Trypanosomes lack many core components of ribonucleoprotein (RNP) granules identified in yeast and humans (e.g., DCP1/2). This study provides evidence for SCD6 being the core RNP granule component in trypanosomes: overexpression induces granules independent of translation, and even when SCD6 is targeted to the nucleus. Granule type and granule number are dependent on the RGG domain.
Abstract
Ribonucleoprotein (RNP) granules are cytoplasmic, microscopically visible structures composed of RNA and protein with proposed functions in mRNA decay and storage. Trypanosomes have several types of RNP granules, but lack most of the granule core components identified in yeast and humans. The exception is SCD6/Rap55, which is essential for processing body (P-body) formation. In this study, we analyzed the role of trypanosome SCD6 in RNP granule formation. Upon overexpression, the majority of SCD6 aggregates to multiple granules enriched at the nuclear periphery that recruit both P-body and stress granule proteins, as well as mRNAs. Granule protein composition depends on granule distance to the nucleus. In contrast to findings in yeast and humans, granule formation does not correlate with translational repression and can also take place in the nucleus after nuclear targeting of SCD6. While the SCD6 Lsm domain alone is both necessary and sufficient for granule induction, the RGG motif determines granule type and number: the absence of an intact RGG motif results in the formation of fewer granules that resemble P-bodies. The differences in granule number remain after nuclear targeting, indicating translation-independent functions of the RGG domain. We propose that, in trypanosomes, a local increase in SCD6 concentration may be sufficient to induce granules by recruiting mRNA. Proteins that bind selectively to the RGG and/or Lsm domain of SCD6 could be responsible for regulating granule type and number.
INTRODUCTION
Ribonucleoprotein (RNP) granules, membrane-free RNP particles, are central to the posttranscriptional regulation of gene expression. The cytoplasmic RNP granules include processing bodies (P-bodies), which are constitutively present and contain mRNA degradation enzymes, as well as various types of stress granules, which contain components of the translation initiation machinery (Anderson and Kedersha, 2009).
Although these two types of RNP granules are broadly classified as foci of mRNA decay machinery or as storage particles, the precise functions and composition of most RNP granules are still poorly defined, and much effort has focused on identifying specific core components as entry to an understanding of RNP granule function. By RNA interference (RNAi) depletion of individual granule components, several proteins have been identified in mammals that are interdependently necessary for correct P-body formation, namely GW182, Ccr4, Lsm1, Lsm4, RCK/p54, eIF4E-T, RAP55, and Ge-1/Hedls (Jakymiw et al., 2007). A similar redundancy of core P-body components was observed in yeast (Teixeira and Parker, 2007). In addition to the identified proteins, micro-RNAs (miRNAs) and mRNAs were found to be necessary for P-body formation (Jakymiw et al., 2007).
However, as P-bodies are formed by translationally repressed RNPs, RNAi experiments do not distinguish between core granule-building components, which are directly necessary for P-body formation, and proteins that could be involved in maintaining the pool of nontranslated mRNAs, for example, by repressing translation. To date, only two proteins have been identified as “true” P-body core proteins, based on the fact that their depletion resulted not only in loss of granule formation but did so without affecting translational repression that occurs concomitantly with P-body–inducing glucose deprivation. One is the Lsm protein EDC3; the other is the Lsm4 protein, a subunit of the cytoplasmic Lsm1-7 complex involved in mRNA decapping (Decker et al., 2007). The Q/N-rich domain of Lsm4 appears to be involved in granule formation by self-aggregation via a prion-like mechanism. The Q/N-rich domain of a yeast prion protein can compensate for the equivalent domain of Lsm4 in granule formation. At least 20 P-body proteins contain Q/N-rich domains (Reijns et al., 2008). Self-aggregating domains are also involved in stress granule formation (Anderson and Kedersha, 2008, 2009); for example, the prion-related Q-rich domain of TIA-1/TIAR (Kedersha et al., 1999; Gilks et al., 2004). Recently, low-complexity domains of RNA-binding proteins were identified to be necessary and sufficient for RNP granule assembly induced in vitro by biotinylated isoxazole (Kato et al., 2012).
One protein shown to be essential for P-body formation in vertebrates (Tanaka et al., 2006; Yang et al., 2006), plants (Xu and Chua, 2009), and trypanosomes (Kramer et al., 2012), although not in Drosophila (Eulalio et al., 2007b) or Caenorhabditis elegans (Audhya et al., 2005), is the Lsm domain containing protein RAP55/SCD6. It was initially discovered as a component of cytoplasmic RNP particles in the salamander Pleurodeles waltl (Lieb et al., 1998). The protein and its localization to P-bodies is highly conserved in eukaryotes (Lieb et al., 1998; Audhya et al., 2005; Boag et al., 2005; Wilhelm et al., 2005; Barbee et al., 2006; Squirrell et al., 2006; Tanaka et al., 2006; Yang et al., 2006; Pepling et al., 2007; Kramer et al., 2008; Xu and Chua, 2009; Mair et al., 2010). In Xenopus and C. elegans, expression of the RAP55/SCD6 orthologues is restricted to germ cells and early embryos (Lieb et al., 1998; Audhya et al., 2005; Boag et al., 2005; Tanaka et al., 2006), while in Drosophila and humans, it is expressed in both germ and somatic cells (Barbee et al., 2006; Yang et al., 2006; Eulalio et al., 2007a, b). RAP55/SCD6 consists of an N-terminal Lsm domain, a C-terminal FDF motif, several RGG motifs (Marnef et al., 2009), and, in some homologues, Q/N-rich sequences (Supplemental Figure S1). RAP55/SCD6 acts as a repressor of translation (Marnef et al., 2009), likely mediated by an interaction between the RGG motif in RAP55/SCD6 and eIF4G (Rajyaguru et al., 2012). In addition to eIF4G, several other proteins have been identified as interacting with RAP55/SCD6, for instance, the RNA helicase DHH1, also known as CGH-1 or p54 (Audhya et al., 2005; Boag et al., 2005; Wilhelm et al., 2005; Tanaka et al., 2006; Tritschler et al., 2009; Nissan et al., 2010; Matsumoto et al., 2012); the decapping enzyme DCP1-DCP2 (Tritschler et al., 2008; Nissan et al., 2010; Fromm et al., 2012); the decapping activator Pat1 (Nissan et al., 2010); and the arginine methyltransferases PRMT1 and PRMT5 (Tanaka et al., 2006; Matsumoto et al., 2012).
The high degree of conservation of SCD6 throughout the eukaryotes, together with its conserved localization to P-bodies and interactions with translation initiation factors and proteins involved in decapping, suggests that SCD6 plays a central role in regulating the formation of RNP granules. A model suggesting that SCD6 might contribute to RNP granule formation by forming a heptameric ring, via interactions between the Lsm domain in a similar manner to the Lsm1-7 or Lsm2-8 complexes, is attractive but currently without supporting evidence. Tra1, the Drosophila orthologue of SCD6, does not form multimers in vitro, and coimmunoprecipitation studies have not revealed interactions of Tra1 with itself or proteins of the Lsm1-7 complex (Tritschler et al., 2008).
Trypanosomes have a large number of different RNP granules, a possible reflection of their almost complete reliance on posttranscriptional regulation of gene expression (De Gaudenzi et al., 2011; Kramer, 2012). At least four different types can be distinguished: P-body–like structures (Cassola et al., 2007; Holetz et al., 2007; Kramer et al., 2008), starvation stress granules (Cassola et al., 2007), heat shock stress granules (Kramer et al., 2008), and nuclear periphery granules (NPGs), which resemble perinuclear germ granules and form after inhibition of trans-splicing (Kramer et al., 2012). Orthologues of many proteins proposed to have core functions in P-body or stress granule formation in metazoan and yeast are not readily identifiable in the trypanosome genome; for example, the P-body components eIF4E-T, EDC3, DCP1-DCP2, GW182, and Ge-1/Hedls or the stress granule components TTP, G3BP, TIAR/TIA-1, and CPEB. The cytoplasmic Lsm1-7 complex is also absent, because trypanosomes lack LSM1 (Liu et al., 2004). The only cytoplasmic Lsm domain protein that can be readily identified in the trypanosome genome is SCD6. The protein localizes to P-bodies (Kramer et al., 2008) and is essential for P-body formation (Kramer et al., 2012).
In this paper, we provide evidence that SCD6 is the P-body protein responsible for granule assembly in trypanosomes, a role possibly related to the absence of other Lsm domain proteins in the cytoplasm. When overexpressed, SCD6-eYFP (enhanced yellow fluorescent protein) aggregates into granules that corecruit other P-body proteins, as well as poly(A)-binding protein, eIF4G5, and mRNAs. Granule formation does not correlate with a repression of translation, and our data provide evidence that SCD6 may recruit its mRNA targets mainly from the nonpolysomal mRNA pool. In fact, granules can also form when SCD6 is targeted to the nucleus. Granule type and number is dependent on the presence of an RGG motif, seemingly in a translation-independent manner. We propose that the formation of P-bodies and P-body–related granules in trypanosomes is largely determined by the local concentration of SCD6 and mRNA.
RESULTS
Overexpressed SCD6 aggregates into cytoplasmic granules enriched at the nuclear periphery
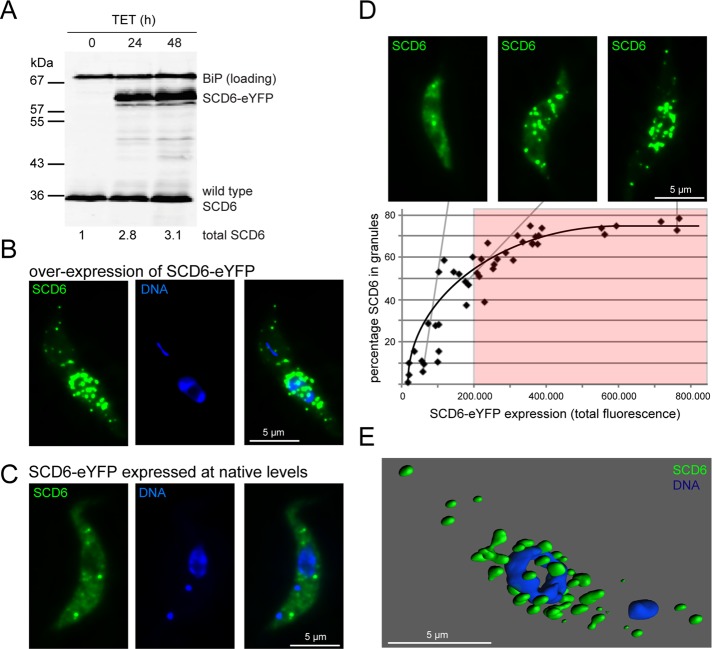
To investigate the role of SCD6 in trypanosome RNP granule formation, we aimed to overexpress an SCD6-eYFP transgene in a procyclic cell line, using the tetracycline-based expression system described by Wirtz et al. (1999) and Sunter et al. (2012). In the absence of tetracycline, no SCD6-eYFP was detectable by Western blotting, while after 24 and 48 h of induction, SCD6-eYFP was present at about twice the wild-type level (Figure 1A), indicating successful overexpression. The majority of SCD6-eYFP was localized to numerous (typically >20) cytoplasmic granules (Figure 1B). The degree of granular localization is a consequence of overexpression, because expression of SCD6-eYFP from its endogenous locus at native expression levels resulted in only a minor fraction of SCD6-eYFP present in a few (<10) P-bodies (Kramer et al., 2008; Figure 1C).
FIGURE 1:
Overexpression of SCD6. (A) Quantitative Western blot over a time course of tetracycline (TET)-induced expression of SCD6-eYFP, probed for SCD6 and, as a loading control, BiP (Bangs et al., 1993). Total SCD6 expression levels are indicated. (B) Localization of SCD6-eYFP after 24 h of overexpression. (C) Localization of SCD6-eYFP expressed from the endogenous locus and thus at native expression levels. (D) Percentage of SCD6-eYFP localized to granules compared with total SCD6-eYFP expression level. Representative images are shown. The threshold fluorescence intensity of 200,000 is indicated, which was used in subsequent experiments to define cells as highly overexpressing SCD6-eYFP (at least threefold). (E) Surface-rendered three-dimensional model of a cell with high SCD6 overexpression (green) and nucleus stained with DAPI (blue). Note that in trypanosomes, DAPI also stains the kinetoplast, the genome of the single mitochondrion. Depending on the cell cycle phase, one or two blue spots can be seen in addition to the larger nucleus.
The expression levels of SCD6-eYFP varied between individual cells more than two orders of magnitude, even though the population was clonal, an effect that had been previously reported (Sunter et al., 2012). This enabled the fraction of SCD6-eYFP localized to granules to be analyzed as a function of expression level (Figure 1D). At low expression levels, only a minor fraction of SCD6-eYFP was localized to granules, and the number and size of the granules were similar to P-bodies marked by expression of fluorescently labeled P-body markers at endogenous levels. With increasing expression, the percentage of SCD6-eYFP in granules increased up to ∼75%, and granule number increased from <10 in cells with low SCD6-eYFP expression up to 45 in cells with high expression. At higher expression levels, granules were not equally distributed throughout the cytoplasm but appeared enriched in regions adjacent to the nucleus. Three-dimensional modeling revealed that all granules were extranuclear (Figure 1E and Supplemental Movie S1).
The total amount of endogenous SCD6 protein present in a trypanosome procyclic cell was estimated to be around 60,000 molecules, using a titration of recombinant SCD6 protein (Figure S4). This is an amount similar to the number of mRNA molecules (Dhalia et al., 2006).
The variation in SCD6-eYFP expression between cells prevented an accurate determination of the expression level necessary for granule induction. However, considering that two thirds of the cells were expressing SCD6-eYFP at concentrations high enough to induce granules, and the mean overexpression level is around threefold the endogenous expression (Figure 1A), we can estimate that a threefold overexpression is sufficient for the induction of granules. Therefore, in all subsequent microscopy experiments, an arbitrary SCD6-eYFP expression threshold corresponding to a total fluorescence intensity of 200,000 (at identical image acquisition conditions), which safely corresponds to at least a threefold overexpression of SCD6, was used to define cells “highly” overexpressing SCD6.
The fusion protein SCD6-eYFP was shown to be functional, as a cell line with one SCD6 allele deleted and the second allele replaced with SCD6-eYFP is viable (Figure S2), whereas the lack of SCD6 is lethal, as previously shown by RNAi depletion (Kramer et al., 2012). Moreover, comparable expression levels of eYFP alone did not result in granule formation (Figure S3A). For further confirmation that induction of granules was not an artifact of the eYFP tag, wild-type SCD6 protein was overexpressed. We were unable to detect SCD6 granules by immunofluorescence. One possible reason is that the granule structure prevents antibody access. We have previously found that some types of trypanosome RNP granules are very difficult to detect with antibodies, for instance, P-bodies (Kramer et al., 2008), while others can be detected easily (Kramer et al., 2012). However, overexpression of wild-type SCD6 in a cell line also expressing the P-body marker protein DHH1 as an mChFP transgene caused localization of mChFP-DHH1 into granules (Figure S3B), similar to that seen with overexpression of SCD6-eYFP did (see results below in Figure 3, A–D). We therefore assume that the phenotypes observed after SCD6-eYFP and SCD6 overexpression are identical.
FIGURE 3:
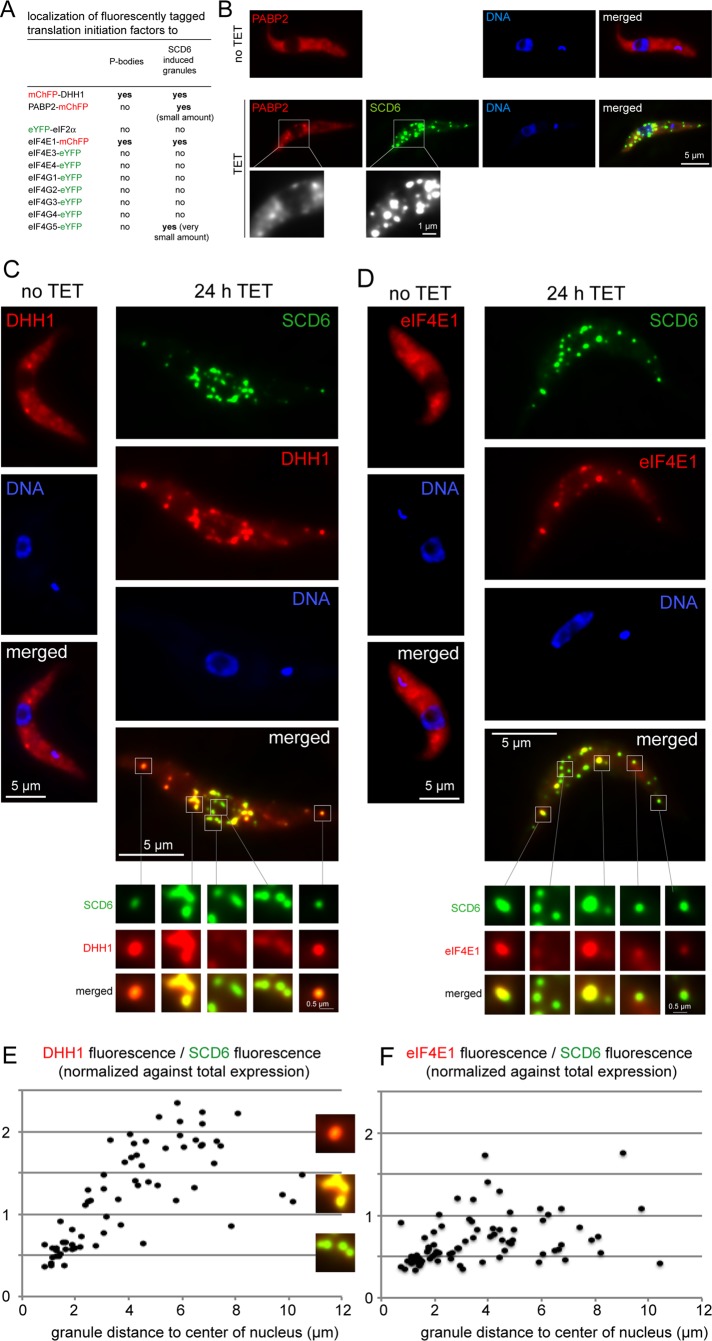
Other components of SCD6-induced granules. (A) List of proteins tested for localization to SCD6-induced granules; localization to P-bodies is also indicated. (B–D) Localization of PABP2-mChFP, mChFP-DHH1, and eIF4E1-mChFP to SCD6-induced granules. Regions of interest are enlarged. (E and F) For an average of 15 randomly selected granules per cell, taken from five individual cells, the content of SCD6-eYFP and mChFP-DHH1 (E) or eIF4E1-mChFP (F) was quantified. The quotient was plotted against granule distance to the center of the nucleus. Cells with an SCD6-eYFP expression level below threshold (see legend for Figure 1D) or more than one nucleus were excluded from the analysis.
Overexpression of SCD6 does not cause major reductions in growth, translation, or global mRNA levels within 24 h
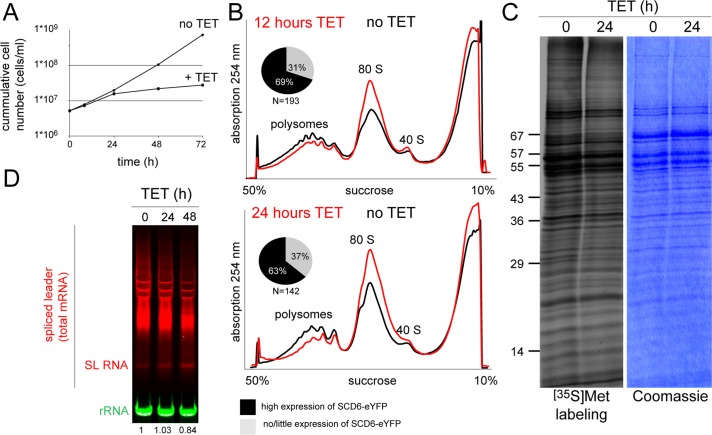
The aggregation of overexpressed SCD6 into granules could be either a direct effect of the increase in protein concentration or a secondary effect caused by translational repression. To distinguish between these two possibilities, we monitored growth, translation, and steady-state mRNA levels over a time course of SCD6-eYFP induction (Figure 2). Cell proliferation was normal for the first 24 h, by which time the SCD6-eYFP granules had formed (Figure 2A), although there was an almost complete growth arrest at later time points. At 24 h after induction, there was only a small reduction in overall translation, monitored by either polysome gradients (Figure 2B) or [35S]methionine incorporation (Figure 2C). In trypanosomes, total mRNA can be quantified by probing a Northern blot for the mini-exon, the part of the spliced leader (SL) RNA that is trans-spliced to the 5′ end of all trypanosome mRNAs. There was no significant change in total steady-state mRNA levels up to 48 h of SCD6 overexpression (Figure 2D).
FIGURE 2:
Effects of SCD6 overexpression on growth, translation, and mRNA levels. Expression of SCD6-eYFP was induced by tetracycline (TET). (A) Growth of cells in the absence and presence of SCD6-eYFP expression. (B) Polysome profiling. The percentage of cells with SCD6-eYFP expression levels high enough for granule induction was determined in parallel by fluorescence microscopy. (C) Autoradiograph and Coomassie-stained gel of cells labeled with [35S]methionine. (D) A Northern blot loaded with total RNA was probed with an oligo antisense to the mini-exon, the part of the SL RNA that is trans-spliced to all trypanosome mRNAs, to detect total mRNA and for rRNA (loading). The mini-exon signal was quantified (numbers at the bottom). All experiments were performed at least three times (twice for D).
The small reduction in translation after 24 h of SCD6 overexpression is not sufficient to account for the formation of the amount of RNP granules observed, even considering that the unequal expression levels of SCD6-eYFP between cells result in a minor underestimation of the effects. In comparison, heat shock granule formation is accompanied by an almost complete dissociation of polysomes (Kramer et al., 2008). Thus the assembly of SCD6-eYFP into multiple granules is likely to be a direct effect of the overexpression and not caused by repression of translation.
Longer induction of SCD6 overexpression caused a proliferation arrest accompanied by a cell cycle phenotype similar to the one obtained after RNAi depletion of the mitotic cyclin CYC6 (Hammarton et al., 2003). Briefly, there is an increase in cells with the abnormal karyotypes 1K2N and 1K0N (K = number of kinetoplasts; N = number of nuclei) that are most likely siblings from a cytokinesis that has occurred in the absence of nuclear division. This phenotype is likely to be a secondary effect of the general proliferation arrest and the possible change in gene expression resulting from the change in RNA granules. The mitotic block may dominate the phenotype, because mitosis may be most sensitive to minor changes in gene expression. This phenotype is described in Figure S5.
Recruitment of other proteins to the SCD6-induced granules is dependent on a granule´s distance from the nucleus
To characterize the granules induced by SCD6 overexpression, we examined whether they recruited proteins associated with translation. Several proteins were expressed as eYFP fusion proteins from their endogenous loci in cell lines containing an inducible SCD6-CerFP transgene; others were expressed as mChFP fusion proteins in cell lines containing an inducible SCD6-eYFP transgene (Figure 3A). Of the proteins tested, only DHH1, eIF4E1, PABP2, and eIF4G5 were detected in SCD6-induced granules (Figures S6 and 3, A–D). It must be noted, however, that nonrecruitment could be due to impaired functionality of the eYFP or mChFP fusion proteins. Only a small fraction of PABP2 (Figure 3B) and eIF4G5 (Figure S6) localized to SCD6-induced granules, while a larger fraction of the two P-body proteins DHH1 and eIF4E1 was present in granules.
A conspicuous difference between the recruitment of the two P-body markers was that DHH1 was enriched in granules distant from the nucleus (Figure 3C and Movie S1), whereas the distribution of eIF4E1 was very similar to that of SCD6-eYFP (Figure 3D). This observation was quantified for individual granules by dividing DHH1 or eIF4E1 content by SCD6 content and relating these values to granule distance to the nucleus (Figure 3, E and F). The amount of DHH1 relative to SCD6 rose significantly with increasing distance from the nucleus. These data indicate that the protein composition of the granules is not homogenous, but gradually changes, depending on distance to the nucleus. Whether this affects other proteins in addition to DHH1 remains to be seen.
SCD6 granules close to the nucleus are sensitive to inhibition of transcription
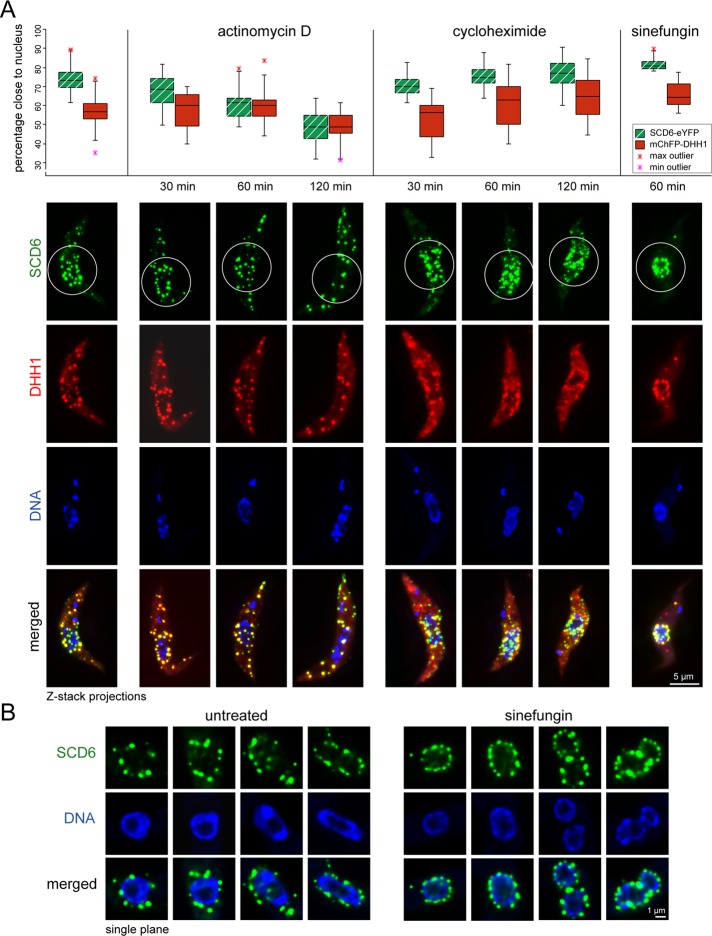
Interference with either translation or transcription affects different types of RNP granules in a specific way, providing some information about their possible functions. To further characterize the granules that formed after induction of SCD6-eYFP expression, we used a cell line that constitutively expressed mChFP-DHH1 from the modified endogenous locus in addition to the inducible SCD6-eYFP transgene. After 24 h of induction, cultures were incubated with either actinomycin D, to inhibit transcription, or cycloheximide, a drug that causes a reduction of P-bodies and an increase of polysomes. Over a time course of 120 min, actinomycin D caused the SCD6-eYFP granule distribution to change from perinuclear enrichment to a more even dispersal throughout the cytoplasm (Figure 4A). This change was less obvious for mChFP-DHH1, as DHH1 was already preferentially present in the distant granules (Figure 3, C and E). The percentage of SCD6-eYFP and mChFP-DHH1 within the perinuclear region was determined in the presence and absence of actinomycin D. The perinuclear region was defined as a 7.1-μm circle around the nuclear center. In untreated cells, an average of 73.3% ± 7% of total SCD6-eYFP and 56.6% ± 8% of mChFP-DHH1 was localized close to the nucleus (Figure 4A). In the presence of actinomycin D, the percentage of SCD6-eYFP in perinuclear granules decreased to 67% ± 10%, 60% ± 9%, and 48% ± 10% at 30, 60, and 120 min respectively, while there was no significant change in the distribution of mChFP-DHH1. After 60 or 120 min of actinomycin D treatment, there was no significant difference between the percentage of SCD6-eYFP and mChFP-DHH1 contained in perinuclear granules (Figure 4A).
FIGURE 4:
Effects of actinomycin D, cycloheximide, and sinefungin on the proximity of SCD6-induced granules to the nucleus. Cells were induced to express SCD6-eYFP for 24 h and were treated with drugs as indicated. (A) The percentage of SCD6-eYFP and mChFP-DHH1 within a 7.1-μm circle centered at the nucleus (= close to nucleus) is shown as box plots (waist is median; box is interquartile range (IQR); whiskers are ±1.5 IQR; only the smallest and largest outliers are shown; n > 10). Cells with an SCD6-eYFP expression level below threshold (see legend to Figure 1C) and cells with two clearly separated nuclei were excluded. Images of representative cells with the 7.1-µm circle indicated are shown. (B) Single-plane images of deconvolved Z-stacks of untreated cells and cells treated with sinefungin (60 min), focused on the plane with the maximum number of granules close to the nucleus.
In contrast, treatment with cycloheximide caused no significant change in the percentage of SCD6-eYFP close to the nucleus and only a minor increase from 57% ± 8% to 64% ± 11% of the perinuclear mChFP-DHH1 fraction after 120 min of incubation. The fraction of SCD6-eYFP in perinuclear granules remained significantly higher than the fraction of mChFP-DHH1 up to 120 min of cycloheximide treatment (Figure 4A).
Taken together, these data indicate that granules close to the nucleus are sensitive to inhibition of transcription but not to an inhibition of translation, and may therefore contain newly transcribed mRNAs.
The induction of transcription-sensitive RNP granules concentrated in a perinuclear compartment is reminiscent of NPGs that form in trypanosomes upon inhibition of trans-splicing (Kramer et al., 2012). In trypanosomes, trans-splicing can be inhibited by sinefungin, an S-adenosylmethionine analogue that prevents methylation of the cap of the spliced leader RNA. In the presence of transcription, inhibition of trans-splicing results in the accumulation of polycistronic mRNAs, which partially leak into the cytoplasm (Kramer et al., 2012). This causes the formation of NPGs. The function of NPGs remains unknown, but one possibility is that they act as a novel cytoplasmic compartment to assess mRNA quality and prevent immature mRNAs from entering translation. NPGs have striking similarities to nucleus-associated germ granules found in gonads of adult animals. Similarities include protein composition (e.g., VASA), localization, dependency on transcription, and stability in the presence of cycloheximide (Eddy and Ito, 1971; Mahowald, 1971; Mahowald and Hennen, 1971; Strome and Wood, 1982, 1983; Hay et al., 1988a, b; Sheth et al., 2010). Both granule types may have a similar evolutionary origin. To investigate whether SCD6-induced granules resemble NPGs, we incubated cells that had been induced for 24 h with the trans-splicing inhibitor sinefungin. There was a significant increase from 73.3% ± 7 to 82% ± 3% of SCD6-eYFP in the perinuclear region (Figure 4A). Single-plane images of deconvolved Z-stacks of nuclei from several untreated and sinefungin-treated cells were compared (Figure 4B). Although NPGs are generally more uniform in size and more evenly distributed around and closer to the nucleus than SCD6-induced granules, the perinuclear subset of SCD6-induced granules resemble NPGs in granule pattern and sensitivity to actinomycin D.
SCD6-induced granules contain mRNA
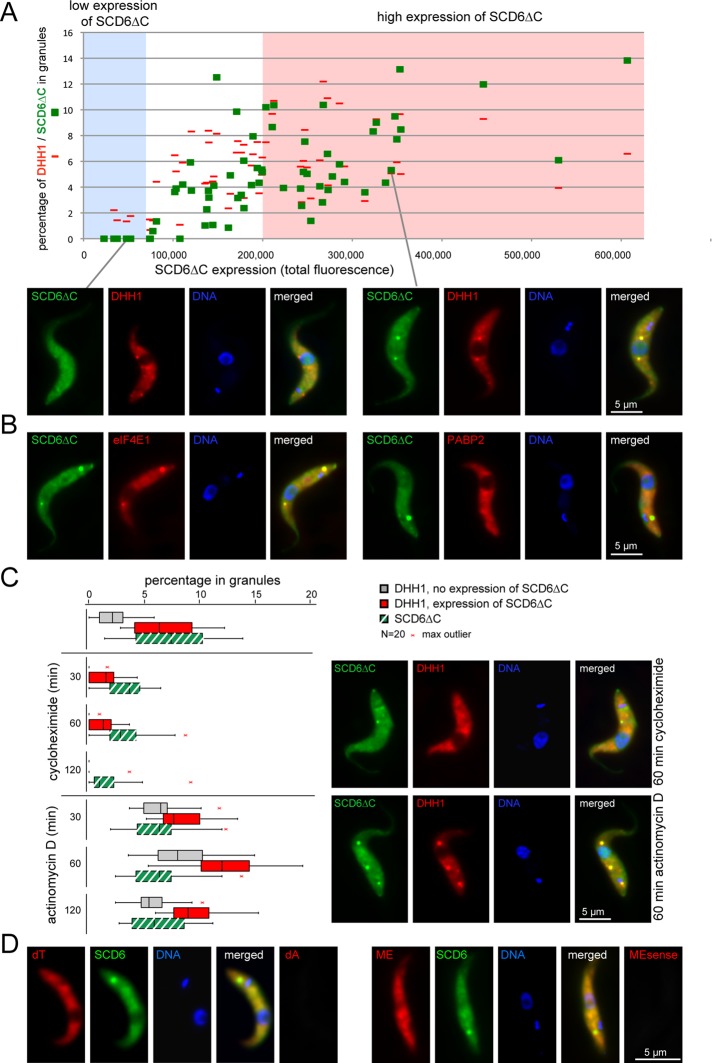
The mRNA content of granules formed after SCD6-eYFP induction was examined by fluorescence in situ hybridization (FISH), using cy3-labeled oligos directed against the poly(A) tail (dT) or the mini-exon sequence (ME) that is trans-spliced to the 5′ end of every trypanosome mRNA. Prior to induction, both oligos were evenly distributed throughout the cytoplasm, whereby the ME oligo also detected an RNA in the nucleus, which is most likely the SL RNA (Kramer et al., 2012). After induction, mRNA was detected in all granules containing SCD6-eYFP, both in the absence and presence of actinomycin D (Figure 5).
FIGURE 5:
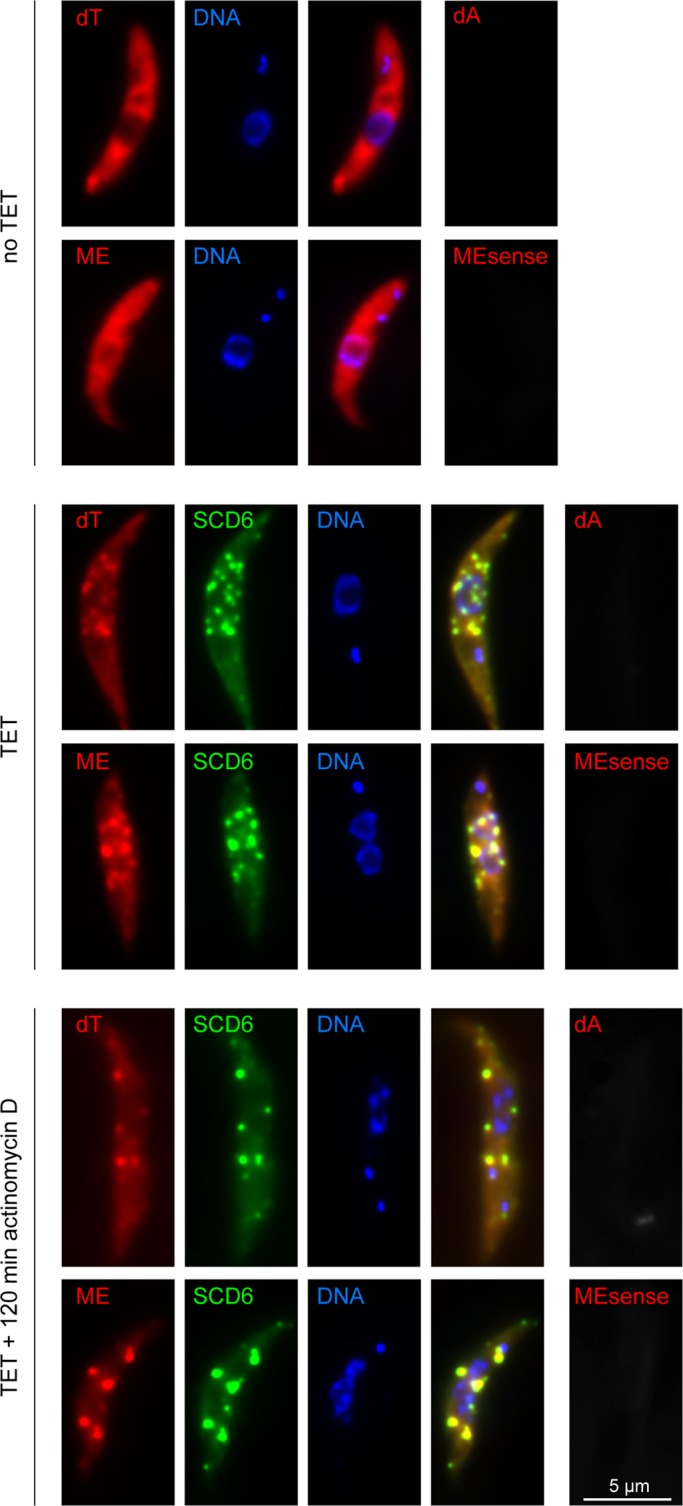
mRNAs in SCD6-induced granules. In situ hybridization of untreated cells (no TET) and cells expressing SCD6-eYFP for 24 h in the absence (TET) and presence of actinomycin D (TET + 120 min actinomycin D). mRNAs were detected by oligo dT or by an oligo antisense to the mini-exon (ME). Sense oligos (dA, MEsense) served as controls and gave significantly weaker signals.
The data suggest that the granules formed after SCD6 induction contain intact mRNAs, although it cannot be ruled out that they contain a mixture of partially 5′–3′ degraded mRNAs and partially 3′–5′ degraded mRNAs. Moreover, mRNA is present in both the transcription- and translation-sensitive populations of SCD6-induced RNP granules. The presence of mRNAs with poly(A) tails and mini-exons indicates that the granules may be involved in storage rather than degradation. This is supported by the observation that total mRNA levels were not altered in SCD6-overexpressing cells (Figure 2D).
The Lsm domain of SCD6 is necessary and sufficient for granule induction, but the RGG motif determines granule number
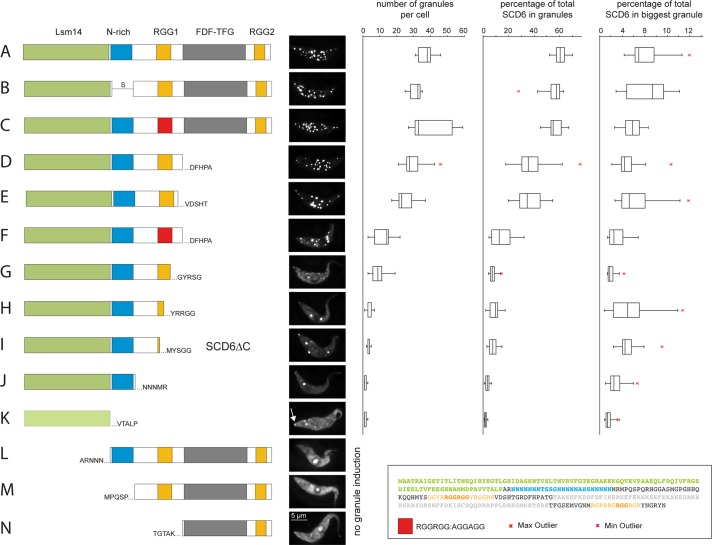
A structure-informed deletion analysis was performed to investigate which part of SCD6 is sufficient for granule induction. In total, transgenes encoding 13 different mutants were inducibly expressed in a cell line containing a mChFP-DHH1 transgene at the endogenous locus, resulting in constitutive expression. The mutants included several truncations, as well as a deletion of the N-rich domain and a mutation of one of the RGG motifs (Figure 6). As already observed for the wild-type protein, the expression levels of the SCD6 mutants were nonuniform, and only cells with the previously defined total expression level of >200,000 were included in the microscopic analysis. First, the ability of each mutant to induce granules at overexpression was tested. This was defined as both the ability of the SCD6-eYFP mutant to localize to granules (Figure 6) and an increase in the percentage of mChFP-DHH1 in granules (data not shown). Granules formed after expression of all transgenes containing the Lsm domain, including the Lsm domain alone (Figure 6K). None of the three mutants lacking the Lsm domain localized to or formed granules. Thus the Lsm domain of SCD6 is necessary and sufficient for the formation of granules.
FIGURE 6:
Deletion analysis of SCD6. The localization of SCD6-eYFP mutant proteins was monitored 24 h after induction of expression. Granule parameters were quantified for all mutants that localized to granules and are presented as box plots (waist is median; box is IQR; whiskers are ±1.5 IQR; only the smallest and largest outliers are shown; n > 10). Cells with subthreshold expression level of SCD6-eYFP were excluded. The arrow points to SCD6-eYFP mutant protein localized at the posterior pole.
The number of granules formed after transgene induction varied significantly between the different SCD6 mutants. This was quantified by measuring 1) the number of granules, 2) the percentage of total SCD6-eYFP mutant protein in granules, and 3) the percentage of total SCD6-eYFP mutant protein in the largest granule. Induction of wild-type SCD6-eYFP expression for 24 h resulted in an average of 38 granules containing ∼60% of the protein (Figure 6A). Neither the removal of the N-rich domain nor the mutation of the N-terminal RGG box (RGG1) caused a significant change in either granule number or the percentage localized to granules (Figure 6, B and C). Deletion of the FDF-TFG domain and the C-terminal RGG box (RGG2) resulted in a minor decrease in the number of granules to ∼25–30, and the percentage of SCD6 localizing to granules decreased to ∼37% (Figure 6, D and E). Deletion of the FDF-TFG domain and the RGG2 box combined with mutation of the RGG1 box resulted in a decrease in granule number to an average of 12, and only ∼15% of SCD6 was localized to granules. Thus absence of the RGG1 box significantly interferes with granule induction, but only in the absence of the C-terminus: the RGG2 box might compensate for the mutated RGG1 box. Successive deletions of RGG1 resulted in a further decrease in both the number of granules and in the percentage of the mutant SCD6-eYFP localizing to granules (Figure 6, G–K). Mutations of SCD6 mainly affected granule number, while granule size was in a similar range compared with wild type. There were two exceptions that had significantly smaller granules than the wild type (Figure 6, G and K). It is possible that the truncation interfered with protein folding in these cases. These data show that the Lsm domain is both necessary and sufficient for SCD6 to localize to and induce granules, but the additional presence of an intact RGG motif is required for the induction of many granules, as opposed to only a few.
We observed that certain SCD6 truncations formed granules located at the posterior pole of the cell (e.g., Figure 6K, arrow); mChFP-DHH1 was not recruited to granules in this position (data not shown). This localization resembles that of XRNA, the trypanosome orthologue to XRN1, during heat shock (Kramer et al., 2008) and suggests an association with the ends of the microtubules in the subpellicular array that underlies the plasma membrane in trypanosomes (Robinson et al., 1995). It is possible that this association with the microtubule cytoskeleton is normally transient but becomes visible under the experimental conditions. This was not further examined in this study, but the possible role of the microtubules’ association of SCD6 in P-body formation is discussed in Discussion.
Granules formed after overexpression of a C-terminally truncated SCD6 resemble P-bodies
The differences between the few large granules formed by overexpression of SCD6 mutants without an RGG domain and the many granules formed by overexpression of wild-type SCD6 were investigated. The “few granules” were induced by the C-terminally truncated SCD6 (Figure 6I), hereafter named SCD6∆C.
While wild-type SCD6-eYFP localized to granules even at endogenous expression levels (Figure 1C; Kramer et al., 2008) and well below an expression level of 50,000 (fluorescence intensity units, Figure 1D), the SCD6∆C-eYFP expression level had to exceed a threshold value of 80,000 before it localized to granules. mChFP-DHH1 was clearly visible in P-bodies at lower expression levels of SCD6∆C-eYFP, indicating that granule formation per se was not disturbed (Figure 7A). As expression levels increased, the percentage of SCD6∆C-eYFP in granules increased, and there was an almost parallel increase in the fraction of mChFP-DHH1 in granules, confirming that SCD6∆C-eYFP not only localized to granules but also recruited other proteins. The P-body protein eIF4E1 was detectable in SCD6∆C-granules but not the stress granule marker protein PABP2, in contrast to wild-type SCD6-induced granules, which include both (Figure 7B). This suggests that the composition of the SCD6∆C granules may resemble that of P-bodies.
FIGURE 7:
Characterization of granules induced by SCD6∆C. SCD6∆C-eYFP expression was induced for 24 h. (A) Percentage of SCD6∆C-eYFP and mChFP-DHH1 localizing to granules as a function of total SCD6∆C-eYFP expression level. Representative images are shown. (B) Localization of eIF4E1-mChFP and PABP2-mChFP upon SCD6∆C-eYFP expression. (C) Localization of SCD6∆C-eYFP and mChFP-DHH1 to granules after treatment with cycloheximide or actinomycin D. mChFP-DHH1 localization to granules was also determined in the absence of SCD6∆C-eYFP expression. Data are shown as box plots (waist is median; box is IQR; whiskers are ±1.5 IQR; only the smallest and largest outliers are shown; n = 20). Cells with a subthreshold expression level of SCD6∆C-eYFP were excluded. Representative images are shown. (D) Detection of total mRNAs in cells expressing SCD6∆C-eYFP for 24 h by in situ hybridization using oligo dT or an oligo antisense to the mini-exon (ME). Sense oligos (dA, MEsense) served as controls and gave significantly weaker signals.
Cells constitutively expressing mChFP-DHH1 were treated with cycloheximide and actinomycin before and after induction of SCD6∆C-eYFP expression (Figure 7C), and the fraction of both proteins in granules was quantified. Before induction, 2% of mChFP-DHH1 was in P-bodies, this percentage increased on incubation with actinomycin D and decreased with cycloheximide, as described previously (Cassola et al., 2007; Holetz et al., 2007; Kramer et al., 2008, 2012). The response of mChFP-DHH1 localization to incubation with actinomycin D or cycloheximide was similar before and after induction of SCD6∆C-eYFP expression, albeit the fraction in granules was generally higher after induction.
The percentage of SCD6∆C-eYFP in granules was reduced by cycloheximide but remained unaffected by actinomycin D treatment, suggesting that the granules are not dependent on active transcription but are in equilibrium with translating polysomes, a behavior that is similar to that of P-bodies. Just as in P-bodies, granules formed after SCD6∆C-eYFP induction did not contain any mRNAs detectable by the same FISH procedure used above (Figure 7D). It is likely that the granules contain little or no mRNA or that any mRNA entering the granules is rapidly degraded. It must also be considered that the granule structure might prevent the penetration of the DNA probe.
In conclusion, the granules formed after induction of SCD6∆C-eYFP resemble P-bodies in number, composition, sensitivity to cycloheximide, and apparent absence of mRNAs.
We further analyzed the localization of SCD6∆C-eYFP to other RNP granule types. After 1 h of sinefungin treatment, only a very small percentage of SCD6∆C-eYFP localized to NPGs (Figure S7A, compare Figure 4A for wild-type SCD6). Similar results were obtained for another SCD6∆C-eYFP truncation (SCD6∆C*; Figure 6J and S7B). Thus the Lsm domain does not mediate localization to NPGs. In contrast, an SCD6 mutant lacking the Lsm domain and the N-rich region (Figure 6M; SCD6∆N) localized to NPGs, although it did not localize to any other cytoplasmic granules (Figure S7C). Thus different requirements exist for the recruitment to NPGs or to other cytoplasmic granules, an observation consistent with P-body but not NPG formation, being dependent on SCD6 (Kramer et al., 2012). Localization to carbon source–starvation stress granules obtained by culturing the cells in phosphate-buffered saline (PBS; Cassola et al., 2007) was similar to wild-type SCD6 for C-terminally truncated SCD6 and only slightly reduced for the N-terminally truncated version (Figure S7). In comparison with localization to other RNP granules, the localization to starvation stress granules appears to be the least restricted, as almost any RNA-binding protein tested has the ability to localize there (unpublished data). It seems probable that the localization to starvation stress granules occurs via many parallel pathways.
SCD6 and SCD6∆C induce nucleoplasmic granules when targeted to the nucleus
Induction of SCD6∆C-eYFP expression caused only a few granules to form, whereas induction of wild-type SCD6-eYFP expression resulted in tens of granules per cell. One possible explanation for this difference could be that wild-type SCD6 is able to induce granules de novo, while SCD6∆C can only increase the size of the few existing P-bodies. If this were true, SCD6∆C would be unable to induce granules in the absence of wild-type SCD6. This was tested by simultaneously inducing expression of SCD6∆C-eYFP and RNAi knockdown of endogenous SCD6. However, even though a major reduction in endogenous SCD6 protein occurred (Figure S8A), SCD6∆C-eYFP still localized to granules (Figure S8B).
Because it is possible that the small fraction of endogenous SCD6 that remains present despite RNAi is sufficient to nucleate P-bodies, a different strategy was used. SCD6∆C-eYFP was targeted to the nucleus, where there is little or no wild-type SCD6 (Kramer et al., 2008). The previously defined NLS (nuclear localization signal) of the La protein (Marchetti et al., 2000) was added to the N-terminus of SCD6-eYFP or SCD6∆C-eYFP, and the fusion proteins were overexpressed in a cell line expressing SCD6-mChFP from the endogenous loci. A similar strategy has been used to study the interactions between mammalian P-body components (Bloch et al., 2011). Both NLS-SCD6-eYFP and NLS-SCD6∆C-eYFP localized to the nucleus, while SCD6-mChFP remained cytoplasmic, indicating the nucleus was free of endogenously expressed SCD6 (Figure 8, A and B). The experiments were repeated in a cell line expressing mChFP-DHH1 instead of SCD6-mChFP from the endogenous locus. After induction, both NLS-SCD6-eYFP and NLS-SCD6∆C-eYFP localized to the nucleus, while mChFP-DHH1 remained cytoplasmic, indicating that no complete P-bodies were coimported into the nucleus (data not shown).
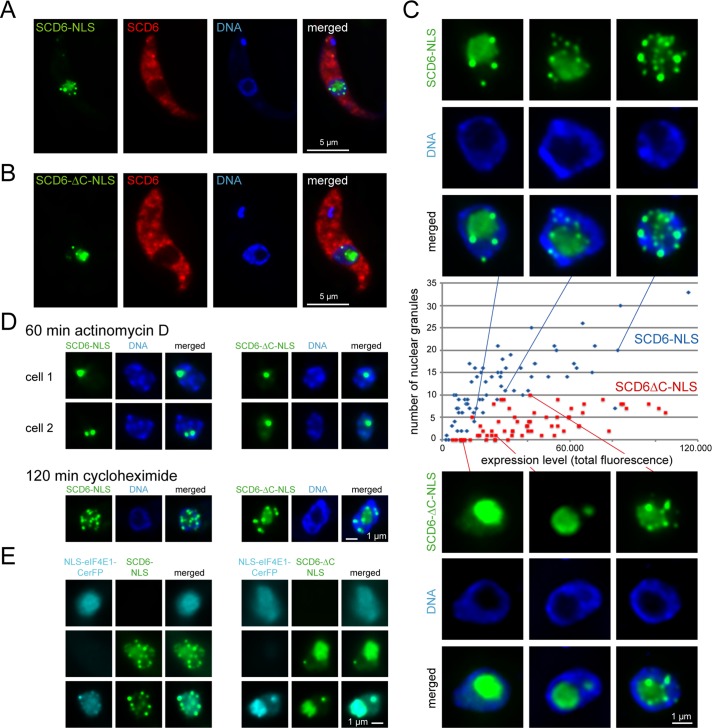
FIGURE 8:
Granule formation after nuclear targeting. NLS-SCD6-eYFP or NLS-SCD6∆C-eYFP was expressed for 4-6 h in cells also expressing SCD6-mChFP from the endogenous locus. (A and B) Representative images. (C) Granule number as a function of expression level (= total fluorescence intensity). Nuclei of representative cells are shown. (D) Nuclei of cells treated with actinomycin D or cycloheximide. Two representative cells (cell 1 and cell 2) are shown for the actinomycin D treatment (E) Expression of NLS-eIF4E1-CerFP together with NLS-SCD6-eYFP (left) or NLS-SCD6∆C-eYFP (right). Nuclei from cells that express either protein alone (top and middle panel) or both proteins together (bottom panel) are shown. The bottom panel is shown as single-plane image rather than as a Z-stack projection for increased clarity.
NLS-SCD6-eYFP and NLS-SCD6∆C-eYFP both accumulated in distinct granules in the nucleoplasm and in the nucleolus (Figure 8, A and B). This finding differs from the equivalent experiment performed with the mammalian SCD6 orthologue, which did not aggregate to nuclear granules and was excluded from the nucleoli (Bloch et al., 2011). Granule formation in the nucleoplasm resembled the cytoplasmic process: the number of granules increased with the increase of transgene expression, and SCD6∆C-eYFP required a higher threshold expression level for aggregation into granules than did the wild-type protein (Figure 8C).
When transcription was inhibited by actinomycin D, nucleoplasmic granules disappeared, and nuclear fluorescence was restricted to one or two small spots that possibly correspond to the remains of the nucleolus (Figure 8D). Cycloheximide did not cause any change in granules (Figure 8D). When a CerFP fusion of eIF4E1 was targeted to the nucleus in a similar way, it did not aggregate into granules, although it is possible that the expression level was too low for granule formation (Figure 8E). When either NLS-SCD6-eYFP or NLS-SCD6∆C-eYFP was expressed at the same time as NLS-eIF4E1-CerFP, part of NLS-eIF4E1-CerFP colocalized with the granules in the nucleoplasm (Figure 8E).
Taken together, the data show that both wild-type SCD6 and SCD6∆C can induce the formation of RNP granules in an “artificial” environment free of wild-type SCD6 and DHH1 and probably other P-body proteins. The granules are dependent on active transcription and are therefore most likely to recruit RNA, and they can recruit at least one other granule protein.
DISCUSSION
To understand RNP granule function, it is essential to understand granule assembly and identify the core granule components. The search for P-body core proteins in yeast and animals has revealed an unexpected redundancy (Jakymiw et al., 2007; Teixeira and Parker, 2007). Trypanosomes seem well suited for the study of RNP granule assembly, as they lack orthologues of many essential P-body core proteins, such as EDC3, DCP1-DCP2, and Lsm1-7. SCD6 is the only cytoplasmic Lsm domain protein, and its depletion by RNAi results in P-body loss (Kramer et al., 2012).
In this work, we have examined the role of trypanosomal SCD6 in RNP granule formation. Upon overexpression, SCD6 aggregated into RNP granules with features of storage granules, in an expression level–dependent way. In contrast to the situation in yeast (Rajyaguru et al., 2012), our data provide evidence that SCD6 recruits mRNAs mainly from nonpolysomal fractions, and in contrast to the situation in human cells (Bloch et al., 2011), SCD6 also aggregates to granules in the nucleoplasm when targeted there. While the Lsm domain is necessary and sufficient for granule induction, the RGG motif determines whether SCD6 aggregates to multiple storage granules or to a few granules resembling P-bodies. Importantly, the differences in granule number remain after targeting SCD6 to the nucleus, indicating that the function of the RGG motif is translation independent. Our work provides evidence that, in the absence of many other P-body proteins, SCD6 may have evolved to be the P-body core protein in trypanosomes.
Preferential recruitment of nontranslated mRNAs to SCD6-induced granules
Overexpression of SCD6 also results in RNP granule induction in human cells and yeast (Matsumoto et al., 2012; Rajyaguru and Parker, 2012), indicating some conservation in SCD6 function. While SCD6 is a well-known translational repressor (Tanaka et al., 2006; Nissan et al., 2010; Rajyaguru and Parker, 2012) and recruits mRNAs at translation initiation via binding to eIF4G (Rajyaguru et al., 2012), our data indicate that the mRNAs recruited to SCD6-induced granules in trypanosomes are not primarily polysomal mRNAs. Granule formation is faster than translational repression; furthermore, the majority of granules are insensitive to cycloheximide. The enrichment of granules close to the nucleus, as well as the sensitivity of these perinuclear granules to inhibition of transcription, suggests that a large fraction of mRNAs in the SCD6 granules are newly transcribed. The ability of SCD6 to aggregate to granules in the nucleus proves that SCD6 can aggregate to granules independent of translation.
We propose that, upon overexpression, trypanosome SCD6 recruits mRNAs nonselectively. Trypanosomes exert almost no transcriptional control, and the majority of cytoplasmic mRNAs are nonpolysomal (Kramer et al., 2010). The preferential recruitment of nontranslated mRNAs to SCD6-induced granules may therefore reflect the total distribution of cytoplasmic mRNAs in trypanosomes. In yeast, transcription is regulated, and the majority of cytoplasmic mRNAs are in translation (Arava et al., 2003), which is a plausible reason for mRNAs mainly being recruited from polysomes.
Spatial differences in granule composition
DHH1 and eIF4E1 are corecruited to the SCD6-induced granules. While eIF4E1 localizes to all granules in amounts similar to SCD6, DHH1 is enriched in granules distant from the nucleus and is almost absent from granules close to the nucleus. Assuming that granules close to the nucleus preferentially contain newly transcribed mRNAs, as is suggested by their sensitivity to actinomycin D, the simplest explanation is that eIF4E1 binds to newly transcribed mRNAs, while DHH1 does not. In fact, eIF4E1 is present in both the cytoplasm and the nucleus (Kramer et al., 2008; Freire et al., 2011), indicating that it may bind its mRNA targets before reaching the cytoplasm, while DHH1, a regulator of translation in trypanosomes (Kramer et al., 2010) and elsewhere (Minshall et al., 2009; Carroll et al., 2011; Sweet et al., 2012), does not localize to the nucleus in trypanosomes (Kramer et al., 2008).
The function of the RGG domain
The Lsm domain of trypanosome SCD6 is fully sufficient for the induction of RNP granules, but one intact RGG domain is required for the induction of many RNP granules with features of storage granules, while the absence of an RGG domain results in the induction of fewer RNP granules with P-body–like features. Thus the RGG domain determines the type of RNP granule. A similar finding has recently been reported in yeast. Saccharomyces cerevisiae SCD6 induces both P-bodies and stress granules upon overexpression, while a truncation lacking the RGG domain (SCD6∆C) induces only P-bodies (Rajyaguru et al., 2012). While SCD6∆C is not impaired in mRNA binding, its ability to repress translation is reduced, and the truncated protein cannot interact with eIF4G (Rajyaguru et al., 2012). Binding of SCD6 to eIF4G is likely to mediate SCD6-induced translational repression by preventing the binding of the 43S initiation complex (Rajyaguru et al., 2012, Nissan et al., 2010). Therefore the RGG domain of SCD6 seems to be essential for translational repression at the initiation stage via eIF4G binding and for mRNA recruitment to storage granules. It may prevent mRNA degradation by hindering access of DCP2 to the cap (Rajyaguru et al., 2012). SCD6∆C must recruit its mRNA targets, if any, from another source, possibly from translating polysomes.
A significant novel finding of this work is that, in trypanosomes, the RGG domain appears to function independent of translation or any other factor restricted to the cytoplasm. The differences between granules induced by either wild-type SCD6 or SCD6∆C remain when the proteins are targeted to the nucleus. The data are not necessarily contradictory to the findings in yeast, if one assumes that SCD6 binds to eIF4G-mRNA complexes already in the nucleus. In fact, eIF4G has important nuclear functions in pre-mRNA processing in humans (McKendrick et al., 2001) and yeast (Kafasla et al., 2009).
The minimal requirement for an RNP granule
There is no evidence for SCD6 forming multimers in Drosophila (Tritschler et al., 2008), and it is likely not do so in trypanosomes, as we could not detect any interactions between SCD6 molecules by either coimmunoprecipitation or FRET (fluorescence resonance energy transfer) (unpublished data). Furthermore, the removal of the N-rich region, a candidate sequence for prion-like aggregation (Decker et al., 2007; Reijns et al., 2008), did not prevent RNP granule induction. Therefore RNP granule aggregation is likely to require other factors in addition to SCD6. The sensitivity of both the perinuclear and nuclear SCD6 granules to actinomycin D indicates a dependency on RNA, possibly mRNA, but other RNA types cannot be excluded. The formation of RNP granules in either the cytoplasm or the nucleus shows that RNP granule formation requires neither exclusively nuclear nor cytoplasmic components. Therefore, it is possible that SCD6 and RNA are fully sufficient for the formation of RNP granules. However, we cannot completely rule out that cytoplasmic proteins are corecruited to the nucleus by SCD6, even though both proteins tested in this work, DHH1 and endogenous SCD6, remained cytoplasmic. There also remains the possibility that nuclear–cytoplasmic shuttling proteins are required.
A model of granule formation
One SCD6 molecule has at least two RNA-binding sites: the Lsm domain and the RGG domain. In addition, it can bind to several mRNA-binding proteins, for instance, eIF4G (Rajyaguru and Parker, 2012), of which trypanosomes have five isoforms. This multivalence enables SCD6 to act as a connector between two mRNA molecules (either directly or via binding to a molecule bound to the mRNA) and can result in the assembly of a network of mRNAs and SCD6. Other proteins may be “passively” corecruited to the granules, because they are bound to the recruited mRNA. At endogenous expression levels, with about one SCD6 molecule per mRNA, the concentration of involved molecules and binding constants result in the formation of only a few small granules. An increase or decrease in the amount of either the available mRNA molecules or the mRNA linker protein SCD6 will shift the equilibrium and result in an increase or decrease of granules. Experimentally, drugs that interfere with translation can produce changes in the concentration of available mRNAs and RNAi, or overexpression can change the concentration of SCD6. Under natural conditions, changes in the concentration of available mRNAs can occur via stress-induced polysomal repression, for example, stress induced by heat shock (Kramer et al., 2008). Changes in SCD6 protein concentration can occur by a change in SCD6 expression or by a subcellular increase of SCD6 mediated by active transport. Evidence for a possible association of RNP granules with the microtubule system comes from the localization of SCD6 truncations (this study) or XRNA (Kramer et al., 2008) to the posterior pole of the cell, which is where the microtubule plus ends are located in trypanosomes (Robinson et al., 1995). The RGG domain of SCD6 determines the type of RNP granule, P-body, or storage granule, possibly by determining the type of mRNA that is recruited. One possible mechanism is the masking of the RGG domain by an unknown factor that normally prevents SCD6 from recruiting intact mRNAs. This would explain why P-bodies or granules induced by SCD6∆C do not contain any detectable mRNAs. On SCD6 overexpression, this factor may become limiting, which would now allow the recruitment of intact mRNAs, possibly by binding to eIF4G (Rajyaguru and Parker, 2012). Under natural conditions, the binding of such factors would be regulated.
Understanding the regulation of RNP granule formation is essential to unraveling gene expression pathways. In yeast and mammals, this is hampered by the complexity and redundancy in RNP granule core proteins. The unique role SCD6 appears to have in the formation of trypanosome RNP granules and the absence of most RNP granule core proteins suggests that trypanosomes may well serve as model organisms for the study of RNP granule formation.
MATERIAL AND METHODS
Trypanosomes
The cell line Lister 427 pSPR2, which expresses a tetracycline repressor, was used for all experiments (Sunter et al., 2012). Transgenic trypanosomes were generated using standard procedures (McCulloch et al., 2004). All experiments were performed with logarithmically growing trypanosomes at a cell density of <1 × 107 cells/ml. The wild-type Lister 427 strain (a kind gift from George Cross, Rockefeller University) was used for Figure S2.
Plasmids used in this work
All plasmids used in this work are summarized in Table S1. Endogenous tagging of proteins was essentially performed as described by Kelly et al. (2007), and inducible overexpression was done based on the 3383 plasmid described by Sunter et al. (2012); in many cases, selectable markers and tags were exchanged/added.
Western blots, Northern blots, polysomes, and [35S]methionine labeling
Western blots were done according to standard protocols. Proteins were detected and quantified by the Odyssey Infrared Imaging System (LI-COR Biosciences, Lincoln, NE). Northern blots, polysome analysis, and [35S]methionine labeling were done as previously described (Kramer et al., 2008).
Drugs
Drugs were used in the following concentrations: cycloheximide, 50 μg/ml; actinomycin D, 10 μg/ml; sinefungin, 2 μg/ml.
RNA FISH
RNA FISH was performed as previously described (Kramer et al., 2012), except that 5% dextran sulphate was included in the hybridization solution, and 1 μM of oligos labeled with Cy3 at both ends were used for detection. Oligo sequences were
dT:45T;
dA:45A;
ME antisense: CAATATAGTACAGAAACTGTTCTAATAATAGCGTT;
ME sense: AACGCTATTATTAGAACAGTTTCTGTACTATATTG.
Microscopic imaging and quantification
Cells were washed with SDM79 without serum and hemin and fixed at a density of 1× 107 cells/ml with 2.4% paraformaldehyde overnight, washed once in PBS, and stained with 4′,6-diamidino-2-phenylindole (DAPI). Z-stacks (100 images, 100-nm spacing) were recorded with a custom-built TILL Photonics iMIC microscope equipped with a 100×, 1.4numerical aperture objective (Olympus, Tokyo, Japan) and a sensicam qe CCD camera (PCO, Kehlheim, Germany); deconvolved using Huygens Essential software (SVI, Hilversum, The Netherlands); and, unless otherwise stated, presented as Z-projections (method sum slices) produced by and quantified with ImageJ. The total expression level of an individual cell was calculated by integration of the total background-subtracted fluorescence of the cell in the 32-bit Z-projections; the percentage of SCD6-eYFP (or mutants) in granules was determined by adding the integrated fluorescence of all granules (manual threshold) and correlating it to the total fluorescence of the cell. For three-dimensional imaging, the recorded stacks were volume- and surface-rendered using Imaris software version 7.6 (Bitplane, Belfast, Northern Ireland).
Supplementary Material
Acknowledgments
The authors thank Mark Carrington (University of Cambridge, UK) for great support, discussions, and help with the manuscript; Markus Engstler (University of Würzburg, Germany) for providing excellent infrastructure and support; Jack Sunter (University of Oxford, UK) for discussions and providing cell lines and plasmids; Jay Bangs (University of Wisconsin–Madison, Madison, WI) for the BiP antibody; Ines Subota for advice on ISHs; and all members of Zoology I (University of Würzburg, Germany) for great support. This work was funded by the DFG (German Research Foundation; grant Kr4017/1-1) and by a Wellcome trust grant awarded to Mark Carrington.
Abbreviations used:
- CerFP
cerulean fluorescent protein
- DAPI
4′,6-diamidino-2-phenylindole
- eYFP
enhanced yellow fluorescent protein
- FISH
fluorescence in situ hybridization
- IQR
interquartile range
- mChFP
mCherry fluorescent protein
- ME
mini-exon
- NPG
nuclear periphery granules
- P-body
processing body
- PBS
phosphate-buffered saline
- RNAi
RNA interference
- RNP
ribonucleoprotein
- SL
spliced leader
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-01-0068) on May 15, 2013.
REFERENCES
- Anderson P, Kedersha N. Stress granules: the Tao of RNA triage. Trends Biochem Sci. 2008;33:141–150. doi: 10.1016/j.tibs.2007.12.003. [DOI] [PubMed] [Google Scholar]
- Anderson P, Kedersha N. RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nature. 2009;10:430–436. doi: 10.1038/nrm2694. [DOI] [PubMed] [Google Scholar]
- Arava Y, Wang Y, Storey JD, Liu CL, Brown PO, Herschlag D. Genome-wide analysis of mRNA translation profiles in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2003;100:3889–3894. doi: 10.1073/pnas.0635171100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audhya A, Hyndman F, McLeod I, Maddox A, Yates J, III, Desai A, Oegema K. A complex containing the Sm protein CAR-1 and the RNA helicase CGH-1 is required for embryonic cytokinesis in Caenorhabditis elegans. J Cell Biol. 2005;171:267. doi: 10.1083/jcb.200506124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bangs JD, Uyetake L, Brickman MJ, Balber AE, Boothroyd JC. Molecular cloning and cellular localization of a BiP homologue in Trypanosoma brucei. Divergent ER retention signals in a lower eukaryote. J Cell Sci. 1993;105:1101–1113. doi: 10.1242/jcs.105.4.1101. [DOI] [PubMed] [Google Scholar]
- Barbee SA, et al. Staufen- and FMRP-containing neuronal RNPs are structurally and functionally related to somatic P bodies. Neuron. 2006;52:997–1009. doi: 10.1016/j.neuron.2006.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloch DB, Nobre RA, Bernstein GA, Yang WH. Identification and characterization of protein interactions in the mammalian mRNA processing body using a novel two-hybrid assay. Exp Cell Res. 2011;317:2183–2199. doi: 10.1016/j.yexcr.2011.05.027. [DOI] [PubMed] [Google Scholar]
- Boag PR, Nakamura A, Blackwell TK. A conserved RNA-protein complex component involved in physiological germline apoptosis regulation in C. elegans. Development. 2005;132:4975–4986. doi: 10.1242/dev.02060. [DOI] [PubMed] [Google Scholar]
- Carroll JS, Munchel SE, Weis K. The DExD/H box ATPase Dhh1 functions in translational repression, mRNA decay, and processing body dynamics. J Cell Biol. 2011;194:527–537. doi: 10.1083/jcb.201007151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassola A, De Gaudenzi JG, Frasch AC. Recruitment of mRNAs to cytoplasmic ribonucleoprotein granules in trypanosomes. Mol Microbiol. 2007;65:655–670. doi: 10.1111/j.1365-2958.2007.05833.x. [DOI] [PubMed] [Google Scholar]
- Decker CJ, Teixeira D, Parker R. Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae. J Cell Biol. 2007;179:437–449. doi: 10.1083/jcb.200704147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Gaudenzi JG, Noe G, Campo VA, Frasch AC, Cassola A. Gene expression regulation in trypanosomatids. Essays Biochem. 2011;51:31–46. doi: 10.1042/bse0510031. [DOI] [PubMed] [Google Scholar]
- Dhalia R, Marinsek N, Reis CRS, Katz R, Muniz JRC, Standart N, Carrington M, de Melo Neto OP. The two eIF4A helicases in Trypanosoma brucei are functionally distinct. Nucleic Acids Res. 2006;34:2495–2507. doi: 10.1093/nar/gkl290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy EM, Ito S. Fine structural and radioautographic observations on dense perinuclear cytoplasmic material in tadpole oocytes. J Cell Biol. 1971;49:90–108. doi: 10.1083/jcb.49.1.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A, Behm-Ansmant I, Izaurralde E. P bodies: at the crossroads of post-transcriptional pathways. Nat Rev Mol Cell Biol. 2007a;8:9–22. doi: 10.1038/nrm2080. [DOI] [PubMed] [Google Scholar]
- Eulalio A, Behm-Ansmant I, Schweizer D, Izaurralde E. P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol Cell Biol. 2007b;27:3970–3981. doi: 10.1128/MCB.00128-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freire ER, et al. The four trypanosomatid eIF4E homologues fall into two separate groups, with distinct features in primary sequence and biological properties. Mol Biochem Parasitol. 2011;176:25–36. doi: 10.1016/j.molbiopara.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromm SA, Truffault V, Kamenz J, Braun JE, Hoffmann NA, Izaurralde E, Sprangers R. The structural basis of Edc3- and Scd6-mediated activation of the Dcp1:Dcp2 mRNA decapping complex. EMBO J. 2012;31:279–290. doi: 10.1038/emboj.2011.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilks N, Kedersha N, Ayodele M, Shen L, Stoecklin G, Dember LM, Anderson P. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol Biol Cell. 2004;15:5383–5398. doi: 10.1091/mbc.E04-08-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammarton TC, Clark J, Douglas F, Boshart M, Mottram JC. Stage-specific differences in cell cycle control in Trypanosoma brucei revealed by RNA interference of a mitotic cyclin. J Biol Chem. 2003;278:22877–22886. doi: 10.1074/jbc.M300813200. [DOI] [PubMed] [Google Scholar]
- Hay B, Ackerman L, Barbel S, Jan LY, Jan YN. Identification of a component of Drosophila polar granules. Development. 1988a;103:625–640. doi: 10.1242/dev.103.4.625. [DOI] [PubMed] [Google Scholar]
- Hay B, Jan LY, Jan YN. A protein component of Drosophila polar granules is encoded by vasa and has extensive sequence similarity to ATP-dependent helicases. Cell. 1988b;55:577–587. doi: 10.1016/0092-8674(88)90216-4. [DOI] [PubMed] [Google Scholar]
- Holetz FB, Correa A, Avila AR, Nakamura CV, Krieger MA, Goldenberg S. Evidence of P-body-like structures in Trypanosoma cruzi. Biochem Biophys Res Commun. 2007;356:1062–1067. doi: 10.1016/j.bbrc.2007.03.104. [DOI] [PubMed] [Google Scholar]
- Jakymiw A, Pauley KM, Li S, Ikeda K, Lian S, Eystathioy T, Satoh M, Fritzler MJ, Chan EKL. The role of GW/P-bodies in RNA processing and silencing. J Cell Sci. 2007;120:1317–1323. doi: 10.1242/jcs.03429. [DOI] [PubMed] [Google Scholar]
- Kafasla P, Barrass JD, Thompson E, Fromont-Racine M, Jacquier A, Beggs JD, Lewis J. Interaction of yeast eIF4G with spliceosome components: implications in pre-mRNA processing events. RNA Biol. 2009;6:563–574. doi: 10.4161/rna.6.5.9861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, et al. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149:753–767. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha NL, Gupta M, Li W, Miller I, Anderson P. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2α to the assembly of mammalian stress granules. J Cell Biol. 1999;147:1431–1442. doi: 10.1083/jcb.147.7.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly S, et al. Functional genomics in Trypanosoma brucei: a collection of vectors for the expression of tagged proteins from endogenous and ectopic gene loci. Mol Biochem Parasitol. 2007;154:103–109. doi: 10.1016/j.molbiopara.2007.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer S. Developmental regulation of gene expression in the absence of transcriptional control: the case of kinetoplastids. Mol Biochem Parasitol. 2012;181:61–72. doi: 10.1016/j.molbiopara.2011.10.002. [DOI] [PubMed] [Google Scholar]
- Kramer S, Marnef A, Standart N, Carrington M. Inhibition of mRNA maturation in trypanosomes causes the formation of novel foci at the nuclear periphery containing cytoplasmic regulators of mRNA fate. J Cell Sci. 2012;125:2896–2909. doi: 10.1242/jcs.099275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer S, Queiroz R, Ellis L, Hoheisel JD, Clayton C, Carrington M. The RNA helicase DHH1 is central to the correct expression of many developmentally regulated mRNAs in trypanosomes. J Cell Sci. 2010;123:699–711. doi: 10.1242/jcs.058511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer S, Queiroz R, Ellis L, Webb H, Hoheisel JD, Clayton C, Carrington M. Heat shock causes a decrease in polysomes and the appearance of stress granules in trypanosomes independently of eIF2α phosphorylation at Thr169. J Cell Sci. 2008;121:3002–3014. doi: 10.1242/jcs.031823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieb B, Carl M, Hock R, Gebauer D, Scheer U. Identification of a novel mRNA-associated protein in oocytes of Pleurodeles waltl and Xenopus laevis. Exp Cell Res. 1998;245:272–281. doi: 10.1006/excr.1998.4249. [DOI] [PubMed] [Google Scholar]
- Liu Q, Liang XH, Uliel S, Belahcen M, Unger R, Michaeli S. Identification and functional characterization of Lsm proteins in Trypanosoma brucei. J Biol Chem. 2004;279:18210–18219. doi: 10.1074/jbc.M400678200. [DOI] [PubMed] [Google Scholar]
- Mahowald AP. Polar granules of Drosophila. IV. Cytochemical studies showing loss of RNA from polar granules during early stages of embryogenesis. J Exp Zool. 1971;176:345–352. doi: 10.1002/jez.1401760309. [DOI] [PubMed] [Google Scholar]
- Mahowald AP, Hennen S. Ultrastructure of the “germ plasm” in eggs and embryos of Rana pipiens. Dev Biol. 1971;24:37–53. doi: 10.1016/0012-1606(71)90045-5. [DOI] [PubMed] [Google Scholar]
- Mair GR, Lasonder E, Garver LS, Franke-Fayard BMD, Carret CK, Wiegant JCAG, Dirks RW, Dimopoulos G, Janse CJ, Waters AP. Universal features of post-transcriptional gene regulation are critical for Plasmodium zygote development. PLoS Pathog. 2010;6:e1000767. doi: 10.1371/journal.ppat.1000767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchetti MA, Tschudi C, Kwon H, Wolin SL, Ullu E. Import of proteins into the trypanosome nucleus and their distribution at karyokinesis. J Cell Sci. 2000;113:899–906. doi: 10.1242/jcs.113.5.899. [DOI] [PubMed] [Google Scholar]
- Marnef A, Sommerville J, Ladomery MR. RAP55: insights into an evolutionarily conserved protein family. Int J Biochem Cell Biol. 2009;41:977–981. doi: 10.1016/j.biocel.2008.06.015. [DOI] [PubMed] [Google Scholar]
- Matsumoto K, Nakayama H, Yoshimura M, Masuda A, Dohmae N, Matsumoto S, Tsujimoto M. PRMT1 is required for RAP55 to localize to processing bodies. RNA Biol. 2012;9:610–623. doi: 10.4161/rna.19527. [DOI] [PubMed] [Google Scholar]
- McCulloch R, Vassella E, Burton P, Boshart M, Barry JD. Transformation of monomorphic and pleomorphic Trypanosoma brucei. Methods Mol Biol. 2004;262:53–86. doi: 10.1385/1-59259-761-0:053. [DOI] [PubMed] [Google Scholar]
- McKendrick L, Thompson E, Ferreira J, Morley SJ, Lewis JD. Interaction of eukaryotic translation initiation factor 4G with the nuclear cap-binding complex provides a link between nuclear and cytoplasmic functions of the m(7) guanosine cap. Mol Cell Biol. 2001;21:3632–3641. doi: 10.1128/MCB.21.11.3632-3641.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minshall N, Kress M, Weil D, Standart N. Role of p54 RNA helicase activity and its C-terminal domain in translational repression, P-body localization and assembly. Mol Biol Cell. 2009;20:2464–2472. doi: 10.1091/mbc.E09-01-0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissan T, Rajyaguru P, She M, Song H, Parker R. Decapping activators in Saccharomyces cerevisiae act by multiple mechanisms. Mol Cell. 2010;39:773–783. doi: 10.1016/j.molcel.2010.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepling ME, Wilhelm JE, O'Hara AL, Gephardt GW, Spradling AC. Mouse oocytes within germ cell cysts and primordial follicles contain a Balbiani body. Proc Natl Acad Sci USA. 2007;104:187–192. doi: 10.1073/pnas.0609923104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajyaguru P, Parker R. RGG motif proteins: modulators of mRNA functional states. Cell Cycle. 2012;11:2594–2599. doi: 10.4161/cc.20716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajyaguru P, She M, Parker R. Scd6 targets eIF4G to repress translation: RGG motif proteins as a class of eIF4G-binding proteins. Mol Cell. 2012;45:244–254. doi: 10.1016/j.molcel.2011.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reijns MA, Alexander RD, Spiller MP, Beggs JD. A role for Q/N-rich aggregation-prone regions in P-body localization. J Cell Sci. 2008;121:2463–2472. doi: 10.1242/jcs.024976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson DR, Sherwin T, Ploubidou A, Byard EH, Gull K. Microtubule polarity and dynamics in the control of organelle positioning, segregation, and cytokinesis in the trypanosome cell cycle. J Cell Biol. 1995;128:1163–1172. doi: 10.1083/jcb.128.6.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth U, Pitt J, Dennis S, Priess JR. Perinuclear P granules are the principal sites of mRNA export in adult C. elegans germ cells. Development. 2010;137:1305–1314. doi: 10.1242/dev.044255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squirrell JM, Eggers ZT, Luedke N, Saari B, Grimson A, Lyons GE, Anderson P, White JG. CAR-1, a protein that localizes with the mRNA decapping component DCAP-1, is required for cytokinesis and ER organization in Caenorhabditis elegans embryos. Mol Biol Cell. 2006;17:336–344. doi: 10.1091/mbc.E05-09-0874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strome S, Wood WB. Immunofluorescence visualization of germ-line-specific cytoplasmic granules in embryos, larvae, and adults of Caenorhabditis elegans. Proc Natl Acad Sci USA. 1982;79:1558–1562. doi: 10.1073/pnas.79.5.1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strome S, Wood WB. Generation of asymmetry and segregation of germ-line granules in early C. elegans embryos. Cell. 1983;35:15–25. doi: 10.1016/0092-8674(83)90203-9. [DOI] [PubMed] [Google Scholar]
- Sunter J, Wickstead B, Gull K, Carrington M. A new generation of T7 RNA polymerase-independent inducible expression plasmids for Trypanosoma brucei. PLoS One. 2012;7:e35167. doi: 10.1371/journal.pone.0035167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweet T, Kovalak C, Coller J. The DEAD-box protein Dhh1 promotes decapping by slowing ribosome movement. PLoS Biol. 2012;10:e1001342. doi: 10.1371/journal.pbio.1001342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka KJ, Ogawa K, Takagi M, Imamoto N, Matsumoto K, Tsujimoto M. RAP55, a cytoplasmic mRNP component, represses translation in Xenopus oocytes. J Biol Chem. 2006;281:40096–40106. doi: 10.1074/jbc.M609059200. [DOI] [PubMed] [Google Scholar]
- Teixeira D, Parker R. Analysis of P-body assembly in Saccharomyces cerevisiae. Mol Biol Cell. 2007;18:2274–2287. doi: 10.1091/mbc.E07-03-0199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tritschler F, Braun JE, Eulalio A, Truffault V, Izaurralde E, Weichenrieder O. Structural basis for the mutually exclusive anchoring of P body components EDC3 and Tral to the DEAD box protein DDX6/Me31B. Mol Cell. 2009;33:661–668. doi: 10.1016/j.molcel.2009.02.014. [DOI] [PubMed] [Google Scholar]
- Tritschler F, Eulalio A, Helms S, Schmidt S, Coles M, Weichenrieder O, Izaurralde E, Truffault V. Similar modes of interaction enable Trailer Hitch and EDC3 to associate with DCP1 and Me31B in distinct protein complexes. Mol Cell Biol. 2008;28:6695–6708. doi: 10.1128/MCB.00759-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm JE, Buszczak M, Sayles S. Efficient protein trafficking requires trailer hitch, a component of a ribonucleoprotein complex localized to the ER in Drosophila. Dev Cell. 2005;9:675–685. doi: 10.1016/j.devcel.2005.09.015. [DOI] [PubMed] [Google Scholar]
- Wirtz E, Leal S, Ochatt C, Cross GA. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol Biochem Parasitol. 1999;99:89–101. doi: 10.1016/s0166-6851(99)00002-x. [DOI] [PubMed] [Google Scholar]
- Xu J, Chua NH. Arabidopsis decapping 5 is required for mRNA decapping, P-body formation, and translational repression during postembryonic development. Plant Cell. 2009;21:3270–3279. doi: 10.1105/tpc.109.070078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W-H, Yu JH, Gulick T, Bloch KD, Bloch DB. RNA-associated protein 55 (RAP55) localizes to mRNA processing bodies and stress granules. RNA. 2006;12:547–554. doi: 10.1261/rna.2302706. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.