Abstract
Penner serotyping has been the principal method for differentiating Campylobacter isolates since its inception. Campylobacter capsule polysaccharide (CPS), the principal serodeterminant on which Penner serotyping is based, is presently of interest as a vaccine component. To determine the required valency of an effective CPS-based vaccine, a comprehensive understanding of CPS distribution is needed. Because of the association between Penner serotype and CPS, we conducted a systematic review to estimate the frequency and distribution of Penner serotypes associated with cases of Campylobacteriosis. In total, more than 21,000 sporadic cases of C. jejuni cases were identified for inclusion. While regional variation exists, distribution estimates indicate that eight serotypes accounted for more than half of all sporadic diarrheal cases globally and three serotypes (HS4 complex, HS2, and HS1/44) were dominant inter-regionally as well as globally. Furthermore, a total of 17 different serotypes reached a representation of 2% or greater in at least one of the five regions sampled. While this review is an important first step in defining CPS distribution, these results make it clear that significant gaps remain in our knowledge. Eliminating these gaps will be critical to future vaccine development efforts.
Introduction
Campylobacter is a gram negative organism that belongs to the class epsilonproteobacteria, and is recognized as a major foodborne pathogen in developed and developing countries alike. Campylobacteriosis, the disease caused by Campylobacter spp., is often a self-limiting disease commonly associated with diarrhea, cramping, headache, and fever. However, severe cases, including those resulting in dysentery, may require medical attention. Furthermore, long-term sequelae, including Guillain-Barré Syndrome (GBS), reactive arthritis, and irritable bowel syndrome (IBS) have been linked to infection [1]. The two major Campylobacter spp. involved in human diarrheal disease are C. jejuni and C. coli. However, for the purpose of this systematic review, analysis will be limited to C. jejuni.
For the past 30 years, epidemiologic studies of C. jejuni have used the Penner serotyping scheme, a passive slide hemaglutination assay developed by Penner and Hennessy [2], to classify differing Campylobacter isolates. This system, which is also called the heat stable or HS serotyping system, was originally thought to be based on lipopolysaccharides (LPS), also referred to as the “O” antigen, hence the frequent designation of “O” serotypes in earlier publications. However, more recent data show that C. jejuni does not express LPS, but instead expresses lipooligosaccharides (LOS) and a capsule polysaccharide (CPS) [3], [4]. The CPS has since been shown to be the primary serodeterminant of the Penner scheme [4], [5]. Today, there are 47 recognized HS serotypes of C. jejuni, which mirror CPS variation within the species [1]. These 47 serotypes, because of similarities in CPS structure, may further be refined into 35 serotypes, serotype cross-reactive pairs or complexes (See Methods). Because CPS is one of the few identified virulence factors of C. jejuni [6] and [7], the potential use of CPS-based vaccines to protect against infection has gained some interest and early evidence appears to support such a strategy. Vaccines based on the CPS of C. jejuni strain 81–176 (HS23/36) and CG8486 (HS4 complex) conjugated to CRM197 (a diptheria toxin mutant) reduced disease in the intranasal mouse model following challenge with the homologous strain. And, vaccines made with CPS of 81–176 also provided 100% protection against diarrheal disease in Aotus nancymaae, a new world monkey, when challenged with 81–176 [8] and 86% protection when challenged with CG8421, another HS23/36 strain which has been used in development of a human challenge model [9] and [Gregory et al. (in preparation)].
These data suggest that CPS based vaccines are a viable strategy to protect against diarrheal disease. However, continued pursuit of such a strategy will require answering questions about the required valency of a broadly effective CPS conjugate vaccine against C. jejuni. A first step in that direction will be determining the prevalence of CPS types circulating globally. Due to the correlation between CPS and Penner types, this review aims to summarize Penner serotyping data of clinical isolates, and by extension C. jejuni CPS type distribution, as reported in the published literature since the advent of Penner’s method.
Methods
Relevant published data were identified from searches of PubMed for research articles containing the keyword “Campylobacter” and the term “Penner” or “serotype”. At the same time, non-English publications, and review articles were excluded. The titles and abstracts of the identified articles were screened for relevance and evaluated independently by two of the study authors for inclusion on the basis of the availability of the article, and whether or not the article had previously unpublished, extractable data. Inclusion was limited to studies of natural sporadic C. jejuni infections in which human isolates were typed by the Penner-serotyping method. Research articles that reported data on fewer than ten isolates, data from outbreaks, or data from collections of isolates with evidence of selection bias (i.e. studies examining isolates from Guillain-Barré Syndrome patients only) were excluded. No further exclusionary restrictions were applied, such as the makeup of the study population, the length of the observation period, or the publication date. Disagreements between reviewers concerning inclusion were resolved by consensus. Data from studies selected for inclusion were extracted by two authors independently. The data were extracted according to a fixed protocol to include, the author, year, and location of the study, the demographics of the study population, the serotyping methodology used, the length of the study period, and the number of serotypes screened, and a count of the serotypes identified. Extracted data were entered independently into a Microsoft Access (Redmond, WA) database. Reported isolates were assigned to 1 of 35 commonly observed serotypes or cross-reactive serotype groups (HS1/44, HS2, HS3, HS4 complex (includes HS4/13/16/43/50/63/64/65), HS5/31, HS6/7, HS8/17, HS9, HS10, HS11, HS12, HS15, HS18, HS19, HS21, HS22, HS23/36, HS27, HS29, HS32, HS33, HS35, HS37, HS38, HS40, HS41, HS42, HS45, HS52, HS53, HS55, HS57, HS58, HS60, HS62, or Non-Typable). Some Penner serotypes were reported as belonging to more than one of the HS types indicated above, and such isolates were distributed across each serotype indicated. For example, an isolate reported as belonging to serotype HS24/55/60 would be distributed equally as HS24 = 0.33, HS55 = 0.33, and HS60 = 0.33. Several research articles also grouped a fraction of isolates into the non-descript category “Other”, when the relative proportion of a given serotype was below a reporting threshold determined by study authors. In these instances, the serotypes of the “Other” isolates were imputed across each study and distributed in the relative global proportions calculated for the 35 C. jejuni serotypes outlined above. C. coli serotypes, when reported, were not included in this analysis. Discrepancies concerning serotype assignment were resolved through discussion amongst all study authors. Serotypes were tallied within each study, and their respective proportions were calculated. Pooled proportional estimates were computed across all studies and within studies grouped by region. The proportional estimates were computed using the DerSimonian & Laird random effects model [10]. Strong evidence of heterogeneity existed across the studies for most of the serotypes examined, the exception being those rarely reported in the literature (HS22, HS29, HS32, HS33, HS35, HS38, HS40, HS41, HS42, HS45, HS52, HS55, HS57, HS60, HS62, and HS66). All statistical analyses were performed using Stata Version 12 (College Station, TX).
Results
A search of the PubMed database identified 596 research articles for possible inclusion. After removing the duplicates, 488 research articles remained for consideration. A review of the titles and abstracts excluded another 410 articles from consideration based on relevance to the topic of interest, leaving 78 studies to be assessed for eligibility for inclusion. The full text of each of the 78 articles was examined in more detail, and data from 54 studies were included for the purpose of this review. Five publications reported stratified data that are included as separate studies for the purpose of this review, bringing the total number of studies to 59 (See Table 1 and Supplementary Figure S1).
Table 1. Included Studies.
| First Author | Countrya | Totalb | Yearc | Duratione | Agef | CatchmentArea g, h | Serotypes Tested i |
| Karmali [13] | Canada | 285 | 1978 | 36 | Children 0 to >10 | Point | 55 |
| Taylor [14] | USA | 46 | 1980 | 6 | Mixed | Regional | NS |
| Skirrow [15] | England | 3400 | 1981 | 132 | Mixed | Country | 43 |
| McMyne [16] | Canada | 153 | 1982d | NS | NS | Regional | 55 |
| Lastovica [17] | South Africa | 258 | 1982 | 6 | Children <10 | Point | 60 |
| Georges-Courbot [18] | CAR | 94 | 1982 | 17 | Children <15 | Point | 56 |
| Neogi [19] | Bangladesh | 102 | 1983 | 12 | Mixed | Point | 42 |
| Patton [20] | USA | 149 | 1985d | NS | NS | Country | 56 |
| Jones [21] | Britain | 406 | 1985d | NS | NS | Unknown | 32 |
| Sjogren [22] | Sweden | 29 | 1985 | 12 | Adults >15 | Point | 23 |
| Sjogren [22] | Mexico | 130 | 1985 | 12 | Infants 0–5 | Point | 23 |
| Nishimura [23] | Japan | 69 | 1985 | NS | NS | Unknown | NS |
| Chatzipanagiotou [24] | Greece | 31 | 1987 | 12 | Children <14 | Point | 25 |
| Albert [25] | Australia | 108 | 1988 | 12 | Mixed | Regional | 66 |
| Albert [26] | Australia | 12 | 1988 | 6 | Mixed | Regional | 66 |
| Sjogren [27] | Kuwait | 47 | 1989 d | NS | Mixed | Point | NS |
| Zaman [28] | Saudi Arabia | 46 | 1989 | 12 | Mixed | Point | NS |
| Prasad [29] | India | 22 | 1989 | 132 | Mixed | Regional | 72 |
| Wareing [30] | England | 754 | 1990 | 7 | NS | Country | 42 |
| Takahashi [31] | Japan | 455 | 1990 | 156 | NS | Country | 25 |
| Owen [32] | UK | 27 | 1992 | 12 | NS | Country | 45 |
| Asrat [33] | Ethiopia | 35 | 1992 | 12 | Mixed | Point | 33 |
| Owen [34] | England | 398 | 1993 | 12 | NS | Country | 47 |
| Marshall [35] | England | 70 | 1994 d | NS | NS | Point | NS |
| Gibson [36] | UK | 27 | 1994 | 2 | NS | Country | 45 |
| Nishimura [23] | China | 85 | 1994 | NS | NS | Regional | NS |
| Fang [37] | Taiwan | 27 | 1994 | 120 | Mixed | Unknown | 25 |
| Nielsen [38] | Denmark | 136 | 1995 | 12 | NS | Country | 49 |
| Nielsen [39] | Denmark | 42 | 1995 | 11 | NS | Country | 47 |
| Poly [5] | Egypt | 142 | 1995 | 43 | Infants 0–5 | Point | 47 |
| Frost [40] | Wales | 2310 | 1996 | 12 | NS | Country | 66 |
| Hudson [41] | New Zealand | 69 | 1996 | 7 | NS | Point | NS |
| Strid [42] | Denmark | 173 | 1996 | NS | Mixed | Country | 47 |
| Petersen [43] | Denmark | 42 | 1996 | 24 | NS | Country | 47 |
| Smith [44] | Nigeria | 17 | 1997 d | NS | NS | Point | 64 |
| Sopwith [45] | England | 2277 | 1997 | 24 | Mixed | Regional | NS |
| McKay [46] | Scotland | 3155 | 1998 | 12 | NS | Country | 66 |
| Moser [47] | Germany | 201 | 1998 | 12 | NS | Regional | 9 |
| Chatzipanagiotou [24] | Greece | 98 | 1998 | 24 | Children <14 | Point | 25 |
| Poly [5] | Thailand | 103 | 1998 | 72 | Adults >15 | Country | 47 |
| Vierikko [48] | Finland | 518 | 1999 | 3 | NS | Country | 25 |
| Saito [49] | Japan | 158 | 2000 | 36 | NS | Regional | 25 |
| Eyles [50] | New Zealand | 54 | 2000 | 12 | Mixed | Regional | NS |
| Ioannidis [51] | Greece | 207 | 2000 | 36 | NS | Regional | 25 |
| Gilpin [52] | New Zealand | 66 | 2000 | 6 | NS | Regional | NS |
| Nielsen [53] | Denmark | 973 | 2001 | 12 | NS | Regional | 47 |
| Fussing [54] | Denmark | 926 | 2001 | 13 | Mixed | Regional | 47 |
| Wierzba [55] | Egypt | 20 | 2001 | 30 | Mixed | Point | NS |
| Oza [56] | England | 414 | 2002 d | NS | NS | Unknown | 66 |
| Cornelius [57] | New Zealand | 106 | 2002 | 2 | NS | Point | NS |
| Gilpin [58] | New Zealand | 168 | 2002 | 6 | NS | Regional | 43 |
| Schonberg-Norio [59] | Finland | 114 | 2002 | 3 | NS | Country | 25 |
| Sonnevend [60] | UAE | 41 | 2002 | 24 | NS | Point | 25 |
| Nakari [61] | Finland | 622 | 2002 | 48 | Mixed | Country | 25 |
| Nakari [61] | Finland | 785 | 2002 | 48 | Mixed | Country | 25 |
| Miljkovic-Selimovic [62] | Serbia | 29 | 2003 | 21 | NS | Regional | NS |
| McTavish [63] | New Zealand | 112 | 2006 | NS | Mixed | Country | 43 |
| Islam [64] | Bangladesh | 31 | 2006 | NS | NS | Point | NS |
| Grozdanova [65] | Macedonia | 20 | 2008 | 11 | NS | Regional | 25 |
Country = Country from which sporadic diarrhea cases were identified;
Total = Total number of isolates analyzed;
Year = Year specimen collection initiated.
When the year in which specimen collection began was not specified, publication year used;
Duration = Length of specimen collection period in months;
Age in years, “Mixed” indicates specimens collected from both children and adults;
Catchment indicates the size of the collection area,
Point = a single collection point (e.g. single hospital or clinic);
Serotypes Tested = number of serotypes included in the panel of screening sera used in each study. A number of studies screened for C. coli serotypes in addition to those for C. jejuni. Therefore, the number of serotypes screened for may exceed the 35 C. jejuni serotypes enumerated in this review. Abbreviations: CAR = Central African Republic, UAE = United Arab Emirates, UK = United Kingdom, USA = United States of America; NS = Not Specified.
In total, the studies were published between 1982 and 2011, reported data on 21,394 individual C. jejuni isolates from sporadic cases of enteric infection collected between 1978 to 2008 from 29 different countries (Table 1). Study size and duration varied considerably. The largest and smallest studies comprised 3,400 and 12 isolates, respectively (mean = 363), while the duration of the studies analyzed ranged from 13 years to 2 months. The included studies also varied in design (i.e. sampling methodology and the size of the catchment area) as well as in their target populations (i.e. age, traveler vs. resident populations). The number of serotypes screened for in each study also differed, ranging from nine to 72 serotypes (including serotypes for C. coli) (See Table 1).
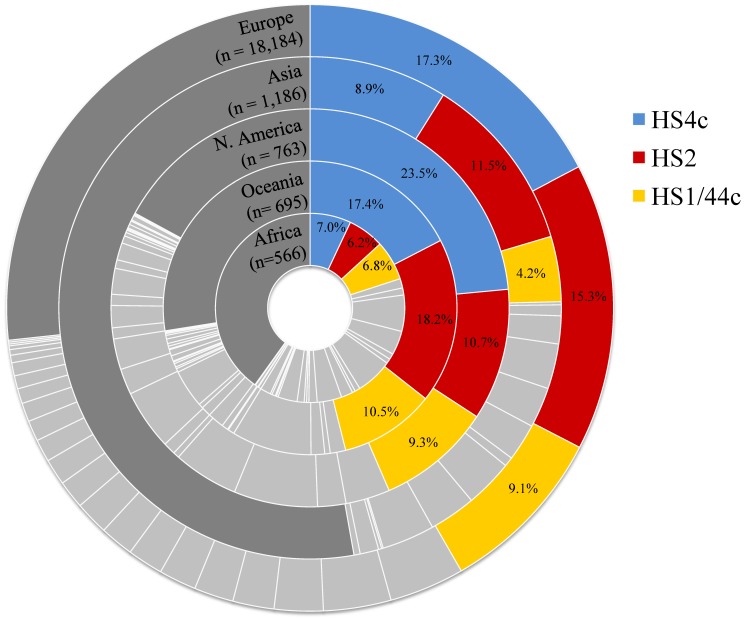
Overall, the studies predominately sampled European populations. Nearly 85% (n = 18,184) of the isolates included in this analysis were from Europe, while 1,186 were from Asia, 763 were from North America, 695 were from the Oceanic Region, and 566 were from Africa (Figure 1). No studies examining South America were identified in the literature search.
Figure 1. Proportional representation of the three most dominant HS serotypes (HS4c, HS2, and HS1/44) by region.
Lightly shaded areas represent the 33 (of 35) HS serotypes not indicated in color on the graph. Darkly shaded areas indicating those isolates not accounted for in the 35 HS serotypes examined were empirically derived by subtracting the sum of the percentages of the 35 serotypes from 100%. The darkly shaded area also includes non-typable isolates.
Globally, eight serotypes (HS4 complex, HS2, HS1/44, HS11, HS5/31, HS8/17, HS6/7, and HS3) accounted for 50.4% of all isolates. The dominant serotypes were those of the HS4 complex (15.3%, CI: 12.9, 17.6), HS2 (13.5%, CI: 11.3, 15.8), and HS1/44 (8.2%, CI: 7.1, 9.3) (See Table 2). Combined, these three serotype categories accounted for nearly 40% of all isolates reported worldwide. HS4 complex, HS2, and HS1/44 were also the three serotypes with the greatest proportional representation across each of the five regions examined (Table 3 and Figure 1). Moreover, these three serotypes remained the most prevalent serotypes when the data were stratified by the economic status of the country in which the study was conducted (Tables 4).
Table 2. Global HS Serotypes with Proportional Estimates of 2% or Greater.
| % | lci | uci | |
| Global (n = 21,394) | |||
| HS4c | 15.3 | 12.9 | 17.6 |
| HS2 | 13.5 | 11.3 | 15.8 |
| HS1/44 | 8.2 | 7.1 | 9.3 |
| HS11 | 3.1 | 2.2 | 4.0 |
| HS5/31 | 2.9 | 2.2 | 3.5 |
| HS8/17 | 2.8 | 2.2 | 3.4 |
| HS6/7 | 2.4 | 1.8 | 3.1 |
| HS3 | 2.2 | 1.7 | 2.7 |
Table 3. HS Serotypes with Proportional Estimates of 2% or Greater by Region.
| % | lci | uci | |
| Africa (n = 566) | |||
| HS4c | 7.0 | 2.8 | 11.2 |
| HS1/44 | 6.8 | 2.8 | 10.8 |
| HS3 | 6.3 | 1.1 | 11.6 |
| HS2 | 6.2 | 2.1 | 10.3 |
| HS5/31 | 6.2 | 2.3 | 10 |
| HS23/36 | 4.2 | 2.3 | 6.1 |
| HS8/17 | 4.1 | 0.1 | 8.1 |
| HS53 | 3.3 | 0.2 | 6.4 |
| HS19 | 2.0 | 0.6 | 3.4 |
| Asia (n = 1,186) | |||
| HS2 | 11.5 | 6.1 | 17 |
| HS4c | 8.9 | 4.3 | 13.5 |
| HS1/44 | 4.2 | 1.9 | 6.5 |
| HS15 | 3.4 | 1.1 | 5.7 |
| HS19 | 3.1 | 0.9 | 5.4 |
| HS23/36 | 3.0 | 0.9 | 5.0 |
| HS8/17 | 2.9 | 1.0 | 4.8 |
| HS3 | 2.6 | 1.1 | 4.2 |
| HS37 | 2.4 | 0.6 | 4.1 |
| Europe (n = 18,184) | |||
| HS4c | 17.3 | 14.6 | 20 |
| HS2 | 15.3 | 12.1 | 18.5 |
| HS1/44 | 9.1 | 7.7 | 10.4 |
| HS11 | 4.0 | 2.8 | 5.2 |
| HS6/7 | 3.6 | 2.7 | 4.5 |
| HS5/31 | 2.6 | 1.9 | 3.4 |
| HS8/17 | 2.2 | 1.5 | 2.9 |
| HS12 | 2.1 | 1.4 | 2.8 |
| HS58 | 2.0 | 1.0 | 3.0 |
| N. America (n = 763) | |||
| HS4c | 23.5 | 15.3 | 31.7 |
| HS2 | 10.7 | 4.3 | 17.1 |
| HS1/44 | 9.3 | 7.1 | 11.5 |
| HS5/31 | 6.8 | 3.0 | 10.5 |
| HS8/17 | 5.3 | 3.4 | 7.2 |
| HS3 | 4.9 | 1.8 | 8.1 |
| HS11 | 3.6 | 1.2 | 5.9 |
| HS21 | 2.5 | 0.8 | 4.2 |
| HS6/7 | 2.3 | 0.7 | 3.9 |
| HS18 | 2.1 | 0.8 | 3.4 |
| HS37 | 2.1 | 0.7 | 3.4 |
| Oceania (n = 695) | |||
| HS2 | 18.2 | 7.9 | 28.5 |
| HS4c | 17.4 | 10.7 | 24.0 |
| HS1/44 | 10.5 | 6.3 | 14.8 |
| HS8/17 | 8.8 | 3.5 | 14.1 |
| HS23/36 | 4.2 | 2.4 | 5.9 |
Table 4. HS Serotypes with Proportional Estimates of 2% or Greater by Economic Development Status.
| % | lci | uci | |
| Developed (n = 1,222) | |||
| HS4c | 17.5 | 15.2 | 19.8 |
| HS2 | 16.5 | 13.8 | 19.1 |
| HS1/44 | 9.0 | 7.8 | 10.1 |
| HS11 | 3.5 | 2.4 | 4.5 |
| HS6/7 | 2.9 | 2.1 | 3.6 |
| HS8/17 | 2.8 | 2.1 | 3.4 |
| HS5/31 | 2.6 | 2.0 | 3.3 |
| HS3 | 2.1 | 1.6 | 2.6 |
| Developing (n = 20,172) | |||
| HS4c | 8.2 | 4.8 | 11.5 |
| HS1/44 | 5.0 | 2.9 | 7.1 |
| HS2 | 5.0 | 2.8 | 7.3 |
| HS5/31 | 4.3 | 2.3 | 6.3 |
| HS3 | 3.7 | 1.7 | 5.7 |
| HS8/17 | 3.5 | 1.5 | 5.5 |
| HS23/36 | 3.3 | 1.5 | 5.1 |
| HS15 | 2.9 | 1.1 | 4.6 |
| HS53 | 2.9 | 1.0 | 4.8 |
Tables 2–4: HS serotypes with a proportional representation of 2% or greater, Globally (Table 2), by Region (Table 3), and by Economic Status (Table 4). Proportional estimates (%) were computed using the DerSimonian & Laird random effects model and include the upper (uci) and lower (lci) 95% confidence intervals. Note: Isolates categorized as a cross-reactive pair HS serotype (e.g. HS1/44, HS5/31, HS6/7, HS8/17, and HS23/36) were originally reported as one of the two serotypes or as the paired serotype itself. Isolates categorized as HS4 complex (or HS4c) represent isolates reported as any combination of the following serotypes HS 4/13/16/43/50/63/64/65.
Beyond the three most dominant serotypes, in all, 17 different serotypes reached a proportional representation of 2% or more in at least one of the five geographic regions considered (Table 5). Nine serotypes reached the 2% threshold in Africa, Asia, and Europe, accounting for 46.1%, 42%, and 58.2% of the total number of isolates in each region, respectively. Combined, the studies from North America had 11 serotypes with a representation of 2% or greater, totaling 73.1% of isolates, while five serotypes met the 2% threshold in Oceania, accounting for 59.1% of isolates in this geographic region. Notably, nearly 14% of isolates were non-typable by the Penner scheme globally (data not shown), a likely consequence of the fact that CPS expression in C. jejuni is known to be phase variable [6] and successful typing in the Penner scheme requires CPS expression. As discussed below, methodological differences amongst the studies may also contribute to an artificially inflated number of non-typable isolates.
Table 5. Comparison of HS Serotypes with Proportional Estimates by Region: Proportions that met or exceeded the 2% threshold are bolded and those that did not are indicated in italics.
| Global % | Africa % | Asia % | Europe % | N. America % | Oceania % | |
| (n = 21,394) | (n = 566) | (n = 1,186) | (n = 18,184) | (n = 763) | (n = 695) | |
| HS4c | 15.3 | 7.0 | 8.9 | 17.3 | 23.5 | 17.4 |
| HS2 | 13.5 | 6.2 | 11.5 | 15.3 | 10.7 | 18.2 |
| HS1/44 | 8.2 | 6.8 | 4.2 | 9.1 | 9.3 | 10.5 |
| HS11 | 3.1 | 1.6 | 0.2 | 4.0 | 3.6 | 1.7 |
| HS5/31 | 2.9 | 6.2 | 1.8 | 2.6 | 6.8 | 1.5 |
| HS8/17 | 2.8 | 4.1 | 2.9 | 2.2 | 5.3 | 8.8 |
| HS6/7 | 2.4 | 1.2 | 0.7 | 3.6 | 2.3 | 0.6 |
| HS3 | 2.2 | 6.3 | 2.6 | 1.9 | 4.9 | 0.7 |
| HS37 | 1.8 | 0.9 | 2.4 | 1.8 | 2.1 | 1.8 |
| HS23/36 | 1.7 | 4.2 | 3.0 | 1.4 | 1.8 | 4.2 |
| HS21 | 1.6 | 0.5 | 0.6 | 1.8 | 2.5 | 1.1 |
| HS19 | 1.5 | 2.0 | 3.1 | 1.5 | 0.9 | 0.5 |
| HS12 | 1.3 | 1.0 | 0.0 | 2.1 | 0.5 | 0.7 |
| HS58 | 1.3 | 0.8 | 0.0 | 2.0 | 1.0 | 0.1 |
| HS15 | 1.1 | 1.4 | 3.4 | 1.2 | 0.9 | 0.4 |
| HS18 | 0.9 | 0.4 | 0.1 | 1.1 | 2.1 | 0.2 |
| HS53 | 0.7 | 3.3 | 1.2 | 0.7 | 0.6 | 0.1 |
Discussion
Since Penner first introduced the method [2], serotyping has been an important means of characterizing Campylobacter isolates. Here, using existing data, we estimate the distribution of C. jejuni serotypes both globally and by geographic region. Estimates were derived from 59 published studies on more than 21,000 cases of sporadic diarrhea. Based on these estimates, eight serotypes account for half of all isolates globally and three serotypes in particular (HS4 complex, HS2, and HS1/44), were consistently represented across all regions.
Although this study is the first of its kind and a significant step forward in understanding the serotype distribution of C. jejuni infections, it is not without limitations. In fact, the estimates presented here are almost certainly imprecise. Data are sparse in every region of the world. No studies reporting extractable data were identified in South America and relatively few studies reported data from Africa and Asia, regions in which enteric infections contribute significantly to morbidity and mortality. The fact that some geographic regions are underrepresented may be partially due to the exclusion of non-English publications. However, the lack of data most probably reflects an absence of surveillance in these regions. With limited data from every region of the world, save Europe, the global estimates presented are biased towards those calculated in Europe. Even in Europe, from which 85% of the isolates in this study originated, there are insufficient data to draw conclusions regarding temporal changes in serotype distribution, geographic variation, and differences across demographic groups (e.g. travelers vs. non-travelers, or children vs. adults, etc.). The estimates presented here are also based on reports of sporadic cases of diarrhea. If an association between serotype and disease severity exists, selection bias has the potential to overestimate serotypes that result in manifest symptoms. Additionally, although a modest number of publications included in this review used a commercially available kit consisting of 25 antisera (Denka Seiken, Co), most studies relied upon custom reagents generated in-house or from another laboratory. The lack of standardized reagents calls into question the comparability of results across individual studies. Similarly, studies varied from one to the next with regards to which and how many serotypes were tested. These methodological differences undoubtedly influenced the estimates calculated here. Studies that did not screen a complete panel of antisera capable of detecting every serotype risked under-reporting certain serotypes, classifying them instead as non-typable. Finally, because C. jejuni is known to be subject to phase variation, assays such as Penner serotyping that depend upon the expression of CPS have the potential to underestimate the prevalence of any given Campylobacter serotype.
If current efforts to develop a CPS-based vaccine are to succeed, robust surveillance systems are needed to address substantial gaps in knowledge surrounding the geographic distribution and temporal stability of serotypes. Future surveillance methods should also aim to reveal demographic differences in serotype distribution (e.g. age, traveler vs. resident populations) and disease/serotype associations (e.g. severity of disease, risk of developing chronic long-term health outcomes such as reactive arthritis, Guillain-Barré syndrome, or gastrointestinal disorders). Combined with investigations into the immunogenic properties of the differing CPS types, addressing these fundamental surveillance-related questions will be important in determining the composition of a future vaccine with regards to valency. Furthermore, the need for surveillance is greatest in developing regions, where diarrheal disease is most prevalent and available data are lacking. Diarrheal episodes amongst children in the developing world are believed to cause millions of deaths annually [11] and, although the estimates are derived from a relatively small number of studies, the proportion of diarrheal cases attributable to Campylobacter infection is believed to be high, ranging between 5–20% of cases [12]. Given this high incidence rate, the potential benefit of a future vaccine is greatest in the developing world. However, realizing this potential will require a significant surveillance effort to inform the development of a multi-valent vaccine that is well-matched to CPS types circulating in these regions. Implementation of such a surveillance program will require a commitment of time and resources that has not been seen to date. Although Penner typing was once considered the gold standard in C. jejuni serotyping, its use has been declining in recent years and, today, the technique is routinely performed by only a small number of reference laboratories in North American and Europe. The limited and declining use of Penner typing is due in part to the complexity and cost of generating polyclonal rabbit sera to the 47 C. jejuni type strains, as well as to the emergence and value of other typing schemes such as Multi Locus Sequence Typing (MLST) and the ever-decreasing cost of direct sequencing. For a surveillance system to be implemented that is sufficiently large enough to address the outstanding questions of CPS distribution and disease association, alternative methodologies for determining the CPS type of C. jejuni isolates will almost certainly need to be employed. Such alternative methodologies will need to be cost-effective, efficient with respect to time, readily transferred to most any laboratory, and have high throughput capacity. Recently, our group offered a method that meets these criteria. Sequencing has revealed that each Penner serotype is unique with regards to the genomic structure of the cassette of genes involved in the biosynthesis of the serodeterminant CPS [5]. We have designed specific PCR primers that exploit these genomic differences and reproduce the original Penner serotypes. The published system covered 14 serotypes, and has recently been extended to 47 serotypes, (Poly et al. in preparation). Standardization and distribution of this CPS typing system offers one potential alternative method for large-scale surveillance. In addition to the already noted benefits this molecular typing system might offer, such a system may also reduce or eliminate the substantial number of non-typable isolates found in previous studies, as the described PCR-based typing system it is not sensitive to CPS expression. Regardless of which method is ultimately used, informed design of a CPS-based vaccine will require a substantial investment of resources to sustain the intensive surveillance needed to move beyond the incomplete and static picture that this review is able to offer.
Supporting Information
Flow diagram of articles search, reviewed, and included in the systematic review.
(DOCX)
Acknowledgments
The authors would like to thank Dr. Chad Porter for his expertise and guidance in statistical analysis.
Disclaimer: The views expressed in this article are those of the author and do not necessarily reflect the official policy or position of the Department of the Navy, Department of Defense, nor the U.S. Government. This is a US Government work. There are no restrictions on its use. There were no financial conflicts of interests among any of the authors. This study was conducted under support of the Military Infectious Disease Research Program.
Copyright Statement: Authors (BP and PG) are employees of the U.S. Government or military service members. This work was prepared as part of official duties. Title 17 U.S.C. §105 provides that ‘Copyright protection under this title is not available for any work of the United States Government.’ Title 17 U.S.C. §101 defines a U.S. Government work as a work prepared by a military service member or employee of the U.S. Government as part of that person’s official duties.
Funding Statement
This work supported by U.S. Navy Work Unit [6000.RAD1.DA3.A0308]. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Guerry P, Poly F, Riddle M, Maue AC, Chen YH, et al. (2012) Campylobacter polysaccharide capsules: virulence and vaccines. Front Cell Infect Microbiol 2: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Penner JL, Hennessy JN (1980) Passive hemagglutination technique for serotyping Campylobacter fetus subsp. jejuni on the basis of soluble heat-stable antigens. J Clin Microbiol 12: 732–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Parkhill J, Wren BW, Mungall K, Ketley JM, Churcher C, et al. (2000) The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403: 665–668. [DOI] [PubMed] [Google Scholar]
- 4. Karlyshev AV, Linton D, Gregson NA, Lastovica AJ, Wren BW (2000) Genetic and biochemical evidence of a Campylobacter jejuni capsular polysaccharide that accounts for Penner serotype specificity. Mol Microbiol 35: 529–541. [DOI] [PubMed] [Google Scholar]
- 5. Poly F, Serichatalergs O, Schulman M, Ju J, Cates CN, et al. (2011) Discrimination of major capsular types of Campylobacter jejuni by multiplex PCR. J Clin Microbiol 49: 1750–1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bacon DJ, Szymanski CM, Burr DH, Silver RP, Alm RA, et al. (2001) A phase-variable capsule is involved in virulence of Campylobacter jejuni 81–176. Mol Microbiol 40: 769–777. [DOI] [PubMed] [Google Scholar]
- 7. Maue AC, Mohawk KL, Giles DK, Poly F, Ewing CP, et al. (2013) The polysaccharide capsule of Campylobacter jejuni modulates the host immune response. Infect Immun 81: 665–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Monteiro MA, Baqar S, Hall ER, Chen YH, Porter CK, et al. (2009) Capsule polysaccharide conjugate vaccine against diarrheal disease caused by Campylobacter jejuni. Infect Immun 77: 1128–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tribble DR, Baqar S, Carmolli MP, Porter C, Pierce KK, et al. (2009) Campylobacter jejuni strain CG8421: a refined model for the study of Campylobacteriosis and evaluation of Campylobacter vaccines in human subjects. Clin Infect Dis 49: 1512–1519. [DOI] [PubMed] [Google Scholar]
- 10. DerSimonian R, Laird N (1986) Meta-analysis in clinical trials. Control Clin Trials 7: 177–188. [DOI] [PubMed] [Google Scholar]
- 11. O'Ryan M, Prado V, Pickering LK (2005) A millennium update on pediatric diarrheal illness in the developing world. Semin Pediatr Infect Dis 16: 125–136. [DOI] [PubMed] [Google Scholar]
- 12. Coker AO, Isokpehi RD, Thomas BN, Amisu KO, Obi CL (2002) Human campylobacteriosis in developing countries. Emerg Infect Dis 8: 237–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Karmali MA, Penner JL, Fleming PC, Williams A, Hennessy JN (1983) The serotype and biotype distribution of clinical isolates of Campylobacter jejuni and Campylobacter coli over a three-year period. J Infect Dis 147: 243–246. [DOI] [PubMed] [Google Scholar]
- 14. Taylor DN, McDermott KT, Little JR, Wells JG, Blaser MJ (1983) Campylobacter enteritis from untreated water in the Rocky Mountains. Ann Intern Med 99: 38–40. [DOI] [PubMed] [Google Scholar]
- 15. Skirrow MB, Jones DM, Sutcliffe E, Benjamin J (1993) Campylobacter bacteraemia in England and Wales, 1981–91. Epidemiol Infect 110: 567–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. McMyne PM, Penner JL, Mathias RG, Black WA, Hennessy JN (1982) Serotyping of Campylobacter jejuni isolated from sporadic cases and outbreaks in British Columbia. J Clin Microbiol 16: 281–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lastovica AJ, Le Roux E, Congi RV, Penner JL (1986) Distribution of sero-biotypes of Campylobacter jejuni and C. coli isolated from paediatric patients. J Med Microbiol 21: 1–5. [DOI] [PubMed] [Google Scholar]
- 18. Georges-Courbot MC, Baya C, Beraud AM, Meunier DM, Georges AJ (1986) Distribution and serotypes of Campylobacter jejuni and Campylobacter coli in enteric Campylobacter strains isolated from children in the Central African Republic. J Clin Microbiol 23: 592–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Neogi PK, Shahid NS (1987) Serotypes of Campylobacter jejuni isolated from patients attending a diarrhoeal disease hospital in urban Bangladesh. J Med Microbiol 24: 303–307. [DOI] [PubMed] [Google Scholar]
- 20. Patton CM, Barrett TJ, Morris GK (1985) Comparison of the Penner and Lior methods for serotyping Campylobacter spp. J Clin Microbiol 22: 558–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jones DM, Sutcliffe EM, Abbott JD (1985) Serotyping of Campylobacter species by combined use of two methods. Eur J Clin Microbiol 4: 562–565. [DOI] [PubMed] [Google Scholar]
- 22. Sjogren E, Ruiz-Palacios G, Kaijser B (1989) Campylobacter jejuni isolations from Mexican and Swedish patients, with repeated symptomatic and/or asymptomatic diarrhoea episodes. Epidemiol Infect 102: 47–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nishimura M, Nukina M, Yuan JM, Shen BQ, Ma JJ, et al. (1996) PCR-based restriction fragment length polymorphism (RFLP) analysis and serotyping of Campylobacter jejuni isolates from diarrheic patients in China and Japan. FEMS Microbiol Lett 142: 133–138. [DOI] [PubMed] [Google Scholar]
- 24. Chatzipanagiotou S, Papavasileiou E, Lakumenta A, Makri A, Nicolaou C, et al. (2003) Heat-stable antigen serotyping of Campylobacter jejuni strains isolated from hospitalized children in Athens, Greece. Eur J Epidemiol 18: 1097–1100. [DOI] [PubMed] [Google Scholar]
- 25. Albert MJ, Leach A, Asche V, Hennessy J, Penner JL (1992) Serotype distribution of Campylobacter jejuni and Campylobacter coli isolated from hospitalized patients with diarrhea in central Australia. J Clin Microbiol 30: 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Albert MJ, Tee W, Leach A, Asche V, Penner JL (1992) Comparison of a blood-free medium and a filtration technique for the isolation of Campylobacter spp. from diarrhoeal stools of hospitalised patients in central Australia. J Med Microbiol 37: 176–179. [DOI] [PubMed] [Google Scholar]
- 27. Sjogren E, Johny M, Kaijser B (1989) The serotype distribution of Campylobacter in patients with diarrhoea in Kuwait. FEMS Microbiol Lett 48: 237–239. [DOI] [PubMed] [Google Scholar]
- 28. Zaman R (1992) Campylobacter enteritis in Saudi Arabia. Epidemiol Infect 108: 51–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Prasad KN, Dixit AK, Ayyagari A (2001) Campylobacter species associated with diarrhoea in patients from a tertiary care centre of north India. Indian J Med Res 114: 12–17. [PubMed] [Google Scholar]
- 30. Wareing DR, Bolton FJ, Fox AJ, Wright PA, Greenway DL (2002) Phenotypic diversity of Campylobacter isolates from sporadic cases of human enteritis in the UK. J Appl Microbiol 92: 502–509. [DOI] [PubMed] [Google Scholar]
- 31. Takahashi M, Koga M, Yokoyama K, Yuki N (2005) Epidemiology of Campylobacter jejuni isolated from patients with Guillain-Barre and Fisher syndromes in Japan. J Clin Microbiol 43: 335–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Owen RJ, Fitzgerald C, Sutherland K, Borman P (1994) Flagellin gene polymorphism analysis of Campylobacter jejuni infecting man and other hosts and comparison with biotyping and somatic antigen serotyping. Epidemiol Infect 113: 221–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Asrat DA, Hathaway A, Sjogren E, Ekwall E, Kaijser B (1997) The serotype distribution of Campylobacter jejuni and C. coli isolated from patients with diarrhoea and controls at Tikur Anbassa Hospital, Addis Ababa, Ethiopia. Epidemiol Infect 118: 91–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Owen RJ, Slater E, Telford D, Donovan T, Barnham M (1997) Subtypes of Campylobacter jejuni from sporadic cases of diarrhoeal disease at different locations in England are highly diverse. Eur J Epidemiol 13: 837–840. [DOI] [PubMed] [Google Scholar]
- 35. Marshall LE, Boswell TC, Kudesia G (1994) False positive legionella serology in campylobacter infection: campylobacter serotypes, duration of antibody response and elimination of cross-reactions in the indirect fluorescent antibody test. Epidemiol Infect 112: 347–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gibson JR, Fitzgerald C, Owen RJ (1995) Comparison of PFGE, ribotyping and phage-typing in the epidemiological analysis of Campylobacter jejuni serotype HS2 infections. Epidemiol Infect 115: 215–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fang SW, Yang CJ, Shih DY, Chou CC, Yu RC (2006) Amplified fragment length polymorphism, serotyping, and quinolone resistance of Campylobacter jejuni and Campylobacter coli strains from chicken-related samples and humans in Taiwan. J Food Prot 69: 775–783. [DOI] [PubMed] [Google Scholar]
- 38. Nielsen EM, Engberg J, Madsen M (1997) Distribution of serotypes of Campylobacter jejuni and C. coli from Danish patients, poultry, cattle and swine. FEMS Immunol Med Microbiol 19: 47–56. [DOI] [PubMed] [Google Scholar]
- 39. Nielsen EM, Engberg J, Fussing V, Petersen L, Brogren CH, et al. (2000) Evaluation of phenotypic and genotypic methods for subtyping Campylobacter jejuni isolates from humans, poultry, and cattle. J Clin Microbiol 38: 3800–3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Frost JA, Oza AN, Thwaites RT, Rowe B (1998) Serotyping scheme for Campylobacter jejuni and Campylobacter coli based on direct agglutination of heat-stable antigens. J Clin Microbiol 36: 335–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hudson JA, Nicol C, Wright J, Whyte R, Hasell SK (1999) Seasonal variation of Campylobacter types from human cases, veterinary cases, raw chicken, milk and water. J Appl Microbiol 87: 115–124. [DOI] [PubMed] [Google Scholar]
- 42. Strid MA, Engberg J, Larsen LB, Begtrup K, Molbak K, et al. (2001) Antibody responses to Campylobacter infections determined by an enzyme-linked immunosorbent assay: 2-year follow-up study of 210 patients. Clin Diagn Lab Immunol 8: 314–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Petersen L, Nielsen EM, Engberg J, On SL, Dietz HH (2001) Comparison of genotypes and serotypes of Campylobacter jejuni isolated from Danish wild mammals and birds and from broiler flocks and humans. Appl Environ Microbiol 67: 3115–3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Smith SI, Coker AO, Olukoya DK (1997) Biotyping of Campylobacter strains isolated in Lagos, Nigeria using the modified Preston biotype. Z Naturforsch C 52: 259–263. [DOI] [PubMed] [Google Scholar]
- 45. Sopwith W, Ashton M, Frost JA, Tocque K, O'Brien S, et al. (2003) Enhanced surveillance of campylobacter infection in the North West of England 1997–1999. J Infect 46: 35–45. [DOI] [PubMed] [Google Scholar]
- 46. McKay D, Fletcher J, Cooper P, Thomson-Carter FM (2001) Comparison of two methods for serotyping Campylobacter spp. J Clin Microbiol 39: 1917–1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Moser I, Lentzsch P, Rieksneuwoehner B, Schwerk P, Wieler LH (2002) High resolution genotyping of Campylobacter jejuni strains by macrorestriction analysis with XhoI and polymerase chain reaction targeting enterobacterial repetitive intergenic consensus sequences: can we predict the zoonotic potential of strains? Epidemiol Infect 129: 435–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Vierikko A, Hanninen ML, Siitonen A, Ruutu P, Rautelin H (2004) Domestically acquired Campylobacter infections in Finland. Emerg Infect Dis 10: 127–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Saito S, Yatsuyanagi J, Harata S, Ito Y, Shinagawa K, et al. (2005) Campylobacter jejuni isolated from retail poultry meat, bovine feces and bile, and human diarrheal samples in Japan: comparison of serotypes and genotypes. FEMS Immunol Med Microbiol 45: 311–319. [DOI] [PubMed] [Google Scholar]
- 50. Eyles RF, Brooks HJ, Townsend CR, Burtenshaw GA, Heng NC, et al. (2006) Comparison of Campylobacter jejuni PFGE and Penner subtypes in human infections and in water samples from the Taieri River catchment of New Zealand. J Appl Microbiol 101: 18–25. [DOI] [PubMed] [Google Scholar]
- 51. Ioannidis A, Nicolaou C, Legakis NJ, Ioannidou V, Chatzipanagiotou S (2006) The first database comprised of flagellin gene (flaA) types of Campylobacter jejuni human clinical isolates from Greece. Eur J Epidemiol 21: 823–829. [DOI] [PubMed] [Google Scholar]
- 52. Gilpin BJ, Thorrold B, Scholes P, Longhurst RD, Devane M, et al. (2008) Comparison of Campylobacter jejuni genotypes from dairy cattle and human sources from the Matamata-Piako District of New Zealand. J Appl Microbiol 105: 1354–1360. [DOI] [PubMed] [Google Scholar]
- 53. Nielsen EM, Fussing V, Engberg J, Nielsen NL, Neimann J (2006) Most Campylobacter subtypes from sporadic infections can be found in retail poultry products and food animals. Epidemiol Infect 134: 758–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Fussing V, Moller Nielsen E, Neimann J, Engberg J (2007) Systematic serotyping and riboprinting of Campylobacter spp. improves surveillance: experiences from two Danish counties. Clin Microbiol Infect 13: 635–642. [DOI] [PubMed] [Google Scholar]
- 55. Wierzba TF, Abdel-Messih IA, Gharib B, Baqar S, Hendaui A, et al. (2008) Campylobacter infection as a trigger for Guillain-Barre syndrome in Egypt. PLoS One 3: e3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Oza AN, Thwaites RT, Wareing DR, Bolton FJ, Frost JA (2002) Detection of heat-stable antigens of Campylobacter jejuni and C. coli by direct agglutination and passive hemagglutination. J Clin Microbiol 40: 996–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Cornelius AJ, Nicol C, Hudson JA (2005) Campylobacter spp. in New Zealand raw sheep liver and human campylobacteriosis cases. Int J Food Microbiol 99: 99–105. [DOI] [PubMed] [Google Scholar]
- 58. Gilpin B, Cornelius A, Robson B, Boxall N, Ferguson A, et al. (2006) Application of pulsed-field gel electrophoresis to identify potential outbreaks of campylobacteriosis in New Zealand. J Clin Microbiol 44: 406–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Schonberg-Norio D, Sarna S, Hanninen ML, Katila ML, Kaukoranta SS, et al. (2006) Strain and host characteristics of Campylobacter jejuni infections in Finland. Clin Microbiol Infect 12: 754–760. [DOI] [PubMed] [Google Scholar]
- 60. Sonnevend A, Rotimi VO, Kolodziejek J, Usmani A, Nowotny N, et al. (2006) High level of ciprofloxacin resistance and its molecular background among Campylobacter jejuni strains isolated in the United Arab Emirates. J Med Microbiol 55: 1533–1538. [DOI] [PubMed] [Google Scholar]
- 61. Nakari UM, Huovinen E, Kuusi M, Siitonen A (2010) Population-based surveillance study of Campylobacter infections in Finland. Epidemiol Infect 138: 1712–1718. [DOI] [PubMed] [Google Scholar]
- 62. Miljkovic-Selimovic B, Ng LK, Price LJ, Kocic B, Babic T (2010) Characterization of Campylobacter jejuni and Campylobacter coli strains isolated in the region of Nis, Serbia. Srp Arh Celok Lek 138: 721–725. [DOI] [PubMed] [Google Scholar]
- 63. McTavish SM, Pope CE, Nicol C, Sexton K, French N, et al. (2008) Wide geographical distribution of internationally rare Campylobacter clones within New Zealand. Epidemiol Infect 136: 1244–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Islam Z, van Belkum A, Wagenaar JA, Cody AJ, de Boer AG, et al. (2009) Comparative genotyping of Campylobacter jejuni strains from patients with Guillain-Barre syndrome in Bangladesh. PLoS One 4: e7257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Grozdanova A, Poceva-Panovska A, Brezovska K, Trajkovska-Dokic E, Dimovski A, et al. (2011) Cross-reactive epitopes present in campylobacter jejuni serotypes isolated from enteritis patients. Prilozi 32: 113–125. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Flow diagram of articles search, reviewed, and included in the systematic review.
(DOCX)