Abstract
Supernumerary chromosomes (B chromosomes) occur in approximately 15% of eukaryote species. Although these chromosomes have been extensively studied, knowledge concerning their specific molecular composition is lacking in most cases. The accumulation of repetitive DNAs is one remarkable characteristic of B chromosomes, and the occurrence of distinct types of multigene families, satellite DNAs and some transposable elements have been reported. Here, we describe the organization of repetitive DNAs in the A complement and B chromosome system in the grasshopper species Abracris flavolineata using classical cytogenetic techniques and FISH analysis using probes for five multigene families, telomeric repeats and repetitive C0t-1 DNA fractions. The 18S rRNA and H3 histone multigene families are highly variable and well distributed in A. flavolineata chromosomes, which contrasts with the conservation of U snRNA genes and less variable distribution of 5S rDNA sequences. The H3 histone gene was an extensively distributed with clusters occurring in all chromosomes. Repetitive DNAs were concentrated in C-positive regions, including the pericentromeric region and small chromosomal arms, with some occurrence in C-negative regions, but abundance was low in the B chromosome. Finally, the first demonstration of the U2 snRNA gene in B chromosomes in A. flavolineata may shed light on its possible origin. These results provide new information regarding chromosomal variability for repetitive DNAs in grasshoppers and the specific molecular composition of B chromosomes.
Introduction
Repetitive DNAs comprise a large portion of eukaryotic genomes, including tandem arrays and scattered repeats. Tandem repeats are represented by microsatellite, minisatellite, and satellite DNAs as well as some multigene families, while dispersed repeats are comprised of transposons and retrotransposons [1]–[3]. Among the multigene families, the ribosomal DNAs (rDNAs), followed by histone genes and to a lesser extent U small nuclear RNA (snRNA) genes, have been mapped cytogenetically, revealing clusters located in one chromosomal loci or dispersed in some chromosomes (see for example [4]–[11]). The spreading of these sequences has been attributed to transposition and ectopic recombination, as well as the possible involvement of extra chromosomal circular DNAs (eccDNA), which have been detected in Drosophila, Arabdopsis thaliana and human (see for example [4], [6]–[8], [12]–[19]).
In grasshoppers, the mapping of multigene families for rDNAs and histone genes has identified distinct patterns of chromosomal distributions. In particular the H3/H4 histone clusters are highly conserved in one chromosomal pair, and the rDNAs are variable due to amplification and dispersion of the clusters in some chromosomes. In some cases these sequences are co-located in the same chromosomal area [4], [5], [8]. Moreover the presence of multigene families in B chromosomes has been reported in some species (see references below).
The B chromosomes, also known as accessory or supernumerary elements, are dispensable chromosomes not required for normal organismal development, constituting a type of selfish DNA element [20], [21]. Since their discovery by Wilson [22], distinct B chromosomes have been described in all eukaryotic groups and occur in approximately 15% of species cytogenetically investigated [20]. Primary characteristics of B chromosomes include their remarkable accumulation due to irregular modes of inheritance; pairing incapacity during meiosis with standard A chromosomes; and accumulation of distinct repetitive DNAs, leading to species-specific evolutionary fates [20], [21], [23]–[25].
Concerning molecular composition of animal B chromosomes, among repetitive sequences the presence of satellite repeats, transposable elements and multigene families, mainly 45S rDNA, have been described [20]. In grasshoppers, the presence of satellite DNA and multigene families such 45S and 5S rDNA and H3/H4 histone genes have been described in distinct species such as Dichroplus pratensis [26], Eyprepocnemis plorans [27], Locusta migratoria [28], and Rhammatocerus brasiliensis [29], [30]. These markers shed light on the possible origin and evolutionary differentiation of B chromosomes in these taxa [28]–[31]. Although Bs have been frequently investigated, knowledge regarding their origin and specific molecular composition is limited.
Eyprepocnemis plorans and L. migratoria are the two model species most commonly used to study B chromosome biology in grasshoppers and animals. Over the years, information has been accumulated in these species regarding B chromosome population dynamics, their possible origin, B chromosome gene activity and the interference in the expression of A complement genes due its presence using distinct cytogenetic and molecular approaches [28], [31]–[39]. In contrast, knowledge regarding molecular composition obtained in other grasshopper species such as D. pratensis [26], Podisma kanoi [40] and R. brasiliensis [29], [30] remains limited.
In an attempt to further understand karyotypes and B chromosome composition and evolution in grasshoppers, we analyzed the karyotypic structure and B chromosomes of the South American grasshopper species Abracris flavolineata (Acrididae, Ommatolampinae). This species exhibits a karyotype composed of 2n = 23,X0 (males) with a distinct biarmed B chromosome in the population of Rio Claro/SP, Brazil [41]. Specifically, general chromosomal characteristics and B chromosome frequency and structure were studied using classical cytogenetic techniques and mapping of multigene families, telomeric repeats and repeated DNA fraction (C0t-1 DNA fraction). Our analyses provide new information regarding chromosomal variability in grasshoppers and the specific molecular composition of B chromosomes.
Materials and Methods
A total of 65 A. flavolineata adult individuals, including 38 males and 27 females, were collected in Rio Claro/SP, Brazil with the authorization of ICMBio SISBIO (process number 16009-1). The animals were anesthetized before dissecting testis follicles and gastric caeca. Chromosomes were obtained from male testis follicles and female gastric caeca, which contain mitotic chromosomes, according to the procedure described by Castillo et al. [42]. The tissues used to obtain chromosomes were fixed in modified Carnoy's solution (3∶1, 100% ethanol:glacial acetic acid), and entire animals were stored in 100% ethanol for DNA extraction.
All individuals were studied using conventional staining with 5% Giemsa to estimate B chromosome presence and frequency. The C-banding was obtained as described by Sumner [43], and to identify G+C or A+T rich regions fluorochrome staining (CMA3/DA/DAPI) was performed as proposed by Schweizer et al. [44].
Genomic DNA was extracted from the posterior legs of animals with and without one B chromosome using phenol-chlorophorm procedure as proposed by Sambrook and Russel [45]. The DNA was used as template to obtain distinct multigene families such as 5S rDNA, H3 histone and U2 small nuclear RNA (snRNA) genes by polymerase chain reaction (PCR) using primers described by Cabral-de-Mello et al. [46] for 5S rDNA, and by Colgan et al. [47] for H3 histone. Moreover, a specific pair of primers designated by sequences deposited in GenBank as U2F (5′-ATC GCT TCT CGG CCT TAT G–3′) and U2R (5′-TCC CGG CGG TAC TGC AAT A-3′) were used for U2 snDNA amplification. The U1 snDNA sequence was obtained from the R. brasiliensis genome using primers described by Cabral-de-Mello et al. [9], and 18S rDNA was obtained from cloned fragments of the Dichotomius semisquamosus (GenBank accession number GQ443313.1) genome [46]. All sequences obtained by PCR were sequenced to confirm the sequence of interest, and were deposited in GenBank under the accession numbers KC936996-5S rDNA, KC896792-H3, KC896793-U1 snDNA and KC896794-U2 snDNA. A BLAST search using these sequences confirmed the isolation of the elements that were used as probes. Telomeric probes were obtained by PCR using complementary primers (TTAGG)5 and (CCTAA)5. Repetitive DNA-enriched samples from individuals with and without one B chromosome were obtained based on the renaturation kinetics of C0t-1 DNA (DNA enriched for highly and moderately repetitive DNA sequences) according to the protocol described by Zwick et al. [48] with modifications later published [46]. DNA samples (200 µl of 100–500 ng/µl genomic DNA in 0.3 M NaCl) were digested with 0.01 U/µl Deoxyribonuclease I (Sigma) for 1 min and 10 sec to 1 min and 45 sec depending on the sample concentration, and the fragmented DNA was separated by 1% agarose gel electrophoresis. The expected DNA fragments ranged in size from 100 to 1,000 base pairs (bp). DNA fragment samples (50 µl) were denatured at 95°C for 10 min, placed on ice for 10 s and transferred into a 65°C water bath for reannealing for 25 min. Subsequently, the samples were incubated at 37°C for 8 min with 1 U of S1 nuclease to permit the digestion of single-stranded DNA, which was then purified/extracted using a traditional phenol–chlorophorm procedure.
The 5S rDNA, telomeric probes and U1 and U2 snRNA gene probes were labeled through PCR with digoxigenin-11-dUTP (Roche, Mannheim, Germany), and the 18S rDNA, H3 histone gene and C0t-1 DNA were labeled using biotin-14-dATP through nick translation (Invitrogen, San Diego, CA, USA). Fluorescent in situ hybridization (FISH) was performed according to the protocol proposed by Pinkel et al. [49] with modifications described by Cabral-de-Mello et al. [46]. Single or two color FISH were performed with the distinct probes and at least 200 ng of each probe was used. Probes labeled with digoxigenin-11-dUTP were detected using anti-digoxigenin rhodamine (Roche), and probes labeled with biotin-14-dATP were detected using Alexa Fluor 488 (Invitrogen). All preparations were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) and mounted in Vectashield (Vector, Burlingame, CA, USA). Chromosomes and signals were observed using an Olympus microscope BX61 equipped with fluorescence lamp and appropriate filters. Photographs were recorded with a DP70 cooled digital camera. Images were merged and optimized for brightness and contrast with Adobe Photoshop CS2.
For the measurement of relative extension occupied by repetitive DNAs (i.e. C-positive blocks, 18S rDNA, H3 histone and C0t-1 DNA) the software ImageJ was used. We comparatively analyzed the extension occupied by the C-positive blocks or FISH signals with repetitive DNAs in relation of the whole chromosomal extension in six metaphases. The analyses were performed using mitotic chromosomes from individuals that presented the rDNA distribution showed in the last row of table 1, considering only the chromosomes harboring these clusters.
Table 1. Intrapopulational polymorphisms of 18S rDNA inA. flavolineata.
| Figure | Chromosomes | Total of elements labeled | Frequency observed | |||||||||||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | X | |||
| S1a | p | p | p* | p | p | p# | p | p | p | 9 | 1 | |||
| S1b | p | p | p* | p | p | p# | p | p | 8 | 1 | ||||
| S1c | p | p | p | p | p | p | 6 | 2 | ||||||
| S1d | p | p* | p | p* | p | p | p | 7 | 2 | |||||
| S1e | p | p* | p | p | p | p | 6 | 1 | ||||||
| S1f | p | p | p | p | p | 5 | 1 | |||||||
| Fig. 2a | p | p | p | p | p | 5 | 1 | |||||||
p: pericentromeric;
heteromorphic pair;
tiny signal.
Results
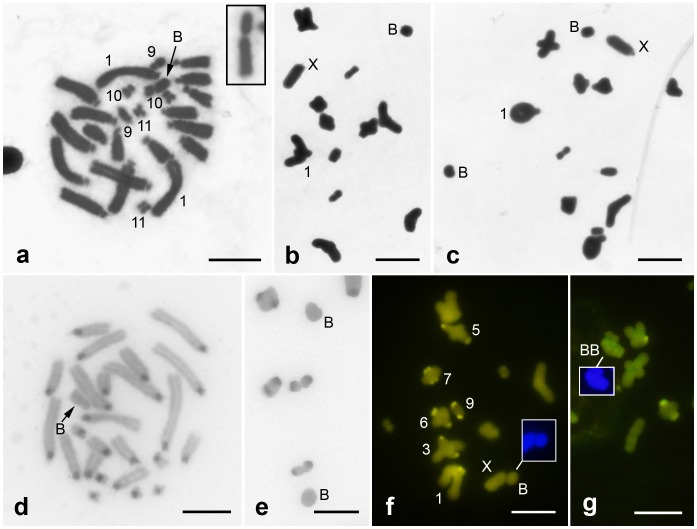
The A. flavolineata karyotype is composed of 2n = 23,X0 (males) and 2n = 24,XX (females). Pairs 1–8 and the X are subtelo-acrocentric, pair 9 is submetacentric, and pairs 10 and 11 are metacentric (Figure 1a). This macro chromosomal structure is similar to that previously reported by Cella and Ferreira [41]. Among the 65 animals studied, 30.7% (12 males and 8 females) carried one or two submetacentric B chromosomes (Figure 1a–c). The frequency of B chromosomes was similar in males (31.5%) and females (29.7%). In males, the B chromosome showed mitotic instability in the germ line since primary spermatocytes with 0–1 or 0–2 B elements were observed in eight individuals (Figure 1b,c).
Figure 1. Classical cytogenetic characterization ofAbracris flavolineata chromosomal complement at mitotic metaphase (a,d) and metaphase I (b,c,e–g) cells.
(a–c) conventional staining, (d,e) C-banding and (f,g) CMA3 fluorochrome staining in cells with one or two B chromosomes. Insets show the B element obtained from another mitotic metaphase (a), and B chromosomes stained with DAPI (f,g). The B and X chromosomes are indicated in all cells, and (f) chromosomes with CMA3-positive blocks are also indicated. The metaphases (e,g) are partial. Bar = 5 µm.
C-positive chromosome regions were concentrated in the pericentromeric region extending to the short chromosomal arms of all chromosomes, while in the B chromosomes no C-positive regions were observed (Figure 1d,e). The heterochromatin blocks of pairs 1, 3, 5, 6, 7 and 9 were G+C-rich, while the other regions were neutral for both fluorochromes (CMA3/DAPI) (Figure 1f). DAPI staining did not reveal any positive blocks, as the chromosomes were homogeneously stained (result not shown). No A+T- or G+C-rich blocks were observed in the B chromosomes (Figure 1f,g).
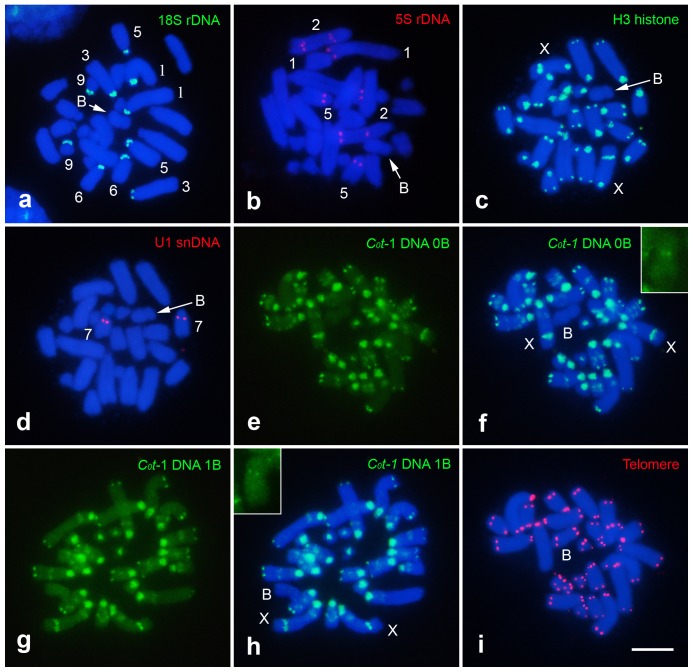
In the nine animals analyzed for FISH with the 18S rDNA probe, we observed extensive variation between individuals in the number of chromosomes (5–9) harboring this kind of repetitive DNA, including autosomes and X chromosomes. In the B chromosome no signal for 18S rDNA was observed. All rDNA blocks were located in the short arm of carrier chromosomes (Figure 2a, Table 1, Figure S1), and heteromorphic pairs were observed in some cases (see Figure S1). Table 1 shows that i) rDNA was found in all A chromosomes except 7 and 11, ii) there was no variation in autosomes 1, 3, 6 and 9, and iii) it was polymorphic in the remaining A chromosomes (2, 4, 5, 8, 10 and X). The 5S rDNA location, however, was highly conserved since all individuals carried a distal cluster in the long arm of autosome 1, two interstitial clusters in autosome 2, and one proximal cluster in autosome 5 (Figure 2b).
Figure 2. Cytogenetic mapping of repetitive DNAs in gastric caeca female mitotic cells bearing one B chromosome.
Each probe used is indicated directly in the images using colors. Insets in (f,h) show the B chromosome with a faint centromeric signal for the C0t-1 DNA probe. Bar = 5 µm.
Remarkably, H3 histone genes were found in all chromosomes, with large pericentromeric clusters in all chromosomes, except the B chromosome. The H3 histone pericentromeric clusters in some chromosomes were extended to the short arms and additional distal clusters in pairs 1–6 and 8, and a large interstitial cluster in the X chromosome was observed (Figure 2c).
U1 snRNA genes were located exclusively in the proximal region of autosome pair 7 (Figure 2d). FISH for highly and moderately repeated DNAs (C0t-1 DNA), with probes obtained from genomic DNA of 0B and 1B individuals, labeled large proximal regions (including short arms) in all A chromosomes, as well as small terminal blocks in all chromosomes but 7, 9–11, and a large interstitial block in the X chromosome. B chromosomes showed, with both C0t-1 probes, a very small hybridization signal in the centromere (Figure 2e–h). This signal was not observed in meiotic cells, most likely due to high chromosome condensation, which limits FISH resolution. FISH with the telomeric DNA probe revealed terminal signals in all chromosomes, including the B elements (Figure 2i). A comparative analysis, using mitotic chromosomes, of repetitive DNAs located near to centromeres (heterochromatin, 18S rDNA, H3 histone and C0t-1 DNA) revealed larger relative region, in size, occupied physically by C0t-1 DNA in comparison to other repeated DNAs (Figure S2, Table S1). All FISH analyses were also performed in individuals harboring two B chromosomes, and identical results described for individuals with one B were observed (Figure S3).
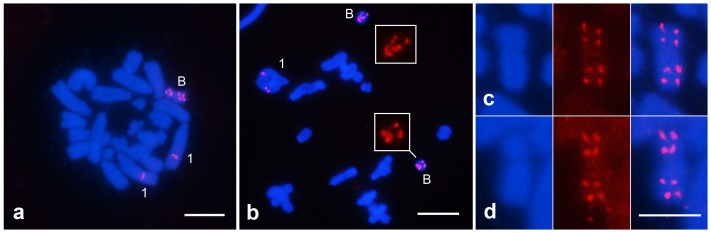
Finally, FISH with the U2 snDNA probe revealed the presence of a single cluster in the A chromosomes, specifically in an interstitial region of the largest autosome. Additionally, in the B chromosome the U2 snDNA presented high copy number with four blocks in this element, two in each arm in a nearly symmetrical location (Figure 3).
Figure 3. FISH with the U2 snRNA gene probe in individuals with one (a) and two (b) B chromosomes.
(a) gastric caeca female mitotic metaphase cell, (b) male metaphase I cell, (c,d) selected B chromosomes showing the symmetrical location of the U2 snDNA clusters in the two arms. Note the double signal in each arm of the B elements. (a,b) Bar = 5 µm, (c,d) Bar = 2.5 µm.
Discussion
The A complement and dynamics of repetitive DNAs in theA. flavolineata genome
The karyotype and heterochromatin distribution observed in A. flavolineata is similar to the previous description provided by Cella and Ferreira [41]. These results mirror the common pattern seen in grasshoppers [50], with the exception of the presence of meta- or submetacentric chromosomes (pairs 9–11), which are less common in acridid karyotypes and may be related to pericentric inversions. The absence of interstitial telomeric DNA indicates that either the breakpoints for chromosomal inversions were located outside of telomeric regions or that these repeats were lost after the inversions took place. Alternatively accumulation of repetitive DNAs in short arms could explain the occurrence of meta-/submetacentric chromosomes [41].
The variability observed in base pair composition for C-positive regions may indicate the dynamics related to their specific composition. The C0t-1 DNA fraction provided additional information concerning repetitive DNAs distribution, revealing that in addition to the high amount of this genome fraction in C-positive regions, repetitive DNAs are also enriched in the terminal and interstitial regions. Additionally, it is noticeable that the regions occupied by C0t-1 DNA signals were larger in physical size if compared to other repetitive DNAs, including C-positive blocks, H3 histone genes and 18S rDNA, indicating that the pericentromeric regions harbor other repetitive elements. In general, the mapping of C0t-1 DNA in animals has been a valuable tool in the understanding of repetitive DNA diversification regarding heterochromatin [51], possible sex chromosomes [52] and the origin/evolution of B chromosomes [46], [53] such as the B of A. flavolineata (see discussion below).
Chromosomal mapping of the five multigene families in A. flavolineata revealed distinct genome dynamics among the sequences studied. This intense difference was observed by contrasting the patterns observed for stable U snRNA genes with only one site, and H3 histone and 18S rRNA genes that are spread and are additionally polymorphic for 18S rDNA. Although the mapping of U1 and U2 has been poorly explored, the cytogenetic mapping of U1 snRNA genes was reported as being highly conserved in cichlid fish and crustaceans, with only one site in the distantly related cichlid fish and more dynamic sites in crustaceans [9], [54], [55]. For U2 snDNA, the occurrence of only one site as observed in A. flavolineata was previously described in fish, such as Halobatrachus didactylus and Plectorhinchus mediterraneus. However, interestingly distinct scenarios were observed in Batrachoididae fish with sites concentrated in one chromosomal pair, sites dispersed in some chromosomes and both organizations in the same genome [56], [57].
The marked genomic dispersal of H3 histone clusters observed in the A. flavolineata genome was also reported in the grasshopper R. brasiliensis [30], although without the occurrence of terminal and interstitial clusters. This surprising chromosomal distribution in A. flavolineata contrasts strongly with the conserved pattern observed in other animals, which possess only a single locus or few loci of histone genes [5], [7], [58]–[60]. Cabrero et al. [5], claimed that, in grasshoppers, the conservation of a number of H3-H4 histone genes dates to the origin of Acrididae, which is reinforced by the analysis of H3 histone clusters in ancient grasshoppers belonging to the Proscopiidae family [60]. The spreading of histone H3 repeats observed in this work suggests greater dynamism of these repeats in grasshoppers. Although deeper analysis is necessary, the role of transposable elements (TE) in histone H3 dispersion must be considered. Furthermore, other mechanisms of repetitive DNA dispersal across the genome may also govern the intense sequence dispersion observed here, such as extrachromosomal circular DNAs (eccDNA), and ectopic recombination, as described for rDNAs [4], [14], [15], [17], [19].
Although less variable than histone H3 genes, 5S rDNA was present at multiple loci, which is a common placement for grasshoppers [8]. Concerning the major rDNA cluster, a more detailed scenario has been described in insects, and high variability has been primarily described at the interspecific level [4], [6], [7], [61]. Variability at the intraspecific level as observed in A. flavolineata is less common and was observed in E. plorans, Orthoptera [62], in some Scarabaeinae beetles, Coleoptera [7] and in Triatoma infestans, Heteroptera [61]. These variations in insects with major rDNA clusters have been frequently attributed to ectopic recombination, transpositions, translocations, structural rearrangements and gene conversion followed by amplifications [4], [6], [7], [61].
The B chromosome
The presence of a B chromosome in A. flavolineata was described for the first time by Cella and Ferreira [41] in the same population analyzed in this study, but without frequency description. The main difference observed in this work with the previous description is the morphology of the B element, which was previously classified as metacentric and in our analysis, it was submetacentric.
The absence of C-positive regions in the B chromosome initially indicated the possible low quantity of repeated DNAs. Additionally, the use of the repetitive DNA fraction (C0t-1 DNA) obtained from genomes with or without one B chromosome as probes indicated a possible low copy number of B-specific repetitive DNAs, such as satellites, or that this element possesses a high number of different repetitive sequences not represented in the C0t-1 DNA fraction. Our results indicate that in addition to the non-accumulation of some repetitive DNAs in the B chromosome, this element does not share the general pool of repetitive DNAs with the A genome, except for less repeated elements such as U2 snRNA, and some transposable elements (unpublished results), possibly not represented in the C0t-1 DNA fraction isolated, besides by the occurrence of a faint and punctual signal in the B centromere. This pattern suggests the non-homogenization of the A complement and B chromosomes in A. flavolineata and it could indicates a recent origin of this chromosome due to the non-accumulation of repetitive DNAs. This non-accumulation of repetitive DNAs in the B chromosome is contrary to a common pattern of B evolution, with accumulation of repetitive DNAs in this element as observed in some species [21], [24], [25]. Similarly, a lack of DNA identity between A and B chromosomes was described in the fish Prochilodus lineatus [63], which contrasts with some common cases of A and B chromosome DNA sharing in animals such as in E. plorans [33], L. migratoria [34], Podisma kanoi [40], P. sapporensis [64], Dichotomius geminatus [46], Astyanax scabripinis [53], Vulpes vulpes [65] and Apodemus peninsulae [66].
Chromosomal mapping of multigene families provided interesting information regarding the genome dynamics of A. flavolineata and the possible autosomal B origin/diversification in this species. The remarkable presence of U2 snRNA genes in the B element is reasonable evidence for its ancestry to the autosomal pair 1, the unique element that harbors a large cluster of this sequence observed by FISH. Autosomal origins for B chromosomes in grasshoppers were also proposed for example in Dichroplus pratensis [26], R. brasiliensis, Xyleus discoideus angulatus [29] and L. migratoria [28] using repetitive DNA mapping.
The presence of other multigene families were described in distinct B chromosomes in grasshoppers, such as 45S rDNA [26], [27], 5S rDNA [30], [31] and H3/H4 histone genes [28], [30]; however, the case of A. flavolineata is the first demonstration of the occurrence of the U2 snRNA gene in B chromosomes among eukaryotes. In contrast to U2 snDNA, the other multigene families used as probes in A. flavolineata, i.e., 18S rRNA and 5S rRNA, H3 histone and U1 snRNA genes, revealed the absence of signals in the B chromosome. The specific cases of H3 histone and 18S rRNA genes are interesting, although highly variable in the number and position for 18S rDNA, and the intense dispersion of H3 histone these sequences were not transposed to the B chromosomes. The dispersion of H3 histone genes could have occurred after the origin of the B chromosome in the genome of A. flavolineata. These aspects reinforce the hypothesis of B origin from pair 1 (with U2 snDNA), which is apparently one sequence without high transposition dynamics in the A. flavolineata genome, considering the occurrence of a single locus. We could not, however, definitively rule out the possibility of a transposition of U2 snDNA sequences to the B element after its origin followed by amplification.
Additionally, the similar distribution of U2 snDNA clusters in the two arms of the B chromosome led us to hypothesize the origin of this chromosome based on isochromosomes, followed by the enlargement of one chromosomal arm or the occurrence of a pericentric inversion, which causes the difference in the size of the arms. The origin of the B chromosome through isochromosomes was observed, for example in the fish Astyanax scabripinis [67] and Prochilodus lineatus [68], other grasshoppers [69]–[73], and plants such as Brachycome dichromosomatica [25] and rye Secale cereale [74].
The classical cytogenetic methods and the mapping of repeated DNAs in individuals harboring two B chromosomes indicated similarity between these elements, suggesting that only one type of B chromosome is present in the population studied. This observation contrasts with the high variability of the molecular composition or the sequence distribution in B elements, as observed in E. plorans [31], [75] and P. lineatus [68]. The variability of B elements has been attributed to the differential amplification/deletion of DNA sequences in addition to the involvement of translocations/transpositions of sequences from the A genome and organellar DNA [20], [25], [53], [76], [77]. Although the B chromosome of A. flavolineata did not present variability, we could not rule out the occurrence of the mechanism cited above, intrinsic for B evolution, and certainly the analysis of other sequences will shed light on this issue.
The data presented provide new information regarding chromosomal evolution of repetitive sequences in grasshoppers and revealed intense dynamics for 18S rDNA and H3 histone genes in A. flavolineata, indicating, in the case of H3 histone genes, that this sequence could be more dynamic than previously reported in Acrididae grasshopper genomes [5]. U2 snDNA can be used as an interesting marker to investigate B chromosome origin/evolution by providing new information concerning B chromosome composition in eukaryotes. The use of this marker in the case of A. flavolineata highlighted the autosomal origin and conservation of the B element, at least in the Rio Claro/SP population. Finally, the analysis of other populations conducted using the U2 snRNA gene will provide information regarding the origin and evolution of this polymorphism in this species, as well as the use of other chromosomal markers through FISH.
Supporting Information
Meiotic cells from distinct individuals of A. flavolineata showing the variable patterns of 18S rDNA distribution. Autosomal bivalents were numbered in order of decreasing size. Bar = 5 µm.
(PDF)
Selected mitotic chromosomes of A. flavolineata after C-banding treatment and FISH with 18S rDNA, H3 histone and C0t -1 DNA as probes. Note the occurrence of large C0t-1 DNA blocks. Blue = DAPI, Green = signals.
(PDF)
Partial metaphases I of A. flavolineata individuals harboring two B chromosomes. The probes used are indicated in colors directly in each cell. Bar = 5 µm.
(PDF)
Relative length occupied by repetitive DNAs in chromosomes 1, 3, 5, 6 and 9 of A. flavolineata . Note that in all chromosomes the region occupied by C0t-1 DNA is larger than for other repetitive DNAs.
(PDF)
Acknowledgments
The authors are grateful to Dr. Juan Pedro M. Camacho for his valuable suggestions and critical review of a previous version of this manuscript and to Dallas Krentzel for English corrections.
Funding Statement
This study was supported by Fundação de Amparo a Pesquisa do Estado de São Paulo-FAPESP (2011/19481-3, 2011/18028-3), Coordenadoria de Aperfeiçoamento de Pessoal de Nível Superior-CAPES and Conselho Nacional de Desenvolvimento Científico e Tecnológico-CNPq (475308/2011-5). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Long EO, Dawid IB (1980) Alternative pathway in processing of ribosomal RNA precursor in Drosophila melanogaster . J Mol Biol 138: 373–378. [DOI] [PubMed] [Google Scholar]
- 2. Charlesworth B, Sniegowski P, Stephan W (1994) The evolutionary dynamics of repetitive DNA in eukaryotes. Nature 371: 215–220. [DOI] [PubMed] [Google Scholar]
- 3. Biémont C, Vieira C (2006) Genetics: Junk DNA as an evolutionary force. Nature 443: 521–524. [DOI] [PubMed] [Google Scholar]
- 4. Cabrero J, Camacho JP (2008) Location and expression of ribosomal RNA genes in grasshoppers: Abundance of silent and cryptic loci. Chromosome Res 16: 595–607. [DOI] [PubMed] [Google Scholar]
- 5. Cabrero J, López-León MD, Teruel M, Camacho JPM (2009) Chromossome mapping of H3 and H4 histone gene clusters in 35 species of acridid grasshoppers. Chromosome Res 17: 397–404. [DOI] [PubMed] [Google Scholar]
- 6. Nguyen P, Sahara K, Yoshido A, Marec F (2010) Evolutionary dynamics of rDNA clusters on chromosomes of moths and butterflies (Lepidoptera). Genetica 138: 343–354. [DOI] [PubMed] [Google Scholar]
- 7. Cabral-de-Mello DC, Oliveira SG, Moura RC, Martins C (2011) Chromosomal organization of the 18S and 5S rRNAs and histone H3 genes in Scarabaeinae coleopterans: insights into the evolutionary dynamics of multigene families and heterochromatin. BMC Genetics 12: 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cabral-de-Mello DC, Cabrero J, López-León MD, Camacho JPM (2011) Evolutionary dynamics of 5S rDNA location in acridid grasshoppers and its relationship with H3 histone gene and 45S rDNA location. Genetica 139: 921–931. [DOI] [PubMed] [Google Scholar]
- 9. Cabral-de-Mello DC, Valente GT, Nakajima RT, Martins C (2012) Genomic organization and comparative chromosome mapping of the U1 snRNA gene in cichlid fish, with an emphasis in Oreochromis niloticus . Chromosome Res 20: 279–292. [DOI] [PubMed] [Google Scholar]
- 10. Roa F, Guerra M (2012) Distribution of 45S rDNA sites in chromosomes of plants: Structural and evolutionary implications. BMC Evol Biol 12: 225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nakajima RT, Cabral-de-Mello DC, Valente GT, Venere PC, Martins C (2012) Evolutionary dynamics of rRNA gene clusters in cichlid fish. BMC Evol Biol 12: 198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schubert I (1984) Mobile nucleolus organizing regions (NORs) in Allium (Liliaceae S-Lat)-inferences from the specifity of silver staining. Plant Syst Evol 144: 291–305. [Google Scholar]
- 13. Hanson RE, Islam-Faridi MN, Percival EA, Crane CF, Ji Y, et al. (1996) Distribution of 5S and 18S–28S rDNA loci in a tetraploid cotton (Gossypium hirsutum L.) and its putative diploid ancestors. Chromosoma 105: 55–61. [DOI] [PubMed] [Google Scholar]
- 14. Raskina O, Belyayev A, Nevo E (2004) Activity of the En/Spm-like transposons in meiosis as a base for chromosome repatterning in a small, isolated, peripheral population of Aegilops speltoides Tausch. Chromosom Res 12: 153–161. [DOI] [PubMed] [Google Scholar]
- 15. Raskina O, Barber JC, Nevo E, Belyayev A (2008) Repetitive DNA and chromosomal rearrangements: speciation-related events in plant genomes. Cytogenet Genome Res 120: 351–357. [DOI] [PubMed] [Google Scholar]
- 16. Pedrosa-Harand A, Almeida CCS, Mosiolek M, Blair MW, Schweizer D, et al. (2006) Extensive ribosomal DNA amplification during Andean common vean (Phaseolus vulgaris L.) evolution. Theor Appl Genet 112: 924–933. [DOI] [PubMed] [Google Scholar]
- 17. Cohen S, Yacobi K, Segal D (2003) Extrachromosomal circular DNA of tandemly repeated genomic sequences in Drosophila. Genome Res 13: 1133–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cohen S, Houben A, Segal D (2008) Extrachromosomal circular DNA derived from tandemly repeated genomic sequences in plants. Plant J 53: 1027–1034. [DOI] [PubMed] [Google Scholar]
- 19. Cohen S, Agmon N, Sobol O, Segal D (2010) Extra chromosomal circles of satellite repeats and 5S ribosomal DNA in human cells. Mobile DNA 1: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Camacho JPM (2005) B chromosomes. In: The evolution of the genome. Gregory TR (ed): 223–286. [Google Scholar]
- 21. Jones RN, Gonzalez-Sanchez M, Gonzalez-Garcia M, Vega JM, Puertas MJ (2008) Chromosomes with a life of their own. Cytogenet Genome Res 120: 265–280. [DOI] [PubMed] [Google Scholar]
- 22. Wilson EB (1907) The supernumerary chromosomes of Hemiptera. Science 26: 870–871. [Google Scholar]
- 23. Jones RN (1995) B chromosomes in plants. New Phytol 131: 411–434. [DOI] [PubMed] [Google Scholar]
- 24. Camacho JPM, Sharbel TF, Beukeboom LW (2000) B chromosome evolution. Phil Trans R SocLond B 355: 163–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jones RN, Houben A (2003) B chromosomes in plants: Escapees from the A chromosome genome? Trends Plant Sci 8: 417–423. [DOI] [PubMed] [Google Scholar]
- 26. Bidau CJ, Rosato M, Martí DA (2004) FISH detection of ribosomal cistrons and assortment-distortion for X and B chromosomes in Dichroplus pratensis (Acrididae). Cytogenet Genome Res 106: 295–301. [DOI] [PubMed] [Google Scholar]
- 27. López-León MD, Neves N, Schwarzacher T, Heslop-Harrison JS, Hewitt GM, et al. (1994) Possible origin of a B chromosome deduced from its DNA composition using double FISH technique. Chromosome Res 2: 87–92. [DOI] [PubMed] [Google Scholar]
- 28. Teruel M, Cabrero J, Perfectti F, Camacho JP (2010) B chromosome ancestry revealed by histone genes in the migratory locust. Chromosoma 119: 217–225. [DOI] [PubMed] [Google Scholar]
- 29. Loreto V, Cabrero J, López-León MD, Camacho JPM, Souza MJ (2008) Possible autosomal origin of macro B chromosomes in two grasshopper species. Chromosome Res 16: 233–241. [DOI] [PubMed] [Google Scholar]
- 30. Oliveira NL, Cabral-de-Mello DC, Rocha MF, Loreto V, Martins C, et al. (2011) Chromosomal mapping of rDNAs and H3 histone sequences in the grasshopper Rhammatocerus brasiliensis (acrididae, gomphocerinae): extensive chromosomal dispersion and co-localization of 5S rDNA/H3 histone clusters in the A complement and B chromosome. Mol Cytogenet 4: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cabrero J, Bakkali M, Bugrov A, Warchalowska-Sliwa E, López-León MD, et al. (2003a) Multiregional origin of B chromosomes in the grasshopper Eyprepocnemis plorans . Chromosoma 112: 207–211. [DOI] [PubMed] [Google Scholar]
- 32. Cabrero J, Teruel M, Carmona FD, Jiménez R, Camacho JPM (2007) Histone H3 lysine 9 acetylation pattern suggests that X and B chromosomes are silenced during entire male meiosis in a grasshopper. Cytogenet Genome Res 119: 135–142. [DOI] [PubMed] [Google Scholar]
- 33. Teruel M, Cabrero J, Perfectti F, Acosta MJ, Sánchez A, et al. (2009a) Microdissection and chromosome painting of X and B chromosomes in the grasshopper Eyprepocnemis plorans. Cytogenet Genome Res 125: 286–291. [DOI] [PubMed] [Google Scholar]
- 34. Teruel M, Cabrero J, Montiel EE, Acosta MJ, Sánchez A, et al. (2009b) Microdissection and chromosome painting of X and B chromosomes in Locusta migratoria . Chromosome Res 17: 11–18. [DOI] [PubMed] [Google Scholar]
- 35. Teruel M, Cabrero J, Perfectti F, Camacho JPM (2009c) Quantitative analysis of NOR expression in a B chromosome of the grasshopper Eyprepocnemis plorans . Chromosoma 118: 291–301. [DOI] [PubMed] [Google Scholar]
- 36. Teruel M, Sørensen JG, Loeschcke V, Cabrero J, Perfectti F, et al. (2011) Level of Heat Shock Proteins Decreases in Individuals Carrying B-Chromosomes in the Grasshopper Eyprepocnemis plorans . Cytogenet Genome Res 132: 94–99. [DOI] [PubMed] [Google Scholar]
- 37. Muñoz-Pajares AJ, Martínez-Rodríguez L, Teruel M, Cabrero J, Camacho JPM, et al. (2011) A Single, Recent Origin of the Accessory B Chromosome of the Grasshopper Eyprepocnemis plorans . Genetics 187: 853–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ruiz-Ruano FJ, Ruiz-Estévez M, Rodríguez-Pérez J, López-Pino JL, Cabrero J, et al. (2011) DNA Amount of X and B Chromosomes in the Grasshoppers Eyprepocnemis plorans and Locusta migratoria . Cytogenet Genome Res 134: 120–126. [DOI] [PubMed] [Google Scholar]
- 39. Ruiz-Estévez M, López-León MD, Cabrero J, Camacho JPM (2012) B-Chromosome Ribosomal DNA Is Functional in the Grasshopper Eyprepocnemis plorans . PLOS ONE 7: e36600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bugrov AG, Karamysheva TV, Perepelov EA, Elisaphenko EA, Rubtsov DN, et al. (2007) DNA content of the B chromosomes in grasshopper Podisma kanoi Storozh. (Orthoptera, Acrididae). Chromosome Res 15: 315–325. [DOI] [PubMed] [Google Scholar]
- 41. Cella DM, Ferreira A (1991) The cytogenetics of Abracris flavolineata (Orthoptera, Caelifera, Ommatolampinae, Abracrini). Brazilian J Genet 14: 315–329. [Google Scholar]
- 42. Castillo ER, Taffarel A, Martí DA (2011) An alternative technique for mitotic grasshoppers karyotype: Fluorescent and C-banding in Adimantus ornatissimus (Orthoptera: Acrididae). Rev Cien Tecnol 16: 31–35. [Google Scholar]
- 43. Sumner AT (1972) A simple technique for demonstrating centromeric heterochromatin. Exp Cell Res 75: 304–306. [DOI] [PubMed] [Google Scholar]
- 44. Schweizer D, Mendelak M, White MJD, Contreras N (1983) Cytogenetics of the parthenogenetic grasshopper Warramaba virgo and its bisexual relatives. X. Pattern of fluorescent banding. Chromosoma 88: 227–236. [Google Scholar]
- 45.Sambrook J, Russel DW (2001) Molecular cloning. A laboratory manual, 3rd edn. Cold Spring Harbor Laboratory Press, New York. [Google Scholar]
- 46. Cabral-de-Mello DC, Moura RC, Martins C (2010) Chromosomal mapping of repetitive DNAs in the beetle Dichotomius geminatus provides the first evidence for an association of 5S rRNA and histone H3 genes in insects, and repetitive DNA similarity between the B chromosome and A complement. Heredity 104: 393–400. [DOI] [PubMed] [Google Scholar]
- 47. Colgan DJ, McLauchlan A, Wilson GDF, Livingston SP, Edgecombe GD, et al. (1998) Histone H3 and U2 snRNA DNA sequences and arthropod molecular evolution. Austral J Zool 46: 419–437. [Google Scholar]
- 48. Zwick MS, Hanson RE, McKnight TD, Nurul-Islam-Faridi M, Stelly DM (1997) A rapid procedure for the isolation of C0t–1 DNA from plants. Genome 40: 138–142. [DOI] [PubMed] [Google Scholar]
- 49. Pinkel D, Straume T, Gray JW (1986) Cytogenetic analysis using quantitative, high-sensitivity, fluorescence hybridization. Proc Natl Acad Sci USA 83: 2934–2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hewitt GH (1979) Orthoptera. Grasshoppers and Crickets. In Animal Citogenetics 3. Insecta 1. John B (ed). Gerbrüder Borntraeger, Berlín-Suttgart, pp 1–170. [Google Scholar]
- 51. Cabral-de-Mello DC, Moura RC, Melo AS, Martins C (2011) Evolutionary dynamics of heterochromatin in the genome of Dichotomius beetles based on chromosomal analysis. Genetica 139: 315–325. [DOI] [PubMed] [Google Scholar]
- 52. Ferreira IA, Martins C (2008) Physical chromosome mapping of repetitive DNA sequences in Nile tilapia Oreochromis niloticus: Evidences for a differential distribution of repetitive elements in the sex chromosomes. Micron 39: 411–418. [DOI] [PubMed] [Google Scholar]
- 53. Vicari MR, Pistune HFM, Castro JP, Almeida MC, Bertollo LAC, et al. (2011) New insights on the origin of B chromosomes in Astyanax scabripinnis obtained by chromosome painting and FISH. Genetica 139: 1073–1081. [DOI] [PubMed] [Google Scholar]
- 54. Pelliccia F, Barzotti R, Bucciarelli E, Rocchi A (2001) 5S ribosomal and U1 small nuclear RNA genes: a new linkage type in the genome of a crustacean that has three different tandemly repeated units containing 5S ribosomal DNA sequences. Genome 44: 331–335. [DOI] [PubMed] [Google Scholar]
- 55. Barzotti R, Pelliccia F, Rocchi A (2003) Identification and characterization of U1 small nuclear RNA genes from two crustacean isopod species. Chromosome Res 11: 365–373. [DOI] [PubMed] [Google Scholar]
- 56. Úbeda-Manzanaro M, Merlo MA, Palazón JL, Cross I, Sarasquete C, et al. (2010) Chromosomal mapping of the major and minor ribosomal genes, (GATA)n and U2 snRNA gene by double-colour FISH in species of the Batrachoididae family. Genetica 138: 787–794. [DOI] [PubMed] [Google Scholar]
- 57. Merlo MA, Cross I, Palazón JL, Úbeda-Manzanaro M, Sarasquete C, et al. (2012) Evidence for 5S rDNA Horizontal Transfer in the toadfish Halobatrachus didactylus (Schneider, 1801) based on the analysis of three multigene families. BMC Evol Biol 12: 202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Pendás AM, Morán P, García-Vázquez E (1994) Organization and chromosomal location of the major histone cluster in brown trout, Atlantic salmon and rainbow trout. Chromosoma 103: 147–152. [DOI] [PubMed] [Google Scholar]
- 59. Zhang L, Bao Z, Wang S, Huang X, Hu J (2007) Chromosome rearrangements in Pectinidae (Bivalvia: Pteriomorphia) implied based on chromosomal localization of histone H3 gene in four scallops. Genetica 130: 193–198. [DOI] [PubMed] [Google Scholar]
- 60. Cabral-de-Mello DC, Martins C, Souza MJ, Moura RC (2011) Cytogenetic Mapping of 5S and 18S rRNAs and H3 Histone Genes in 4 Ancient Proscopiidae Grasshopper Species: Contribution to Understanding the Evolutionary Dynamics of Multigene Families. Cytogenet Genome Res 132: 89–93. [DOI] [PubMed] [Google Scholar]
- 61. Panzera Y, Pita S, Ferreiro MJ, Ferrandis I, Lages C, et al. (2012) High Dynamics of rDNA Cluster Location in Kissing Bug Holocentric Chromosomes (Triatominae, Heteroptera). Cytogenet Genome Res 138: 56–67. [DOI] [PubMed] [Google Scholar]
- 62. Cabrero J, Perfectti F, Gómez R, Camacho JPM, López-León MD (2003b) Population variation in the A chromosome distribution of satellite DNA and ribosomal DNA in the grasshopper Eyprepocnemis plorans . Chromosome Res 11: 375–381. [DOI] [PubMed] [Google Scholar]
- 63. Voltolin TA, Laudicina A, Senhorini JA, Bortolozzi J, Oliveira C, et al. (2010) Origin and molecular organization of supernumerary chromosomes of Prochilodus lineatus (Characiformes, Prochilodontidae) obtained by DNA probes. Genetica 138: 1133–1139. [DOI] [PubMed] [Google Scholar]
- 64. Bugrov AG, Karamysheva TV, Rubtsov DN, Andreenkova OV, Rubtsov NB (2004) Comparative FISH analysis of distribution of B chromosome repetitive DNA in A and B chromosomes in two subspecies of Podisma sapporensis (Orthoptera, Acrididae). Cytogenet Genome Res 106: 284–288. [DOI] [PubMed] [Google Scholar]
- 65. Yang F, O'Brien PCM, Milne BS, Graphodatsky AS, Solanky N, et al. (1999) A complete comparative chromosome map for the dog, red fox, and human and its integration with canine genetic maps. Genomics 62: 189–202. [DOI] [PubMed] [Google Scholar]
- 66. Rubtsov NB, Karamysheva TV, Andreenkova OV, Bochkaerev MN, Kartavtseva IV, et al. (2004) Comparative analysis of micro and macro B chromosomes in the Korean field mouse Apodemus peninsulae (Rodentia, Murinae) performed by chromosome microdissection and FISH. Cytogenet Genome Res 106: 289–294. [DOI] [PubMed] [Google Scholar]
- 67. Mestriner CA, Galetti PM Jr, Valentini SR, Ruiz IRG, Abel LDS, et al. (2000) Structural and functional evidence that a B chromosome in the characid fish Astyanax scabripinnis is an isochromosome. Heredity 85: 1–9. [DOI] [PubMed] [Google Scholar]
- 68. Artoni RF, Vicari MR, Endler AL, Cavallaro ZI, Jesus CM, et al. (2006) Banding pattern of A and B chromosome of Prochilodus lineatus (Characiformes, Prochilodontidae), with comments on B chromosome evolution. Genetica 127: 277–284. [DOI] [PubMed] [Google Scholar]
- 69. John B, Hewitt GM (1965) The B-chromosome system of Myrmeleotettix maculatus (Thunb.) I. The mechanics. Chromosoma 16: 548–578. [DOI] [PubMed] [Google Scholar]
- 70. Gallagher A, Hewitt G, Gibson I (1973) Differential giemsa staining of heterochromatic B-chromosomes in Myrmeleotettix maculatus (Thunb.) (Orthoptera: Acrididae). Chromosoma 40: 167–172. [DOI] [PubMed] [Google Scholar]
- 71. Confalonieri VA, Bidau CJ (1986) The B-chromosomes of two species of Cylindrotettix (Leptysminae, Acrididae). Genetica 68: 87–95. [Google Scholar]
- 72. Vilardi JC (1986) Isocromosomas B e irregularidades meióticas en dos especies de Euplectrotettix (Orthoptera: Acrididae). Mendeliana 7: 125–137. [Google Scholar]
- 73. Grieco ML, Bidau C (2000) The dicentric nature of the metacentric B chromosome of Metaleptea brevicornis adspersa (Acridinae, acrididae). Heredity 84: 639–646. [DOI] [PubMed] [Google Scholar]
- 74.Jones RN, Puertas MJ (1993) The B chromosomes of rye (Secale cereale L.). In Frontiers in Plant Science Research. Ed: Dhir KK, Sareen TS. Delhi: Bhagwati Enterprises, pp 81–112. [Google Scholar]
- 75. López-León MD, Cabrero J, Dzyubenko VV, Bugrov AG, Karamysheva TV, et al. (2008) Differences in ribosomal DNA distribution on A and B chromosomes between eastern and western populations of the grasshopper Eyprepocnemis plorans plorans . Cytogenet Gen Res 121: 260–265. [DOI] [PubMed] [Google Scholar]
- 76. Dhar MK, Friebe B, Koul AK, Gill BS (2002) Origin of an apparent B chromosome by mutation, chromosome fragmentation and specific DNA sequence amplification. Chromosoma 111: 332–340. [DOI] [PubMed] [Google Scholar]
- 77. Martis MM, Klemme S, Banaei-Moghaddam AM, Blattner FR, Macas J, et al. (2012) Selfish supernumerary chromosome reveals its origin as a mosaic of host genome and organellar sequences. Proc Natl Acad Sci U S A 109: 13343–13346. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Meiotic cells from distinct individuals of A. flavolineata showing the variable patterns of 18S rDNA distribution. Autosomal bivalents were numbered in order of decreasing size. Bar = 5 µm.
(PDF)
Selected mitotic chromosomes of A. flavolineata after C-banding treatment and FISH with 18S rDNA, H3 histone and C0t -1 DNA as probes. Note the occurrence of large C0t-1 DNA blocks. Blue = DAPI, Green = signals.
(PDF)
Partial metaphases I of A. flavolineata individuals harboring two B chromosomes. The probes used are indicated in colors directly in each cell. Bar = 5 µm.
(PDF)
Relative length occupied by repetitive DNAs in chromosomes 1, 3, 5, 6 and 9 of A. flavolineata . Note that in all chromosomes the region occupied by C0t-1 DNA is larger than for other repetitive DNAs.
(PDF)