Abstract
The induction of humoral response in ducks by DNA-based immunization against duck hepatitis B virus (DHBV) core protein (DHBc) was investigated. In addition, the amino acid specificity of the induced response was compared by using peptide scanning to that elicited either by protein immunization or during chronic DHBV infection. Immunization of ducks with a plasmid expressing DHBc protein led to the induction of a long-lasting antibody response able to specifically recognize viral protein in chronically infected duck livers. Peptide scanning analysis of anti-DHBc response induced during chronic DHBV infection allowed us to identify six major antigenic regions (AR1 to AR6). The reactivity spectrum of duck sera elicited by protein immunization appeared narrower and was restricted to only four of these antigenic regions in spite of higher anti-DHBc antibody titers. Interestingly, anti-DHBc antibodies induced by DNA-based immunization recognized five of six antigenic regions, and the epitope pattern was broader and more closely related to that observed in chronic viral infections. To gain more insight into the location of antigenic regions, we built a three-dimensional (3-D) model of DHBc protein based on human and duck core sequence alignment data and the HBc 3-D crystal structure. The results suggest that two identified antigenic regions (AR2, amino acids [aa] 64T-P84, and AR5, aa 183A-R210) are located at positions on the protein surface equivalent to those of the two HBc major epitopes. Moreover, we identified another antigenic region (AR3, aa 99I-I112) that was recognized by all sera from chronically infected, DNA- or protein-immunized ducks within the large 45-aa insertion in DHBc protein, suggesting that this region, which lacks HBc, is externally exposed.
DNA vaccine is a powerful new approach to immunization and immunotherapy that has been shown to induce specific humoral and cellular responses against variety of viral antigens (13). In a genetic vaccine, direct inoculation of plasmid DNA results in the in situ synthesis of antigen by the immunized host; therefore, it has a potential advantage over a recombinant protein immunization in that the resulting antigen would have the conformation and posttranslational modifications identical to those occurring during viral infection (13, 27). Genetic vaccination is also an efficient tool for defining immunogenic epitopes of selected viral antigens. For example, this approach allowed the successful characterization of B- and T-cell epitopes of human immunodeficiency virus Nef, Rev, and Tat proteins (9).
The effectiveness of DNA vaccine to Hepatitis B Virus (HBV) envelope proteins has been extensively studied in a murine model and showed induction of high-titer and long-lasting humoral responses (19). Studies with a transgenic mouse, lineage E36, which constitutively expresses the HBsAg, strongly suggest the therapeutic efficacy of DNA vaccine encoding viral envelope proteins (17). Moreover, DNA-based vaccination allowed better insight into the murine immune response to selected T-cell or antibody-defined determinants of HBV envelope proteins (19, 25).
Pekin duck infected with duck hepatitis B virus (DHBV), which is closely related to the human virus, is a reference model for in vivo studies of hepadnaviral replication and for antiviral approaches testing, since the chronic infection of ducks mimics aspects of what occurs in the HBV long-term carrier state. It is also a pertinent model for studying the efficacy of DNA vaccine against hepadnaviral structural proteins. The DNA vaccination of ducks with plasmid expressing DHBV small or large envelope proteins elicited a potent, specific, and protective humoral response (21, 22, 31). The first attempts of therapeutic immunization of chronic DHBV carrier ducks with this plasmid showed promise, since a dramatic and sustained decrease in viral replication was observed that was further enhanced by association of DNA vaccine with antiviral drugs (15, 22, 30).
Hepadnaviral core antigen represents another interesting target for DNA-based vaccine since it plays a central role in nucleocapsid formation and pregenomic viral RNA packaging, although less data are available on DNA immunization to core compared to the envelope protein. DNA-based immunization of mouse to the HBV core has been shown to efficiently prime specific antibody and cytotoxic-T-lymphocyte responses (12). In the American woodchuck model the DNA vaccination to Woodchuck Hepatitis Virus core was shown to induce a potent immune response that was further enhanced by coadminstration of a plasmid expressing woodchuck gamma interferon (16, 26).
However, the effectiveness of naked DNA immunization to DHBV core protein (DHBc) has not yet been investigated. Moreover, in contrast to DHBV envelope protein, for which the humoral response of ducks has been extensively studied and major antigenic regions (ARs) have been precisely mapped by us and others (3, 4, 28, 35), data on the humoral response of ducks to DHBV core protein are scarce. Similarly to HBV, the DHBV core gene encodes two polypeptides: the core protein that forms particles and the secreted e-antigen. The DHBV core protein is a basic phosphoprotein of 32 to 35 kDa (20) that is assembled from a polypeptide species of 226 amino acids and forms nucleocapsids of a three-dimensional structure similar to that of HBV (11, 34). However, neither crystal structure nor ARs of DHBc protein have yet been described.
In the present study, we have investigated the induction of humoral response after DNA-based immunization of ducks with a plasmid expressing DHBc protein. In addition, we have taken advantage of naked DNA immunization to identify for the first time the ARs of DHBc and compare the amino acid specificity of the induced response with that elicited either by protein immunization or during chronic DHBV infection. Moreover, we have localized the identified ARs on a three-dimensional (3-D) model DHBc structure and compared their positions with previously described major HBc epitopes recognized by monoclonal antibodies and chronically infected patient sera.
MATERIALS AND METHODS
Virus.
A pool of viremic sera from ducklings infected with the cloned and sequenced DHBV (18) was used as an inoculum. This inoculum was quantified into virus genome equivalents by quantitative dot blot hybridization as described previously (1, 28).
Animals.
Three-day-old DHBV-free Pekin ducklings (Anas domesticus) were purchased from a commercial supplier. Ducks were maintained outdoors in accordance with the guidelines for animal care at the facilities of the École Nationale Veterinaire de Lyon, Lyon, France. The chronically infected animals were obtained by inoculation of 3-day-old ducklings with DHBV-positive serum (2 × 108 virus genome equivalents/duck) as described previously (14).
Construction of vectors for DNA immunization.
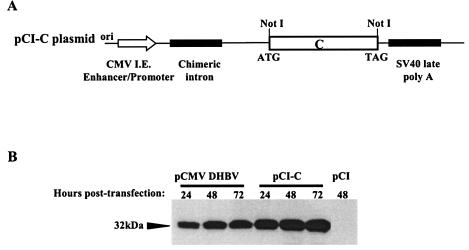
For DNA immunization, the DHBV DNA fragment encoding core protein (nucleotides 2647 to 412) was cloned into pCI plasmid (Promega, Charboniere, France), leading to the pCI-C vector (Fig. 1A). Empty pCI plasmid was used as a negative control. The plasmids were grown in Escherichia coli, followed by purification using Qiagen anion-exchange chromatography columns (Qiagen, Hilden, Germany) and quantification by determining the optical density at 260 nm and ethidium bromide staining as previously described (22).
FIG. 1.
DNA vaccination vector and DHBc protein expression. (A) Schematic representation of pCI-C plasmid encoding DHBV core protein. (B) Expression of the DHBV core protein from pCI-C and pCMV-DHBV plasmid (control) in transiently transfected LMH cells. Proteins from cells lysates were probed in an immunoblotting analysis with rabbit anti-DHBc antibody. The black arrowhead indicates the 32-kDa DHBc protein.
In vitro transient expression of DHBc protein.
The avian hepatoma LMH cell line was transiently transfected with plasmids by using Transfection-Fugene 6 Reagent (Roche Diagnostics, Basel, Switzerland) according to the manufacturer's protocol. As a positive control, pCMV-DHBV plasmid (kindly provided by C. Seeger, Fox Chase Center, Philadelphia, Pa.) encoding the entire DHBV pregenomic sequence under the control of the cytomegalovirus (CMV) promoter was used (22). At 24, 48, and 72 h posttransfection, LMH cells were lysed and then analyzed for core protein expression by immunoblotting.
Immunizations of ducks.
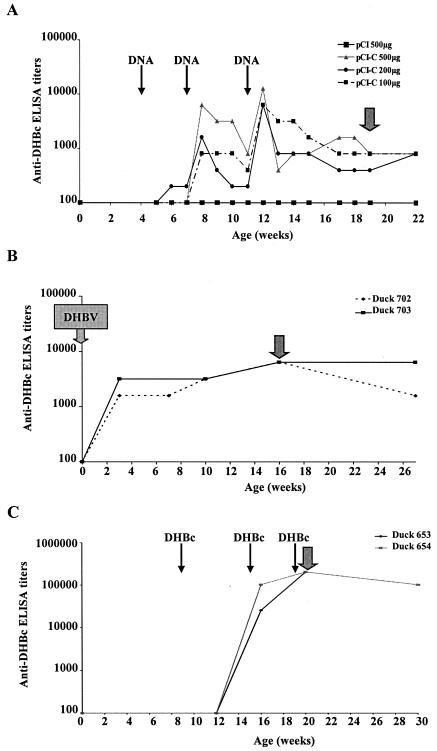
Four-week-old uninfected ducks were DNA immunized with 100 μg (three animals), 200 μg (three animals), and 500 μg (two animals) of pCI-C plasmids diluted in phosphate-buffered saline given by intramuscular injection in the anterior quadriceps of both legs and breast as previously described (22). Booster doses were administered at the same sites 3 and 7 weeks later. The control duck group was immunized under the same protocol with the empty pCI plasmid (500 μg). Two other animals were immunized by 50 μg/injection of recombinant DHBc protein mixed with an equal volume of complete Freund adjuvant at weeks 9, 14, and 19. The anti-DHBc humoral response was monitored for 22 and 30 weeks by enzyme-linked immunosorbent assay (ELISA).
ELISA test for anti-DHBc antibodies detection.
The anti-DHBc response was determined by a direct ELISA with microtiter plates coated with 100 ng of recombinant DHBc protein/well in 0.1 M carbonate buffer (pH 9.6). The recombinant DHBc protein, purified as previously described (36), was kindly provided by F. Schödel. The plates were sealed and incubated overnight at 4°C. Thereafter, the plates coated with DHBc protein were washed and treated as previously described (4). Endpoint titers were taken as the reciprocal of highest serum dilution, which gave an optical density signal >3 standard deviations above the mean signal of duplicates of control sera.
Immunoblotting.
Protein from DHBV-infected or uninfected duck livers were extracted as previously described (3). The immunoblotting was performed with either rabbit polyclonal anti-DHBc antiserum (kindly provided by A. Kay and diluted 1:4,000) or immunized duck sera (diluted 1:10) (1, 22). After washes, filters were incubated with either anti-rabbit (diluted 1:2,000; Kirkegaard & Perry Laboratories, Inc., Gaithersburg, Md.) or anti-duck (diluted 1:500; Kirkegaard & Perry) immunoglobulin G-peroxidase conjugates, and the reaction was developed with the ECL chemoluminescence detection kit (Amersham, Courtaboeuf, France).
Pepscan analysis.
A set of 37 overlapping peptides corresponding to 14-mers spanning the entire amino acid sequence of the DHBV core protein was synthesized in duplicates on pins (Chiron Mimotyopes, Clayton, Victoria, Australia) according to the Pepscan method (7, 8). The DHBc sequence was scanned by using a 14-amino-acid (aa) window moved by 7 aa at each step (Table 1). The pin-bound peptides were held on rack, which had a 96-well format compatible with conventional ELISA plates. The peptide recognition was tested with sera (dilution 1:200) from DNA- or DHBc protein-immunized ducks or with sera from chronically infected animals. Direct ELISA was performed according to the manufacturer's protocol (Chiron) with anti-duck immunoglobulin G-peroxidase conjugate and ABTS [2,2′azinobis(3-ethylbenzthiazolinesulfonic acid)] substrate (Boehringer Ingelheim, Ingelheim am Rhein, Germany). The test was repeated at least two times for each serum and, between each test, the bound antibodies were removed from the pins according to the manufacturer's protocol (Chiron) by sonication for 10 min in disruption buffer at 65°C (0.1 M phosphate buffer, 1% sodium dodecyl sulfate, 0.1% β-mercaptoethanol [pH 7.2]). After a wash with water at 60°C (30 min), pins were plunged into methanol (15 min at 60°C) and then air dried for 15 min before storage at 4°C. A peptide was considered significantly reactive when its absorbance was higher than a mean of 10 lowest absorbance values plus two standard deviations determined for each assay (29).
TABLE 1.
Amino acid sequences of the overlapping DHBc synthetic peptides used for Pepscan analysis
| Peptide no. | Peptide location (aa) | Peptide sequence |
|---|---|---|
| 1 | 1-14 | MDINASRALANVYD |
| 2 | 8-21 | ALANVYDLPDDFFP |
| 3 | 15-28 | LPDDFFPKIDDLVR |
| 4 | 22-35 | KIDDLVRDAKDALE |
| 5 | 29-42 | DAKDALEPYWKSDS |
| 6 | 36-49 | PYWKSDSIKKHVLI |
| 7 | 43-56 | IKKHVLIATHFVDL |
| 8 | 50-63 | ATHFVDLIEDFWQT |
| 9 | 57-70 | IEDFWQTTQGMHEI |
| 10 | 64-77 | TQGMHEIAESLRAV |
| 11 | 71-84 | AESLRAVIPPTTTP |
| 12 | 78-91 | IPPTTTPVPPGYLI |
| 13 | 85-98 | VPPGYLIQHEEAEE |
| 14 | 92-105 | QHEEAEEIPLGDLF |
| 15 | 99-112 | IPLGDLFKHQEERI |
| 16 | 106-119 | KHQEERIVSFQPDY |
| 17 | 113-126 | VSFQPDYPITARIH |
| 18 | 120-133 | PITARIHAHLKAYA |
| 19 | 127-140 | AHLKAYAKINEESL |
| 20 | 134-147 | KINEESLDRARRLL |
| 21 | 141-154 | DRARRLLWWHYNCL |
| 22 | 148-161 | WWHYNCLLWGEAQV |
| 23 | 155-168 | LWGEAQVTNYISRL |
| 24 | 162-175 | TNYISRLRTWLSTP |
| 25 | 169-182 | RTWLSTPEKYRGRD |
| 26 | 176-189 | EKYRGRDAPTIEAI |
| 27 | 183-196 | APTIEAITRPIQVA |
| 28 | 190-203 | TRPIQVAQGGRKTT |
| 29 | 197-210 | QGGRKTTTGTRKPR |
| 30 | 204-217 | TGTRKPRGLEPRRR |
| 31 | 211-224 | GLEPRRRKVKTTVV |
| 32 | 218-231 | KVKTTVVYGRRRSK |
| 33 | 225-238 | YGRRRSKSRERRAP |
| 34 | 232-245 | SRERRAPTPQRAGS |
| 35 | 239-252 | TPQRAGSPLPRSSS |
| 36 | 246-259 | PLPRSSSSHHRSPS |
| 37 | 249-262 | RSSSSHHRSPSPRK |
Molecular modeling.
The 3-D model for the DHBV core protein was built by using Geno 3D, a comparative molecular modeling system for proteins (5). The HBc 3-D structure (pdb code 1GQT) (33) was taken as the template on the basis of sequence identity after sequence alignment with DHBc protein as described by Bringas (2). Distance restraints and dihedral angles were calculated on the template structure. These measures were performed for all common atoms revealed by the alignment between the DHBc and HBc proteins. The CNS 1.0 program was used to generate the model by a distance geometry approach similar to that used in modeling from nuclear magnetic resonance experiments. Mirror images were eliminated on the basis of energy calculation. Ten models were generated and were superimposed with the ANTHEPROT 3D package (6) by minimizing the root mean square deviation between alpha carbons. The visualization of the molecule was performed with the help of a WebLab ViewerLite 3.2 (Accelrys).
RESULTS
In vitro expression of DHBc protein by DNA-immunization vector.
To test the capacity of pCI-C vector to express DHBV core proteins in eukaryotic cells, the LMH cells were transiently transfected with this plasmid. As illustrated in Fig. 1B, high levels of 32-kDa protein, which was reactive with a specific anti-DHBc rabbit antibody, were observed in these cells beginning already at 24 h and accumulating in the cells up to 72 h posttransfection. This protein comigrates with the 32-kDa DHBc protein from cells transfected with the pCMV-DHBV plasmid encoding the entire viral genome, indicating that pCI-C plasmid expresses the entire DHBc protein of correct size. No DHBV core gene products were detected in the control cells transfected with empty pCI plasmid.
Anti-DHBc humoral response elicited in ducks after DNA or protein immunization and during chronic infection.
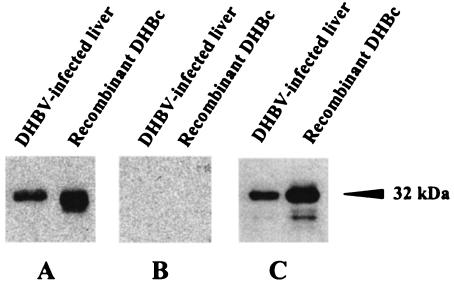
To test the ability of pCI-C plasmid to induce DHBc-specific immune response, uninfected ducks were DNA immunized with this vector at weeks 4, 7, and 11. Different amounts of pCI-C plasmid (ranging from 100 to 500 μg) were injected to evaluate the DNA dose effect. As shown in Fig. 2A, a control duck immunized with the empty pCI plasmid (500 μg) did not develop any detectable anti-DHBc response. In contrast, beginning after the first DNA boost, a humoral response was detected in the ducks immunized with pCI-C plasmid, which was further enhanced by the second boost (titers peaking to 104.1), followed by a decrease (to ca. 103), and was maintained during 3 months of follow-up without additional boosts (Fig. 2A and data not shown). Importantly, all eight immunized ducks responded to this DNA vaccine. Interestingly, even the lowest plasmid dose (100 μg) resulted in the induction of high anti-DHBc response, although three injections of this plasmid dose were required to elicit antibody titers comparable to those elicited by only two injections of high plasmid dose (500 μg). We have confirmed the specificity of these antibodies, since they were able to recognize in immunoblotting the recombinant 32-kDa DHBc protein and, more importantly, the DHBc protein in the chronically infected duck livers (Fig. 3A), similarly to anti-DHBc control serum from a rabbit immunized with the recombinant DHBc protein (Fig. 3C). Antibodies elicited by empty pCI plasmid immunization were not reactive in this test (Fig. 3B).
FIG. 2.
Humoral response induction by DNA or protein immunization and during DHBV chronic infection. The kinetics of humoral response induced in ducks immunized with pCI-C plasmid (100 μg, 200 μg, 500 μg) and empty pCI (500 μg) plasmid (A), in chronic DHBV carrier ducks (B), and in ducks immunized with recombinant DHBc protein (C) are shown. Detection of anti-DHBc, endpoint titer antibodies, expressed as the log10, was performed by using a direct ELISA. The graphs show mean anti-DHBc responses for each duck group immunized with pCI-C plasmid at doses of 100 μg (three ducks), 200 μg (three ducks), or 500 μg (two ducks) (A) and individual values for chronically infected (B) or DHBc protein-immunized animals (C). Thin black arrows represent DNA or protein injections, and large gray arrows indicate the serum sample used for the Pepscan analysis.
FIG. 3.
Recognition in immunoblotting assay of recombinant DHBc proteins and proteins from chronically infected duck liver with serum from pCI-C immunized animals (A), control duck serum from empty pCI plasmid-immunized ducks (B), and polyclonal anti-DHBc rabbit antiserum (C). The DHBV core protein is indicated.
To compare the humoral response induced by genetic vaccination with the response elicited during chronic DHBV, we evaluated the anti-DHBc response of DHBV carrier ducks, which had been infected with DHBV as neonates. As illustrated in Fig. 2B, chronically infected ducks mounted rapidly an anti-DHBc response, reaching a plateau with anti-DHBc titers of ∼103.8. Immunization with the recombinant DHBc protein led to the induction of an anti-DHBc response, which peaked to higher ELISA titers (105.3) compared to the responses observed during chronic viral infection or specific DNA vaccination.
Identification of ARs of DHBc protein recognized by antibodies induced by DNA or protein immunizations or during chronic infection.
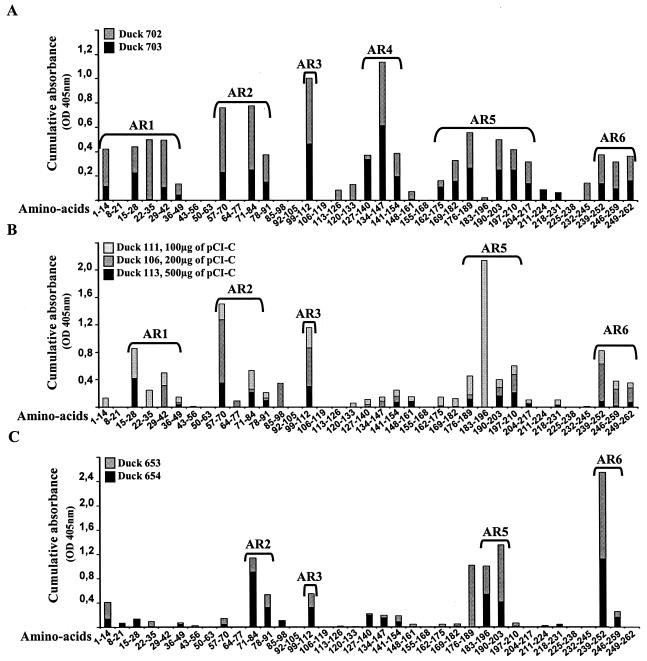
To determine the ARs of DHBc protein recognized by antibodies induced by either DNA immunization, DHBc immunization, or chronic viral infection, the corresponding duck sera were taken at the plateau of anti-DHBc response (weeks 16 to 20) (Fig. 2, arrow) and were screened for their reactivity with the overlapping 14-mer peptides covering the entire DHBc region (peptides 1 to 37) (Table 1). Figure 4 illustrates the accumulated optical density values in ELISAs for the sera from chronically infected animals, ducks immunized with plasmid DNA (100, 200, and 500 μg, respectively), or recombinant DHBc protein (panels A, B, and C, respectively). Despite of the individual variation in peptide recognition and in the intensity of peptide reactivity, six ARs (AR1 to AR6) were reactive with the sera from chronically infected ducks (Fig. 4A). Thus, chronically infected duck sera recognized AR1 (aa 1M-S42), AR2 (aa 57I-P84), AR4 (aa 134K-L147), AR5 (aa 169R-R210), and AR6 (aa 246P-K262). Moreover, these sera were highly reactive with a single peptide corresponding to AR3 (aa 99I-I112) (Fig. 4A). Despite the differences in the reactivity intensities and individual bird-to-bird responses, sera from DNA-immunized ducks recognized these ARs except for AR4 (Fig. 4B). The recognition of some ARs by the pCI-C plasmid-immunized duck sera was less broad compared to the sera elicited during chronic infections as illustrated for AR1, which was restricted to aa 22K-S42. No marked differences in reaction intensity were observed with antibodies elicited by different amounts of pCI-C vector (100, 200, and 500 μg) used for the DNA immunization (Fig. 4B). Importantly, no reactivity was detected with the two control sera from empty pCI plasmid-immunized ducks (data not shown).
FIG. 4.
Screening by Pepscan of duck antibodies binding to overlapping 14-mer peptides spanning the entire sequence of DHBV core protein. Anti-DHBc duck antibodies induced either by chronic viral infection or by DNA or DHBc protein immunization were taken at the plateau of anti-DHBc response (weeks 16 to 20) (Fig. 2) and were screened for their reactivity with the overlapping 14-mer peptides covering the entire DHBc region (peptides 1 to 37). Cumulative optical density values as determined by ELISA for the sera from two chronically infected ducks (A), three DNA-immunized ducks (100, 200, and 500 μg, respectively) (B), or two recombinant DHBc protein-immunized ducks (C) are shown. AR1 to AR6 indicate the positions of identified ARs.
The antibodies induced by immunization with the recombinant DHBc protein were reactive only with four (AR2, AR3, AR5, AR6) of six ARs recognized by the sera from chronically infected ducks (Fig. 4C). Moreover, in spite of higher anti-HBc titers in ELISA, the reactivity spectrum of these sera appeared narrower and was restricted to few peptides within AR2 (aa 78I-P85), AR5 (aa 190T-A197), and AR6 (aa 246P-S252) compared to the spectrum of response induced by DNA-immunization or during chronic infection. Interestingly, AR3 (aa 99I-I112) was reactive with all duck sera elicited as well during chronic infection as by DNA or protein immunization.
Comparison of AR localization on DHBc and HBc proteins.
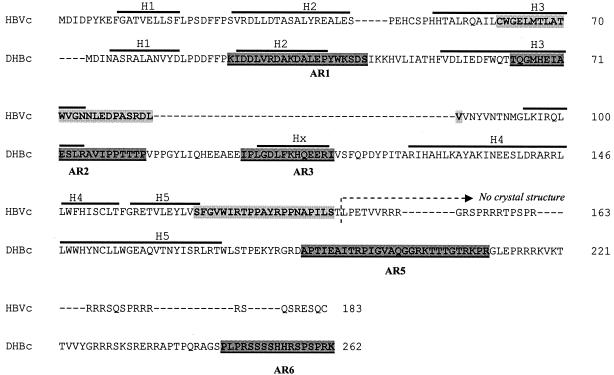
Next, we localized the experimentally defined ARs on the DHBc amino acid sequence and compared to the known HBc protein antigenic sites. The alignment of duck and human viral capsid proteins used for molecular modeling, mainly based on that established by Bringas (2) is shown in Fig. 5. The two major HBc protein ARs (aa 61 to 85 and aa 121 to 140), recognized by the antibodies in chronically infected patient sera (24), were highlighted on the HBc sequence. This alignment shows a large amino-acid insertion of 45 aa (from aa 85 to 130) in DHBc protein within amino-acid 85 and 86 of HBc protein.
FIG. 5.
Sequence alignment of DHBV and HBV core proteins showing localization of identified DHBc ARs. The alignment of duck and human viral capsid proteins was mainly based on that established by Bringas (2). The large dotted line shows a 45-aa insertion (from aa 85 to 130) in DHBc protein within aa 85 and 86 of the HBc protein. Five ARs (AR1, AR2, AR3, AR5, and AR6) that were recognized by Pepscan analysis of all five duck sera from the three pCI-C-immunized animals and the two chronically infected animals are highlighted in gray and underlined in the DHBc sequence. Two major HBV core protein ARs (aa 61 to 85 and aa 121 to 140) were highlighted in light gray on the HBc sequence. Numbers on the right side of sequences denote amino acid position of human and duck sequences. H1 to H5 and Hx indicate predicted α-helical conformation by Bringas (2).
A total of five ARs (AR1, AR2, AR3, AR5, and AR6) that were recognized in Pepscan by all five duck sera from the three pCI-C immunized animals and the two chronically infected ducks were localized on this DHBV sequence (Fig. 5). Three ARs, AR1, AR3, and AR6, defined on the DHBc protein were not located near known HBc ARs. Interestingly, the positions of AR2 and AR5 partly overlap the two major immunodominant epitopes of HBc protein (aa 61 to 85 and aa 121 to 140) (24). The alignment data suggest that these immunodominant regions are located on the surface of both proteins and on the equivalent α-helixes (Fig. 5). Moreover, the AR3 corresponding to the residues 99 to 112 was localized within the 45-aa DHBc large insertion.
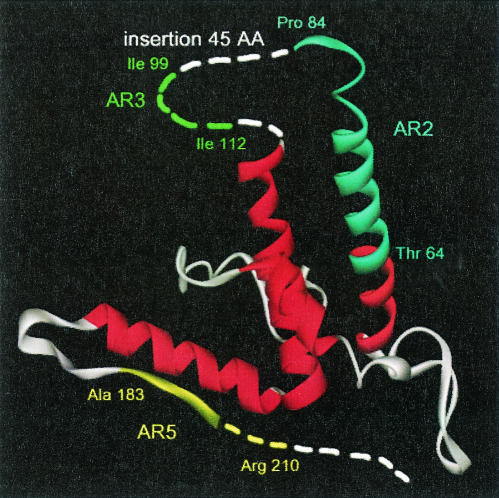
To gain more insight into AR locations on DHBc, we built a 3-D model of virus core protein based on the HBc 3-D crystal structure (33) and alignment data (2) by using Geno 3D (5) (Fig. 6). Importantly, the two regions of interest, AR2 (aa 64T-P84) and AR5 (aa 183A-R210), were localized within the modelized α-helix 3 and on the loop following the α-helix 5, respectively (Fig. 6). These regions were rather exposed at the surface of the DHBc protein structure and were localized on α-helices equivalent to those of HBc protein. For AR3, localized within the 45-aa insertion, no 3-D information is available and therefore this portion of protein (shown as dotted line) cannot be modelized since there is no corresponding region within HBc protein (Fig. 6). Interestingly, AR3 (aa 99I-I112), identified by us, was localized within this large insertion that could correspond to a short α-helix (from aa 101L to R111, Hx) as predicted by Bringas (2) (Fig. 5).
FIG. 6.
3-D model of DHBV core protein. The model was built as described in Materials and Methods with 1GQT as the molecular template (33). All helices have been stained in red. Three identified ARs have been positioned onto the 3-D model (AR2 from amino acids 64T to 84P is indicated in blue; AR3 from aa 99I to 112I is indicated in green in the region with unavailable atomic coordinates, corresponding to the 45-aa insertion; and AR5 from aa 183A to 210R is indicated in yellow). The dotted lines indicate regions in DHBc for which no atomic coordinates are available.
DISCUSSION
The aim of this study was to induce and analyze the humoral response elicited by DNA immunization toward the DHBV core protein. We report that immunization of ducks with a plasmid expressing DHBc protein induces a specific and long-lasting humoral response. The antibody response elicited by plasmid vaccination appears to be dependent on multiple factors such as antigen immunogenicity, plasmid DNA dose, animal species, and body weight (13). In the duck model, DNA immunization with only 100 μg of plasmid encoding DHBV capsid protein induced an anti-DHBc response in DNA-immunized ducks weighing from 2 to 3 kg, although the response was weaker than the anti-pre-S response induced by DNA immunization to DHBV L envelope protein with the same plasmid dose (22). The comparison of DNA immunization with different amounts of plasmid expressing DHBc protein (100, 200, or 500 μg) showed that immunization with higher amounts of plasmid DNA was more efficient, since three injections of 100 μg of plasmid DNA were required to reach the same magnitude of antibody response obtained with only two injections of 500 μg of DNA. Importantly, antibodies elicited by DNA immunization that we have developed were able to specifically recognize not only the recombinant DHBV core protein but also the native viral protein in the livers of chronically infected ducks.
An important aspect of the present study concerns DHBc ARs recognized by antibodies elicited by DNA immunization in comparison to chronic viral infection. In contrast to DHBV envelope protein, for which a number of studies have analyzed humoral response and mapped ARs, detailed analysis of anti-DHBc response of ducks to viral core protein had not yet been performed. To identify DHBc ARs, we used the overlapping peptide scanning technique, an approach initially described by Geysen et al. (7, 8), which has been successful in the determination of the ARs of different viruses, including the DHBV large envelope protein major antigenic domains (3, 4). In the experimentally infected DHBV carrier ducks, anti-DHBc antibodies were demonstrated to be present early after infection, depending of the dose of inoculated virus, and persisted throughout the course of infection, similarly to anti-HBc antibodies in serum of patients with chronic HBV infection (10). We have reproduced such chronic DHBV infection and demonstrated by overlapping peptides analysis that the elicited anti-DHBc antibodies recognize six major ARs (AR1 to AR6). We observed some bird-to-bird differences in the reactivity of individual duck sera with peptides, a finding similar to what was previously observed by us in a Pepscan analysis of an anti-DHB pre-S response in DHBV-infected ducks (4). Interestingly, five of six identified ARs were also recognized by anti-DHBc antibodies induced by DNA-based immunization. Although immunization with the recombinant DHBc protein increased the antibody titer, the epitope pattern appeared to be broader after DNA immunization and closer to the one observed in chronic viral infection. Broader recognition of epitopes by antisera elicited by DNA compared to protein immunization was also demonstrated in mice immunized against human immunodeficiency virus regulatory proteins (9) or against Plasmodium falciparum merosite surface protein (32). This is not surprising since, in contrast to protein immunization, plasmid DNA inoculation should result in the intracellular synthesis of a protein in which posttranslational modifications lead to the induction of immune responses mimicking those observed during viral infection (13). Our results support these findings and demonstrate for the first time in the DHBV infection model that the humoral response induced by genetic immunization to hepadnaviral capsid protein is closely related to that induced during natural infection. Further studies are needed to identify and analyze cellular immune response to DHBc induced by DNA immunization, although tools for investigation of the cellular response in duck model are still under development.
It was of interest to compare the localization of identified ARs on the DHBc amino acid sequence with the known HBc antigenic sites. In spite of the sequence divergence between mammalian and duck hepadnaviral capsid proteins, the alignment of these sequences and secondary structure prediction showed a good correlation, suggesting the existence of five helical segments common to both mammalian and avian core proteins (2). Interestingly, the localization of ARs identified by us on this alignment showed that of five ARs identified, two (AR2 and AR5) are localized at the equivalent positions to HBc major antigenic epitopes (Fig. 5). Thus, the AR2 of DHBc protein (aa 64T-P84) was defined in the α-helix 3 of DHBc protein. This localization is similar to that of the major immunodominant epitope (residues 61 to 85) of HBc protein recognized by monoclonal antibodies and chronically infected patient sera (23, 24), which was identified within an immunodominant loop located at about residue 80 (2, 11). In addition, AR5 region (aa 183A-R210) has been localized within the loop between α-helix 5 and the C-terminal extended strand of DHBc protein, similarly to the secondary epitope of HBc, which has been identified in the equivalent position (residues 121 to 140) (24). Our 3-D molecular modeling data have further indicated that these two ARs, i.e., AR2 and AR5, were rather exposed at the surface of the DHBc protein structure and were localized on equivalent position to the two major immunodominant epitopes of HBc. Moreover, another AR, AR3 (aa 99I-I112), which was recognized by all sera from chronically infected, DNA- or protein-immunized ducks, was identified by us within the large 45-aa insertion in the DHBc protein, suggesting that this region, lacking in HBc, is externally exposed. The previously described prediction of DHBc protein secondary structure (2) has suggested that this 45-residue insertion may be a protruding domain located within a predicted α-helix. Our localization of an immunodominant region within this domain provides experimental data which strengthen this hypothesis. Giving the absence of 3-D information on DHBc protein, these data may be extremely useful and should be taken into account in further models of DHBc structure.
The AR identified by us with the help of synthetic peptides thus represented more likely linear sites. As a consequence, there could be partial epitopes that may be part of much larger conformational sites. However, since there is an unresolved region where AR3 was localized, it is difficult to hypothesize about conformational epitopes involving this region.
Taken together, the present results, which for the first time demonstrate the humoral response induction by DNA immunization to the DHBV core protein and identify the major ARs on a 3-D protein model, can be valuable for further studies on hepadnaviral capsid structure and morphogenesis and for the design of new antiviral strategies in the DHBV infection model.
Acknowledgments
A. Thermet was the recipient of a fellowship from the ARC.
We thank Alan Kay for stimulating discussions. We are grateful to Catherine Jamard for excellent assistance with the animals.
REFERENCES
- 1.Borel, C., C. Sunyach, O. Hantz, C. Trepo, and A. Kay. 1998. Phosphorylation of DHBV pre-S: identification of the major site of phosphorylation and effects of mutations on the virus life cycle. Virology 242:90-98. [DOI] [PubMed] [Google Scholar]
- 2.Bringas, R. 1997. Folding and assembly of hepatitis B virus core protein: a new model proposal. J. Struct. Biol. 118:189-196. [DOI] [PubMed] [Google Scholar]
- 3.Chassot, S., V. Lambert, A. Kay, C. Godinot, B. Roux, C. Trepo, and L. Cova. 1993. Fine mapping of neutralization epitopes on duck hepatitis B virus (DHBV) pre-S protein using monoclonal antibodies and overlapping peptides. Virology 192:217-223. [DOI] [PubMed] [Google Scholar]
- 4.Chassot, S., V. Lambert, A. Kay, C. Godinot, C. Trepo, and L. Cova. 1994. Identification of major antigenic domains of duck hepatitis B virus pre-S protein by peptide scanning. Virology 200:72-78. [DOI] [PubMed] [Google Scholar]
- 5.Combet, C., M. Jambon, G. Deleage, and C. Geourjon. 2002. Geno3D: automatic comparative molecular modeling of protein. Bioinformatics 18:213-214. [DOI] [PubMed] [Google Scholar]
- 6.Geourjon, C., and G. Deleage. 1995. ANTHEPROT 2.0: a three-dimensional module fully coupled with protein sequence analysis methods. J. Mol. Graph. 13:199-212. [DOI] [PubMed] [Google Scholar]
- 7.Geysen, H. M., R. H. Meloen, and S. J. Barteling. 1984. Use of peptide synthesis to probe viral antigens for epitopes to a resolution of a single amino acid. Proc. Natl. Acad. Sci. USA 81:3998-4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Geysen, H. M., S. J. Rodda, T. J. Mason, G. Tribbick, and P. G. Schoofs. 1987. Strategies for epitope analysis using peptide synthesis. J. Immunol. Methods 102:259-274. [DOI] [PubMed] [Google Scholar]
- 9.Hinkula, J., C. Svanholm, S. Schwartz, P. Lundholm, M. Brytting, G. Engstrom, R. Benthin, H. Glaser, G. Sutter, B. Kohleisen, V. Erfle, K. Okuda, H. Wigzell, and B. Wahren. 1997. Recognition of prominent viral epitopes induced by immunization with human immunodeficiency virus type 1 regulatory genes. J. Virol. 71:5528-5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jilbert, A. R., and I. Kotlarski. 2000. Immune responses to duck hepatitis B virus infection. Dev. Comp. Immunol. 24:285-302. [DOI] [PubMed] [Google Scholar]
- 11.Kenney, J. M., C. H. von Bonsdorff, M. Nassal, and S. D. Fuller. 1995. Evolutionary conservation in the hepatitis B virus core structure: comparison of human and duck cores. Structure 3:1009-1019. [DOI] [PubMed] [Google Scholar]
- 12.Kuhober, A., H. P. Pudollek, K. Reifenberg, F. V. Chisari, H. J. Schlicht, J. Reimann, and R. Schirmbeck. 1996. DNA immunization induces antibody and cytotoxic T-cell responses to hepatitis B core antigen in H-2b mice. J. Immunol. 156:3687-3695. [PubMed] [Google Scholar]
- 13.Lai, W. C., and M. Bennett. 1998. DNA vaccines. Crit. Rev. Immunol. 18:449-484. [DOI] [PubMed] [Google Scholar]
- 14.Lambert, V., S. Chassot, A. Kay, C. Trépo, and L. Cova. 1991. In vivo neutralization of duck hepatitis B virus by antibodies specific to the N-terminal portion of pre-S protein. Virology 185:446-450. [DOI] [PubMed] [Google Scholar]
- 15.Le Guerhier, F., A. Thermet, S. Guerret, M. Chevallier, C. Jamard, C. S. Gibbs, C. Trepo, L. Cova, and F. Zoulim. 2003. Antiviral effect of adefovir in combination with a DNA vaccine in the duck hepatitis B virus infection model. J. Hepatol. 38:328-334. [DOI] [PubMed] [Google Scholar]
- 16.Lu, M., G. Hilken, J. Kruppenbacher, T. Kemper, R. Schirmbeck, J. Reimann, and M. Roggendorf. 1999. Immunization of woodchucks with plasmids expressing woodchuck hepatitis virus (WHV) core antigen and surface antigen suppresses WHV infection. J. Virol. 73:281-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mancini, M., M. Hadchouel, H. L. Davis, R. G. Whalen, P. Tiollais, and M. L. Michel. 1996. DNA-mediated immunization in a transgenic mouse model of the hepatitis B surface antigen chronic carrier state. Proc. Natl. Acad. Sci. USA 93:12496-12501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mandart, E., A. Kay, and F. Galibert. 1984. Nucleotide sequence of a cloned duck hepatitis B virus genome: comparison with woodchuck and human hepatitis B virus sequences. J. Virol. 49:782-792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Michel, M. L., and D. Loirat. 2001. DNA vaccines for prophylactic or therapeutic immunization against hepatitis B. Intervirology 44:78-87. [DOI] [PubMed] [Google Scholar]
- 20.Pugh, J., A. Zweidler, and J. Summers. 1989. Characterization of the major duck hepatitis B virus core particle protein. J. Virol. 63:1371-1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rollier, C., C. Charollois, C. Jamard, C. Trepo, and L. Cova. 2000. Maternally transferred antibodies from DNA-immunized avians protect offspring against hepadnavirus infection. J. Virol. 74:4908-4911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rollier, C., C. Sunyach, L. Barraud, N. Madani, C. Jamard, C. Trepo, and L. Cova. 1999. Protective and therapeutic effect of DNA-based immunization against hepadnavirus large envelope protein. Gastroenterology 116:658-665. [DOI] [PubMed] [Google Scholar]
- 23.Sallberg, M., U. Ruden, L. O. Magnius, H. P. Harthus, M. Noah, and B. Wahren. 1991. Characterisation of a linear binding site for a monoclonal antibody to hepatitis B core antigen. J. Med. Virol. 33:248-252. [DOI] [PubMed] [Google Scholar]
- 24.Sallberg, M., U. Ruden, B. Wahren, and L. O. Magnius. 1994. Immune recognition of linear antigenic regions within the hepatitis B pre-C and C-gene translation products using synthetic peptides. J. Med. Virol. 42:7-15. [DOI] [PubMed] [Google Scholar]
- 25.Schirmbeck, R., X. Zheng, M. Roggendorf, M. Geissler, F. V. Chisari, J. Reimann, and M. Lu. 2001. Targeting murine immune responses to selected T cell- or antibody-defined determinants of the hepatitis B surface antigen by plasmid DNA vaccines encoding chimeric antigen. J. Immunol. 166:1405-1413. [DOI] [PubMed] [Google Scholar]
- 26.Siegel, F., M. Lu, and M. Roggendorf. 2001. Coadministration of gamma interferon with DNA vaccine expressing woodchuck hepatitis virus (WHV) core antigen enhances the specific immune response and protects against WHV infection. J. Virol. 75:5036-5042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Srivastava, I. K., and M. A. Liu. 2003. Gene vaccines. Ann. Intern. Med. 138:550-559. [DOI] [PubMed] [Google Scholar]
- 28.Sunyach, C., C. Rollier, M. Robaczewska, C. Borel, L. Barraud, A. Kay, C. Trepo, H. Will, and L. Cova. 1999. Residues critical for duck hepatitis B virus neutralization are involved in host cell interaction. J. Virol. 73:2569-2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tahtinen, M., A. Ranki, S. L. Valle, V. Ovod, and K. Krohn. 1997. B-cell epitopes in HIV-1 Tat and Rev proteins colocalize with T-cell epitopes and with functional domains. Biomed. Pharmacother. 51:480-487. [DOI] [PubMed] [Google Scholar]
- 30.Thermet, A., C. Rollier, F. Zoulim, C. Trepo, and L. Cova. 2003. Progress in DNA vaccine for prophylaxis and therapy of hepatitis B. Vaccine 21:659-662. [DOI] [PubMed] [Google Scholar]
- 31.Triyatni, M., A. R. Jilbert, M. Qiao, D. S. Miller, and C. J. Burrell. 1998. Protective efficacy of DNA vaccines against duck hepatitis B virus infection. J. Virol. 72:84-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang, L., J. G. Menting, C. G. Black, A. Stowers, D. C. Kaslow, S. L. Hoffman, and R. L. Coppel. 2000. Differences in epitope recognition, isotype and titer of antisera to Plasmodium falciparum merozoite surface protein 4 raised by different modes of DNA or protein immunization. Vaccine 19:816-824. [DOI] [PubMed] [Google Scholar]
- 33.Wynne, S. A., R. A. Crowther, and A. G. Leslie. 1999. The crystal structure of the human hepatitis B virus capsid. Mol. Cell 3:771-780. [DOI] [PubMed] [Google Scholar]
- 34.Yang, W., J. Guo, Z. Ying, S. Hua, W. Dong, and H. Chen. 1994. Capsid assembly and involved function analysis of twelve core protein mutants of duck hepatitis B virus. J. Virol. 68:338-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yuasa, S., R. C. Cheung, Q. Pham, W. S. Robinson, and P. L. Marion. 1991. Peptide mapping of neutralizing and nonneutralizing epitopes of duck hepatitis B virus pre-S polypeptide. Virology 181:14-21. [DOI] [PubMed] [Google Scholar]
- 36.Zheng, J., F. Schodel, and D. L. Peterson. 1992. The structure of hepadnaviral core antigens. Identification of free thiols and determination of the disulfide bonding pattern. J. Biol. Chem. 267:9422-9429. [PubMed] [Google Scholar]