Abstract
In order to elucidate the function of Myc in the maintenance of pluripotency and self-renewal in mouse embryonic stem cells (mESCs), we screened for novel embryonic stem cell (ESC)-specific interactors of Myc by mass spectrometry. Undifferentiated embryonic cell transcription factor 1 (Utf1) was identified in the screen as a putative Myc binding protein in mESCs. We found that Myc and Utf1 directly interact. Utf1 is a chromatin-associated factor required for maintaining pluripotency and self-renewal in mESCs. It can also replace c-myc during induced pluripotent stem cell (iPSC) generation with relatively high efficiency, and shares target genes with Myc in mESCs highlighting a potentially redundant functional role between Myc and Utf1. A large region of Utf1 was found to be necessary for direct interaction with N-Myc, while the basic helix-loop-helix leucine zipper domain of N-Myc is necessary for direct interaction with Utf1.1
Keywords: mESCs, Transcription, Myc, Utf1
INTRODUCTION
Myc proteins are basic-helix-loop-helix leucine zipper (bHLHZ) transcription factors that dimerize with a related, small bHLHZ protein called Max [1]. Myc-Max binds DNA at E-box sequences at both canonical (CACGTG) and non-canonical sites (CANNTG) resulting in recruitment of multiple coactivator complexes and gene activation [2,3]. Myc can also function as a transcriptional repressor at least in part through the interaction with the transcriptional activator Miz-1 [4,5]. Myc regulates a wide variety of genes including those important in cell growth, ribosome biogenesis, protein synthesis, cell cycling, and metabolism [6,7] at least in part through direct regulation of such genes, but also potentially through global chromatin regulation [8,9,10,11]. Myc also contributes to such cellular processes as proliferation, differentiation, and apoptosis (reviewed in [12]).
Elucidating the mechanism by which Myc controls self-renewal and pluripotency in ESCs is an emerging area of interest within the Myc field. myc genes are expressed during early embryogenesis with N-myc most highly expressed in ESCs and c-myc in extraembryonic cells [13,14,15]. N- or c-myc knockout results in embryonic lethality at E11.5 or E10.5, respectively [16,17]. The embryonic lethality resulting from a c-myc knockout is rescued when c-myc is replaced with N-myc [18]. These results highlight the importance of N- and c-Myc in early embryo development as well as their substantially redundant roles. Simultaneous disruption of both N- and c-myc expression by conditional knockout in mESCs leads to loss of self-renewal and pluripotency [19].
Myc is also a potent driver of cellular reprogramming, enhancing the production of iPSCs by one-to-two orders of magnitude when combined with Oct4, Sox2, and Klf4. While Myc has subsequently been shown not to be formally required for the iPSC production process [20], it consistently enhances the efficiency of production dramatically and a role for endogenous Myc in iPSC formation is clear [21]. Utf1 can substitute for Myc in the iPSC production process by an unknown mechanism [22] suggesting links between the functions of Myc and Utf1.
Utf1 is highly expressed and restricted to cells of the inner cell mass (ICM), ESCs, and germ line tissue [23]. Knockdown of Utf1 in mESCs results in a delay or block of differentiation [24]. It functions as a translational and epigenetic factor that binds to approximately 75,000 different sites on mESC chromatin [25]. Over 1,700 Utf1 target genes overlap with other factors that maintain ESC pluripotency including Nanog, Oct4, Klf4, and in particular c-Myc [26], supporting the notion that Myc and Utf1 may have related or overlapping functions in ESCs. Utf1 expression is also an early stage indicator of successful reprogramming [27].
To address the molecular mechanisms of Myc function in maintaining distinctive ESC characteristics, we conducted a proteomics screen to identify pluripotency and self-renewal-related Myc cofactors in mESCs. Both known and putative novel cofactors of Myc were identified. Amongst the candidate novel Myc interactors identified in this screen was Utf1. We focused on the putative interaction between Myc and Utf1 due to Utf1’s restricted expression pattern to ICM cells, ESCs, and germ line tissue and potentially redundant functional roles with Myc in ESCs and reprogramming.
MATERIALS AND METHODS
Myc Co-Immunoprecipitation (co-IP)
Lysis buffer (50mM Hepes pH 7.9, 1mM MgCl2, 20mM NaCl, 10mM EDTA pH 8.0, 10% glycerol, 0.2% NP-40, and 1× protease inhibitor) was added onto mouse embryonic fibroblast-depleted F1A mESCs on 3×15cm plates in order to lyse the cells and maintain Myc protein complexes. Myc complexes were immunoprecipitated with either antimouse N-Myc (Abcam Inc., Cambridge, MA) or anti-rabbit c-Myc (Santa Cruz Biotechnology, Santa Cruz, CA) antibody and either G or A sepharose beads (GE Healthcare Life Sciences, Piscataway, NJ), respectively. Cell lysates without antibody added were used as negative controls. Beads were washed 4 times with 450μL lysis buffer. Samples were then washed 5 times and resuspended with 450μL of 55mM ammonium bicarbonate in preparation for mass spectrometry analysis.
Mass Spectrometry
The UC Davis Genome Center Proteomics Core Facility carried out tandem mass spectrometry on Myc immunoprecipitated samples. Mass spectrometry results were viewed with Scaffold Viewer 3 Software (Proteome Software Inc., Portland, OR).
GST-Fusion Protein Expression
pGEX plasmids with full length mouse Utf1, Max, truncated/mutated Utf1, or truncated/mutated N-myc were transformed into BL21 cells (Sigma-Aldrich, St. Louis, MO). Induced cells were sonicated with a Bioruptor (Diagenode Inc., Denville, NJ) at medium setting for 15 bursts. Samples were centrifuged at 14,000×g for 5 minutes at 4°C. Supernatants were aliquoted and stored at −80°C. Protein samples were run out on SDSPAGE and stained with Coomassie to ensure proper protein size expression. Fusion protein expression was further validated by SDS-PAGE followed by western blot with rabbit anti-GST antibody (Cell Signaling Technology Inc., Boston, MA).
GST-Pull Down
20μL of GST fusion proteins were incubated with glutathione agarose beads (Sigma-Aldrich) for 1 hour rocking at 4°C. Beads were washed 3 times with 1mL PBST buffer followed by 2 washes with 1mL HEMG buffer (100mM HEPES PH 7.8, 1M KCl, 0.5M EDTA, 1M MgCl2, 10%NP-40, 50% Glycerol, 1M DTT, and 1× protease inhibitor). Plasmids containing either full-length mouse N-myc, c-myc, or Utf1 under the Sp6 promoter were expressed according to supplier protocol of the Sp6 TNT Quick Coupled Transcription/Translation System (Promega Corp., Madison, WI). 8μL of in vitro transcribed/translated protein was incubated with the GST protein:glutathione beads for 2 hours rocking at 4°C. Samples were washed 4 times with 1mL HEMG buffer. Proteins were boiled from the beads and loaded onto SDS-PAGE for further investigation via western blot. Membranes were blotted with either mouse anti-N-Myc (Abcam) or rabbit ant-Utf1 (Abcam) antibody.
Luciferase Assay
293FT cells (Invitrogen, Grand Island, NY) were transfected utilizing X-tremeGENE HP DNA transfection reagent (Roche) with plasmids containing full length N-myc( 500ng), Utf1 (250ng, 500ng, 1µg, or 2.5µg), renilla luciferase (10ng) and a reporter consisting of 4 tandem E-box sites upstream of the promoter for firefly luciferase (2µg). All cells were transfected with 5.01µg total DNA by adding appropriate amounts of pMX-empty plasmid (2.25µg, 2µg, 1.5µg or 0µg). In parallel, 293FT cells were transfected with a GFP plasmid to ensure a 70% or higher successful transfection rate. 48 hours post-transfection experimental cells were lysed and subjected to luciferase activation utilizing the dual-luciferase reporter assay system (Promega) and measured by the 1450 Microbeta TriLux luminescence counter (Perkin Elmer, Waltham, MA). Fold induction of the reporter was calculated as a ratio of firefly over renilla luciferase relative to the N-Myc, renilla, and reporter sample set normalized to 100, n=3. Student’s t-test was carried out to identify p-values amongst two specified fold induction data sets. Results shown as mean +/− standard deviation.
RESULTS
Screen for pluripotency and self-renewal-related Myc cofactors
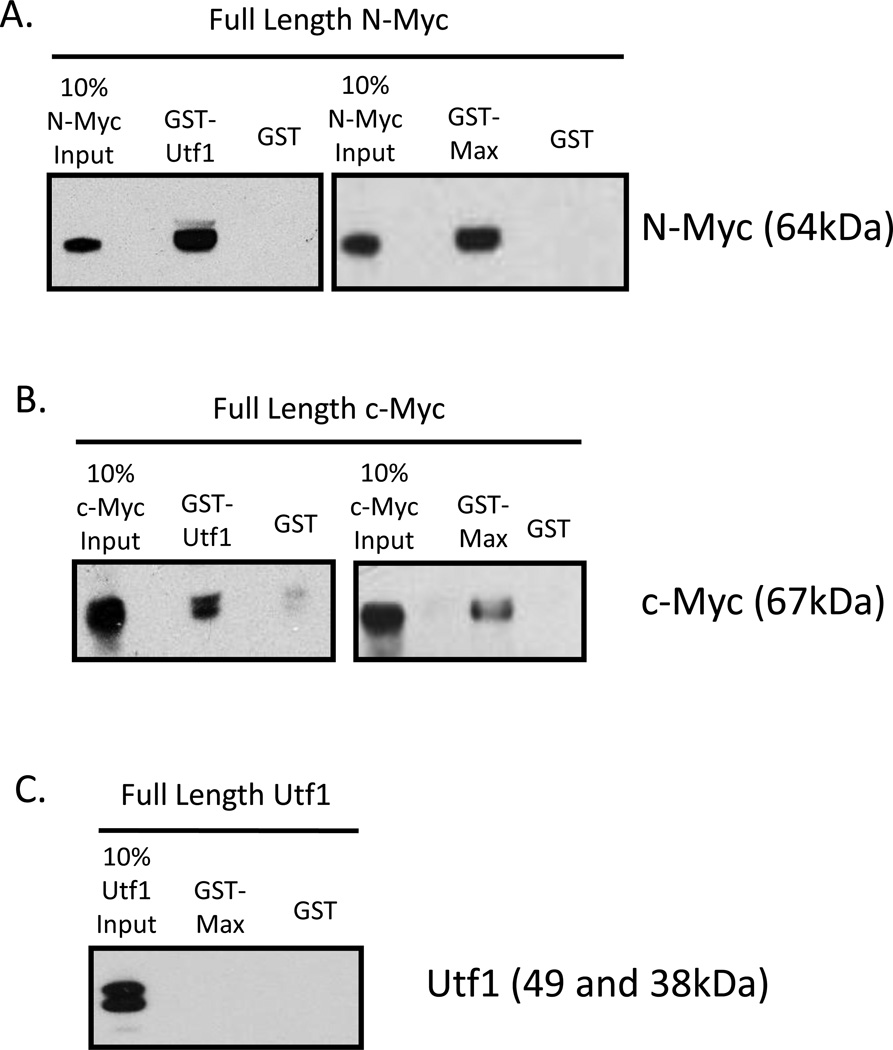
We hypothesized that Myc forms ESC-specific multi-protein complexes that target genes important for maintaining ESC characteristics. To test this notion and in order to delineate the role of Myc in maintaining ESC characteristics of pluripotency and self-renewal from its regular upkeep processes, we screened for Myc protein cofactors in mESCs. F1A mESCs were lysed under low stringency conditions to isolate Myc complexes. Lysates were subjected to either N- or c-Myc immunoprecipitation followed by tandem mass spectrometry analysis. Isolated protein complexes are listed in order of highest total peptide number and total percent peptide coverage (Table 1). The c-Myc antibody showed cross-reactivity with N-Myc protein, increasing the likelihood that cofactors isolated by c-Myc antibody are N-Myc cofactors. Our screen identified previously reported mESC-specific Myc cofactors Ddx20 [28], Ruvbl2 [29,30], and Ybx1 [28]. Also a mouse embryonic fibroblast cell-specific Myc cofactor, Nucleophosmin 1 [31] and an osteosarcoma cell-specific Myc cofactor, Dnmt3a were identified [32]. Other proteins not previously identified as Myc cofactors but that have similar number of peptide hits in our screen as previously identified Myc cofactors include Eef1a1 (Eukaryotic translation elongation factor 1 alpha 1), Hist1h2be (Histone cluster 1, H2be) and H3f3b (H3 histone, family 3B). Utf1 was also identified in the screen. Relatively strong direct interaction of Utf1 with both N-Myc and c-Myc were consistently evident (Figure 1A and 1B). N-Myc and c-Myc heterodimerizing partner, Max was used as a positive control. Interestingly, Utf1 did not directly interact with bHLHZ protein Max (Figure 1C).
Table 1. Identification of Myc Cofactors in mESCs by Co-Immunoprecipitation and Tandem Mass Spectrometry.
mESCs were lysed and immunoprecipitated two times with N- or c-Myc antibody. Isolated protein complexes identified by tandem mass spectrometry are listed in order of highest total peptide number and total percent peptide coverage. Known Myc cofactors are designated by cell type in which the interaction was previously found.
| Gene Symbol | Gene Name | Peptide # (N-N-c-c- Myc co-IP) |
Peptide # Control (No Antibody) |
Total Percent Peptide Coverage |
Range of Percent Probability |
Known Myc Cofactors (Cell Type) |
|---|---|---|---|---|---|---|
| Eef1a1 | Eukaryotic translation elongation factor 1 alpha 1 | 0-0-6-5 | 0-0 | 16 | 60–95 | |
| Ddx20 | DEAD box polypeptide 20 | 0-0-4-3 | 0-0 | 10 | 50–95 | mESCs30 |
| Hist1h2be | Histone cluster 1, H2be | 0-0-4-2 | 0-0 | 37 | 77–95 | |
| Ruvbl2 | Ruvb-like protein 2 | 0-0-4-2 | 0-0 | 7 | 75–95 | mESCs31,32 |
| H3f3b | H3 histone, family 3b | 0-0-3-3 | 0-0 | 25 | 65–95 | |
| Npm1 | Nucleophosmin 1 | 0-3-3-0 | 0-0 | 11 | 81–95 | MEFs33 |
| Ybx1 | V box protein 1 | 1-1-1-2 | 0-1 | 17 | 70–95 | mESCs30 |
| hnrnpk | Heterogeneous nuclear ribonucleoprotein K |
0-0-5-0 | 0-0 | 8 | 61–95 | |
| Hist1h1b | Histone cluster 1, H1b | 0-0-3-2 | 1-0 | 20 | 95 | |
| Dppa4 | Developmental pluripotency associated 4 | 0-2-0-0 | 0-0 | 11 | 52–69 | |
| Nono | Non-POU-domain- containing, octamer binding protein |
1-0-1-0 | 0-0 | 7 | 95 | |
| Ctrnb1 | Catenin, beta 1 | 0-2-0-0 | 0-0 | 6 | 95 | |
| Mycn | V-myc myelocytomatosis viral related oncogene, neuroblastoma derived |
0-0-1-1 | 0-0 | 10 | 80–90 | Control |
| Hist1h1a | Histone cluster 1, H1a | 0-0-0-1 | 0-0 | 15 | 95 | |
| Hist4h4 | Histone cluster 4, H4 | 0-0-1-0 | 0-0 | 12 | 94–95 | |
| Utf1 | Undifferentiated embryonic cell transcription factor 1 |
0-0-0-1 | 0-0 | 5 | 95 | 95 |
| Dnmt3a | DNA methyltransferase 3a | 1-0-0-0 | 0-0 | 2 | 76 | U2OS34 |
Figure 1.
Direct Interaction of Utf1 with N- and c-Myc but not Max.
(A) Pull down with in vitro transcribed/translated full-length N-Myc and Utf1 fusion protein identified a direct interaction between N-Myc and Utf1. N-Myc and Max were used as a positive control.
(B) Pull down with in vitro transcribed/translated full-length c-Myc and Utf1 fusion protein identified a direct interaction between c-Myc and Utf1. c-Myc and Max were used as a positive control.
(C) Pull down of in vitro transcribed/translated Utf1 and Max fusion did not identify a direct interaction between Utf1 and Max.
The bHLHZ portion of Myc is necessary for binding Utf1
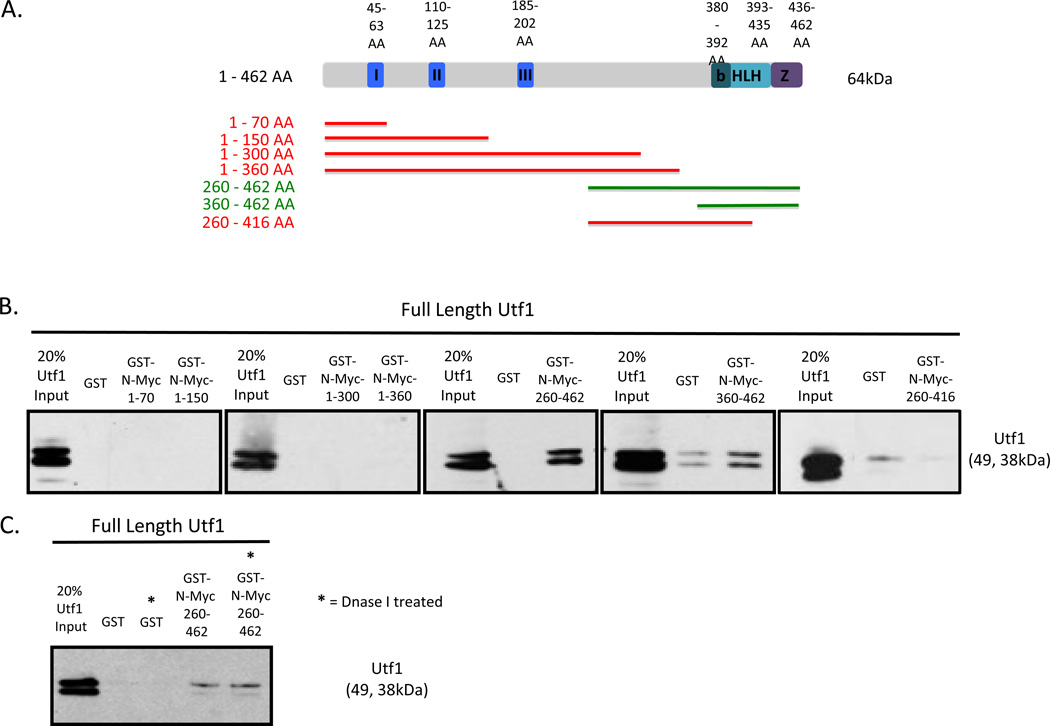
To identify the interaction domains within Myc responsible for direct Myc and Utf1 interaction, we created and tested N-Myc truncation proteins. Both N-Myc and c-Myc have six highly conserved domains in the N-terminal: Myc Box I, Myc Box II, and Myc Box III and in the C-terminal: the basic region, helix-loop-helix region, and the leucine zipper domain. Due to the substantially conserved domains of N- and c-Myc as well as the highly redundant biological roles of the proteins during early embryonic development we decided to map the interaction domains of Myc by focusing on N-Myc truncation proteins. Using previously published c-Myc amino acid sequence designations for Myc Box I, Myc Box II, basic region, helix-loop-helix, and leucine zipper [33] and Myc Box III [34] the amino acid regions of these domains were designated for N-Myc through amino acid sequence alignment (Figure 2A).
Figure 2.
The bHLHZ of N-Myc is Necessary for Direct Interaction with Utf1.
(A) Schematic diagram outlining the conserved domains of the N-Myc protein as well as N-Myc truncation proteins generated for pull down. The following bars summarize the pull down results. I = Myc Box I, II = Myc Box II, III = Myc Box III, B = Basic Region, HLH = Helix-loop-helix, Z = Leucine zipper domain, Red bar = negative pull down interaction, Green bar = positive pull down interaction.
(B) Pull down results showing positive interaction of N-Myc 260-462 AA and N-Myc-360-462 AA with both isoforms of Utf1.
(C) Pull down results after treating N-Myc 260-460 AA with DNase I.
N-Myc truncation proteins were designed and generated to determine which regions of Myc are necessary for direct interaction with Utf1 with specific regard to the well-established conserved domains. Colored bars correlating to the size of the truncated proteins are drawn as a schematic representation (Figure 2A). Red bars indicate lack of interaction with full length Utf1, and the green bars indicate positive interaction with full length Utf1. N-Myc truncation proteins expressed well and were detected by western blot prior to pull down (Suppl. Figure 1A).
The C-terminal containing the bHLHZ domain of N-Myc was found to be necessary for direct interaction with both isoforms of Utf1 (Figure 2A and 2B). The only N-Myc truncation proteins capable of binding both isoforms of Utf1 have the bHLHZ domain intact. Two isoforms of Utf1 are expressed in mESCs [35]. The second smaller isoform is due to an alternative start site at the 43rd amino acid position resulting in a smaller Utf1 protein (Figure 3A). The loss of detection with N-Myc 260–416 AA may be the result of either the requirement of the helix-loop-helix and leucine zipper domain to interact with Utf1 or the improper folding of the remaining half of the helix-loop-helix domain which then subsequently blocks Utf1 binding to the basic region of N-Myc. DNase I treatment did not abolish the interaction between N-Myc bHLHZ domain from interacting with Utf1 (Figure 2C), validating that the interaction between the two proteins is the result of the proteins directly interacting with one another through the C-terminal region of Myc and not due to DNA contamination.
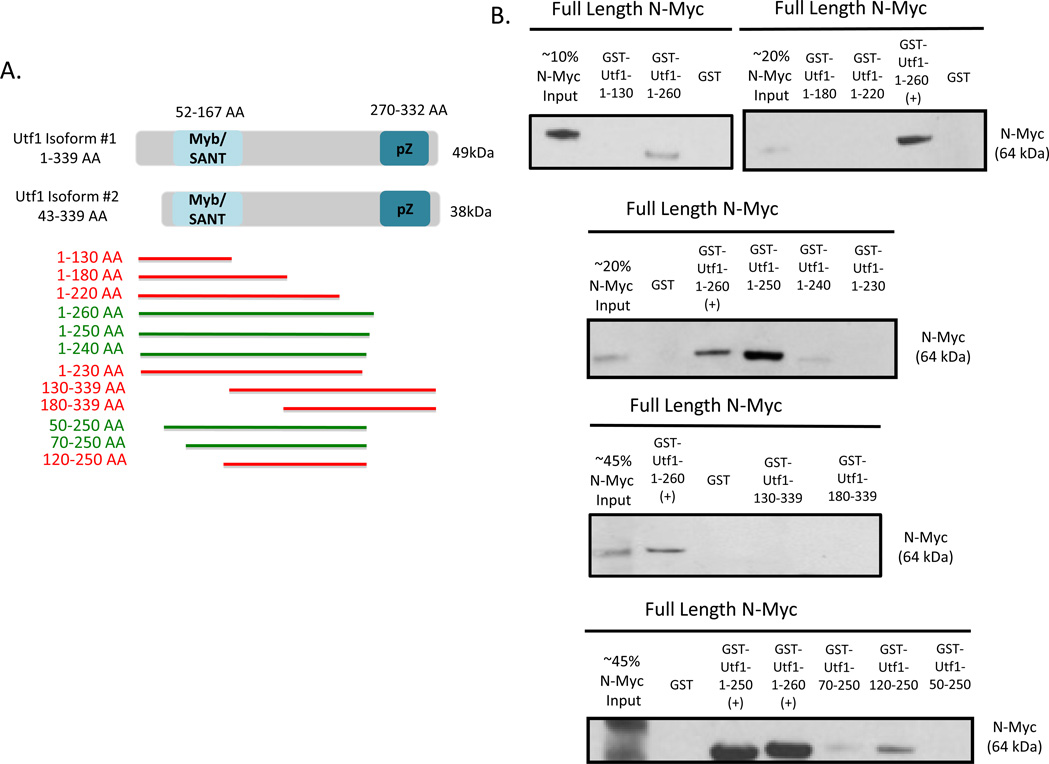
Figure 3.
One Large Region of Utf1 Required for Direct Interaction with N-Myc.
(A) Schematic diagram outlining the two endogenous isoforms of Utf1 (1–339 AA and 43–339 AA) expressed in mESCs. The smaller isoform is due to an alternative start site. Regions highlighted in blue (57–167 AA and 270–332 AA) indicate the two highly conserved domains of Utf1. Utf1 truncation proteins designed and generated for pull down are designated by subsequent bars. pZ = putative leucine zipper, Red bar = negative pull down interaction, Green bar = positive pull down interaction.
(B) Pull down results showing positive interaction of Utf1 1–260 AA, 1–250 AA, 1–240 AA, 50–250 AA, and 70–250 AA with full length N-Myc.
A large region of Utf1 is required for Myc binding
Two highly conserved domains are present within Utf1, the Myb/SANT domain near the N-terminus and a putative leucine zipper near the C-terminus [24,36] (Figure 3A). Utf1 truncations were designed and generated in regard to the highly conserved domains of Utf1 in order to determine the regions necessary for direct interaction with full length N-Myc. Utf1 truncation proteins are indicated by colored bars of approximate truncation protein lengths (Figure 3A). Green bars indicate positive direct interaction and red bars indicate lack of interaction with full length N-Myc. Proper expression of expected Utf1 truncation proteins were detected by western blot prior to carrying out pull down (Suppl. Figure 1B). The region required for interaction with full length N-Myc consists of the Myb/SANT domain, a domain known to bind DNA. Also the region required for N-Myc interaction extends towards the C-terminus of Utf1 just outside the putative leucine zipper motif (Figure 3A and 3B).
Utf1 does not influence Myc transcriptional activation at E-box promoters
We performed a luciferase assay to determine whether the direct interaction between N-Myc and Utf1 affects Myc activity at E-box containing gene targets. 293FT cells were transfected with four different plasmids to overexpress N-myc, renilla luciferase, a reporter construct consisting of 4 tandem E-box sites upstream of the firefly luciferase, and increasing amounts of Utf1 (Suppl. Figure 2). As seen previously with this reporter construct [37,38] Myc increases firefly luciferase expression levels approximately 5-fold (Suppl. Figure 2). The presence of Utf1 as well as increasing amounts of Utf1 does not have a significant effect on N-Myc transcriptional activation activity at E-box promoter sites (Suppl. Figure 2).
DISCUSSION
Here we have identified the direct interaction of the pluripotency and self-renewal factors Utf1 and N-Myc/c-Myc as well as designated the regions required for this interaction. N-Myc and c-Myc are highly conserved within the bHLHZ region strongly supporting the requirement of bHLHZ domain for Utf1 interaction for both c-and N-Myc. Due to the requirement of the bHLHZ domain of N-Myc and the Myb/SANT domain of Utf1 for their direct interaction suggest that the proteins may bind specific regions of DNA together.
Other Myc cofactors have been known to interact with Myc through the C-terminal domain including Miz-1 [4] and Skp2 [39]. Most importantly Myc binds its heterodimeric binding partner, Max through the helix-loop-helix leucine zipper domain making Myc capable of binding DNA at E-box sites. Also, Utf1 was not found to directly interact with Max. These results suggest that Utf1 and Max may either competitively bind Myc due to their common binding interface or are able to bind Myc simultaneously.
The direct interaction of Utf1 and Myc could not be recapitulated in vivo, in both an ESC context and overexpressed in 293FT cells (data not shown). There are several possible explanations for these negative findings. Myc-Utf1 complexes may require an ESC context and be present at relatively low abundance making detection particularly challenging. Secondly, Myc is a highly unstable protein [40] with an average half-life of 20 minutes, which may contribute to the difficulty in copurifying its cofactors.
Previous studies have suggested a redundant functional role between Myc and Utf1 in both reprogramming and maintenance of self-renewal and pluripotency in ESCs. First the relative efficient generation of iPSCs in which Myc is replaced with Utf1 as one of the four factors for reprogramming [22] as well as the 415 genes co-bound by Utf1 and c-Myc in mESCs [26]. From gene target data sets it can be conservatively estimated that Myc binds ~20% of Utf1 target genes in mESCs [26,41].
Utf1 functions as an epigenetic factor and translational factor in mESCs by blocking PRC2 binding and recruiting the mRNA decapping complex [25]. Myc on the other hand is a well-established transcription and epigenetic factor, functioning by the recruitment of a wide variety of cofactors affecting the transcriptional machinery and acetylation of histone H3 and H4 (reviewed in [42]). Further studies of comparative global chromatin binding sites as well as expression levels of target genes in the absence and presence of either protein in mESCs will increase our understanding of the combinatorial roles of Myc and Utf1 in maintenance of ESC characteristics.
Supplementary Material
Supplemental Figure 1. Expression of GST-N-Myc and GST-Utf1 truncation proteins.
(A) Expression and expected sizes of all GST-N-Myc fusion truncation proteins, visualized by GST western blot.
(B) Expression and expected sizes of all GST-Utf1 fusion truncation proteins, visualized by GST western blot.
Supplemental Figure 2. Utf1 Does Not Influence N-Myc Activation Activity at E-Box Promoter Sites.
Fold induction level of E-Box luciferase reporter relative to N-Myc with increasing amounts of Utf1. Error bars indicate mean +/− standard deviation. ** = p-value < .005 relative to fold induction set of N-Myc alone.
Highlights.
Identification of known and novel Myc cofactors in mESCs.
Myc and Utf1 directly interact.
The bHLHZ region of Myc is required for direct interaction with Utf1.
Large region of Utf1 including Myb/SANT domain is required for interaction with Myc.
ACKNOWLEDGMENTS
We thank the UCD Genome Center Proteomics Core for the mass spectrometric work and Bonnie Barrilleaux for feedback on the manuscript. The work was supported by NIH Grant 5K01CA114400-05 and CIRM Grant RN2-00922-1 (both to PSK).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
bHLHZ, basic-helix-loop-helix leucine zipper; co-IP, co-immunoprecipitation; ESC, embryonic stem cell; GST, glutathione-S-transferase; ICM, inner cell mass cell; iPSC, induced pluripotent stem cell; mESC, mouse embryonic stem cell; Utf1, Undifferentiated embryonic cell transcription factor 1.
REFERENCES
- 1.Blackwood EM, Eisenman RN. Max: a helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science. 1991;251:1211–1217. doi: 10.1126/science.2006410. [DOI] [PubMed] [Google Scholar]
- 2.Cheng SW, Davies KP, Yung E, Beltran RJ, Yu J, Kalpana GV. c-MYC interacts with INI1/hSNF5 and requires the SWI/SNF complex for transactivation function. Nat Genet. 1999;22:102–105. doi: 10.1038/8811. [DOI] [PubMed] [Google Scholar]
- 3.Cheng AS, Jin VX, Fan M, Smith LT, Liyanarachchi S, Yan PS, Leu YW, Chan MW, Plass C, Nephew KP, Davuluri RV, Huang TH. Combinatorial analysis of transcription factor partners reveals recruitment of c-MYC to estrogen receptor-alpha responsive promoters. Mol Cell. 2006;21:393–404. doi: 10.1016/j.molcel.2005.12.016. [DOI] [PubMed] [Google Scholar]
- 4.Herold S, Wanzel M, Beuger V, Frohme C, Beul D, Hillukkala T, Syvaoja J, Saluz HP, Haenel F, Eilers M. Negative regulation of the mammalian UV response by Myc through association with Miz-1. Mol Cell. 2002;10:509–521. doi: 10.1016/s1097-2765(02)00633-0. [DOI] [PubMed] [Google Scholar]
- 5.Varlakhanova N, Cotterman R, Bradnam K, Korf I, Knoepfler PS. Myc and Miz-1 have coordinate genomic functions including targeting Hox genes in human embryonic stem cells. Epigenetics Chromatin. 2011;4:20. doi: 10.1186/1756-8935-4-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fernandez PC, Frank SR, Wang L, Schroeder M, Liu S, Greene J, Cocito A, Amati B. Genomic targets of the human c-Myc protein. Genes Dev. 2003;17:1115–1129. doi: 10.1101/gad.1067003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zeller KI, Zhao X, Lee CW, Chiu KP, Yao F, Yustein JT, Ooi HS, Orlov YL, Shahab A, Yong HC, Fu Y, Weng Z, Kuznetsov VA, Sung WK, Ruan Y, Dang CV, Wei CL. Global mapping of c-Myc binding sites and target gene networks in human B cells. Proc Natl Acad Sci U S A. 2006;103:17834–17839. doi: 10.1073/pnas.0604129103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frank SR, Schroeder M, Fernandez P, Taubert S, Amati B. Binding of c-Myc to chromatin mediates mitogen-induced acetylation of histone H4 and gene activation. Genes Dev. 2001;15:2069–2082. doi: 10.1101/gad.906601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bieda M, Xu X, Singer MA, Green R, Farnham PJ. Unbiased location analysis of E2F1-binding sites suggests a widespread role for E2F1 in the human genome. Genome Res. 2006;16:595–605. doi: 10.1101/gr.4887606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Knoepfler PS, Zhang XY, Cheng PF, Gafken PR, McMahon SB, Eisenman RN. Myc influences global chromatin structure. EMBO J. 2006;25:2723–2734. doi: 10.1038/sj.emboj.7601152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cotterman R, Jin VX, Krig SR, Lemen JM, Wey A, Farnham PJ, Knoepfler PS. N-Myc regulates a widespread euchromatic program in the human genome partially independent of its role as a classical transcription factor. Cancer Res. 2008;68:9654–9662. doi: 10.1158/0008-5472.CAN-08-1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meyer N, Penn LZ. Reflecting on 25 years with MYC. Nat Rev Cancer. 2008;8:976–990. doi: 10.1038/nrc2231. [DOI] [PubMed] [Google Scholar]
- 13.Downs KM, Martin GR, Bishop JM. Contrasting patterns of myc and N-myc expression during gastrulation of the mouse embryo. Genes Dev. 1989;3:860–869. doi: 10.1101/gad.3.6.860. [DOI] [PubMed] [Google Scholar]
- 14.Kelly DL, Rizzino A. DNA microarray analyses of genes regulated during the differentiation of embryonic stem cells. Mol Reprod Dev. 2000;56:113–123. doi: 10.1002/(SICI)1098-2795(200006)56:2<113::AID-MRD1>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 15.Ramalho-Santos M, Yoon S, Matsuzaki Y, Mulligan RC, Melton DA. "Stemness": transcriptional profiling of embryonic and adult stem cells. Science. 2002;298:597–600. doi: 10.1126/science.1072530. [DOI] [PubMed] [Google Scholar]
- 16.Sawai S, Shimono A, Hanaoka K, Kondoh H. Embryonic lethality resulting from disruption of both N-myc alleles in mouse zygotes. New Biol. 1991;3:861–869. [PubMed] [Google Scholar]
- 17.Davis AC, Wims M, Spotts GD, Hann SR, Bradley A. A null c-myc mutation causes lethality before 10.5 days of gestation in homozygotes and reduced fertility in heterozygous female mice. Genes Dev. 1993;7:671–682. doi: 10.1101/gad.7.4.671. [DOI] [PubMed] [Google Scholar]
- 18.Malynn BA, de Alboran IM, O'Hagan RC, Bronson R, Davidson L, DePinho RA, Alt FW. N-myc can functionally replace c-myc in murine development, cellular growth, and differentiation. Genes Dev. 2000;14:1390–1399. [PMC free article] [PubMed] [Google Scholar]
- 19.Varlakhanova NV, Cotterman RF, deVries WN, Morgan J, Donahue LR, Murray S, Knowles BB, Knoepfler PS. myc maintains embryonic stem cell pluripotency and self-renewal. Differentiation. 2010;80:9–19. doi: 10.1016/j.diff.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakagawa M, Koyanagi M, Tanabe K, Takahashi K, Ichisaka T, Aoi T, Okita K, Mochiduki Y, Takizawa N, Yamanaka S. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat Biotechnol. 2008;26:101–106. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- 21.Smith KN, Singh AM, Dalton S. Myc represses primitive endoderm differentiation in pluripotent stem cells. Cell Stem Cell. 2010;7:343–354. doi: 10.1016/j.stem.2010.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao Y, Yin X, Qin H, Zhu F, Liu H, Yang W, Zhang Q, Xiang C, Hou P, Song Z, Liu Y, Yong J, Zhang P, Cai J, Liu M, Li H, Li Y, Qu X, Cui K, Zhang W, Xiang T, Wu Y, Zhao Y, Liu C, Yu C, Yuan K, Lou J, Ding M, Deng H. Two supporting factors greatly improve the efficiency of human iPSC generation. Cell Stem Cell. 2008;3:475–479. doi: 10.1016/j.stem.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 23.Okuda A, Fukushima A, Nishimoto M, Orimo A, Yamagishi T, Nabeshima Y, Kuro-o M, Nabeshima Y, Boon K, Keaveney M, Stunnenberg HG, Muramatsu M. UTF1, a novel transcriptional coactivator expressed in pluripotent embryonic stem cells and extra-embryonic cells. EMBO J. 1998;17:2019–2032. doi: 10.1093/emboj/17.7.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van den Boom V, Kooistra SM, Boesjes M, Geverts B, Houtsmuller AB, Monzen K, Komuro I, Essers J, Drenth-Diephuis LJ, Eggen BJ. UTF1 is a chromatin-associated protein involved in ES cell differentiation. J Cell Biol. 2007;178:913–924. doi: 10.1083/jcb.200702058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jia J, Zheng X, Hu G, Cui K, Zhang J, Zhang A, Jiang H, Lu B, Yates J, 3rd, Liu C, Zhao K, Zheng Y. Regulation of pluripotency and self-renewal of ESCs through epigenetic-threshold modulation and mRNA pruning. Cell. 2012;151:576–589. doi: 10.1016/j.cell.2012.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kooistra SM, van den Boom V, Thummer RP, Johannes F, Wardenaar R, Tesson BM, Veenhoff LM, Fusetti F, O'Neill LP, Turner BM, de Haan G, Eggen BJ. Undifferentiated embryonic cell transcription factor 1 regulates ESC chromatin organization and gene expression. Stem Cells. 2010;28:1703–1714. doi: 10.1002/stem.497. [DOI] [PubMed] [Google Scholar]
- 27.Buganim Y, Faddah DA, Cheng AW, Itskovich E, Markoulaki S, Ganz K, Klemm SL, van Oudenaarden A, Jaenisch R. Single-cell expression analyses during cellular reprogramming reveal an early stochastic and a late hierarchic phase. Cell. 150(2012):1209–1222. doi: 10.1016/j.cell.2012.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Agrawal P, Yu K, Salomon AR, Sedivy JM. Proteomic profiling of Myc-associated proteins. Cell Cycle. 2010;9:4908–4921. doi: 10.4161/cc.9.24.14199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim J, Woo AJ, Chu J, Snow JW, Fujiwara Y, Kim CG, Cantor AB, Orkin SH. A Myc network accounts for similarities between embryonic stem and cancer cell transcription programs. Cell. 2010;143:313–324. doi: 10.1016/j.cell.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith KN, Lim JM, Wells L, Dalton S. Myc orchestrates a regulatory network required for the establishment and maintenance of pluripotency. Cell Cycle. 2011;10:592–597. doi: 10.4161/cc.10.4.14792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Z, Boone D, Hann SR. Nucleophosmin interacts directly with c-Myc and controls c-Myc-induced hyperproliferation and transformation. Proc Natl Acad Sci U S A. 2008;105:18794–18799. doi: 10.1073/pnas.0806879105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brenner C, Deplus R, Didelot C, Loriot A, Vire E, De Smet C, Gutierrez A, Danovi D, Bernard D, Boon T, Pelicci PG, Amati B, Kouzarides T, de Launoit Y, Di Croce L, Fuks F. Myc represses transcription through recruitment of DNA methyltransferase corepressor. EMBO J. 2005;24:336–346. doi: 10.1038/sj.emboj.7600509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oster SK, Mao DY, Kennedy J, Penn LZ. Functional analysis of the N-terminal domain of the Myc oncoprotein. Oncogene. 2003;22:1998–2010. doi: 10.1038/sj.onc.1206228. [DOI] [PubMed] [Google Scholar]
- 34.Atchley WR, Fitch WM. Myc and Max: molecular evolution of a family of proto-oncogene products and their dimerization partner. Proc Natl Acad Sci U S A. 1995;92:10217–10221. doi: 10.1073/pnas.92.22.10217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nishimoto M, Fukushima A, Miyagi S, Suzuki Y, Sugano S, Matsuda Y, Hori T, Muramatsu M, Okuda A. Structural analyses of the UTF1 gene encoding a transcriptional coactivator expressed in pluripotent embryonic stem cells. Biochem Biophys Res Commun. 2001;285:945–953. doi: 10.1006/bbrc.2001.5265. [DOI] [PubMed] [Google Scholar]
- 36.Fukushima A, Okuda A, Nishimoto M, Seki N, Hori TA, Muramatsu M. Characterization of functional domains of an embryonic stem cell coactivator UTF1 which are conserved and essential for potentiation of ATF-2 activity. J Biol Chem. 1998;273:25840–25849. doi: 10.1074/jbc.273.40.25840. [DOI] [PubMed] [Google Scholar]
- 37.Hurlin PJ, Steingrimsson E, Copeland NG, Jenkins NA, Eisenman RN. Mga, a dual-specificity transcription factor that interacts with Max and contains a T-domain DNA-binding motif. EMBO J. 1999;18:7019–7028. doi: 10.1093/emboj/18.24.7019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.James L, Eisenman RN. Myc and Mad bHLHZ domains possess identical DNA-binding specificities but only partially overlapping functions in vivo. Proc Natl Acad Sci U S A. 2002;99:10429–10434. doi: 10.1073/pnas.162369299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim SY, Herbst A, Tworkowski KA, Salghetti SE, Tansey WP. Skp2 regulates Myc protein stability and activity. Mol Cell. 2003;11:1177–1188. doi: 10.1016/s1097-2765(03)00173-4. [DOI] [PubMed] [Google Scholar]
- 40.Ramsay G, Evan GI, Bishop JM. The protein encoded by the human proto-oncogene c-myc. Proc Natl Acad Sci U S A. 1984;81:7742–7746. doi: 10.1073/pnas.81.24.7742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim J, Chu J, Shen X, Wang J, Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132:1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eilers M, Eisenman RN. Myc's broad reach. Genes Dev. 2008;22:2755–2766. doi: 10.1101/gad.1712408. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Expression of GST-N-Myc and GST-Utf1 truncation proteins.
(A) Expression and expected sizes of all GST-N-Myc fusion truncation proteins, visualized by GST western blot.
(B) Expression and expected sizes of all GST-Utf1 fusion truncation proteins, visualized by GST western blot.
Supplemental Figure 2. Utf1 Does Not Influence N-Myc Activation Activity at E-Box Promoter Sites.
Fold induction level of E-Box luciferase reporter relative to N-Myc with increasing amounts of Utf1. Error bars indicate mean +/− standard deviation. ** = p-value < .005 relative to fold induction set of N-Myc alone.