Abstract
Rigorous phylogenetic analyses were used to compare the nucleotide sequences of feline immunodeficiency virus strains isolated from Texas and throughout the world. The envelope V3-V4 sequences and capsid gene of the Texas isolates formed a cluster between subtypes B and E. Statistical comparisons with other published sequences confirmed that the Texas group is a unique cluster, possibly a new subtype, arising from subtype B.
Feline immunodeficiency virus (FIV) was initially isolated in 1987 from a cat in California with severe immunodeficiency and has been recognized as a common worldwide feline pathogen (11, 14, 19, 20, 33). FIV-infected cats exhibit a progressive impairment of cellular immunity, leaving the animals susceptible to opportunistic infections (17, 23, 24). Similar to other lentiviruses, genetic variations may occur as a result of point mutations or recombination (1, 2). Since genetic variation and associated disease characteristics are similar between FIV and human immunodeficiency virus, FIV is a practical model for T-tropic lentiviral infection, especially for vaccine development and the design of antiviral therapies (3, 4, 11, 12, 14, 17, 21, 27, 29, 33, 34).
A number of FIV strains have been identified throughout the world, and their corresponding genomes have been partially sequenced (1, 5, 6, 10, 13, 15, 18, 19, 22, 27, 32). Studies developed on the basis of the nucleotide sequences from the envelope gene separated FIV strains into five distinct phylogenetic subtypes designated A to E (1, 2, 10, 13, 22, 27). Although geographic isolation could be a major factor in the evolution of FIV, individual subtypes are found in more than one part of the world (1, 2, 5, 13, 19, 27). For example, subtypes A and B have been isolated from cats in the United States, Europe, Japan, and Australia, and subtype C-infected cats have been identified in North America, Europe, and Taiwan. Consequently, recombinant strains have also been described (notably between subtype A and B subtypes and between B and D subtypes) (1, 2). The isolation and characterization of new FIV isolates with a broad feline representation, including high-risk feral cats, are critical for better understanding of ongoing genetic diversity.
In the present study, FIV was isolated from 11 feral cats that were trapped by the Aggie Feral Cat Alliance in Brazos County, Tex., and surrounding areas. Rigorous phylogenetic analyses were used to determine nucleotide sequences from the V3-V4 region of the envelope gene and from the whole capsid gene of the new Texas FIV strains and to compare those sequences with existing sequences of other FIV strains.
Feline peripheral blood mononuclear cells (PBMCs) were isolated from EDTA (K3)-treated whole-blood samples by Histopaque-1077 (Sigma Chemicals, St. Louis, Mo.) density gradient centrifugation. PBMCs were grown ex vivo for 7 to 10 days as previously described (27). Cells were grown at 37°C in a humidified atmosphere containing 5% CO2, with a change of medium every 3 to 4 days. FIV replication in the culture was determined by detection of capsid antigen in the supernatants with an antigen capture enzyme-linked immunosorbent assay (IDEXX, Portland, Maine) as described elsewhere (25).
Total RNA was extracted (using TRIzol reagent [Invitrogen, Carlsbad, Calif.]) from virus-infected PBMCs and stored at −70°C. Viral cDNA was synthesized using SuperScript II RNase H reverse transcriptase (Invitrogen) primed with oligo(dT). The V3-V4 sequence and capsid gene were separately amplified by nested PCR. For the V3-V4 sequence, the primary PCR used primers 6785F (5′-GCG CAA GTA GTG TGG AG-3′) and 8842R (5′-GCT TCA TCA TTC CTC CTC TT-3′); the nested, secondary amplification employed primers 7316F (5′-ATA CCA AAA TGT GGA TGG TG-3′) and 7866R (5′-CAA GAC CAA TTT CCA GCA AT-3′). The following two sets of primers were used for amplifying the capsid genes: 1053F (5′-GAA GAT CTC ATG GGG AAC GGA CAG GGG CGA-3′) and 2403R (5′-GTC GAC TTA TAA ATC CAA TAG TCT CTC CTC-3′) for the primary PCR and 1458F (5′-GAA GAT CTC CCT ATT CAA ACA GCA AAT GGA GCA-3′) and 2127R (5′-CCA TCG ATG GCA AGA GTT GCA TTT TAT ATC CTG G-3′) for the nested, secondary PCR. Primary reactions (50 μl) contained 100 ng of cDNA, a 100 nM concentration of each primer, 200 μM concentrations of each of the four deoxynucleoside triphosphates, and 1.5 U of TaqDNA polymerase. After incubating at 94°C for 5 min, reactions were subjected to five cycles of amplification at 94°C for 60 s, 53°C for 60 s, and 72°C for 2 min, an additional 30 cycles at 94°C for 15 s and 53°C for 15 s (with a 0.1°C increase after each cycle), and 72°C for 2 min, which was followed by 72°C for 15 min of incubation to ensure the completion of elongation. The same conditions and parameters were applied to the second set of amplifications, except that 10 μl of each of the primary PCR products was used as a template. The PCR products were purified using a Genelute PCR clean-up kit (Sigma Chemical Co.) and directly used as templates for DNA sequencing using a dye terminator method (Gene Technologies Core Facility, Texas A&M University).
Phylogenetic relationships of these new isolates were inferred from analyses of the V3-V4 and capsid sequences (7). To determine the relationships of the V3-V4 region, nucleotide sequences from 11 Texas isolates were aligned with 43 homologues retrieved (using a MacVector v7.0 program [Genetics Computer Group, Madison, Wis]) from the GenBank database. All aligned sequences were also inspected manually. Positions containing gaps or ambiguously aligned positions were removed from the data set. Finally, a total of 501 nucleotide (nt) positions from 54 taxa that can be unambiguously aligned by codons were included in the final phylogenetic analyses. Similarly, DNA sequences of capsid genes from 27 isolates (including 7 Texas isolates) were aligned; a total of 606 nt positions were used for phylogenetic reconstructions.
Phylogenetic analyses were performed using Bayesian inference (BI), maximum likelihood (ML), and maximum parsimony (MP) approaches for both V3-V4 and capsid datasets. For BI phylogeny, nucleotide sequences were partitioned by codons. The MrBayes v3.0 program (9) was used to perform a total of 1,000,000 generations of searches with a Markov chain Monte Carlo sampling method with four chains simultaneously running. Start trees for all chains were randomly generated by the MrBayes program. Posterior possibility (PP) values were obtained using an extended majority ruling method to calculate the consensus tree from the best BI trees. ML analysis employed a quartet-puzzling method using a HKY85 nucleotide substitution model with the consideration of two rates of heterogeneity (one invariable plus one variable). Best quartet-likelihood trees were constructed using the Treepuzzle v5.0 program with 1,000 puzzling steps (30). All parameters, including transition-transversion ratio, nucleotide frequency, and fraction of invariable sites, were estimated from the datasets on the basis of neighbor-joining topologies. For MP bootstrapping analysis, a full heuristic algorithm and the PAUP* v4.0b10 program (31) were used to search for the best trees from 1,000 replicates. Sequences were added randomly, and tree-bisection reconnection was applied for branch swapping during the MP tree searches.
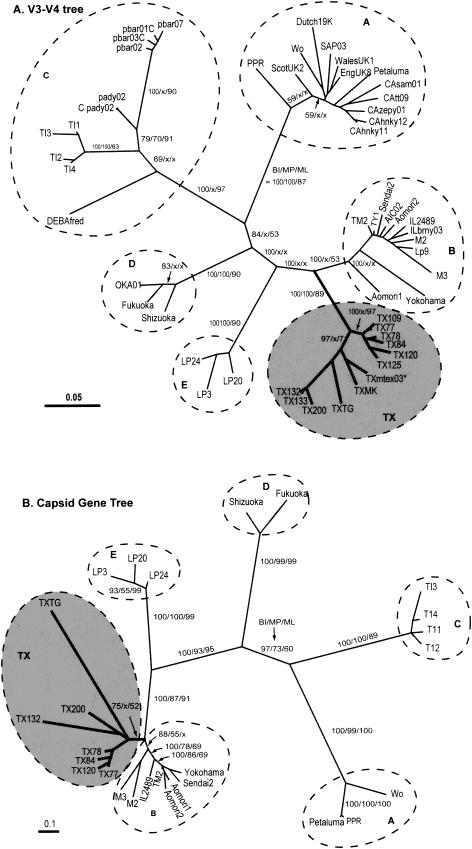
During BI analysis, the best-likelihood values were reached before 40,000 generations of Markov chain Monte Carlo searches for both ENV-V3-V4 and the capsid datasets, indicating that 1,000,000 generations were sufficient for the chains. The BI consensus trees inferred from all alignments clearly separated FIV viral strains into six major clusters, five of which were the previously established subtypes (i.e., A, B, C, D, and E); the sixth cluster consisted of the 11 newly cloned Texas (TX) isolates plus the previously reported Txmtex03 strain (Fig. 1 and 2). The separation of these major subtypes and the cluster was robustly supported by the PP values (i.e., PP in all major nodes = 97 to 100%). The branch lengths between the B subtype and the Texas cluster were better separated in V3-V4 trees than in the capsid gene trees. The B subtype and Texas cluster are clearly related to each other, indicating that these two clusters evolved from a common FIV viral ancestor. When the two alignments were subjected to the quartet-puzzling ML and MP analyses, both methods again divided all viral isolates into six major clusters. However, relationships of isolates among these major FIV clusters were not well resolved by quartet likelihood and MP analyses (Fig. 1 and 2; collapsed branches in ML/MP analyses are marked by an x). In all trees, the ML puzzling indices and MP bootstrapping supporting values were lower than the BI PP values at corresponding nodes with both puzzling and bootstrapping methods (indicative of rapidly evolving genomes), suggesting an overconservative nature.
FIG.1.
(A) Phylogenetic relationships among 54 FIV isolates determined (using BI, MP, and quartet-puzzling methods) on the basis of envelope V3-V4 region nucleotide sequences. A total of 501 nt positions were included for the tree reconstructions. The tree shown here is a consensus tree calculated from the best 9,000 BI trees. Statistical supporting values shown at major branches are PP (BI [from 9,000 best trees]), bootstrapping proportions (MP [from 1,000 replicates]), and puzzling indices (ML [with 10,000 puzzling steps]). Collapsed (unsupported) branches (as determined by MP and/or ML analysis) are marked by an x at corresponding positions. The strain names (GenBank accession numbers) for the V3-V4 sequences are as follows: EngUK8 (X69496); ScotUK2 (X69494); Wales UK14 (X69497); Wo (L06312); Dutch19K32 (M73965); USCAzepy01A (U02417); USCAhnky12A (U02403); USCAsam01A (U02410); USCAtt09A (U02413); SAP03 (AB010404); Sendai2 (D37814); Aomori 2 (D37817); Aomori 1 (D37816); Yokohama (D37812); AICO2 (AB010397); TY1 (D67064); USMAboy03B (U02419); USILbrny03B (U02418); USMOglwd03B (U02420); USTXmtex03B (U02422); M88 (Y13868); M2 (X69501); M3 (X69502); Lp9 (D84497); CABCpady02C (U02392); CABCpbar02C (U02394); CABCpbar03C (U02395); CABCpbar01C (U02393); CABCpbar07C (U02397); DEBAfredC (U57020); TI1 (AB016025); TI2 (AB016026); TI3 (AB016027); TI4 (AB016028); Fukuoka (D37815); Shizuoka (D37811); OKA01D (AB010400); Lp3 (D84496); Lp20 (D84498); Lp24 (D84500); TM2 (M59418); Usil2489_7B (U11820); Petaluma (M25381); PPR (M36968): TX125 (AY139094); TX120 (AY139095); TX200 (AY139096); TXMK (AY139097); TX133 (AY139098); TX132(AY139099); TX84 (AY139100); TXTG (AY139101); TX78 (AY139102); TX109 (AY139102); and TX77 (AY139103). (B) Consensus BI tree inferred from 27 capsid gene sequences with a total of 606 nt positions. PP values (BI [from 9,000 best trees]), bootstrapping proportions (MP [from 1,000 replicates]), and puzzling indices (ML [with 10,000 puzzling steps]) are shown at major branches. Collapsed branches (as determined by MP and/or ML analysis) are marked by an x at corresponding positions. The strain names (GenBank accession numbers) for the capsid sequences are as follows: Wo (L06136); Sendai1 (D37820); Z1 (X57001); Aomori2 (D37824); Sendai2 (D37821); Yokohama (D37819); Aomori1 (D37820); ItM3 (Y13866); ItM2 (Y13867); TI3 (AB027300); TI4 (AB027301); TI2 (AB027299); TI1 (AB027298); Shizuoka (D37822); Fukuoka (D37818); Lp24 (AB027304); Lp20 (AB027303); Lp3 (AB027302); TM2 (M59418); Usil2489_7B (U11820); Petaluma (M25381); PPR (M36968); TX120 (AY139105); TX77 (AY139106); TX78 (AY139107); TX84 (AY139108); TX53 (AY139109); TX200 (AY139110); TXTG (AY139111); and TX132 (AY139112).
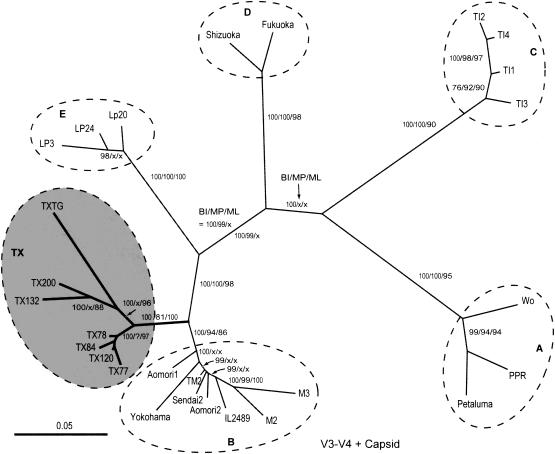
FIG. 2.
Phylogenetic tree determined on the basis of the concatenated V3-V4 and capsid sequences (total positions = 1,107 nt). Statistically supporting values, which included PP values (BI [from 9,000 best trees]), bootstrapping proportions (MP [from 1,000 replicates]), and puzzling indices (ML [with 10,000 puzzling steps]), are again shown at major branches. Collapsed branches determined by MP and/or ML analysis are marked by an x at corresponding positions. The GenBank accession numbers are listed in the legend for Fig. 1.
To further validate these observations with a more extensive region, the capsid data set was fused with corresponding V3-V4 sequences to form a large data set containing 1,107 nt positions. Such concatenated sequences included more phylogenetic informative positions for tree constructions than individual sequences. Upon phylogenetic analysis using the BI method, the same six clusters were formed as in the previous trees (Fig. 2). Again, Texas isolates clustered to a unique branch between the B and E clades. All major clade findings were 100% supported by the PP analysis. The fused data set also improved the resolution of the tree and statistically supported values for the quartet likelihood and MP analyses, though a few major nodes were still unsolved by ML and/or MP analyses (marked by an x in Fig. 2).
The divergence of Texas strains (as a unique cluster) from subtype B viruses was also confirmed by the genetic distances of the V3-V4 regions. The means of the distances of the FIV isolates within and among various subtypes were calculated using a Jukes-Cantor method (Table 1). The results revealed that the Texas cluster was most divergent from subtype A and least divergent from subtype B. The genetic distances between sequences of isolates within the Texas group (mean of distances = 6.4) were similar to those of subtype B (i.e., 6.2), which were less than those within subtypes A and C (i.e., 10.1) but more than those within subtypes D and E (i.e., 4.4 to 4.9). Although double-crossover events could have occurred in regions not sequenced to maintain the observed coevolution of V3-V4 and the capsid sequences, genetic shifts were not identified and the similarities were maintained. Therefore, observed divergence would seem to have occurred through point mutations rather than recombination (1, 2).
TABLE 1.
Means of Jukes-Cantor distances between isolates within and among various subtypesa
| Subtype | Results (genetic distance) for subtypeb:
|
|||||
|---|---|---|---|---|---|---|
| A | B | C | D | E | TX | |
| A | 10.10 | |||||
| B | 23.16 | 6.19 | ||||
| C | 23.95 | 22.82 | 10.33 | |||
| D | 22.60 | 18.29 | 23.36 | 4.88 | ||
| E | 23.37 | 17.78 | 23.97 | 18.78 | 4.39 | |
| TX | 23.83 | 15.07 | 23.40 | 21.58 | 18.04 | 6.37 |
Kimura three-parameter and general time-reversal analyses provided similar results and confirmed the relationships.
The similar comparison results represent distances within each subtype or cluster.
Our data clearly indicate that FIV strains within the United States are much more divergent than initially thought. The Texas cluster is apparently distinct from subtype B viruses, suggesting the presence or emergence of a possible new FIV subtype in the United States. The Texas cluster and the B subtype were more closely related than the others, indicating that the two could have evolved from a common ancestor. The original isolate from Texas (TXmtex03) was, in fact, classified as a subtype B isolate in the absence of other isolates. The sequences of additional isolates allowed for more accurate phylogenetic analysis of this group of FIV strains. Conclusive evidence that the Texas cluster indeed represents a new FIV subtype may depend on immunological studies and sequencing analyses of additional genes and additional isolates. Nonetheless, the divergence of this Texas cluster from subtype B was evident. At the least, a parsimonious explanation is that these Texas FIV isolates may represent a distinct subgroup within the subtype B.
The divergence of FIV in the United States raises broader questions relative to vaccine design (1, 4). Although the only commercial vaccine for FIV is comprised of subtypes A and D viruses, this vaccine may not be effective against natural challenge strains (12, 29). While there are no reports of significant differences in the biological impact of FIV isolated from Texas, the phylogenetic analyses presented in this study suggested that a possible new subtype (or subtype cluster) should be taken into consideration, especially with regard to vaccine design and testing. The amount of variation within the United States warrants additional ongoing comprehensive analyses to truly understand the breadth of FIV diversity throughout the world.
Acknowledgments
We express our appreciation to the Veterinary Medical Teaching Hospital at Texas A&M University and the students, staff, and faculty involved in the AFCAT project.
We thank the Department of Veterinary Pathobiology (department head, Ann Kier) for financial support. This work was further funded by the National Institute of Allergy and Infectious Diseases (grants AI32360 [E.W.C.] and AI44594 [G.Z.]), by the Center for AIDS Research at Baylor (grant 502342), and by the Morris Animal Health Foundation (grant 96FE-09).
REFERENCES
- 1.Bachmann, M. H., C. Mathiason-Dubard, G. H. Learn, A. G. Rodrigo, D. L. Sodora, P. Mazzetti, E. A. Hoover, and J. I. Mullins. 1997. Genetic diversity of feline immunodeficiency virus: dual infection, recombination, and distinct evolutionary rates among envelope sequence subtypes. J. Virol. 71:4241-4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carpenter, M.A., E. W. Brown, D. W. MacDonald, and S. J. O'Brien. 1998. Phylogeographic patterns of feline immunodeficiency virus genetic diversity in the domestic cat. Virology 251:234-243. [DOI] [PubMed] [Google Scholar]
- 3.Elder, J. H., G. A. Dean, E. A. Hoover, J. A. Hoxie, M. H. Malim, L. Mathes, J. C. Neil, T. W. North, E. Sparger, M. B. Tompkins, W. A. Tompkins, J. Yamamoto, N. Yuhki, N. C. Pedersen, and R. H. Miller. 1998. Lessons from the cat: feline immunodeficiency virus as a tool to develop intervention strategies against human immunodeficiency virus type 1. AIDS Res. Hum. Retrovir. 14:797-801. [DOI] [PubMed] [Google Scholar]
- 4.Elyar, J. S., M. C. Tellier, J. M. Soos, and J. K. Yamamoto. 1997. Perspectives on FIV vaccine development. Vaccine 15:1437-1444. [DOI] [PubMed] [Google Scholar]
- 5.Greene, W. K., J. Meers, B. Chadwick, P. R. Carnegie, and W. F. Robinson. 1993. Nucleotide sequences of Australian isolates of the feline immunodeficiency virus: comparison with other feline lentiviruses. Arch. Virol. 132:369-379. [DOI] [PubMed] [Google Scholar]
- 6.Greene, W. K., J. Meers, G. del Fierro, P. R. Carnegie, and W. F. Robinson. 1993. Extensive sequence variation of feline immunodeficiency virus env genes in isolates from naturally infected cats. Arch. Virol. 133:51-62. [DOI] [PubMed] [Google Scholar]
- 7.Hall, B. G. 2001. Phylogenetic trees made easy: a how-to manual for molecular biologists. Sinauer Associates, Inc., Sunderland, Mass.
- 8.Hosie, M. J., R. Osborne, J. K. Yamamoto, J. C. Neil, and O. Jarrett. 1995. Protection against homologous but not heterologous challenge induced by inactivated feline immunodeficiency virus vaccines. J. Virol. 69:1253-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huelsenbeck, J. P., and F. Ronquist. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17:754-755. [DOI] [PubMed] [Google Scholar]
- 10.Inada, G., T. Miyazawa, Y. Inoshima, M. Kohmoto, Y. Ikeda, C. H. Liu, J. A. Lin, T. F. Kuo, and T. Mikami. 1997. Phylogenetic analysis of feline immunodeficiency virus isolated from cats in Taiwan. Arch. Virol. 142:1459-1467. [PubMed] [Google Scholar]
- 11.Ishida, T., and I. Tomoda. 1990. Clinical staging of feline immunodeficiency virus infection. Nippon Juigaku Zasshi-Japanese J. Vet. Sci. 52:645-648. [DOI] [PubMed] [Google Scholar]
- 12.Kakemizu, M., K. A. Ito, A. Yamamoto, Y. Yoshida, M. Sugiyama, N. Minamoto, and R. Pu. 2001. Dual-subtype FIV vaccine protects cats against in vivo swarms of both homologous and heterologous subtype FIV isolates. Microbiol. Immunol. 45:51-58. [DOI] [PubMed] [Google Scholar]
- 13.Kakinuma, S., K. Motokawa, T. Hohdatsu, J. K. Yamamoto, H. Koyama, and H. Hashimoto. 1995. Nucleotide sequence of feline immunodeficiency virus: classification of Japanese isolates into two subtypes which are distinct from non-Japanese subtypes. J. Virol. 69:3639-3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kawaguchi, Y., M. Kohmoto, J. Sakuragi, A. Adachi, M. Fukasawa, T. Mikami, and A. Moraillon. 1992. In vitro properties and experimental pathogenic effect of three strains of feline immunodeficiency viruses (FIV) isolated from cats with terminal disease. J. Gen. Virol. 73:1543-1546.1318947 [Google Scholar]
- 15.Maki, N., T. Miyazawa, M. Fukasawa, A. Hasegawa, M. Hayami, K. Miki, and T. Mikami. 1992. Molecular characterization and heterogeneity of feline immunodeficiency virus isolates. Arch. Virol. 123:29-45. [DOI] [PubMed] [Google Scholar]
- 16.Matteucci, D., A. Poli, P. Mazzetti, S. Sozzi, F. Bonci, P. Isola, L. Zaccaro, S. Giannecchini, M. Calandrella, M. Pistello, S. Specter, and M. Bendinelli. 2000. Immunogenicity of an anti-subtype B feline immunodeficiency fixed-cell virus vaccine in field cats. J. Virol. 74:10911-10919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Novotney, C., R. V. English, J. Housman, M. G. Davidson, M. P. Nasisse, C. R. Jeng, W. C. Davis, and M. B. Tompkins. 1990. Lymphocyte population changes in cats naturally infected with feline immunodeficiency virus. AIDS 4:1213-1218. [DOI] [PubMed] [Google Scholar]
- 18.Olmsted, R. A., R. Langley, M. E. Roelke, R. M. Goeken, D. Adger-Johnson, J. P. Goff, J. P. Albert, C. Packer, M. K. Laurenson, and T. M. Caro. 1992. Worldwide prevalence of lentivirus infection in wild feline species: epidemiologic and phylogenetic aspects. J. Virol. 66:6008-6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pecoraro, M. R., K. Tomonaga, T. Miyazawa, Y. Kawaguchi, S. Sugita, Y. Tohya, C. Kai, M. E. Etcheverrigaray, and T. Mikami. 1996. Genetic diversity of Argentine isolates of feline immunodeficiency virus. J. Gen. Virol. 77:2031-2035. [DOI] [PubMed] [Google Scholar]
- 20.Pedersen, N. C., E. W. Ho, M. L. Brown, and J. K. Yamamoto. 1987. Isolation of a T-lymphotropic virus from domestic cats with an immunodeficiency-like syndrome. Science 235:790-793. [DOI] [PubMed] [Google Scholar]
- 21.Pedersen, N. C., M. Torten, B. Rideout, and N. Levy. 1990. Asymptomatic feline leukemia virus carrier cats have an enhanced susceptibility to feline immunodeficiency virus-induced disease. Devel. Biol. Standard 72:173-184. [PubMed] [Google Scholar]
- 22.Pistello, M., G. Cammarota, E. Nicoletti, D. Matteucci, M. Curcio, D. Del Mauro, and M. Bendinelli. 1997. Analysis of the genetic diversity and phylogenetic relationship of Italian isolates of feline immunodeficiency virus indicates a high prevalence and heterogeneity of subtype B. J. Gen. Virol. 78:2247-2257. [DOI] [PubMed] [Google Scholar]
- 23.Podell, M., K. Hayes, M. Oglesbee, and L. Mathes. 1997. Progressive encephalopathy associated with CD4/CD8 inversion in adult FIV-infected cats. J. Acquir. Immune. Defic. Syndr. Hum. Retrovirol. 15:332-340. [DOI] [PubMed] [Google Scholar]
- 24.Podell, M., P. A. March, W. R. Buck, and L. E. Mathes. 2000. The feline model of neuroAIDS: understanding the progression towards AIDS dementia. J. Psychopharm. 14:205-213. [DOI] [PubMed] [Google Scholar]
- 25.Rigby, M. A., E. C. Holmes, M. Pistello, A. Mackay, A. J. Brown, and J. C. Neil. 1993. Evolution of structural proteins of feline immunodeficiency virus: molecular epidemiology and evidence of selection for change. J. Gen. Virol. 74:425-436. [DOI] [PubMed] [Google Scholar]
- 26.Siebelink, C. H., R. W. Windrich, I. Chu, J. Groen, K. Weijer, F. G. UytdeHaag, and A. D. Osterhaus. 1990. An enzyme linked immunosorbent assay (ELISA) for the detection of feline immunodeficiency virus (FIV) antigen in cell culture and FIV specific antibodies in feline serum. Devel. Biol. Standard 72:189-196. [PubMed] [Google Scholar]
- 27.Sodora, D. L., E. G. Shpaer, B. E. Kitchell, S. W. Dow, E. A. Hoover, and J. I. Mullins. 1994. Identification of three feline immunodeficiency virus (FIV) env gene subtypes and comparison of the FIV and human immunodeficiency virus type 1 evolutionary patterns. J. Virol. 68:2230-2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Song, W., E. Collisson, P. Billingsley, and W. Brown. 1992. Induction of feline immunodeficiency virus-specific cytotoxic T-cell responses from experimentally infected cats. J. Virol. 66:5407-5417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Soos, J., R. Pu, D. Pollock, J. K. Yamamoto, and J. S. Elyar. 1997. Perspectives on FIV vaccine development. Vet. Microbiol. 57:1-11. [DOI] [PubMed] [Google Scholar]
- 30.Strimmer, K., and A. Vonhaeseler. 1996. Quartet puzzling—a quartet maximum-likelihood method for reconstructing tree topologies. Mol. Biol. Evol. 13:964-969. [Google Scholar]
- 31.Swofford, D. L. 2000. PAUP*. Phylogenetic analysis using parsimony (*and other methods). Sinauer Associates, Sunderland, Mass.
- 32.Yamada, H., T. Miyazawa, K. Tomonaga, Y. Kawaguchi, K. Maeda, M. C. Castellano, C. Kai, Y. Tohya, and T. Mikami. 1995. Phylogenetic analysis of the long terminal repeat of feline immunodeficiency viruses from Japan, Argentina and Australia. Arch. Virol. 140:41-52. [DOI] [PubMed] [Google Scholar]
- 33.Yamamoto, J. K., H. Hansen, E. W. Ho, T. Y. Morishita, T. Okuda, T. R. Sawa, R. M. Nakamura, N. C. Pedersen, and T. Ishida. 1989. Epidemiologic and clinical aspects of feline immunodeficiency virus infection in cats from the continental United States and Canada and possible mode of transmission of feline immunodeficiency virus infection in cats of Japan. J. Am. Vet. Med. Assoc. 194:213-220. [PubMed] [Google Scholar]
- 34.Yamamoto, J. K., E. Sparger, E. W. Ho, P. R. Andersen, T. P. O'Connor, C. P. Mandell, L. Lowenstine, R. Munn, and N. C. Pedersen. 1988. Pathogenesis of experimentally induced feline immunodeficiency virus infection in cats. Am. J. Vet. Res. 49:1246-1258. [PubMed] [Google Scholar]