Ubiquitylation of partially misfolded proteins by the yeast Doa10 E3 ligase requires the Hsp40 cochaperone Sis1, whereas the Hsp70 chaperones Ssa1 and Ssa2 are dispensable. Elimination of the Hsp70 chaperones prevents proteasomal degradation, resulting in ubiquitin-dependent sequestration of the misfolded proteins in Hsp42-positive foci.
Abstract
Ubiquitin accumulation in amyloid plaques is a pathological marker observed in the vast majority of neurodegenerative diseases, yet ubiquitin function in these inclusions is controversial. It has been suggested that ubiquitylated proteins are directed to inclusion bodies under stress conditions, when both chaperone-mediated refolding and proteasomal degradation are compromised or overwhelmed. Alternatively, ubiquitin and chaperones may be recruited to preformed inclusions to promote their elimination. We address this issue using a yeast model system, based on expression of several mildly misfolded degradation substrates in cells with altered chaperone content. We find that the heat shock protein 70 (Hsp70) chaperone pair Ssa1/Ssa2 and the Hsp40 cochaperone Sis1 are essential for degradation. Substrate ubiquitylation is strictly dependent on Sis1, whereas Ssa1 and Ssa2 are dispensable. Remarkably, in Ssa1/Ssa2-depleted cells, ubiquitylated substrates are sequestered into detergent-insoluble, Hsp42-positive inclusion bodies. Unexpectedly, sequestration is abolished by preventing substrate ubiquitylation. We conclude that Hsp40 is required for the targeting of misfolded proteins to the ubiquitylation machinery, whereas the decision to degrade or sequester ubiquitylated proteins is mediated by the Hsp70s. Accordingly, diminished Hsp70 levels, as observed in aging or certain pathological conditions, might be sufficient to trigger ubiquitin-dependent sequestration of partially misfolded proteins into inclusion bodies.
INTRODUCTION
Cell viability depends on the cell's ability to maintain the balance of the proteome in the face of constant challenges. A major challenge to cell homeostasis is protein misfolding, which may occur frequently during its lifespan as a result of changes in environmental conditions, stochastic fluctuations, metabolic challenges, or mutations (Hartl and Hayer-Hartl, 2009). Cells have developed specialized protective mechanisms to cope with protein misfolding that show a high degree of evolutionary conservation. Molecular chaperones are believed to function as the first line of defense, initially recognizing exposed hydrophobic patches on protein surfaces, protecting them from aggregation and assisting refolding (Muchowski and Wacker, 2005). If refolding to the native structure fails, the protein may be delivered to ubiquitin (Ub) ligases for tagging and then shuttled to the 26S proteasome, the major proteolytic system in eukaryotic cells (Ross and Pickart, 2004).
Failure to remove misfolded proteins may also lead to their sequestration in designated protein quality control (PQC) foci or inclusion bodies (IBs). Intracellular accumulation of amyloidogenic protein IBs is a pathological hallmark of various neurodegenerative diseases (Ross and Poirier, 2004). Although a link between accumulation of IBs and neurodegenerative diseases has been known for more than a century (Holdorff, 2002; Cipriani et al., 2011), the mechanism by which they are formed is poorly understood and under constant controversy. A key, unresolved question is whether these IBs are “burial grounds” for terminally aggregated proteins or serve as functional protein PQC sites. The fact that Ub conjugates, molecular chaperones, and 26S proteasome subunits often accumulate together with misfolded proteins within these IBs appears to support the PQC site hypothesis (Cummings et al., 1998; Stenoien et al., 1999; Wang et al., 2009). In agreement with this hypothesis is the observation that under stress conditions, ubiquitylated misfolded proteins are sequestered in a dynamic juxtanuclear PQC compartment, whereas nonubiquitylated, terminally misfolded proteins accumulate in an inactive peripheral compartment (Kaganovich et al., 2008). In addition, studies of mutant polyglutamine (PolyQ)-expended huntingtin protein demonstrated that Ub, molecular chaperones, and other critical cellular factors associate irreversibly with existing IBs, leading to their depletion, eventually resulting in proteostasis collapse and cell death (Wang et al., 2009; Hipp et al., 2012).
Molecular chaperones play a key role in PQC by mediating the triage decision of whether to refold, degrade, or sequester a misfolded protein (Chen et al., 2011). Chaperones, such as members of the heat shock protein 70 (Hsp70) family, bind to short, hydrophobic polypeptide motifs, enabling proteins to fold to their native, functional state (Bukau et al., 2006). An important, but less-studied, role of the Hsp70 chaperones is the escorting of misfolded proteins to E3 Ub-ligases (Nishikawa et al., 2005; Buchberger et al., 2010), although not all protein quality control–associated degradation (PQCD) E3 ligases depend on chaperones for executing the ubiquitylation reaction (Fang et al., 2011; Rosenbaum et al., 2011). Finally, Hsp70s have also been implicated as functioning downstream of ubiquitylation (Park et al., 2007; Furth et al., 2011), although their precise role in assisting the degradation of ubiquitylated proteins and their mechanism of action is unclear.
Hsp70s participation in PQCD depends on the cooperation of obligatory cochaperones, the 40-kDa Hsps, also termed J-domain proteins. Hsp40s show a large degree of sequence and structural divergence, consistent with the idea that they play a major part in the highly diverse Hsp70 functions (Kampinga and Craig, 2010). A major function of Hsp40s is to stimulate the ATPase activity of Hsp70, thereby facilitating substrate capture (Liberek et al., 1991). Hsp40s have also been implicated in the targeting of misfolded substrates to PQCD factors (Metzger et al., 2008; Nakatsukasa et al., 2008). The yeast Hsp40, Sis1, has another important PQC function: protection from prion cytotoxicity by maintaining them in nontoxic conformations and facilitating their disaggregation, in conjunction with the protein-remodeling factor Hsp104 (Shorter and Lindquist, 2008). Paradoxically, these pathways also result in prion maintenance and propagation, perhaps due to their preservation in a soluble, nontoxic state (Tipton et al., 2008). Finally, recent studies show that Sis1 is engaged in targeting misfolded proteins to cytosolic, stress-induced PQC compartments (Malinovska et al., 2012), as well as to proteasomal degradation (Summers et al., 2013).
To gain insights into the PQC-related function of the different Hsp chaperones/cochaperones in PQCD, we investigated their role in the proteolysis of substrates of the Doa10 PQCD ligase, which contain a well-defined degradation signal (degron) derived from the yeast protein Ndc10 (Ravid et al., 2006). Ndc10 is an essential kinetochore protein and a key component of the CBF3 multisubunit complex that binds to the centromere (Doheny et al., 1993). We previously identified a bipartite hydrophobic region in this protein, confined to the last 100 amino acids (aa) of Ndc10, which, upon exposure, triggers its chaperone- and Ub-dependent degradation (Furth et al., 2011). This region contains two distinct degron elements, DegA and DegB, which are necessary for substrate ubiquitylation and initiation of proteasomal degradation, respectively (Furth et al., 2011; Alfassy et al., 2013). The entire Ndc10 degron was therefore named DegAB (Alfassy et al., 2013). Of importance, DegAB functioned autonomously when attached to various protein reporters (Furth et al., 2011). The novelty of this model degron, in comparison to substrates that undergo rapid and terminal misfolding, such as PolyQ-extended huntingtin protein or the von Hippel–Lindau tumor suppressor, is that it does not aggregate spontaneously and hence may be defined as representative of mildly misfolded PQCD substrates (Furth et al., 2011). Similar combinations of hydrophobic motifs are predicted to be present in multiple proteins. Thus DegAB-mediated degradation is likely to represent a general pathway for the elimination of mildly misfolded proteins before they undergo irreversible misfolding that can lead to cytotoxicity.
Using these PQCD model substrates, we show that the yeast Hsp70 chaperones Ssa1 and Ssa2 (Ssa1/2), as well as the Hsp40 cochaperone Sis1, are required for degradation of substrates carrying DegAB. We further provide evidence that Sis1 acts as an essential factor for ubiquitylation, whereas Ssa1/2 function is essential for facilitating proteasomal degradation subsequent to ubiquitylation. When Hsp70 chaperone levels become limiting, ubiquitylated substrates accumulate in PQC aggregation foci, whereas nonubiquitylated substrates remain soluble. Thus, this study demonstrates a direct link between misfolded PQCD substrate ubiquitylation and sequestration.
RESULTS
Ssa1 and Ssa2 are essential for the degradation of proteins containing the degron domain of Ndc10
The degradation of a temperature-sensitive (ts) mutant of Ndc10, termed Ndc10-2, and its endoplasmic reticulum (ER)–anchored derivatives displayed strong dependence on the activity of Ssa-chaperone family members (Furth et al., 2011). To identify specific Ssa protein(s) required for the degradation of Ndc10-2, we expressed a FLAG-tagged version of the protein in strains with individual knockouts of each of the four SSA family members. The rate of protein degradation in each strain was determined after treatment with the translation inhibitor cycloheximide (CHX). There was no apparent difference in the intensity of the Ndc10-2 protein band in these single-knockout strains (Supplemental Figure S1). We next examined Ndc10-2 degradation in a double-knockout ssa1Δ ssa2Δ strain (ssa1/2Δ), considering that these chaperones are highly homologous (96% aa sequence identity) and hence carry out overlapping functions (Werner-Washburne et al., 1987). Strains with SSA1 and SSA2 double knockout are less tolerant to stress conditions (Craig and Jacobsen, 1984). Therefore, to allow efficient delivery of the plasmid DNA containing Ndc10-2 into yeast and reduce the risk of spontaneous reverse mutations, we performed cell transformations in ssa1/2Δ cells expressing a covering plasmid containing wild-type SSA1 and the URA3 gene as a selectable marker. This plasmid was removed 2–3 d before experiments by treating the cells with 5-fluoroorotic acid, which is cleaved into a toxic substance in cells containing the URA3 gene product orotidine-5′-phosphate decarboxylase. As shown in Figure 1A, Ndc10-2 was rapidly degraded in ssa1/2Δ cells expressing Ssa1 from a plasmid, but was substantially stabilized after the removal of Ssa1. The stabilizing effect of Ssa1/2 knockout was not associated with localization of Ndc10-2 at the kinetochore, since it similarly stabilized an ER-anchored substrate composed of a fusion between the ER-anchored protein Vma12 and DegAB, as well as a soluble substrate composed of green fluorescent protein (GFP) fused to DegAB (Figure 1, B– D). These results indicate that either Ssa1 or Ssa2 is strictly required for the degradation of proteins containing DegAB and that they are functionally interchangeable. See Supplemental Table S1 for a concise description of the different features of the various DegAB-containing substrates used in the present study.
FIGURE 1:
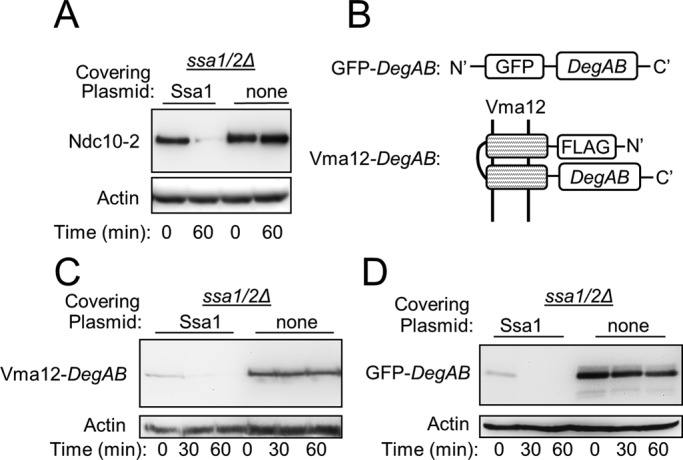
Ssa1 and Ssa2 are essential for the degradation of proteins containing the degron domain of Ndc10. (A) Degradation of Ndc10-2 requires either of the Hsp70 chaperones Ssa1 or Ssa2 (Ssa1/2). (B) Schematic presentation of the topology of substrates used in this study. Top, fusion between GFP and the 100-aa Ndc10 degron (DegAB). Bottom, fusion between the ER-anchored protein Vma12 and DegAB. (C, D) Degradation of Vma12-DegAB and GFP- DegAB, respectively, requires intact Ssa1/2.
The cochaperone Sis1 is required for the degradation of several misfolded Doa10 substrates
Ydj1 is the most common Hsp40 cochaperone that operates together with Ssa chaperones in the degradation of Doa10 substrates (McClellan, 2012). However, the inactivation of Ydj1 did not stabilize Ndc10-2 (Furth et al., 2011). Thus, to identify Hsp40 chaperone(s) involved in the degradation of DegAB-expressing substrates, we compared Ndc10-2 protein levels in a set of yeast strains with individual Hsp40 cochaperone deletions from a yeast knockout library. Ndc10-2 was not stabilized in any of the Hsp40-deleted strains, as judged by the intensity of the Ndc10-2 protein band in the various deletions compared with that in doa10Δ cells (Supplemental Figure S2a).
The degradation of Ndc10-2 in cells with conditional depletion of Hsp40s that are essential for cell viability was next examined. We used mutant strains in which the Hsp40 activity was inhibited by various means, such as the deletion of an essential domain, controlled regulation of protein levels, or temperature-dependent inhibition of catalytic activity (Figure 2A and Supplemental Figure S2, b and c). In these analyses, only the abrogation of Sis1 activity in sis1-85 cells showed some effect on Ndc10-2 degradation kinetics, mildly stabilizing Ndc10-2 (Figure 2A). We next tested the degradation of Vma12-DegAB in strains carrying sis1-85 or ydj1-151 ts alleles, using temperature shift as a robust on/off switch of chaperone activity (Figure 2B). At the restrictive temperature of 37°C, Vma12-DegAB was rapidly degraded in both Hsp40 wild-type and ydj1-151 cells. However, it was substantially stabilized in cells expressing sis1-85. Similarly, GFP-DegAB showed marked stabilization in sis1-85 cells (Figure 2C, top). Of note, the degradation of CPY*, an endoplasmic reticulum–associated degradation (ERAD) substrate of the Hrd1 ubiquitylation pathway (Bordallo et al., 1998), was not affected in sis1-85 cells (Figure 2C, middle), emphasizing Sis1’s specific role in the Doa10 pathway of ERAD. The selectivity of Sis1 in PQCD in the Doa10 pathway was further demonstrated by following the degradation kinetics of a fusion between the Mat alpha2 Deg1 degradation signal, Vma12, and protein A (Deg1-VP; Ravid et al., 2006) and of the E2 enzyme, Ubc6 (Figure 2, D and E, respectively). Whereas Deg1-VP containing a misfolded degron was markedly stabilized, the degradation of Ubc6, a native Doa10 substrate (Swanson et al., 2001), was unaffected in sis1-85 cells.
FIGURE 2:
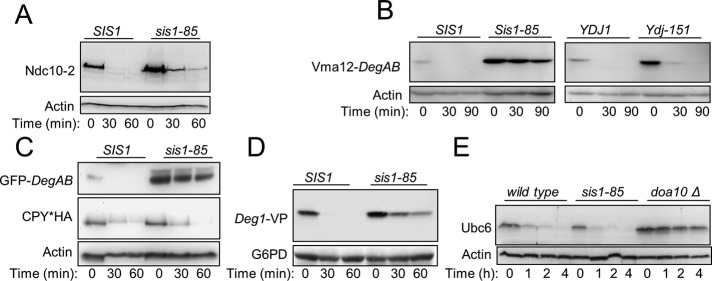
Sis1 is essential for the degradation of several misfolded Doa10 substrates. (A) Degradation of Ndc10-2 in sis1-85 cells. Cells incubated at 24°C were shifted to 37°C at 30 min before the CHX-chase experiment and kept at 37°C thereafter. (B) Vma12-DegAB is rapidly degraded in ydj1-151 cells but stabilized in sis1-85 cells. (C) Sis1 activity is required for the elimination of GFP-DegAB but not of mutant carboxypeptidase Y (CPY*). (D) Degradation of Deg1-Vma12-protein A (Deg1-VP) requires intact Sis1. (E) Degradation of the Doa10 native substrate, Ubc6, does not require Sis1 activity.
As previously indicated, the maintenance and propagation of prions is an important cellular function of Sis1. Because sis1-85 is in the W303 background, which harbors the [RNQ+] prion (Lopez et al., 2003), it is conceivable that the stabilization of Doa10 substrates observed in sis1-85 cells was due to sequestration of Ssa1/2 by aggregated [RNQ+] prions, thereby inhibiting degradation indirectly. To test this possibility, we examined the degradation kinetics of Vma12-DegAB in a W303 strain in which the protein aggregation-remodeling factor HSP104 was deleted, thereby disabling prion propagation (Chernoff et al., 1995). As shown in Supplemental Figure S3, Vma12-DegAB was rapidly degraded in the prion-free Hsp104Δ but was stabilized in sis1-85 Hsp104Δ cells. Overall these findings indicate a direct role for Sis1 in the degradation of DegAB-harboring PQCD substrates, independent of prion infection.
Ssa1/2 and Sis1 contribute differently to ubiquitylation in PQCD
To gain further insights into the precise roles of Ssa1/2 and Sis1 in PQCD, we next tested whether they are required for Doa10-dependent ubiquitylation of Vma12-DegAB. To this end, we used a reporter protein expressing a mutated degron in which two Leu residues at the extreme C-terminal region of DegAB were replaced with negatively charged Asp residues (Vma12-DegABDD). This mutation selectively prevents proteasomal degradation without impairing the ubiquitylation of substrate proteins (Alfassy et al., 2013).
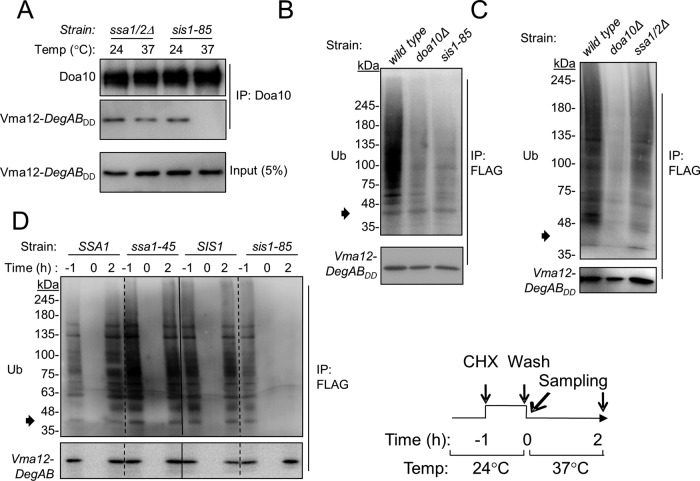
Initially we tested whether Ssa1/2 and Sis1 are required for substrate binding to Doa10 (Figure 3A). Cells expressing Vma12-DegABDD, were incubated at either 24 or 37°C for 2 h, and proteins were extracted under mild lysis conditions. Doa10 complexes were purified by immunoprecipitation (IP) using anti-Doa10 antibodies, and Doa10-bound substrate was subsequently detected by immunoblotting using anti-FLAG antibodies. Remarkably, when incubated at the restrictive temperature, Vma12-DegABDD copurified with Doa10 in ssa1/2Δ cells but not in sis1-85 cells. Thus Sis1 activity is required for stable interaction between Doa10 and the substrate before substrate ubiquitylation, whereas Ssa1 activity is largely dispensable.
FIGURE 3:
Ssa1/2 and Sis1 contribute differently to the ubiquitylation of the misfolded ER-anchored substrate Vma12-DegAB. (A) Sis1 is required for Vma12-DegAB binding to Doa10, whereas Ssa1 is dispensable. Microsomes were prepared from cells incubated at either 24 or 37°C under mild lysis conditions. Doa10 complexes were purified after treatment with digitonin using anti Doa10 antibodies. (B, C) IP of Vma12-DegABDD using anti-FLAG affinity gel, followed by immunoblotting with anti-Ub antibodies. Arrowhead indicates the approximate molecular weight of the substrate. (B) Vma12- DegABDD is not ubiquitylated in sis1-85 cells incubated at the restrictive temperature. (C) Vma12- DegABDD is partially ubiquitylated in ssa1/2Δ cells. (D) Newly synthesized Vma12- DegAB is ubiquitylated in ssa1-45 cells but remains unubiquitylated in sis1-85 cells. Cells expressing Vma12-DegAB were treated with CHX for 1 h at 24°C. CHX was washed, and the cells were shifted to grow at 37°C for 2 h. Ub conjugation was assayed as illustrated in B and C.
To test ubiquitylation directly, we next isolated Vma12-DegABDD from cell extracts by IP with anti-FLAG, followed by immunoblotting with anti-Ub antibodies (Figure 3, B and C). PolyUb-Vma12-DegABDD conjugates were detected in wild-type cells but were essentially absent in doa10Δ cells. In agreement with the substrate-binding experiment (Figure 3A), polyubiquitylation was abolished in sis1-85 cells but retained in ssa1/2Δ cells, albeit at reduced levels (∼40–50% compared with wild-type cells; Figure 3, B and C). To confirm that Vma12-DegAB was still ubiquitylated in the absence of Ssa chaperones, we examined the ubiquitylation of newly synthesized Vma12-DegAB in ssa1-45 cells and compared it to that in sis1-85 cells, using a CHX washout-chase protocol. Because members of the SSA chaperone family have overlapping functions, we examined the specific activity of Ssa1 in a strain lacking three of the four SSA genes (ssa2Δ ssa3Δ ssa4Δ) and carrying a ts ssa1 allele (ssa1-45; Becker et al., 1996). Initially, cells were treated with CHX at the permissive temperature (24°C) for 1 h, allowing degradation of the substrate. Next, CHX was washed away, and cells were further incubated at 37°C for 2 h, after which ubiquitylation was analyzed. As shown in Figure 3D, newly formed protein-polyUb conjugates were detected in ssa1-45 cells 2 h after washing the CHX, independent of the incubation temperature. In contrast, incubating sis1-85 cells at 37°C during the wash period considerably impaired formation of Vma12-DegAB-poly-Ub conjugates. Overall these results confirm that Sis1 is essential, whereas Ssa chaperones are largely dispensable for the ubiquitylation of Vma12-DegAB. It is thus likely that the reduced degradation in the absence of SSA chaperones (Figure 1) is due to a requirement for chaperone activity downstream of protein ubiquitylation.
Ub-dependent sequestration of PQCD substrates into foci in the absence of SSA1/2
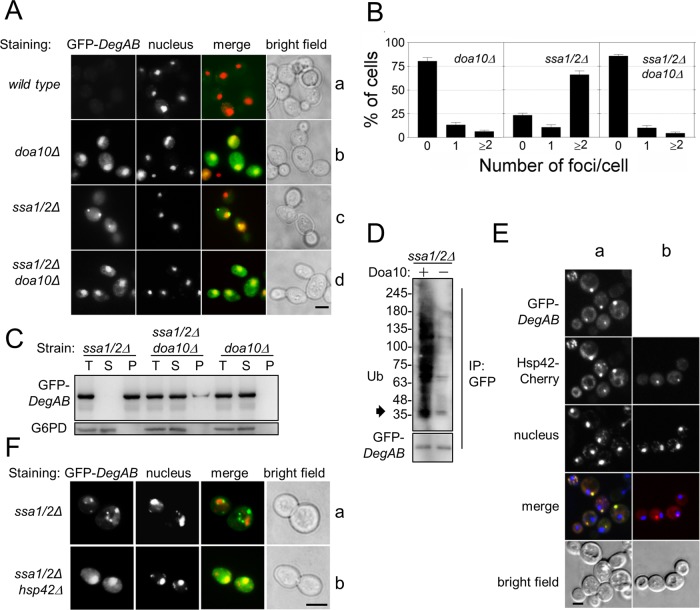
Given that the principal role of Hsp70 chaperones is to prevent aggregation of misfolded proteins, we speculated that Ssa1/2 might likewise prevent deposition of proteins expressing the Ndc10 degron in aggregation foci. Consequently we examined the spatial localization of Vma12-DegAB harboring an internal GFP (Vma12-GFP-DegAB) in ssa1/2Δ cells by fluorescence microscopy. Vma12-GFP-DegAB was hardly detected in wild-type cells but was clearly visible in doa10Δ cells, where it was mostly localized to the ER (Figure 4A, a and b). In ssa1/2Δ cells, the protein exhibited punctate staining, typical of substrate deposition in aggregation foci. This punctate staining was hardly detectable in a triple-mutant strain lacking both DOA10 and SSA1/2 (compare Figure 4A, c and d). Quantitative image analysis revealed that ∼80% of ssa1/2Δ cells expressing Vma12-GFP-DegAB contained aggregation foci, whereas aggregates were present in only ~20% of the doa10Δ cells and the doa10Δ and ssa1/2Δ triple-mutant cells (Figure 4B).
FIGURE 4:
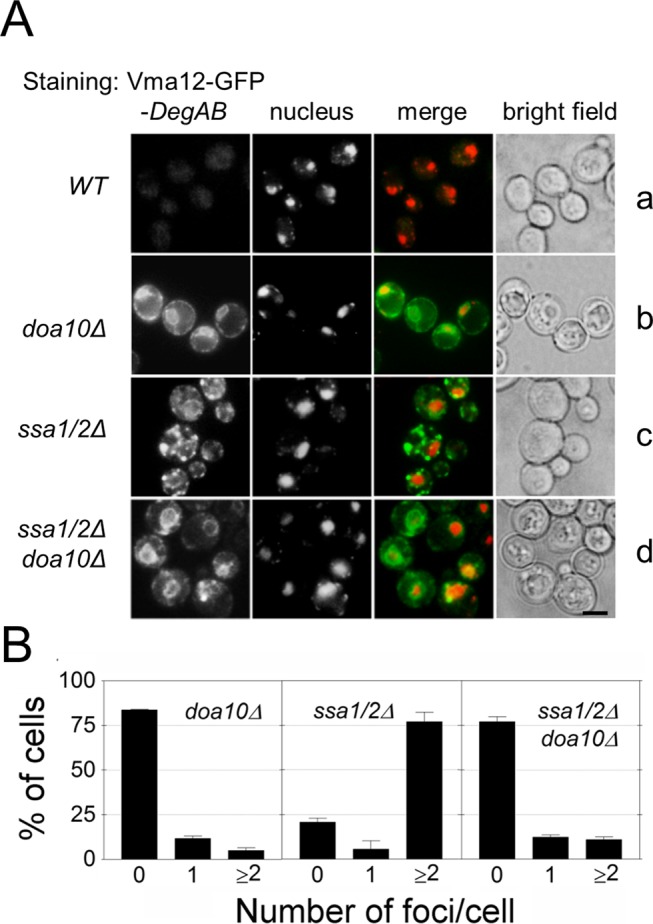
Ubiquitin conjugation is required for triggering sequestration of Vma12-GFP-DegAB into foci in the absence of Ssa1/2. (A) Vma12-GFP-DegAB was expressed in wild-type, doa10Δ, ssa1/2Δ, and ssa1/2Δ doa10Δ cells. Cells were grown to log phase at 30°C. Nuclei were visualized by Hoechst staining (red). Cells were fixed in 3.7% paraformaldehyde and visualized by fluorescence microscopy. Green channel, GFP; red channel, Hoechst. Scale bar, 5 μm. (B) Quantitative assessment of Vma12-GFP-DegAB–positive foci in each strain. The total number of foci per cell in GFP-expressing strains was counted manually. Quantifications are based on the analysis of 150–200 cells. n ≥ 3.
Aggregation of Vma12-GFP-DegAB in ssa1/2Δ cells could be attributed to the exposure of hydrophobic and/or misfolded regions within DegAB or, alternatively, within membrane-spanning regions of Vma12. To distinguish between the two possibilities, we performed a similar fluorescence analysis using the soluble Doa10 substrate GFP-DegAB. In doa10Δ cells GFP-DegAB showed diffused staining that was mainly nuclear (Figure 5A, b). Given that Ndc10 localizes to the kinetochore and the spindle pole body (Doheny et al., 1993), our findings point to the existence of a nuclear localization signal within DegAB. Furthermore, GFP-DegAB accumulated in punctate foci in ssa1/2Δ cells, but showed mainly diffused staining in doa10Δ ssa1/2Δ cells (Figure 5A, c and d). Quantitatively, the percentile of punctate foci in the various strains was similar to that observed for the ER-embedded substrate (Figure 5B).
FIGURE 5:
Ubiquitin conjugation is required for triggering sequestration of GFP-DegAB into detergent-insoluble, Hsp42-positive punctate foci. (A) Cells expressing GFP-DegAB were treated and visualized as illustrated in Figure 4A. Green channel, GFP; red channel, Hoechst. Scale bar, 5 μm. (B) Quantitative assessment of GFP-DegAB–positive foci in each strain. The total number of foci per cell in GFP-expressing strains was counted manually. Quantifications are based on the analysis of 150–200 cells. n ≥ 3. (C) GFP-DegAB is detergent insoluble in ssa1/2Δ cells but remains soluble in doa10Δ and ssa1/2Δ doa10Δ cells. Proteins extracted with 0.5% Triton X-100 were separated by centrifugation at 17,000 × g. T, total; S, supernatant; P, pellet. (D) Detergent-insoluble GFP-DegAB is ubiquitylated in ssa1/2Δ cells. (E) ssa1/2Δ cells coexpressing GFP-DegAB and mCherry-Hsp42 were visualized by confocal microscopy. The cells were prepared as illustrated in Figure 4A. (a) GFP-DegAB colocalizes with mCherry-Hsp42 in punctate foci, (b) mCherry-Hsp42 localization in punctate foci is independent of the presence of GFP-DegAB in ssa1/2Δ cells. Green channel, GFP; red channel, Cherry; blue channel, Hoechst. Scale bar, 5 μm. (F) Hsp42 is required for sequestration of GFP-DegAB in ssa1/2Δ cells. Green channel, GFP; red channel, Hoechst. Scale bars, 5 μm.
Given that the accumulation of detergent-insoluble aggregates is tightly associated with the pathology of misfolded protein diseases, we next tested the solubility of GFP-DegAB in 0.5% Triton X-100 (Figure 5C). In ssa1/2Δ cells, GFP-DegAB was strictly confined to the Triton X-100–insoluble fraction while remaining largely soluble in DOA10-knockout strains, regardless of SSA1/2 presence. That the detergent-insoluble substrate is indeed ubiquitylated was next confirmed by IP using anti-GFP antibodies followed by immunoblotting with anti-Ub antibodies (Figure 5D).
To characterize the GFP-DegAB foci, we used Hsp42, an established marker of peripheral aggregates in yeast, essential for sequestration of misfolded proteins into aggregation foci (Specht et al., 2011; Malinovska et al., 2012). In agreement with localization in aggregation foci, GFP-DegAB expressed in ssa1/2Δ cells colocalized with Hsp42 fused to monomeric cherry fluorescence protein (mCherry; Figure 5E, a). Hsp42 punctate foci were also present in ssa1/2Δ cells that did not express GFP-DegAB, suggesting that in the absence of Ssa1/2 additional proteins are being sequestered (Figure 5E, b). Furthermore, in agreement with the proposed role of Hsp42 as a recruiting factor (Specht et al., 2011), its elimination from ssa1/2Δ cells resulted in enhanced solubility of GFP-DegAB (Figure 5F, b). Together these results demonstrate that polyUb conjugation plays a significant role in Hsp42-mediated sequestration of misfolded proteins into detergent-insoluble PQC aggregation foci. Because both ER-localized and cytosolic proteins were targeted to aggregation foci, we concluded that sequestration is independent of the protein's spatial localization.
To verify that polyUb conjugation is the primary cause for the sequestration of GFP-DegAB in ssa1/2Δ cells and that reduced levels of Hsp70 alone are insufficient to trigger aggregation, we tested the cellular distribution of a stable mutant of GFP-DegAB devoid of the last 10 aa of Ndc10 (GFP-DegABΔ10). This mutant protein did not interact with Doa10 (Supplemental Figure S4) and therefore remained unconjugated to Ub (Furth et al., 2011). In line with the data shown in Figure 5, when GFP-DegABΔ10 was expressed in ssa1/2Δ cells, it did not aggregate (Supplemental Figure S5). This is likely due to inability of GFP-DegABΔ10 to undergo ubiquitylation, although we cannot exclude the possibility that the mutant affected some structural elements of the DegAB degron. Accordingly, we hypothesize that SSA1/2 depletion by itself is insufficient to promote sequestration of DegAB-expressing substrates.
In a complementary experiment, we tested a DegABDD mutant, which is ubiquitylated but cannot be degraded (Alfassy et al., 2013; Figure 3, B and C). The results show that GFP-DegABDD remained soluble in wild-type cells, whereas it was sequestered in ssa1/2Δ cells (Supplemental Figure S6), indicating that polyubiquitylation per se is unlikely to be the driving force for sequestration when Ssa1/2 levels are sufficient.
The collective results indicate that the sequestration of the misfolded substrate into PQC aggregation foci requires both polyUb conjugation and Hsp70 depletion and that each condition by itself is insufficient to induce aggregation.
Given that the appearance of cellular aggregates is commonly associated with reduced cell viability, we wanted to assess the consequences of protein aggregation observed in the ssa1/2Δ strain on cell growth. Previous studies demonstrated that the growth rate of ssa1/2Δ cells is slightly decreased at 24°C and 30°C compared with wild-type cells (Sanchez et al., 1993), and we observed no additional growth defect upon expression of GFP-DegAB in these cells (Supplemental Figure S7a). However, spontaneous mutants evading aggregation appear with high frequency in ssa1/2Δ cells (Meriin et al., 2002). Thus we used flow cytometry to analyze the number of GFP-DegAB–expressing cells within the total population. Results presented in Supplemental Figure S7, b and c, show a substantial decrease in the number of florescent ssa1/2Δ cells measured 3 d after transformation compared with strains with lower aggregate levels, doa10Δ and ssa1/2Δ doa10Δ. As a consequence, as a technical note, we took care that studies of protein sequestration were done soon after the removal of the ectopically expressed SSA1, followed by enrichment of fluorescent cells by fluorescence-activated cell sorting (FACS).
GFP-DegAB accumulates in a soluble form in cells with impaired Sis1 activity
Because Sis1 was required for the ubiquitylation of Vma12-DegAB (Figure 3, B and D), we hypothesized that GFP-DegAB will remain soluble in sis1-85 cells under the restrictive temperature. We therefore followed the cellular distribution of GFP-DegAB in sis1-85 cells and found that in agreement with the requirement for ubiquitylation, GFP-DegAB levels increased in sis1-85 cells at the restrictive temperature, yet the protein remained soluble (Figure 6).
FIGURE 6:
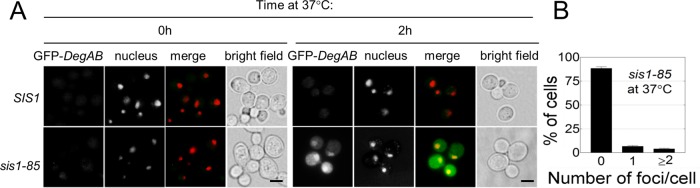
GFP-DegAB accumulates in a soluble form in cells with impaired Sis1 activity. (A) The indicated GFP-DegAB–expressing strains were grown to log phase at 24°C. Cells were kept at 24°C or shifted to 37°C for 2 h before fixation in paraformaldehyde. Nuclei were visualized by Hoechst staining (red). Green channel, GFP; red channel, Hoechst. (B) Quantitative assessment of GFP-DegAB–positive foci in sis1-85 cells, as illustrated in Figure 4B. n ≥ 3. Scale bar, 5 μm.
The formation of DegAB foci in ssa1/2Δ cells is dose dependent and displays an asymmetrical inheritance pattern during bud formation
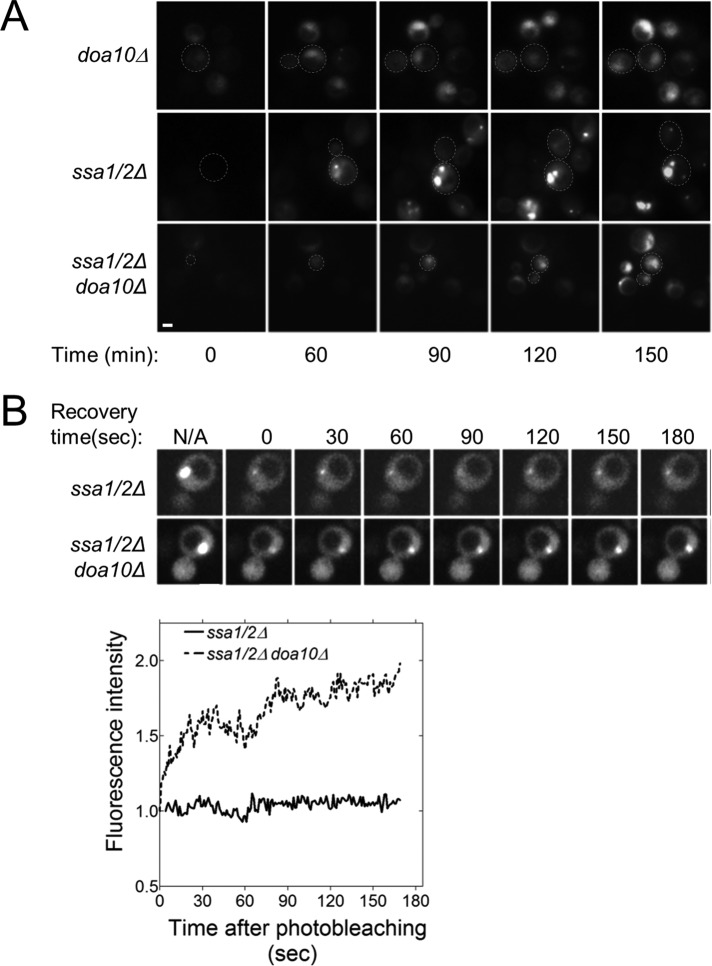
To investigate the dynamics of aggregate formation, we used time-lapse imaging of ssa1/2Δ cells expressing GFP-DegAB. Cells grown to stationary phase were diluted and subsequently followed by time-lapse microscopy. During the stationary phase TDH3 promoter activity, which controls GFP-DegAB transcription, is reduced, whereas shifting the cells to a logarithmic growth phase relieves this inhibition (Werner-Washburne et al., 1993). This mode of regulation of the TDH3 promoter enabled us to follow the kinetics of formation of nascent aggregates. Figure 7A and Supplemental Videos S1 and S2 show that the stationary population (time 0) displayed only a small portion of GFP-DegAB–derived aggregates and that aggregation increased when cells reached logarithmic phase. This accumulation of aggregates occurred in a dose-dependent manner. The substrate accumulated initially in the nucleus in a soluble form, whereas aggregates began to emerge only after a critical concentration was reached. Initially a single aggregate formed in a perinuclear inclusion, similar to the juxtanuclear quality control compartment (JUNQ; Kaganovich et al., 2008; Specht et al., 2011). Next peripheral inclusions started to appear. These “seeds” continued to grow and occasionally divided until multiple punctate foci were established. Yet these protein aggregates rarely moved to the daughter cells. In doa10Δ cells, substantially smaller fraction of the cells accumulated aggregates. The apparent punctate foci rarely expanded above a critical size and ultimately disappeared. The triple mutant lacking both DOA10 and SSA1/2 displayed similar distribution of GFP-DegAB as in doa10Δ cells.
FIGURE 7:
GFP-DegAB foci display asymmetric inheritance and differential mobility in ssa1/2Δ cells. (A) Time-dependent changes in the localization of GFP-DegAB. GFP-DegAB was visualized in the indicated strains by fluorescence microscopy. Images were collected from logarithmically growing cells at 5-min intervals. (B) Aggregates containing GFP-DegAB exhibit low mobility. Prebleach and postbleach images of a representative FRAP experiment and subsequent recovery of GFP-DegAB in the indicated strains are shown. Scale bars, 5 μm.
To gain additional insights into the dynamic properties of these aggregates we used the FRAP technique. Although no recovery of the GFP signal was observed in ssa1/2Δ cells at the indicated time intervals up to 3 min after photobleaching, about twofold increase in fluorescence intensity was detected in the sporadic aggregates formed in the triple-knockout cells (Figure 7B). Thus aggregates obtained in the absence of the E3 ligase show higher dynamics, which is in agreement with our working hypothesis that the capacity of these cells to dismantle preformed aggregates is enhanced (Supplemental Videos S1 and S2).
DISCUSSION
Sis1 functions as a misfolded protein–sorting factor
We show that Hsp40 cochaperone Sis1 is required for the ubiquitylation of proteins carrying the DegAB degron and for their subsequent degradation or sequestration (Figures 2, 3, and 6). This is the first time that Sis1 is directly implicated in PQCD by the Doa10 pathway. Previously Sis1 was largely associated with prion recognition (Sondheimer et al., 2001; Douglas et al., 2008; Tipton et al., 2008). It was suggested that it facilitates sorting of prions to stress-inducible PQC compartments containing other aggregation-prone model substrates, such as von Hippel–Lindau (Malinovska et al., 2012). Similarly, Sis1 and Ssa1 are associated with aggresomes of expanded PolyQ huntingtin domains in a yeast model (Wang et al., 2009). Our novel finding that Sis1 is specifically required for ubiquitylation of PQCD substrates by facilitating their binding to Doa10 E3 ligase is coincident with a recent report indicating a direct role for Sis1 in proteasomal degradation of a cytosolic substrate of the yeast PQCD E3 ligases Ubr1 and San1 (Summers et al., 2013).
Hsp70 levels determine the fate of ubiquitylated proteins
The constitutively expressed Ssa1 and Ssa2 chaperones were required for PQCD of Ndc10 and its derivatives, whereas the stress-inducible members of the SSA family, Ssa3 and Ssa4, were dispensable. This finding highlights the constitutive role of Hsp70s in maintaining cell proteostasis under physiological conditions.
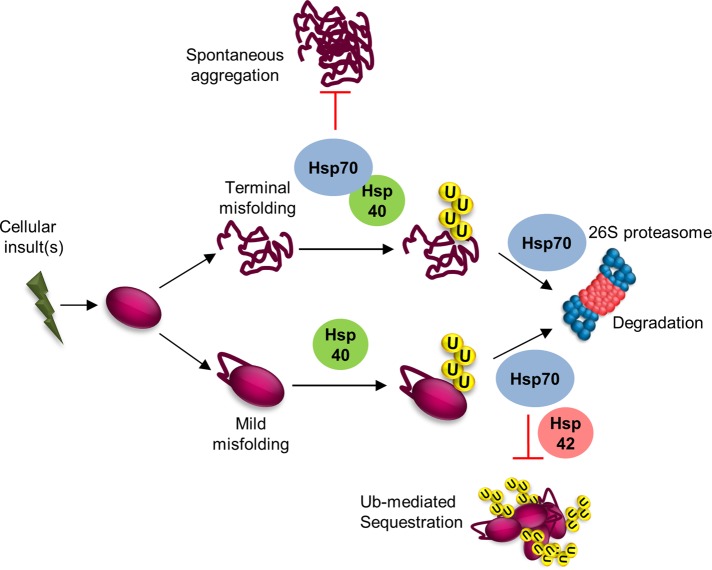
We found that Hsp70s were largely dispensable for the ubiquitylation of substrates harboring the DegAB degron (Figure 3). Our findings are different from various studies reporting requirement of Hsp70s for ubiquitylation of PQCD substrates. For example, the association of the E3 ubiquitin-ligase C-terminus of Hsc70-interacting protein with Hsp70 triggers ubiquitylation of its substrates (Murata et al., 2001), and the ubiquitylation of certain misfolded Doa10 substrates similarly requires Hsp70s (Han et al., 2007; Metzger et al., 2008; Nakatsukasa et al., 2008). In accordance with our findings, the ubiquitylation of the cytosolic model PQC substrate ΔssCPY* (signal sequence–deleted carboxypeptidase YGly255Arg) did not require the SSA family, whereas its aggregation was prevented by Ssa1 (Park et al., 2007). However, the connection between ubiquitylation and aggregation was not clarified. ΔssCPY* is likely to be defective mainly in its tertiary structure due to impaired disulfide bond formation (Endrizzi et al., 1994; Jamsa et al., 1994). Similarly, DegAB undergoes a mild perturbation that does not affect its secondary structure (Furth et al., 2011). It is therefore possible that the requirement for chaperones in the ubiquitylation pathway is determined by the severity of the lesion (Figure 8): severely misfolded substrates engage Ssa chaperones rapidly, before ubiquitylation, in order to maintain their solubility, whereas substrates with mild structural perturbation remain soluble, requiring chaperones activity only after ubiquitylation.
FIGURE 8:
A proposed model for the sorting of misfolded proteins to distinct PQCD pathways, depending on their folding and ubiquitylation states and the cellular levels of Hsp40/70 chaperones. Hsp40/70 activities are required for keeping terminally misfolded proteins in a soluble form, enabling their ubiquitylation. Presence of these chaperones in sufficient amounts prevents spontaneous aggregation. On the other hand, mildly/partially misfolded proteins do not require Hsp70 activity for maintaining their solubility. Still, Hsp40s are required for targeting these substrates to the ubiquitylation complex. Mildly misfolded ubiquitylated substrates still require Hsp70s for escorting them to the 26S proteasome, thereby preventing Ub-mediated, Hsp42-assisted sequestration.
Lack of Hsp70 requirement for Ub conjugation, although calling for further clarification, provided us with an unprecedented opportunity to assess the roles of Hsp70 downstream to ubiquitylation. We found that in ssa1/2Δ cells, ubiquitylated substrates were sequestered into distinct PQC foci (Figures 4, 5, and 7). This sequestration was dependent on ubiquitylation, since inhibition of ubiquitylation, whether by eliminating Sis1 or Doa10, resulted in the accumulation of the substrate in a soluble form, regardless of Ssa1/2 activity (Figures 5–7).
The specific role of Ssa1/2 downstream to ubiquitylation is obscure. It is unlikely that Hsp70s are required to specifically increase the solubility of the Ub-conjugated substrate, since, a priori, both conjugate moieties (the substrate and polyUb) are soluble, and, furthermore, Ub conjugation increases protein solubility (Catanzariti et al., 2004; Wang et al., 2012). Instead, it is plausible that Hsp70s are required for shuttling the ubiquitylated substrate to the proteasome. According to this hypothesis, by binding to the substrate moiety of the conjugate, Hsp70s prevent binding of sequestration factors and thus ensure delivery to the proteasome for degradation. Hence, in their absence, conjugates are sequestered. The findings of a physical interaction between Hsp70s and proteasomal Ub receptors are in accordance with this hypothesis (Guerrero et al., 2008; Gong et al., 2009).
Roles of ubiquitin in IBs formation
Ubiquitin is a prominent constituent of IBs in the vast majority of neurodegenerative diseases (Lehman, 2009). Previous studies suggest that ubiquitylation of misfolded proteins can lead to their aggregation in distinct quality control foci (Johnston et al., 1998; Kaganovich et al., 2008). In yeast, several misfolded proteins that undergo ubiquitylation are sequestered in a juxtanuclear PQC compartment, termed JUNQ, whereas nonubiquitylated, terminally misfolded proteins accumulated in a peripheral compartment termed IPOD (Kaganovich et al., 2008). However, the pattern of protein aggregation is likely to be more complex, as in many cases, the number of distinct aggregation foci emerging from misfolded protein accumulation is larger than these two compartments (Specht et al., 2011).
It is important to note that in the present study we used a model protein with structural properties distinct from those used for characterizing the IPOD and JUNQ compartments (Kaganovich et al., 2008; Specht et al., 2011). Whereas the previous studies largely relied on heterologously expressed human von Hippel–Lindau and PolyQ-expanded huntingtin proteins, both insoluble in yeast, we used a soluble, mildly misfolded substrate derived from a native yeast protein. This substrate colocalized in an Ub-dependent manner to Hsp42-positive foci in both juxtanuclear and peripheral aggregation compartments, where it showed low mobility. Furthermore, whereas the previous studies were carried out under environmental stress conditions, the present study was performed in cells with intact proteolytic activity, subjected to Hsp70 depletion. The novel findings emerging from our study can therefore be attributed to the different experimental conditions and substrates used. These findings imply that the depletion of Hsp70 chaperones directs mildly misfolded proteins into an alternative Ub-dependent sequestration pathway.
Recognition of ubiquitylated proteins by the proteasome is mediated by Ub receptors (Deveraux et al., 1994; Husnjak et al., 2008). Several studies show proteasome-unbound Ub-receptors, implying that they may act as proteasome-targeting factors (Matiuhin et al., 2008). Furthermore, Ub receptors are found in aggresomes of polyQ huntingtin domains (Wang et al., 2009), and several Ub-binding proteins, such as PLIC, ataxin3, and sequestosome1/p62, are implicated in misfolded-protein sequestration pathways (Donaldson et al., 2003; Burnett and Pittman, 2005; Heir et al., 2006). Moreover, the expression of isolated Ub-interacting motifs inhibits aggregation of PolyQ huntingtin in a cell model of Huntington disease (Miller et al., 2007), suggesting that Ub-binding proteins are actively involved in huntingtin sequestration and that overexpression of isolated Ub-interacting motifs can compete for this interaction. Thus our findings that ubiquitylation of misfolded PQCD substrates leads to their sequestration when Hsp70 levels limit degradation (Figures 3–5 and 7) may be explained by either active targeting via sequestration factors (as described earlier) or by competing interactions with alternative Ub receptors localized in PQC foci.
Implications for neurodegenerative diseases and aging
The misfolded-protein substrates used in this study are not cytotoxic, as indicated by the roughly similar growth rates of mutant and wild-type cell cultures, where the substrate is stable and rapidly degraded, respectively. Nonetheless, the ubiquitylated substrate aggregates observed in ssa1/2Δ cells are retained in the mother cell, as shown by time-lapse imaging of budding cells (Figure 7 and Supplemental Movie S1). This asymmetrical distribution between mother and daughter cells is characteristic of damaged and potentially toxic protein species, facilitating superior damaged-protein management in the progeny of Saccharomyces cerevisiae (Erjavec and Nystrom, 2007). This situation is analogous, albeit at a much longer time scale, to the time-dependent cytotoxicity typical of most neurodegenerative disorders. Because the most common risk factor for neurodegeneration is aging (Hebert et al., 1995; Cao et al., 2010), the time-dependent gain of cytotoxicity may be the result of gradual depletion of critical cellular factors, such as molecular chaperones, as well as proteasome subunits (Cummings et al., 1998; Chai et al., 1999; Stenoien et al., 1999; Waelter et al., 2001), which ensure removal of deleterious proteins rather than their accumulation in the form of IBs. Consistent with such a mechanism are the findings in Caenorhabditis elegans and Drosophila melanogaster of an age-dependent reduction of Hsp70 expression associated with diminished proteostasic capacity (Sørensen and Loeschcke, 2002; Ben-Zvi et al., 2009) and of decreased proteasome activity in aged mammalian brains (Keller et al., 2002).
Here we established an effective model system to study the molecular mechanism of misfolded-protein PQCD aggregation by limiting chaperone expression. Our findings strongly support the hypothesis that Ub is directly involved in the sequestration of mildly misfolded proteins (a working model is presented in Figure 8). Identifying Ub-dependent interactions and characterizing their interplay with the Hsp70/40 machinery may facilitate the elucidation of the mechanisms by which cellular decisions are made on whether to degrade or divert defective proteins to IBs.
MATERIALS AND METHODS
Yeast and bacterial methods
Yeast extract–peptone–dextrose (YPD)-rich media, synthetic dextrose (SD) minimal media, and lysogeny broth bacterial media were prepared using standard protocols. Standard methods were used for genomic manipulation of yeast and for recombinant DNA work.
Yeast strains
S. cerevisiae strains used in this study are listed in Supplemental Table S2. Unless indicated otherwise, the genetic background for yeast strains used in this study was that of W303 (α, his3-11,15, leu2-3,112, ura3-52, trp1-1, ade2-1, can 1-100). Deletion strains constructed in this study contain the KanMX cassette that was used to disrupt the ORF of the appropriate genes and are derived from the yeast knockout collection (Open Biosystems, Huntsville, AL).
Plasmids
Plasmids used in this study are listed in Supplemental Table S3. The plasmids were constructed using standard molecular biology techniques as described below. Primers for constructing plasmids and strains using PCR amplification techniques are available upon request.
To obtain plasmid pRS414 FLAG-Vma12-GFP-DegAB, a GFP fragment from plasmid pFa6a-GFP was PCR amplified and cloned into pJET, then cloned to pRS414GPDp-FLAG-Vma12-6His-DegAB at the AgeI and StuI sites. To obtain plasmid pRS414GPDp-GFP-DegAB, GFP-DegAB was generated by PCR amplification of pRS414-FLAG-Vma12-GFP-DegAB and cloned to pRS414GPDp CEN/TRP1 at the PstI and SalI sites.
GFP-DegABD10 and GFP-DegABDD were created by ligation of the corresponding fragments from pRS414-FLAG-Vma12-GFP-N90 and pRS414-FLAG-Vma12-GFP-DegABDD, respectively (Furth et al., 2011), into pRS414GPDp-GFP-DegAB at the AgeI and SalI sites.
Cycloheximide chase and immunoblotting
Unless otherwise indicated, all experiments were done at 30°C. Yeast cells were grown to log phase, then cycloheximide (0.5 mg/ml) was added, and aliquots from each time point were taken. Proteins were extracted by incubating cells with 0.1 N NaOH for 5 min at 23°C and spinning down the cells. The pellets were then dissolved in sample buffer and boiled at 95°C for 5 min. Samples were separated on SDS–PAGE gels (5–15% gradient), transferred to polyvinylidene fluoride membranes, and immunoblotted. The following antibodies were used: anti-Ndc10 (a gift from P. Meluh, Johns Hopkins University, Baltimore, MD), anti-GFP (Roche, Indianapolis, IN), anti–glucose-6-phosphate dehydrogenase (G6PD; Sigma-Aldrich, St. Louis, MO), anti-FLAG (Sigma-Aldrich), anti-actin (MP Biomedicals, Solon, OH), anti-hemagglutinin (anti-HA; Roche), anti-Doa10 antiserum from rabbit raised against a hexahistidine (His6)-tagged N-terminal 128-residue fragment of Doa10 (Kreft et al., 2006), and a rabbit polyclonal anti-Ubc6 antiserum. The rabbit antiserum was raised against the N-terminal 225-residue fragment of Ubc6 tagged by His6 at the C-terminus (1–225 aa). Proteins were visualized by enhanced chemiluminescence reaction.
Fluorescence imaging
Yeast cells transformed with indicated plasmids were grown to early log phase at 30°C (unless indicated otherwise) and fixed with 3.7% paraformaldehyde. Images were obtained by confocal microscopy using a Bio-Rad (Hercules, CA) MRC-1024 workstation attached to an Axiovert 135M microscope equipped with a 63×/1.4 objective (Carl Zeiss, Jena, Germany) or a DP71 camera mounted on an IX70 microscope with a 40×/1.6 oil objective (Olympus, Tokyo, Japan).
For live-cell imaging cells were immobilized using concanavalin A on glass-bottom tissue culture dishes (MatTek, Ashland, MA) and filmed using a charge-coupled device camera (Roper, Tucson, AZ) mounted on an Olympus IX70 microscope with a 60× oil objective. Imaging of cells was started immediately after dilution of stationary-phase cells. The entire setup was controlled by MetaMorph software (Molecular Devices, Sunnyvale, CA). ImageJ (National Institutes of Health, Bethesda, MD) was used for image processing, analysis, and assembly.
FRAP analysis
For FRAP analysis, yeast cells were grown to log phase and immobilized on concanavalin A–coated, glass-bottom culture dishes. Five individual cells for each strain were measured. Measurements were performed at 30°C by confocal microscopy using a Bio-Rad MRC-1024 workstation attached to a Zeiss Axiovert 135M microscope equipped with a 63×/1.4 objective. The resulting loss of fluorescence in the region of interest as a function of time provides a measure of the relative exchange rate with the bleached aggregate fraction of molecules.
Ubiquitylation analysis
Ubiquitylation assays were performed according to Loayza and Michaelis (1998). Yeast cells (∼20 A600 eq) coexpressing the indicated plasmids, together with a plasmid containing copper-induced Ub, were lysed by addition of 1.5 ml of 2 N NaOH/1 M β-mercaptoethanol. The lysates were incubated with 5% trichloroacetic acid. Proteins were separated by centrifugation at 17,000 × g for 10 min at 4°C, and the pellet was resuspended in 100 μl of sample buffer. Cell extracts were diluted 30-fold with buffer supplemented with protease inhibitors (Sigma-Aldrich) and 5 mM N-ethylmaleimide. Extracted proteins were immunoprecipitated with anti-GFP (Roche), followed by precipitation with rProtein A beads (RepliGen, Waltham, MA). Immunoblotting was done as described. Ubiquitylation was also tested in microsomes, which were prepared precisely as described by Bazirgan et al. (2006). Briefly, yeast cells (∼5 A600 eq) expressing the indicated plasmids were harvested and resuspended in 400 μl of ice-cold membrane fractionation buffer (20 mM Tris, pH 7.5, 0.1 M NaCl, 0.3 M sorbitol) with protease inhibitors. Glass beads were added, and lysis was conducted by high-speed vortexing. The resulting lysates were cleared by repeated 10-s microcentrifuge pulses to remove unlysed cells and large debris. The cleared supernatant contained microsome membranes, which were harvested by centrifugation at 17,000 × g for 30 min. Microsomal pellets were resuspended in SDS-sample buffer with 50 mM dithiothreitol (DTT) and diluted ∼30-fold in buffer supplemented with protease inhibitors (Sigma-Aldrich) and 5 mM N-ethylmaleimide. Extracted proteins were immunoprecipitated with anti-FLAG M2 affinity gel (Sigma-Aldrich), and immunoblotting was done as described.
Solubility analysis
Detergent solubility assay was adapted from Theodoraki et al. (2012). Yeast cells (∼5 A600 eq) at late logarithmic phase were harvested and lysed in 200 μl of lysis buffer (100 mM Tris, pH 7.5, 200 mM NaCl, 1 mM EDTA, 1 mM DTT, 5% glycerol, and 0.5% Triton X-100) with protease inhibitors (Sigma-Aldrich) by vortexing for 5 min at 4°C with glass beads. Repeated 10-s microcentrifuge pulses cleared the resulting lysates. A 50-μl amount of lysate, representing the “total lysate,” was removed and added to 50 μl of SUMEB (8 M urea, 1% SDS, 10 mM 3-(N-morpholino)propanesulfonic acid, pH 6.8, 10 mM EDTA, 0.01% bromphenol blue). The remaining lysate was centrifuged at 17,000 × g for 15 min. A 100-μl amount of supernatant was added to 100 μl of SUMEB. The pellet was resuspended in 100 μl of lysis buffer plus 100 μl of SUMEB. Proteins were immunoblotted as described.
Coimmunoprecipitation analysis
Coimmunoprecipitation analysis was done precisely as described by Kreft and Hochstrasser (2011). Briefly, yeast cells (∼20 A600 eq) were lysed in extraction buffer (50 mM Tris-HCl, pH 7.5, with protease inhibitors phenylmethylsulfonyl fluoride and aprotinin [Sigma-Aldrich]) by vortexing 5 min at 4°C with glass beads. The cleared, crude microsomal fraction was collected by centrifugation at 17,000 × g for 10 min and resuspended in 0.5 ml of resuspension buffer (RB; 50 mM Tris-HCl, pH 7.5, 200 mM NaAc, 10% glycerol with protease inhibitors [Sigma-Aldrich]). Membranes were solubilized by addition of digitonin (Calbiochem, La Jolla, CA) to 1%. The supernatant after centrifugation (16,000 × g for 10 min) was diluted 1:1 with RB. Extracted proteins were immunoprecipitated and immunoblotted as described.
Cell sorting and analysis by flow cytometry
Flow cytometry analysis and sorting were performed using a FACS Aria III cell sorter equipped with 488- and 531-nm lasers (BD Biosciences, San Jose, CA). Sorting criteria used were set to select the 5–10% highest percentile of fluorescent cells, with a minimum collection of 1 × 105 cells.
Supplementary Material
Acknowledgments
We thank O. S. Alfassy for the data in Figure 2B. We thank Y. Reiss for helpful discussions during this study and for critically reviewing the manuscript. We also thank Drora Zenvirth for technical assistance with time-lapse imaging. We are grateful to R. Kulka, D. Kaganovich, E. Craig, and J. L. Brodsky for strains or plasmids. This work was supported by Israel Science Foundation Grant 786/08, Marie Curie International Reintegration Grant MIRG-CT-2007-205425, and the Lejwa Fund for Biochemistry (T.R.).
Abbreviations used:
- CHX
cycloheximide
- ER
endoplasmic reticulum
- ERAD
endoplasmic reticulum–associated degradation
- FACS
fluorescence-activated cell sorting
- GFP
green fluorescent protein
- Hsp
heat shock protein
- IBs
inclusion bodies
- IP
immunoprecipitation
- PolyQ
polyglutamine
- PQC
protein quality control
- PQCD
protein quality control–associated degradation
- Ub
ubiquitin
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-01-0010) on May 1, 2013.
REFERENCES
- Alfassy OS, Cohen I, Reiss Y, Tirosh B, Ravid T. Placing a disrupted degradation motif at the C-terminus of proteasome substrates attenuates degradation without impairing ubiquitylation. J Biol Chem 288, 12645–12653. 2013 doi: 10.1074/jbc.M113.453027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazirgan OA, Garza RM, Hampton RY. Determinants of RING-E2 fidelity for Hrd1p, a membrane-anchored ubiquitin ligase. J Biol Chem. 2006;281:38989–39001. doi: 10.1074/jbc.M608174200. [DOI] [PubMed] [Google Scholar]
- Becker J, Walter W, Yan W, Craig EA. Functional interaction of cytosolic hsp70 and a DnaJ-related protein, Ydj1p, in protein translocation in vivo. Mol Cell Biol. 1996;16:4378–4386. doi: 10.1128/mcb.16.8.4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Zvi A, Miller EA, Morimoto RI. Collapse of proteostasis represents an early molecular event in Caenorhabditis elegans aging. Proc Natl Acad Sci USA. 2009;106:14914–14919. doi: 10.1073/pnas.0902882106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordallo J, Plemper RK, Finger A, Wolf DH. Der3p/Hrd1p is required for endoplasmic reticulum-associated degradation of misfolded lumenal and integral membrane proteins. Mol Biol Cell. 1998;9:209–222. doi: 10.1091/mbc.9.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchberger A, Bukau B, Sommer T. Protein quality control in the cytosol and the endoplasmic reticulum: brothers in arms. Mol Cell. 2010;40:238–252. doi: 10.1016/j.molcel.2010.10.001. [DOI] [PubMed] [Google Scholar]
- Bukau B, Weissman J, Horwich A. Molecular chaperones and protein quality control. Cell. 2006;125:443–451. doi: 10.1016/j.cell.2006.04.014. [DOI] [PubMed] [Google Scholar]
- Burnett BG, Pittman RN. The polyglutamine neurodegenerative protein ataxin 3 regulates aggresome formation. Proc Natl Acad Sci USA. 2005;102:4330–4335. doi: 10.1073/pnas.0407252102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao K, Chen-Plotkin AS, Plotkin JB, Wang L-S. Age-correlated gene expression in normal and neurodegenerative human brain tissues. PLoS One. 2010;5:e13098. doi: 10.1371/journal.pone.0013098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catanzariti AM, Soboleva TA, Jans DA, Board PG, Baker RT. An efficient system for high-level expression and easy purification of authentic recombinant proteins. Protein Sci. 2004;13:1331–1339. doi: 10.1110/ps.04618904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai Y, Koppenhafer SL, Shoesmith SJ, Perez MK, Paulson HL. Evidence for proteasome involvement in polyglutamine disease: localization to nuclear inclusions in SCA3/MJD and suppression of polyglutamine aggregation in vitro. Hum Mol Genet. 1999;8:673–682. doi: 10.1093/hmg/8.4.673. [DOI] [PubMed] [Google Scholar]
- Chen B, Retzlaff M, Roos T, Frydman J. Cellular strategies of protein quality control. Cold Spring Harb Perspect Biol. 2011;3:a004374. doi: 10.1101/cshperspect.a004374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernoff YO, Lindquist SL, Ono B, Inge-Vechtomov SG, Liebman SW. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+] Science. 1995;268:880–884. doi: 10.1126/science.7754373. [DOI] [PubMed] [Google Scholar]
- Cipriani G, Dolciotti C, Picchi L, Bonuccelli U. Alzheimer and his disease: a brief history. Neurol Sci. 2011;32:275–279. doi: 10.1007/s10072-010-0454-7. [DOI] [PubMed] [Google Scholar]
- Craig EA, Jacobsen K. Mutations of the heat inducible 70 kilodalton genes of yeast confer temperature sensitive growth. Cell. 1984;38:841–849. doi: 10.1016/0092-8674(84)90279-4. [DOI] [PubMed] [Google Scholar]
- Cummings CJ, Mancini MA, Antalffy B, DeFranco DB, Orr HT, Zoghbi HY. Chaperone suppression of aggregation and altered subcellular proteasome localization imply protein misfolding in SCA1. Nat Genet. 1998;19:148–154. doi: 10.1038/502. [DOI] [PubMed] [Google Scholar]
- Deveraux Q, Ustrell V, Pickart C, Rechsteiner M. A 26 S protease subunit that binds ubiquitin conjugates. J Biol Chem. 1994;269:7059–7061. [PubMed] [Google Scholar]
- Doheny KF, Sorger PK, Hyman AA, Tugendreich S, Spencer F, Hieter P. Identification of essential components of the S. cerevisiae kinetochore. Cell. 1993;73:761–774. doi: 10.1016/0092-8674(93)90255-O. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson KM, Li W, Ching KA, Batalov S, Tsai CC, Joazeiro CA. Ubiquitin-mediated sequestration of normal cellular proteins into polyglutamine aggregates. Proc Natl Acad Sci USA. 2003;100:8892–8897. doi: 10.1073/pnas.1530212100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas PM, Treusch S, Ren HY, Halfmann R, Duennwald ML, Lindquist S, Cyr DM. Chaperone-dependent amyloid assembly protects cells from prion toxicity. Proc Natl Acad Sci USA. 2008;105:7206–7211. doi: 10.1073/pnas.0802593105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endrizzi JA, Breddam K, Remington SJ. 2.8-A structure of yeast serine carboxypeptidase. Biochemistry. 1994;33:11106–11120. doi: 10.1021/bi00203a007. [DOI] [PubMed] [Google Scholar]
- Erjavec N, Nystrom T. Sir2p-dependent protein segregation gives rise to a superior reactive oxygen species management in the progeny of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2007;104:10877–10881. doi: 10.1073/pnas.0701634104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang NN, Ng AH, Measday V, Mayor T. Hul5 HECT ubiquitin ligase plays a major role in the ubiquitylation and turnover of cytosolic misfolded proteins. Nat Cell Biol. 2011;13:1344–1352. doi: 10.1038/ncb2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furth N, Gertman O, Shiber A, Alfassy OS, Cohen I, Rosenberg MM, Doron NK, Friedler A, Ravid T. Exposure of bipartite hydrophobic signal triggers nuclear quality control of Ndc10 at the endoplasmic reticulum/nuclear envelope. Mol Biol Cell. 2011;22:4726–4739. doi: 10.1091/mbc.E11-05-0463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Y, Kakihara Y, Krogan N, Greenblatt J, Emili A, Zhang Z, Houry WA. An atlas of chaperone-protein interactions in Saccharomyces cerevisiae: implications to protein folding pathways in the cell. Mol Syst Biol. 2009;5:275. doi: 10.1038/msb.2009.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero C, Milenkovic T, Przulj N, Kaiser P, Huang L. Characterization of the proteasome interaction network using a QTAX-based tag-team strategy and protein interaction network analysis. Proc Natl Acad Sci USA. 2008;105:13333–13338. doi: 10.1073/pnas.0801870105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S, Liu Y, Chang A. Cytoplasmic Hsp70 promotes ubiquitination for endoplasmic reticulum-associated degradation of a misfolded mutant of the yeast plasma membrane ATPase, PMA1. J Biol Chem. 2007;282:26140–26149. doi: 10.1074/jbc.M701969200. [DOI] [PubMed] [Google Scholar]
- Hartl FU, Hayer-Hartl M. Converging concepts of protein folding in vitro and in vivo. Nat Struct Mol Biol. 2009;16:574–581. doi: 10.1038/nsmb.1591. [DOI] [PubMed] [Google Scholar]
- Hebert LE, Scherr PA, Beckett LA, Albert MS, Pilgrim DM, Chown MJ, Funkenstein HH, Evans DA. Age-specific incidence of Alzheimer's disease in a community population. J Am Med Assoc. 1995;273:1354–1359. [PubMed] [Google Scholar]
- Heir R, Ablasou C, Dumontier E, Elliott M, Fagotto-Kaufmann C, Bedford FK. The UBL domain of PLIC-1 regulates aggresome formation. EMBO Rep. 2006;7:1252–1258. doi: 10.1038/sj.embor.7400823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hipp MS, Patel CN, Bersuker K, Riley BE, Kaiser SE, Shaler TA, Brandeis M, Kopito RR. Indirect inhibition of 26S proteasome activity in a cellular model of Huntington's disease. J Cell Biol. 2012;196:573–587. doi: 10.1083/jcb.201110093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holdorff B. Friedrich Heinrich Lewy (1885–1950) and his work. J Hist Neurosci. 2002;11:19–28. doi: 10.1076/jhin.11.1.19.9106. [DOI] [PubMed] [Google Scholar]
- Husnjak K, Elsasser S, Zhang N, Chen X, Randles L, Shi Y, Hofmann K, Walters KJ, Finley D, Dikic I. Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature. 2008;453:481–488. doi: 10.1038/nature06926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamsa E, Simonen M, Makarow M. Selective retention of secretory proteins in the yeast endoplasmic reticulum by treatment of cells with a reducing agent. Yeast. 1994;10:355–370. doi: 10.1002/yea.320100308. [DOI] [PubMed] [Google Scholar]
- Johnston JA, Ward CL, Kopito RR. Aggresomes: a cellular response to misfolded proteins. J Cell Biol. 1998;143:1883–1898. doi: 10.1083/jcb.143.7.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaganovich D, Kopito R, Frydman J. Misfolded proteins partition between two distinct quality control compartments. Nature. 2008;454:1088–1095. doi: 10.1038/nature07195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampinga HH, Craig EA. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat Rev. Mol Cell Biol. 2010;11:579–592. doi: 10.1038/nrm2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller JN, Gee J, Ding Q. The proteasome in brain aging. Ageing Res Rev. 2002;1:279–293. doi: 10.1016/s1568-1637(01)00006-x. [DOI] [PubMed] [Google Scholar]
- Kreft SG, Hochstrasser M. An unusual transmembrane helix in the Doa10 ERAD ubiquitin ligase modulates degradation of its cognate E2. J Biol Chem. 2011;286:20163–20174. doi: 10.1074/jbc.M110.196360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreft SG, Wang L, Hochstrasser M. Membrane topology of the yeast endoplasmic reticulum-localized ubiquitin ligase Doa10 and comparison with its human ortholog TEB4 (MARCH-VI) J Biol Chem. 2006;281:4646–4653. doi: 10.1074/jbc.M512215200. [DOI] [PubMed] [Google Scholar]
- Lehman NL. The ubiquitin proteasome system in neuropathology. Acta Neuropathol. 2009;118:329–347. doi: 10.1007/s00401-009-0560-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberek K, Marszalek J, Ang D, Georgopoulos C, Zylicz M. Escherichia coli DnaJ and GrpE heat shock proteins jointly stimulate ATPase activity of DnaK. Proc Natl Acad Sci USA. 1991;88:2874–2878. doi: 10.1073/pnas.88.7.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loayza D, Michaelis S. Role for the ubiquitin-proteasome system in the vacuolar degradation of Ste6p, the a-factor transporter in Saccharomyces cerevisiae. Mol Cell Biol. 1998;18:779–789. doi: 10.1128/mcb.18.2.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez N, Aron R, Craig EA. Specificity of class II Hsp40 Sis1 in maintenance of yeast prion [RNQ+] Mol Biol Cell. 2003;14:1172–1181. doi: 10.1091/mbc.E02-09-0593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinovska L, Kroschwald S, Munder MC, Richter D, Alberti S. Molecular chaperones and stress-inducible protein sorting factors coordinate the spatio-temporal distribution of protein aggregates. Mol Biol Cell. 2012;23:3041–3056. doi: 10.1091/mbc.E12-03-0194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matiuhin Y, Kirkpatrick DS, Ziv I, Kim W, Dakshinamurthy A, Kleifeld O, Gygi SP, Reis N, Glickman MH. Extraproteasomal Rpn10 restricts access of the polyubiquitin-binding protein Dsk2 to proteasome. Mol Cell. 2008;32:415–425. doi: 10.1016/j.molcel.2008.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClellan AJ. eLS. DOI: 10.1002/9780470015902.a0020886.pub2; 2012. Quality control of protein folding in the cytosol. [Google Scholar]
- Meriin AB, Zhang X, He X, Newnam GP, Chernoff YO, Sherman MY. Huntingtin toxicity in yeast model depends on polyglutamine aggregation mediated by a prion-like protein Rnq1. J Cell Biol. 2002;157:997–1004. doi: 10.1083/jcb.200112104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger MB, Maurer MJ, Dancy BM, Michaelis S. Degradation of a cytosolic protein requires endoplasmic reticulum-associated degradation machinery. J Biol Chem. 2008;283:32302–32316. doi: 10.1074/jbc.M806424200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SL, Scappini EL, O'Bryan J. Ubiquitin-interacting motifs inhibit aggregation of polyQ-expanded huntingtin. J Biol Chem. 2007;282:10096–10103. doi: 10.1074/jbc.M611151200. [DOI] [PubMed] [Google Scholar]
- Muchowski PJ, Wacker JL. Modulation of neurodegeneration by molecular chaperones. Nat Rev Neurosci. 2005;6:11–22. doi: 10.1038/nrn1587. [DOI] [PubMed] [Google Scholar]
- Murata S, Minami Y, Minami M, Chiba T, Tanaka K. CHIP is a chaperone-dependent E3 ligase that ubiquitylates unfolded protein. EMBO Rep. 2001;2:1133–1138. doi: 10.1093/embo-reports/kve246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsukasa K, Huyer G, Michaelis S, Brodsky JL. Dissecting the ER-associated degradation of a misfolded polytopic membrane protein. Cell. 2008;132:101–112. doi: 10.1016/j.cell.2007.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa S, Brodsky JL, Nakatsukasa K. Roles of molecular chaperones in endoplasmic reticulum (ER) quality control and ER-associated degradation (ERAD) J Biochem. 2005;137:551–555. doi: 10.1093/jb/mvi068. [DOI] [PubMed] [Google Scholar]
- Park S-H, Bolender N, Eisele F, Kostova Z, Takeuchi J, Coffino P, Wolf DH. The cytoplasmic Hsp70 chaperone machinery subjects misfolded and endoplasmic reticulum import-incompetent proteins to degradation via the ubiquitin-proteasome system. Mol Biol Cell. 2007;18:153–165. doi: 10.1091/mbc.E06-04-0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravid T, Kreft SG, Hochstrasser M. Membrane and soluble substrates of the Doa10 ubiquitin ligase are degraded by distinct pathways. EMBO J. 2006;25:533–543. doi: 10.1038/sj.emboj.7600946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum JC, et al. Disorder targets misorder in nuclear quality control degradation: a disordered ubiquitin ligase directly recognizes its misfolded substrates. Mol Cell. 2011;41:93–106. doi: 10.1016/j.molcel.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross CA, Pickart CM. The ubiquitin-proteasome pathway in Parkinson's disease and other neurodegenerative diseases. Trends Cell Biol. 2004;14:703–711. doi: 10.1016/j.tcb.2004.10.006. [DOI] [PubMed] [Google Scholar]
- Ross CA, Poirier MA. Protein aggregation and neurodegenerative disease. Nat Med. 2004;10(Suppl):S10–S17. doi: 10.1038/nm1066. [DOI] [PubMed] [Google Scholar]
- Sanchez Y, Parsell DA, Taulien J, Vogel JL, Craig EA, Lindquist S. Genetic evidence for a functional relationship between Hsp104 and Hsp70. J Bacteriol. 1993;175:6484–6491. doi: 10.1128/jb.175.20.6484-6491.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorter J, Lindquist S. Hsp104, Hsp70 and Hsp40 interplay regulates formation, growth and elimination of Sup35 prions. EMBO J. 2008;27:2712–2724. doi: 10.1038/emboj.2008.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sondheimer N, Lopez N, Craig EA, Lindquist S. The role of Sis1 in the maintenance of the [RNQ+] prion. EMBO J. 2001;20:2435–2442. doi: 10.1093/emboj/20.10.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Specht S, Miller SB, Mogk A, Bukau B. Hsp42 is required for sequestration of protein aggregates into deposition sites in Saccharomyces cerevisiae. J Cell Biol. 2011;195:617–629. doi: 10.1083/jcb.201106037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenoien DL, Cummings CJ, Adams HP, Mancini MG, Patel K, DeMartino GN, Marcelli M, Weigel NL, Mancini MA. Polyglutamine-expanded androgen receptors form aggregates that sequester heat shock proteins, proteasome components and SRC-1, and are suppressed by the HDJ-2 chaperone. Hum Mol Genet. 1999;8:731–741. doi: 10.1093/hmg/8.5.731. [DOI] [PubMed] [Google Scholar]
- Summers DW, Wolfe KJ, Ren HY, Cyr DM. The Type II Hsp40 Sis1 cooperates with Hsp70 and the E3 ligase Ubr1 to promote degradation of terminally misfolded cytosolic protein. PLoS One. 2013;8:e52099. doi: 10.1371/journal.pone.0052099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson R, Locher M, Hochstrasser M. A conserved ubiquitin ligase of the nuclear envelope/endoplasmic reticulum that functions in both ER-associated and Matalpha2 repressor degradation. Genes Dev. 2001;15:2660–2674. doi: 10.1101/gad.933301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen JG, Loeschcke V. Decreased heat-shock resistance and down-regulation of Hsp70 expression with increasing age in adult Drosophila melanogaster. Funct Ecol. 2002;16:379–384. [Google Scholar]
- Theodoraki MA, Nillegoda NB, Saini J, Caplan AJ. A network of ubiquitin ligases is important for the dynamics of misfolded protein aggregates in yeast. J Biol Chem. 2012;287:23911–23922. doi: 10.1074/jbc.M112.341164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tipton KA, Verges KJ, Weissman JS. In vivo monitoring of the prion replication cycle reveals a critical role for Sis1 in delivering substrates to Hsp104. Mol Cell. 2008;32:584–591. doi: 10.1016/j.molcel.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waelter S, Boeddrich A, Lurz R, Scherzinger E, Lueder G, Lehrach H, Wanker EE. Accumulation of mutant huntingtin fragments in aggresome-like inclusion bodies as a result of insufficient protein degradation. Mol Biol Cell. 2001;12:1393–1407. doi: 10.1091/mbc.12.5.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Li N, Wang Y, Wu Y, Mu T, Zheng Y, Huang L, Fang X. Ubiquitin-intein and SUMO2-intein fusion systems for enhanced protein production and purification. Protein Expr Purif. 2012;82:174–178. doi: 10.1016/j.pep.2011.11.017. [DOI] [PubMed] [Google Scholar]
- Wang Y, Meriin AB, Zaarur N, Romanova NV, Chernoff YO, Costello CE, Sherman MY. Abnormal proteins can form aggresome in yeast: aggresome-targeting signals and components of the machinery. FASEB J. 2009;23:451–463. doi: 10.1096/fj.08-117614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner-Washburne M, Braun E, Johnston GC, Singer RA. Stationary phase in the yeast Saccharomyces cerevisiae. Microbiol Rev. 1993;57:383–401. doi: 10.1128/mr.57.2.383-401.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner-Washburne M, Stone DE, Craig EA. Complex interactions among members of an essential subfamily of hsp70 genes in Saccharomyces cerevisiae. Mol Cell Biol. 1987;7:2568–2577. doi: 10.1128/mcb.7.7.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.