Skp2 in human breast cancer. The Journal of clinical investigation. 2002;110:633–641. doi: 10.1172/JCI15795. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [143].Latres E, Chiarle R, Schulman BA, Pavletich NP, Pellicer A, Inghirami G, Pagano M. Role of the F-box protein Skp2 in lymphomagenesis. Proc Natl Acad Sci U S A. 2001;98:2515–2520. doi: 10.1073/pnas.041475098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Shim EH, Johnson L, Noh HL, Kim YJ, Sun H, Zeiss C, Zhang H. Expression of the F-box protein SKP2 induces hyperplasia, dysplasia, and low-grade carcinoma in the mouse prostate. Cancer Res. 2003;63:1583–1588. [PubMed] [Google Scholar]
- [145].Timmerbeul I, Garrett-Engele CM, Kossatz U, Chen X, Firpo E, Grunwald V, Kamino K, Wilkens L, Lehmann U, Buer J, Geffers R, Kubicka S, Manns MP, Porter PL, Roberts JM, Malek NP. Testing the importance of p27 degradation by the SCFskp2 pathway in murine models of lung and colon cancer. Proc Natl Acad Sci U S A. 2006;103:14009–14014. doi: 10.1073/pnas.0606316103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Lin HK, Chen Z, Wang G, Nardella C, Lee SW, Chan CH, Yang WL, Wang J, Egia A, Nakayama KI, Cordon-Cardo C, Teruya-Feldstein J, Pandolfi PP. Skp2 targeting suppresses tumorigenesis by Arf-p53-independent cellular senescence. Nature. 2010;464:374–379. doi: 10.1038/nature08815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Lee SH, McCormick F. Downregulation of Skp2 and p27/Kip1 synergistically induces apoptosis in T98G glioblastoma cells. J Mol Med (Berl) 2005;83:296–307. doi: 10.1007/s00109-004-0611-7. [DOI] [PubMed] [Google Scholar]
- [148].Katagiri Y, Hozumi Y, Kondo S. Knockdown of Skp2 by siRNA inhibits melanoma cell growth in vitro and in vivo. Journal of dermatological science. 2006;42:215–224. doi: 10.1016/j.jdermsci.2005.12.016. [DOI] [PubMed] [Google Scholar]
- [149].Kudo Y, Kitajima S, Ogawa I, Kitagawa M, Miyauchi M, Takata T. Small interfering RNA targeting of S phase kinase-interacting protein 2 inhibits cell growth of oral cancer cells by inhibiting p27 degradation. Molecular cancer therapeutics. 2005;4:471–476. doi: 10.1158/1535-7163.MCT-04-0232. [DOI] [PubMed] [Google Scholar]
- [150].Barry M, Fruh K. Viral modulators of cullin RING ubiquitin ligases: culling the host defense. Sci STKE. 2006;2006:pe21. doi: 10.1126/stke.3352006pe21. [DOI] [PubMed] [Google Scholar]
- [151].Frescas D, Pagano M. Deregulated proteolysis by the F-box proteins SKP2 and beta-TrCP: tipping the scales of cancer. Nat Rev Cancer. 2008;8:438–449. doi: 10.1038/nrc2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Winston JT, Strack P, Beer-Romero P, Chu CY, Elledge SJ, Harper JW. The SCFbeta-TRCP-ubiquitin ligase complex associates specifically with phosphorylated destruction motifs in IkappaBalpha and beta-catenin and stimulates IkappaBalpha ubiquitination in vitro. Genes Dev. 1999;13:270–283. doi: 10.1101/gad.13.3.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Tang W, Li Y, Yu D, Thomas-Tikhonenko A, Spiegelman VS, Fuchs SY. Targeting beta-transducin repeat-containing protein E3 ubiquitin ligase augments the effects of antitumor drugs on breast cancer cells. Cancer Res. 2005;65:1904–1908. doi: 10.1158/0008-5472.CAN-04-2597. [DOI] [PubMed] [Google Scholar]
- [154].Kudo Y, Guardavaccaro D, Santamaria PG, Koyama-Nasu R, Latres E, Bronson R, Yamasaki L, Pagano M. Role of F-box protein betaTrcp1 in mammary gland development and tumorigenesis. Mol Cell Biol. 2004;24:8184–8194. doi: 10.1128/MCB.24.18.8184-8194.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Ougolkov A, Zhang B, Yamashita K, Bilim V, Mai M, Fuchs SY, Minamoto T. Associations among beta-TrCP, an E3 ubiquitin ligase receptor, beta-catenin, and NF-kappaB in colorectal cancer. Journal of the National Cancer Institute. 2004;96:1161–1170. doi: 10.1093/jnci/djh219. [DOI] [PubMed] [Google Scholar]
- [156].Muerkoster S, Arlt A, Sipos B, Witt M, Grossmann M, Kloppel G, Kalthoff H, Folsch UR, Schafer H. Increased expression of the E3-ubiquitin ligase receptor subunit betaTRCP1 relates to constitutive nuclear factor-kappaB activation and chemoresistance in pancreatic carcinoma cells. Cancer Res. 2005;65:1316–1324. doi: 10.1158/0008-5472.CAN-04-1626. [DOI] [PubMed] [Google Scholar]
- [157].Belaidouni N, Peuchmaur M, Perret C, Florentin A, Benarous R, Besnard-Guerin C. Overexpression of human beta TrCP1 deleted of its F box induces tumorigenesis in transgenic mice. Oncogene. 2005;24:2271–2276. doi: 10.1038/sj.onc.1208418. [DOI] [PubMed] [Google Scholar]
- [158].Besnard-Guerin C, Belaidouni N, Lassot I, Segeral E, Jobart A, Marchal C, Benarous R. HIV-1 Vpu sequesters beta-transducin repeat-containing protein (betaTrCP) in the cytoplasm and provokes the accumulation of beta-catenin and other SCFbetaTrCP substrates. J Biol Chem. 2004;279:788–795. doi: 10.1074/jbc.M308068200. [DOI] [PubMed] [Google Scholar]
- [159].Palumbo A, Anderson K. Multiple myeloma. N Engl J Med. 2011;364:1046–1060. doi: 10.1056/NEJMra1011442. [DOI] [PubMed] [Google Scholar]
- [160].Ruschak AM, Slassi M, Kay LE, Schimmer AD. Novel proteasome inhibitors to overcome bortezomib resistance. Journal of the National Cancer Institute. 2011;103:1007–1017. doi: 10.1093/jnci/djr160. [DOI] [PubMed] [Google Scholar]
- [161].Battistutta R. Protein kinase CK2 in health and disease: Structural bases of protein kinase CK2 inhibition. Cellular and molecular life sciences : CMLS. 2009;66:1868–1889. doi: 10.1007/s00018-009-9155-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Busino L, Millman SE, Scotto L, Kyratsous CA, Basrur V, O'Connor O, Hoffmann A, Elenitoba-Johnson KS, Pagano M. Fbxw7alpha- and GSK3-mediated degradation of p100 is a pro-survival mechanism in multiple myeloma. Nat Cell Biol. 2012;14:375–385. doi: 10.1038/ncb2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Thompson BJ, Buonamici S, Sulis ML, Palomero T, Vilimas T, Basso G, Ferrando A, Aifantis I. The SCFFBW7 ubiquitin ligase complex as a tumor suppressor in T cell leukemia. The Journal of experimental medicine. 2007;204:1825–1835. doi: 10.1084/jem.20070872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Rajagopalan H, Jallepalli PV, Rago C, Velculescu VE, Kinzler KW, Vogelstein B, Lengauer C. Inactivation of hCDC4 can cause chromosomal instability. Nature. 2004;428:77–81. doi: 10.1038/nature02313. [DOI] [PubMed] [Google Scholar]
- [165].O'Neil J, Grim J, Strack P, Rao S, Tibbitts D, Winter C, Hardwick J, Welcker M, Meijerink JP, Pieters R, Draetta G, Sears R, Clurman BE, Look AT. FBW7 mutations in leukemic cells mediate NOTCH pathway activation and resistance to gamma-secretase inhibitors. The Journal of experimental medicine. 2007;204:1813–1824. doi: 10.1084/jem.20070876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Chapman MA, Lawrence MS, Keats JJ, Cibulskis K, Sougnez C, Schinzel AC, Harview CL, Brunet JP, Ahmann GJ, Adli M, Anderson KC, Ardlie KG, Auclair D, Baker A, Bergsagel PL, Bernstein BE, Drier Y, Fonseca R, Gabriel SB, Hofmeister CC, Jagannath S, Jakubowiak AJ, Krishnan A, Levy J, Liefeld T, Lonial S, Mahan S, Mfuko B, Monti S, Perkins LM, Onofrio R, Pugh TJ, Rajkumar SV, Ramos AH, Siegel DS, Sivachenko A, Stewart AK, Trudel S, Vij R, Voet D, Winckler W, Zimmerman T, Carpten J, Trent J, Hahn WC, Garraway LA, Meyerson M, Lander ES, Getz G, Golub TR. Initial genome sequencing and analysis of multiple myeloma. Nature. 2011;471:467–472. doi: 10.1038/nature09837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Duan S, Cermak L, Pagan JK, Rossi M, Martinengo C, di Celle PF, Chapuy B, Shipp M, Chiarle R, Pagano M. FBXO11 targets BCL6 for degradation and is inactivated in diffuse large B-cell lymphomas. Nature. 2012;481:90–93. doi: 10.1038/nature10688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Lohr JG, Stojanov P, Lawrence MS, Auclair D, Chapuy B, Sougnez C, Cruz-Gordillo P, Knoechel B, Asmann YW, Slager SL, Novak AJ, Dogan A, Ansell SM, Link BK, Zou L, Gould J, Saksena G, Stransky N, Rangel-Escareno C, Fernandez-Lopez JC, Hidalgo-Miranda A, Melendez-Zajgla J, Hernandez-Lemus E, Schwarz-Cruz y Celis A, Imaz-Rosshandler I, Ojesina AI, Jung J, Pedamallu CS, Lander ES, Habermann TM, Cerhan JR, Shipp MA, Getz G, Golub TR. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc Natl Acad Sci U S A. 2012;109:3879–3884. doi: 10.1073/pnas.1121343109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, Kryukov GV, Lawrence MS, Sougnez C, McKenna A, Shefler E, Ramos AH, Stojanov P, Carter SL, Voet D, Cortes ML, Auclair D, Berger MF, Saksena G, Guiducci C, Onofrio RC, Parkin M, Romkes M, Weissfeld JL, Seethala RR, Wang L, Rangel-Escareno C, Fernandez-Lopez JC, Hidalgo-Miranda A, Melendez-Zajgla J, Winckler W, Ardlie K, Gabriel SB, Meyerson M, Lander ES, Getz G, Golub TR, Garraway LA, Grandis JR. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333:1157–1160. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Kan Z, Jaiswal BS, Stinson J, Janakiraman V, Bhatt D, Stern HM, Yue P, Haverty PM, Bourgon R, Zheng J, Moorhead M, Chaudhuri S, Tomsho LP, Peters BA, Pujara K, Cordes S, Davis DP, Carlton VE, Yuan W, Li L, Wang W, Eigenbrot C, Kaminker JS, Eberhard DA, Waring P, Schuster SC, Modrusan Z, Zhang Z, Stokoe D, de Sauvage FJ, Faham M, Seshagiri S. Diverse somatic mutation patterns and pathway alterations in human cancers. Nature. 2010;466:869–873. doi: 10.1038/nature09208. [DOI] [PubMed] [Google Scholar]
- [172].Barbash O, Zamfirova P, Lin DI, Chen X, Yang K, Nakagawa H, Lu F, Rustgi AK, Diehl JA. Mutations in Fbx4 inhibit dimerization of the SCF(Fbx4) ligase and contribute to cyclin D1 overexpression in human cancer. Cancer Cell. 2008;14:68–78. doi: 10.1016/j.ccr.2008.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Barbash O, Diehl JA. SCF(Fbx4/alphaB-crystallin) E3 ligase: when one is not enough. Cell Cycle. 2008;7:2983–2986. doi: 10.4161/cc.7.19.6775. [DOI] [PubMed] [Google Scholar]
- [174].Fu J, Qiu H, Cai M, Pan Y, Cao Y, Liu L, Yun J, Zhang CZ. Low cyclin F expression in hepatocellular carcinoma associates with poor differentiation and unfavorable prognosis. Cancer Sci. 2013 doi: 10.1111/cas.12100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Yang Y, Kitagaki J, Dai RM, Tsai YC, Lorick KL, Ludwig RL, Pierre SA, Jensen JP, Davydov IV, Oberoi P, Li CC, Kenten JH, Beutler JA, Vousden KH, Weissman AM. Inhibitors of ubiquitin-activating enzyme (E1), a new class of potential cancer therapeutics. Cancer Res. 2007;67:9472–9481. doi: 10.1158/0008-5472.CAN-07-0568. [DOI] [PubMed] [Google Scholar]
- [176].Soucy TA, Smith PG, Milhollen MA, Berger AJ, Gavin JM, Adhikari S, Brownell JE, Burke KE, Cardin DP, Critchley S, Cullis CA, Doucette A, Garnsey JJ, Gaulin JL, Gershman RE, Lublinsky AR, McDonald A, Mizutani H, Narayanan U, Olhava EJ, Peluso S, Rezaei M, Sintchak MD, Talreja T, Thomas MP, Traore T, Vyskocil S, Weatherhead GS, Yu J, Zhang J, Dick LR, Claiborne CF, Rolfe M, Bolen JB, Langston SP. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature. 2009;458:732–736. doi: 10.1038/nature07884. [DOI] [PubMed] [Google Scholar]
- [177].Goldenberg SJ, Cascio TC, Shumway SD, Garbutt KC, Liu J, Xiong Y, Zheng N. Structure of the Cand1-Cul1-Roc1 complex reveals regulatory mechanisms for the assembly of the multisubunit cullin-dependent ubiquitin ligases. Cell. 2004;119:517–528. doi: 10.1016/j.cell.2004.10.019. [DOI] [PubMed] [Google Scholar]
- [178].Duda DM, Borg LA, Scott DC, Hunt HW, Hammel M, Schulman BA. Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation. Cell. 2008;134:995–1006. doi: 10.1016/j.cell.2008.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Soucy TA, Smith PG, Rolfe M. Targeting NEDD8-activated cullin-RING ligases for the treatment of cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2009;15:3912–3916. doi: 10.1158/1078-0432.CCR-09-0343. [DOI] [PubMed] [Google Scholar]
- [180].Swords RT, Kelly KR, Smith PG, Garnsey JJ, Mahalingam D, Medina E, Oberheu K, Padmanabhan S, O'Dwyer M, Nawrocki ST, Giles FJ, Carew JS. Inhibition of NEDD8-activating enzyme: a novel approach for the treatment of acute myeloid leukemia. Blood. 2010;115:3796–3800. doi: 10.1182/blood-2009-11-254862. [DOI] [PubMed] [Google Scholar]
- [181].Ye Y, Rape M. Building ubiquitin chains: E2 enzymes at work. Nat Rev Mol Cell Biol. 2009;10:755–764. doi: 10.1038/nrm2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [182].Ceccarelli DF, Tang X, Pelletier B, Orlicky S, Xie W, Plantevin V, Neculai D, Chou YC, Ogunjimi A, Al-Hakim A, Varelas X, Koszela J, Wasney GA, Vedadi M, Dhe-Paganon S, Cox S, Xu S, Lopez-Girona A, Mercurio F, Wrana J, Durocher D, Meloche S, Webb DR, Tyers M, Sicheri F. An allosteric inhibitor of the human Cdc34 ubiquitin-conjugating enzyme. Cell. 2011;145:1075–1087. doi: 10.1016/j.cell.2011.05.039. [DOI] [PubMed] [Google Scholar]
- [183].Gonzalez-Ruiz D, Gohlke H. Targeting protein-protein interactions with small molecules: challenges and perspectives for computational binding epitope detection and ligand finding. Current medicinal chemistry. 2006;13:2607–2625. doi: 10.2174/092986706778201530. [DOI] [PubMed] [Google Scholar]
- [184].Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, Kong N, Kammlott U, Lukacs C, Klein C, Fotouhi N, Liu EA. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- [185].Wiman KG. Strategies for therapeutic targeting of the p53 pathway in cancer. Cell Death Differ. 2006;13:921–926. doi: 10.1038/sj.cdd.4401921. [DOI] [PubMed] [Google Scholar]
- [186].Shangary S, Qin D, McEachern D, Liu M, Miller RS, Qiu S, Nikolovska-Coleska Z, Ding K, Wang G, Chen J, Bernard D, Zhang J, Lu Y, Gu Q, Shah RB, Pienta KJ, Ling X, Kang S, Guo M, Sun Y, Yang D, Wang S. Temporal activation of p53 by a specific MDM2 inhibitor is selectively toxic to tumors and leads to complete tumor growth inhibition. Proc Natl Acad Sci U S A. 2008;105:3933–3938. doi: 10.1073/pnas.0708917105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [187].Chen Q, Xie W, Kuhn DJ, Voorhees PM, Lopez-Girona A, Mendy D, Corral LG, Krenitsky VP, Xu W, Moutouh-de Parseval L, Webb DR, Mercurio F, Nakayama KI, Nakayama K, Orlowski RZ. Targeting the p27 E3 ligase SCF(Skp2) results in p27- and Skp2-mediated cell-cycle arrest and activation of autophagy. Blood. 2008;111:4690–4699. doi: 10.1182/blood-2007-09-112904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [188].Orlicky S, Tang X, Neduva V, Elowe N, Brown ED, Sicheri F, Tyers M. An allosteric inhibitor of substrate recognition by the SCF(Cdc4) ubiquitin ligase. Nat Biotechnol. 2010;28:733–737. doi: 10.1038/nbt.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [189].Wu L, Grigoryan AV, Li Y, Hao B, Pagano M, Cardozo T. Specific Small Molecule Inhibitors of Skp2-Mediated p27 Degradation. Chemistry and Biology. 2013 doi: 10.1016/j.chembiol.2012.09.015. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [190].Aghajan M, Jonai N, Flick K, Fu F, Luo M, Cai X, Ouni I, Pierce N, Tang X, Lomenick B, Damoiseaux R, Hao R, Del Moral PM, Verma R, Li Y, Li C, Houk KN, Jung ME, Zheng N, Huang L, Deshaies RJ, Kaiser P, Huang J. Chemical genetics screen for enhancers of rapamycin identifies a specific inhibitor of an SCF family E3 ubiquitin ligase. Nat Biotechnol. 2010;28:738–742. doi: 10.1038/nbt.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [191].Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- [192].Ma WW, Messersmith WA, Dy GK, Weekes CD, Whitworth A, Ren C, Maniar M, Wilhelm F, Eckhardt SG, Adjei AA, Jimeno A. Phase I study of Rigosertib, an inhibitor of the phosphatidylinositol 3-kinase and Polo-like kinase 1 pathways, combined with gemcitabine in patients with solid tumors and pancreatic cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2012;18:2048–2055. doi: 10.1158/1078-0432.CCR-11-2813. [DOI] [PubMed] [Google Scholar]