Abstract
Mammalian reoviruses are thought to assemble and replicate within cytoplasmic, nonmembranous structures called viral factories. The viral nonstructural protein μNS forms factory-like globular inclusions when expressed in the absence of other viral proteins and binds to the surfaces of the viral core particles in vitro. Given these previous observations, we hypothesized that one or more of the core surface proteins may be recruited to viral factories through specific associations with μNS. We found that all three of these proteins—λ1, λ2, and σ2—localized to factories in infected cells but were diffusely distributed through the cytoplasm and nucleus when each was separately expressed in the absence of other viral proteins. When separately coexpressed with μNS, on the other hand, each core surface protein colocalized with μNS in globular inclusions, supporting the initial hypothesis. We also found that λ1, λ2, and σ2 each localized to filamentous inclusions formed upon the coexpression of μNS and μ2, a structurally minor core protein that associates with microtubules. The first 40 residues of μNS, which are required for association with μ2 and the RNA-binding nonstructural protein σNS, were not required for association with any of the three core surface proteins. When coexpressed with μ2 in the absence of μNS, each of the core surface proteins was diffusely distributed and displayed only sporadic, weak associations with μ2 on filaments. Many of the core particles that entered the cytoplasm of cycloheximide-treated cells following entry and partial uncoating were recruited to inclusions of μNS that had been preformed in those cells, providing evidence that μNS can bind to the surfaces of cores in vivo. These findings expand a model for how viral and cellular components are recruited to the viral factories in infected cells and provide further evidence for the central but distinct roles of viral proteins μNS and μ2 in this process.
The molecular machinery used for viral replication in the cytoplasm or nucleus of infected cells is commonly concentrated and organized in distinct sites or structures (reviewed in references 22 and 27). Schwartz et al. (44) recently proposed that all viruses that replicate through mRNA intermediates, including double-stranded RNA (dsRNA) viruses, may sequester their mRNA templates within a multiprotein complex that either is attached to cellular membranes or forms a distinct core-like structure. By doing so, these viruses may concentrate the minus-strand RNA products for use as templates, while limiting the exposure of dsRNA intermediates or products to host cell defense mechanisms, such as protein kinase R, RNase L, and the factors that mediate RNA interference (13, 43, 44).
The nonfusogenic mammalian orthoreoviruses (reoviruses) sequester their segmented dsRNA genomes, together with the viral polymerase molecules and capping enzymes for mRNA synthesis, within a 52-MDa core particle (38, 41). This core displays T=1 icosahedral symmetry and is composed of the following five viral proteins: λ1 and σ2, which form the core shell and decorating nodules (41); λ2, the mRNA capping guanylyltransferase and methyltransferase, which forms a turret on the exterior of the core shell around each fivefold axis (12, 32, 41, 54); and λ3 and μ2, the RNA-dependent RNA polymerase (14, 51, 52) and its cofactor (57), respectively, which are situated internal to the shell near the fivefold axes (15). The 10 dsRNA genome segments are also packaged inside the shell, where they can be used as templates for mRNA synthesis by the viral transcriptases (2, 5, 46). Over the course of reovirus infection, many new core particles are assembled and presumably then coated with the three remaining viral outer capsid proteins to produce infectious progeny virions (36, 47). In addition, some or all of the newly assembled cores synthesize more of the viral mRNAs, thereby amplifying the production of viral genes, gene products, and particles (23, 25, 29).
How the core is assembled remains poorly understood. It is a seemingly complex process that involves multiple events as follows (relative timing is not implied by the listed order): (i) formation of an icosahedral protein shell from λ1 and σ2, (ii) addition of the λ2 mRNA capping enzyme turret outside this shell, (iii) addition of the λ3 polymerase and μ2 cofactor inside the shell, (iv) assortment and packaging of the 10 distinct mRNA molecules, and (v) one round of minus-strand synthesis from each of the mRNA templates to regenerate the 10 dsRNA genome segments (reviewed in references 39 and 58). Packaging and minus-strand synthesis may be linked (1, 59), and the manner by which the internal components are placed inside the shell or by which the shell forms around the internal components remains a mystery. Despite these uncertainties, the assembly of cores and the replication of viral RNA are believed to occur within distinct structures that form in the cytoplasm of reovirus-infected cells and are commonly referred to as viral inclusions or factories (3, 4, 8, 33, 35, 40, 42, 45, 48, 49).
We and others have recently identified several determinants of reovirus factory formation and morphology. Nearly all reovirus strains examined to date form microtubule-associated filamentous factories that are similar to those of the type 1 prototype strain, Lang (T1L) (40). In contrast, the type 3 prototype strain, Dearing, in use in our lab (T3DN) forms globular factories that lack filaments (40). Analysis of a panel of T1L/T3D reassortants identified the M1 genome segment, which encodes the structurally minor core protein μ2, as the genetic determinant of this difference in factory morphology (40). The μ2 protein from reovirus strains that form filamentous factories was identified as a microtubule-associated protein, which determines this morphology (40). Another recent study identified μ2 as the primary determinant of a difference in the rates of formation of viral factories in T1L- versus T3D-infected cells (33). We have further found that when the reovirus nonstructural protein μNS is expressed in cells in the absence of other viral proteins, it forms large globular inclusions that are indistinguishable by phase-contrast microscopy from the globular factories formed during T3DN infection (8). From this result, we have concluded that the matrix of the viral factories is largely composed of μNS. When coexpressed with μ2, however, μNS is not present in globular inclusions but instead colocalizes with μ2 in filamentous inclusions associated with microtubules (8). The N-terminal 40 to 41 residues of μNS have been shown to be both necessary and sufficient for colocalization with μ2 (8). In more recent experiments, we have found that the RNA-binding nonstructural protein σNS, which is diffusely distributed in the cytoplasm and nucleus when expressed alone, is specifically recruited to μNS globular inclusions in a manner that requires the N-terminal 40 amino acids of μNS and is promoted by RNA (35). Other recent studies identified the S3 genome segment and its encoded protein σNS as secondary determinants of a difference in the rates of formation of viral factories in T1L- versus T3D-infected cells (33) and provided independent evidence that σNS is recruited to viral factories through its association with μNS (3, 4). Based on these observations, we have proposed a model in which the μNS matrix acts to sequester and concentrate viral proteins, viral RNAs, and possibly host factors in distinct locations within the cytoplasm, thus building the factories in which viral replication and assembly occur (8, 35).
For this report, we subjected our model to further scrutiny by using immunofluorescence (IF) microscopy to examine the subcellular locations of the three surface proteins (λ1, λ2, and σ2) of the viral core particle in infected cells when expressed in cells in the absence of other viral proteins or when coexpressed with viral proteins μNS and/or μ2. Based on previous evidence that μNS binds to the surfaces of cores in vitro (7), we specifically hypothesized that one or more of the core surface proteins are recruited to viral factories through an association(s) with μNS. We also tested whether entering viral cores may be recruited to preformed μNS inclusions inside cells, which may have relevance for understanding how the factories initially form. These observations provide further evidence for the central but distinct roles of μNS and μ2 in recruiting viral and cellular components that are required for the assembly of cores and for replication of the genome within viral factories.
MATERIALS AND METHODS
Cells and viruses.
CV-1 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) (Invitrogen) containing 10% fetal bovine serum (FBS) (HyClone) and 10 μg of gentamicin (Invitrogen) per ml. Reovirus strains T1L and T3D were lab stocks. The isolate of T3D in general use in our lab is designated T3DN to differentiate it from another lab's isolate (T3DC) that differs in viral factory morphology and the M1 sequence, as described previously (40). Third-passage L-cell-lysate stocks of twice plaque-purified reovirus clones were used for cell infections.
Antibodies.
Goat anti-mouse immunoglobulin G (IgG) and goat anti-rabbit IgG conjugated to Alexa 488 or Alexa 594 were obtained from Molecular Probes. Rabbit polyclonal antisera against the μNS and μ2 proteins have been described previously (7, 8). A rabbit polyclonal antiserum against heat-inactivated whole T1L core particles (11, 26; S. Noble and M. L. Nibert, unpublished data) was used to detect λ1, λ2, and σ2. Mouse monoclonal antibody (MAb) 7F4, which is specific for λ2, was described previously (53). All antibodies were titrated to optimize signal-to-noise ratios. For some experiments, we used Alexa 488, Alexa 594, or Texas Red conjugates of μNS and μ2 antibodies that were prepared from protein A-purified IgG conjugated to fluorophores by use of kits obtained from Molecular Probes.
Mammalian expression vectors.
All reovirus proteins examined in this study were expressed from genes cloned into pCI-neo (Promega). The plasmids pCI-M1(T1L) to express μ2 (40), pCI-M3(T1L) to express μNS (8), and pCI-M3(41-721) to express μNS(41-721) (8) were previously described. For expression of the λ1 protein, the T1L L3 gene was excised from pFbD-L3LS2L (26) with EcoRI and was ligated to pCI-neo cut with EcoRI to generate pCI-L3(T1L). For expression of the σ2 protein, the T1L S2 gene was excised from pBS-S2L (26) with EcoRV and NotI and was ligated to pCI-neo cut with NotI and XhoI after the cut XhoI site was blunt ended by treatment with the Klenow fragment of DNA polymerase I. This procedure generated pCI-S2(T1L). For expression of the λ2 protein, the T1L L2 gene was excised from pPCR-Script-L2 (31) with BamHI and EcoRI and was ligated to pBluescript (Stratagene) cut with the same enzymes to generate pBS-L2(T1L). When sequenced, the subcloned T1L L2 gene was found to encode two amino acid changes relative to the published T1L L2 sequence (6), namely Phe129→Ser and Thr534→Ala. These changes were corrected by using the QuikChange site-directed mutagenesis protocol (Stratagene), and the new pBS-L2(T1L) construct was sequenced again to confirm that the encoded amino acid sequence matched that of the published T1L L2 gene. The T1L L2 gene was then removed from pBS-L2(T1L) with SpeI and EcoRI and was ligated to pCI-neo cut with NheI and EcoRI to generate pCI-L2(T1L). All enzymes were obtained from New England Biolabs unless otherwise stated.
Transfections and infections.
CV-1 cells were seeded the day before infection or transfection at a density of 1.5 × 104 per cm2 in 6-well plates (9.6 cm2 per well) containing round glass coverslips (18-mm diameter). Cells were transfected with 2 μg of DNA by use of 6 μl of Lipofectamine (Invitrogen) or with 1.5 μg of DNA by use of 10 μl of Polyfect (Qiagen) according to the manufacturers' directions. Cells on coverslips were inoculated with third-passage lysates of T1L or T3D virus at 5 PFU/cell in phosphate-buffered saline (PBS) (137 mM NaCl, 3 mM KCl, 8 mM Na2HPO4, pH 7.5) supplemented with 2 mM MgCl2 (PBS-MC), and the virus was allowed to adsorb to cells for 1 h at room temperature before fresh medium was added. Cells were further incubated for 18 to 24 h at 37°C before being processed for IF microscopy.
Immunoblot analysis.
CV-1 cells were transfected as described above in plates without coverslips, and whole-cell lysates were collected at 24 h posttransfection (p.t.). CV-1 cells (8 × 105) were washed briefly in PBS and then were scraped into 1 ml of PBS and pelleted. The pelleted cells were resuspended in 30 μl of PBS containing protease inhibitors (Roche), lysed in sample buffer, boiled for 10 min, and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Approximately 1.3 × 105 cell equivalents were analyzed per lane. Proteins were electroblotted from the gels onto nitrocellulose in 25 mM Tris-192 mM glycine, pH 8.3. The binding of antibodies was detected with alkaline phosphatase-coupled goat anti-rabbit IgG (Bio-Rad) and colorimetric reagents p-nitroblue tetrazolium chloride and 5-bromo-4-chloro-3-indolyl phosphate p-toluidine salt (Bio-Rad).
Immunostaining and IF microscopy.
Cells to be processed for IF microscopy were fixed for 10 min at room temperature in 2% paraformaldehyde or for 3 min at −20°C in 100% methanol unless otherwise stated. Cells fixed in paraformaldehyde were washed with PBS and then permeabilized and blocked in PBS containing 1% bovine serum albumin and 0.1% Triton X-100 (PBSAT). After methanol fixation, cells were incubated in PBSAT three times for 5 min at room temperature prior to incubation in primary antibody. Primary antibodies were diluted in PBSAT and incubated with cells for 25 to 40 min at room temperature. After three washes in PBS, secondary antibodies diluted in PBSAT were added and incubated with cells for 25 min at room temperature. Coverslips were incubated with 300 nM 4,6-diamidino-2-phenylindole (Molecular Probes) in PBS for 5 min to counterstain cell nuclei, briefly washed in PBS, and then mounted on glass slides with Prolong (Molecular Probes). Samples were examined under a Nikon TE-300 inverted microscope equipped with phase-constrast and fluorescence optics. Images were collected as described elsewhere (40). All images were processed and prepared for presentation by using Adobe Photoshop.
Expression of μNS and infection with top-component ISVPs.
CV-1 cells seeded on 18-mm-diameter coverslips as described above were transfected with 1.5 μg of pCI-M3(T1L) by use of 10 μl of Polyfect per the manufacturer's directions. After 5.5 h, the cells were washed with PBS-MC and then incubated with 100 μg of cycloheximide (Sigma) per ml in DMEM containing 10% FBS for 30 min at 37°C. Top-component infectious subvirion particles (ISVPs) were prepared by digesting purified top-component virions (1013 particles/ml) with 200 μg of α-chymotrypsin (Sigma) per ml for 10 min at 32°C, followed by quenching with 2 mM phenylmethylsulfonyl fluoride (Sigma). Each coverslip of transfected cells was incubated with 2 × 1010 top-component ISVPs and 100 μg of cycloheximide/ml in 50 μl of PBS-MC on ice for 30 min and then was placed in DMEM containing 10% FBS and 100-μg/ml cycloheximide at 37°C for 90 min before the cells were fixed and processed for IF microscopy.
RESULTS
Core surface proteins localize to viral factories in reovirus-infected cells.
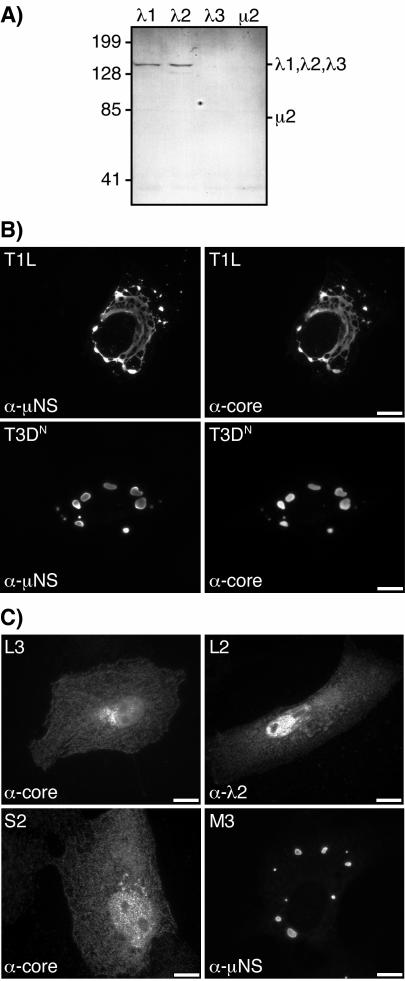
We and others have shown that the viral core proteins λ2, λ3, and μ2 colocalize with μNS in viral factories in reovirus-infected cells (9, 33, 40). To examine the distributions of the remaining two core proteins, λ1 and σ2, we performed IF microscopy with a polyclonal antiserum raised against whole core particles (11, 26; Noble and Nibert, unpublished data). This antiserum specifically recognizes the three core surface proteins—λ1, λ2, and σ2—by both immunoblotting (Fig. 1A) (see reference 26 for σ2 data) and immunostaining of protein-expressing cells (Fig. 1C). It does not, however, recognize the two structurally minor core proteins—λ3 and μ2—by either method (Fig. 1A and C; also data not shown). Viral factories were identified in this study by using antibodies derived from a μNS-specific polyclonal antiserum as previously described (8, 40). At 18 h postinfection (hpi), staining for the three core surface proteins showed that they colocalized with μNS in both the filamentous factories of reovirus T1L and the globular factories of reovirus T3DN (Fig. 1B). There was little detectable staining for these core proteins either in the nucleus or in the cytoplasm outside the factories (Fig. 1B), suggesting that all three core surface proteins were largely localized to the factories. Similarly strong colocalization of the core surface proteins and μNS was observed at 6 and 24 hpi (data not shown). Based on these and previous findings, we conclude that all three core surface proteins are concentrated in the viral factories throughout much of the infection cycle.
FIG. 1.
Specificity of core antiserum and distribution of reovirus core surface proteins λ1, λ2, and σ2 and nonstructural protein μNS in infected and singly transfected cells. (A) CV-1 cells were transfected with pCI-L3(T1L) encoding λ1, pCI-L2(T1L) encoding λ2, pCI-L1(T3D) encoding λ3, or pCI-M1(T1L) encoding μ2. At 24 h p.t., whole-cell lysates were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred to nitrocellulose, and immunoblotted with a core-specific rabbit polyclonal antiserum. Positions of molecular weight markers (Bio-Rad Laboratories) are indicated (in kilodaltons) to the left. Positions of the core proteins are indicated to the right; the presence of λ3 and μ2 in the samples was confirmed by using other antisera in parallel (data not shown). (B) CV-1 cells were infected with T1L (top row) or T3DN (bottom row) at 5 PFU/cell, fixed at 18 hpi, and coimmunostained with the core-specific antiserum followed by goat anti-rabbit IgG conjugated to Alexa 488 (right column) or with μNS-specific rabbit IgG conjugated to Texas Red (left column). Bars, 10 μm. (C) CV-1 cells were singly transfected with pCI-L3(T1L) (top left), pCI-L2(T1L) (top right), pCI-S2(T1L) encoding σ2 (bottom left), or pCI-M3(T1L) encoding μNS (bottom right) and were fixed at 18 h p.t. Samples were immunostained with the core-specific antiserum followed by goat anti-rabbit IgG conjugated to Alexa 488 (left column), λ2-specific mouse MAb 7F4 followed by goat anti-mouse IgG conjugated to Alexa 488 (top right), or μNS-specific rabbit IgG conjugated to Texas Red (bottom right). Bars, 10 μm.
Core surface proteins are diffusely distributed in cells in the absence of other viral proteins.
Although localized to viral factories in infected cells, the RNA-binding nonstructural protein σNS is diffusely distributed through the cytoplasm and nucleus when it is expressed in the absence of other viral proteins (4, 35). Given this observation, we investigated the distribution of core surface proteins λ1, λ2, and σ2 when each was separately expressed. To express these proteins, we cloned their genes (L3, L2, and S2, respectively) into the mammalian expression vector pCI-neo (Promega). Each expression construct was then separately transfected into CV-1 cells, and the distribution of each protein was determined by IF microscopy. The core-specific antiserum was used to detect λ1 and σ2, and λ2-specific MAb 7F4 (53) was used to detect λ2. In this manner, we found that each of the core surface proteins was diffusely distributed through the cytoplasm and nuclei of the transfected cells (Fig. 1C). The significance of the nuclear staining is unknown and requires further study, but as noted above, significant staining for λ1, λ2, and σ2 was not seen in the nuclei of infected cells (Fig. 1B). The diffuse distributions of λ1, λ2, and σ2 in transfected cells (Fig. 1C) contrasted with their localization to viral factories in infected cells (Fig. 1B), suggesting that a mechanism exists in infected cells to recruit the core surface proteins to the factories.
Core surface proteins localize to globular inclusions when coexpressed with μNS.
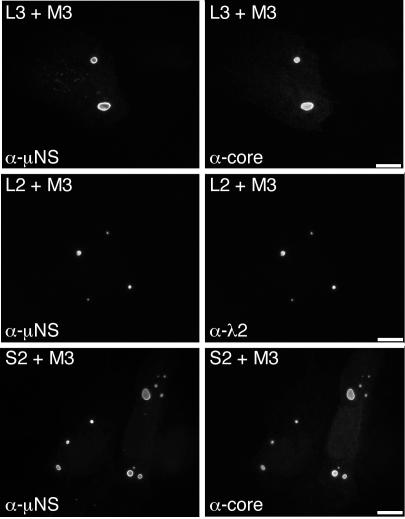
The structurally minor core protein μ2 and the RNA-binding nonstructural protein σNS localize to μNS inclusions when either of these proteins is coexpressed with μNS (8, 35). Given these observations and our findings that these proteins, as well as the core surface proteins λ1, λ2, and σ2 (Fig. 1B), are concentrated within viral factories in infected cells, we hypothesized that μNS may also recruit and concentrate the core surface proteins within μNS inclusions in transfected cells. To address this hypothesis, we separately coexpressed each of the core surface proteins with μNS and examined the distributions of the proteins by IF microscopy using the μNS-specific antibodies and the same reagents as those used to detect each core protein as described above. The core-specific antiserum and λ2-specific MAb did not cross-react with μNS inclusions in transfected cells expressing μNS in the absence of other viral proteins (data not shown). When λ1, λ2, and σ2 were each separately coexpressed with μNS, each was concentrated in globular inclusions that colocalized with μNS (Fig. 2). The distribution of μNS was not detectably changed from that of the protein expressed in the absence of other viral proteins (8). The localization of λ1, λ2, and σ2 to globular inclusions (Fig. 2) contrasted with their diffuse distribution in the cytoplasm and nucleus when each was expressed in the absence of μNS (Fig. 1C). We conclude from these results that μNS inclusions recruit and concentrate the core surface proteins in transfected cells.
FIG. 2.
Distribution of core surface proteins and μNS in doubly transfected cells. CV-1 cells were cotransfected with 1 μg of pCI-M3(T1L) encoding μNS and 1 μg of either pCI-L3(T1L) encoding λ1 (top row) or pCI-L2(T1L) encoding λ2 (middle row) and were fixed at 18 h p.t. Other CV-1 cells were cotransfected with 1.8 μg of pCI-M3(T1L) encoding μNS and 0.2 μg of pCI-S2(T1L) encoding σ2 (bottom row) and were fixed at 18 h p.t. Cells were coimmunostained for nonstructural protein μNS (left column) and for core surface protein λ1, λ2, or σ2 (right column) as for Fig. 1C. Bars, 10 μm.
Core surface proteins also localize to μNS(41-721) globular inclusions.
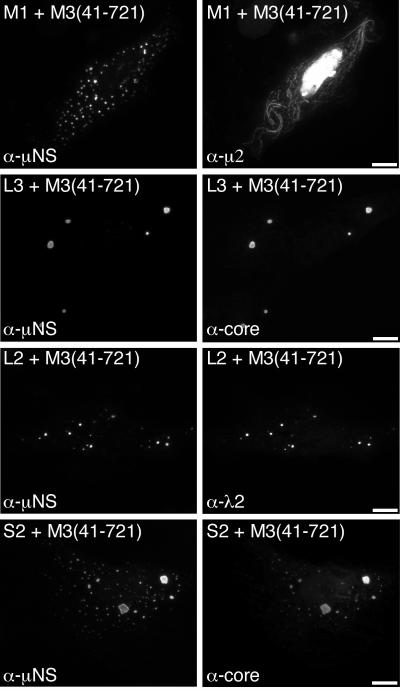
A form of μNS that lacks the N-terminal 40 residues, μNS(41-721), also forms globular inclusions when expressed in the absence of other viral proteins but does not associate with either μ2 (8) (Fig. 3) or σNS upon coexpression (35). These findings may be relevant for understanding the role(s) of these proteins in infection because a smaller form of μNS, μNSC, which lacks ∼5 kDa from the N terminus of μNS and is thus similar or identical to μNS(41-721), is expressed during infection (30, 55). We therefore examined the distribution of core surface proteins λ1, λ2, and σ2 when each was separately coexpressed with μNS(41-721). We found that each of the core surface proteins colocalized in globular inclusions with μNS(41-721) (Fig. 3). The distribution of μNS(41-721) was not detectably changed from that of the protein expressed in the absence of other viral proteins (8). The findings for λ1, λ2, and σ2 thus strongly contrasted with those for both μ2 (8) (Fig. 3) and σNS (35) and suggested that the first 40 residues of μNS are not required for association of any of the three core surface proteins with μNS inclusions.
FIG. 3.
Distribution of core surface proteins and μNS(41-721) in doubly transfected cells. CV-1 cells were cotransfected with 1 μg of pCI-M3(41-721) encoding μNS(41-721) and 1 μg of pCI-M1(T1L) encoding μ2 (first row), pCI-L3(T1L) encoding λ1 (second row), or pCI-L2(T1L) encoding λ2 (third row). Other CV-1 cells were cotransfected with 1.8 μg of pCI-M3(41-721) and 0.2 μg of pCI-S2(T1L) encoding σ2 (fourth row). Cells were fixed at 18 h p.t. and coimmunostained for μNS (left column) as for Fig. 1C and for either minor core protein μ2 by using a μ2-specific rabbit IgG conjugated to Texas Red or core surface protein λ1, λ2, or σ2 as for Fig. 1C (right column). Bars, 10 μm.
Core surface proteins localize to μ2(T1L)-μNS filamentous inclusions.
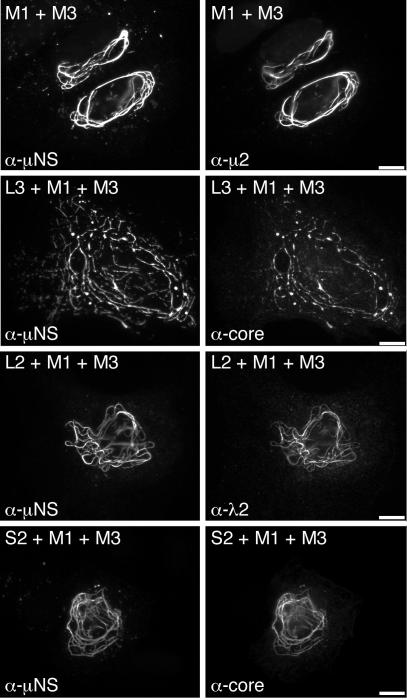
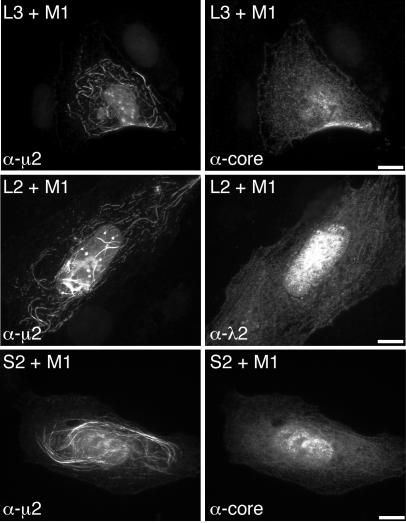
The μNS protein can be redistributed from globular inclusions to microtubule-associated filamentous inclusions when coexpressed with the minor core protein μ2(T1L) (8). To determine if the λ1, λ2, and σ2 proteins may be redistributed to filamentous inclusions along with μNS when coexpressed with μ2(T1L), we triply coexpressed these protein combinations in transfected cells and examined their distributions by IF microscopy. In all samples, μNS had a filamentous distribution (Fig. 4), similar to that observed when μNS and μ2(T1L) are coexpressed (8). Each of the core surface proteins displayed a filamentous distribution as well and colocalized with μNS (Fig. 4). We did not stain for μ2 in these experiments, but the filamentous distribution of μNS depends on its association with microtubule-associated μ2(T1L) (8), thus suggesting that μ2(T1L) colocalized with μNS and each of the core surface proteins as well. These results also provide evidence that the association of λ1, λ2, and σ2 with μNS is specific and not due to nonspecific recruitment to globular inclusions.
FIG. 4.
Distribution of core surface proteins coexpressed with μ2(T1L) and μNS in triply transfected cells. CV-1 cells were cotransfected with 1 μg of pCI-M1(T1L) encoding μ2 and 1 μg of pCI-M3(T1L) encoding μNS (first row) to illustrate their colocalization. For determination of the distribution of core surface proteins, CV-1 cells were transfected with 0.67 μg of pCI-M1(T1L) and 0.67 μg of pCI-M3(T1L) along with 0.67 μg of either pCI-L3(T1L) encoding λ1 (second row) or pCI-L2(T1L) encoding λ2 (third row). Other CV-1 cells were cotransfected with 0.9 μg of pCI-M1(T1L), 0.9 μg of pCI-M3(T1L), and 0.2 μg of pCI-S2(T1L) encoding σ2 (fourth row). Cells were fixed at 18 h p.t. and coimmunostained for major nonstructural protein μNS (left column) as for Fig. 1C and for either minor core protein μ2 or core surface protein λ1, λ2, or σ2 (right column) as for Fig. 3. Bars, 10 μm.
Core surface proteins separately coexpressed with μ2(T1L) are diffusely distributed and exhibit only sporadic, weak association with filaments.
We considered the possibility that the recruitment of core proteins to μ2(T1L)-μNS filamentous inclusions may be due to an association of the core surface proteins with μ2 rather than with μNS. To determine if λ1, λ2, and σ2 are recruited to microtubules when coexpressed with μ2(T1L) in the absence of μNS, we separately expressed each of the core proteins along with μ2(T1L) and examined their distributions by IF microscopy using antibodies derived from a μ2-specific polyclonal antiserum, as previously described (8, 40), and either an anti-core serum or MAb 7F4. The μ2(T1L) protein showed a primarily filamentous distribution, but with some diffuse cytoplasmic staining and diffuse and punctate nuclear staining, as previously described (8, 40) (Fig. 5). In most cells, we found that λ1, λ2, and σ2 were diffusely distributed through the cytoplasm and nuclei of the cells (Fig. 5). In occasional cells, however, each of the core surface proteins exhibited a weak filamentous distribution, which colocalized with μ2(T1L), in addition to the diffuse cytoplasmic and nuclear distribution (Fig. 5). These results suggest that μNS is required for the strong recruitment of λ1, λ2, and σ2 to filamentous inclusions, as shown in Fig. 4, but that λ1, λ2, and σ2 may be capable of weak associations with μ2(T1L) on filaments in the absence of μNS. We have previously reported a similarly weak association of the RNA-binding nonstructural protein σNS with the μ2(T1L) protein on filaments (35).
FIG. 5.
Distribution of core surface proteins coexpressed with μ2(T1L) in doubly transfected cells. CV-1 cells were cotransfected with 1 μg of pCI-M1(T1L) encoding μ2 and 1 μg of either pCI-L3(T1L) encoding λ1 (first row) or pCI-L2(T1L) encoding λ2 (second row). Other CV-1 cells were cotransfected with 1.8 μg of pCI-M1(T1L) and 0.2 μg of pCI-S2(T1L) encoding σ2 (third row). Cells were fixed at 18 h p.t. and coimmunostained for minor core protein μ2 (left column) and for core surface protein λ1, λ2, or σ2 (right column) as for Fig. 3. Bars, 10 μm.
Core-like particles released into the cytoplasm during entry can also be recruited to μNS globular inclusions.
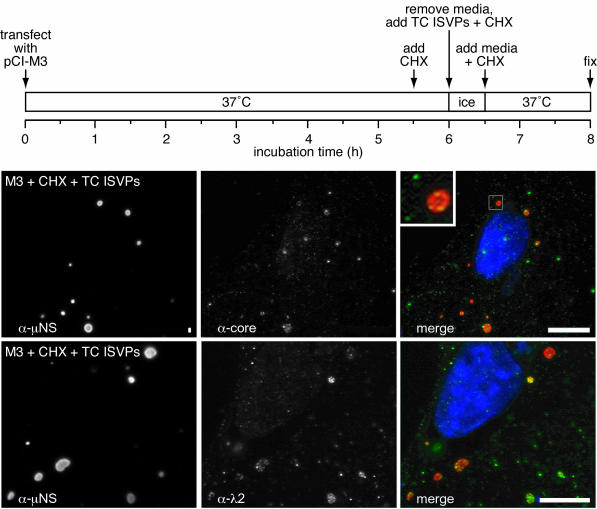
Recent findings indicated that viral particles similar to cores, lacking most or all copies of outer capsid proteins μ1, σ1, and σ3, are released into the cytoplasm during the course of cell entry by reoviruses (10). Based on previous evidence that μNS binds to the surfaces of cores in vitro (7) as well as on the new evidence in this study for associations between μNS and each of the core surface proteins, we hypothesized that core particles released into the cytoplasm during entry may be capable of binding to preformed μNS inclusions in cells. To test this hypothesis, we first expressed μNS in transfected cells at 37°C. At 5.5 h p.t., we added cycloheximide to block further protein synthesis and 30 min later exposed the cells to either ISVPs or top-component ISVPs of reovirus T1L (see Materials and Methods). After a 30-min absorption period at 4°C, the infection was allowed to proceed in the presence of cycloheximide for 90 min at 37°C, and the cells were then fixed and processed for IF microscopy. The results with ISVPs (data not shown) and top-component ISVPs (Fig. 6) were essentially identical, namely that core surface protein staining with either an anti-core serum or λ2 MAb 7F4 was restricted to punctate structures in the cytoplasm. Additionally, many of these punctate structures colocalized with μNS in globular inclusions (Fig. 6). Because new protein synthesis was inhibited in this experiment, we conclude that the core surface protein staining represents intact core particles introduced into the cytoplasm (10), some of which then bound to μNS globular inclusions. The cytoplasmic core particles produced during entry by either ISVPs or top-component ISVPs of reovirus T1L showed a similar pattern of binding to μNS(41-721) globular inclusions in parallel experiments (data not shown). These findings provide strong evidence for the binding of μNS and μNS(41-721) to the surfaces of cores inside cells, consistent with previous in vitro results (7). They further suggest that μNS and perhaps μNSC as well (30, 55), which themselves are first translated from RNA transcripts produced by parental cores (primary transcriptase particles), may then bind to these same newly arrived, transcriptionally active articles, allowing them to become embedded within nascent viral factories (see Discussion for further considerations).
FIG. 6.
Distribution of μNS and newly arrived core particles in cells transfected with pCI-M3(T1L) and then infected with T1L top-component (TC) ISVPs. CV-1 cells were transfected with 2 μg of pCI-M3(T1L) encoding μNS, and then at 5.5 h p.t., 100 μg of cycloheximide (CHX)/ml was added to the cells. At 6 h p.t., the cells were incubated (in the presence of cycloheximide) with T1L top-component ISVPs (5,000 particles per cell) at 4°C for 30 min and then were warmed to 37°C for 90 min and fixed. (The preceding steps are summarized in the time line at the top.) Cells were immunostained either with core-specific rabbit polyclonal antiserum followed by goat anti-rabbit IgG conjugated to Alexa 488 and μNS-specific IgG conjugated to Texas Red (top row) or with λ2-specific mouse MAb 7F4 followed by goat anti-mouse IgG conjugated to Alexa 488 and μNS-specific IgG conjugated to Texas Red (bottom row). Signals from individual antibodies are shown in the left and middle columns, and merged images are shown in the right column, as indicated. Bars, 10 μm. The boxed region in the merged image of the top row is magnified ×4 in the inset in order to show more clearly the punctate core staining that does (yellow) or does not (green) colocalize with μNS.
DISCUSSION
Several labs have reported recent progress in understanding how the cytoplasmic factories in reovirus-infected cells are formed, how different viral and cellular components are recruited to them (or vice versa), and how some differences in the factories observed between viral strains are genetically determined (3, 4, 8, 33, 35, 40). To date, including the results of this study, the network of protein-protein and protein-RNA interactions schematized in Fig. 7 has been identified. Other viral and cellular components that may be specifically recruited and play important roles in viral genome packaging, replication, and particle assembly remain to be determined. A general hypothesis that guides these studies is that the complexly structured, 130-MDa reovirus virion, including its 10 different RNA genome segments, is likely to require an organized intracellular environment in which to be assembled with high fidelity and efficiency. Another general hypothesis is that the large and complex milieu of the infected cytoplasm has compelled reoviruses to evolve signals in their various components that specify how they are recruited to and arranged within the factories. We view the viral factories as specialized organelles, and our long-range goal is to understand their genesis, structure, and functions. The factories may be related in some ways to cellular aggresomes (24, 28, 40), and thus our studies of the factories may reveal some new aspects of cellular functions as well.
FIG. 7.
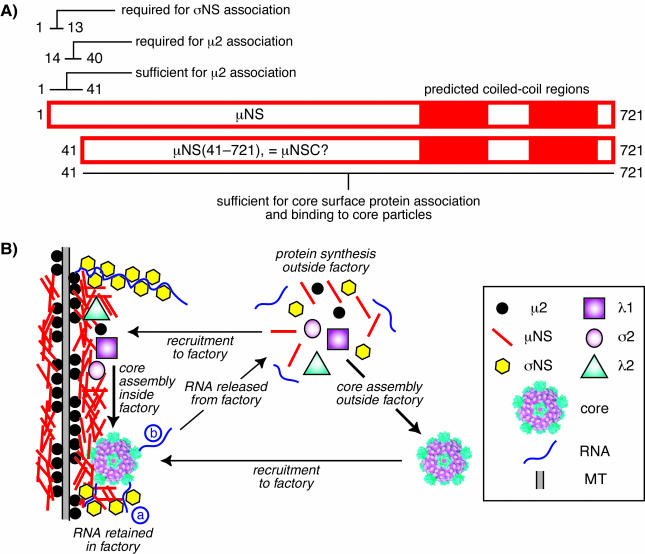
Summary model of μNS associations with other components in viral factories. (A) Bar diagram of μNS primary sequences (residues 1 to 721) indicating known features and regions shown to be required for protein associations. It remains to be demonstrated that μNS(41-721) and μNSC are equivalent. (B) Cartoon depicting the factory of a reovirus strain, such as T1L, whose μ2 protein recruits μNS to microtubules (MT) (8, 40). Ribosomes are excluded from the factories (42, 45), so protein synthesis must occur in surrounding regions of the cytoplasm. Core surface proteins λ1, λ2, and σ2 are recruited to the factory through association with μNS (this study). The single-stranded RNA-binding protein σNS is also recruited to the factory through association with μNS (4, 35). Core assembly, including the 10 genomic RNA segments and polymerase λ3 which are not shown here, is proposed to occur within the factory but might also occur in surrounding regions of the cytoplasm as also shown in the cartoon. Cores assembled in the cytoplasm may then be recruited to the factory through association with μNS (this study). New plus-strand RNA transcripts produced by μNS-associated cores (36, 37) within the factory may be largely retained there, possibly by binding to σNS (a), which may promote their assembly into progeny particles (sponge model). Some newly produced viral transcripts, however, must be released into the surrounding cytoplasm (b) to promote ongoing viral protein synthesis. The precise role of μNSC remains unclear and is therefore not shown. The mechanism of outer capsid assembly is also unclear and therefore not shown. Although the various protein associations are shown as direct interactions, there may be unidentified intermediaries or promoting agents in one or more cases.
Reovirus core surface proteins are separately recruited to μNS factory-like inclusions: evidence for core assembly within the factories?
Previously, the reovirus nonstructural protein μNS has been shown to form inclusions that are similar in morphology to viral factories (8) and to recruit the viral proteins μ2 (8) and σNS (4, 35) to those inclusions. In addition, μNS has been found attached to progeny cores in transcriptionally active particle assembly intermediates isolated from infected cells (36, 37), and these are thought to represent the secondary transcriptase particles that produce 80 to 95% of the viral transcripts during an infection (reviewed in reference 58). Lastly, recombinant μNS has been shown to bind to the surfaces of viral core particles in vitro (7). These data identify μNS as a strong candidate for recruiting additional viral components, especially the viral core surface proteins, to factories. Thus, the starting hypothesis of this study was that one or more of these proteins may be separately recruited to viral factories through a specific association with μNS. Our new results indicate that in fact each of the three core surface proteins can be independently recruited to μNS inclusions (Fig. 2).
We were unable to distinguish between assembled cores and unassembled proteins in infected cells since the available antibodies recognize core surface proteins that are both free and bound in core particles. In transfected cells, however, the individually expressed core surface proteins are not assembled into particles because the necessary companion proteins are not coexpressed (26, 56). Under these conditions, each of the core surface proteins was separately recruited to μNS inclusions. These results suggest that λ1, λ2, and σ2 may each be recruited to viral factories in infected cells prior to assembly into cores, which is consistent with the hypothesis that core assembly occurs within the factories. However, since the expression of core proteins from either vaccinia or baculovirus vectors allows the assembly of core-like particles in the absence of μNS coexpression (26, 56), it appears likely that particles can assemble in the cytoplasm of infected cells outside the factories as well. Since the vaccinia- or baculovirus-generated core-like particles lack genomic RNA (26, 56), an interesting hypothesis is that core assembly in association with μNS-containing factories may be necessary to promote packaging of the viral genome (see below).
Reovirus nonstructural protein σNS is a single-stranded RNA-binding protein (17, 19, 21) that localizes to μNS inclusions (4, 35) through RNA-enhanced associations requiring the N termini of both μNS and σNS (35). We have proposed that σNS may recruit or retain viral mRNAs within the factories (35). Consistent with this hypothesis, we have recently obtained evidence that large amounts of newly transcribed viral RNAs localize to factories in infected cells (C. L. Miller and M. L. Nibert, unpublished data). A high concentration of viral RNAs within factories may promote their assortment and packaging into newly assembling core particles. We propose that top-component virions (50), which largely lack the viral genome (15, 16, 50), might be derived from cores that assemble in the cytoplasm and therefore fail to package the viral RNAs, whereas genome-containing virions might be derived from cores that assemble in the factories, where both the viral RNAs and core proteins are concentrated. In any case, based on evidence in this and previous studies (Fig. 1B and 6) (7), cores with or without a genome that may assemble in the cytoplasm are likely recruited to μNS-containing factories through associations of the core surface proteins with μNS.
μNS recruits different components to factories through different regions of its primary sequence: unique roles for μNSC in the viral life cycle?
Previous studies have shown that the N-terminal 40 residues of μNS are required for association with two different viral proteins, μ2 and σNS (8, 35). The exact residues in μNS that determine these associations remain to be identified; however, μNS residues 1 to 13 are dispensable for association with μ2 but are required for association with σNS (35) (Fig. 7A). Moreover, μNS residues 1 to 41 are sufficient for association with μ2 (8) (Fig. 7A). The region of μNS that is sufficient for association with σNS has not been reported. In this study, we determined that residues 1 to 40 are dispensable for associations between μNS and each of the core surface proteins (Fig. 7A). Further dissection of the region(s) of μNS residues 41 to 721 involved in the associations with λ1, λ2, and σ2 awaits future studies, but the present evidence is sufficient to conclude that different regions of μNS have evolved to promote associations with different viral proteins. We hypothesize that these associations are important not only for recruiting these proteins to the viral factories, but also for arranging them properly within the factories for viral genome replication and assembly.
An N-terminally truncated form of μNS, called μNSC, is also found in infected cells (30, 55). μNSC has been proposed to be a product of alternative translation initiation from a second in-frame AUG codon 41 codons downstream of the full-length μNS start site (34, 55). The μNS(41-721) protein expressed in this and previous studies (8, 35) is therefore believed to represent this second form of μNS. The failure of μNS(41-721) to associate with either μ2 or σNS in transfected cells (8, 35) is consistent with the hypothesis that μNS and μNSC each performs some distinct function(s) in infected cells. Previous evidence that both μNS and μNS(41-721) form factory-like inclusions (8) and new evidence that both μNS and μNS(41-721) can recruit the core surface proteins to these inclusions (Fig. 2 and 3) suggest that some functions of μNS and μNSC are overlapping. The distinguishable functions could conceivably be important at several different steps in the viral replication cycle and are the subject of ongoing studies in our lab.
The data in this report do not demonstrate whether the recruitment of each core surface protein to μNS inclusions is through a direct two-component interaction, an indirect association involving one or more RNAs or a cellular protein intermediate, or some undefined mechanism whereby μNS determines that certain proteins are shuttled to the inclusions. The recruitment clearly has some specificity, however, in that core surface proteins are recruited to μNS(41-721) inclusions (Fig. 3), whereas μ2 and σNS are not (8, 35). Further evidence for specificity is that at least one of the three reovirus outer capsid proteins is not recruited to inclusions when coexpressed with μNS (4; J. S. L. Parker, K. S. Myers, and M. L. Nibert, unpublished data).
Do entering parental core particles seed viral factories?
New evidence in this study demonstrates that many of the core particles released into the cytoplasm following entry and partial uncoating (10) become bound to preformed μNS factory-like inclusions within 90 min after infection of cells that have previously expressed μNS or μNS(41-721) from an expression plasmid (Fig. 6). Thus, one model for viral factory formation is that an incoming core particle transcribes and releases the viral mRNAs, which are translated by cellular machinery in adjacent regions of the cytoplasm to yield the viral proteins, including μNS, which then binds to the adjacent core and seeds a factory. Alternatively, the newly synthesized μNS could first form a small inclusion to which the nearby core is then recruited. In either case, the μNS protein is likely to recruit other viral proteins, including λ1, λ2, σ2, σNS, and μ2, to enlarge the nascent factory (4, 8, 35). We and others have previously noted similarities between viral factories and cellular aggresomes (8, 20, 40), structures that develop in the cytoplasm of cells overexpressing misfolded proteins (24). The two preceding hypotheses for reovirus factory formation are both similar to a seeding hypothesis proposed for the development of aggresomes (28).
If cores are embedded within a matrix of μNS that also recruits the viral RNA-binding protein σNS, some of the plus-strand transcripts that are produced and released by cores might be bound up by the σNS protein recruited to the factory (Fig. 7B). In fact, the ongoing production of new viral plus-strand RNA molecules by both parental and progeny cores in situ and the ongoing addition of μNS and σNS to the developing factories could create a sponge-like environment in which the viral plus-strand RNAs remain largely trapped within the factory. Excess transcripts could be released into the surrounding cytoplasm for translation (the factories lack ribosomes [42, 45]) when the “sponge” is saturated. In this scenario, no specificity for the viral RNAs would be required for the enrichment of viral over cellular transcripts within the factories, consistent with evidence that σNS has a nonspecific RNA-binding activity (18, 21). As a result, the viral plus-strand RNAs would be concentrated within the factories for assortment and packaging into progeny cores. In addition, because of their larger numbers of σNS-binding sites per molecule (18) and lower rates of diffusion, larger RNAs may be retained in the factories at higher frequencies than smaller ones. Since the viral plus-strand RNAs are transcribed by cores in nonequimolar amounts that are proportional to size (small > medium > large RNAs) (60), a sponge-like environment that favors the retention of larger over smaller RNAs in the factories would tend to equalize the concentrations of the 10 different transcripts. This could, in turn, promote equimolar assortment and packaging of the 10 RNAs into newly assembling cores and virions, as is thought to occur in the factories.
There are, of course, many other possible scenarios for the complex events in the reovirus life cycle that encompass transcription, translation, replication, packaging, and assembly. For example, instead of only transcripts being released from the factories as in the preceding model (Fig. 7B), some whole cores could be released to produce the transcripts for translation, whereas other cores could be retained in the factories to produce the transcripts for replication and packaging. Only a small number of released cores might be needed to generate sufficient mRNAs and proteins to fuel the viral factories, consistent with our IF microscopy evidence that the core proteins are in fact highly concentrated within the factories in infected cells (Fig. 1B). It is also important to note that the preceding model does not yet incorporate the events in outer capsid assembly that are required to complete the production of infectious virions. Further studies of the localization of reovirus RNA and protein molecules within cells are clearly needed to test these and other hypotheses relating to RNA assortment and packaging and particle assembly within reovirus-infected cells.
Acknowledgments
We express our sincere thanks to Elaine Freimont for lab maintenance and technical assistance and to other members of our lab for helpful discussions. We also thank Cindy Luongo for providing pPCR-Script-L2.
This work was supported by NIH grants R01 AI47904 (to M.L.N.) and K08 AI52209 (to J.S.L.P.) and by a junior faculty research grant from the Giovanni Armenise-Harvard Foundation (to M.L.N.). C.L.M. and T.J.B. received additional support, respectively, from NIH grants T32 AI07061 to the Combined Infectious Diseases Training Program at Harvard Medical School and T32 AI07245 to the Viral Infectivity Training Program at Harvard Medical School. C.D.S.P. received additional support from the Harvard College Research Program.
REFERENCES
- 1.Antczak, J. B., and W. K. Joklik. 1992. Reovirus genome segment assortment into progeny genomes studied by the use of monoclonal antibodies directed against reovirus proteins. Virology 187:760-776. [DOI] [PubMed] [Google Scholar]
- 2.Banerjee, A. K., and A. J. Shatkin. 1970. Transcription in vitro by reovirus-associated ribonucleic acid-dependent polymerase. J. Virol. 6:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Becker, M. M., M. I. Goral, P. R. Hazelton, G. S. Baer, S. E. Rodgers, E. G. Brown, K. M. Coombs, and T. S. Dermody. 2001. Reovirus σNS protein is required for nucleation of viral assembly complexes and formation of viral inclusions. J. Virol. 75:1459-1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Becker, M. M., T. R. Peters, and T. S. Dermody. 2003. Reovirus σNS and μNS proteins form cytoplasmic inclusion structures in the absence of viral infection. J. Virol. 77:5948-5963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borsa, J., and A. F. Graham. 1968. Reovirus: RNA polymerase activity in purified virions. Biochem. Biophys. Res. Commun. 33:895-901. [DOI] [PubMed] [Google Scholar]
- 6.Breun, L. A., T. J. Broering, A. M. McCutcheon, S. J. Harrison, C. L. Luongo, and M. L. Nibert. 2001. Mammalian reovirus L2 gene and λ2 core spike protein sequences and whole-genome comparisons of reoviruses type 1 Lang, type 2 Jones, and type 3 Dearing. Virology 287:333-348. [DOI] [PubMed] [Google Scholar]
- 7.Broering, T. J., A. M. McCutcheon, V. E. Centonze, and M. L. Nibert. 2000. Reovirus nonstructural protein μNS binds to core particles but does not inhibit their transcription and capping activities. J. Virol. 74:5516-5524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Broering, T. J., J. S. L. Parker, P. L. Joyce, J. Kim, and M. L. Nibert. 2002. Mammalian reovirus nonstructural protein μNS forms large inclusions and colocalizes with reovirus microtubule-associated protein μ2 in transfected cells. J. Virol. 76:8285-8297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cashdollar, L. W. 1994. Characterization and structural localization of the reovirus λ3 protein. Res. Virol. 145:277-285. [DOI] [PubMed] [Google Scholar]
- 10.Chandran, K., J. S. L. Parker, M. Ehrlich, T. Kirchhausen, and M. L. Nibert. 2003. The δ region of outer-capsid protein μ1 undergoes conformational change and release from reovirus particles during cell entry. J. Virol. 77:13361-13375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chandran, K., S. B. Walker, Y. Chen, C. M. Contreras, L. A. Schiff, T. S. Baker, and M. L. Nibert. 1999. In vitro recoating of reovirus cores with baculovirus-expressed outer-capsid proteins μ1 and σ3. J. Virol. 73:3941-3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cleveland, D. R., H. Zarbl, and S. Millward. 1986. Reovirus guanylyltransferase is L2 gene product lambda 2. J. Virol. 60:307-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cullen, B. R. 2002. RNA interference: antiviral defense and genetic tool. Nat. Immunol. 3:597-599. [DOI] [PubMed] [Google Scholar]
- 14.Drayna, D., and B. N. Fields. 1982. Activation and characterization of the reovirus transcriptase: genetic analysis. J. Virol. 41:110-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dryden, K. A., D. L. Farsetta, G.-J. Wang, J. M. Keegan, B. N. Fields, T. S. Baker, and M. L. Nibert. 1998. Internal structures containing transcriptase-related proteins in top component particles of mammalian orthoreovirus. Virology 225:33-46. [DOI] [PubMed] [Google Scholar]
- 16.Furlong, D. B., M. L. Nibert, and B. N. Fields. 1988. Sigma 1 protein of mammalian reoviruses extends from the surfaces of viral particles. J. Virol. 62:246-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gillian, A. L., and M. L. Nibert. 1998. Amino terminus of reovirus nonstructural protein σNS is important for ssRNA binding and nucleoprotein complex formation. Virology 240:1-11. [DOI] [PubMed] [Google Scholar]
- 18.Gillian, A. L., S. C. Schmechel, J. Livny, L. A. Schiff, and M. L. Nibert. 2000. Reovirus nonstructural protein σNS binds in multiple copies to single-stranded RNA and shares properties with single-stranded DNA binding proteins. J. Virol. 74:5939-5948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gomatos, P. J., O. Prakash, and N. M. Stamatos. 1981. Small reovirus particle composed solely of sigma NS with specificity for binding different nucleic acids. J. Virol. 39:115-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heath, C. M., M. Windsor, and T. Wileman. 2001. Aggresomes resemble sites specialized for virus assembly. J. Cell Biol. 153:449-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huismans, H., and W. K. Joklik. 1976. Reovirus-coded polypeptides in infected cells: isolation of two native monomeric polypeptides with affinity for single-stranded and double-stranded RNA, respectively. Virology 70:411-424. [DOI] [PubMed] [Google Scholar]
- 22.Hunter, E. 2001. Virus assembly, p. 171-197. In D. M. Knipe et al. (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 23.Ito, Y., and W. K. Joklik. 1972. Temperature-sensitive mutants of reovirus. I. Patterns of gene expression by mutants of groups C, D, and E. Virology 50:189-201. [DOI] [PubMed] [Google Scholar]
- 24.Johnston, J. A., C. L. Ward, and R. R. Kopito. 1998. Aggresomes: a cellular response to misfolded proteins. J. Cell Biol. 143:1883-1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Joklik, W. K. 1980. The structure and function of the reovirus genome. Ann. N. Y. Acad. Sci. 80:107-124. [DOI] [PubMed] [Google Scholar]
- 26.Kim, J., X. Zhang, V. E. Centonze, V. D. Bowman, S. Noble, T. S. Baker, and M. L. Nibert. 2002. The hydrophilic amino-terminal arm of reovirus core shell protein λ1 is dispensable for particle assembly. J. Virol. 76:12211-12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Knipe, D. M., C. E. Samuel, and P. Palese. 2001. Virus-host cell interactions, p. 133-170. In D. M. Knipe et al. (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 28.Kopito, R. R. 2000. Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol. 10:524-530. [DOI] [PubMed] [Google Scholar]
- 29.Lai, M. H., and W. K. Joklik. 1973. The induction of interferon by temperature-sensitive mutants of reovirus, UV-irradiated reovirus, and subviral reovirus particles. Virology 51:191-204. [DOI] [PubMed] [Google Scholar]
- 30.Lee, P. W. K., E. C. Hayes, and W. K. Joklik. 1981. Characterization of anti-reovirus immunoglobulins secreted by cloned hybridoma cell lines. Virology 108:134-146. [DOI] [PubMed] [Google Scholar]
- 31.Luongo, C. L. 2002. Mutational analysis of a mammalian reovirus mRNA capping enzyme. Biochem. Biophys. Res. Commun. 291:932-938. [DOI] [PubMed] [Google Scholar]
- 32.Luongo, C. L., C. M. Contreras, D. L. Farsetta, and M. L. Nibert. 1998. Binding site for S-adenosyl-L-methionine in a central region of mammalian reovirus λ2 protein. J. Biol. Chem. 273:23773-23780. [DOI] [PubMed] [Google Scholar]
- 33.Mbisa, J. L., M. M. Becker, S. Zou, T. S. Dermody, and E. G. Brown. 2000. Reovirus μ2 protein determines strain-specific differences in the rate of viral inclusion formation in L929 cells. Virology 272:16-26. [DOI] [PubMed] [Google Scholar]
- 34.McCutcheon, A. M., T. J. Broering, and M. L. Nibert. 1999. Mammalian reovirus M3 gene sequences and conservation of coiled-coil motifs near the carboxyl terminus of the μNS protein. Virology 264:16-24. [DOI] [PubMed] [Google Scholar]
- 35.Miller, C. L., T. J. Broering, J. S. L. Parker, M. M. Arnold, and M. L. Nibert. 2003. Reovirus σNS protein localizes to inclusions through an association requiring the μNS amino-terminus. J. Virol. 77:4566-4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morgan, E. M., and H. J. Zweerink. 1974. Reovirus morphogenesis. Corelike particles in cells infected at 39° with wild-type reovirus and temperature-sensitive mutants of groups B and G. Virology 59:556-565. [DOI] [PubMed] [Google Scholar]
- 37.Morgan, E. M., and H. J. Zweerink. 1975. Characterization of transcriptase and replicase particles isolated from reovirus-infected cells. Virology 68:455-466. [DOI] [PubMed] [Google Scholar]
- 38.Nibert, M. L. 1998. Structure of mammalian orthoreovirus particles. Curr. Top. Microbiol. Immunol. 233:1-30. [DOI] [PubMed] [Google Scholar]
- 39.Nibert, M. L., and L. A. Schiff. 2001. Reoviruses and their replication, p. 1679-1728. In D. M. Knipe et al. (ed.), Fields virology, 4th ed. Lippincott Willams & Wilkins, Philadelphia, Pa.
- 40.Parker, J. S. L., T. J. Broering, J. Kim, D. E. Higgins, and M. L. Nibert. 2002. Reovirus core protein μ2 determines the filamentous morphology of viral inclusion bodies by interacting with and stabilizing microtubules. J. Virol. 76:4483-4496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reinisch, K. M., M. L. Nibert, and S. C. Harrison. 2000. Structure of the reovirus core at 3.6 Å resolution. Nature 404:960-967. [DOI] [PubMed] [Google Scholar]
- 42.Rhim, J. S., L. E. Jordan, and H. D. Mayor. 1962. Cytochemical, fluorescent-antibody, and electron microscopic studies on the growth of reovirus (ECHO 10) in tissue culture. Virology 17:342-355. [DOI] [PubMed] [Google Scholar]
- 43.Samuel, C. E. 1998. Reoviruses and the interferon system. Curr. Top. Microbiol. Immunol. 233:125-145. [DOI] [PubMed] [Google Scholar]
- 44.Schwartz, M., J. Chen, M. Janda, M. Sullivan, J. den Boon, and P. Ahlquist. 2002. A positive-strand RNA virus replication complex parallels form and function of retrovirus capsids. Mol. Cell 9:505-514. [DOI] [PubMed] [Google Scholar]
- 45.Sharpe, A. H., L. B. Chen, and B. N. Fields. 1982. The interaction of mammalian reoviruses with the cytoskeleton of monkey kidney CV-1 cells. Virology 120:399-411. [DOI] [PubMed] [Google Scholar]
- 46.Shatkin, A. J., and J. D. Sipe. 1968. RNA polymerase activity in purified reoviruses. Proc. Natl. Acad. Sci. USA 61:1462-1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shing, M., and K. M. Coombs. 1996. Assembly of the reovirus outer capsid requires μ1/σ3 interactions which are prevented by misfolded σ3 protein in temperature-sensitive mutant tsG453. Virus Res. 46:19-29. [DOI] [PubMed] [Google Scholar]
- 48.Silverstein, S. C., and S. Dales. 1968. The penetration of reovirus RNA and initiation of its genetic function in L-strain fibroblasts. J. Cell Biol. 36:197-230. [PubMed] [Google Scholar]
- 49.Silverstein, S. C., and P. H. Schur. 1970. Immunofluorescent localization of double-stranded RNA in reovirus-infected cells. Virology 41:564-566. [DOI] [PubMed] [Google Scholar]
- 50.Smith, R. E., H. J. Zweerink, and W. K. Joklik. 1969. Polypeptide components of virions, top component, and cores of reovirus type 3. Virology 39:791-810. [DOI] [PubMed] [Google Scholar]
- 51.Starnes, M. C., and W. K. Joklik. 1993. Reovirus protein λ3 is a poly(C)-dependent poly(G) polymerase. Virology 193:356-366. [DOI] [PubMed] [Google Scholar]
- 52.Tao, Y., D. L. Farsetta, M. L. Nibert, and S. C. Harrison. 2002. RNA synthesis in a cage—structural studies of reovirus polymerase λ3. Cell 111:733-745. [DOI] [PubMed] [Google Scholar]
- 53.Virgin, H. W., IV, M. A. Mann, B. N. Fields, and K. L. Tyler. 1991. Monoclonal antibodies to reovirus reveal structure/function relationships between capsid proteins and genetics of susceptibility to antibody action. J. Virol. 65:6772-6781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.White, C. K., and H. J. Zweerink. 1976. Studies on the structure of reovirus cores: selective removal of polypeptide λ2. Virology 70:171-180. [DOI] [PubMed] [Google Scholar]
- 55.Wiener, J. R., J. A. Bartlett, and W. K. Joklik. 1989. The sequences of reovirus serotype 3 genome segments M1 and M3 encoding the minor protein μ2 and the major nonstructural protein μNS, respectively. Virology 169:293-304. [DOI] [PubMed] [Google Scholar]
- 56.Xu, P., S. E. Miller, and W. K. Joklik. 1993. Generation of reovirus core-like particles in cells infected with hybrid vaccinia viruses that express genome segments L1, L2, L3, and S2. Virology 197:726-731. [DOI] [PubMed] [Google Scholar]
- 57.Yin, P., M. Cheang, and K. M. Coombs. 1996. The M1 gene is associated with differences in the temperature optimum of the transcriptase activity in reovirus core particles. J. Virol. 70:1223-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zarbl, H. J., and S. Millward. 1983. The reovirus multiplication cycle, p. 107-196. In W. K. Joklik (ed.), The reoviridae. Plenum Press, New York, N.Y.
- 59.Zweerink, H. J. 1974. Multiple forms of ss leads to dsRNA polymerase activity in reovirus-infected cells. Nature 247:313-315. [DOI] [PubMed] [Google Scholar]
- 60.Zweerink, H. J., and W. K. Joklik. 1970. Studies on the intracellular synthesis of reovirus-specified proteins. Virology 41:501-518. [DOI] [PubMed] [Google Scholar]