Binding and dephosphorylation of the yeast lipin Pah1p by its phosphatase Nem1p-Spo7p is essential for its membrane targeting and is mediated by a C-terminal acidic stretch on Pah1p. This results in the recruitment of Pah1p to the vicinity of lipid droplets, where it controls triglyceride and droplet biogenesis in an acidic tail–dependent manner.
Abstract
Lipins are evolutionarily conserved phosphatidate phosphatases that perform key functions in phospholipid, triglyceride, and membrane biogenesis. Translocation of lipins on membranes requires their dephosphorylation by the Nem1p-Spo7p transmembrane phosphatase complex through a poorly understood mechanism. Here we identify the carboxy-terminal acidic tail of the yeast lipin Pah1p as an important regulator of this step. Deletion or mutations of the tail disrupt binding of Pah1p to the Nem1p-Spo7p complex and Pah1p membrane translocation. Overexpression of Nem1p-Spo7p drives the recruitment of Pah1p in the vicinity of lipid droplets in an acidic tail–dependent manner and induces lipid droplet biogenesis. Genetic analysis shows that the acidic tail is essential for the Nem1p-Spo7p–dependent activation of Pah1p but not for the function of Pah1p itself once it is dephosphorylated. Loss of the tail disrupts nuclear structure, INO1 gene expression, and triglyceride synthesis. Similar acidic sequences are present in the carboxy-terminal ends of all yeast lipin orthologues. We propose that acidic tail–dependent binding and dephosphorylation of Pah1p by the Nem1p-Spo7p complex is an important determinant of its function in lipid and membrane biogenesis.
INTRODUCTION
In eukaryotes, the central hub for lipid synthesis is the endoplasmic reticulum (ER), where cells coordinate the expression, localization, and activity of a multitude of lipid-modifying enzymes (Nohturff and Zhang, 2009). Phosphatidate (PA) is a key precursor in lipid metabolism: its dephosphorylation gives rise to diacylglycerol (DAG), which can be acylated to generate triacylglycerol (TAG), an important form of energy and fatty acid storage that is deposited in lipid droplets (Figure 1A). In the presence of choline and ethanolamine, DAG can be also used to generate phospholipids through the Kennedy pathway. Alternatively, PA can be converted to cytidine diphosphate diacylglycerol (CDP-DAG), from which all membrane phospholipids are produced (Henry et al., 2012).
FIGURE 1:
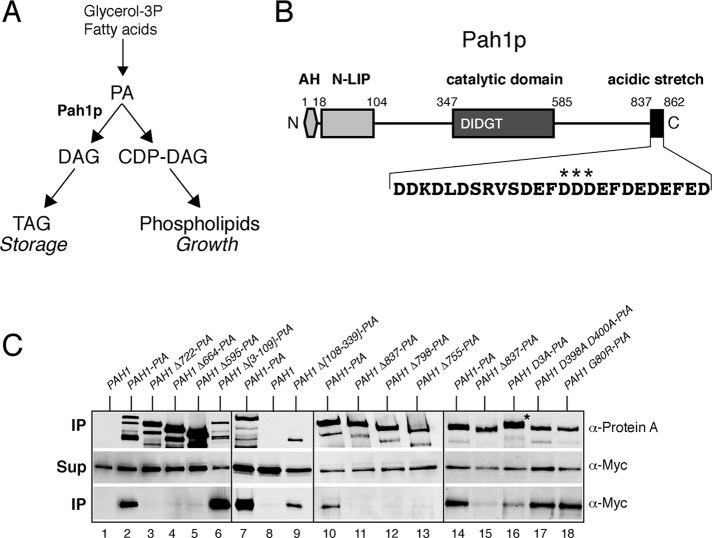
The C-terminal acidic stretch of Pah1p is required for binding to the Nem1p-Spo7p complex in vivo. (A) Simplified schematic of the role of Pah1p in lipid biosynthesis. (B) Schematic of the primary structure of Pah1p with the position of key motifs/domains shown. AH, amphipathic helix. The sequence of the acidic stretch is shown, with the three aspartates mutated in Pah1D3A-PtA highlighted by stars. (C) Mutational analysis of the interaction of Pah1p and Nem1p-Spo7p. Control untagged PAH1 or the indicated wild-type or mutant PAH1-PtA fusions were expressed in pah1Δ cells carrying GAL-NEM1-Myc and GAL-SPO7. Cells were transferred from raffinose- into galactose-containing medium and grown for 8 h before lysis and affinity purification over IgG–Sepharose columns. Each of the four boxes corresponds to one independent representative set of experiments. Top and bottom (IP) correspond to equivalent amounts of affinity purified eluates, analyzed by SDS–PAGE (8%) and Western blotting using anti–protein A and anti-Myc antibodies, respectively. Middle (Sup) corresponds to equal amounts of clarified cell extracts used for the affinity purifications, analyzed by Western blotting using anti-Myc antibodies.
In the budding yeast Saccharomyces cerevisiae, PA homeostasis is controlled by Pah1p, which dephosphorylates PA to DAG (Han et al., 2006). Consistent with the important roles of these two lipids in cell physiology Pah1p is required for cell surface biosynthesis (Lussier et al., 1997), nuclear/ER membrane biogenesis (Santos-Rosa et al., 2005), vacuole function (Sasser et al., 2012), and lipid droplet biogenesis (Adeyo et al., 2011). In addition to its lipid metabolic roles on cytosolic membranes, Pah1p can also translocate to the nucleus, where it binds to chromatin (Santos-Rosa et al., 2005). How the intranuclear localization of Pah1p relates to its ER biosynthetic roles is not known.
Pah1p is a member of the conserved lipin family (Peterfy et al., 2001). Mammals express three lipin paralogues, lipins 1–3, which are PA phosphatases (Han et al., 2006; Donkor et al., 2007) and also have functions as transcriptional regulators (reviewed in Harris and Finck, 2011). The important roles of lipins in metabolism are underscored by the fact that lipin 1–deficient mice display features of generalized lipodystrophy, characterized by significant reduction in fat mass and lack of adipocyte differentiation (Peterfy et al., 2001; Phan et al., 2004).
Similar to its mammalian orthologues, Pah1p lacks transmembrane domains, and therefore an essential step during its function is its translocation onto membranes. Multisite phosphorylation controls the activity and membrane translocation of Pah1p (O'Hara et al., 2006; Karanasios et al., 2010; Choi et al., 2011, 2012). Genetic screens in yeast have identified a transmembrane phosphatase complex consisting of two subunits, Nem1p and Spo7p, required for the dephosphorylation of Pah1p (Siniossoglou et al., 1998; Santos-Rosa et al., 2005). Nem1p belongs to the CPD phosphatase family and is the catalytic subunit, whereas Spo7p binds to the catalytic domain of Nem1p and is required for the activity of the holoenzyme (Siniossoglou et al., 1998; Santos-Rosa et al., 2005). Dephosphorylation of Pah1p by Nem1p-Spo7p leads to its membrane anchoring via an amino-terminal amphipathic helix (Karanasios et al., 2010). The mammalian orthologue complex of Nem1p-Spo7p has been identified as the Dullard-TMEM188 transmembrane complex, recently renamed as CTDNEP1-NEP1R1 (Kim et al., 2007; Han et al., 2012), raising the possibility that aspects of the Pah1p recruitment pathway are conserved in higher eukaryotes.
The association of Pah1p with ER membranes will influence the partitioning of lipid biosynthetic precursors between storage and membrane phospholipid biogenesis (Figure 1A). In addition, it may affect local PA and DAG pools and organelle function through downstream signaling pathways. Despite these important roles, the mechanisms by which Pah1p translocates onto membranes and their spatial regulation are poorly understood. Here we identify a short carboxy-terminal acidic peptide on Pah1p that mediates its interaction with the Nem1p-Spo7p complex and is consequently important for Pah1p membrane translocation, production of DAG and TAG, lipid droplet biogenesis, and transcriptional regulation of INO1. This requirement can be bypassed if Pah1p is dephosphorylated and thus membrane anchored in an Nem1p-Spo7p–independent manner. Similar carboxy-terminal tails are found in other yeast lipins, suggesting that this represents an ancient mechanism for the activation of the lipin PA phosphatases.
RESULTS
The acidic tail of Pah1p mediates its interaction with the Nem1p-Spo7p phosphatase complex
To identify proteins interacting with the membrane-bound Pah1p, we expressed a Pah1p protein A fusion (Pah1p-PtA) in a pah1Δ strain expressing NEM1 and SPO7 under the control of the inducible GAL1/10 promoter. Pah1p-PtA from cells grown in galactose was affinity purified over an immunoglobulin G (IgG)–Sepharose column. Mass spectrometric analysis identified a number of candidate interacting proteins that will be characterized in a separate study. Both Nem1p and Spo7p were identified among the interacting proteins. To confirm this result, we expressed galactose (Gal)-inducible NEM1-Myc and SPO7 in pah1Δ strains expressing either PAH1 or PAH1-PtA and isolated the Pah1p proteins as described. Western blot confirmed that Nem1p-Myc can be recovered in association with Pah1p-PtA but not untagged Pah1p (Figure 1C, lanes 1 and 2). Moreover, Gal-inducible Spo7p-Myc also can be coprecipitated with Pah1p-PtA (data not shown). We were unable to detect an interaction between Nem1p and Pah1p when the former was expressed at endogenous levels (data not shown), however, implying a more transient binding, which is typical of enzyme and substrate. Taken together, these data show that overexpressed Nem1p-Spo7p complex interacts with Pah1p.
To validate this interaction and explore its functional significance in vivo, we sought to identify Pah1p mutants that are compromised in their binding to Nem1p-Spo7p. Pah1p contains two conserved domains (N-LIP and C-LIP/catalytic), a linker domain in between, and a long carboxy-terminal tail (Figure 1B). Partial or complete deletions of the carboxy-terminal tail led to loss of binding to Nem1p-Spo7p, whereas deletion of the N-LIP domain did not (Figure 1C). Pah1p lacking the linker domain was less stable than the full-length version but still able to retain the interaction with Nem1p-Spo7p (Figure 1C, compare lanes 7–9). Therefore the C-terminal domain of Pah1p is essential for the interaction with Nem1p-Spo7p. Shorter deletions identified the 25 extreme C-terminal residues as the sequence necessary for this binding (Figure 1C, lanes 10–13). This sequence is highly enriched in acidic residues (61% aspartate and glutamates; Figure 1B). The acidic nature of this sequence is important for the interaction with Nem1p-Spo7p because mutation of only three central aspartates to alanines (Pah1pD851A/D852A/D853A-PtA, referred here to as Pah1pD3A-PtA) decreased the interaction without affecting the stability of Pah1p (Figure 1C, lanes 14–16). Of importance, Pah1pD3A-PtA displays a mobility shift when compared with the wild-type Pah1p-PtA, consistent with the decreased interaction and therefore dephosphorylation by Nem1p-Spo7p in vivo (Figure 1C, lanes 14 and 16). On the other hand, mutations within its catalytic site (D398A D400A) or the N-LIP domain (the lipodystrophy mutation G80R; Peterfy et al., 2001) did not disrupt the binding of Pah1p to Nem1p-Spo7p (Figure 1C, lanes 17 and 18). Taken together, these data show that the C-terminal acidic stretch of Pah1p is required for its binding to the Nem1p-Spo7p phosphatase complex.
The acidic tail is conserved and functions as an autonomous unit
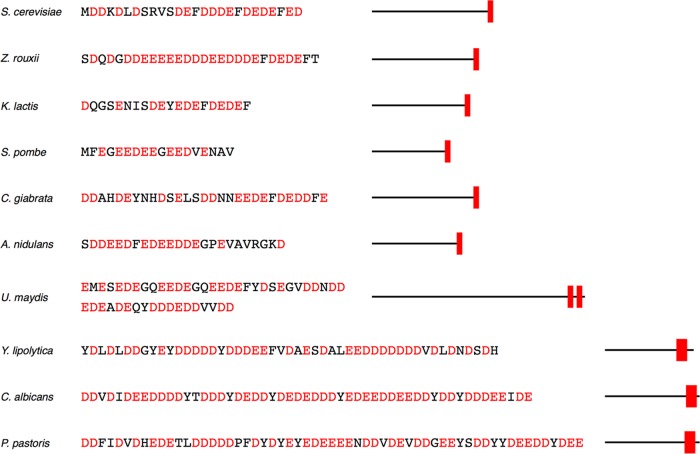
Acidic domains similar to the one found in Pah1p are also present in lipins from many other evolutionarily divergent yeast species. In all cases, these domains are positioned at their C-terminal ends and range from short clusters of few aspartates/glutamates, as in S. cerevisiae or Schizosaccharomyces pombe, to longer sequences of seven to 10 residues that repeat over larger domains, as in Pichia pastoris or Candida albicans—the latter containing a tail of 59 residues with 80% glutamates or aspartates (Figure 2). A common feature in both short and long acidic domains is the presence of hydrophobic residues, mostly F, Y, and V, that are inserted in between the aspartate/glutamate clusters.
FIGURE 2:
The Pah1p acidic tail is evolutionarily conserved. Sequences and schematics of similar Pah1p-like aspartate/glutamate-rich sequences found in other yeasts. Species, amino acid sequence (with aspartate and glutamate residues in red), and the position of the acidic domains (in red) within the primary structure of the proteins are shown.
To confirm the function of the acidic tail as a Nem1p-Spo7p–binding platform in budding yeast, we fused either the entire C-terminal domain of Pah1p (residues S586–D862; see also Figure 1B, lipin 2-Pah1pLongC) or only the 26-residue-long acidic stretch (residues M837–D862, lipin 2-Pah1pShortC) to the C-terminal end of mouse lipin 2 and asked whether these fusions would now bind to the Nem1p-Spo7p complex in yeast (Figure 3A). We choose lipin 2 because, in contrast to lipin 1, its full-length PtA fusion is readily expressed in yeast extracts (Grimsey et al., 2008; our unpublished observations). Whereas lipin 2-PtA could specifically bind to the mammalian lipin phosphatase complex CTDNEP1-NEP1R1 when coexpressed in yeast, we could not detect a stable interaction with Nem1p-Spo7p (Figure 3, B and C). On the other hand, once fused to either the long or short acidic tail domains derived from Pah1p, lipin 2 could bind very efficiently to Nem1p-Spo7p (Figure 3C). Therefore the acidic stretch of Pah1p functions as an autonomous unit and can promote stable binding to Nem1p-Spo7p even when expressed in the context of an evolutionarily distant lipin enzyme.
FIGURE 3:
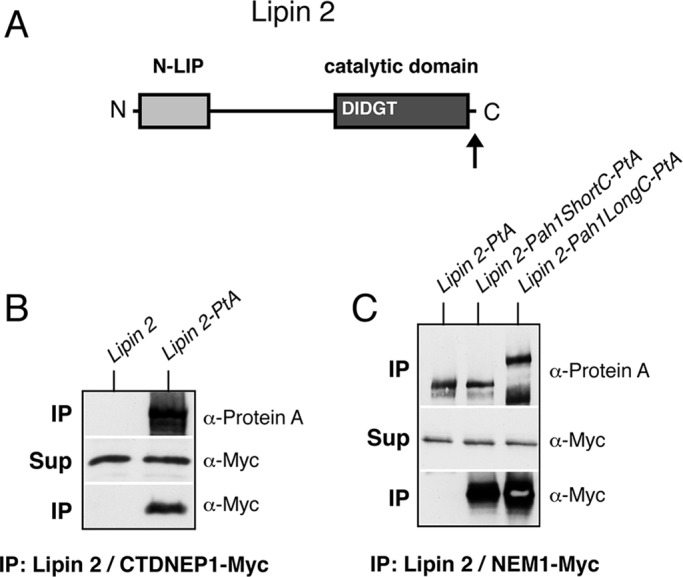
Pah1p acidic stretch–dependent interaction of mouse lipin 2 with the Nem1p-Spo7p complex. (A) Schematic of the primary structure of lipin 2, with the position of key motifs/domains shown. The arrow indicates the position where the Pah1p acidic stretch was fused. (B) Control untagged lipin 2 or lipin 2-PtA was affinity purified from pah1Δ cells expressing CTDNEP1-MyC and NEP1R1. IP and Sup samples were processed as in Figure 1C. (C) Affinity purification of PtA-tagged lipin 2, lipin 2-Pah1ShortC, or lipin 2-Pah1LongC. The PtA fusions were expressed under the control of the PAH1 promoter and low-copy plasmids, except for lipin 2-Pah1LongC, which was less stable and expressed from a high-copy (2 μ) vector. All fusions were expressed in pah1Δ cells carrying GAL-NEM1-Myc and GAL-SPO7. IP and Sup samples were processed as in Figure 1C.
The acidic tail is necessary and sufficient for Nem1p-Spo7p–dependent recruitment of Pah1p close to lipid droplets and is required for droplet biogenesis
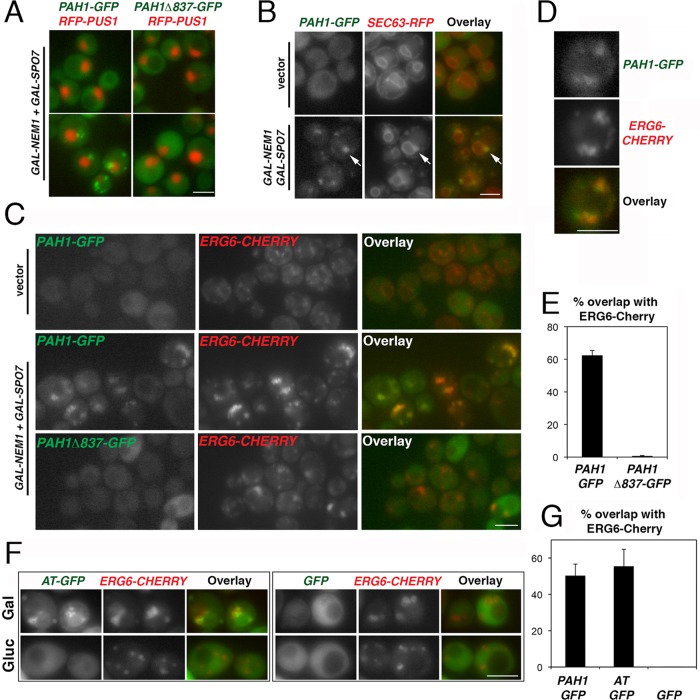
To address the role of the acidic stretch of Pah1p on its intracellular distribution, we examined the localization of Pah1p–green fluorescent protein (GFP) or Pah1pΔ837-GFP in cells carrying Gal-inducible NEM1-SPO7. In cells grown in medium containing raffinose, Pah1p-GFP showed a diffused cytosolic distribution (data not shown). After the addition of galactose, Pah1p-GFP localized to discrete punctuate structures that are in contact with the ER membrane, whereas Pah1pΔ837-GFP maintained a diffused cytosolic localization similar to the one seen under noninducible NEM1-SPO7 conditions (Figure 4, A and B). Because a recent study from the Goodman group reported that Nem1p localizes in close proximity to lipid droplets (Adeyo et al., 2011), we asked whether high levels of Nem1p-Spo7p could target Pah1p-GFP close to lipid droplets visualized by a chromosomally tagged ERG6-Cherry fusion (Jacquier et al., 2011). Pah1p-GFP overlapped with the Erg6p-Cherry–labeled droplets in 63% of the cells examined (Figure 4, C and E), whereas the remaining cells with no overlap had no detectable membrane-bound pool of Pah1p-GFP at all (see, for example, PAH1-GFP in Figure 4C, middle). Often Pah1p-GFP appeared to be in contact, but not entirely colocalizing, with Erg6p-Cherry (Figure 4D), consistent with recruitment to the ER site of lipid droplet formation, whereas Pah1pΔ837-GFP remained cytosolic. To test whether the acidic tail alone is sufficient for targeting, we fused the 25 C-terminal residues of Pah1p to GFP and examined its localization in response to Nem1p-Spo7p levels. After GAL-NEM1/SPO7 overexpression, the acidic tail–GFP fusion showed similar levels of lipid droplet targeting as Pah1p-GFP (Figure 4, F and G). Thus the acidic tail of Pah1p is necessary and sufficient for the recruitment of Pah1p by Nem1p-Spo7p.
FIGURE 4:
The acidic stretch is necessary and sufficient for the Nem1p-Spo7p–directed recruitment of Pah1p-GFP to lipid droplets. (A) pah1Δ cells expressing the intranuclear reporter RFP-PUS1 and either PAH1-GFP (left) or PAH1Δ837-GFP (right) were transformed with GAL-NEM1 and GAL-SPO7, transferred from raffinose- to galactose-containing medium, grown for 6 h, and imaged by epifluorescence microscopy. Bar, 5 μm. (B) pah1Δ cells expressing PAH1-GFP and SEC63-RFP were transformed with either empty vectors (top) or GAL-NEM1 and GAL-SPO7 (bottom). Cells were grown in galactose-containing medium for 6 h as in A. Arrows point to typical ER-associated Pah1p-GFP labeling. Bar, 5 μm. (C) pah1Δ cells with chromosomally integrated ERG6-Cherry expressing PAH1-GFP (top and middle) or PAH1Δ837-GFP (bottom) were transformed with either empty vectors (top) or GAL-NEM1 and GAL-SPO7 (middle and bottom). Cells were grown in galactose for 6 h as in A. (D) Representative PAH1-GFP cell overexpressing GAL-NEM1 and GAL-SPO7 from C. (E) The percentage of cells in which Pah1p-GFP or Pah1pΔ837-GFP overlaps with more than half of Erg6p-Cherry puncta per cell is given. Values represent the mean ± SD of three independent experiments. At least 300 cells per experiment and strain were scored. (F) Nem1p-Spo7p–dependent targeting of the Pah1p acidic stretch to lipid droplets. The acidic tail fusion to GFP (AT-GFP, left) or GFP alone (right) was expressed under the control of the NOP1 promoter in RS453 cells coexpressing chromosomally integrated ERG6-Cherry. Cells were grown in galactose as in A (Gal) or glucose (Gluc) and imaged by epifluorescence microscopy. Bar, 5 μm. (G) RS453 ERG6-Cherry cells expressing PAH1-GFP, AT-GFP, or GFP, all under the control of the NOP1 promoter, were grown in galactose as in F, and the percentage of overlap between the GFP fusions and Erg6p-Cherry was quantified as described in E.
Of interest, we noticed that GAL-NEM1/SPO7 cells contained what appeared to be larger and irregularly distributed lipid droplets when compared with the vector controls (Figure 4C, compare top and middle). Lack of Pah1pΔ837-GFP recruitment coincided with a decrease of the Erg6p-Cherry labeling compared with that seen in Pah1p-GFP cells, indicating that the acidic stretch is required for droplet biogenesis (Figure 4C, middle and bottom). Droplets in many Pah1pΔ837-GFP cells appeared to be still modestly larger when compared with the vector control (Figure 4C, top and bottom), however, suggesting that an acidic-stretch independent mechanism contributes also to lipid droplet biogenesis. Labeling lipid droplets with the lipophilic dye BODIPY 493/503 confirmed the results obtained with Erg6p-Cherry (Supplemental Figure S1A). Of importance, droplets in pah1Δ cells were not affected by GAL-NEM1/SPO7, confirming that the observed effects are mediated by Pah1p and not some other factor that could be dephosphorylated by Nem1p-Spo7p (Supplemental Figure S1B).
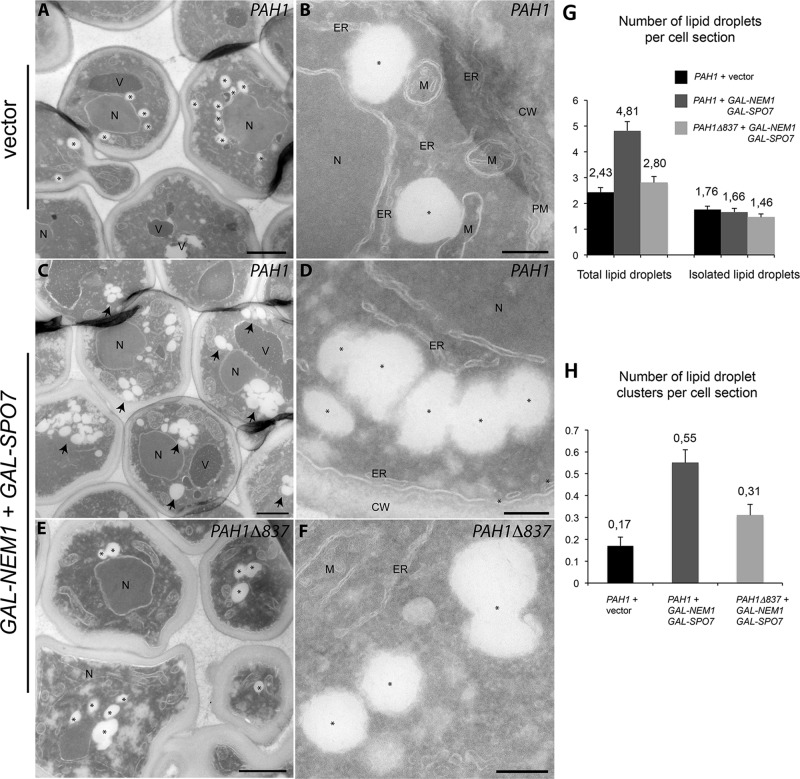
To investigate the role of the acidic tail in lipid droplet biogenesis and quantify the changes at the ultrastructural level, we examined PAH1 or PAH1Δ837 strains after Nem1p-Spo7p overexpression using a cryosectioning procedure for the morphological examination of yeast (Griffith et al., 2008). Whereas wild-type cells containing the vector controls displayed a homogeneous distribution of lipid droplets (Figure 5, A and B), cells expressing full-length Pah1p with GAL-NEM1/SPO7 showed a striking accumulation of tightly packed and possibly connected lipid droplets forming clusters (Figure 5, C and D). The total number of lipid droplets per cell section showed an almost 100% increase after Nem1p-Spo7p overproduction but only in the presence of the acidic tail: the Pah1pΔ837 cells displayed only a minor increase in lipid droplet number (Figure 5, E and F). Of interest, the number of isolated lipid droplets does not increase in GAL-NEM1/SPO7 cells; the difference in total lipid droplet number is due to the increase of droplet cluster formation, which is again partly dependent on the acidic tail (Figure 5, G and H). The formation of these closely associated droplets in clusters explains the increased Erg6p-Cherry and BODIPY labeling of GAL-NEM1/SPO7 cells, which cannot be resolved by fluorescence microscopy (Figures 4C and Supplemental Figure S1). Taken together, these data show that the acidic tail of Pah1p is required for lipid droplet biogenesis in response to Nem1p-Spo7p and that the partial rescue of the Δ837 mutant is likely due to remaining Pah1p activity in droplet biogenesis.
FIGURE 5:
Pah1p acidic tail–dependent formation of lipid droplet clusters. (A–F) pah1Δ cells expressing either PAH1 and vector controls (A, B), PAH1 and GAL-NEM1/GAL-SPO7 (C, D), or PAH1Δ837 and GAL-NEM1/GAL-SPO7 (E, F) were grown in a galactose-containing medium for 5 h as in Figure 4C. Cells were then processed for electron microscopy as described in Materials and Methods. Cryosections were picked up with 0.47% uranyl acetate (mild uranyl pickup) before being immediately stained and viewed with an electron microscope. CW, cell wall; ER, endoplasmic reticulum; M, mitochondria; N, nucleus; PM, plasma membrane; V, vacuole. Lipid droplets are marked with an asterisk (A, B, D–F) or, when in clusters, an arrow. Bars, 1 μm (A, C, E), 0.2 μm (B, D, F). (G) Number of total lipid droplets increases upon induction of Nem1p and Spo7p overexpression in a Pah1p-acidic tail–dependent manner. The total number of lipid droplets (left) or only isolated lipid droplets (right) per cell section was determined over 200 cell profiles randomly selected. (H) The number of lipid droplet clusters (more than three adjacent lipid droplets) per cell section increases when Nem1p and Spo7p are overexpressed in the presence of full-length Pah1p and is partly dependent on the acidic tail. Values represent the mean ± SEM of measurements from at least three different grids.
The acidic stretch mediates the Nem1p-Spo7p–dependent function of Pah1p in cell growth
We next sought further evidence for the function of the acidic tail as a Nem1p-Spo7p–binding domain of Pah1p using two genetic assays. First, overexpression of NEM1-SPO7 causes lethality that is mediated by Pah1p, as it is rescued in pah1Δ cells (Santos-Rosa et al., 2005). If the acidic stretch of Pah1p is important for binding to Nem1p-Spo7p in vivo, then its removal should rescue this lethality. Indeed, in contrast to PAH1, both PAH1Δ837 and PAH1D3A restore growth in cells overexpressing NEM1-SPO7 (Figure 6A). Of importance, however, the same tail deletion could not rescue the lethality caused by the overexpression of a phosphorylation-deficient mutant (PAH1 7A) in which seven phosphoresidues were replaced by alanines (O'Hara et al., 2006; Figure 6B). Thus mutation of the acidic stretch rescues the lethality of dephosphorylated Pah1p only if the latter is generated by the Nem1p-Spo7p phosphatase complex. If, on the other hand, NEM1-SPO7 is bypassed by PAH1 7A, then the acidic tail is no longer required.
FIGURE 6:
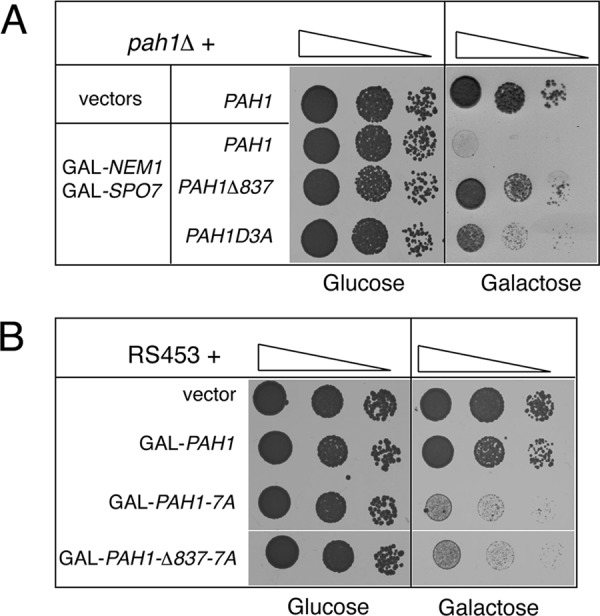
Mutation of the Pah1p acidic stretch rescues the NEM1-SPO7-dependent but not the PAH1 7A -dependent lethality. (A) Serial dilutions of pah1Δ cells carrying YCpLac33-URA3-GAL-NEM1 and pRS313-HIS3-GAL-SPO7 or the corresponding empty vectors, and the indicated YCplac111-LEU2-based PAH1 constructs, were spotted onto synthetic plates supplemented with either glucose (left) or galactose (right) and grown for 1.5 or 3 d, respectively, at 30°C. (B) Serial dilutions of RS453 wild-type cells expressing the indicated YEplac181-LEU2-GAL–inducible constructs were spotted onto synthetic plates lacking inositol and supplemented with either glucose (left) or galactose (right). Plates were grown for 1.5 or 3 d, respectively, at 30°C.
Second, overexpression of PAH1 from either a high- or a low-copy plasmid suppresses the slow growth of nem1Δ, which could be accounted for by a low level of dephosphorylated Pah1p generated spontaneously or by the action of some other phosphatase (Santos-Rosa et al., 2005; Figure 7). If the main function of the acidic stretch is to mediate the interaction of Pah1p with the Nem1p-Spo7p complex, then removing it should not influence the ability of PAH1 to rescue cells lacking NEM1. Indeed, as shown in Figure 7, similar to wild-type PAH1, both PAH1Δ837 and PAH1D3A are able to rescue equally well the growth defect of nem1Δ at 37°C. Taken together, these data strongly support the notion that the main function of the acidic tail is to mediate the Nem1p-Spo7p–dependent activation of Pah1p.
FIGURE 7:
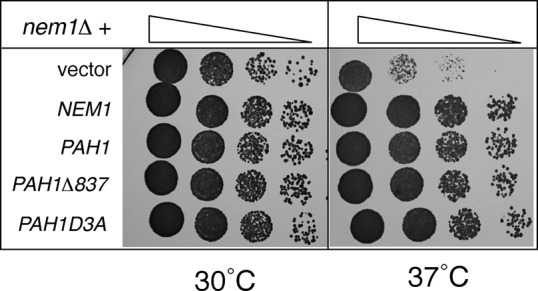
The acidic stretch is not required for the suppression of the nem1Δ growth defect by PAH1. Serial dilutions of nem1Δ cells transformed with an empty YEplac181 vector or the same vector containing PAH1, PAH1Δ837, and PAH1D3A, and pUN100-NEM1, were spotted onto synthetic plates and grown for 2 d at 30 or 37°C.
The acidic tail is required for the functions of Pah1p in nuclear membrane biogenesis, INO1 gene expression, and neutral lipid metabolism
We next reasoned that if the Pah1p acidic stretch is important for its binding to the Nem1p-Spo7p complex, then acidic stretch mutants should display defects due to the lack of Pah1p dephosphorylation and membrane recruitment, such as those described in the nem1Δ or spo7Δ cells. To test this, we integrated the Δ837 or D3A mutation into the endogenous PAH1 chromosomal locus. Because nem1Δ cells display nuclear/ER membrane expansion (Siniossoglou et al., 1998), we first examined nuclear morphology. In contrast to the round nuclei of wild-type cells, both PAH1Δ837 and PAH1D3A integrants displayed irregularly shaped nuclei (Figure 8A), although these were less severe when compared with the pah1Δ cells (Santos-Rosa et al., 2005; data not shown). A PAH1Δ837 mutant expressed by a centromeric plasmid in a pah1Δ strain displayed a similar phenotype, which was rescued when combined with the 7A mutations (Figure 8B). Therefore, and consistent with GAL-PAH1Δ837-7A lethality (Figure 6B), deletion of the acidic tail does not affect the function of Pah1p once the latter is dephosphorylated.
FIGURE 8:
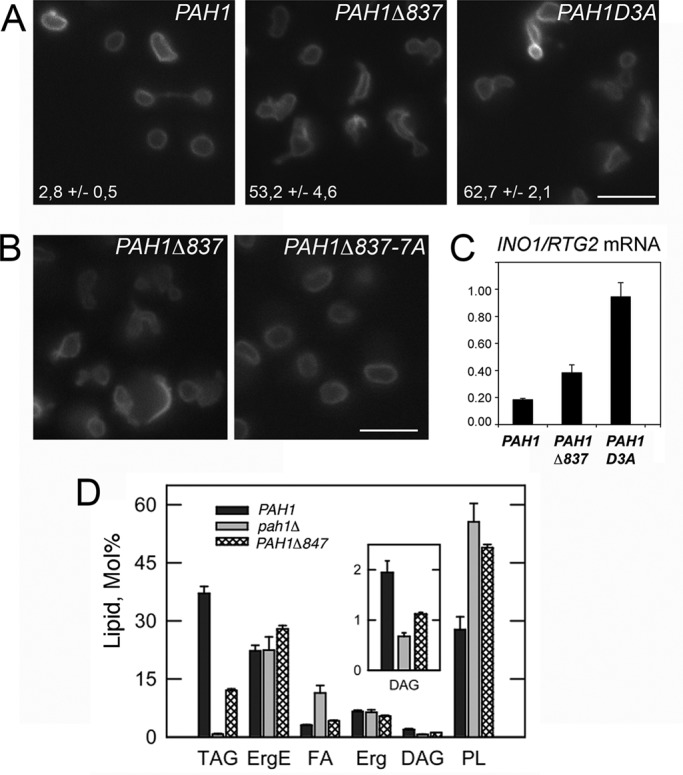
The Pah1p acidic stretch is required for nuclear structure, INO1 gene expression, and neutral lipid synthesis. (A) Cells with the PAH1-KanMX4, PAH1Δ837-KanMX4, or PAH1D3A-KanMX4 alleles integrated into the PAH1 locus were transformed with the nuclear envelope reporter HEH2-GFP and examined by fluorescence microscopy. The percentage of cells displaying irregular nuclear morphology (excluding mitotic cells) is given. Values represent the mean ± SD of three experiments for each strain, with >100 cells counted for each. Bar, 5 μm. (B) pah1Δ cells expressing the indicated PAH1 mutants from a YCplac111 vector were transformed with HEH2-GFP and examined by fluorescence microscopy. Bar, 5 μm. (C) The mRNA levels of INO1 were analyzed in the indicated strains grown in yeast extract/peptone/dextrose medium by quantitative real-time PCR. Amplification of each sample was performed in duplicate and normalized to RTG2. Values and ± SDs were calculated from three experiments. (D) Neutral lipid composition of the indicated strains in the stationary phase of growth. Cells were grown to stationary phase in the presence of [2-14C]acetate, and lipids were extracted, separated, and quantified by one-dimensional TLC as described in Materials and Methods. Each data point represents the mean ± SD of three experiments. DAG, diacylglycerol; Erg, ergosterol; ErgE, ergosterol ester; FA, fatty acid; PL, phospholipid; TAG, triacylglycerol.
Because lack of Pah1p dephosphorylation leads to derepression of the INO1 gene expression (Santos-Rosa et al., 2005), we asked whether the acidic stretch mutants could display a similar defect. Indeed, INO1 mRNA levels increase in both PAH1Δ837 and PAH1D3A cells twofold and fivefold, respectively (Figure 8C). We next tested the functional significance of the acidic tail in lipid production. Lack of PAH1 decreases both DAG levels, blocks TAG production, and increases total phospholipid levels (Han et al., 2006; Figure 8D). Chromosomal integration of a PAH1 allele lacking the 15 carboxy-terminal residues of the acidic tail also led to significant decrease of both DAG and TAG levels. The tail mutant also displayed an increase in total phospholipids (Figure 8D), consistent with the derepression of INO1 transcription and membrane biogenesis defects. Taken together, these data show that the acidic tail of Pah1p is required for the functions of Pah1p in lipid and membrane biogenesis.
DISCUSSION
In this study we showed that a prime role for the acidic C-terminal tail of Pah1p is to mediate its activation through its physical interaction with the transmembrane phosphatase Nem1p-Spo7p. Several lines of evidence support the conclusion that the Pah1p tail is required for this function in vivo. First, the tail is necessary and sufficient for the Nem1p-Spo7p–driven membrane targeting of Pah1p. Second, mutation of the tail disrupts the binding of Pah1p to Nem1p-Spo7p. This binding may be direct or indirect, mediated by a third, as-yet-unidentified partner. Third, the same tail mutations rescue the lethality caused by the overexpression of Nem1p-Spo7p. Fourth, the tail mutations phenocopy nem1Δ and spo7Δ mutants. We also showed that the acidic tail is dispensable once Pah1p is dephosphorylated, as shown by the function of PAH1Δ837-7A. Therefore we propose that the acidic stretch functions at an initial step during which Pah1p interacts with Nem1p-Spo7p. Once dephosphorylated by the latter, Pah1p can associate with membranes independently of the Nem1p-Spo7p complex via an N-terminal amphipathic helix (Karanasios et al., 2010), and therefore the tail is no longer needed for the function of Pah1p. These data are consistent with a model in which the C-terminal acidic tail and the N-terminal amphipathic helix cooperate sequentially during the membrane binding of Pah1p.
We also demonstrate here that overexpression of Nem1p-Spo7p induces lipid droplet biogenesis, manifested as an increase in droplet numbers, in a pathway that is mediated by Pah1p. Because Pah1p regulates triglyceride and phospholipid synthesis, both of which affect droplet biogenesis, multiple, non–mutually exclusive pathways could be responsible for this phenotype. Constitutive membrane anchoring of Pah1p by Nem1p-Spo7p would increase production of DAG and thus drive TAG synthesis, which may be sufficient to increase lipid droplet numbers (Yang et al., 2012). Alternatively, given that DAG generated by Pah1p has been proposed to be also important for the initial steps of lipid droplet formation at the ER membrane (Adeyo et al., 2011), Nem1p-Spo7p–anchored Pah1p could act as a platform for derepressed lipid droplet biogenesis. The formation of tightly associated droplets in clusters could be due to altered phospholipid composition either on lipid droplet monolayers (Fei et al., 2011) or on ER membranes, affecting droplet formation and dynamics (Jacquier et al., 2011). In this regard, it will be interesting to examine the effects of Nem1p-Spo7p overexepression in other lipid droplet assembly mutants. In either case, disrupting the interaction between Pah1p and Nem1p-Spo7p by mutating the acidic tail of Pah1p inhibits the recruitment of Pah1p-GFP and consequently decreases lipid droplet numbers. Because removal of the tail does not completely abrogate the effect of Nem1p-Spo7p on droplets, it is likely that Pah1pΔ837 retains some affinity for Nem1p-Spo7p through a second binding site. Interaction with Nem1p-Spo7p under these conditions may not be readily detectable by fluorescence microscopy with a Pah1pΔ837-GFP fusion, as is the case of wild-type cells and non-NEM1/SPO7 induction conditions. Nevertheless, that the Δ837 mutation alone can suppress GAL-NEM1/SPO7 lethality and significantly decrease lipid droplet cluster formation strongly supports an important functional role for the Pah1p acidic tail in Nem1p-Spo7p–dependent activation in vivo.
Of interest, in addition to the effects on lipid droplet biogenesis, cells overproducing Nem1p-Spo7p also arrest with an undivided nucleus and a short spindle at the bud neck (Santos-Rosa et al., 2005). It is tempting to speculate that these two events may be linked. Dephosphorylation of PA by Pah1p determines whether the glycerol backbones and fatty acids will be channeled toward TAG for storage or phospholipids for membrane biogenesis and growth through the CDP-DAG pathway. It is conceivable that driving the lipid biosynthetic precursors toward storage by anchoring Pah1p onto the membrane may restrict the amount of lipids available for nuclear membrane expansion during anaphase or activate a checkpoint that monitors availability of lipids for membrane biogenesis and growth. According to an alternative possibility, cell cycle arrest by membrane-bound Pah1p causes triglyceride and lipid droplets to gradually accumulate over membrane phospholipid growth that would normally be needed for cell division.
Ectopic expression of the mammalian Nem1p-Spo7p orthologue complex CTDNEP1-NEP1R1 leads to an increase of the nuclear fraction of lipin 1b (Han et al., 2012). We did not detect any significant change in its soluble intranuclear pool after GAL-NEM1/SPO7 overexpression, suggesting that Pah1p-GFP is stably bound to membrane-anchored Nem1p-Spo7p. Because of the very low levels of PAH1-GFP expression (our experiments were performed with the endogenous PAH1 promoter and centromeric plasmid), however, we cannot exclude more subtle changes in the distribution of Pah1p-GFP. More detailed studies will be required to address this possibility.
Highly acidic sequences of variable length are always found at the C-terminal tails of all other yeast lipin orthologues, and our results suggest that they may also be required for interaction to their respective Nem1p-like phosphatases. Mammalian lipins differ from their yeast counterparts in that they lack the long C-terminal tails attached next to their C-LIP domains (Figure 1B). Of interest, the C-terminus of mouse lipin 2 ends with the peptide DLDLDDLA, which is reminiscent of the longer acidic repeats, interrupted by hydrophobic residues, seen in yeasts. On the other hand, lipin 1 lacks acidic residues in its C-terminal end and is still a well-known target of CTDNEP1-NEP1R1 (Kim et al., 2007; Han et al., 2012), suggesting the presence of different and/or multiple modes of interaction with the mammalian phosphatase.
Stretches of 15–25 contiguous acidic residues are not that uncommon in the yeast proteome (Kleiger et al., 2009a). Such negatively charged sequences can promote very special types of interactions with ligands through ionic bonds. One important property of electrostatically driven bonding is the very fast on-rate for binding, like those described for the assembly of the Cdc34p-SCF complex, which catalyzes the transfer of polyubiquitin chains on substrates and depends on the highly acidic tail of Cdc34p (Kleiger et al., 2009b). A similar dynamic interaction may allow fast cycles of Pah1p recruitment to the Nem1p-Spo7p complex and could be important for the dynamics of PA dephosphorylation in vivo and the processivity of the enzyme. Future work will address these possibilities.
MATERIALS AND METHODS
Yeast strains, media, and growth conditions
Yeast strains used in this study are listed in Table 1. PAH1 mutants expressed from centromeric plasmids were first transformed in a pah1Δ strain carrying a pURA-PAH1 plasmid (SS1746), followed by growth on 5-fluoroorotic acid (5-FOA)–containing media to select for loss of the pURA-PAH1 plasmid. Yeast cells were grown in synthetic medium (SC) containing 2% glucose, lacking the appropriate amino acids for plasmid selection. For the Nem1p-Spo7p-Pah1p interaction experiments, pah1Δ cells expressing the appropriate PAH1-PtA fusion and carrying the GAL-NEM1-Myc and GAL-SPO7 plasmids were grown in SC medium containing 2% raffinose, pelleted, resuspended into SC medium containing 2% galactose to a final OD600 of 0.1, and grown for further 8 h. For growth assays on plates, yeast cells were grown in the corresponding SC liquid medium to early logarithmic phase. Ten microliters of serial 10-fold dilutions was spotted onto the appropriate SC plates and incubated at 30°C for 2–4 d. To assay growth of yeast cells in media lacking inositol, synthetic medium was prepared using yeast nitrogen base lacking inositol (Bio101). To integrate the PAH1Δ837 and PAH1D3A into the endogenous chromosomal locus of PAH1, the PAH1-KanMX4 cassettes, carrying 0.7 kb upstream of the ATG codon and 0.3 kb downstream of KanMX4, were excised as XbaI fragments and transformed in wild-type RS453 cells. Integrants were selected by resistance to G418. To construct control strains, the wild-type PAH1-KanMX4 cassette was also integrated in parallel. To integrate the pah1::URA3, the cassette was released by XbaI/HindIII digest from pGH338 and transformed in wild-type W303-1A cells, and transformants showing uracil prototrophy were selected. To integrate PAH1Δ847, the cassette was released by XbaI/SphI digest from pGH315-CD15 and transformed in the pah1::URA3 strain, and transformants showing resistance to 5-FOA were selected. In all cases, correct integrants were verified by sequencing of genomic DNA.
TABLE 1:
Yeast strains used in this study.
| Strain | Genotype | Reference/source |
|---|---|---|
| RS453 | MATα ade2-1 his3-11,15 ura3-52 leu2-3112 trp1-1 | Wimmer et al. (1992) |
| SS1746 | RS453 pah1::TRP1 + YCplac33-URA3-PAH1 | This study |
| SS2017 | RS453 pah1::TRP1 ERG6-Cherry::KANMX4 + YCplac33-URA3-PAH1 | This study |
| SS1423 | RS453 pah1::TRP1 Cherry-PUS1::URA3 | This study |
| SS1328 | RS453 PAH1::KANMX4 | This study |
| SS1829 | RS453 PAH1 Δ837::KANMX4 | This study |
| SS1363 | RS453 PAH1 D3A::KANMX4 | This study |
| W303-1A | MATa ade2-1 can1-100 his3-11,15 leu2-3112 trp1-1 ura3-1 | Thomas and Rothstein (1989) |
| GHY67 | W303-1A PAH1 Δ847 | This study |
Plasmid constructs
Plasmids used in this study are listed in Supplemental Table S1. PAH1 carboxy-terminal truncation mutants were constructed by introducing a BamHI or XhoI site by PCR just after the codon corresponding to the last amino acid of the respective mutant, followed by the stop codon and PAH1 terminator. Internal truncation mutants were constructed by introducing a BamHI site at the site of the deletion. Point mutations at the carboxy-terminal acidic stretch of Pah1p were introduced by PCR-mediated mutagenesis. For tagging of PAH1 mutants, two IgG-binding domains of protein A or the S65T/V163A variant of GFP were introduced in-frame before the stop codon. All PAH1 mutants were expressed under the control of the PAH1 promoter and centromeric vectors. To introduce PAH1Δ837 and PAH1D3A into the endogenous chromosomal locus of PAH1, the mutant alleles were first cloned into a pBluescript vector and the KanMX4 gene was introduced 170 nucleotides downstream of the PAH1 stop codon and integrated as described later. Plasmids pGH338 and pGH315-CD15 carrying the pah1::URA3 and PAH1Δ847 cassettes, respectively, were used to integrate them as described later. Mouse lipin 2, human CTDNEP1, and yeast NEM1 were all tagged by introducing a BamHI site before the stop codon and inserting in-frame the protein A, GFP. or 3xMyc tags. Lipin 2-PtA fusion was expressed under the control of the PAH1 promoter, CTDNEP1 and NEP1R1 under the control of the NOP1 promoter, and NEM1 and SPO7 under the control of the inducible GAL1/10 promoter. The fusions of the Pah1p carboxy-terminal domain to lipin 2 were constructed by introducing an XbaI site before the stop codon of lipin 2 and inserting the Pah1p sequences corresponding to Pah1p S586-D862 or M837-D862 as XbaI fragments followed by the protein A tag. The fusion of the Pah1p acidic tail to GFP was constructed by introducing a NotI site after the start codon and inserting the Pah1p sequences corresponding to Pah1p D838-D862 followed by GFP. All constructs were verified by DNA sequencing.
Affinity purification of Pah1p-PtA and Lipin 2-PtA
Cells were grown in galactose to induce GAL-NEM1 and GAL-SPO7, as described, washed once with water, and incubated for 10 min in 100 mM Tris-HCl, pH 9.4, and 10 mM dithiothreitol at room temperature. Cells were then spheroplasted with Zymolyase 20T (5 mg/g of cells) for 15 min at room temperature in spheroplasting buffer (1.2 M sorbitol, 20 mM K2HPO4/KH2PO4, pH 7.4). Cells were washed once with spheroplasting buffer, and the cell pellet was frozen in liquid nitrogen. Affinity purification of the PtA fusions with IgG-Sepharose was done as described (Siniossoglou et al., 2000). PtA fusions were eluted with acetic acid, pH 3.4, freeze-dried, and resuspended in SDS-sample buffer. Protein identification by mass spectrometry of tryptic fragments was performed as previously described (O'Hara et al., 2006). To assay binding to CTDNEP1-NEP1R1, pull-downs were performed identically, with the exception that cells were grown in glucose. The anti–protein A antibody was from Dako (Z0113; Carpinteria, CA) and the anti–cMyc-peroxidase antibody from Roche (11814150001; Indianapolis, IN). Blot signals were developed using enhanced chemiluminescence (GE Healthcare, Piscataway, NJ).
RNA extractions and quantitative PCR
Total RNA was isolated from yeast cells using the hot phenol method as described previously (O'Hara et al., 2006). RNA was subsequently purified using the RNeasy Mini Kit (Qiagen, Valencia, CA) following the manufacturer's instructions. Quantitative real-time PCR was performed on total RNA using an ABI 7900HT system and a SYBR Green Mastermix (Applied Biosystems, Foster City, CA) as described previously (O'Hara et al., 2006).
Labeling and analysis of lipids
Steady-state labeling of neutral lipids with [2-14C]acetate was performed as described previously (Han et al., 2006). Neutral lipids were separated by one-dimensional TLC on silica gel plates using a solvent system of hexane/diethyl ether/glacial acetic acid (40:10:1, vol/vol/vol). The identity of the labeled lipids on TLC plates was confirmed by comparison with standards after exposure to iodine vapor. Radiolabeled lipids were visualized by phosphorimaging and subjected to ImageQuant analysis.
Microscopy
Cells grown at 30°C in synthetic medium according to the selection marker of the plasmids were pelleted when reached mid–log phase, resuspended in the same medium, and immediately imaged live at room temperature. For GAL-NEM1 and GAL-SPO7 inductions, cells were grown in galactose as described. Images were acquired with an epifluorescence microscope (Axioplan; Carl Zeiss, Jena, Germany) using a 100× Plan-Apochromatic 1.4 numerical aperture objective lens (Carl Zeiss) connected to an Orca R2 charge-coupled device camera and controlled by Simple PCI6 software (Hamamatsu, Hamamatsu, Japan). The brightness and contrast of the resulting images were adjusted using Photoshop (Adobe, San Jose, CA). For lipid droplet labeling, cells were stained for 10 min with 1.25 μg/ml of BODIPY 493/503 (D-3922; Invitrogen, Carlsbad, CA) at room temperature, mounted on glass slides, covered with cover glasses, and sealed with nail polish and imaged with a Zeiss LSM710 confocal microscope.
Electron microscopy
For galactose inductions, cells were grown in galactose-containing medium for 5 h, as described. Cells were then collected by centrifugation, chemically fixed, embedded in 12% gelatin, and cryosectioned as described previously (Griffith et al., 2008). For better preservation of lipid droplets, cryosections were picked up with 0.47% uranyl acetate (Jacquier et al., 2011) before being immediately stained and viewed in a JEOL1010 electron microscope (JEOL, Tokyo, Japan). For statistical analyses, the numbers of structures, for example, lipid droplets or clusters of lipid droplets, per cell section were determined by counting 200 randomly selected profiles. The clusters of lipid droplets were defined as a concentration of more than three lipid droplets in close proximity, and each cluster was counted as one individual structure when determining the average number of clusters of lipid droplets per cell section. SDs were used to perform a t test, confirming the significance of the data (p < 0.05).
Supplementary Material
Acknowledgments
This work was supported by grants from the Medical Research Council (G0701446) to S.S., the National Institutes of Health (GM-50679) to G.M.C., and the ECHO (700.59.003), ALW Open Program (821.02.017), Deutsche Forschungsgemeinschaft–Nederlands Organisatie voor Wetenschappelijk Onderzoek cooperation (DN82-303), and Netherlands Organisation for Health Research and Development (Vici) to F.R. We thank Sew Peak Chew for help with protein identification, Michael Wilson for bioinformatic advice, and Helena Santos-Rosa for critical reading of the manuscript.
Abbreviations used:
- CDP-DAG
cytidine diphosphate diacylglycerol
- DAG
diacylglycerol
- ER
endoplasmic reticulum
- PA
phosphatidate
- TAG
triacylglycerol
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-01-0021) on May 8, 2013.
*These authors contributed equally to this work.
†Present address: Signaling Program, Babraham Institute, Cambridge CB22 3AT, United Kingdom.
REFERENCES
- Adeyo O, Horn PJ, Lee S, Binns DD, Chandrahas A, Chapman KD, Goodman JM. The yeast lipin orthologue Pah1p is important for biogenesis of lipid droplets. J Cell Biol. 2011;192:1043–1055. doi: 10.1083/jcb.201010111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi HS, Su WM, Han GS, Plote D, Xu Z, Carman GM. Pho85p-Pho80p phosphorylation of yeast Pah1p phosphatidate phosphatase regulates its activity, location, abundance, and function in lipid metabolism. J Biol Chem. 2012;287:11290–11301. doi: 10.1074/jbc.M112.346023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi HS, Su WM, Morgan JM, Han GS, Xu Z, Karanasios E, Siniossoglou S, Carman GM. Phosphorylation of phosphatidate phosphatase regulates its membrane association and physiological functions in Saccharomyces cerevisiae: identification of SER(602), THR(723), AND SER(744) as the sites phosphorylated by CDC28 (CDK1)-encoded cyclin-dependent kinase. J Biol Chem. 2011;286:1486–1498. doi: 10.1074/jbc.M110.155598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donkor J, Sariahmetoglu M, Dewald J, Brindley DN, Reue K. Three mammalian lipins act as phosphatidate phosphatases with distinct tissue expression patterns. J Biol Chem. 2007;282:3450–3457. doi: 10.1074/jbc.M610745200. [DOI] [PubMed] [Google Scholar]
- Fei W, et al. A role for phosphatidic acid in the formation of “supersized” lipid droplets. PLoS Genet. 2011;7:e1002201. doi: 10.1371/journal.pgen.1002201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith J, Mari M, De Maziere A, Reggiori F. A cryosectioning procedure for the ultrastructural analysis and the immunogold labelling of yeast Saccharomyces cerevisiae. Traffic. 2008;9:1060–1072. doi: 10.1111/j.1600-0854.2008.00753.x. [DOI] [PubMed] [Google Scholar]
- Grimsey N, Han GS, O'Hara L, Rochford JJ, Carman GM, Siniossoglou S. Temporal and spatial regulation of the phosphatidate phosphatases lipin 1 and 2. J Biol Chem. 2008;283:29166–29174. doi: 10.1074/jbc.M804278200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han GS, Wu WI, Carman GM. The Saccharomyces cerevisiae Lipin homolog is a Mg2+-dependent phosphatidate phosphatase enzyme. J Biol Chem. 2006;281:9210–9218. doi: 10.1074/jbc.M600425200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S, Bahmanyar S, Zhang P, Grishin N, Oegema K, Crooke R, Graham M, Reue K, Dixon JE, Goodman JM. Nuclear envelope phosphatase 1-regulatory subunit 1 (formerly TMEM188) is the metazoan Spo7p ortholog and functions in the lipin activation pathway. J Biol Chem. 2012;287:3123–3137. doi: 10.1074/jbc.M111.324350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris TE, Finck BN. Dual function lipin proteins and glycerolipid metabolism. Trends Endocrinol Metab. 2011;22:226–233. doi: 10.1016/j.tem.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry SA, Kohlwein SD, Carman GM. Metabolism and regulation of glycerolipids in the yeast Saccharomyces cerevisiae. Genetics. 2012;190:317–349. doi: 10.1534/genetics.111.130286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquier N, Choudhary V, Mari M, Toulmay A, Reggiori F, Schneiter R. Lipid droplets are functionally connected to the endoplasmic reticulum in Saccharomyces cerevisiae. J Cell Sci. 2011;124:2424–2437. doi: 10.1242/jcs.076836. [DOI] [PubMed] [Google Scholar]
- Karanasios E, Han GS, Xu X, Carman GM, Siniossoglou S. A phosphorylation-regulated amphipathic helix controls the membrane translocation and function of the yeast phosphatidate phosphatase. Proc Natl Acad Sci USA. 2010;107:17539–17544. doi: 10.1073/pnas.1007974107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Gentry MS, Harris TE, Wiley SE, Lawrence JC, Jr, Dixon JE. A conserved phosphatase cascade that regulates nuclear membrane biogenesis. Proc Natl Acad Sci USA. 2007;104:6596–6601. doi: 10.1073/pnas.0702099104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleiger G, Hao B, Mohl DA, Deshaies RJ. The acidic tail of the Cdc34 ubiquitin-conjugating enzyme functions in both binding to and catalysis with ubiquitin ligase SCFCdc4. J Biol Chem. 2009a;284:36012–36023. doi: 10.1074/jbc.M109.058529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleiger G, Saha A, Lewis S, Kuhlman B, Deshaies RJ. Rapid E2-E3 assembly and disassembly enable processive ubiquitylation of cullin-RING ubiquitin ligase substrates. Cell. 2009b;139:957–968. doi: 10.1016/j.cell.2009.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lussier M, et al. Large scale identification of genes involved in cell surface biosynthesis and architecture in Saccharomyces cerevisiae. Genetics. 1997;147:435–450. doi: 10.1093/genetics/147.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nohturfft A, Zhang SC. Coordination of lipid metabolism in membrane biogenesis. Annu Rev Cell Dev Biol. 2009;25:539–566. doi: 10.1146/annurev.cellbio.24.110707.175344. [DOI] [PubMed] [Google Scholar]
- O'Hara L, Han GS, Peak-Chew S, Grimsey N, Carman GM, Siniossoglou S. Control of phospholipid synthesis by phosphorylation of the yeast lipin Pah1p/Smp2p Mg2+-dependent phosphatidate phosphatase. J Biol Chem. 2006;281:34537–34548. doi: 10.1074/jbc.M606654200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterfy M, Phan J, Xu P, Reue K. Lipodystrophy in the fld mouse results from mutation of a new gene encoding a nuclear protein, lipin. Nat Genet. 2001;27:121–124. doi: 10.1038/83685. [DOI] [PubMed] [Google Scholar]
- Phan J, Péterfy M, Reue K. Lipin expression preceding peroxisome proliferator-activated receptor-gamma is critical for adipogenesis in vivo and in vitro. J Biol Chem. 2004;279:29558–29564. doi: 10.1074/jbc.M403506200. [DOI] [PubMed] [Google Scholar]
- Santos-Rosa H, Leung J, Grimsey N, Peak-Chew S, Siniossoglou S. The yeast lipin Smp2 couples phospholipid biosynthesis to nuclear membrane growth. EMBO J. 2005;24:1931–1941. doi: 10.1038/sj.emboj.7600672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasser T, Qiu QS, Karunakaran S, Padolina M, Reyes A, Flood B, Smith S, Gonzales C, Fratti RA. Yeast lipin 1 orthologue pah1p regulates vacuole homeostasis and membrane fusion. J Biol Chem. 2012;287:2221–2236. doi: 10.1074/jbc.M111.317420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniossoglou S, Peak-Chew SY, Pelham HR. Ric1p and Rgp1p form a complex that catalyses nucleotide exchange on Ypt6p. EMBO J. 2000;19:4885–4894. doi: 10.1093/emboj/19.18.4885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniossoglou S, Santos-Rosa H, Rappsilber J, Mann M, Hurt E. A novel complex of membrane proteins required for formation of a spherical nucleus. EMBO J. 1998;17:6449–6464. doi: 10.1093/emboj/17.22.6449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas BJ, Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989;56:619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- Wimmer C, Doye V, Grandi P, Nehrbass U, Hurt EC. A new subclass of nucleoporins that functionally interact with nuclear pore protein NSP1. EMBO J. 1992;11:5051–5061. doi: 10.1002/j.1460-2075.1992.tb05612.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Galea A, Sytnyk V, Crossley M. Controlling the size of lipid droplets: lipid and protein factors. Curr Opin Cell Biol. 2012;24:509–516. doi: 10.1016/j.ceb.2012.05.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.