Abstract
The cellular immune response to respiratory syncytial virus (RSV) is important in both protection and immunopathogenesis. In contrast to HLA class I, HLA class II-restricted RSV-specific T-cell epitopes have not been identified. Here, we describe the generation and characterization of two human RSV-specific CD4+-T-cell clones (TCCs) associated with type 0-like cytokine profiles. TCC 1 was specific for the matrix protein and restricted over HLA-DPB1*1601, while TCC 2 was specific for the attachment protein G and restricted over either HLA-DPB1*0401 or -0402. Interestingly, the latter epitope is conserved in both RSV type A and B viruses. Given the high allele frequencies of HLA-DPB1*0401 and -0402 worldwide, this epitope could be widely recognized and boosted by recurrent RSV infections. Indeed, peptide stimulation of peripheral blood mononuclear cells from healthy adults resulted in the detection of specific responses in 8 of 13 donors. Additional G-specific TCCs were generated from three of these cultures, which recognized the identical (n = 2) or almost identical (n = 1) HLA-DP4-restricted epitope as TCC 2. No significant differences were found between the capacities of cell lines obtained from infants with severe (n = 41) or mild (n = 46) RSV lower respiratory tract infections to function as antigen-presenting cells to the G-specific TCCs, suggesting that the severity of RSV disease is not linked to the allelic frequency of HLA-DP4. In conclusion, we have identified an RSV G-specific human T helper cell epitope restricted by the widely expressed HLA class II alleles DPB1*0401 and -0402. Its putative role in protection and/or immunopathogenesis remains to be determined.
Respiratory syncytial virus (RSV), a member of the genus Pneumovirus of the family Paramyxoviridae, is a major cause of severe lower respiratory tract disease in infants, immunocompromised individuals, and the elderly (7, 29). RSV infections cause yearly epidemics in the winter season of moderate climate zones and are most often associated with relatively mild upper respiratory tract infections (29). In general, specific immunity is insufficient for protection, and RSV infections continue to occur throughout life.
At present, no licensed RSV vaccine is available. During vaccine trials in the 1960s, vaccination with a formalin-inactivated whole-virus preparation (FI-RSV) was found to predispose for enhanced clinical disease following natural infection with RSV (17). Although the exact mechanism of this apparently immunopathological phenomenon remains unclear, studies of both rodent and nonhuman primate models have suggested that a skewed RSV-specific T helper type 2 (Th2) response was a key factor in this process (11, 23). Several studies have suggested that primary infections in young infants resulting in severe RSV bronchiolitis are also associated with Th2 responses (24, 28). However, in two other cohort studies of infants with either severe RSV bronchiolitis or relatively mild RSV upper respiratory tract infection, this observation was not confirmed (6, 12).
Few studies have described the RSV-specific T-cell response at the epitope level. In rodents, four T-cell epitopes have been described, of which three were MHC class I and one was class II-restricted. The MHC class I-restricted epitopes were located in the RSV F protein (10, 16) and the RSV M2 protein (19), whereas the MHC class II-restricted epitope was located in the RSV G protein (33). In humans, class I-restricted epitopes have been identified in the RSV F (5, 27) and NP (14, 34) proteins. However, no human class II-restricted T-cell epitopes have been described. Recently, van Bleek et al. (32) described the human CD4 response to the RSV F protein. Using a set of overlapping peptides, they were able to demonstrate ex vivo F-specific CD4 memory T-cell responses. Similar RSV F-specific CD4 responses were also described earlier by Levely et al. (20).
In the present study, we describe two RSV-specific CD4+-T-cell clones (TCCs) generated from clinical materials collected from infants during the acute or convalescent phase of a laboratory-confirmed RSV infection. Gamma interferon (IFN-γ) enzyme-linked immunospot (ELISPOT) assays were used to determine the protein specificity, minimal epitope, and HLA restriction element.
MATERIALS AND METHODS
Antigen-presenting cell (APC) lines.
Autologous B-lymphoblastic cell lines (BLCL) were established by transformation of peripheral blood mononuclear cells (PBMC) with Epstein-Barr virus as described previously (31). For the generation of target and/or stimulator cells, BLCL were infected with RSV-A2 (ATCC VR1322) at a multiplicity of infection of 100, resulting in persistently infected BLCL (BLCL-RSV) as previously described (3). The percentage of RSV-positive cells was checked by FACScan with fluorescein isothiocyanate-labeled RSV-specific monoclonal antibodies (MAbs) (Imagen; DAKO, Glostrup, Denmark). All BLCL were maintained in RPMI 1640 medium (BioWhittaker, Verviers, Belgium) containing penicillin (100 U/ml; BioWhittaker), streptomycin (100 μg/ml; BioWhittaker), l-glutamine (2 mM; BioWhittaker), β-mercaptoethanol (10−5 M; Merck, Darmstadt, Germany) (referred to below as culture medium) supplemented with 10% heat-inactivated (30 min; 56°C) fetal bovine serum (Greiner, Frickenhausen, Germany). BLCL used for restriction element analysis were obtained from the European Collection of Cell Cultures (cells lines represented by “I” followed by the International Histocompatibility Workshop number) or were in-house generated (represented by “E” followed by a unique number). For autologous and in-house-generated BLCL, molecular typing of the HLA-DRB1*, DQB1*, and DPB1* loci was performed using a commercial typing system (GenoVision, Vienna, Austria).
Generation of RSV-specific T-cell clones.
TCC 1 was generated from nasal-brush cells collected from an infant (age, 20 months) during the acute phase of a laboratory-confirmed RSV-mediated upper respiratory tract infection. The nasal-brush T cells were stimulated in vitro with autologous γ-irradiated (3,000 rad) BLCL-RSV, and TCCs were generated by limiting dilution as described before (35). In short, T cells were seeded in 60-well Terasaki plates (Greiner Bio-One, Frickenhausen, Germany) at concentrations of 3, 1, and 0.3 per well and stimulated with allogeneic feeder cells and recombinant human interleukin-2 (rhIL-2; Red Swan, Utrecht, The Netherlands). After 2 weeks of expansion, positive cultures were restimulated specifically using γ-irradiated autologous BLCL-RSV, expanded with rhIL-2, and tested for RSV specificity in an IFN-γ ELISPOT assay (see below) after stimulation with BLCL or BLCL-RSV. RSV-specific TCCs were phenotyped by fluorescence using MAbs against CD3, CD4, and CD8 (DAKO).
TCC 2 was generated from PBMC collected from another infant (age, 2 months) during the convalescent phase of an RSV-mediated lower respiratory tract infection. PBMC were stimulated in vitro with γ-irradiated autologous BLCL-RSV, and TCCs were generated by limiting dilution as described above.
TCCs P1, P2, and P3 were generated from PBMC collected from healthy adult donors. PBMC were stimulated in vitro with peptide G158-189 (0.01 μM), and TCCs were generated by limiting dilution as described above.
T cells and T-cell lines were maintained in culture medium supplemented with pooled 10% heat-inactivated human serum (referred to below as R10H) and rhIL-2.
ELISPOT assay.
The reactivities of TCCs with APCs were determined in an IFN-γ ELISPOT assay as described previously (12). In short, TCCs were seeded in concentrations ranging from 4,000 to 10,000 per well in a 96-well V-bottom plate (Greiner Bio-One), and APCs were added in an effector-to-target ratio of 1:5 and incubated for 1 h at 37°C. The cells were transferred to nylon bottom plates (Nalge Nunc, Rochester, N.Y.) precoated with a MAb against IFN-γ (1-D1K; Mabtech AB, Stockholm, Sweden), incubated for 4 h at 37°C, and subsequently washed with phosphate-buffered saline containing 0.05% Tween 20 (Merck). The spots were visualized by incubation with a secondary biotinylated MAb against IFN-γ (7-B6-1; Mabtech AB), streptavidin alkaline phosphatase (Mabtech AB), and nitroblue tetrazolium-5-bromo-4-chloro-3-indolylphosphate (Kirkegaard & Perry Laboratories, Gaithersburg, Md.). Finally, the color reaction was stopped by washing the plates with distilled water, and the spots were counted with a stereomicroscope at 25-fold magnification. IFN-γ ELISPOT results are shown in figures and tables as IFN-γ spot-forming cells (SFC) per well.
RSV protein specificity.
Recombinant vaccinia virus (rVV) constructs mediating the expression of the individual proteins of RSV (rVV-F, -G, -M, -P, -N, -1A, -1B, -1C, -L, and -22K) and a wild-type VV were used to infect autologous BLCL at a multiplicity of infection of 10. rVV-infected BLCL were used as APCs in IFN-γ ELISPOT assays to determine protein specificities.
Peptide-specific T-cell responses.
For evaluation of the responses to different peptides, BLCL were pulsed overnight with peptides at different concentrations (1 to 0.001 μM) and subsequently used as APCs in IFN-γ ELISPOT assays.
For TCC 1, 15-mer peptides (n = 49) with 5-amino-acid overlaps spanning the M protein (GenBank accession no. P03419) were tested (last peptide, 16 amino acids). For fine T-cell epitope mapping, peptides were constructed with one or more deletions on either the N-terminal or C-terminal side. Peptides with free N and C termini were synthesized as described before (13), dissolved in dimethyl sulfoxide at a concentration of 10 mg/ml, and diluted to 100 μM in RPMI 1640 (BioWhittaker).
For TCC 2, a 101-mer peptide spanning the conserved region of the RSV G protein (G2Na; amino acids 130 to 230; kindly provided by U. F. Power, Centre d'Immunologie Pierre-Fabre, Saint-Julien-en-Genevois, France) and smaller peptides spanning different regions of G2Na (G170-187, G187-223, G187-198, G174-189, and G158-189) were tested. For fine epitope mapping, overlapping 15-mers with 14-amino-acid overlaps or deletion mutants were used.
The protein preparations used to pulse BLCL overnight at a concentration of 1 μg/ml were β-propiolactone (Sigma Aldrich, St. Louis, Mo.) inactivated RSV (BPL-RSV cultured in Vero cells) and Vero cell antigen (BPL-Vero), as described by De Swart et al. (11).
RSV-specific T-cell responses in PBMC or CBMC.
PBMC collected from healthy adults or cord blood mononuclear cells (CBMC) were stimulated with peptide G158-189 (0.1 or 0.01 μM) in R10H and expanded in the presence of rhIL-2. After 2.5 weeks of culture, the outgrowth of specific cells was analyzed in a CD69 expression assay as described before (30). In short, residual CD69 molecules were enzymatically removed (0.1% chymotrypsin type II [Sigma Aldrich] in PBS; 10 min; 37°C), and subsequently the cells were stimulated with autologous BLCL pulsed with peptide G158-189 (positive) or G174-189 (negative). After 6 h, the cells were washed; incubated with αCD3-FITC (DAKO), αCD69-PE (BD Pharmingen), αCD8-PerCP (BD Pharmingen), and αCD4-APC (BD Pharmingen); and analyzed in a FACSCalibur (Becton-Dickinson, Erembodegem, Belgium).
Cytokine enzyme-linked immunosorbent assays.
To determine cytokine profiles, TCCs (105 per well) were stimulated in vitro with autologous BLCL either uninfected or infected with RSV or were pulsed overnight with peptides (0.1 μM) M245-256 and M241-251 (TCC 1 positive and negative peptides, respectively) or G161-175 and G167-181 (TCC P3 positive and negative peptides, respectively), at an effector-to-target ratio of 2:1. After 5 days, the culture supernatants were harvested, and cytokine levels were determined according to the manufacturer's instructions for IFN-γ, IL-2, IL-4, and IL-5 (Biosource, Fleurus, Belgium) and IL-13 (U-Cytech, Utrecht, The Netherlands).
RESULTS
Generation and characterization of human RSV-specific T-cell clones.
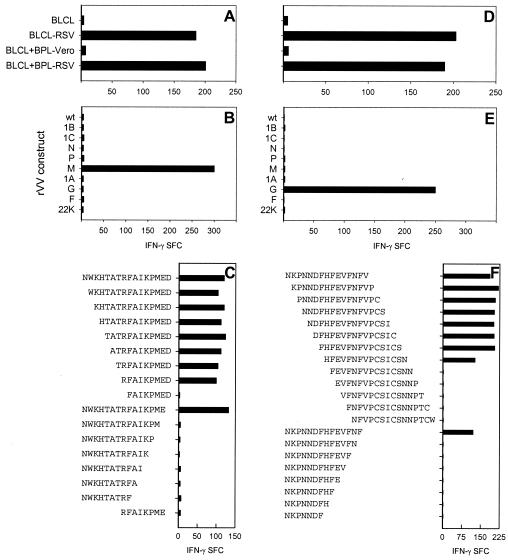
TCC 1 was cloned from nasal-brush cells collected from an infant during the acute phase of a laboratory-confirmed RSV upper respiratory tract infection. It was RSV specific (Fig. 1A), of the CD3+ CD4+ phenotype, and it recognized the RSV M protein (Fig. 1B). Of the overlapping peptides tested, peptide N241 WKHTATRFAIKPMED256 was recognized, and the minimal epitope was R248FAIKPME255 with an additional amino acid on either side (Fig. 1C).
FIG. 1.
RSV specificities of TCC 1 (A) and TCC 2 (D) were determined in an IFN-γ ELISPOT assay, using autologous BLCL either uninfected (BLCL) or infected with RSV (BLCL-RSV) or pulsed with antigen (BLCL+BPL-Vero and BLCL+BPL-RSV). The protein specificities of TCC 1 (B) and TCC 2 (E) were determined using autologous BLCL infected with different rVV constructs. The minimal epitopes recognized by TCC 1 (C) and TCC 2 (F) were determined by using autologous BLCL pulsed overnight with 0.1 (C) or 0.01 (F) μM peptide. The results are shown as SFC per well.
TCC 2 was cloned from PBMC collected from an infant during the convalescent phase of an RSV lower respiratory tract infection. It was RSV specific (Fig. 1D), of the CD3+ CD4+ phenotype, and it recognized the RSV G protein (Fig. 1E). The clone was also found to recognize BLCL pulsed with G2Na or with G158-189 (data not shown). The minimal epitope was F163HFEVFNFV171 (Fig. 1F).
Analysis of published sequences suggested that the M epitope is conserved in RSV A but not in RSV B, whereas the G epitope is conserved in both subgroups (results not shown). This was confirmed in an IFN-γ ELISPOT assay using autologous BLCL pulsed with RSV A (Long strain; ATCC VR26) or RSV B (B1 strain; ATCC 1400) antigens as APCs. Whereas the G-specific TCC P2 was able to recognize both RSV A- and B-pulsed APCs, the M-specific TCC 1 was capable of recognizing only RSV A-pulsed APCs (data not shown).
Determination of HLA restriction elements of TCCs 1 and 2.
The class II genotype of the donor from whom TCC 1 originated was HLA-DRB1*0301,0701;DQB1*0201,0202;DPB1*1101,1601; that of the donor of TCC 2 was HLA-DRB1*0102,0701;DQB1*0303,0501;DPB1*0201,0401. Bytesting reactivity with peptide-pulsed BLCL matched or mismatched for HLA-DR or -DQ, we were unable to determine the restriction pattern for either of these TCCs: for TCC 1, no responses were found, while for TCC 2, the majority of the APCs resulted in positive responses. When the HLA-DP alleles were included, it was found that TCC 1 was restricted by HLA-DPB1*1601 (Table 1), while TCC 2 recognized peptide in the context of either HLA-DPB1*0401 or -0402 (Table 2).
TABLE 1.
Determination of HLA restriction elements of TCC 1
| BLCL | Restriction elementsc
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Aut.a | E-0002 | I-9019 | I-9023 | I-9050 | I-9095 | I-9041 | I-9063 | |
| DRB1* | 0301 | 0102 | 0301 | 0301 | 1104 | 1302 | ||
| DRB1* | 0701 | 0701 | 0701 | 0701 | ||||
| DQB1* | 0201 | 0303 | 0201 | 0201 | 0201 | 0301 | 0604 | |
| DQB1* | 0202 | 0501 | 0202 | 030302 | ||||
| DPB1* | 1101 | 0201 | 0402 | 0101 | 020102 | 0401 | 1101 | |
| DPB1* | 1601 | 0401 | 1301 | 0402 | 1601 | |||
| IFN-γ SFCb | 168 | 3 | 2 | 0 | 2 | 1 | 2 | 188 |
Aut., autologous BLCL.
IFN-γ SFC per well were obtained by using BLCL pulsed with M241-256.
HLA class II phenotypes that BLCL have in common with the autologous BLCL are in boldface.
TABLE 2.
Determination of HLA restriction elements of TCC 2
| BLCL | Restriction elementc
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Aut.a | E-0004 | E-0001 | E-7366 | I-9003 | I-9105 | I-9026 | E-0003 | I-9013 | |
| DRB1* | 0102 | 0102 | 0301 | 1501 | 0101 | 11041 | 0402 | 1301 | 1501 |
| DRB1* | 0701 | 0302 | 0701 | 0701 | 1501 | ||||
| DQB1* | 0303 | 0402 | 0201 | 0202 | 0603 | 0302 | 0602 | 0602 | |
| DQB1* | 0501 | 0501 | 0202 | 0602 | 0501 | 0603 | |||
| DPB1* | 0201 | 0101 | 1101 | 0301 | 1301 | 0201 | 0402 | 0402 | |
| DPB1* | 0401 | 0301 | 1601 | 1001 | 0401 | 0401 | |||
| IFN-γ SFCb | 126 | 1 | 0 | 3 | 1 | 3 | 145 | 130 | 122 |
Aut., autologous BLCL.
IFN-γ SFC per well were obtained by using BLCL pulsed with G158-189.
HLA class II phenotypes that BLCL have in common with the autologous BLCL are in boldface.
Peptide G158-189 responses in the human population.
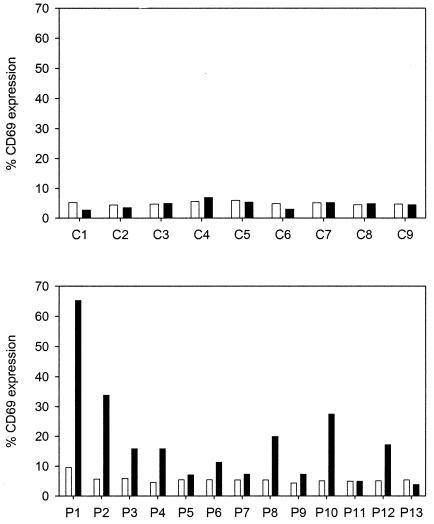
Since HLA-DPB1*0401 and -0402 are the most frequent HLA class II alleles in the human population (8), we investigated the response to peptide G158-189 in PBMC obtained from healthy adult donors and in CBMC. No specific T cells were found in the CBMC cultures, while G158-189-specific T cells were detected in 8 of 13 PBMC cultures (Fig. 2).
FIG. 2.
Detection of G158-189-specific T cells in CBMC (C1 to C9) or PBMC (P1 to P13) stimulated for 2.5 weeks with 0.01 μM peptide G158-189. The percentage of CD69+ cells in the CD3+ CD4+ fraction of the expanded bulk cultures was determined after 6 h of stimulation with autologous BLCL pulsed with G174-189 (negative; open bars) or G158-189 (positive; black bars). Responses were considered positive when the ratio of the percentages of CD69+ cells after stimulation with the positive and negative peptides was >2.
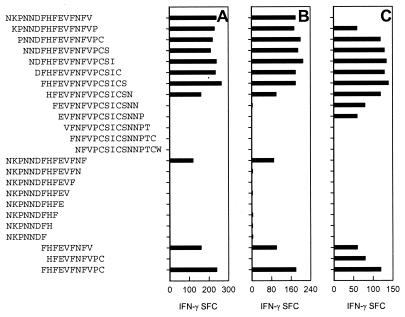
In order to confirm that the G158-189-expanded cells were indeed RSV-specific T cells, TCCs were generated from three of these cultures (P1, P2, and P3), and the minimal epitopes of these TCCs were determined. As shown in Fig. 3, TCC P1 and TCC P2 recognized the same minimal epitope as TCC 2, while TCC P3 recognized a 10-mer peptide shifted 1 amino acid in the C-terminal direction, H162FEVFNFVPC173.
FIG. 3.
Determination of the minimal epitopes of TCC-P1, -P2, and -P3 in an IFN-γ ELISPOT assay using autologous BLCL pulsed with 0.01 μM G158-189 as APCs. The results are shown as SFC per well.
The class II genotype of donor P1 was HLA-DRB1*1301,1501;DQB1*0602,0603;DPB1*0401,0402, that of donor P2 wasHLA-DRB1*0401,1201;DQB1*0301;DPB1*0301,0402, andthat of donor P3 was HLA-DRB1*1201,1501;DQB1*0301,0601;DPB1*0201,0401. Similarly to TCC 2, both TCCs P1 and P2 recognized peptide in the context of HLA-DPB1*0401 or -0402 (Tables 3 and 4). Interestingly, TCC P3 was also able to recognize peptide in the context of HLA-DPB1*0201 and -02012 (Table 5).
TABLE 3.
Determination of HLA restriction elements of TCC P1
| BLCL | Restriction elementc
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Aut.a | I-9060 | E-7366 | I-9013 | I-9105 | I-9016 | I-9002 | I-9026 | |
| DRB1* | 1301 | 1301 | 0701 | 11041 | 1602 | 0102 | 0402 | |
| DRB1* | 1501 | 1501 | 1501 | |||||
| DQB1* | 0602 | 0602 | 0602 | 0301 | 0501 | 0302 | ||
| DQB1* | 0603 | 0603 | 0202 | 0603 | ||||
| DPB1* | 0401 | 0301 | 0201 | 0401 | 0401 | |||
| DPB1* | 0402 | 1001 | 0402 | 0402 | ||||
| IFN-γ SFCb | 200 | 0 | 1 | 140 | 10 | 126 | 190 | 180 |
Aut., autologous BLCL.
IFN-γ SFC per well were obtained by using BLCL pulsed with G158-189.
HLA class II phenotypes that BLCL have in common with the autologous BLCL are in boldface.
TABLE 4.
Determination of HLA restriction elements of TCC P2
| BLCL | Restriction elementc
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Aut.a | I-9032 | I-9038 | I-9016 | I-9043 | E-1519 | I-9013 | I-9002 | |
| DRB1* | 0401 | 0401 | 1602 | 1101 | 0801 | 0102 | ||
| DRB1* | 1201 | 1201 | 1401 | 1501 | ||||
| DQB1* | 0301 | 0302 | 0301 | 0301 | 0301 | 0402 | 0602 | 0501 |
| DQB1* | 0503 | |||||||
| DPB1* | 0301 | 02012 | 02012 | 1001 | 0301 | 0401 | ||
| DPB1* | 0402 | 0402 | 0501 | 0402 | ||||
| IFN-γ SFCb | 180 | 3 | 2 | 165 | 0 | 6 | 175 | 185 |
Aut., autologous BLCL.
IFN-γ SFC per well were obtained by using BLCL pulsed with G158-189.
HLA class II phenotypes that BLCL have in common with the autologous BLCL are in boldface.
TABLE 5.
Determination of HLA restriction elements of TCC P3
| BLCL | Restriction elementc
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Aut.a | I-9038 | E-7366 | I-9016 | I-9043 | E-0005 | I-9105 | I-9050 | I-9002 | I-9013 | |
| DRB1* | 1201 | 1201 | 0701 | 1602 | 1101 | 1502 | 11041 | 0701 | 0102 | |
| DRB1* | 1501 | 1501 | 1501 | |||||||
| DQB1* | 0301 | 0301 | 0602 | 0301 | 0301 | 0202 | 0501 | 0602 | ||
| DQB1* | 0601 | 0202 | 0601 | 0603 | ||||||
| DPB1* | 0201 | 02012 | 0301 | 0402 | 1001 | 0901 | 0201 | 02012 | 0402 | |
| DPB1* | 0401 | 1001 | 0401 | |||||||
| IFN-γ SFCb | 125 | 80 | 1 | 104 | 2 | 1 | 130 | 80 | 124 | 112 |
Aut., autologous BLCL.
IFN-γ SFC per well were obtained by using BLCL pulsed with G158-189.
HLA class II phenotypes that BLCL have in common with the autologous BLCL are in boldface.
All TCCs produce both type 1 and 2 cytokines.
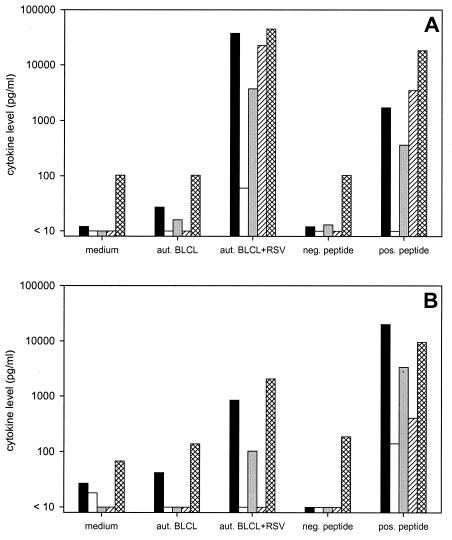
TCCs 1 (two subclones), P2, and P3 were tested for the ability to produce cytokines after stimulation with different antigens. All of the TCCs showed specific cytokine production after stimulation with RSV-infected or peptide-pulsed autologous BLCL, with IFN-γ and IL-13 predominating. Interestingly, while TCC 1 (both subclones) produced higher levels of IL-5 than IL-4 upon stimulation, TCCs P2 and P3 produced more IL-4 than IL-5 (Fig. 4 and data not shown).
FIG.4.
Cytokine levels in culture supernatants of TCC 1 (A) and TCC P3 (B) 5 days after stimulation with medium, autologous (aut.) BLCL, autologous BLCL-RSV, autologous BLCL pulsed with negative (neg.) peptide (M241-251 for TCC 1 and G167-181 for TCC P3), or autologous BLCL pulsed with positive (pos.) peptide (M245-256 for TCC 1 and G161-175 for TCC P3). The detection limits were 10 pg/ml for IFN-γ (solid bars), IL-2 (open bars), IL-4 (shaded bars), and IL-5 (hatched bars) and 100 pg/ml for IL-13 (cross-hatched bars).
HLA-DP4 phenotype in infants with RSV infections of different clinical severities.
To determine if the T-cell response to the HLA-DP4-restricted G epitope plays a role in the pathogenesis of severe RSV disease, the allelic frequency of HLA-DP4 or the precursor frequency of the G-specific T cells could be compared among infants with RSV disease of different severities. As an alternative to the determination of HLA-DP4 allelic frequencies, specimens from a previous cohort study (6) allowed us to test the functional capacities of BLCL obtained from infants with severe or mild RSV lower respiratory tract infections to function as APCs to the G-specific TCCs. Positive responses of the TCCs were found after stimulation by 35 of 41 (85%) and 50 of 64 (78%) peptide-pulsed BLCL, respectively (no significant difference; P > 0.1; Fisher's exact test, two sided).
DISCUSSION
We have identified two HLA-DP-restricted T helper cell epitopes in the RSV M and G proteins, conserved in subgroup A and in both subgroups A and B, respectively. The M epitope was recognized by a TCC restricted by HLA-DPB1*1601, while the G epitope was recognized in the context of either HLA-DPB1*0401 or -0402. The ubiquitous distribution of the last alleles and the results of our peptide PBMC bulk stimulations suggest that responses to the G epitope are generated in a large part of the human population.
The majority of studies of human T helper cell responses have focused on HLA-DR- and -DQ-restricted T cells, to a large extent because HLA-DP appeared less important in contributing to the risk of graft-versus-host disease (21). However, several HLA-DP-restricted T-cell epitopes have now been described (2), including viral epitopes (9, 18). To our knowledge, the RSV M-specific TCC described here is the first HLA-DPB1*1601-restricted epitope identified. Recently, a number of TCCs have been described as recognizing their epitopes in the context of HLA-DPB1*0401 and/or -0402, which both belong to the serologically defined HLA-DPw4 antigenic group (4). HLA-DPw4 is the most prevalent HLA class II antigen, with an allelic frequency of 78% in the Caucasian population (1). The RSV G epitope identified in the present paper is consistent with the suggested motif for HLA-DP4-restricted epitopes as previously described (8).
HLA-DPB1*0401 and -0402 differ by only 3 amino acids, and TCCs raised in subjects with HLA-DPB1*0401 were in some cases also able to recognize their antigens in the context of HLA-DPB1*0402 (4). We have found the same in the donor of TCC 2 and in donor P3, but have also raised a TCC in a donor who was HLA-DPB1*0402 positive (donor P3) that could recognize its epitope in the context of HLA-DPB1*0401. This TCC, which recognized an epitope that was shifted 1 amino acid in the C-terminal direction compared to TCCs 2, P1, and P2, could also recognize its epitope in the context of HLA-DPB1*0201 and -02012. Interestingly, HLA-DPB1*0201 and HLA-DPB1*0402 differ by only 1 amino acid in the P4 pocket of the peptide-binding groove, and HLA-DP2 and -DP4 were previously suggested to form a supertype of class II molecules on the basis of homology in the peptide-binding pockets P1 and P6 (8).
The functional studies using BLCL from infants with mild or severe RSV-mediated lower respiratory tract disease as APCs to the G-specific TCCs suggested that the severity of RSV disease is not linked to the allelic frequency of HLA-DP4. However, frequency studies of epitope-specific T cells in infants with different disease severities will have to be performed to determine whether they play a role in RSV pathogenesis. The fact that the conserved HLA-DP4-restricted epitope was found in the G protein brings another dimension to this question, since G-specific responses have often been suggested to be involved in natural or vaccine-mediated enhanced disease (15, 22). However, in our study, similar Th0-like cytokine production profiles were found for both the M- and G-specific TCCs.
Castelli et al. suggested that specific HLA-DP4-restricted epitopes could be used as peptide vaccines, because of the high frequency of HLA-DP4 worldwide (8). However, vaccination with nonreplicating RSV vaccines that induce only HLA class II-restricted T-cell responses has been associated with immunopathology in humans (17), nonhuman primates (11, 25), and rodents (23, 26).
In conclusion, we have identified the first HLA-DPB1*1601-restricted T-cell epitope and a conserved HLA-DP4-restricted T-cell epitope in the RSV M and G proteins, respectively. Whether immune responses to the latter epitope are involved in RSV-mediated immunopathogenesis remains to be determined.
Acknowledgments
We thank A. C. M. Boon, L. P. Koopman, P. G. H. Mulder, G. van der Net, and H. H. Timmerman for their contributions to this study and the Department of Immunohaematology and Blood Transfusion, Leiden University Medical Center, for the kind gift of BLCL E-0005 (SAS).
This work was sponsored by The Netherlands Asthma Foundation (grant 93.96.1), The Netherlands Organization for Health Sciences (grant 940-35-025), and the Sophia Foundation for Medical Research (grant 214).
REFERENCES
- 1.al Daccak, R., F. Q. Wang, D. Theophille, P. Lethielleux, J. Colombani, and P. Loiseau. 1991. Gene polymorphism of HLA-DPB1 and DPA1 loci in caucasoid population: frequencies and DPB1-DPA1 associations. Hum. Immunol. 31:277-285. [DOI] [PubMed] [Google Scholar]
- 2.Austin, P., J. Trowsdale, C. Rudd, W. Bodmer, M. Feldmann, and J. Lamb. 1985. Functional expression of HLA-DP genes transfected into mouse fibroblasts. Nature 313:61-64. [DOI] [PubMed] [Google Scholar]
- 3.Bangham, C. R. M., and A. J. McMichael. 1986. Specific human cytotoxic T cells recognize B-cell lines persistently infected with respiratory syncytial virus. Proc. Natl. Acad. Sci. USA 83:9183-9187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baselmans, P. J., E. Pollabauer, F. C. van Reijsen, H. C. Heystek, A. Hren, P. Stumptner, M. G. Tilanus, W. C. Vooijs, and G. C. Mudde. 2000. IgE production after antigen-specific and cognate activation of HLA-DPw4-restricted T-cell clones, by 78% of randomly selected B-cell donors. Hum. Immunol. 61:789-798. [DOI] [PubMed] [Google Scholar]
- 5.Brandenburg, A. H., L. de Waal, H. H. Timmerman, P. Hoogerhout, R. L. De Swart, and A. D. M. E. Osterhaus. 2000. HLA class-I restricted cytotoxic T-cell epitopes of the respiratory syncytial virus fusion protein. J. Virol. 74:10240-10244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brandenburg, A. H., A. Kleinjan, B. van het Land, H. A. Moll, H. H. Timmerman, R. L. De Swart, H. J. Neijens, W. Fokkens, and A. D. M. E. Osterhaus. 2000. Type 1-like immune response is found in children with respiratory syncytial virus infection regardless of clinical severity. J. Med. Virol. 62:267-277. [PubMed] [Google Scholar]
- 7.Carbonell-Estrany, X., and J. Quero. 2001. Hospitalization rates for respiratory syncytial virus infection in premature infants born during two consecutive seasons. Pediatr. Infect. Dis. J. 20:874-879. [DOI] [PubMed] [Google Scholar]
- 8.Castelli, F. A., C. Buhot, A. Sanson, H. Zarour, S. Pouvelle-Moratille, C. Nonn, H. Gahery-Segard, J. G. Guillet, A. Menez, B. Georges, and B. Maillere. 2002. HLA-DP4, the most frequent HLA II molecule, defines a new supertype of peptide-binding specificity. J. Immunol. 169:6928-6934. [DOI] [PubMed] [Google Scholar]
- 9.Celis, E., J. Larson, L. Otvos, Jr., and W. H. Wunner. 1990. Identification of a rabies virus T cell epitope on the basis of its similarity with a hepatitis B surface antigen peptide presented to T cells by the same MHC molecule (HLA-DPw4). J. Immunol. 145:305-310. [PubMed] [Google Scholar]
- 10.Chang, J., A. Srikiatkhachorn, and T. J. Braciale. 2001. Visualization and characterization of respiratory syncytial virus F-specific CD8+ T cells during experimental virus infection. J. Immunol. 167:4254-4260. [DOI] [PubMed] [Google Scholar]
- 11.De Swart, R. L., T. Kuiken, H. H. Timmerman, G. Van Amerongen, B. G. van den Hoogen, H. W. Vos, H. J. Neijens, A. C. Andeweg, and A. D. M. E. Osterhaus. 2002. Immunization of macaques with formalin-inactivated respiratory syncytial virus (RSV) induces interleukin-13-associated hypersensitivity to subsequent RSV infection. J. Virol. 76:11561-11569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Waal, L., L. P. Koopman, I. J. van Benten, A. H. Brandenburg, P. G. H. Mulder, R. L. De Swart, W. J. Fokkens, H. J. Neijens, and A. D. M. E. Osterhaus. 2003. Moderate local and systemic respiratory syncytial virus-specific T-cell responses upon mild or subclinical RSV infection. J. Med. Virol. 70:309-318. [DOI] [PubMed] [Google Scholar]
- 13.Fields, C. G., D. H. Lloyd, R. L. MacDonald, K. M. Otteson, and R. L. Noble. 1991. HBTU activation for automated Fmoc solid phase peptide synthesis. Peptide Res. 4:95-101. [PubMed] [Google Scholar]
- 14.Goulder, P. J. R., F. Lechner, P. Klenerman, K. Mcintosh, and B. D. Walker. 2000. Characterization of a novel respiratory syncytial virus-specific human cytotoxic T-lymphocyte epitope. J. Virol. 74:7694-7697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Graham, B. S., J. A. Rutigliano, and T. R. Johnson. 2002. Respiratory syncytial virus immunobiology and pathogenesis. Virology 297:1-7. [DOI] [PubMed] [Google Scholar]
- 16.Jiang, S., N. J. Borthwick, P. Morrison, G. F. Gao, and M. W. Steward. 2002. Virus-specific CTL responses induced by an H-2Kd-restricted, motif-negative 15-mer peptide from the fusion protein of respiratory syncytial virus. J. Gen. Virol. 83:429-438. [DOI] [PubMed] [Google Scholar]
- 17.Kim, H. W., J. G. Canchola, C. D. Brandt, G. Pyles, R. M. Chanock, K. Jensen, and R. H. Parrott. 1969. Respiratory syncytial virus disease in infants despite prior administration of antigenic inactivated vaccine. Am. J. Epidemiol. 89:422-434. [DOI] [PubMed] [Google Scholar]
- 18.Koelle, D. M., S. N. Reymond, H. Chen, W. W. Kwok, C. McClurkan, T. Gyaltsong, E. W. Petersdorf, W. Rotkis, A. R. Talley, and D. A. Harrison. 2000. Tegument-specific, virus-reactive CD4 T cells localize to the cornea in herpes simplex virus interstitial keratitis in humans. J. Virol. 74:10930-10938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kulkarni, A. B., H. C. Morse III, J. R. Bennink, J. W. Yewdell, and B. R. Murphy. 1993. Immunization of mice with vaccinia virus-M2 recombinant induces epitope-specific and cross-reactive Kd-restricted CD8+ cytotoxic T cells. J. Virol. 67:4086-4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Levely, M. E., C. A. Bannow, C. W. Smith, and J. A. Nicholas. 1991. Immunodominant T-cell epitope on the F protein of respiratory syncytial virus recognized by human lymphocytes. J. Virol. 65:3789-3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moreau, P., and A. Cesbron. 1994. HLA-DP and allogeneic bone marrow transplantation. Bone Marrow Transplant. 13:675-681. [PubMed] [Google Scholar]
- 22.Openshaw, P. J., S. L. Clarke, and F. M. Record. 1992. Pulmonary eosinophilic response to respiratory syncytial virus in mice sensitized to the major surface glycoprotein G. Int. Immunol. 4:493-500. [DOI] [PubMed] [Google Scholar]
- 23.Openshaw, P. J. M., F. J. Culley, and W. Olszewska. 2001. Immunopathogenesis of vaccine-enhanced RSV disease. Vaccine 20:S27-S31. [DOI] [PubMed] [Google Scholar]
- 24.Pala, P., R. Bjarnason, F. Sigurbergsson, C. Metcalfe, N. Sigurs, and P. J. Openshaw. 2002. Enhanced IL-4 responses in children with a history of respiratory syncytial virus bronchiolitis in infancy. Eur. Respir. J. 20:376-382. [DOI] [PubMed] [Google Scholar]
- 25.Ponnuraj, E. M., A. R. Hayward, A. Raj, H. Wilson, and E. A. F. Simoes. 2001. Increased replication of respiratory syncytial virus (RSV) in pulmonary infiltrates is associated with enhanced histopathological disease in bonnet monkeys (Macaca radiata) pre-immunized with a formalin-inactivated RSV vaccine. J. Gen. Virol. 82:2663-2674. [DOI] [PubMed] [Google Scholar]
- 26.Prince, G. A., S. J. Curtis, K. C. Yim, and D. D. Porter. 2001. Vaccine-enhanced respiratory syncytial virus disease in cotton rats following immunization with Lot 100 or a newly prepared reference vaccine. J. Gen. Virol. 82:2881-2888. [DOI] [PubMed] [Google Scholar]
- 27.Rock, M. T., and J. E. Crowe, Jr. 2003. Identification of a novel human leucocyte antigen-A*01-restricted cytotoxic T-lymphocyte epitope in the respiratory syncytial virus fusion protein. Immunology 108:474-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roman, M., W. J. Calhoun, K. L. Hinton, L. F. Avendano, V. Simon, A. M. Escobar, A. Gaggero, and P. V. Diaz. 1997. Respiratory syncytial virus infection in infants is associated with predominant Th-2-like response. Am. J. Respir. Crit. Care Med. 156:190-195. [DOI] [PubMed] [Google Scholar]
- 29.Simoes, E. A. F. 1999. Respiratory syncytial virus infection. Lancet 354:847-852. [DOI] [PubMed] [Google Scholar]
- 30.Stittelaar, K. J., L. S. Wyatt, R. L. De Swart, H. W. Vos, J. Groen, G. Van Amerongen, R. S. Van Binnendijk, S. Rozenblatt, B. Moss, and A. D. M. E. Osterhaus. 2000. Protective immunity in macaques vaccinated with a modified vaccinia virus Ankara-based measles vaccine in the presence of passively acquired antibodies. J. Virol. 74:4236-4243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van Binnendijk, R. S., M. C. M. Poelen, P. De Vries, H. O. Voorma, A. D. M. E. Osterhaus, and F. G. C. M. UytdeHaag. 1989. Measles virus-specific human T cell clones. Characterization of specificity and function of CD4+ helper/cytotoxic and CD8+ cytotoxic T cell clones. J. Immunol. 142:2847-2854. [PubMed] [Google Scholar]
- 32.Van Bleek, G. M., M. C. Poelen, R. Van der Most, H. F. Brugghe, H. A. M. Timmermans, C. J. Boog, P. Hoogerhout, H. G. Otten, and C. A. C. M. Van Els. 2003. Identification of immunodominant epitopes derived from the respiratory syncytial virus fusion protein that are recognized by human CD4 T cells. J. Virol. 77:980-988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Varga, S. M., E. L. Wissinger, and T. J. Braciale. 2000. The attachment (G) glycoprotein of respiratory syncytial virus contains a single immunodominant epitope that elicits both Th1 and Th2 CD4+ T cell responses. J. Immunol. 165:6487-6495. [DOI] [PubMed] [Google Scholar]
- 34.Venter, M., M. Rock, A. J. Puren, C. T. Tiemessen, and J. E. Crowe, Jr. 2003. Respiratory syncytial virus nucleoprotein-specific cytotoxic T-cell epitopes in a South African population of diverse HLA types are conserved in circulating field strains. J. Virol. 77:7319-7329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Verjans, G. M., R. Janssen, F. G. UytdeHaag, C. E. van Doornik, and J. Tommassen. 1995. Intracellular processing and presentation of T cell epitopes, expressed by recombinant Escherichia coli and Salmonella typhimurium, to human T cells. Eur. J. Immunol. 25:405-410. [DOI] [PubMed] [Google Scholar]