Abstract
The circadian clock in mammals is driven by an autoregulatory transcriptional feedback mechanism that takes about 24 hours to complete. A key component of this mechanism is a heterodimeric transcriptional activator consisting of two bHLH-PAS domain protein subunits, CLOCK and BMAL1. Here we report the crystal structure of a complex containing the mouse CLOCK:BMAL1 bHLH-PAS domains at 2.3Å resolution. The structure reveals an unusual asymmetric heterodimer with the three domains in each of the two subunits, bHLH, PAS-A and PAS-B tightly intertwined and involved in dimerization interactions, resulting in three distinct protein interfaces. Mutations that perturb the observed heterodimer interfaces affect the stability and activity of the CLOCK:BMAL1 complex as well as the periodicity of the circadian oscillator. The structure of the CLOCK:BMAL1 complex is a starting point for understanding at an atomic level the mechanism driving the mammalian circadian clock.
The basic helix-loop-helix PER-ARNT-SIM (bHLH-PAS) proteins, CLOCK and BMAL1 (ARNTL), are the primary transcriptional activators within the circadian clock mechanism of mammals. Since the molecular identification of the Clock gene fifteen years ago (1, 2), the transcriptional network that drives circadian oscillations has been systematically identified (3–5). CLOCK and BMAL1 heterodimerize and interact with E-box regulatory elements in the Period (Per1, Per2) and Cryptochrome (Cry1, Cry2) genes to activate their transcription during the daytime (6, 7). Their protein products, PER and CRY, accumulate, dimerize and translocate into the nucleus at night where they interact directly with CLOCK:BMAL1 to repress their own transcription (7–10). As the PER:CRY repressor complex is degraded by specific E3 ubiquitin ligase complexes (11–14), repression is relieved, and CLOCK:BMAL1 then activate a new round of transcription to begin the circadian cycle anew. This cell-autonomous, autoregulatory transcriptional feedback loop takes approximately 24 hours to complete and forms the core mechanism of the circadian clock in mammals (5).
CLOCK and BMAL1 belong to a family of transcriptional regulators that contain bHLH and PAS domains. In mammals, these bHLH-PAS transcription factors participate in a wide array of functions including responses to environmental contaminants (Aryl hydrocarbon receptor, AHR), hypoxia (Hypoxia inducible factor, HIF), neurogenesis (SIM1), synaptic plasticity (NPAS4) and circadian regulation (CLOCK, NPAS2, BMAL1) (15) and most of them remain poorly characterized at the structural level. By contrast, the structures of individual PAS domains and their interactions with small molecule ligands such as heme and flavin cofactors are well understood, especially among microorganisms and plants, where PAS domains serve important roles in two-component signaling and blue-light detection (16, 17). Although the PAS fold is widely conserved, it has intrinsic flexibility and can adapt to different conformations depending on bound ligands or interacting partners (16). Here we present the three-dimensional structure of the bHLH-PAS domains from the mouse CLOCK:BMAL1 heterodimer at 2.3 Å resolution.
Overall structure of CLOCK:BMAL1
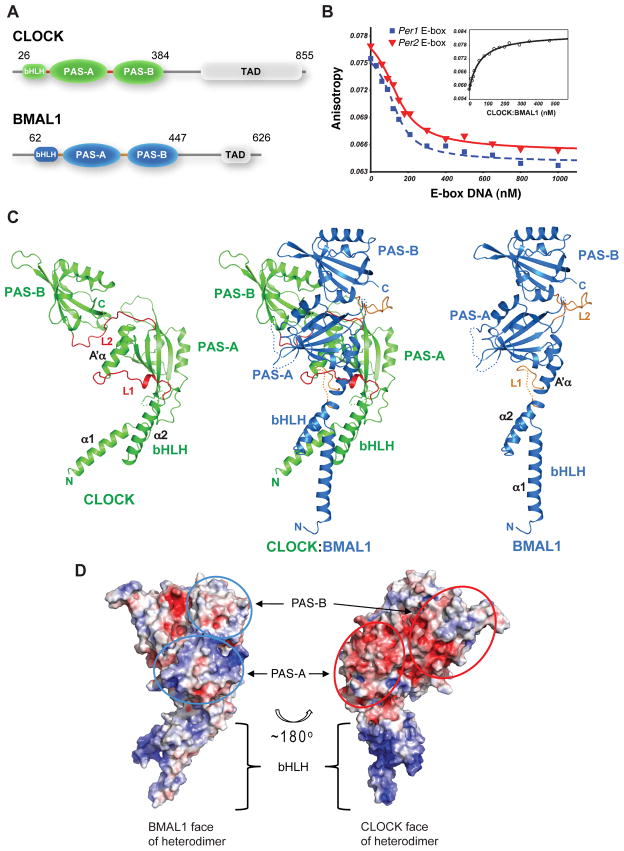
To obtain stable CLOCK:BMAL1 complexes suitable for crystallographic analysis, we used protein constructs containing the bHLH and the two tandem PAS-A and PAS-B domains (Fig. 1A). N-terminal His tagged mouse CLOCK (residues 26–384) and native mouse BMAL1 (residues 62–447) constructs were co-expressed in Sf9 insect cells and co-purified (See Supplementary Materials and Methods). To confirm that that the resulting heterodimeric protein binds DNA, we assayed the affinity of binding to oligonucleotides containing the canonical E-box sequence (CACGTG) from the mPer1 and mPer2 promoters and observed Kd’s of ~10 nM (Fig. 1B, S1). Crystals of CLOCK:BMAL1 were obtained that diffracted to 2.3Å at synchrotron sources. The phases of CLOCK:BMAL1 were determined by the single wavelength anomalous dispersion (SAD) method using selenomethionine-labeled CLOCK:BMAL1 crystals (Fig. S2). Data collection and refinement statistics are shown in Supplementary Table S1.
Fig. 1.
Overall structure of mouse CLOCK:BMAL1. (A) Domain organization of CLOCK and BMAL1. Crystals were obtained from the truncated proteins (indicated by the amino acid residue number) encompassing the bHLH-PAS-AB domains. (B) DNA-binding affinity of the truncated CLOCK:BMAL1 complex measured by fluorescence anisotropy. Dissociation constant (Kd) of the fluorescein labeled mPer2 E2-box DNA was 59 ± 7.3 nM by direct binding to CLOCK:BMAL1 (inset). Using unlabeled DNA probes as competitor, the Kd’s of unlabeled 18-mer mPer1 E1-box DNA (blue) and mPer2 E2-box DNA (red) (40) were 9.0 ± 2.3 nM and 13 ± 2.0 nM, respectively. (See Materials and Method for details). (C) Ribbon diagram of CLOCK:BMAL1 heterodimer (center). The CLOCK subunit is colored green, BMAL1 blue. Each individual domain is labeled. The CLOCK (left) and BMAL1 (right) subunits are also shown separately to illustrate their different spatial domain arrangements. The linker regions between domains in the two subunits (L1 and L2) are highlighted in red or orange color. Flexible loops lacking density are indicated by dotted lines. (D) Electrostatic potentials of CLOCK:BMAL1 heterodimer showing that the surfaces composed of CLOCK PAS domains (red ovals, right) have mostly negative potentials while the surfaces of BMAL1 PAS domains (blue ovals, left) are mostly positive or neutral. The colors are ramped from negative potential −5 kT/q (red) to positive 5 kT/q (blue).
The three-dimensional structure of CLOCK:BMAL1 reveals a tightly intertwined heterodimer (Fig. 1C, center) with all three domains, the N-terminal bHLH domain and two tandem PAS domains (PAS-A and PAS-B), involved in dimerization interactions. Each domain interacts primarily with the corresponding domain of its partner subunit so that CLOCK bHLH interacts with BMAL1 bHLH, and CLOCK PAS-A (or PAS-B) with BMAL1 PAS-A (or PAS-B). Although the primary sequences of these three domains are similar in CLOCK and BMAL1 (Fig. S3, S4), the spatial arrangement of these domains with respect to one another is strikingly different in the two subunits (Fig. 1C). In BMAL1, the second helix of the bHLH domain (α2) is nearly continuous with the N-terminal flanking helix (A′α) of the PAS-A domain despite insertion of a ~15 residue flexible loop (L1) (Fig. 1C, right panel). By contrast, in CLOCK there is a ~23Å displacement between the end of α2 and the beginning of the PAS-A A′α helix (Fig. 1C, left panel). As a consequence, the CLOCK PAS-A domain is in direct contact with the α2 helix of its bHLH domain while there are no direct contacts between the BMAL1 PAS-A and bHLH domains.
The asymmetry of the CLOCK:BMAL1 complex is also reflected in the divergent electrostatic potential distributions on the two subunits. The BMAL1 subunit has an overall positive electrostatic potential with a pI of 9.01 (and 8.55 for the PAS-A/B domains). The CLOCK subunit, on the other hand, has an overall negative electrostatic potential, with a pI of 5.86 for the bHLH-PAS domains (and 5.28 for the PAS-A/B domain only). In the three-dimensional CLOCK:BMAL1 complex structure, the exposed CLOCK PAS domain surfaces have a largely negative electrostatic potential, whereas the exposed BMAL1 PAS domains are mostly positively charged or neutral (Fig. 1D). These electrostatic features of the CLOCK:BMAL1 heterodimer produce an interesting dichotomy in the potential interaction interfaces of the complex and are consistent with prior work suggesting that the PER1, PER2, CRY1 and CRY2 proteins differentially interact with CLOCK and BMAL1 (8, 18–20).
The PAS-A domains
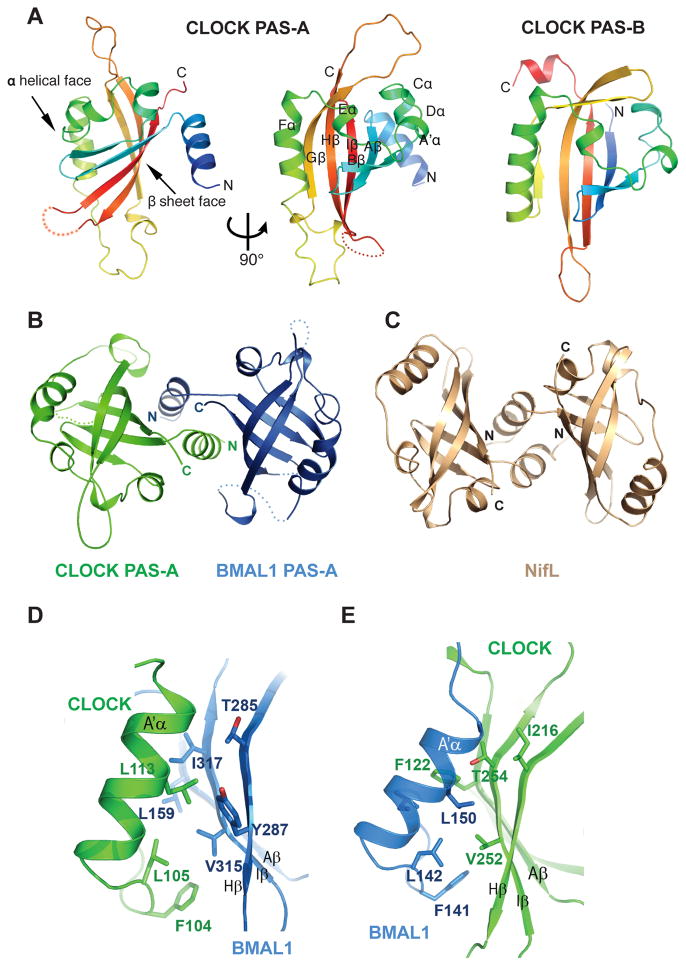
While all three domains of the CLOCK and BMAL1 subunits are involved in intermolecular interactions, the heterodimeric interfaces between the individual PAS domains are particularly interesting. The CLOCK:BMAL1 heterodimer has long flexible loops interspersed with canonical PAS secondary structure elements in both PAS-A domains. In the BMAL1 PAS-A domain, a total of ~60 residues in three loop regions are disordered in the crystal structure, whereas in the CLOCK PAS-A domain, a single loop of ~25 residues is disordered. In spite of this high degree of flexibility, the two PAS-A domains adopt a typical PAS fold (Fig. 2A) and are structurally similar to each other with a root mean square deviation (rmsd) of only 0.62Å over 79 Cα atoms. As seen in typical PAS domains, the core of CLOCK and BMAL1 PAS-A domains contains a five-stranded antiparallel β sheet (AβBβGβHβIβ) and several α helices (Cα, DαEαFα) flanking the concave surface of the sheet. In contrast to the PAS-B domain (Fig. 2A), both PAS-A domains in the two subunits contain an N-terminal flanking helix (A′α) external to the canonical PAS-domain fold. The A′α helices of the CLOCK and BMAL1 PAS-A domains pack in between the β-sheet faces of the two domains to mediate the heterodimeric PAS-A interactions (Fig. 2B). The A′αhelix of CLOCK PAS-A makes extensive contacts with the β-sheet face of BMAL1 PAS-A, while the A′αhelix of BMAL1 interacts with the β-sheet face of CLOCK PAS-A (Fig. 2B). This domain-swapped helical interface is reminiscent of a number of other PAS domain proteins, such as the N-terminal PAS domain of the nitrogen fixation regulatory protein NifL from Azotobacter vinelandii (21) (Fig. 2C) and the N-terminal domain of the transcriptional factor TyrR from E. coli (22).
Fig. 2.
Structure and interaction of the PAS-A domains of CLOCK:BMAL1. (A) Ribbon representations of CLOCK PAS-A domain. Secondary structures are color ramped from blue to red and labeled from the A′α helix located N-terminal to the canonical PAS domain fold, in an alphabetical progression through the whole domain.. The CLOCK PAS-B domain is also shown for comparison. (B) Dimerization of the two PAS-A domains in CLOCK:BMAL1, looking down the approximate two-fold symmetry axis. (C) Similar domain-swapped structure of the redox sensing PAS domain of NifL from A. vinelandii (pdb: 2GJ3). (D) Left panel, detailed interface between A′α helix of CLOCK PAS-A (green) and the β sheet face of BMAL1 PAS-A (blue). Right panel, the corresponding interface between A′α helix of BMAL1 PAS-A and the β sheet face of CLOCK PAS-A.
The CLOCK:BMAL1 PAS-A dimer interface is largely mediated by conserved hydrophobic interactions. Specifically, Phe104, Leu105, and Leu113 on the A′α helix of CLOCK (corresponding residues on BMAL1 PAS-A are Phe141, Leu142 and Leu150) contact a hydrophobic region on the β-sheet face of BMAL1 PAS-A comprised of residues Leu159 on strand Aβ, Thr285 and Tyr287 on Hβ, Val315 and Ile317 on strand Iβ (Fig. 2B, left). A similar interface can be found between the A′α helix of BMAL1 and CLOCK PAS-A domain (Fig. 2B, right). As a result, the two PAS-A domains in CLOCK:BMAL1 form a parallel dimer related by an approximate two-fold symmetry, with an extensive buried surface area (~1950 Å2) and topologically complex interface between the two subunits. Many of the residues observed in the CLOCK:BMAL1 PAS-A dimer interface are conserved among bHLH-PAS transcription factors (Fig. S4), suggesting that these proteins may share a common PAS-A domain dimerization mode.
The PAS-B domains
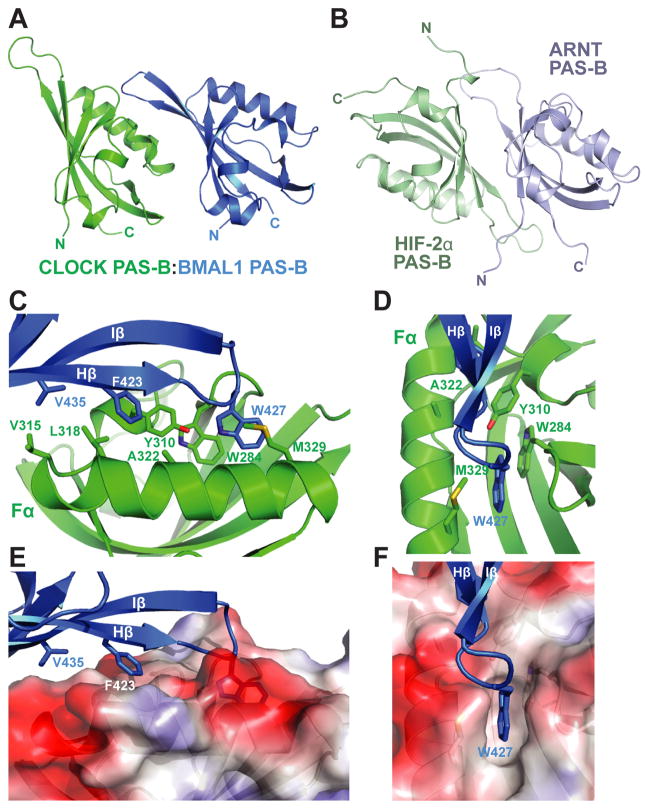
A ~15 residue linker (L2) connects the PAS-A and PAS-B domains in each of the CLOCK and BMAL1 subunits, though the linker conformation and the relative spatial arrangement of the two PAS domains in the two subunits are different (Fig. 1C). In CLOCK, a large portion of L2 is buried between the interface of CLOCK and BMAL1 and is well ordered (Fig. 1C). By contrast, in BMAL1, L2 between PAS-A and PAS-B is solvent exposed and very flexible as indicated by high atomic displacement parameters (B-factors). The PAS-B domains of CLOCK and BMAL1 are related predominantly by a ~26Å translation and are stacked in a roughly parallel fashion (Fig. 3A), different from the antiparallel β-sheet interface seen for the isolated PAS-B domain complex of HIF-2α:ARNT (Fig. 3B) (23). The β sheet of BMAL1 PAS-B makes contacts with the helical face of CLOCK PAS-B, burying a patch of hydrophobic residues on both subunits. Several surface-exposed hydrophobic residues on both CLOCK and BMALl PAS-B become mostly or partially buried upon dimerization, including Tyr310, Val315, Leu318 of CLOCK and Phe423, Trp427 and Val435 of BMAL1, resulting in ~700 Å2 of buried surface area (Fig. 3C, 3E). Most prominently within these hydrophobic interactions, BMAL1 Trp427, located on the short HI loop (connecting the Hβ and Iβ strands), intrudes into a hydrophobic cleft created between the Fα helix and the AB loop of CLOCK PAS-B (Fig. 3D, 3F), and partially stacks against the indole ring of CLOCK Trp284.
Fig. 3.
Interface between CLOCK:BMAL1 PAS-B domains. (A) The spatial arrangement of the two PAS-B domains in CLOCK:BMAL1. (B) Antiparallel orientation of β sheet-mediated interaction between isolated HIF-2α:ARNT PAS-B domains (pdb: 3F1P). (C) Detailed interface between CLOCK:BMAL1 PAS-B domains. (D) Front facing view of CLOCK:BMAL1 PAS-B interface highlighting role of BMAL1 Trp427and CLOCK Trp284 interaction. (E) Side view of PAS-B interface displaying surface electrostatic potential of CLOCK PAS-B. (F) Front facing view of CLOCK surface electrostatic potential displaying the binding pocket for BMAL1 Trp427. The color scheme used is the same as in Fig. 1D.
CLOCK:BMAL1 heterodimer conformation and transactivation function
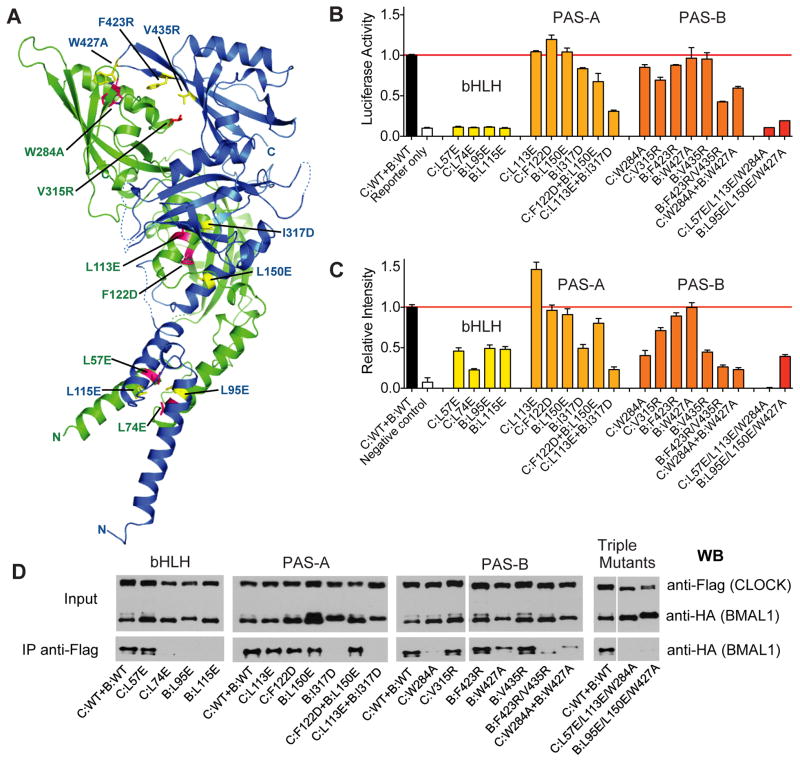
To probe the relationship between the observed conformation of the CLOCK:BMAL1 heterodimer and its function, we generated a series of mutations predicted to perturb the interfaces between each of the three domains (Fig. 4A). For the bHLH domains, the C-terminal halves of the α1 helices, together with the α2 helices of both CLOCK and BMAL1 participate in the formation of a canonical four-helical bHLH bundle in the heterodimer similar to that observed in USF1 and MYC:MAX (24–26) (Fig. S3A). As seen in other bHLH proteins, the core of this four-helical bundle is highly hydrophobic (26, 27), indicating that dimerization of the bHLH domains should help stabilize the CLOCK:BMAL1 complex. The proper conformation of the bHLH domain is also critical for E-box DNA recognition as the DNA binding α1 helices need to be positioned precisely to interact with the major groove sites of the E-box DNA duplex (24, 26, 27). Indeed, when the bHLH hydrophobic core residues Leu57 and Leu74 of CLOCK, and Leu95 and Leu115 of BMAL1 are mutated to glutamate, the transactivation activity of these full-length CLOCK:BMAL1 mutants are eliminated, as demonstrated by measuring E-box-driven luciferase reporter gene activity in human kidney cells (HEK293T) with transiently transfected CLOCK and BMAL1 mutant proteins (Fig. 4B).
Fig. 4.
Functional analysis of CLOCK:BMAL1 mutants. (A) Locations of domain interface mutants in CLOCK (green) and BMAL1 (blue). (B) Per2 promoter:Luciferase reporter assays to evaluate the effects of structure-based mutations on transactivation by full-length CLOCK:BMAL1. Data are average of 2 independent experiments performed in duplicate. (C) Bimolecular fluorescence complementation (BiFC) experiments on the same set of mutants in CLOCK:BMAL1 truncated constructs. The fluorescent intensities of WT and mutant CLOCK:BMAL1 bHLH-PAS-AB constructs (for details, see Supplementary Materials and Methods) were quantified using data from 3 independent experiments. (E) Coimmunoprecipitation experiments assessing the association of CLOCK and BMAL1 in full-length WT and mutant proteins. Anti-FLAG affinity gel was used to precipitate FLAG tagged CLOCK along with the tightly associated BMAL1, which is HA tagged. The Western blots using an anti-HA antibody were then performed to detect the association of WT and mutant BMAL1 with CLOCK constructs. The Co-IP data are representative of at least 3 independent replicates, with the exception of C:W284A which had stronger co-IP interaction in other experiments, but on average was weaker than WT CLOCK.
We examined the dimerization of these mutants in living cells through a bimolecular fluorescence complementation (BiFC) assay, in which the N- and C-terminal fragments of the fluorescent protein Venus (Ven-N and Ven-C) were fused to the C-termini of truncated bHLH-PAS domain constructs of CLOCK and BMAL1, respectively (See Supplementary Online Material). Dimerization of CLOCK and BMAL1 brings the two Venus fragments into close proximity to facilitate the formation of an intact fluorescent protein, thus providing a fluorescent readout for protein-protein interactions in living cells (28). The BiFC data showed that mutation at the bHLH hydrophobic core reduced formation of a stable heterodimeric complex (Fig. 4C, Fig. S5). Furthermore, three of these bHLH domain mutations, CLOCK L74E (C:L74E), BMAL1 L95E and L115E (B:L95E and B:L115E) also destabilized the full-length CLOCK:BMAL1 heterodimer as shown by co-immunoprecipitation (co-IP) assays (Fig 4D).
Next, we mutated residues involved in PAS-A and PAS-B domain interfaces to test their effects on transactivation activity and CLOCK:BMAL1 heterodimer formation. To examine the PAS-A domain interface, we made following mutations: L113E (on A′α) and F122D (on Aβ of CLOCK, and L150E (on A′α) and I317D (on Iβ of BMAL1, and performed transactivation, BiFC, and co-IP assays (Fig. 4). Single mutations of C:L113E, C:F122D or B:L150E were not sufficient to reduce dimerization or transactivation activity (Fig. 4B–D). However, BMAL1 mutant I317D had decreased transcriptional activity (~80% of wild type control) (Fig. 4B) and decreased affinity for CLOCK as demonstrated by the BiFC and co-IP experiments (Fig. 4C, 4D, SOM Fig. S5). Furthermore, when opposing CLOCK and BMAL1 PAS-A domain interface residues were doubly mutated as in C:L113E+B:I317D, the association between full-length CLOCK and BMAL1 subunits was not detectable under the assay conditions and transactivation activity was reduced to ~25% of the control (Fig. 4B–D).
To examine the unusual interface between the PAS-B domains of the CLOCK:BMAL1 heterodimer, we made the following PAS-B domain mutations: W284A and V315R on the CLOCK helical face, and W427A, F423R, and V435R on the β-sheet face of BMAL1. Single mutations in either CLOCK or BMAL1 PAS-B domains had a limited effect on the transactivation activity by the full-length mutant protein (Fig. 4B), although the activities of C:W284A, C:V315R and B:F423R were reduced by ~20–30% compared to the wild-type (WT) protein (Fig. 4B). Additionally, the BiFC signal of mutants C:W284A, C:V315R and B:V435R decreased dramatically compared to the WT protein (Fig. 4C, SOM Fig. S5), indicating that the PAS-B domain interactions of these mutants may be altered. The effect of these single mutations on the interactions of the full-length proteins, as measured by co-IP, was more subtle, with partially weakened interactions for mutants C:W284A and B:W427A (Fig 4D). Notably, the double BMAL1 PAS-B domain mutant, B:F423R/V435R and the combined CLOCK:BMAL1 mutant C:W284A+B:W427A showed a decreased interaction in both co-IP and BiFC assays, as well as a reduction in transactivation activity (Fig. 4B–D). These data support the unusual PAS-B domain interface observed in the crystal structure involving the helical face of CLOCK and the β-sheet face of BMAL1, and specifically indicate that contact between CLOCK Trp284 and BMAL1 Trp427 is important for PAS-B interaction.
CLOCK:BMAL1 mutants alter circadian cycling in cells
To examine the functional consequences of mutations that compromise CLOCK:BMAL1 heterodimer formation and transactivation potential, we assessed circadian rhythmns in mouse Per2Luc fibroblasts overexpressing mutant CLOCK or BMAL1 constructs introduced by lentiviral vectors (see Supplemental Materials and Methods) (SOM Fig. S6). On the basis of in vivo transgenic experiments, we can infer that CLOCK levels are rate limiting and that overexpression of CLOCK leads to a shortening of circadian period in both constitutively expressed or conditionally expressed transgenic mice (2, 29). In contrast, overexpression of BMAL1 can have no effect or can lengthen circadian period (30), and these effects of BMAL1 overexpression are consistent with the hypothesis that BMAL1 is normally in excess of CLOCK. Higher overexpression of BMAL1 can lead to period lengthening, possibly by sequestering of CLOCK via a squelching mechanism (31). Thus, we can assay the WT function of CLOCK and BMAL1 by overexpression in PER2::luciferase cycling cell assays (32), and by extension, infer loss-of-function mutations by their inability to mimic WT function, or in contrast, dominant-negative mutations by their disruption of normal rhythms.
Control Per2Luc fibroblasts overexpressing GFP had robust luciferase rhythms with a period of 23.1 hours (SOM Fig. S6). Cells overexpressing WT CLOCK or BMAL1 exhibited rhythms with either shorter (~22.0 hr) or longer (24.6 hr) periods, respectively (SOM Fig. S6A). Both the CLOCK mutants tested (C:L57E and C:W284A) failed to mimic WT CLOCK and had period values similar to the GFP control cells (~23 hr), and therefore behaved as loss-of-function mutations. Notably, the C:L57E mutant abolished transactivation by full-length CLOCK:BMAL1 and reduced dimerization of the truncated heterodimer (Fig. 4B, 4C). Although the C:W284A PAS-B mutant had only a 20% reduction in activty in transactivation assays (Fig. 4B), it weakened CLOCK:BMAL1 dimerization significantly (Fig. 4C–D) and failed to mimic the WT function of CLOCK on circadian periodicity. Interestingly, overexpression of BMAL1 mutants within the bHLH and PAS-A domains had different effects on the period of circadian rhythms (SOM Fig. S6). Overexpression of the bHLH mutant B:L95E led initially to a longer period (25.1 hrs) for the first 3 days followed by disruption of circadian rhythmicity, whereas overexpression of the PAS-A mutant B:I317D led to a shorter period (~23.8 hr) compared to the cells overexpressing WT BMAL1 (24.6 hrs) and thus behaved as a partial loss-of-function mutation.
Discussion
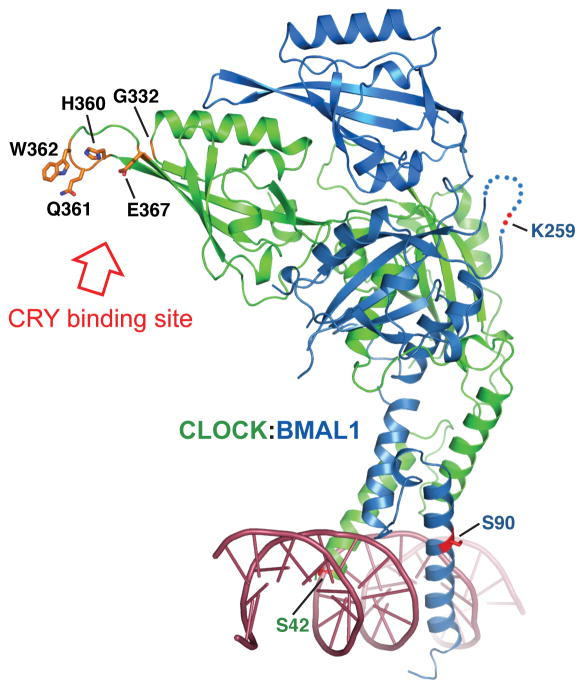
Here we present the X-ray structure of the mouse CLOCK:BMAL1 transcriptional activator complex, a central regulator in the circadian clock. With the CLOCK:BMAL1 complex structure in hand, it will now be possible to analyze the multiprotein complexes involved in mammalian circadian clock mechanisms at an atomic level. Existing genetic and biochemical data indicate that the negative regulators CRY and PER physically interact with CLOCK:BMAL1 to form the major repressive clock complex containing CLOCK:BMAL1 and PER:CRY (9, 10, 18, 33). Although the structural details of these interactions have not been elucidated, the binding of CRY and/or PER to CLOCK:BMAL1 could affect DNA-binding, modulate transactivation potential, or modify interactions with co-activators and co-repressors. Previous work suggests that CRY interacts with the PAS-B domain of CLOCK near its β-sheet face and also with a C-terminal region of BMAL1 (18, 19, 33). Specifically, mutations of residues Gln332, His360, Gln361, Trp362 and Glu367 of the CLOCK PAS-B domain interfere with repression by CRY. In the crystal structure, these residues are located on the HI loop of the solvent exposed β-sheet face of the CLOCK PAS-B domain fully accessible for interaction with CRY (Fig. 5). The electrostatic distribution of CLOCK PAS domains is also consistent with the idea that CLOCK is the site for CRY binding because CRY is a highly positively charge protein (pI = 8.24 for CRY1) and would complement the negative surface charge on CLOCK (Fig 1D). Thus, the unusual spatial arrangement of the PAS-B domains of CLOCK:BMAL1 observed in the crystal structure is consistent with the earlier biochemical data on the PAS-B domain function. Interestingly the tandem PAS domains in BMAL1 have a spatial arrangement similar to that observed in crystal structures of the mouse and Drosophila PER tandem PAS domains (Fig. S7) (34–37). This suggests that the tandem PAS domains in BMAL1 and PER may have a deeper degree of structural and/or functional conservation than was previously appreciated, which may have implications for how the PAS-A and PAS-B domains of PER2 interact with either CLOCK or BMAL1 (10).
Fig. 5.
Mutations that reduce repression of CLOCK:BMAL1 transactivation by CRY localize to CLOCK PAS-B HI loop. CRY-derepressing mutations arising from a random mutagenesis screen: G332E, H360Y, E367K (18) or directed mutagenesis study, Q361P/W362R (19) are predominantly found on the β-sheet face of CLOCK PAS-B domain and are fully solvent accessible. Residues mutated in these studies are in orange. The locations of the SUMOylation site on BMAL1 PAS-A (K259) (41), the Casein kinase 2 phosphorylation site on BMAL1 (S90) (42), and the phosphorylation site on CLOCK (S42) (43) are also indicated. A double strand DNA is modeled based on the superposition with USF-DNA complex structure (25).
Interestingly, Trp362 of CLOCK, implicated in an interaction with CRY, corresponds to Trp427 of BMAL1, which was shown here to interact with CLOCK (Fig. 3D, 3F). Moreover, a tryptophan residue at the same position is also conserved in the Drosophila and mouse PER proteins (i.e., Trp482 of dPER and Trp419 of mPER2), and they are shown in the crystal structures to be involved in the interaction with a second PER protein to form homodimers (35). These observations highlight a potentially conserved functional role for the tryptophan residue located at the HI loop of the PAS-B domains of these clock proteins.
The CLOCK:BMAL1 PAS-B domain interface reveals details of a mode of PAS protein-protein interaction involving the α-helical face of CLOCK PAS-B and the β-sheet face of BMAL1 PAS-B (Fig. 3C–3F and Fig S8A). Notably, the same region (between Fα and the AB loop) on the helical face of PER PAS-B is utilized for intramolecular interactions with a C-terminal α helix (αE) containing nuclear exporting signal (NES) residues (37) (Note: helix αE is equivalent to Jα in canonical PAS nomenclature.) (Fig. S8B). Similarly, ARNT has been shown by NMR studies to use the same helical region for interacting with a family of helical coactivator proteins that are required for transactivation by the heterodimeric HIF complex (38, 39). Moreover, the same region of many bacterial and plant PAS proteins binds to small molecule ligands such as flavin cofactors, flavin adenine dinucleotide (FAD) and flavin mononucleotide (FMN) (16) (Fig. S8C). Overall, these data highlight the remarkable structural plasticity and adaptability of PAS domains. Because CLOCK:BMAL1 is a prototypical bHLH-PAS protein family member, the structural features of the CLOCK:BMAL1 complex may be shared by other bHLH-PAS proteins. It will be important in future work to determine the structures of additional heterodimeric bHLH-PAS proteins such as HIF:ARNT and AHR:ARNT and observe the structural basis by which these homologous proteins confer their distinct and pathway specific functions.
In summary, the structure of CLOCK:BMAL1 has revealed the locations of previously identified sites on these proteins that affect their inhibition by CRY. It has also revealed an unexpected similarity in the orientation of the tandem PAS-A and PAS-B domains of BMAL1 to that found in the PERIOD proteins. These observations provide a starting point for the determination of how the CRY and PER proteins interact with and repress CLOCK:BMAL1, which in turn should yield insight into the detailed biochemical mechanism by which this transcriptional feedback loop drives the circadian clock.
Supplementary Material
Acknowledgments
This work was supported by the Howard Hughes Medical Institute (J.S.T.), an American Heart Association grant 10GRNT4310090 (H.Z.), and NIH R01 GM090247 (C.B.G.). We thank N. Grishin and K. Gardner for helpful discussions; S. Padrick for help with the fluorescence polarization assay; M. Rosen for use of his fluorometer; D. Tomchick and H. Aronovich for technical assistance; and C. Ralston, L. Steinhour and C. Brautigan for help with data collection. The Berkeley Center for Structural Biology is supported in part by the National Institutes of Health, National Institute of General Medical Sciences, and the Howard Hughes Medical Institute. The Advanced Light Source is supported by the Director, Office of Science, Office of Basic Energy Sciences, of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231. J.S.T. is an Investigator, Y.C. is a Research Specialist 3 and C.P. was an Associate in the Howard Hughes Medical Institute.
Footnotes
Atomic coordinates for the reported crystal structures have been deposited with the Protein Data Bank under accession code 4F3L.
References and Notes
- 1.King DP, et al. Cell. 1997;89:641. doi: 10.1016/s0092-8674(00)80245-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antoch MP, et al. Cell. 1997;89:655. doi: 10.1016/s0092-8674(00)80246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reppert SM, Weaver DR. Nature. 2002;418:935. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- 4.Lowrey PL, Takahashi JS. Annu Rev Genom Hum Genet. 2004;5:407. doi: 10.1146/annurev.genom.5.061903.175925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lowrey PL, Takahashi JS. Adv Genet. 2011;74:175. doi: 10.1016/B978-0-12-387690-4.00006-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gekakis N, et al. Science. 1998;280:1564. doi: 10.1126/science.280.5369.1564. [DOI] [PubMed] [Google Scholar]
- 7.Kume K, et al. Cell. 1999;98:193. doi: 10.1016/s0092-8674(00)81014-4. [DOI] [PubMed] [Google Scholar]
- 8.Griffin EA, Jr, Staknis D, Weitz CJ. Science. 1999;286:768. doi: 10.1126/science.286.5440.768. [DOI] [PubMed] [Google Scholar]
- 9.Lee C, Etchegaray JP, Cagampang FR, Loudon AS, Reppert SM. Cell. 2001;107:855. doi: 10.1016/s0092-8674(01)00610-9. [DOI] [PubMed] [Google Scholar]
- 10.Chen R, et al. Mol Cell. 2009;36:417. doi: 10.1016/j.molcel.2009.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shirogane T, Jin J, Ang XL, Harper JW. J Biol Chem. 2005;280:26863. doi: 10.1074/jbc.M502862200. [DOI] [PubMed] [Google Scholar]
- 12.Reischl S, et al. J Biol Rhythms. 2007;22:375. doi: 10.1177/0748730407303926. [DOI] [PubMed] [Google Scholar]
- 13.Busino L, et al. Science. 2007;316:900. doi: 10.1126/science.1141194. [DOI] [PubMed] [Google Scholar]
- 14.Siepka SM, et al. Cell. 2007;129:1011. doi: 10.1016/j.cell.2007.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McIntosh BE, Hogenesch JB, Bradfield CA. Annu Rev Physiol. 2010;72:625. doi: 10.1146/annurev-physiol-021909-135922. [DOI] [PubMed] [Google Scholar]
- 16.Möglich A, Ayers RA, Moffat K. Structure. 2009;17:1282. doi: 10.1016/j.str.2009.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henry JT, Crosson S. Annu Rev Microbiol. 2011;65:261. doi: 10.1146/annurev-micro-121809-151631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sato TK, et al. Nat Genet. 2006;38:312. doi: 10.1038/ng1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao WN, et al. Nature cell biology. 2007;9:268. doi: 10.1038/ncb1539. [DOI] [PubMed] [Google Scholar]
- 20.Ye R, Selby CP, Ozturk N, Annayev Y, Sancar A. J Biol Chem. 2011;286:25891. doi: 10.1074/jbc.M111.254680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Key J, Hefti M, Purcell EB, Moffat K. Biochemistry. 2007;46:3614. doi: 10.1021/bi0620407. [DOI] [PubMed] [Google Scholar]
- 22.Verger D, Carr PD, Kwok T, Ollis DL. J Mol Biol. 2007;367:102. doi: 10.1016/j.jmb.2006.12.018. [DOI] [PubMed] [Google Scholar]
- 23.Scheuermann TH, et al. Proc Natl Acad Sci USA. 2009;106:450. doi: 10.1073/pnas.0808092106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ferré-D’ Amaré AR, Prendergast GC, Ziff EB, Burley SK. Nature. 1993;363:38. doi: 10.1038/363038a0. [DOI] [PubMed] [Google Scholar]
- 25.Ferré-D’ Amaré AR, Pognonec P, Roeder RG, Burley SK. EMBO J. 1994;13:180. doi: 10.1002/j.1460-2075.1994.tb06247.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nair SK, Burley SK. Cell. 2003;112:193. doi: 10.1016/s0092-8674(02)01284-9. [DOI] [PubMed] [Google Scholar]
- 27.Ma PC, Rould MA, Weintraub H, Pabo CO. Cell. 1994;77:451. doi: 10.1016/0092-8674(94)90159-7. [DOI] [PubMed] [Google Scholar]
- 28.Kerppola TK. Nat Rev Mol Cell Biol. 2006;7:449. doi: 10.1038/nrm1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hong HK, et al. PLoS Genet. 2007;3:e33. doi: 10.1371/journal.pgen.0030033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McDearmon EL, et al. Science. 2006;314:1304. doi: 10.1126/science.1132430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ptashne M. Nature. 1988;335:683. doi: 10.1038/335683a0. [DOI] [PubMed] [Google Scholar]
- 32.Yoo SH, et al. Proc Natl Acad Sci USA. 2004;101:5339. [Google Scholar]
- 33.Kiyohara YB, et al. Proc Natl Acad Sci USA. 2006;103:10074. doi: 10.1073/pnas.0601416103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yildiz O, et al. Mol Cell. 2005;17:69. doi: 10.1016/j.molcel.2004.11.022. [DOI] [PubMed] [Google Scholar]
- 35.Hennig S, et al. PLoS Biol. 2009;7:e94. doi: 10.1371/journal.pbio.1000094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.King HA, Hoelz A, Crane BR, Young MW. J Mol Biol. 2011;413:561. doi: 10.1016/j.jmb.2011.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kucera N, et al. Proc Natl Acad Sci USA. 2012;109:3311. doi: 10.1073/pnas.1113280109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Partch CL, Card PB, Amezcua CA, Gardner KH. J Biol Chem. 2009;284:15184. doi: 10.1074/jbc.M808479200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Partch CL, Gardner KH. Proc Natl Acad Sci USA. 2011;108:7739. doi: 10.1073/pnas.1101357108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoo SH, et al. Proc Natl Acad Sci USA. 2005;102:2608. [Google Scholar]
- 41.Cardone L, et al. Science (New York, NY) 2005;309:1390. doi: 10.1126/science.1110689. [DOI] [PubMed] [Google Scholar]
- 42.Tamaru T, et al. Nature Structural & Molecular Biology. 2009;16:446. doi: 10.1038/nsmb.1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yoshitane H, et al. Mol Cell Biol. 2009;29:3675. doi: 10.1128/MCB.01864-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou T, et al. Structure. 2004;12:1891. doi: 10.1016/j.str.2004.07.021. [DOI] [PubMed] [Google Scholar]
- 45.Otwinowski Z, Minor W. Method Enzymol. 1997;276:307. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 46.Abrahams JP, Leslie AG. Acta Crystallogr D Biol Crystallogr. 1996;52:30. doi: 10.1107/S0907444995008754. [DOI] [PubMed] [Google Scholar]
- 47.Sheldrick GM. Acta Crystallogr A. 2008;64:112. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 48.Vonrhein C, Blanc E, Roversi P, Bricogne G. Methods Mol Biol. 2007;364:215. doi: 10.1385/1-59745-266-1:215. [DOI] [PubMed] [Google Scholar]
- 49.Perrakis A, Morris R, Lamzin VS. Nat Struct Biol. 1999;6:458. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 50.Cowtan K. Acta Crystallogr D Biol Crystallogr. 2006;62:1002. doi: 10.1107/S0907444906022116. [DOI] [PubMed] [Google Scholar]
- 51.Emsley P, Lohkamp B, Scott WG, Cowtan K. Acta Cryst. 2010;66:486. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.N. Collaborative Computational Project. Acta Crystallogr D Biol Crystallogr. 1994;50:760. [Google Scholar]
- 53.Murshudov GN, Vagin AA, Dodson EJ. Acta Crystallogr D Biol Crystallogr. 1997;53:240. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 54.Adams PD, et al. Acta Crystallogr D Biol Crystallogr. 2010;66:213. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Baker NA. Proceedings of the National Academy of Sciences. 2001;98:10037. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Anderson BJ, Larkin C, Guja K, Schildbach JF. Methods Enzymol. 2008;450:253. doi: 10.1016/S0076-6879(08)03412-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hida A, et al. Genomics. 2000;65:224. doi: 10.1006/geno.2000.6166. [DOI] [PubMed] [Google Scholar]
- 58.Wang ZX. FEBS Lett. 1995;360:111. doi: 10.1016/0014-5793(95)00062-e. [DOI] [PubMed] [Google Scholar]
- 59.Liu H, Naismith JH. BMC Biotechnol. 2008;8:91. doi: 10.1186/1472-6750-8-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hu CD, Chinenov Y, Kerppola TK. Mol Cell. 2002;9:789. doi: 10.1016/s1097-2765(02)00496-3. [DOI] [PubMed] [Google Scholar]
- 61.Hu CD, Kerppola TK. Nat Biotechnol. 2003;21:539. doi: 10.1038/nbt816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Meyer P, Saez L, Young MW. Science. 2006;311:226. doi: 10.1126/science.1118126. [DOI] [PubMed] [Google Scholar]
- 63.Li Y, Yu W, Liang Y, Zhu X. Cell Res. 2007;17:701. doi: 10.1038/cr.2007.65. [DOI] [PubMed] [Google Scholar]
- 64.Wang F, Zhang Q, Cao J, Huang Q, Zhu X. Exp Cell Res. 2008;314:213. doi: 10.1016/j.yexcr.2007.09.022. [DOI] [PubMed] [Google Scholar]
- 65.Welsh DK, Yoo SH, Liu AC, Takahashi JS, Kay SA. Curr Biol. 2004;14:2289. doi: 10.1016/j.cub.2004.11.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Buhr ED, Yoo SH, Takahashi JS. Science. 2010;330:379. doi: 10.1126/science.1195262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen VB, et al. Acta Crystallogr D Biol Crystallogr. 2009;66:12. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.