Abstract
The peptidoglycan (PG) cell wall is a unique macromolecule responsible for both shape determination and cellular integrity under osmotic stress in virtually all bacteria. A quantitative understanding of the relationships between PG architecture, morphogenesis, immune system activation, and pathogenesis can provide molecular-scale insights into the function of proteins involved in cell-wall synthesis and cell growth. High Performance Liquid Chromatography (HPLC) has played an important role in our understanding of the structural and chemical complexity of the cell wall by providing an analytical method to quantify differences in chemical composition. Here, we present a primer on the basic chemical features of wall structure that can be revealed through HPLC, along with a description of the applications of HPLC PG analyses for interpreting the effects of genetic and chemical perturbations to a variety of bacterial species in different environments. We describe the physical consequences of different PG compositions on cell shape, and review complementary experimental and computational methodologies for PG analysis. Finally, we present a partial list of future targets of development for HPLC and related techniques.
Introduction
A bacterial cell must maintain positive turgor pressure in order to drive growth and division (Whatmore et al., 1990). This turgor pressure can be several atmospheres in magnitude (Sochacki et al., 2011), leading to mechanical stresses that are withstood in virtually all bacterial species by the cell wall, a rigid exoskeleton surrounding the cytoplasmic membrane. The cell wall is both necessary and sufficient for cell shape determination; cells become round when the cell wall is removed (Harold,1990), and isolated cell walls (sacculi) maintain the shape of the intact cell (Fig. 1A) (Heidrich et al., 2001; Priyadarshini et al., 2007; Goley et al., 2010). Throughout the great diversity of shapes and sizes across the bacterial domain (Young,2006), the cell wall is universally composed of peptidoglycan (PG), a macromolecular network of sugar strands (glycans) cross-linked by short peptides (Holtje,1998). The general importance of PG for bacterial physiology (Scheffers et al., 2005) and as an antibiotic target (Kong et al., 2010) has motivated genetic, structural, and cell biological studies of how it is robustly assembled during growth and division. Critical to our understanding of cell shape determination are the identification of key enzymatic activities in the PG synthesis pathway and the chemical composition of the resulting network. High Pressure Liquid Chromatography (HPLC) has played, and continues to play, an important role in both of these aims by providing an analytical method to quantify differences in the chemical composition of the wall between strains, growth conditions, and during chemical perturbations.
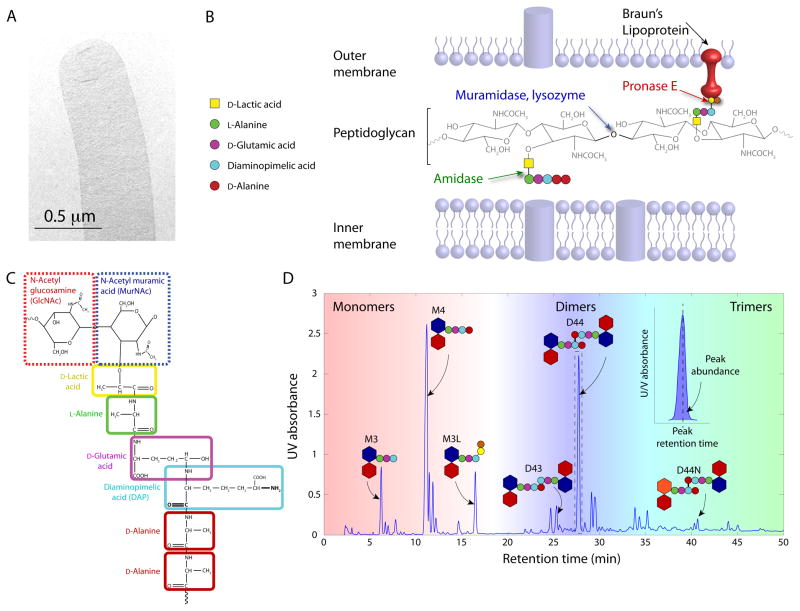
Figure 1. Basic features of peptidoglycan muropeptide chemistry and analysis.
(A) Electron micrograph of a sacculus from the rod-shaped bacterium E. coli. (B) After sacculus isolation, the preparation protocol for HPLC analyses includes several enzymatic treatments that digest sacculi into muropeptide monomers or cross-linked oligomers. Colored arrows designate the site of action of the respective enzyme. Yellow and orange circles denote an oligopeptide from Braun’s lipoprotein that remains bound to the muropeptide after cleavage by Pronase E. (C) Each muropeptide is composed of a disaccharide and its associated peptide stem. The disaccharide consists of the sugars MurNAc and GlcNAc, while the peptide stem consists of up to five amino acids (shown in colored rectangles) connected to the MurNAc through a D-Lactic acid. The bolded atoms on DAP indicate potential cross-linking locations to other muropeptides, which would result in either a dimer or other oligomer. (D) Example of an HPLC PG analysis, with characteristic peaks labeled with cartoons of their muropeptide structure (disaccharides: hexagons; amino acids in peptide stem: circles; both are colored as in (C)). Lipoprotein amino acids on M3L are shown with orange and yellow circles; anhydro modification on D44N in orange). Inset shows a zoomed-in version of the D44 peak, with shading indicating the species abundance and dashed line indicating the retention time.
The basic subunit of the cell wall is a disaccharide with a peptide stem that can be cross-linked to neighboring subunits. These “muropeptide” subunits are manufactured with pentapeptide stems within the cytoplasm and then flipped across the cytoplasmic membrane, whereupon they are integrated into the existing network through covalent bonds, causing expansion of the wall (den Blaauwen et al., 2008). Most of the proteins in this synthesis pathway are part of the murein (Mur) or Penicillin Binding Protein (PBP) families (Typas et al., 2011). The standard substrate for the PBPs is the terminal D-Ala-D-Ala residues of the peptide stems (Sauvage et al., 2008; Chen et al., 2009). The structural similarity between D-Ala-D-Ala and β-lactam antibiotics facilitates binding of these antibiotics to the active site of PBPs, with the β-lactam nucleus of the molecule irreversibly binding to the catalytic serine residue of the PBP active site. This inhibition typically leads to loss of cell shape, and often cell death (Dalhoff et al., 2003; Joseleau-Petit et al., 2007; Legaree et al., 2007a; Legaree et al., 2007b).
Enzymes that modify the PG have been implicated in cell shape determination; for example, the helical shape in Helicobacter pylori requires an enzyme that cleaves cross-links (Sycuro et al., 2012), while transcription factors and lipoprotein cofactors that regulate the expression (Santos et al., 2002) and activity (Paradis-Bleau et al., 2010; Typas et al., 2010), respectively, of PBPs in E. coli play a role in both cell wall morphology and integrity. In addition to the proteins directly involved in cell-wall synthesis, several cytoskeletal proteins that contribute to cell wall production have been identified, and their disruption often alters cellular morphology. The actin homolog MreB has been implicated in rod-shaped growth; depolymerization of MreB using the small molecule A22 leads to a round morphology (Karczmarek et al., 2007; Jiang et al., 2011) and point mutations in MreB can result in changes in the width of rod-shaped Escherichia coli cells (Wachi et al., 1989). The tubulin homolog FtsZ is essential for cytokinesis and localizes the machinery required for constructing the septal wall (Strauss et al., 2012). Finally, a wide variety of bacteria incorporate non-canonical D-amino acids into their cell walls to alter PG density and strength (Lam et al., 2009; Cava et al., 2011b; Horcajo et al., 2012), and their environmental release can act as a developmental signal, for example by disrupting biofilms in Bacillus subtilis (Kolodkin-Gal et al., 2010).
A major challenge in bacterial morphogenesis is to understand how cells enlarge the cell wall under stress while maintaining a defined shape. As our knowledge of the repertoire of shape-related genes and their complex, systems-level interactions has expanded due to advances in genetics, imaging, and structural modeling, HPLC has provided an increasingly valuable tool for assaying how molecular components impact the construction of the cell wall. In concert with the recent development of tools for rapid quantification of cell morphology from imaging data (Guberman et al., 2008; Sliusarenko et al., 2011) and genetic libraries such as the KEIO collection of single-gene knockouts in E. coli (Baba et al., 2006), we are positioned to deconstruct cell-wall growth through multiple, complementary lines of inquiry.
Here, we present a primer on the basic chemical features of wall structure that can be revealed through HPLC, along with a description of the applications of HPLC PG analyses for interpreting the effects of genetic and chemical perturbations to a variety of species in different environments. We then provide a physical framework for dissecting how different PG compositions can impact cell shape and mechanics, and review emerging tools that can provide spatial, mechanical, and dynamic information about wall ultrastructure to complement future advances in HPLC. Our goal is to provide the reader with enough of the details to appreciate how the technique has been refined to answer a variety of questions about PG structure and assembly. In the interests of space and conciseness, we omit some information that is less critical for the non-expert on PG biochemistry, sacculus preparation, and HPLC techniques; more information can be found in several excellent reviews (Holtje,1998; den Blaauwen et al., 2008; Vollmer et al., 2008; Typas et al., 2011). Moreover, we have compiled information on the scope of organisms whose PG has been quantified by HPLC, and the relevant data collected, from a large fraction of the existing studies (Supp. Tables 1 and 2) to serve as a comprehensive resource and to highlight unexplored species and questions.
Chemical composition of PG
Knowledge of the chemical composition of PG provides key information regarding the structural and metabolic properties of wall synthesis. To isolate sacculi (Fig. 1A), bacteria are lysed, digested with enzymes targeted to the PG, and prepared for HPLC analysis. Although this process has been performed in several ways by different researchers, common features include lysing in boiling SDS, purifying away bound protein (e.g. Braun’s lipoprotein) in enterobacteria such as E. coli with protease treatment, digesting down to disaccharide subunits with muramidase (Fig. 1B), reducing muropeptides with sodium borohydride, setting pH to the isoelectric point of the muropeptides, and filtering before injection onto an HPLC system.
The glycan component of each muropeptide is composed of two sugar moieties, an N-acetylglucosamine (GlcNAc) and an N-acetylmuramic acid (MurNAc). Together, these sugars are commonly referred to as the disaccharide subunit of a muropeptide (not to be confused with a muropeptide dimer, described below). MurNAc is a unique sugar that appears only in the chemical make-up of bacteria, and is the location where the peptides are covalently bonded (Fig. 1C). The peptide stem is typically composed of five amino acids, including D-amino acids that are not commonly used in nature. This stereochemistry confers resistance from typical peptidases that would otherwise cleave the cross-links and thereby lead to lysis (Cava et al., 2011a). Peptide stems are bound to MurNAc through its lactic acid component, thus ensuring resistance to chemical degradation through a strong ether bond to the sugar.
Most of the variation in PG composition between species occurs in the peptide stem and the method by which the glycan strands are cross-linked. In Gram-negative bacteria, a meso-diaminopimelic acid (DAP) participates in the cross-linking bridge between muropeptides, which leads to the formation of muropeptide dimers (two disaccharide subunits covalently linked through their peptide stems), and larger oligomers upon further cross-linking. This non-protein amino acid is the third residue of the peptide stem and is an achiral carboxy derivative of lysine, which gives the muropeptide more opportunities to cross-link, thereby strengthening the PG network. In E. coli, DAP cross-links 30–50% of the available sites for cross-linking, depending on the strain (Vollmer et al., 2010). Muramidase digestion of sacculi produces uncross-linked monomers, and cross-linked dimers, trimers, and tetramers of muropeptides whose combined abundance indicates the level of cross-linking. Some bacteria instead use a peptide bridge such as a pentaglycine to cross-link muropeptides (Koyama et al., 2012).
Glycan strands are formed by linking muropeptide subunits together into a chain. These strands are then cross-linked to the peptides of adjacent strands, thereby forming a two- or three-dimensional network. For all bacteria, each peptide stem initially terminates with two D-alanine residues, which is the site of action for D,D-transpeptidases that catalyze covalent cross-linking between muropeptides. This transpeptidase reaction is energized by the hydrolysis of the D-Ala-D-Ala bond. Most Gram-negative bacteria form cross-links by bonding the carboxyl group of the terminal D-Ala at position 4 on one muropeptide directly to the D-amino group of DAP in position 3 of an adjacent muropeptide, forming a characteristic DD, 4–3 crossbridge linkage. In addition, E. coli and many other species can form 3-3 cross-links through an alternate reaction in which the L-carboxyl group of DAP in one muropeptide is bonded to the D-amino group of DAP in a neighboring muropeptide (Magnet et al., 2007; Lavollay et al., 2008; Magnet et al., 2008; Cava et al., 2011b; Lavollay et al., 2011; Peltier et al., 2011). The enzymes that mediate this cross-linking are known as L,D-transpeptidases and are also responsible for the covalent attachment of muropeptides to Braun’s lipoprotein (Lpp) in the outer membrane of some Gram-negative bacteria (Dramsi et al., 2008; Piek et al., 2012). The relative abundance of PG-bound Lpp is an indication of the strength of the OM/PG interaction; the proportion of PG subunits bound to Lpp can be quantified since protease cleavage of Lpp from PG during sacculus preparation leaves a C-terminal oligopeptide from the lipoprotein attached to DAP, giving these muropeptides added hydrophobicity to distinguish them chemically (Fig. 1D) (Magnet et al., 2008). A glycan strand can be released from its membrane anchor by an internal transglycosylation reaction. This results in the formation of a terminating 1,6-anhydro-MurNAc, which prevents further polymerization and thus signals the end of a glycan strand. Therefore, the “anhydro” muropeptide fraction provides an indirect measurement of average glycan strand length, an important parameter for dictating the structure of the PG and the resulting cell shape (Furchtgott et al., 2011).
Linking cell-wall composition to physical aspects of morphology
Cell shape is ultimately dictated by the balance between the force exerted on the membrane by turgor pressure and the elastic extension of the PG network. From a physical perspective, the cell wall can be thought of as a network of springs whose stiffness dictates the amount of stretching for a given force (Boulbitch et al., 2000). In the rod-shaped E. coli, the peptides are thought to be oriented roughly along the longitudinal direction; since peptide cross-links are likely more flexible than the glycan strands (Huang et al., 2008), the density of cross-links largely dictates the expansion of the cell along its longitudinal axis due to turgor pressure. Simulations based on a biophysical model of PG growth have shown that the distribution of glycan strand lengths can affect cell width and the ordering of the PG (Furchtgott et al., 2011). Finally, the number of layers within the wall dictates its thickness and thus affects the overall stiffness of the PG. In E. coli, recent cryo-electron microscopy tomograms indicate that the wall is mostly single layered, although other Gram-negative organisms exhibit patterns of cross-linking that suggest a non-planar organization in which strands may cross over each other at multiple sites (Brown et al., 2012). There is some evidence that Gram-positive organisms such as B. subtilis can tolerate higher turgor pressures (Brill et al., 2011); this capacity may be linked to the increased wall thickness. Therefore, quantification of the cross-linking densities, glycan strand lengths, and layering within the wall provides direct information about the physical properties of cells.
Historical overview of applications of chromatography to PG structural analysis
Methods for PG purification and muropeptide solubilization by enzymatic digestion were established soon after discovery of the cell wall itself (Weidel et al., 1960). However, development of an appropriate methodology for muropeptide analysis was not possible until the advent of advanced chromatographic techniques in the early 1980s. Prior to this, most information about wall components was gleaned from amino acid analysis (Sutow et al., 1967; Boylen et al., 1968; Wang et al., 1968) or paper chromatography (Kandler et al., 1968; de la Rosa et al., 1985) of lysozyme-digested cell walls. These tedious, low-resolution techniques allowed for the detection of at most six different muropeptide species. The initial breakthroughs applying HPLC techniques to PG analysis were carried out by Bernd Glauner and the late Uli Schwarz (Hakenbeck et al., 1983). They obtained the first comprehensive, high-resolution analysis of E. coli PG composition (Glauner et al., 1988). This led to the identification of new families of muropeptides, in particular cross-linked trimmers and tetramers and L,D-cross-linked muropeptides, indicating a far more complicated wall structure and metabolism than previously appreciated.
Development of PG analysis by HPLC
High Performance Liquid Chromatography (HPLC) is a separation technique whereby a liquid sample is injected onto a column tightly packed with porous particles (the “stationary phase”). The individual components of the sample are transported by a liquid (the “mobile phase”). Separation of the different individual components of the sample is based upon their relative solubility between the two phases. Typically in PG analyses, reverse phase C18 columns that provide a strongly hydrophobic matrix are used with sodium phosphate/methanol-buffered mobile phases to separate the different muropeptide species. However, varying the type of column and mobile phase can be used to tune the purification of PG components in different organisms. High-pressure pumps affect the elution of all components off the column, whereupon detection occurs to produce a chromatogram with the detector output (UV absorbance at 202–208 nm) as a function of time. The time at which a particular chemical species elutes off the column is called its “retention time,” which varies with buffer composition and gradient (Fig. 1D). Standard protocols currently require about three days to prepare and analyze a PG sample, but many samples can be processed in parallel. The ability to resolve and quantify all the individual muropeptides in large sets of samples provides the opportunity to link wall composition with specific enzymatic activities, environmental conditions, and shape determination (Fig. 2).
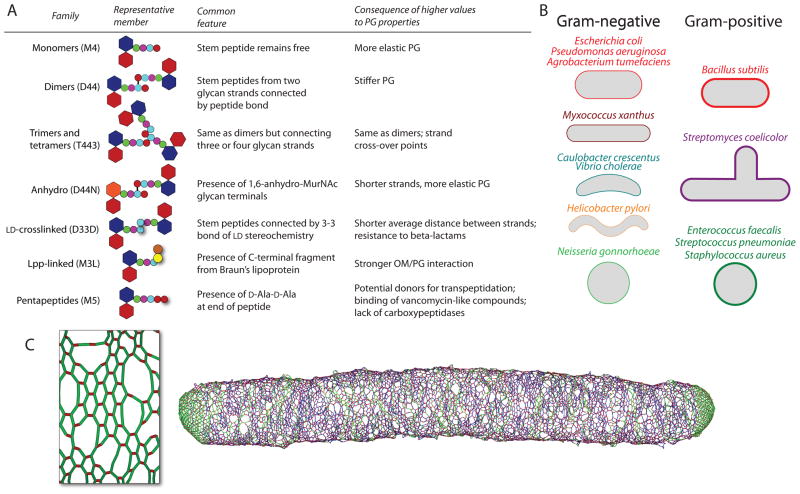
Figure 2. Mapping PG composition to morphology.
(A) For each family of muropeptides, the table describes the salient chemical features and the physical or chemical consequences of having a high proportion of each family. (B) Species for which at least two HPLC PG analyses have been published, grouped by shape. A majority of studies have focused on Gram-negative PG, although recent efforts have expanded the diversity of species studied (see Supp. Table 2). (C) Connecting differences in PG composition to morphology can be facilitated by biophysical simulations of cell growth, using HPLC quantification as a constraint on growth. The inset on the left is zoomed in on the architecture of a single-layered cell wall from a simulation of rod-shaped growth, with the results of one such simulation shown on the right (modified from (Furchtgott et al., 2011)). Each green or blue cylinder represents a disaccharide in an old or new glycan strand, respectively, while each red cylinder represents a peptide cross-link.
One of the first reported cases of PG analysis by HPLC was performed on the Gram-negative bacterium Caulobacter crescentus (Markiewicz et al., 1983), revealing the absence of PG carboxypeptidase activity through the high abundance of pentapeptides. Direct comparisons of HPLC PG analyses between wild-type and mutant strains can be used to infer important differences in wall composition, for instance between penicillin-resistant and susceptible strains of the round, Gram-negative Neisseria gonorrhoeae (Dougherty,1985); these studies indicated that antibiotic resistance is correlated to a change in wall composition (Pisabarro et al. 1985). Although most enzymes involved in cell wall synthesis are membrane-associated and hence are difficult to purify for in vitro studies, they can be genetically manipulated to produce knockout and depletion strains whose muropeptide profile is an indicator of function.
An important advantage of HPLC has been the capability to easily identify individual PG components by collecting the eluate from the column. The advent of Matrix Assisted Laser Desorption/Ionization – Time Of Flight Mass Spectrometry (MALDI-TOF MS) has greatly accelerated muropeptide characterization in comparison with previous chemical and sequencing methods. Unlike previous MS techniques, MALDI-TOF has a mass range appropriate for muropeptides, is relatively tolerant to salt contamination, requires only minute amounts of sample, and is carried out at many research institutions by core facilities (Wieser et al., 2012). This has allowed for the rapid and systematic analysis of PG composition from new species, reducing the time to discovery by over an order of magnitude relative to Schwarz and Glauner’s original work.
E. coli as a paradigm for PG analysis
By far the species that has served as the focus of the largest number of PG analyses his E. coli (Supp. Table 1) (Holtje,1998), making it the reference for PG studies. High-resolution chromatograms have revealed up to 80 different muropeptide species (Glauner,1988), though the 30 most abundant muropeptides account for >96–98% of total abundance. From a chemical and structural point of view, muropeptides are often classified into a small number of groups that affect certain properties of the sacculus (Fig. 2A). For example, a higher fraction of muropeptide monomers indicates a lower density of cross-links, likely leading to more mechanically flexible PG. For specific HPLC conditions, there is a strong correlation between peak retention times and the chemical structure of the muropeptides (Glauner et al., 1988); with some exceptions, polar, low molecular weight muropeptides (i.e. monomers) elute first, and apolar, high molecular weight muropeptides (dimers, trimers, tetramers, anhydromuropeptides) elute last (Fig. 1D).
Extensive analysis of E. coli PG has revealed specific changes to wall composition that occur in different growth conditions and during cell division. Stationary phase PG has increased cross-linking, higher proportions of lipoprotein-bound muropeptides, and shorter glycan strands relative to exponentially growing cells (Pisabarro et al., 1985; Prats et al., 1989); the biological implications of these changes have yet to be elucidated. HPLC was also instrumental in defining the mechanism of incorporation of D-amino acids (Caparros et al., 1991; Horcajo et al., 2012), a reaction once thought to be artificial, but now considered part of a potential sensing mechanism of environmental stress (Caparros et al., 1991; Lam et al., 2009; Cava et al., 2011a; Cava et al., 2011b; Horcajo et al., 2012). Inspired by the universality of D-amino acid incorporation across bacteria (Cava et al., 2011b), new approaches based on the incorporation of fluorescent (Kuru et al., 2012) or azide/alkyne derivatives of D-amino acids (Siegrist et al., 2013) have launched direct methods for imaging PG dynamics in vivo. In all cases, HPLC has been fundamental for determining the specificity of the PG modification.
Connecting cell shape and peptidoglycan synthesis
Given the importance of PG in cell-shape determination, the relationship between PG make-up and cell morphology has been a topic of growing interest. This was initially investigated by purifying sacculi from E. coli minicells (Goodell et al., 1977), which are nonviable round cells produced through aberrantly placed polar division events (Markiewicz et al., 1992). Because minicells are made of two poles, they provide a mechanism to compare the PG compositions of the poles and cylindrical cell body. HPLC analysis showed that minicell PG consists of shorter glycan strands, suggesting that in rod-shaped cells, the poles have shorter strands than the cylindrical body (Obermann et al., 1994). E. coli grown in the presence of β-lactams and an osmoprotectant shed most of their wall and enter a spherical, “L-form”-like state; HPLC was used to confirm that they still retain approximately 7% of their PG that is perhaps used as a template for division and proliferation (Joseleau-Petit et al., 2007). This study also established that HPLC can detect low levels of PG from normal preparations (Supp. Table 1). HPLC has similarly demonstrated roles for PG modification in organisms with non-rod-like morphology. HPLC studies of a straight H. pylori mutant led to the characterization of a DL-carboxypeptidase that cleaves tripeptides to produce a helical shape in wild-type cells (Sycuro et al., 2012). In C. crescentus, HPLC revealed that the cross-linking density is unchanged despite the curvature of the cell body, suggesting that other factors such as spatially dependent synthesis are responsible for their morphology (Cabeen et al., 2009).
PG variability across bacterial species
Using the extensive knowledge framework derived from E. coli PG analyses, chromatographic outputs in comparative studies with other bacteria have revealed significant biological differences in PG makeup (a summary of these studies can be found in Supp. Table 2). A study to develop a consensus of PG make-up in Gram-negative bacteria, spanning Aeromonas sp., Acinetobacter acetoaceticus, Agrobacterium tumefaciens, Enterobacter cloacae, Proteus morganii, Pseudomonas aeruginosa, Pseudomonas putida, Vibrio parahaemolyticus, and Yersinia enterocolitica, unveiled that although the collection of muropeptide identities was almost identical across organisms, abundances of muropeptides varied considerably (Quintela et al., 1995). To date, over 50 strains of more than 20 different bacterial species have been analyzed for PG composition; this represents a small fraction of both the prokaryotic kingdom and the morphological heterogeneity exhibited by bacteria (Young,2006). A more comprehensive set of analyses will allow for determinations of the variations in PG composition due to shape, phylogeny, and growth environment.
Not all bacterial species offer PG that is easily purified, due to differences in how PG is synthesized from organism to organism. PG purification from Gram-positive bacteria can be more challenging, likely due to teichoic acids and/or other PG-associated polymers that may shield chemical bonds from cleavage (Tipper et al., 1969; Garcia-Bustos et al., 1987). Nevertheless, the use of specific amidases has proved effective for purification of soluble peptide stems from the Gram-positive species Streptococcus pneumoniae (Garcia-Bustos et al., 1988; Garcia-Bustos et al., 1990) and Staphylococcus aureus (Snowden et al., 1990). Although HPLC separation of S. aureus PG does not have the resolution necessary to definitively identify all muropeptide species, characteristic peaks can be assigned to a fraction of the muropeptide species (Snowden et al., 1990; Snowden et al., 1991). In methicillin resistant S. aureus, the high abundance of D-Ala-D-Ala termini was indicative of low D,D-carboxypeptidase activity (de Jonge et al., 1992).
The analysis of “exotic” bacteria has also revealed other PG components. The wall of the extreme thermophile Thermus thermophilus contains ornithine in place of DAP, with crossbridges consisting of Gly-Gly peptides, leading to a far more complex HPLC spectrum (Quintela et al., 1995). Deinococcus radiodurans, a species closely related to T. thermophilus, has similar muropeptide composition, with the exception of the absence of phenylacetate acylated muropeptides found in T. thermophilus (Quintela et al., 1999). However, T. thermophilus is Gram-negative while D. radiodurans is Gram-positive and has a much thicker wall, illustrating that similarities in chemical composition can occur even if the structure is substantially different. The Gram-negative species Thermotoga maritima and Myxococcus xanthus also have similarly peculiar PG properties: both have two DAP enantiomers, L- and D-Lys in T. maritima and meso- and LL-DAP in M. xanthus (Boniface et al., 2009; Bui et al., 2009). The significance of this chemical diversity remains to be determined.
Revealing the functions of PG-related proteins
In addition to PG characterization, HPLC has found further application in the study of enzymes involved in PG metabolism and modification. Differences in muropeptide abundances have been used to link proteins to enzymatic activities, such as the role of LD-carboxypeptidases in hydrolyzing peptide stems (Leguina et al., 1994), DD-endopeptidases in cleaving peptide cross-bridges (Singh et al., 2012), and amidase activity in cleaving peptide stems during cell separation (Heidrich et al., 2001). Two lytic transglycosylases that cleave between the two sugars in the muropeptide subunit were discovered in P. aeruginosa using a modified HPLC method that utilizes anion exchange to resolve the muropeptide products of digestion (Blackburn et al., 2002). By separating soluble muropeptides that are freed during growth, a lytic transglycosylase from the pathogen N. gonorrhoeae was identified that removes PG strands to aid in cell division (Cloud et al., 2004). These studies demonstrate the utility of HPLC to discover novel players in PG synthesis and degradation.
Another control knob for cell wall synthesis and structure lies in non-PG synthesis morphogenetic proteins; HPLC can be used to discover their indirect effects on PG. The outer membrane proteins LpoA and LpoB aid PBP1A and PBP1B, respectively, by stimulating their transpeptidase activity to result in higher cross-linking percentages (Typas et al., 2010). In C. crescentus, depolymerization of MreB by A22 resulted in decreased average glycan strand length, indicating that MreB promotes transglycosylase activity(Takacs et al., 2010). In S. aureus, teichoic acids regulate PG metabolism by positioning PBP4 at the division septum to catalyze a high level of cross-linking (Atilano et al., 2010). Combining HPLC analysis with gel filtration chromatography has even enabled the quantification of the enzymatic rates of PG precursor synthesis (Mengin-Lecreulx et al., 1982; Barreteau et al., 2009).
Biological functions specific to bacteria can also be discovered from HPLC PG analyses. Amycolatopsis balhimycina produces the antibiotic balhimycin that inhibits cell-wall synthesis in other Gram-positive bacteria; using HPLC it was found that A. balhimycina has few pentapeptides ending in D-Ala-D-Ala, the target for balhimycin, thereby protecting itself from the inhibitory effects of its own synthesis (Schaberle et al., 2011). HPLC was used to identify muropeptide degradation products of bacteriocins in a number of pathogenic Pseudomonas species that target PG metabolism (Barreteau et al., 2009), to discover type-VI secretion-driven endopeptidases (Russell et al., 2011), and to categorize the bacterial toxin pesticin from Yersinia pestis as a muramidase (Vollmer et al., 1997).
PG-mediated activation of the human immune system
Toll-like receptors comprise a complex system of receptors on host cells that sense a wide array of microbial ligands, and play a central role in innate immunity by recognizing pathogens (Yano et al., 2011). HPLC studies of S. aureus and S. pneumoniae were used to determine that a muropeptide dimer is the minimal unit needed to trigger the Toll pathway (Filipe et al., 2005). Side reactions that chemically alter the structure of PG can promote infectivity, such as the acetylation of MurNAc in N. gonorrhoeae and N. meningitidis (Dillard et al., 2005). HPLC was also used to elucidate mechanisms of pathogenesis involving PG degradation by the type-IV secretion apparatus (Kohler et al., 2007); a virulence factor protects N. gonorrhoeae from host attack through DD-carboxypeptidase and endopeptidase activity, resulting in subtle, localized changes in PG structure and smaller colonies that may protect against the host immune response (Stohl et al., 2012). In S. aureus, the amidation of D-Glu in the stem peptides to D-Gln was discovered to be important for drug resistance (Figueiredo et al., 2012). Therefore, HPLC PG analyses can provide novel information about mechanisms of pathogenesis and drug resistance.
UPLC: HPLC reloaded
There are a few drawbacks that limit the widespread utility of HPLC. Slow flow-through, even with advances in column design and injectors, limits the number of samples processed. In addition, accurate detection of the minor PG components requires samples taken from109 – 1010 cells, which may be a limiting constraint for the study of intracellular pathogens or endosymbionts. Application of HPLC to the detection of PG in natural samples, such as blood, would also likely be limited by sensitivity. One potential avenue for addressing these limitations is the recent introduction of Ultra Performance Liquid Chromatography (UPLC). UPLC was recently developed as an extension of the HPLC method that utilizes new stationary phases with sub-2-μm particle size able to withstand very high pressures, which increases resolution, speed, and sensitivity (Wilson et al., 2005; Kumar et al., 2012). Coupled with detectors that have high sampling rates and low-volume sample injectors, this allows for the use of smaller sample volumes (1–10 μl, compared with 100–200 μl for a typical HPLC PG analysis) and shorter run times (5–20 minutes, compared with 1–2 hours for HPLC). Although microbiologists have yet to harness the power of UPLC for PG analyses, they are likely forthcoming and have the potential to leverage higher resolution and throughput for novel discoveries.
Complementary methodologies for PG analysis
To provide additional information about cell wall ultrastructure, Atomic Force Microscopy and TEM tomography have been used to probe the mechanics and spatial architecture of sacculi from E. coli (Yao et al., 1999; Gan et al., 2008), C. crescentus (Gan et al., 2008), B. subtilis (Hayhurst et al., 2008), S. aureus (Turner et al., 2010), and ovococci (Wheeler et al., 2011). Solid-state nuclear magnetic resonance spectroscopy has been used to resolve the 3D atomic structure of the PG polymer, and determined that the PG layer is very flexible (Renault et al., 2012). These methods are destructive, but chemical tools to label PG have enabled real-time tracking of bacterial growth using fluorescent vancomycin (Turner et al., 2010; Wheeler et al., 2011) or fluorescent D-amino acids (Kuru et al., 2012; Siegrist et al., 2013) and promise further insight into the coordination of PG synthesis.
Finally, an understanding of the molecular mechanisms underpinning cell wall growth will require bridging the molecular scale of the synthesis machinery and the cellular scales that can involve spatial and mechanical feedback on growth. Given the wealth of quantitative information regarding wall biochemistry and mechanics, computational modeling provides a tool that can assay the consequences of hypothesized growth mechanisms on morphogenesis (Fig. 2C). In concert with fluorescence imaging of protein dynamics, the wall composition parameters provided by HPLC PG analyses, such as cross-linking density and glycan strand lengths, can be directly incorporated into simulations to predict wall ultrastructure and map this to the ensuing shape (Huang et al., 2008; Furchtgott et al., 2011; van Teeffelen et al., 2011; Wang et al., 2012). Future approaches could involve exhaustive generation of architectures that satisfy the properties constrained by HPLC analyses, and mapping wall networks to morphologies can be compared to shapes determined from light microscopy and high-resolution electron microscopy. Importantly, ongoing advances in HPLC, modeling, and complementary experimental tools should drive further developments in each area.
Outstanding questions and challenges
Although the union of HPLC with other assays has produced much insight, several hurdles remain to be overcome for achieving a mechanistic understanding of bacterial morphogenesis. It remains to be seen if all enzymes relating to PG synthesis have been discovered; recent chemical screens using the KEIO library have identified new PG-related proteins as well as connections to known factors such as the PBPs (Typas et al., 2010), motivating the generation of similar genetic libraries in other organisms. Moreover, the full functionality of known proteins may not be fully elucidated, both from an in vitro perspective and in the context of a spatially heterogeneous, complex cell. Clearly, it will be critical to expand the application of HPLC to a greater scope of species, with the ability to compare compositions between samples while keeping as many variables as possible constant. Given that HPLC is economically and technically accessible to many microbiology labs, an expansion in the number of scientists utilizing this technology would be a natural route to expand the biological diversity of PG analyses.
Thus far, the majority of PG analyses have focused on the cross-linking diversity, with glycan strand length distributions known only for E. coli and S. aureus (Boneca et al., 2000), and only in a few growth conditions (Mollner et al., 1984; Harz et al., 1990; Obermann et al., 1994). Glycan strand quantification would provide a complementary dimension with which to map wall ultrastructure. The glycan strands can be purified using amidases to cleave the peptide stem from muropeptides (Harz et al., 1990; Bui et al., 2012). Most glycan strand analyses have resolution limited to 30 disaccharide units by reverse-phase HPLC (Vollmer et al., 2008), although recent applications of size-exclusion-based chromatography have enabled resolution of strands of up to 200 disaccharides (Hayhurst et al., 2008; Wheeler et al., 2011). A greater shortcoming is the difficulty of obtaining the amidase enzymes necessary to cleave stem peptides from muropeptides, which are not available commercially and take considerable effort to purify in-house (Hash et al., 1967). Until more streamlined techniques emerge for amidase purification, accurately quantifying differences in strand length will remain challenging.
HPLC PG analysis provides information on muropeptide identity, chemical composition, cross-linking and glycan strand lengths. However, there are several other potentially important variables that merit further development, such as the spatial distribution of structural features in the sacculus. Finer details such as where crossbridging occurs along a glycan chain, where Lpp binds spatially, or where DAP-DAP cross-links appear cannot be gleaned from HPLC analyses. In addition, differences in PG make-up within a population cannot be discerned from HPLC analysis. Finally, even though HPLC methods provide data on glycan strand termini, that evidence is not highly dependable.
Conclusion
Given recent advances in our understanding of the role of the cell wall in bacterial morphogenesis and physiology, the utility of HPLC for the analysis of peptidoglycan composition is of increasing importance to the microbiological community as a complement to cell biological and genetic studies. Historically, HPLC analyses have been critical for revealing peptidoglycan structural and chemical modifications through muropeptide identification and quantification, and for linking cell-wall composition to morphogenesis, pathogenesis, and diverse essential enzymatic functions. Building upon the HPLC PG data collected from E. coli, we look forward to expanding our knowledge of the diversity of PG architectures across the prokaryotic kingdom. HPLC has proven to be the tool of choice in identifying nuances in PG composition, with advantages such as affordability and ease of use, and can be used in combination with other technologies in the quest to discover the architecture of a bug’s life. Recent advances in HPLC column materials and the introduction of UPLC should keep these technologies at the forefront of cell wall research for years to come.
Supplementary Material
Acknowledgments
This work was supported by NIH Director’s New Innovator Award DP2OD006466 (to K.C.H.). The authors thank Gabriel Billings and Carolina Tropini for helpful discussions.
References
- Atilano ML, Pereira PM, Yates J, Reed P, Veiga H, Pinho MG, Filipe SR. Teichoic acids are temporal and spatial regulators of peptidoglycan cross-linking inStaphylococcus aureus. Proc Natl Acad Sci U S A. 2010;107:18991–18996. doi: 10.1073/pnas.1004304107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The keio collection. Mol Syst Biol. 2006;2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreteau H, Bouhss A, Fourgeaud M, Mainardi JL, Touze T, Gerard F, et al. Human- and plant-pathogenic Pseudomonas species produce bacteriocins exhibiting colicin M-like hydrolase activity towards peptidoglycan precursors. J Bacteriol. 2009;191:3657–3664. doi: 10.1128/JB.01824-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn NT, Clarke AJ. Characterization of soluble and membrane-bound family 3 lytic transglycosylases from Pseudomonas aeruginosa. Biochemistry. 2002;41:1001–1013. doi: 10.1021/bi011833k. [DOI] [PubMed] [Google Scholar]
- Boneca IG, Huang ZH, Gage DA, Tomasz A. Characterization of Staphylococcus aureus cell wall glycan strands, evidence for a new beta-N-acetylglucosaminidase activity. J Biol Chem. 2000;275:9910–9918. doi: 10.1074/jbc.275.14.9910. [DOI] [PubMed] [Google Scholar]
- Boniface A, Parquet C, Arthur M, Mengin-Lecreulx D, Blanot D. The elucidation of the structure of Thermotoga maritima peptidoglycan reveals two novel types of cross-link. J Biol Chem. 2009;284:21856–21862. doi: 10.1074/jbc.M109.034363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulbitch A, Quinn B, Pink D. Elasticity of the rod-shaped gram-negative eubacteria. Phys Rev Lett. 2000;85:5246–5249. doi: 10.1103/PhysRevLett.85.5246. [DOI] [PubMed] [Google Scholar]
- Boylen CW, Ensign JC. Ratio of teichoic acid and peptidoglycan in cell walls of Bacillus subtilis following spire germination and during vegetative growth. J Bacteriol. 1968;96:421–427. doi: 10.1128/jb.96.2.421-427.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brill J, Hoffmann T, Bleisteiner M, Bremer E. Osmotically controlled synthesis of the compatible solute proline is critical for cellular defense of Bacillus subtilis against high osmolarity. J Bacteriol. 2011;193:5335–5346. doi: 10.1128/JB.05490-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown PJ, de Pedro MA, Kysela DT, Van der Henst C, Kim J, De Bolle X, et al. Polar growth in the alphaproteobacterial order rhizobiales. Proc Natl Acad Sci U S A. 2012;109:1697–1701. doi: 10.1073/pnas.1114476109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui NK, Gray J, Schwarz H, Schumann P, Blanot D, Vollmer W. The peptidoglycan sacculus of Myxococcus xanthus has unusual structural features and is degraded during glycerol-induced myxospore development. J Bacteriol. 2009;191:494–505. doi: 10.1128/JB.00608-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui NK, Eberhardt A, Vollmer D, Kern T, Bougault C, Tomasz A, et al. Isolation and analysis of cell wall components from Streptococcus pneumoniae. Anal Biochem. 2012;421:657–666. doi: 10.1016/j.ab.2011.11.026. [DOI] [PubMed] [Google Scholar]
- Cabeen MT, Charbon G, Vollmer W, Born P, Ausmees N, Weibel DB, Jacobs-Wagner C. Bacterial cell curvature through mechanical control of cell growth. EMBO J. 2009;28:1208–1219. doi: 10.1038/emboj.2009.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caparros M, Torrecuadrada JL, de Pedro MA. Effect of D-amino acids on Escherichia coli strains with impaired penicillin-binding proteins. Res Microbiol. 1991;142:345–350. doi: 10.1016/0923-2508(91)90050-k. [DOI] [PubMed] [Google Scholar]
- Cava F, Lam H, de Pedro MA, Waldor MK. Emerging knowledge of regulatory roles of D-amino acids in bacteria. Cell Mol Life Sci. 2011a;68:817–831. doi: 10.1007/s00018-010-0571-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cava F, de Pedro MA, Lam H, Davis BM, Waldor MK. Distinct pathways for modification of the bacterial cell wall by non-canonical D-amino acids. EMBO J. 2011b;30:3442–3453. doi: 10.1038/emboj.2011.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Zhang W, Shi Q, Hesek D, Lee M, Mobashery S, Shoichet BK. Crystal structures of penicillin-binding protein 6 from Escherichia coli. J Am Chem Soc. 2009;131:14345–14354. doi: 10.1021/ja903773f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloud KA, Dillard JP. Mutation of a single lytic transglycosylase causes aberrant septation and inhibits cell separation of Neisseria gonorrhoeae. J Bacteriol. 2004;186:7811–7814. doi: 10.1128/JB.186.22.7811-7814.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalhoff A, Nasu T, Okamoto K. Target affinities of faropenem to and its impact on the morphology of Gram-positive and Gram-negative bacteria. Chemotherapy. 2003;49:172–183. doi: 10.1159/000071141. [DOI] [PubMed] [Google Scholar]
- de Jonge BL, Chang YS, Gage D, Tomasz A. Peptidoglycan composition of a highly methicillin-resistant Staphylococcus aureus strain. The role of penicillin binding protein 2A. J Biol Chem. 1992;267:11248–11254. [PubMed] [Google Scholar]
- de la Rosa EJ, de Pedro MA, Vazquez D. Penicillin binding proteins: Role in initiation of murein synthesis in Escherichia coli. Proc Natl Acad Sci U S A. 1985;82:5632–5635. doi: 10.1073/pnas.82.17.5632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Blaauwen T, de Pedro MA, Nguyen-Disteche M, Ayala JA. Morphogenesis of rod-shaped sacculi. FEMS Microbiol Rev. 2008;32:321–344. doi: 10.1111/j.1574-6976.2007.00090.x. [DOI] [PubMed] [Google Scholar]
- Dillard JP, Hackett KT. Mutations affecting peptidoglycan acetylation inNeisseria gonorrhoeae and Neisseria meningitidis. Infect Immun. 2005;73:5697–5705. doi: 10.1128/IAI.73.9.5697-5705.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougherty TJ. Analysis of Neisseria gonorrhoeae peptidoglycan by reverse-phase, high-pressure liquid chromatography. J Bacteriol. 1985;163:69–74. doi: 10.1128/jb.163.1.69-74.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dramsi S, Magnet S, Davison S, Arthur M. Covalent attachment of proteins to peptidoglycan. FEMS Microbiol Rev. 2008;32:307–320. doi: 10.1111/j.1574-6976.2008.00102.x. [DOI] [PubMed] [Google Scholar]
- Figueiredo TA, Sobral RG, Ludovice AM, Almeida JM, Bui NK, Vollmer W, et al. Identification of genetic determinants and enzymes involved with the amidation of glutamic acid residues in the peptidoglycan of Staphylococcus aureus. PLoS Pathog. 2012;8:e1002508. doi: 10.1371/journal.ppat.1002508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipe SR, Tomasz A, Ligoxygakis P. Requirements of peptidoglycan structure that allow detection by the Drosophila Toll pathway. EMBO Rep. 2005;6:327–333. doi: 10.1038/sj.embor.7400371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furchtgott L, Wingreen NS, Huang KC. Mechanisms for maintaining cell shape in rod-shaped Gram-negative bacteria. Mol Microbiol. 2011;81:340–353. doi: 10.1111/j.1365-2958.2011.07616.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan L, Chen S, Jensen GJ. Molecular organization of Gram-negative peptidoglycan. Proc Natl Acad Sci U S A. 2008;105:18953–7. doi: 10.1073/pnas.0808035105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Bustos J, Tomasz A. A biological price of antibiotic resistance: Major changes in the peptidoglycan structure of penicillin-resistant pneumococci. Proc Natl Acad Sci U S A. 1990;87:5415–5419. doi: 10.1073/pnas.87.14.5415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Bustos JF, Chait BT, Tomasz A. Altered peptidoglycan structure in a pneumococcal transformant resistant to penicillin. J Bacteriol. 1988;170:2143–2147. doi: 10.1128/jb.170.5.2143-2147.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Bustos JF, Chait BT, Tomasz A. Structure of the peptide network of pneumococcal peptidoglycan. J Biol Chem. 1987;262:15400–15405. [PubMed] [Google Scholar]
- Glauner B. Separation and quantification of muropeptides with high-performance liquid chromatography. Anal Biochem. 1988;172(2):451–464. doi: 10.1016/0003-2697(88)90468-x. [DOI] [PubMed] [Google Scholar]
- Glauner B, Holtje J, Schwarz U. The composition of the murein of Escherichia coli. The Journal of Biological Chemistry. 1988;263:10088–10095. [PubMed] [Google Scholar]
- Goley ED, Comolli LR, Fero KE, Downing KH, Shapiro L. DipM links peptidoglycan remodelling to outer membrane organization in Caulobacter. Mol Microbiol. 2010;77:56–73. doi: 10.1111/j.1365-2958.2010.07222.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodell EW, Schwarz U. Enzymes synthesizing and hydrolyzing murein in Escherichia coli. Topographical distribution over the cell envelope. Eur J Biochem. 1977;81:205–210. doi: 10.1111/j.1432-1033.1977.tb11942.x. [DOI] [PubMed] [Google Scholar]
- Guberman JM, Fay A, Dworkin J, Wingreen NS, Gitai Z. PSICIC: Noise and asymmetry in bacterial division revealed by computational image analysis at sub-pixel resolution. PLoS Comput Biol. 2008;4:e1000233. doi: 10.1371/journal.pcbi.1000233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakenbeck R, Holtje J, Labischinski H, editors. The target of penicillin: The murein sacculus of bacterial cell walls architecture and growth. Walter De Gruyter Incorporated; 1983. [Google Scholar]
- Harold FM. To shape a cell: An inquiry into the causes of morphogenesis of microorganisms. Microbiol Rev. 1990;54:381–431. doi: 10.1128/mr.54.4.381-431.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harz H, Burgdorf K, Holtje JV. Isolation and separation of the glycan strands from murein of Escherichia coli by reversed-phase high-performance liquid chromatography. Anal Biochem. 1990;190:120–128. doi: 10.1016/0003-2697(90)90144-x. [DOI] [PubMed] [Google Scholar]
- Hash JH, Rothlauf MV. The N,O-diacetylmuramidase of Chalaropsis species. I. purification and crystallization. J Biol Chem. 1967;242:5586–5590. [PubMed] [Google Scholar]
- Hayhurst EJ, Kailas L, Hobbs JK, Foster SJ. Cell wall peptidoglycan architecture in Bacillus subtilis. Proc Natl Acad Sci U S A. 2008;105:14603–14608. doi: 10.1073/pnas.0804138105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidrich C, Templin MF, Ursinus A, Merdanovic M, Berger J, Schwarz H, et al. Involvement of N-acetylmuramyl-L-alanine amidases in cell separation and antibiotic-induced autolysis of Escherichia coli. Mol Microbiol. 2001;41:167–178. doi: 10.1046/j.1365-2958.2001.02499.x. [DOI] [PubMed] [Google Scholar]
- Holtje JV. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol Mol Biol Rev. 1998;62:181–203. doi: 10.1128/mmbr.62.1.181-203.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horcajo P, dePedro MA, Cava F. Peptidoglycan plasticity in bacteria: Stress-induced peptidoglycan editing by noncanonical D-amino acids. Microb Drug Resist. 2012;18:306–313. doi: 10.1089/mdr.2012.0009. [DOI] [PubMed] [Google Scholar]
- Huang KC, Mukhopadhyay R, Wen B, Gitai Z, Wingreen NS. Cell shape and cell-wall organization in Gram-negative bacteria. Proc Natl Acad Sci U S A. 2008;105:19282–19287. doi: 10.1073/pnas.0805309105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Si F, Margolin W, Sun SX. Mechanical control of bacterial cell shape. Biophys J. 2011;101:327–335. doi: 10.1016/j.bpj.2011.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseleau-Petit D, Liebart JC, Ayala JA, D’Ari R. Unstable Escherichia coli L forms revisited: Growth requires peptidoglycan synthesis. J Bacteriol. 2007;189:6512–6520. doi: 10.1128/JB.00273-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandler O, Schleifer KH, Dandl R. Differentiation of Streptococcus faecalis Andrewes and Horder and Streptococcus faecium Orla-jensen based on the amino acid composition of their murein. J Bacteriol. 1968;96:1935–1939. doi: 10.1128/jb.96.6.1935-1939.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karczmarek A, Martinez-Arteaga R, Alexeeva S, Hansen FG, Vicente M, Nanninga N, den Blaauwen T. DNA and origin region segregation are not affected by the transition from rod to sphere after inhibition of Escherichia coli MreB by A22. Mol Microbiol. 2007;65:51–63. doi: 10.1111/j.1365-2958.2007.05777.x. [DOI] [PubMed] [Google Scholar]
- Kohler PL, Hamilton HL, Cloud-Hansen K, Dillard JP. AtlA functions as a peptidoglycan lytic transglycosylase in the Neisseria gonorrhoeae type IV secretion system. J Bacteriol. 2007;189:5421–5428. doi: 10.1128/JB.00531-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodkin-Gal I, Romero D, Cao S, Clardy J, Kolter R, Losick R. D-amino acids trigger biofilm disassembly. Science. 2010;328:627–629. doi: 10.1126/science.1188628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong KF, Schneper L, Mathee K. Beta-lactam antibiotics: From antibiosis to resistance and bacteriology. APMIS. 2010;118:1–36. doi: 10.1111/j.1600-0463.2009.02563.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama N, Tokura Y, Munch D, Sahl HG, Schneider T, Shibagaki Y, et al. The nonantibiotic small molecule cyslabdan enhances the potency of beta-lactams against MRSA by inhibiting pentaglycine interpeptide bridge synthesis. PLoS One. 2012;7:e48981. doi: 10.1371/journal.pone.0048981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Saini G, Nair A, Sharma R. UPLC: A preeminent technique in pharmaceutical analysis. Acta Pol Pharm. 2012;69:371–380. [PubMed] [Google Scholar]
- Kuru E, Hughes HV, Brown PJ, Hall E, Tekkam S, Cava F, et al. In situ probing of newly synthesized peptidoglycan in live bacteria with fluorescent D-amino acids. Angew Chem Int Ed Engl. 2012;51(50):12519–2. doi: 10.1002/anie.201206749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam H, Oh DC, Cava F, Takacs CN, Clardy J, dePedro MA, Waldor MK. D-amino acids govern stationary phase cell wall remodeling in bacteria. Science. 2009;325:1552–1555. doi: 10.1126/science.1178123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavollay M, Fourgeaud M, Herrmann JL, Dubost L, Marie A, Gutmann L, et al. The peptidoglycan of Mycobacterium abscessus is predominantly cross-linked by L,D-transpeptidases. J Bacteriol. 2011;193:778–782. doi: 10.1128/JB.00606-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavollay M, Arthur M, Fourgeaud M, Dubost L, Marie A, Veziris N, et al. The peptidoglycan of stationary-phase Mycobacterium tuberculosis predominantly contains cross-links generated by L,D-transpeptidation. J Bacteriol. 2008;190:4360–4366. doi: 10.1128/JB.00239-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legaree BA, Adams CB, Clarke AJ. Overproduction of penicillin-binding protein 2 and its inactive variants causes morphological changes and lysis in Escherichia coli. J Bacteriol. 2007a;189:4975–4983. doi: 10.1128/JB.00207-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legaree BA, Daniels K, Weadge JT, Cockburn D, Clarke AJ. Function of penicillin-binding protein 2 in viability and morphology of Pseudomonas aeruginosa. J Antimicrob Chemother. 2007b;59:411–424. doi: 10.1093/jac/dkl536. [DOI] [PubMed] [Google Scholar]
- Leguina JI, Quintela JC, de Pedro MA. Substrate specificity of Escherichia coli LD-carboxypeptidase on biosynthetically modified muropeptides. FEBS Lett. 1994;339:249–252. doi: 10.1016/0014-5793(94)80425-7. [DOI] [PubMed] [Google Scholar]
- Magnet S, Dubost L, Marie A, Arthur M, Gutmann L. Identification of the L,D-transpeptidases for peptidoglycan cross-linking in Escherichia coli. J Bacteriol. 2008;190:4782–4785. doi: 10.1128/JB.00025-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnet S, Bellais S, Dubost L, Fourgeaud M, Mainardi JL, Petit-Frere S, et al. Identification of the L,D-transpeptidases responsible for attachment of the Braun lipoprotein to Escherichia coli peptidoglycan. J Bacteriol. 2007;189:3927–3931. doi: 10.1128/JB.00084-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markiewicz Z, Holtje JV. Failure to trigger the autolytic enzymes in minicells of Escherichia coli. FEMS Microbiol Lett. 1992;70:119–123. doi: 10.1016/0378-1097(92)90670-j. [DOI] [PubMed] [Google Scholar]
- Markiewicz Z, Glauner B, Schwarz U. Murein structure and lack of DD- and LD-carboxypeptidase activities in Caulobacter crescentus. J Bacteriol. 1983;156:649–655. doi: 10.1128/jb.156.2.649-655.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengin-Lecreulx D, Flouret B, van Heijenoort J. Cytoplasmic steps of peptidoglycan synthesis in Escherichia coli. J Bacteriol. 1982;151:1109–1117. doi: 10.1128/jb.151.3.1109-1117.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollner S, Braun V. Murein hydrolase (N-acetyl-muramyl-L-alanine amidase) in human serum. Arch Microbiol. 1984;140:171–177. doi: 10.1007/BF00454921. [DOI] [PubMed] [Google Scholar]
- Obermann W, Holtje JV. Alterations of murein structure and of penicillin-binding proteins in minicells from Escherichia coli. Microbiology. 1994;140 ( Pt 1):79–87. doi: 10.1099/13500872-140-1-79. [DOI] [PubMed] [Google Scholar]
- Paradis-Bleau C, Markovski M, Uehara T, Lupoli TJ, Walker S, Kahne DE, Bernhardt TG. Lipoprotein cofactors located in the outer membrane activate bacterial cell wall polymerases. Cell. 2010;143:1110–1120. doi: 10.1016/j.cell.2010.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltier J, Courtin P, ElMeouche I, Lemee L, Chapot-Chartier MP, Pons JL. Clostridium difficile has an original peptidoglycan structure with a high level of N-acetylglucosamine deacetylation and mainly 3-3 cross-links. J Biol Chem. 2011;286:29053–29062. doi: 10.1074/jbc.M111.259150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piek S, Kahler CM. A comparison of the endotoxin biosynthesis and protein oxidation pathways in the biogenesis of the outer membrane of Escherichia coli and Neisseria meningitidis. Front Cell Infect Microbiol. 2012;2:162. doi: 10.3389/fcimb.2012.00162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisabarro AG, dePedro MA, Vazquez D. Structural modifications in the peptidoglycan of Escherichia coli associated with changes in the state of growth of the culture. J Bacteriol. 1985;161:238–242. doi: 10.1128/jb.161.1.238-242.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prats R, de Pedro MA. Normal growth and division of escherichia coli with a reduced amount of murein. J Bacteriol. 1989;171:3740–3745. doi: 10.1128/jb.171.7.3740-3745.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priyadarshini R, dePedro MA, Young KD. Role of peptidoglycan amidases in the development and morphology of the division septum in Escherichia coli. J Bacteriol. 2007;189:5334–5347. doi: 10.1128/JB.00415-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintela JC, Caparros M, de Pedro MA. Variability of peptidoglycan structural parameters in Gram-negative bacteria. FEMS Microbiol Lett. 1995;125:95–100. doi: 10.1111/j.1574-6968.1995.tb07341.x. [DOI] [PubMed] [Google Scholar]
- Quintela JC, Garcia-del Portillo F, Pittenauer E, Allmaier G, de Pedro MA. Peptidoglycan fine structure of the radiotolerant bacterium Deinococcus radiodurans Sark. J Bacteriol. 1999;181:334–337. doi: 10.1128/jb.181.1.334-337.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintela JC, Pittenauer E, Allmaier G, Aran V, de Pedro MA. Structure of peptidoglycan from Thermus thermophilus HB8. J Bacteriol. 1995;177:4947–4962. doi: 10.1128/jb.177.17.4947-4962.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renault M, Tommassen-van Boxtel R, Bos MP, Post JA, Tommassen J, Baldus M. Cellular solid-state nuclear magnetic resonance spectroscopy. Proc Natl Acad Sci U S A. 2012;109:4863–4868. doi: 10.1073/pnas.1116478109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell AB, Hood RD, Bui NK, LeRoux M, Vollmer W, Mougous JD. Type VI secretion delivers bacteriolytic effectors to target cells. Nature. 2011;475:343–347. doi: 10.1038/nature10244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos JM, Lobo M, Matos AP, De Pedro MA, Arraiano CM. The gene bolA regulates DacA (PBP5), DacC (PBP6) and ampC (AmpC), promoting normal morphology in Escherichia coli. Mol Microbiol. 2002;45:1729–1740. doi: 10.1046/j.1365-2958.2002.03131.x. [DOI] [PubMed] [Google Scholar]
- Sauvage E, Powell AJ, Heilemann J, Josephine HR, Charlier P, Davies C, Pratt RF. Crystal structures of complexes of bacterial DD-peptidases with peptidoglycan-mimetic ligands: The substrate specificity puzzle. J Mol Biol. 2008;381:383–393. doi: 10.1016/j.jmb.2008.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaberle TF, Vollmer W, Frasch HJ, Huttel S, Kulik A, Rottgen M, et al. Self-resistance and cell wall composition in the glycopeptide producer Amycolatopsis balhimycina. Antimicrob Agents Chemother. 2011;55:4283–4289. doi: 10.1128/AAC.01372-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffers DJ, Pinho MG. Bacterial cell wall synthesis: New insights from localization studies. Microbiol Mol Biol Rev. 2005;69:585–607. doi: 10.1128/MMBR.69.4.585-607.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegrist MS, Whiteside S, Jewett JC, Aditham A, Cava F, Bertozzi CR. D-amino acid chemical reporters reveal peptidoglycan dynamics of an intracellular pathogen. ACS Chem Biol. 2013;8(3):500–5. doi: 10.1021/cb3004995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SK, SaiSree L, Amrutha RN, Reddy M. Three redundant murein endopeptidases catalyse an essential cleavage step in peptidoglycan synthesis of Escherichia coli K12. Mol Microbiol. 2012;86:1036–1051. doi: 10.1111/mmi.12058. [DOI] [PubMed] [Google Scholar]
- Sliusarenko O, Heinritz J, Emonet T, Jacobs-Wagner C. High-throughput, subpixel precision analysis of bacterial morphogenesis and intracellular spatio-temporal dynamics. Molecular Microbiology. 2011;80:612–27. doi: 10.1111/j.1365-2958.2011.07579.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snowden MA, Perkins HR. Cross-linking and O-acetylation of peptidoglycan in Staphylococcus aureus (strains H and MR-1) grown in the presence of sub-growth-inhibitory concentrations of beta-lactam antibiotics. J Gen Microbiol. 1991;137:1661–1666. doi: 10.1099/00221287-137-7-1661. [DOI] [PubMed] [Google Scholar]
- Snowden MA, Perkins HR. Peptidoglycan cross-linking in Staphylococcus aureus. An apparent random polymerisation process. Eur J Biochem. 1990;191:373–377. doi: 10.1111/j.1432-1033.1990.tb19132.x. [DOI] [PubMed] [Google Scholar]
- Sochacki KA, Shkel IA, Record MT, Weisshaar JC. Protein diffusion in the periplasm of E. coli under osmotic stress. Biophys J. 2011;100:22–31. doi: 10.1016/j.bpj.2010.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stohl EA, Chan YA, Hackett KT, Kohler PL, Dillard JP, Seifert HS. Neisseria gonorrhoeae virulence factor NG1686 is a bifunctional M23B family metallopeptidase that influences resistance to hydrogen peroxide and colony morphology. J Biol Chem. 2012;287:11222–11233. doi: 10.1074/jbc.M111.338830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss MP, Liew AT, Turnbull L, Whitchurch CB, Monahan LG, Harry EJ. 3D-SIM super resolution microscopy reveals a bead-like arrangement for FtsZ and the division machinery: Implications for triggering cytokinesis. PLoS Biol. 2012;10:e1001389. doi: 10.1371/journal.pbio.1001389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutow AB, Welker NE. Chemical composition of the cell walls of Bacillus stearothermophilus. J Bacteriol. 1967;93:1452–1457. doi: 10.1128/jb.93.4.1452-1457.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sycuro LK, Wyckoff TJ, Biboy J, Born P, Pincus Z, Vollmer W, Salama NR. Multiple peptidoglycan modification networks modulate Helicobacter pylori’s cell shape, motility, and colonization potential. PLoS Pathog. 2012;8:e1002603. doi: 10.1371/journal.ppat.1002603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takacs CN, Poggio S, Charbon G, Pucheault M, Vollmer W, Jacobs-Wagner C. MreB drives de novo rod morphogenesis in Caulobacter crescentus via remodeling of the cell wall. J Bacteriol. 2010;192:1671–1684. doi: 10.1128/JB.01311-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tipper DJ, Berman MF. Structures of the cell wall peptidoglycans of Staphylococcus epidermidis Texas 26 and Staphylococcus aureus Copenhagen. I. Chain length and average sequence of cross-bridge peptides. Biochemistry. 1969;8:2183–2192. doi: 10.1021/bi00833a060. [DOI] [PubMed] [Google Scholar]
- Turner RD, Ratcliffe EC, Wheeler R, Golestanian R, Hobbs JK, Foster SJ. Peptidoglycan architecture can specify division planes in Staphylococcus aureus. Nat Commun. 2010;1:26. doi: 10.1038/ncomms1025. [DOI] [PubMed] [Google Scholar]
- Typas A, Banzhaf M, Gross CA, Vollmer W. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat Rev Microbiol. 2011;10:123–136. doi: 10.1038/nrmicro2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Typas A, Banzhaf M, van den Berg van Saparoea B, Verheul J, Biboy J, Nichols RJ, et al. Regulation of peptidoglycan synthesis by outer-membrane proteins. Cell. 2010;143:1097–1109. doi: 10.1016/j.cell.2010.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Teeffelen S, Wang S, Furchtgott L, Huang KC, Wingreen NS, Shaevitz JW, Gitai Z. The bacterial actin MreB rotates, and rotation depends on cell-wall assembly. Proc Natl Acad Sci U S A. 2011;108:15822–15827. doi: 10.1073/pnas.1108999108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmer W, Seligman SJ. Architecture of peptidoglycan: More data and more models. Trends Microbiol. 2010;18:59–66. doi: 10.1016/j.tim.2009.12.004. [DOI] [PubMed] [Google Scholar]
- Vollmer W, Blanot D, de Pedro MA. Peptidoglycan structure and architecture. FEMS Microbiol Rev. 2008;32:149–167. doi: 10.1111/j.1574-6976.2007.00094.x. [DOI] [PubMed] [Google Scholar]
- Vollmer W, Pilsl H, Hantke K, Holtje JV, Braun V. Pesticin displays muramidase activity. J Bacteriol. 1997;179:1580–1583. doi: 10.1128/jb.179.5.1580-1583.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachi M, Matsuhashi M. Negative control of cell division by mreB, a gene that functions in determining the rod shape of Escherichia coli cells. J Bacteriol. 1989;171:3123–3127. doi: 10.1128/jb.171.6.3123-3127.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Furchtgott L, Huang KC, Shaevitz JW. Helical insertion of peptidoglycan produces chiral ordering of the bacterial cell wall. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E595–604. doi: 10.1073/pnas.1117132109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WS, Lundgren DG. Peptidoglycan of a chemolithotrophic bacterium,Ferrobacillus ferrooxidans. J Bacteriol. 1968;95:1851–1856. doi: 10.1128/jb.95.5.1851-1856.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidel W, Frank H, Martin HH. The rigid layer of the cell wall of Escherichia coli strain B. J Gen Microbiol. 1960;22:158–166. doi: 10.1099/00221287-22-1-158. [DOI] [PubMed] [Google Scholar]
- Whatmore AM, Reed RH. Determination of turgor pressure in Bacillus subtilis: A possible role for K+ in turgor regulation. J Gen Microbiol. 1990;136:2521–2526. doi: 10.1099/00221287-136-12-2521. [DOI] [PubMed] [Google Scholar]
- Wheeler R, Mesnage S, Boneca IG, Hobbs JK, Foster SJ. Super-resolution microscopy reveals cell wall dynamics and peptidoglycan architecture in ovococcal bacteria. Mol Microbiol. 2011;82:1096–1109. doi: 10.1111/j.1365-2958.2011.07871.x. [DOI] [PubMed] [Google Scholar]
- Wieser A, Schneider L, Jung J, Schubert S. MALDI-TOF MS in microbiological diagnostics-identification of microorganisms and beyond (mini review) Appl Microbiol Biotechnol. 2012;93:965–974. doi: 10.1007/s00253-011-3783-4. [DOI] [PubMed] [Google Scholar]
- Wilson ID, Nicholson JK, Castro-Perez J, Granger JH, Johnson KA, Smith BW, Plumb RS. High resolution “ultra performance” liquid chromatography coupled to oa-TOF mass spectrometry as a tool for differential metabolic pathway profiling in functional genomic studies. J Proteome Res. 2005;4:591–598. doi: 10.1021/pr049769r. [DOI] [PubMed] [Google Scholar]
- Yano T, Kurata S. Intracellular recognition of pathogens and autophagy as an innate immune host defence. J Biochem. 2011;150:143–149. doi: 10.1093/jb/mvr083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao X, Jericho M, Pink D, Beveridge T. Thickness and elasticity of Gram-negative murein sacculi measured by atomic force microscopy. J Bacteriol. 1999;181:6865–6875. doi: 10.1128/jb.181.22.6865-6875.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young KD. The selective value of bacterial shape. Microbiol Mol Biol Rev. 2006;70:660–703. doi: 10.1128/MMBR.00001-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.