SUMMARY
Period determination in the mammalian circadian clock involves the turnover rate of the repressors, CRY and PER. Here we show that CRY ubiquitination engages two competing E3 ligase complexes that either lengthen or shorten circadian period in mice. Cloning of a short-period circadian mutant, Past-time, revealed a glycine to glutamate (G149E) missense mutation in Fbxl21, an F-box protein gene that is a paralog of Fbxl3 that targets the CRY proteins for degradation. While loss-of-function of FBXL3 leads to period lengthening, mutation of Fbxl21 causes period shortening. FBXL21 forms an SCF E3 ligase complex that slowly degrades CRY in the cytoplasm, but antagonizes the stronger E3 ligase activity of FBXL3 in the nucleus. FBXL21 plays a dual role: protecting CRY from FBXL3 degradation in the nucleus and promoting CRY degradation within the cytoplasm. Thus, the balance and cellular compartmentalization of competing E3 ligases for CRY determine circadian period of the clock in mammals.
INTRODUCTION
In mammals, the circadian clock regulates daily oscillations in behavior, physiology and metabolism (Bass and Takahashi, 2010; Lowrey and Takahashi, 2011; Mohawk et al., 2012). The mechanism of the circadian clock is composed of an autoregulatory transcriptional network (Lowrey and Takahashi, 2011). At the core, the heterodimeric transcription factors, CLOCK:BMAL1 (and NPAS2:BMAL1), drive expression of the Period (Per) 1/2 and Cryptochrome (Cry) 1/2 genes. PER1/2 and CRY1/2 proteins in turn interact, translocate into the nucleus and repress the activity of CLOCK:BMAL1 to inhibit their own transcription (King et al., 1997; Gekakis et al., 1998; Lee et al., 2001). Bmal1 expression is also regulated by a secondary feedback loop comprised of the nuclear hormone receptors, REV-ERBα/β, and the retinoid-related orphan receptors (RORs) (Preitner et al., 2002; Cho et al., 2012).
Although the majority of the core clock components have been identified, the molecular mechanisms that define the periodicity or rate of the circadian clock are still not well understood (Zheng and Sehgal, 2012). PER and CRY are essential negative feedback elements of the clock, and the turnover rates of PER and CRY are correlated with the period length of circadian rhythms in mutant strains. For example, the Casein kinase 1ε (Csnk1e) tau mutation shortens period and destabilizes the PER proteins by hyperphosphorylation of a phosphodegron site that targets the PER proteins for ubiquitination by β-TrCP and proteasomal degradation (Meng et al., 2008). Conversely, mutations in the F-box protein, FBXL3, lengthen period and stabilize the CRY proteins by a reduction in ubiquitination-mediated proteosomal degradation involving the SCFFbxl3 ubiquitin ligase complex (Busino et al., 2007; Godinho et al., 2007; Siepka et al., 2007). Recent work shows that CRY ubiquitination by SCFFbxl3 is triggered by AMPK-mediated phosphorylation (Lamia et al., 2009) and reversed by the deubiquitinating enzyme USP2a (Tong et al., 2012).
Genetic analysis of PER and CRY in period determination suggests that these two pathways act independently and additively to regulate circadian period in vivo (Maywood et al., 2011). Here, we report the isolation and positional cloning of a novel circadian mutant, Past-time (Psttm), that shortens circadian period and destabilizes CRY proteins during the circadian cycle. We describe a newly discovered SCF ubiquitin ligase complex (SCFFbxl21) for the CRY proteins that plays a dual role to attenuate CRY degradation by SCFFbxl3 in the nucleus and to promote CRY degradation in the cytoplasm.
RESULTS
Identification of the Past-time mutation
In an ENU mutagenesis recessive screen using C57BL/6J mice (Figure S1), we identified a short-period mutant with a period length of 22.91 hr (Figure 1, A and B). Crosses with C3H/HeJ mice produced animals of 3 phenotypic classes in the F2 generation, indicating that the mutant, named Past-time (Psttm), was semi-dominant (Figure 1C). Mice heterozygous or homozygous for the Psttm mutation showed a mean free-running period of 23.22 hr or 22.92 hr, respectively. We initially mapped the Psttm mutation to a 40-Mb region on chromosome 13 (Figure 1D). Production of additional recombinants further reduced the interval to 11.4 Mb. This smaller interval contains 167 open reading frames and no known circadian clock genes. However, one candidate, Fbxl21, was of particular interest. Its predicted protein product shares 85% amino acid sequence similarity with the F-box protein FBXL3, indicating that FBXL21 is a paralog of FBXL3 (Jin et al., 2004). FBXL3 has been shown to form an SCF (Skp1/Cullin/F-box protein) E3 ligase complex that polyubiquitinates the CRY proteins for degradation by the proteasome (Busino et al., 2007; Godinho et al., 2007; Siepka et al., 2007), and there is evidence suggesting FBXL21 is a clock-controlled E3 ligase involved in ovine CRY1 degradation (Dardente et al., 2008). We sequenced all of the annotated exons of Fbxl21 from Psttm mice and found a single G to A nucleotide change at nucleotide 787 in exon 5. This mutation co-segregated perfectly with the short period phenotype of Psttm/Psttm mice (0/156 recombinants; Fig. 1D). The point mutation converts amino acid 149 from a highly conserved glycine to a glutamic acid residue within the third leucine-rich repeat (LRR) of the protein. This G149E mutation is expected to create a charged protrusion, likely destabilizing the LRR structure (Figure 1E).
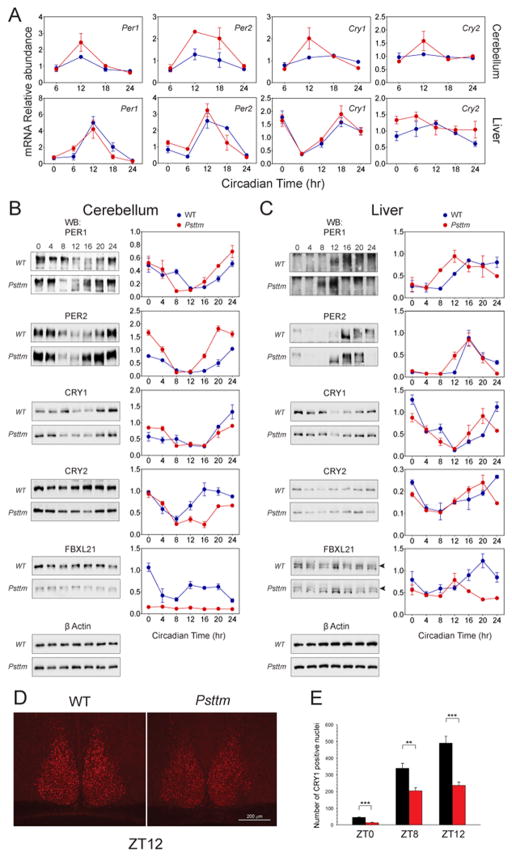
Figure 1. Positional cloning of Past-time (Psttm) and identification of the Fbxl21 mutation.
A. Histogram showing circadian period distribution of ENU mutagenized generation 3 (G3) mutant mice in a recessive screen. The shaded area represents the average period for WT C57BL/6J mice ± 3 standard deviations. The Past-time founder mouse is indicated by the arrow.
B. Representative actogram of a WT C57BL/6J mouse (upper). The actogram is double plotted where each horizontal line represents 48 hrs of activity. The mice were kept on an LD12:12 cycle (represented in the bar above) for the first 7 days and then released into constant darkness for 21 days (indicated by the arrowhead on the right). Actogram of the Past-time founder G3 mouse (period = 22.91 hours) (lower).
C. Period distribution of F2 intercross mice used for genetic mapping. The 3 panels from top to bottom represent WT, Psttm/+ and Psttm/Psttm mice, respectively. ANOVA of the period was performed on the 3 populations of F2 mice (grouped by genotype) (DF = 2; F= 78.80; p = 3.983 ×10−29).
D. Psttm was initially mapped to a 40 Mb region on chromosome 13 between rs13481788 and rs367922 (left). The chromosome 13 schematic lists the markers used to map Psttm to a smaller genetic interval. Haplotypes of the 78 Psttm/Psttm F2 intercross progeny (156 meioses) are shown on the right. Black boxes represent C57BL/6J WT alleles, and white boxes represent C3H/HeJ alleles. The number of recombinants per total meioses is indicated to the right of the haplotype map. Sequencing of Fbxl21 where a single-base change from G to A is indicated by the asterisk (right).
E. Structural modeling of the FBXL21 LRR3 motif based on the Skp2 structure in WT and mutant proteins. See also Figure S1.
The Psttm Mutation Causes Period Shortening
To confirm that Fbxl21 is the causative gene, we generated transgenic mice expressing the Psttm allele using the tetracycline-transactivator (tTA) system (Hong et al., 2007). Three independent tetO::Psttm transgenic lines were crossed to Scg2::tTA mice in which the Secretogranin-2 promoter drives tTA expression in the SCN and other brain areas to evaluate whether the Psttm allele affects circadian behavior. Western blotting analysis using cerebellum extracts confirmed that only double transgenic mice showed mutant protein expression (Figure S2A). To determine whether Scg2::tTA-driven PSTTM expression in the brain also shortens free-running period as seen in Psttm/+ mice (Figure S2B), we measured their circadian activity rhythms in constant darkness. Double transgenic mice displayed short circadian periods compared with single transgenic lines (Figure 2A, S2C). The period shortening was conditional because doxycycline (Dox) treatment, which represses tTA transgene expression, restored the period to wild-type (WT) values (Figures 2A, S2C). Removal of Dox from the drinking water after 4 wks of treatment caused the double transgenic mice to return to their previous, short free-running periods, confirming that inducible PSTTM expression was the cause of the short free-running periods (Figures 2B, S2D). In contrast, overexpression of WT FBXL21 under the control of the Scg2::tTA driver did not cause period changes in Scg2::tTA/tetO::Fbxl21 #38 mice (Figure S2, E, F and G). Together, these observations show that conditional expression of the Psttm allele in the SCN and brain shortens circadian period in vivo, thus, verifying that Fbxl21 is the causative gene. Because transgenic expression of Psttm in a WT background shortens period and because the Psttm allele is semidominant, this suggests that Psttm can act as an antimorphic mutation.
Figure 2. The Psttm mutation shortens period and antagonizes the period lengthening effect of the Ovtm mutation in FBXL3.
A. Representative actogram of tetO::Psttm #11 single transgenic mouse (left). Representative actogram of tetO::Psttm #11 transgenic mouse with the Scg2::tTA driver (right). Arrowheads indicate LD to DD transition. Doxycycline-containing (10 μg/ml) water was administered during the interval indicated by yellow shading on the actogram.
B. Free-running periods of single and double transgenic mice during constant darkness with water (black), Dox (yellow), and water after Dox treatment (grey). Error bars show SEM (tetO::Psttm #11 n = 3, Scg2::tTA/tetO::Psttm #11 n=5; *p < 0.05, Bonferroni corrected pair-wise comparison).
C. Representative PER2::LUC bioluminescence recording of SCN and pituitary explants from WT and homozygous Psttm mice. Blue and red traces represent PER2 rhythm from WT and Psttm mice, respectively. WT SCN mean circadian period: 24.76 hr (n=6), Psttm SCN mean circadian period: 23.36 hr (n=6). t test: *p-value 0.0027. WT pituitary mean circadian period: 23.2hr, N=6; Psttm pituitary mean circadian period: 22.61hr, N=6. t test: **p-value 0.0004.
D. Representative actograms of WT, Ovtm/Ovtm, Psttm/Psttm and Ovtm/Ovtm Psttm/Psttm mice subjected to the same experimental schedule as in Fig. 1B.
E. Period distribution. The four panels from top to bottom represent WT, Ovtm/Ovtm, Psstm/Psstm and Ovtm/Ovtm Psttm/Psttm mice, respectively. ANOVA of the period was performed among the genotype groups. Results: DF = 3; F= 388.507; p = 2.295 × 10−51. See also Figure S2 and Table S1.
To investigate the effects of Psttm at the cellular and molecular level, we crossed the Psttm mice with mice carrying the Per2::luc reporter (Yoo et al., 2004), and examined the circadian rhythmicity in tissue explants from WT and Psttm homozygous mice (Figure 2C). SCN organotypic cultures from Psttm mice showed robust rhythms, yet with significantly shorter periods compared with those of WT SCNs (Figure 2C, lower right). Similarly, pituitary explants from Psttm mice also showed shorter periods than WT explants (Figure 2C, lower). Thus, the primary effect of the Psttm mutation on the circadian clock is period shortening at both behavioral and molecular levels.
The FBXL3 I364T mutation in Overtime (Ovtm) mutant mice lengthens period by about 2.5 hrs (Siepka et al., 2007), providing evidence that CRY stability is important for setting the pace of the clock (Busino et al., 2007; Godinho et al., 2007; Maywood et al., 2011). Given that both Ovtm and Psttm correspond to mutations in F-box genes related to CRY degradation, we created Psttm/Psttm Ovtm/Ovtm double-mutant mice to assess the genetic interaction of these mutant alleles (Figure 2, D and E). While WT mice showed a mean period of 23.34 hr (SD=0.30, N=32), Ovtm mice displayed a long period of 26.00 hr (SD=0.46, N=13). In contrast, Psttm mice exhibited a short period of 22.77 hr. Interestingly, the Ovtm/Psttm double-homozygous mutant mice displayed a free-running period of 23.16 hr (SD=0.27, N=29), similar to that of WT mice, and different from the predicted value of 24.4 hr for an additive effect (Table S1). Thus, the Psttm and Ovtm alleles appear to act in an antagonistic rather than additive manner.
Effects of the Psttm Mutation on Core Clock Gene Expression
To explore the effects of the Psttm mutation on clock gene expression, we collected liver and cerebellum samples from WT and Psttm mice maintained in constant darkness and measured cycling clock gene mRNA expression patterns. The Psttm mutation caused significant elevation of Per1, Per2, Cry1 and Cry2 transcripts in cerebellum and a moderate elevation of clock gene transcript levels in liver (Figure 3A, S3A), perhaps due to lower Fbxl21 expression in liver relative to cerebellum (mean Ct value of WT cerebellum and liver: 27.1 and 31.2, respectively; Figure S3D shows different levels of endogenous FBXL21 expression in various mouse tissues). Interestingly, the levels of several clock-controlled genes, including Rev-erbα, Dec2 and Dbp, were significantly increased in liver (Figure S3A).
Figure 3. Psttm alters circadian clock gene expression in mice.
A. Real-time RT-PCR analysis of clock gene expression in WT and Psttm mice. Blue and red circles represent WT and Psttm mice, respectively. Error bars represent SEM for each time point from four independent replicates. 2-way ANOVA shows significant statistical differences between WT and Psttm mutants for Per1 (p < 0.001) and Per2 (p < 0.0001). Cry1 expression shows statistically significant difference at CT12 (p < 0.01) between WT and Psttm mice. Clock protein oscillation in cerebellum. (B) and liver (C). Western blotting was performed using total protein extracts with the indicated antibodies. Representative blots from 3 independent experiments are shown. Quantification of 3 independent experiments are shown to the right of each representative blot. Blue and red circles represent values from WT and Psttm mice. Error bars represent SEM for each time point from 3 independent repeats. In B, 2-way ANOVA shows significant statistical differences between WT and Psttm for PER2 (p < 0.0001), CRY2 (p < 0.0001), and FBXL21 (p < 0.0001). In C, 2-way ANOVA shows significant statistical differences between WT and Psttm for FBXL21 (p < 0.0001).
D. Immunohistochemical staining of CRY1 in SCN sections from WT and Psttm mice. Representative images from ZT12 are shown.
E. Number of CRY1-positive nuclei in the SCN sections collected from WT and Psttm mice at ZT0, 8, and 12. Student t-test shows statistically significant difference between WT and Psttm mice at ZT0 and ZT12 (*** p < 0.001) and ZT8 (* p < 0.01). See also Figure S3.
Next we examined the effects of the Psttm mutation on core clock protein expression in cerebellum and liver. PER1 and PER2 levels were elevated in the cerebellum of Psttm mice (Figure 3B), consistent with the observed increase in transcript levels. Furthermore, Psttm also appeared to advance the phase of the PER protein rhythm. In contrast, CRY1 protein levels were not elevated, and CRY2 levels were significantly reduced (Figure 3B). Consistent with the previously reported rhythmic Fbxl21 mRNA accumulation (Dardente et al., 2008), we observed a circadian oscillation of FBXL21 protein in the cerebellum of WT mice. In Psttm mutants, FBXL21 protein levels in cerebellum were significantly reduced throughout the circadian cycle compared with WT (Figure 3B), suggesting the Psttm mutation impaired FBXL21 protein stability. In addition, in the liver of Psttm mutants, the phase of PER1/2 and CRY1/2 accumulation was advanced relative to WT (Figure 3C). FBXL21 protein accumulation was also both reduced and phase-advanced in Psttm liver. The reduction in CRY, despite enhanced transcript levels, suggests that the Psttm mutation leads to the destabilization of CRY protein. In addition, because the expression of FBXL21 is rhythmic and in phase with that of CRY1/2 (Figure 3B), it is possible that FBXL21 could exert phase-dependent effects.
To determine whether the Psttm mutation affects CRY expression in the SCN, we used immunocytochemical quantitation of SCN sections from WT and Psttm mice. As shown in Figure 3D, the number of CRY1 positive nuclei was significantly reduced in SCN from Psttm mice at ZT0, 8, 12 compared with WT controls (Figure 3E, S3B,C). Taken together, these experiments show that the Psttm mutation leads to a general increase in the RNA expression of CLOCK:BMAL1 target genes, while at the same time altering the expression of CRY1 and CRY2 proteins and substantially reducing FBXL21 protein levels. The reduction in CRY, despite enhanced transcript levels, suggests that the Psttm mutation leads to destabilization of CRY protein.
FBXL21 Stabilizes CRY by Competing Against FBXL3
The concomitant reduction of FBXL21 and CRY proteins caused by the Psttm mutation prompted us to investigate the biochemical interaction between CRY and FBXL21. Co-immunoprecipitation assays showed that among the 6 core clock proteins in the primary loop (CRY1/2, PER1/2, CLOCK, BMAL1), only CRY1 and CRY2 were able to interact with both FBXL21 and PSTTM (Figure S4A). All CRY1 mutants with C-terminal domain truncation were co-immunoprecipitated with both FBXL3 and FBXL21, indicating that FBXL binds to CRY in the photolyase homology domain of CRY1 (amino acids 1–530) (Figure S4). Next we expressed the F-box proteins in NIH3T3 cells and the cell lysates were immunoprecipitated with anti-FLAG (FBXL3, FBXL21, PSTTM) or anti-V5 (β-TRCP1) antibodies. All the F-box proteins including PSTTM were found to interact strongly with native CULLIN1 and SKP1 (the other components of the SCF complex), indicating FBXL21 and PSTTM can form SCF E3 ligase complexes (Figure 4A). Only low levels of CRY1 co-immunoprecipitated with ectopically expressed FBXL3, likely due to robust CRY1 degradation. To our surprise, FBXL21 or PSTTM co-immunoprecipitated much higher levels of CRY1 compared with FBXL3. The stable binding between CRY1 and FBXL21/PSTTM differs from the transient E3 ligase-substrate interaction commonly associated with rapid degradation (e.g., FBXL3-CRY interaction). In addition, ectopically expressed PSTTM and FBXL21 both showed strong affinity toward CRY, indicating that the mutation does not significantly affect CRY binding.
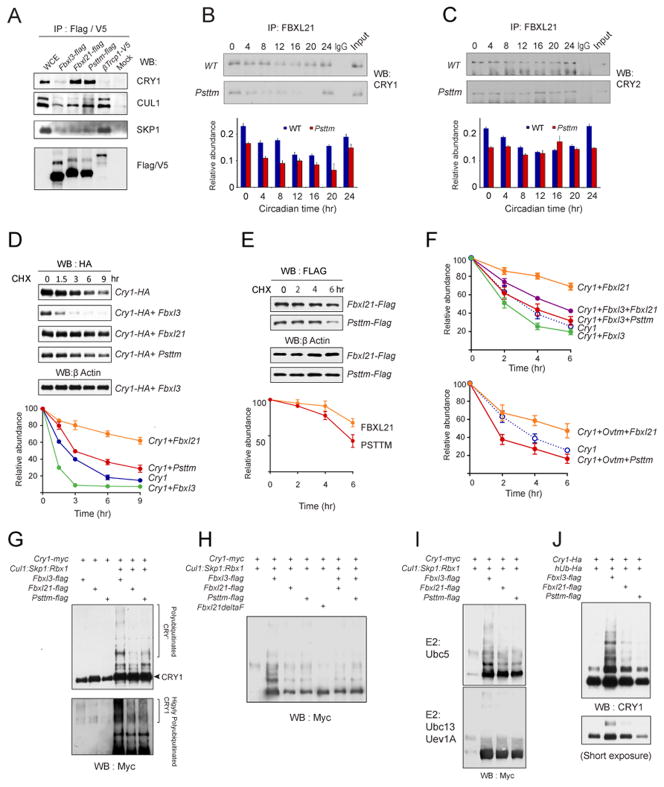
Figure 4. FBXL21 forms an SCF E3 ligase complex that slowly ubiquitinates CRY1 and antagonizes the activity of SCFFbxl3.
A. Interaction of FBXL proteins with Cullin1 and Skp1. NIH3T3 cells were transfected with Flag-Fbxl3, Flag-Fbxl21, Flag-Psttm and V5-βTrcp1 expression constructs. Co-immunoprecipitated proteins were analyzed by western blotting with anti-CRY1, anti-CUL1, and anti-SKP1 antibodies. Representative blots from 3 independent experiments are shown.
B and C. FBXL21 interacts with CRY1 (B) and CRY2 (C) in native extracts in a circadian manner. Liver extracts from CT0 to CT24 were immunoprecipitated (IP) with an FBXL21 antibody, and Western blotting was performed with CRY1 and CRY2 antibodies. Lower: quantification of co-immunoprecipitated CRY1 and CRY2 amounts. Data are taken from 2 independent experiments. Error bars show ± range (n = 2). 2-way ANOVA shows statistically significant differences between WT and Psttm for the amount of CRY1 co-precipitated with FBXL21 throughout the circadian cycle (p < 0.0001).
D. Differential effects of FBXL21 and PSTTM on CRY1 stability. 293A cells were co-transfected with indicated constructs. 32 hrs after transfection, cells were treated with 20 μg/ml cycloheximide and incubated for the indicated time before harvest. Western blotting was performed to monitor CRY1 levels using an anti-HA antibody. Lower: Quantification of the effects of FBXL3, FBXL21, PSTTM on CRY1 stability. Data are taken from 3 independent experiments. Error bars show ± SEM (n = 3). Half life was determined by using nonlinear, one phase exponential decay analysis (the half-life parameter, K, is significantly different in all four conditions: p<0.0001)
E. The Psttm mutation destabilizes FBXL21. 293A cells were transfected with Fbxl21-Flag or Psttm-Flag constructs. Cycloheximide treatment was performed as in D. Data are taken from 3 independent experiments. Error bars show ± SEM (n = 3). The half-life parameter, K, is significantly different: p=0.0436.
F. Competition between FBXL3 and FBXL21/PSTTM modulates CRY1 degradation. Cycloheximide treatment and CRY1 western blotting were performed as in D. Data for quantification represent 3 independent experiments (Figure S4F). The half-life parameter, K, is significantly different: p<0.0001. Data are taken from 3 independent experiments. Error bars show ± SEM (n = 3).
Lower: Competition between Ovtm and Fbxl21/Psttm in CRY1 degradation. Cycloheximide treatment and CRY1 western blotting were performed as in D. Data for quantification represent 3 independent experiments (representative western blots are shown in Figure S4G). The half-life parameter, K, is significantly different: p<0.0001. Error bars show ± SEM (n = 3).
G. In vitro ubiquitination assay. Sf9 cells were infected with the indicated baculovirus constructs. Samples were analyzed by western blotting with an anti-Myc antibody. Upper shows a short exposure image, and the bracket to the right marks polyubiquitinated CRY1. Lower shows a long exposure image, and the bracket to the right marks highly polyubiquitinated CRY1. Results are representative of more than 3 replicates.
H. In vitro ubiquitination was performed as indicated in G. Sf9 cells were infected with the indicated baculovirus. Co-infection of Fbxl baculovirus (Fbxl3+Fbxl21, Fbxl3+Psttm) attenuated the E3 ligase activity of FBXL3. Results are representative of 3 replicates.
I. FBXL3/FBXL21 mediated CRY1 ubiquitination requires Ubc5 as an E2 ligase. Top: Ubc5-mediated robust ubiquitination by FBXL3-SCF complexes and multi ubiquitination by FBXL21/PSTTM SCF complex. Lower: lack of CRY1 ubiquitination in the presence of Ubc13/Uev1A as the E2 ligase. Results are representative of 3 replicates.
J. 293A cells were co-transfected with Cry1-HA, ubiquitin (hUb-HA) and the indicated F-box constructs. Cells were treated with MG132 (10μg/ml) 6 hr before harvest. Whole-cell lysates were analyzed by western blotting with an anti-CRY1 antibody. Results are representative of 3 replicates. See also Figure S4.
To verify the interactions between FBXL21 and CRY1/2 in vivo, we assessed native protein interactions over the circadian cycle by immunoprecipitating endogenous FBXL21 proteins from WT and Psttm liver tissue lysates collected at different circadian times (Figure 4, B and C). Levels of co-immunoprecipitated CRY1/2 showed a circadian rhythm correlated with their periodic accumulation (Figure 3C) in WT tissue; on the other hand, lower levels of CRY1/2 were pulled down in Psttm liver, presumably due to reduced FBXL21 expression (Figure 3C). Next we compared the potential E3 ligase activity of FBXL3 with that of FBXL21 and PSTTM in CRY-degradation assays in 293A cells (Figure 4D, and Figure S4, B and C). In the absence of ectopic FBXL3 expression, transfected CRY1 had a half-life of 2.1 hr (Figure 4D and Figure S4C for CRY2). This rapid degradation was reversed when Cry1 was co-transfected with hFbxl3si RNA, indicating that endogenous hFBXL3 regulates CRY1-HA degradation in 293A cells (Figure S4D). As expected, co-transfection of FBXL3 accelerated CRY1 degradation (0.75 hr). In contrast, co-transfection of FBXL21 significantly decelerated CRY1 degradation (12.3 hr). PSTTM also appeared to slow down CRY1 degradation, but to a lesser extent than WT FBXL21 (3.7 hr). Because endogenous hFBXL3 is present, exogenously expressed FBXL21 likely antagonizes endogenous hFBXL3 to reduce CRY1 degradation. Since FBXL21 is a less efficient E3 ligase for CRY1 than FBXL3 (Fig. 4D), FBXL21 can act antagonistically as a partial agonist in a manner analogous to classic pharmacological experiments in which a partial agonist (FBXL21) can act as a competitive antagonist in the presence of a full agonist (FBXL3). In addition, because PSTTM itself is less stable than WT FBXL21 (Figure 4E), the unstable PSTTM (4 hr) would be expected to antagonize SCFFbxl3 less efficiently than WT FBXL21 (8.6 hr). Next we introduced the corresponding Psttm mutation into Fbxl3 (G143E) and measured the effect of the mutation on CRY1 degradation and FBXL3 stability (Figure S4B, lower). The G143E mutation in Fbxl3 did not alter protein stability or E3 ligase activity of FBXL3. Importantly, we rule out the possibility of FBXL21 or PSTTM as an E3 ligase for FBXL3: co-expression of FBXL21 or PSTTM showed no effects on FBXL3 stability independent of CRY, indicating that FBXL21 does not decelerate CRY degradation by serving as an E3 ligase for FBXL3 (Figure S4E).
To test the hypothesis that FBXL21 antagonizes FBXL3, we performed competition assays for CRY1 degradation in 293A cells (Figures 4F and S4F). In support of an antagonistic relationship between FBXL3 and FBXL21, co-expression of FBXL3 and FBXL21 resulted in an intermediate CRY1 half-life (4.4 hr) relative to expression of CRY1 with either FBXL3 (1.9 hr) or FBXL21 alone (11 hr). Interestingly, in the presence of FBXL3, PSTTM was less effective in decelerating CRY1 degradation (3 hr) than FBXL21 (4.4 hr). This supports the conclusion that PSSTM is a less effective antagonist of FBXL3-mediated CRY degradation than WT FBXL21. Next, we carried out similar assays using the hypomorphic Ovtm mutant of Fbxl3 to investigate whether the WT-like circadian periodicity in Ovtm/Psttm compound homozygous mice is associated with restored CRY degradation rate (Figure 2, D and E). Specifically, 293A cells were co-transfected with Cry1 and F-box pairs, Ovtm+Fbxl21, and Ovtm+Psttm. CRY1 degradation was attenuated in the presence of OVTM (4.9 hr; Figures 4F, lower, and S4G), whereas PSTTM expression in place of FBXL21 restored CRY1 degradation in the presence of OVTM (1.6 hr). These changes in CRY1 degradation rate are directly correlated with the period shortening or lengthening in the Psttm and Ovtm mouse mutants (Figure 2E), suggesting that the WT periods of Ovtm/Psttm mice resulted from an antagonistic interaction of the two mutations on CRY degradation in which impaired CRY degradation in OVTM was rescued (or increased) in the presence of PSTTM.
To verify definitively that FBXL21 forms an SCF E3 ligase complex, we performed in vitro ubiquitination assays to examine the ubiquitination activity of SCFFbxl3, SCFFbxl21 and SCFPsttm. CRY1 was efficiently polyubiquitinated in the presence of SCFFbxl3 complex. Both FBXL21 and PSTTM were able to carry out CRY1 polyubiquitination, but were less active in generating highly polyubiquitinated forms of CRY compared with FBXL3 (Figure 4G, lower: long exposure). The observed differences in CRY ubiquitination processivity by FBXLs paralleled their effects on CRY degradation (Figure 4D). CRY1 ubiquitination by SCFFbxl3 was significantly reduced in the presence of SCFFbxl21 or SCFPsttm (Figure 4H), suggesting that attenuated CRY1 degradation in the presence of both FBXL3 and FBXL21 (Figure 4F upper) was due to direct competition between FBXL3 and FBXL21 as CRY1 E3 ligases (Figure 4H). As shown in the end-point competition assay (Figure S4H), FBXL21 and PSTTM (as a lesser extent than WT) were able to protect CRY1 from FBXL3 mediated degradation in a dose-dependent manner, suggesting direct competition between FBXL3 and FBXL21.
No activity was detected from in vitro ubiquitination assays using the UBC13/Uev1A E2 ligase required for K63 ubiquitination (Figure 4I), indicating that FBXL21 is not involved in non-proteolytic K63 ubiquitination, and that CRY stabilization (Figure 4D) does not involve K63 ubiquitination by FBXL21. To confirm these results in intact cells, we performed ubiquitination assays by expressing CRY1, the F-box proteins, and ubiquitin (hUb) in 293A cells in the presence of the proteasome inhibitor MG132 (Figure 4J). Consistent with the in vitro assay results, FBXL21 and PSTTM showed significantly attenuated polyubiquitination activity relative to FBXL3.
Psttm and Fbxl21 Knockdown Differentially Alter Nuclear and Cytoplasmic CRY Degradation
To determine the allelic nature of the Psttm mutant, we compared the effects of the Psttm mutation with loss-of-function of Fbxl21 using shRNA knockdown of Fbxl21 (shFbxl21) on mPER2::LUC rhythms in mouse embryonic fibroblasts (MEFs). Psttm MEFs showed advanced phases with significantly shorter periods relative to WT MEFs (Figure 5, A and D). PSTTM was less abundant than WT FBXL21 in MEFs from the mPER2::LUC genetic background (Figure 5C left), consistent with what we observed in the tissue samples (Figure 3B,C), and knockdown efficiently depleted FBXL21 protein (Figure 5C right). Importantly, Fbxl21 knockdown also elicited phase advance and period shortening phenotypes compared with shRNA control MEFs (Figure 5, B and D), indicating that the Psttm allele behaves as either a loss-of-function hypomorphic allele or a dominant negative antimorphic allele. This is consistent with the in vitro ubiquitination assays that show that SCFPsttm also does not show a significant gain-of-function in ubiquitination activity relative to SCFFbxl21.
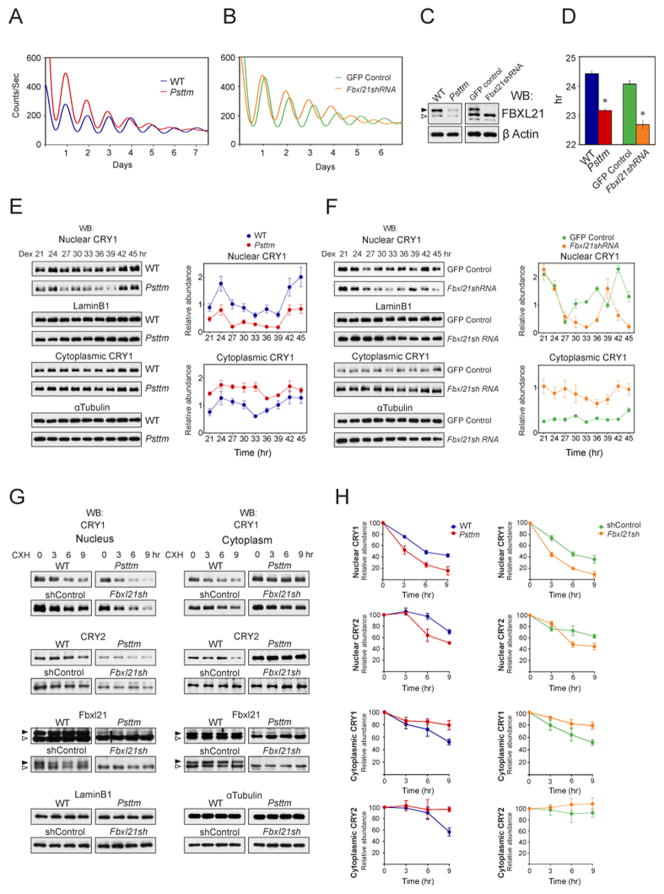
Figure 5. The Psttm mutation is mimicked by Fbxl21 knockdown and that promotes accelerated CRY1 degradation in the nucleus and stabilization in the cytoplasm.
A. Psttm shortens circadian period in MEFs. Representative PER2::LUC bioluminescence recording from WT (blue trace) and Psttm (red trace) MEFs.
B. Fbxl21 knockdown shortens circadian period in MEFs. Representative PER2::LUC bioluminescence recording from Lenti-GFP infected (green trace) and Lenti-Fbxl21sh infected (orange trace) PER2::LUC MEFs.
C. Western blotting of WT, Psttm, Lenti-GFP and Lenti-Fbxl21sh cell lysates showing that FBXL21 is low in Psttm MEFs and depleted in Fbxl21 knockdown MEFs. Filled and open arrows indicate FBXL21 and a nonspecific band respectively.
D. Average period values of the experimental groups in A and B. Statistically significant difference in the mean period was detected between WT (24.4h) and Psttm (23.1hr) MEFs (t test: * p < 0.001,). Likewise, statistically significant difference in the mean period was detected between Lenti-GFP (24.06 h) and Lenti-Fbxl21si (22.68 hr) MEFs (t test: * p < 0.001).
E. The Psttm mutation decreases nuclear CRY1 levels and oscillation amplitude. Western blotting for CRY1 was performed using nuclear and cytosolic fractions. Right: Quantification from triplicates including examples shown here. CRY1 levels from Psttm nuclei (red) were significantly reduced (upper, 2-way ANOVA, p < 0.001) relative to CRY1 from WT nuclei (blue). In contrast, cytoplasmic CRY1 levels from Psttm (red) were significantly elevated throughout the circadian cycle (lower, 2-way ANOVA, p < 0.001). Error bars show ± SEM (n = 3).
F. Fbxl21 depletion reveals that Psttm behaves as a loss-of-function mutation relative to WT Fbxl21. Lenti-GFP and Lenti-Fbxl21sh infected mPER2::LUC MEFs were treated the same as in E. Right: Quantification from triplicates including example shown in F. Error bars show ± SEM (n = 3). CRY1 levels in Lenti-Fbxl21sh nuclei were significantly reduced (orange, upper, p < 0.001), whereas cytoplasmic CRY1 levels were significantly increased throughout the circadian cycle (orange, lower, p < 0.001).
G. Accelerated nuclear CRY degradation and decelerated cytoplasmic CRY degradation in Psttm and Fbxl21 knockdown MEFs. Western blotting was performed using anti-CRY1, -CRY2, -FBXL21 antibodies for nuclear and cytoplasmic fractions. Representative blots from 3 independent experiments are shown.
H. Quantification of nuclear and cytoplasmic CRY1 and CRY2 degradation in Psttm and Fbxl21sh MEFs from triplicate experiments including the representative blots shown in G. Error bars show ± SEM (n = 3). See also Figure S5.
To investigate the cellular localization of Fbxl21 and Psttm action on CRY abundance, we prepared nuclear and cytoplasmic fractions from samples collected over the circadian cycle and assessed CRY levels (Figures 5E, 5F, S5A, and S5B). In Psttm MEFs (red circles), nuclear CRY1 and CRY2 levels were significantly lower than in WT (blue circles) (Figures 5E, right upper, and CRY2 results are shown in Figure S5A). In contrast, cytoplasmic CRY levels were significantly elevated in Psttm MEFs (red circles) throughout the cycle (Figure 5E, right lower, and Figure S5A). In a similar manner, Fbxl21 knockdown (orange circles) reduced nuclear CRY1/2 (Figure 5F, right upper, and Figure S5B) compared to GFP control (green circles) yet enhanced cytoplasmic CRY1/2 levels (Figure 5F, right lower, and Figure S5B). To understand the mechanism underlying the differential effects of FBXL21/PSTTM on nuclear and cytoplamic CRY levels, we compared the rate of CRY degradation in nuclear and cytoplasmic fractions from WT vs. Psttm MEFs and shControl vs. shFbxl21 cell lines (Figure 5, G and H). Both the Psttm mutation and Fbxl21 knockdown accelerated nuclear CRY1/2 degradation compared with controls (Figure 5, G and H), with opposite effects on cytoplasmic CRY degradation. Endogenous FBXL21 was detected in both nuclear and cytoplasmic fractions, and the Psttm mutation impaired FBXL21 accumulation in both sub-cellular compartments (Figure 5G). Neither the Psttm mutation nor depletion of endogenous FBXL21 had any effect on nuclear FBXL3 protein levels (Figure S5C). Together, these findings demonstrate that reduced levels of FBXL21 elicited opposing effects on CRY degradation in the nucleus and the cytoplasm, leading to CRY accumulation in the cytoplasm but accelerated turnover in the nucleus.
FBXL21 Forms SCF Complexes in the Cytoplasm
To determine the subcellular localization of the FBXL proteins, we generated fluorescent protein fusions of the FBXL proteins. Consistent with previous results (Cenciarelli et al., 1999), Venus-FBXL3 localized primarily to the nucleus (Figure 6A). On the other hand, Venus-FBXL21 was found in both the nucleus and the cytoplasm (Figure 6A), in accordance with MEF fractionation experiments (Figure 5G). Venus-PSTTM showed a subcellular distribution similar to WT FBXL21, and Venus-CRY1 localized primarily in the nucleus as reported previously (Yagita et al., 2002). Next we used bimolecular fluorescence complementation (BiFC) to investigate the subcellular localization of FBXL-CRY1 complexes. Specifically, CRY1-VenC was co-expressed with the indicated VenN-FBXLs (Figure 6B), and protein interaction was measured at the Venus emission wavelength (528 nm). The CRY1-FBXL3 complex was observed primarily in the nucleus (~90%), whereas CRY1-FBXL21 or CRY1-PSTTM complexes were found in both the nucleus and cytoplasm. Furthermore, in a 3-way competitive BiFC assay (Kerppola, 2006), in which C-terminal Cerulean fused to CRY1 (CRY1-CerC) was allowed to interact with FBXL proteins fused with either Ven-N (VenN-FBXLs) or Cer-N (CerN-FBXLs), demonstrated that FBXL21, and to a lesser degree PSTTM, interact with CRY1 more strongly than FBXL3 (Figure 6C, lower: quantification). The stronger interaction of FBXL21 with CRY1 relative to FBXL3 likely accounts for how FBXL21 is capable of antagonizing FBXL3 ubiquitination in the nucleus to balance CRY stabilization in the WT situation.
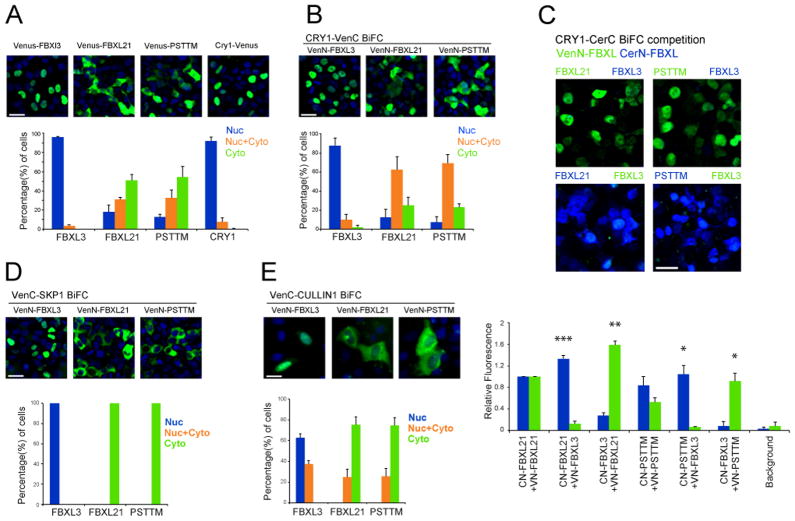
Figure 6. Differential localization of FBXL3 and FBXL21 SCF complexes in the nucleus and cytoplasm.
A. Differential subcellular localization of FBXL3, FBXL21 and PSTTM. Full-length Venus tagged FBXL3, FBXL21, PSTTM, and CRY1 were expressed in 293A cells, and percentages of cells with fluorescence signals in nuclei only, cytoplasm only, or both were measured. Whereas FBXL3 (96%) and CRY1 (92%) were predominantly localized to the nucleus, FBXL21 and PATTM were found in the both nucleus and cytoplasm with 51% and 54% of cells in the cytoplasm, respectively. Smaller percentages of cells showed nuclei only localization (FBXL21: 18%, PSTTM: 12%) or both (FBXL21: 30%, PSTTM: 36%). Green: Venus; Blue: DAPI. Bar graphs show the mean ± SD of 3 replicate experiments. 2-way ANOVA shows the localization is significantly different, p < 0.0001.
B. Differential subcellular localization of CRY-FBXL complexes. VenN tagged FBXL3, FBXL21 and PSTTM formed BiFC complexes with VenC tagged CRY1 in 293A cells. FBXL3-CRY1 complexes were primarily localized to the nuclei (88%); however, FBXL21-CRY1 (62%) and PSTTM-CRY1 (70%) were distributed in both nuclei and cytoplasm. Green: Venus; Blue: DAPI. Bar graphs show the mean ± SD of 3 replicate experiments. 2-way ANOVA shows the localization is significantly different, p < 0.0001.
C. Reciprocal 2-color, 3-way BiFC competition assays in 293A cells using CRY1-CerC complementation with CerN-FBXL3 + VenN-FBXL21 or VenN-PSTTM (upper), VenN-FBXL3 + CerN-FBXL21 or CerN-PSTTM (lower). FBXL21, and to a lesser degree PSTTM, interact with CRY1 more strongly than FBXL3 (Bottom: quantification). Green: Venus; Blue: Cerulean. Bar graphs show the mean ± SEM of 3 replicate experiments; * p < 0.01, ** p < 0.001, *** p < 0.0001.
D. FBXL3 and SKP1 form complexes predominantly in the nuclei of 293A cells, whereas FBXL21 and PSTTM interact with SKP1 in the cytoplasm. Bar graphs show the mean ± SD of 3 replicate experiments. 2-way ANOVA shows the localization is significantly different, p < 0.0001.
E. FBXL3 and CULLIN form complexes mainly in the nuclei of U2OS cells (63%), whereas FBXL21 (75%) and FBXL21Psttm (74%) bind to CULLIN mainly in the cytoplasm. All scale bars are 30 μm. Bar graphs show the mean ± SD of 3 replicate experiments. 2-way ANOVA shows the localization is significantly different, p < 0.0001.
To investigate the cause of the differential effects of FBXL21 depletion on subcellular CRY degradation, we examined SCF complex formation involving FBXL3, FBXL21 and PSTTM by using BiFC assays. Ven-C and Ven-N were fused to the N-termini of Cullin1 or SKP1 and the N-termini of FBXL3, FBXL21 or PSTTM, respectively. SKP1-FBXL3 and Cullin1-FBXL3 complexes were observed primarily in the nucleus (Figure 6D). In contrast, SKP1-FBXL21, SKP1-PSTTM, Cullin1-FBXL21, and Cullin1-PSTTM complexes were abundant in the cytoplasm (Figure 6D, E). Together, these results demonstrate that FBXL21 is primarily involved in CRY degradation in the cytoplasm, and the unstable Psttm allele or Fbxl21 knockdown lead to cytoplasmic CRY accumulation. On the other hand, reduced nuclear FBXL21 levels in Psttm or Fbxl21 knockdown MEFs reduce the antagonistic effects of FBXL21 against FBXL3, leading to accelerated CRY degradation in the nucleus.
FBXL3 and FBXL21 Ubiquitinate Distinct Lysine Residues to Regulate CRY1 Degradation
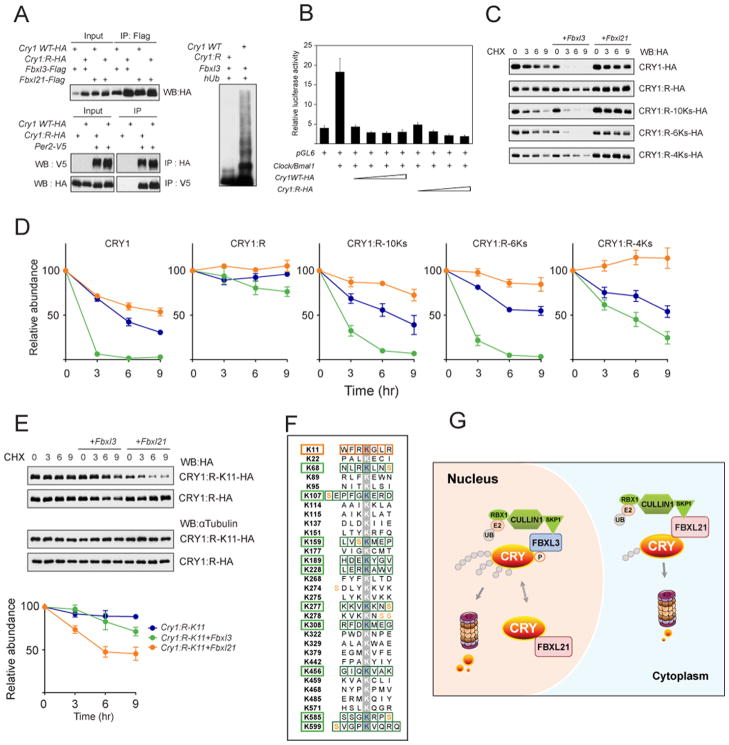
In order to delineate the mechanism of CRY1 degradation by FBXL3 and FBXL21, we sought to identify the ubiquitination sites required for CRY1 degradation by these two F-box proteins. Initially we chose 18 conserved lysine residues in CRY1 and CRY2 and converted them individually to arginine to test for loss of function. No single lysine residue was found to be critically required for CRY1 degradation in the presence of FBXL3 or FBXL21. Of these individual CRY1 mutants, K107R and K228R showed mildly impaired degradation by FBXL3. Although K159R and K308R are not conserved between CRY1 and 2, these sites showed similarity to the consensus sequence of ubiquitination sites (Catic et al., 2004) and the K159R/K308R double mutation caused mild CRY1 stabilization. These initial experiments suggested that multiple lysine residues likely play redundant roles for CRY1 degradation. Thus we also employed a reverse approach, starting from a degradation-incompetent mutant CRY1 in which all 31 lysine residues were mutated to arginine (CRY1:R). To determine whether CRY1:R retained protein-protein interactions comparable to WT CRY1, we first performed co-immunoprecipitation (IP) with FBXL3 and FBXL21. As shown in Figure 7A, co-transfected FBXL3 immunoprecipitated low levels of WT CRY1-HA in the absence of MG132, likely due to active CRY1 degradation. In contrast, CRY1:R efficiently co-immunoprecipitated with FBXL3; concordantly, the in vivo ubiquitination assay showed that this mutant was resistant to FBXL3-mediated ubiquitination (Figure 7A, right). Furthermore, co-IP with PER2 confirmed that CRY1:R could also interact with PER2 (Figure 7A, lower). In addition, reporter assays using a mPer2 promoter construct, pGL6 (Yoo et al., 2005) demonstrated that CRY1:R was able to repress CLOCK/BMAL1 transactivation of E-box dependent transcription to a similar extent as WT CRY1 (Figure 7B). Thus, mutation of all 31 lysine residues in CRY1 renders it resistant to ubiquitination and degradation, but surprisingly functional in protein interaction and transcriptional repression assays.
Figure 7. Identification of CRY1 ubiquitination sites for FBXL3 and FBXL21.
A. Left: CRY1:R co-immunoprecipited with FBXL3, FBXL21 and PER2. Upper: Co-immunoprecipitation of CRY1WT and CRY1:R with FBXL proteins. Lower: Reciprocal co-IP of WT and CRY1:R with PER2. WT or Cry1:R expression constructs were transfected into 293A cells as indicated (+). Right: 293A cells were co-transfected with indicated constructs. Whole-cell lysates were analyzed by western blotting with an anti-CRY1 antibody. Representative blots from 3 independent experiments are shown.
B. CRY1:R repressed CLOCK/BMAL1-mediated transcriptional activation of the Per2 promoter. pGL6 was transfected into 293A cells with the indicated constructs (+). Luciferase reporter assay showed similar repression efficiency for CRY1:R and CRY1 WT. Results are mean ± SEM for 3 independent experiments in duplicate.
C. Representative western blots of HA-tagged CRY1 (WT, R, R-10Ks, R-6Ks, or R-4Ks). The Cry1 construct was transfected alone or with Fbxl3 or Fbxl21 into 293A cells. Cells were treated with CHX (100 μg/ml) and collected after 0, 3, 6, or 9 hrs.
D. Quantification of CRY1 and CRY1 mutant degradation by Fbxl3 or Fbxl21 from 2 independent experiments in duplicate (n = 4 western blots; representative blots shown in C). Data represent mean ± SEM for expression of Cry1 alone (blue), Cry1 + Fbxl3 (green), or Cry1 + Fbxl21 (orange). Half life was determined by nonlinear, one phase exponential decay analysis. CRY1 half life: alone 5 hr; + Fbxl3 0.7 hr; + Fbxl21 9 hr. CRY1:R half life: alone >24 hr; + Fbxl3 21 hr; + Fbxl21 >24 hr. CRY1:R-10Ks half life: alone 6 hr; + Fbxl3 1.8 hr; + Fbxl21 21 hr. CRY1:R-6Ks half life: alone 8.9 hr; + Fbxl3 1.3 hr; + Fbxl21 >24 hr. CRY1:4Ks half life: alone 10.6 hr; + Fbxl3 4.6 hr; + Fbxl21 >24 hr. The half-life parameter, K, is significantly different in all conditions, p<0.0001, with the exception of the CRY1:R group.
E. FBXL21-mediated CRY1 degradation via the K11 residue. Representative western blots of Cry1:R-K11-HA, Cry1:R-HA are shown. The experiment was performed as in C. The K11 revertant underwent moderate degradation by FBXL21. K11 alone half life: >24 hr, + Fbxl3 18.4 hr, + Fbxl21 6.7 hr. The half-life parameter, K, for Fbxl21 is significantly different: p<0.0001).
F. 11 candidate CRY1 lysines subject to ubiquitination. All 31 lysine residues of CRY1 are shown, and candidate lysines for FBXL3- and FBXL21-mediated degradation are indicated with green and orange boxes, respectively.
G. Differential roles of FBXL21 in nuclear and cytoplasmic CRY turnover. FBXL21 appears to form SCF complexes only in the cytoplasm and functions as a cytoplasmic-specific, weak E3 ligase for CRY degradation. In contrast, nuclear FBXL21 antagonizes FBXL3-mediated CRY degradation, thus conferring a CRY-protective function. In the absence of FBXL21, cytoplasmic CRY is stabilized, whereas in the nucleus CRY is destabilized because FBXL21 cannot antagonize the action of FBXL3. See also Figure S6, S7.
We then used site-directed mutagenesis to generate 31 individual lysine revertants from Cry1:R to test for gain of function. Individual clones were transfected, either alone or co-transfected with Fbxl3 or Fbxl21 expression constructs, into 293A cells, and degradation was monitored as above (Figures S6 and S7). An initial gain-of-function screen revealed 6 putative FBXL3 target residues: K68, K189, K277, K456, K585, and K599 (Figure 7F). Combining the four aforementioned residues from the initial loss-of-function screen, we generated a panel of 10 candidate lysine revertants from Cry1:R, and degradation of these mutants in the presence of FBXL3 and FBXL21 is shown in Figure 7C. We further divided the candidate lysine sites into two groups according to their proximity to known CRY1 phosphorylation sites (Lamia et al., 2009). Cry1:R-6Ks is a 6-lysine revertant where the lysine residues are located close to phosphorylation sites, whereas Cry1:R-4Ks contains 4 lysine residues without nearby phosphorylation sites. As shown in Figure 7C, CRY1–10K regained FBXL3 mediated degradation, displaying a degradation rate similar to that of WT CRY1 (Figure 7D). Interestingly, Cry1:R-6Ks displayed a similar degradation rate as Cry1:R-10K, whereas Cry1:R-4Ks was much more stable. Multiple CRY1 mutants with fewer lysine combinations were more resistant to FBXL3-mediated degradation than Cry1:R-6Ks. Thus, there are multiple and redundant potential CRY1 degron sites for FBXL3. FBXL21 expression showed no effects on the half-life of these revertant CRYs, indicating that FBXL3 and FBXL21 do not share ubiquitination sites. Remarkably, similar site mapping for FBXL21 revealed a single candidate residue, K11 (Figure 7E), as the preferred ubiquitination site for FBXL21 (Figure S7). Figure 7F shows the summary of results from degron screening. As indicated by the green boxes, FBXL3 appears to utilize multiple lysine residues for CRY1 degradation. In contrast, FBXL21 showed a highly restricted preference for CRY1 ubiquitination, perhaps explaining why FBXL21 displayed low activity in CRY degradation.
DISCUSSION
CRY proteins are the major negative regulators in the core loop of the mammalian circadian clock. Multiple lines of evidence suggest a key role for CRY1 in regulating circadian period length (Sato et al., 2006; Liu et al., 2007). For example, impaired CRY degradation in the Ovtm mutant caused significant repression of CLOCK:BMAL1-driven transcription and lengthening of period (Siepka et al., 2007). In addition, it has been known for more than a decade that the Cry1 knockout mouse has a short period while the Cry2 knockout has a long period (van der Horst et al., 1999; Vitaterna et al., 1999). Recent work has provided insight into the transcriptional regulation of Cry1 (Ukai-Tadenuma et al., 2011), as well as, identification of a domain in CRY1 that is required for the maintenance of circadian rhythms (Khan et al., 2012). Despite these advances, how CRY stability and turnover is regulated and how CRY stability is related to circadian period remains to be understood. In the work presented here, we identified a short period mutant, Past-time, that is caused by a G149E missense mutation in FBXL21, a leucine-rich repeat F-box protein. FBXL21 is a paralog of FBXL3, an E3 ligase responsible for CRY protein turnover. Contrary to working in an additive manner with FBXL3, as might be expected of a paralog, we find instead that FBXL21 antagonizes FBXL3 in genetic interaction experiments in vivo, in cellular assays of CRY stability, and in biochemical ubiquitination assays of the reconstituted SCF E3 ligase complex in vitro. Taken together these experiments show definitively that FBXL21 interacts with FBXL3 to counterbalance the E3 ligase activity of SCFFbxl3 on CRY1 and CRY2 to stabilize them. The mechanism by which FBXL21 protects CRY is quite unusual and novel for an E3 ligase. Although FBXL21 has intrinsic E3 ligase activity as an SCFFbxl21 complex, it is much less efficient than SCFFbxl3 in polyubiquination of CRY, but has greater affinity for CRY as measured in vivo with competitive BiFC assays. Thus, FBXL21 interacts more strongly with CRY than FBXL3, and as a consequence can protect CRY from SCFFbxl3 activity.
From subcellular localization experiments, we found that the distribution of FBXL21 and FBXL3 is strikingly different: FBXL3 is found almost exclusively in the nucleus, while FBXL21 is distributed in both nucleus and cytoplasm. The distribution of the FBXL proteins was also mirrored by their interactions with SKP1 and Cullin1 so that SCFFbxl3 complexes were primarily nuclear whereas SCFFbxl21 complexes were mainly cytoplasmic. Based on the subcellular distribution of FBXL21 and FBXL3, we infer that their interactions likely occur in the nucleus, and this hypothesis is supported by two types of evidence. First, in the Psttm mutant and in Fbxl21 shRNA knockdown experiments, the levels of CRY1 and CRY2 are lower in the nucleus under conditions where either PSTTM is destabilized or FBXL21 is depleted. Second, the half-lives of CRY1 and CRY2 are shorter in the nucleus. These results are consistent with a model in which FBXL3 is a primary E3 ligase for CRY in the nucleus and FBXL21 protects CRY from rapid polyubiquitination by SCFFbxl3 by competing with FBXL3. In the absence of FBXL21, CRY would then be more susceptible to SCFFbxl3 and would turn over faster to shorten circadian periodicity. Interestingly, FBXL21 does have modest E3 ligase activity so that the SCFFbxl21 complex is not inactive, but is significantly lower than that of SCFFbxl3. Thus, the antagonistic activity of FBXL21 is most likely to occur mechanistically in a manner analogous to a “partial agonist” in classical pharmacology where a partial agonist can act as a competitive antagonist in the presence of a full agonist. In this case the full agonist is FBXL3. The evidence that FBXL21 and the SCFFbxl21 complex display “partial agonist” activity can be seen in the cytoplasmic effects of loss-of-function mutations of either PSTTM or FBXL21 knockdown where the levels of CRY1 and CRY2 are elevated significantly. Because there is essentially no FBXL3 in the cytoplasm, the SCFFbxl21 complex is the primary E3 ligase for CRY in the cytoplasm.
Here, identification of the E3 ligase FBXL21 reveals a novel mechanism involving two paralogous E3 ligases regulating nuclear CRY protein turnover in an antagonistic manner. Previous work has provided examples of multiple E3 ligases co-regulating a single target in the cell cycle (Reed, 2003; Pines, 2011). For example, turnover of the key cell cycle regulator MYC has been shown to be regulated by distinct E3 ligases, such as SCFFbw7, SCFSkp2 and SCFβTrCP, at various cell cycle phases (Popov et al., 2010). Additionally, p53 protein degradation also appears to involve several E3 ligases including Mdm2, ARF-BP1, Pirh1 and the component of the signalosome, Cop9, and WWP1 (Dai and Gu, 2010). Importantly, SCFβTrCP and WWP1 have been shown to confer substrate-protective functions for MYC and p53, respectively. However, unlike any of these precedents for interacting E3 ligases in the cell cycle, where distinct E3 ligases from different protein families interact, the antagonism of two paralogous SCF complexes (SCFFbxl3 and SCFFbxl21) in the same pathway is unique.
Translocation between the cytoplasm and the nucleus is a key step in the circadian cycles of both the positive factors (CLOCK:BMAL1) (Kondratov et al., 2003) and the negative factors (CRY/PER) of the core circadian clock loop. The subcellular distribution and relative E3 ligase activity of FBXL3 and FBXL21 create an entirely novel regulatory mode for CRY stability in the cell, analogous to that seen in the cell cycle where the ubiquitin-proteasomal system can degrade a specific protein at a specific time and in a specific place (Pines and Lindon, 2005). Thus, the discovery of FBXL21 and the differential regulation of CRY stability in the nucleus and cytoplasm reveal a prominent and sensitive role of the nucleus in regulating circadian period. That such a model would be expected can be easily dismissed given the countless models or diagrams of the circadian clock mechanism that uniformly place the SCF complexes for both CRY and PER in the cytoplasm! Clearly, a revision in our thinking is in order. The Psttm mutation accelerates nuclear CRY degradation, consequently speeding up clock progression and shortening the circadian period. Thus, in the WT condition we can infer that FBXL21 must antagonize FBXL3 in the nucleus, and furthermore that nuclear degradation of CRY is primarily instructive for determination of the speed of the clock in mammals.
EXPERIMENTAL PROCEDURES
ENU Mutagenesis Screen
ENU mutagenesis screen were performed as described previously (Siepka et al., 2007).
In Vitro Ubiquitination Assays
CRY1 containing SCF complex were purified from Sf9 cells (Invitrogen) using M2 FLAG agarose. Ubiquitination assays for CRY1 were carried out as described previously (Xu et al., 2009).
Supplementary Material
Highlights.
Ashort period circadian mouse mutant is caused by a missense mutation in FBXL21
FBXL21 forms an SCF E3 ligase complex for CRY degradation in the cytoplasm
FBXL21 antagonizes the E3 ligase activity of FBXL3 to protect CRY in the nucleus
The balance of competing E3 ligases for CRY determines circadian period of the clock
Acknowledgments
We thank Choogon Lee for generously providing the anti-PER1 antibody, Homin Kim for LRR3 modeling, Junghea Park and Weimin Song for excellent technical support, and Yi Liu for editorial comments on the paper. Research was supported by NIH grants (U01 MH61915, P50 MH074924, and R01 MH078024) to J.S.T., (P50 MH074924 and R01 GM090247) to C.B.G., (F32 DA024556) to V.K., the Howard Hughes Medical Institute to J.S.T. and Z.J.C., and the American Heart Association (11SDG7600045) to Z.C. J.S.T. and Z.J.C. are Investigators, J.A.M. is an Associate, I.K. is a Laboratory Manager, and S.-H.Y., S.M.S., H.-K.H., V.K. were Associates in the Howard Hughes Medical Institute.
Footnotes
Supplemental information includes Extended Experimental Procedures and seven supplemental figures and one table.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bass J, Takahashi JS. Circadian integration of metabolism and energetics. Science. 2010;330:1349–1354. doi: 10.1126/science.1195027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busino L, Bassermann F, Maiolica A, Lee C, Nolan PM, Godinho SI, Draetta GF, Pagano M. SCFFbxl3 controls the oscillation of the circadian clock by directing the degradation of cryptochrome proteins. Science. 2007;316:900–904. doi: 10.1126/science.1141194. [DOI] [PubMed] [Google Scholar]
- Catic A, Collins C, Church GM, Ploegh HL. Preferred in vivo ubiquitination sites. Bioinformatics. 2004;20:3302–3307. doi: 10.1093/bioinformatics/bth407. [DOI] [PubMed] [Google Scholar]
- Cenciarelli C, Chiaur DS, Guardavaccaro D, Parks W, Vidal M, Pagano M. Identification of a family of human F-box proteins. Curr Biol. 1999;9:1177–1179. doi: 10.1016/S0960-9822(00)80020-2. [DOI] [PubMed] [Google Scholar]
- Cho H, Zhao X, Hatori M, Yu RT, Barish GD, Lam MT, Chong LW, Ditacchio L, Atkins AR, Glass CK, Liddle C, Auwerx J, Downes M, Panda S, Evans RM. Regulation of circadian behaviour and metabolism by REV-ERB-α and REV-ERB-β. Nature. 2012;485:123–127. doi: 10.1038/nature11048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai C, Gu W. p53 post-translational modification: deregulated in tumorigenesis. Trends Mol Med. 2010;16:528–536. doi: 10.1016/j.molmed.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dardente H, Mendoza J, Fustin JM, Challet E, Hazlerigg DG. Implication of the F-Box Protein FBXL21 in circadian pacemaker function in mammals. PLoS One. 2008;3:e3530. doi: 10.1371/journal.pone.0003530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gekakis N, Staknis D, Nguyen HB, Davis FC, Wilsbacher LD, King DP, Takahashi JS, Weitz CJ. Role of the CLOCK protein in the mammalian circadian mechanism. Science. 1998;280:1564–1569. doi: 10.1126/science.280.5369.1564. [DOI] [PubMed] [Google Scholar]
- Godinho SI, Maywood ES, Shaw L, Tucci V, Barnard AR, Busino L, Pagano M, Kendall R, Quwailid MM, Romero MR, O’Neill J, Chesham JE, Brooker D, Lalanne Z, Hastings MH, Nolan PM. The after-hours mutant reveals a role for Fbxl3 in determining mammalian circadian period. Science. 2007;316:897–900. doi: 10.1126/science.1141138. [DOI] [PubMed] [Google Scholar]
- Hong HK, Chong JL, Song W, Song EJ, Jyawook AA, Schook AC, Ko CH, Takahashi JS. Inducible and reversible Clock gene expression in brain using the tTA system for the study of circadian behavior. PLoS Genetics. 2007;3:e33. doi: 10.1371/journal.pgen.0030033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Cardozo T, Lovering RC, Elledge SJ, Pagano M, Harper JW. Systematic analysis and nomenclature of mammalian F-box proteins. Genes Dev. 2004;18:2573–2580. doi: 10.1101/gad.1255304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerppola TK. Visualization of molecular interactions by fluorescence complementation. Nat Rev Mol Cell Biol. 2006;7:449–456. doi: 10.1038/nrm1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan SK, Xu H, Ukai-Tadenuma M, Burton B, Wang Y, Ueda HR, Liu AC. Identification of a novel cryptochrome differentiating domain required for feedback repression in circadian clock function. J Biol Chem. 2012;287:25917–25926. doi: 10.1074/jbc.M112.368001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King DP, Zhao Y, Sangoram AM, Wilsbacher LD, Tanaka M, Antoch MP, Steeves TD, Vitaterna MH, Kornhauser JM, Lowrey PL, Turek FW, Takahashi JS. Positional cloning of the mouse circadian clock gene. Cell. 1997;89:641–653. doi: 10.1016/s0092-8674(00)80245-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondratov RV, Chernov MV, Kondratova AA, Gorbacheva VY, Gudkov AV, Antoch MP. BMAL1-dependent circadian oscillation of nuclear CLOCK: posttranslational events induced by dimerization of transcriptional activators of the mammalian clock system. Genes Dev. 2003;17:1921–1932. doi: 10.1101/gad.1099503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamia KA, Sachdeva UM, DiTacchio L, Williams EC, Alvarez JG, Egan DF, Vasquez DS, Juguilon H, Panda S, Shaw RJ, Thompson CB, Evans RM. AMPK regulates the circadian clock by cryptochrome phosphorylation and degradation. Science. 2009;326:437–440. doi: 10.1126/science.1172156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Etchegaray JP, Cagampang FR, Loudon AS, Reppert SM. Posttranslational mechanisms regulate the mammalian circadian clock. Cell. 2001;107:855–867. doi: 10.1016/s0092-8674(01)00610-9. [DOI] [PubMed] [Google Scholar]
- Liu AC, Welsh DK, Ko CH, Tran HG, Zhang EE, Priest AA, Buhr ED, Singer O, Meeker K, Verma IM, Doyle FJ, 3rd, Takahashi JS, Kay SA. Intercellular coupling confers robustness against mutations in the SCN circadian clock network. Cell. 2007;129:605–616. doi: 10.1016/j.cell.2007.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowrey PL, Takahashi JS. Genetics of circadian rhythms in Mammalian model organisms. Adv Genet. 2011;74:175–230. doi: 10.1016/B978-0-12-387690-4.00006-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maywood ES, Chesham JE, Meng QJ, Nolan PM, Loudon AS, Hastings MH. Tuning the period of the mammalian circadian clock: additive and independent effects of CK1epsilonTau and Fbxl3Afh mutations on mouse circadian behavior and molecular pacemaking. J Neurosci. 2011;31:1539–1544. doi: 10.1523/JNEUROSCI.4107-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng QJ, Logunova L, Maywood ES, Gallego M, Lebiecki J, Brown TM, Sladek M, Semikhodskii AS, Glossop NR, Piggins HD, Chesham JE, Bechtold DA, Yoo SH, Takahashi JS, Virshup DM, Boot-Handford RP, Hastings MH, Loudon AS. Setting clock speed in mammals: the CK1 epsilon tau mutation in mice accelerates circadian pacemakers by selectively destabilizing PERIOD proteins. Neuron. 2008;58:78–88. doi: 10.1016/j.neuron.2008.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohawk JA, Green CB, Takahashi JS. Central and Peripheral Circadian Clocks in Mammals. Annu Rev Neurosci. 2012 doi: 10.1146/annurev-neuro-060909-153128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pines J. Cubism and the cell cycle: the many faces of the APC/C. Nat Rev Mol Cell Biol. 2011;12:427–438. doi: 10.1038/nrm3132. [DOI] [PubMed] [Google Scholar]
- Pines J, Lindon C. Proteolysis: anytime, any place, anywhere? Nat Cell Biol. 2005;7:731–735. doi: 10.1038/ncb0805-731. [DOI] [PubMed] [Google Scholar]
- Popov N, Schulein C, Jaenicke LA, Eilers M. Ubiquitylation of the amino terminus of Myc by SCF(beta-TrCP) antagonizes SCF(Fbw7)-mediated turnover. Nat Cell Biol. 2010;12:973–981. doi: 10.1038/ncb2104. [DOI] [PubMed] [Google Scholar]
- Preitner N, Damiola F, Lopez-Molina L, Zakany J, Duboule D, Albrecht U, Schibler U. The orphan nuclear receptor REV-ERBα controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110:251–260. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- Reed SI. Ratchets and clocks: the cell cycle, ubiquitylation and protein turnover. Nat Rev Mol Cell Biol. 2003;4:855–864. doi: 10.1038/nrm1246. [DOI] [PubMed] [Google Scholar]
- Sato TK, Yamada RG, Ukai H, Baggs JE, Miraglia LJ, Kobayashi TJ, Welsh DK, Kay SA, Ueda HR, Hogenesch JB. Feedback repression is required for mammalian circadian clock function. Nat Genet. 2006;38:312–319. doi: 10.1038/ng1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepka SM, Yoo SH, Park J, Song W, Kumar V, Hu Y, Lee C, Takahashi JS. Circadian mutant Overtime reveals F-box protein FBXL3 regulation of cryptochrome and period gene expression. Cell. 2007;129:1011–1023. doi: 10.1016/j.cell.2007.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong X, Buelow K, Guha A, Rausch R, Yin L. USP2a Protein Deubiquitinates and Stabilizes the Circadian Protein CRY1 in Response to Inflammatory Signals. J Biol Chem. 2012;287:25280–25291. doi: 10.1074/jbc.M112.340786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ukai-Tadenuma M, Yamada RG, Xu H, Ripperger JA, Liu AC, Ueda HR. Delay in feedback repression by cryptochrome 1 is required for circadian clock function. Cell. 2011;144:268–281. doi: 10.1016/j.cell.2010.12.019. [DOI] [PubMed] [Google Scholar]
- van der Horst GT, Muijtjens M, Kobayashi K, Takano R, Kanno S, Takao M, de Wit J, Verkerk A, Eker AP, van Leenen D, Buijs R, Bootsma D, Hoeijmakers JH, Yasui A. Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature. 1999;398:627–630. doi: 10.1038/19323. [DOI] [PubMed] [Google Scholar]
- Vitaterna MH, Selby CP, Todo T, Niwa H, Thompson C, Fruechte EM, Hitomi K, Thresher RJ, Ishikawa T, Miyazaki J, Takahashi JS, Sancar A. Differential regulation of mammalian period genes and circadian rhythmicity by cryptochromes 1 and 2. Proc Natl Acad Sci U S A. 1999;96:12114–12119. doi: 10.1073/pnas.96.21.12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Skaug B, Zeng W, Chen ZJ. A ubiquitin replacement strategy in human cells reveals distinct mechanisms of IKK activation by TNFalpha and IL-1beta. Mol Cell. 2009;36:302–314. doi: 10.1016/j.molcel.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagita K, Tamanini F, Yasuda M, Hoeijmakers JH, van der Horst GT, Okamura H. Nucleocytoplasmic shuttling and mCRY-dependent inhibition of ubiquitylation of the mPER2 clock protein. EMBO J. 2002;21:1301–1314. doi: 10.1093/emboj/21.6.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SH, Ko CH, Lowrey PL, Buhr ED, Song EJ, Chang S, Yoo OJ, Yamazaki S, Lee C, Takahashi JS. A noncanonical E-box enhancer drives mouse Period2 circadian oscillations in vivo. Proc Natl Acad Sci U S A. 2005;102:2608–2613. doi: 10.1073/pnas.0409763102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SH, Yamazaki S, Lowrey PL, Shimomura K, Ko CH, Buhr ED, Siepka SM, Hong HK, Oh WJ, Yoo OJ, Menaker M, Takahashi JS. PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci U S A. 2004;101:5339–5346. doi: 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Sehgal A. Speed control: cogs and gears that drive the circadian clock. Trends Neurosci. 2012 doi: 10.1016/j.tins.2012.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.