Abstract
Reactive metabolites from carcinogens and oxidative stress can drive genetic mutations, genomic instability, neoplastic transformation, and ultimately carcinogenesis. Numerous dietary phytochemicals in vegetables/fruits have been shown to possess cancer chemopreventive effects in both preclinical animal models and human epidemiological studies. These phytochemicals could prevent the initiation of carcinogenesis via either direct scavenging of reactive oxygen species/reactive nitrogen species (ROS/RNS) or, more importantly, the induction of cellular defense detoxifying/antioxidant enzymes. These defense enzymes mediated by Nrf2-antioxidative stress and anti-inflammatory signaling pathways can contribute to cellular protection against ROS/RNS and reactive metabolites of carcinogens. In addition, these compounds would kill initiated/transformed cancer cells in vitro and in in vivo xenografts via diverse anti-cancer mechanisms. These mechanisms include the activation of signaling kinases (e.g., JNK), caspases and the mitochondria damage/cytochrome c pathways. Phytochemicals may also have anti-cancer effects by inhibiting the IKK/NF-κB pathway, inhibiting STAT3, and causing cell cycle arrest. In addition, other mechanisms may include epigenetic alterations (e.g., inhibition of HDACs, miRNAs, and the modification of the CpG methylation of cancer-related genes). In this review, we will discuss: the current advances in the study of Nrf2 signaling; Nrf2-deficient tumor mouse models; the epigenetic control of Nrf2 in tumorigenesis and chemoprevention; Nrf2-mediated cancer chemoprevention by naturally occurring dietary phytochemicals; and the mutation or hyper-expression of the Nrf2–Keap1 signaling pathway in advanced tumor cells. The future development of dietary phytochemicals for chemoprevention must integrate in vitro signaling mechanisms, relevant biomarkers of human diseases, and combinations of different phytochemicals and/or non-toxic therapeutic drugs, including NSAIDs.
Keywords: Dietary phytochemical, Nrf2, Antioxidant response, Inflammation, Epigenetics, Cancer stem cell
1. Introduction
The recent explosion of research on dietary phytochemicals has contributed much to our overall understanding of these compounds in terms of their chemical and biological functions and beneficial effects on human health. With the advent of cellular, molecular, and genomic experimental systems and in vivo transgenic and knockout animal models, a tremendous understanding of the mechanisms involved in the action of dietary phytochemicals has been achieved. Upon entering cells, these phytochemicals can directly scavenge free radicals and can also generate “chemical or electrophilic stress signals” that trigger proteins related to various cellular signaling pathways (Kong et al., 1998, 2001a; Finley et al., 2011). This ability includes the activation of the nuclear factor erythroid-2 (NF-E2)-related factor 2 (Nrf2)-Kelch-like ECH associated protein 1 (Keap1) complex. Activation of the Nrf2-Keap1 complex results in the induction of cellular defense mechanisms, including phase II detoxifying enzymes, phase III transporters, anti-oxidative stress proteins, and other stress-defense molecules that protect normal cells from reactive oxygen species (ROS) and/or reactive nitrogen species (RNS) and reactive metabolites of carcinogenic species. These protective mechanisms that block the initiation of carcinogenesis can be classically defined as chemoprevention, a concept that was originally introduced by Wattenberg (1966). The regulation of the growth, survival, and proliferative signaling pathways in initiated tumor cells would surpass that of normal cells because the cell-growth and cell-death machinery in tumor cells are dysregulated (Jacks & Weinberg, 2002). Interestingly, dietary phytochemicals could induce apoptotic cell death in pre-neoplastic or neoplastic cells through various growth inhibitory mechanisms, including the activation of cytochrome c (Cyt c)/caspases and cell cycle arrest and the inhibition of the nuclear factor-κB (NF-κB) and Janus kinase (JAK)-signal transducer and activator of transcription (STAT) signaling pathways, which results in the inhibition of tumor progression (Keum et al., 2004; Chen & Kong, 2005). However, advanced metastatic cancers have many genetic mutations, loss of heterozygosity, and/or epigenetic changes would be too resistant to radiation or chemotherapeutic drugs and likely would not respond to dietary phytochemicals alone.
Increasing data from both cancer epidemiology and experimental attempts support the bright future of dietary phytochemicals in chemoprevention and suggest that daily consumption is a promising new approach to prevent carcinogenesis. In this review, we will discuss the current progress in the study of dietary phytochemicals that block tumor initiation in normal cells and inhibit tumor progression in initiated/tumor cells, as well as the recent findings on epigenetic mechanisms and the existence of cancer stem cells (CSCs).
2. Oxidative stress and the defense system are mediated by dietary phytochemicals that inhibit cancer initiation
2.1. Oxidative stress and cancer
The generation of ROS/RNS is an essential metabolic process for maintaining homeostasis in our body. A small amount of ROS is released as a byproduct of mitochondrial respiratory electron-transportation (Harman, 1956). Various types of cells secrete ROS/RNS as a physiological signaling molecule used to communicate with other vital cells or to trigger certain signaling pathways (Sarti et al., 2002). Furthermore, ROS/RNS generated by immune cells are used to eliminate invading pathogens (Babior, 2000; Nathan & Ding, 2010). In contrast to the endogenous production discussed above, ROS/RNS is also generated by exogenous processes, such as UV radiation, inflammatory cytokines, lipid peroxidation, and environmental pollutants (Ma, 2010). The antioxidant defense system, such as superoxide dismutase (SOD), converts or conjugates these potentially deleterious ROS/RNS into less toxic molecules (Yu, 1994; Chae et al., 1999). Oxidative stress is an imbalance between the generation of ROS/RNS and the anti-oxidative stress defense systems in the body (Beckman & Ames, 1998; Halliwell & Gutteridge, 1999). In the context of oxidative stress, if acute inflammation fails to remove the exogenous stimuli or lasts too long, it will develop into chronic inflammation, which is believed to be the cause of many human diseases, including cancer (Coussens & Werb, 2002; Hussain & Harris, 2007). In chronically inflamed cells, the secretion of a large amount of ROS/RNS recruits more activated immune cells, which leads to the amplification of malfunction processes. If this crosstalk between inflammation and oxidative stress is prolonged, excessive cellular ROS/RNS will be produced, thereby exacerbating the oxidation of intracellular proteins, lipids, and nucleic acids (Kong et al., 1999; Ma, 2010; Schetter et al., 2010). This exacerbation could result in either genetic changes and/or epigenetic alterations leading to the dysregulation of oncogenes and tumor suppressor genes. Hence, the oxidative stress and chronic inflammation processes are tightly coupled, and the failure to stop these processes could result in genetic/epigenetic changes that drive the initiation of carcinogenesis (Chen & Kong, 2004; Hussain & Harris, 2007; Schetter et al., 2010) (Fig. 1). In addition, non-neoplastic stem/progenitor cells that have epigenetic disruptions are likely targets for further progression of carcinogenesis when challenged with chronic oxidative stress/inflammation (Feinberg et al., 2006). These initiated tumor cells could potentially be an important subject for cancer risk assessment and chemoprevention by dietary phytochemicals.
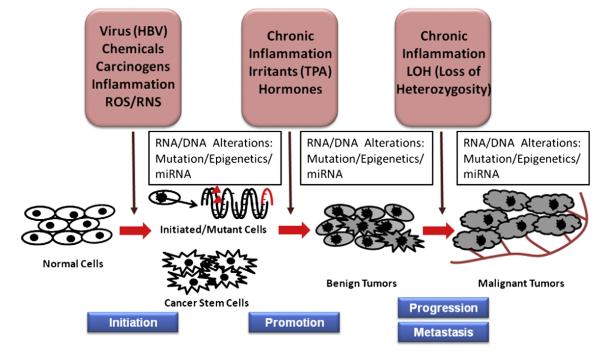
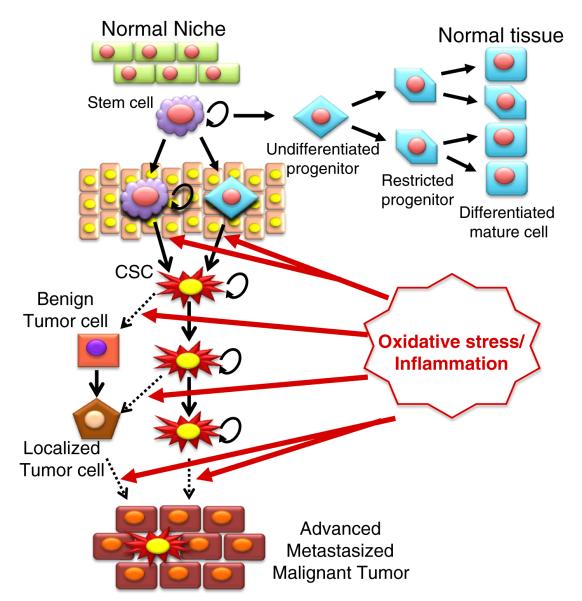
Fig. 1.
Schematic representation of multi-stage carcinogenesis. Exposure to intrinsic/extrinsic factors, including various toxic chemicals, oncogenes, viruses (e.g., HBV, hepatitis B virus), ROS/RNS, and inflammation, can resulting in genetic mutations and/or epigenetic alterations that cause the initiation of carcinogenesis in normal cells. The initiated cells and non-neoplastic cancer stem/progenitor cells can first progress to benign tumors, which would be amendable with surgery, radiation and or chemotherapy, if detected early, with subsequent progression to advanced/metastasized/malignant/drug-resistant tumors due to the prolonged effects of chronic inflammation, various irritants and aberrant hormones.
2.2. Nuclear factor erythroid-2 (NF-E2)-related factor 2–Kelch-like ECH associated protein 1 complex and phase II detoxifying/antioxidant enzymes
Phase II detoxifying/antioxidant enzymes provide a cytoprotective mechanism by reducing the toxicity of reactive intermediates, which results in cellular defense against oxidative or electrophilic challenges, and maintaining chemical homeostasis in cells (Kong et al., 1999; Kwak et al., 2001). The induction of phase II detoxifying and antioxidant enzymes, such as glutathione S-transferase (GST), UDP-glucuronosyltransferase (UGT), heme oxygenase-1 (HO-1), NADP(H):quinone oxidoreductase (NQO), glutamate cysteine ligase (GCL) and gamma glutamylcysteine synthetase (γGCS), among others, has been proposed as an effective chemopreventive approach because they can potentially block toxic and neoplastic processes in carcinogenesis (Motohashi & Yamamoto, 2004; Wakabayashi et al., 2004; Chen & Kong, 2005; Jeong et al., 2006). These phase II detoxifying/antioxidant enzymes conjugate oxidants or electrophilic xenobiotics with endogenous ligands, such as glutathione and glucuronide, to reduce their toxicity and reactivity by increasing the solubility of these conjugations and excreting them (Maheo et al., 1997; Talalay, 2000; Zhao et al., 2001; Paul et al., 2005; Pool-Zobel et al., 2005; Jeong et al., 2006). The genes encoding the detoxifying/antioxidant enzymes are typically regulated through a consensus cis-element at the 5′-flanking promoter region, known as the antioxidant responsive element (ARE) or electrophile response element (EpRE), which are located in their promoter region. ARE-mediated gene expression plays a central role in the cellular defense against oxidants and electrophiles (C. Chen & Kong, 2004). The transcriptional activation of ARE-mediated genes is mainly regulated by Nrf2, a member of the NF-E2 family of nuclear basic leucine zipper (bZIP) transcription factors and the Keap1 complex (Itoh et al., 1997, 1999; Motohashi & Yamamoto, 2004). Extensive research has been conducted both in vitro and in vivo using an Nrf2 (−/−) knockout (Nrf2 KO) mouse model, and clinical trials have demonstrated the pivotal role of Nrf2 in the regulation of ARE-mediated gene expression (Kong et al., 2001b; Balogun et al., 2003; Jeong et al., 2005; Saw et al., 2011). Many studies have reported that Nrf2, as a key player in cytoprotection, induced ARE-mediated type II detoxifying/antioxidant enzymes in response to antioxidative/electrophilic stimuli and therapeutic signals (Itoh et al., 1997; Alam et al., 2000; Jeong et al., 2005; He et al., 2007). Nrf2 KO mice showed increased susceptibility to chemical carcinogens and inflammatory inducers, confirming that the Nrf2 protective mechanism is applied to the whole body system (Khor et al., 2006; Osburn et al., 2007; Khor et al., 2008). A blinded clinical trial using oltipraz, an Nrf2 activator, was conducted recently in Qidong, P.R. China. Study participants who were exposed to dietary carcinogenic aflatoxins and were at high risk for the development of liver cancer received an oral administration of oltipraz daily. Oltipraz significantly enhanced the excretion in the urine of a non-toxic conjugated metabolite aflatoxin-glutathione catalyzed by an Nrf2-mediated GST phase II detoxifying enzyme (Kensler et al., 2000). This study demonstrated the role of Nrf2 in detoxifying metabolism and addressed the importance of utilizing food-based chemopreventive agents.
2.3. Mitogen-activated protein kinases signaling cascade
Mitogen-activated protein kinases (MAPKs) are characterized as proline directed serine/threonine kinases that convert various extracellular signals into intracellular responses through serial phosphorylation cascades and the activation of transcription factors. MAPKs are involved in the regulation of proliferation, differentiation, stress-reduction, and apoptosis (Cobb & Goldsmith, 1995; Chang & Karin, 2001). In yeast and mammalian cells, three distinct but parallel MAP cascades have been identified: extracellular signal-regulated kinase (ERK), c-JUN NH2-terminal protein kinase (JNK), and p38 (Kong et al., 1998). Each cascade consists of a module of three kinases. Phosphorylation is mediated by a MAPK kinase (MAP2K) activated through phosphorylation by its MAPKK kinase (MAP3K) (Marshall, 1994). The ERK pathway plays an important role in controlling cell growth and differentiation. In response to growth factors, tyrosine-phosphorylated transmembrane receptors recruit the exchange factor SOS and adaptor protein Grb2 complex. This complex activates the membrane bound small G-protein Rat sarcoma (Ras) (Egan et al., 1993). Activated Ras recruits c-Raf (a MAP3K) to the membrane and activates it. c-Raf phosphorylates and activates MEK (a MAP2K), which in turn leads to ERK activation (Stokoe et al., 1994). In addition to tyrosine kinase receptors, ERK can also be activated through certain G-protein-coupled receptors and protein kinase c (PKC) (Burgering & Bos, 1995). JNK and p38 are known as stress-activated MAP kinases, because both are only modestly activated by growth factors and phorbol esters but potently activated by stress signals, such as UV light, inflammatory cytokines, protein synthesis inhibitors, DNA damaging agents and chemopreventive agents. Especially, JNK is called stress-activated protein kinases (SAPK). Two MAP2Ks (MKK4 and MKK7) that activate JNK have been identified (Hibi et al., 1993; Kyriakis et al., 1994; Lin, 2003). Several MAP3Ks, including members of the MEKK family ASK1, MLK, TAK1 and TPL-2, have been reported to act as MAP3Ks in JNK activation (Busciglio & Yankner, 1995; Nakahara et al., 1999; Raman et al., 2007). Similar to JNK, several MAP2Ks, such as MKK3, MKK6, and SEK, activate p38 MAPK by phosphorylation at Thr180 and Tyr182 (Lee et al., 1994). Both JNK and p38 are activated by ASK1 and lead to cytokine- and stress-induced apoptosis. Redox regulatory protein thioredoxin (Trx) associates with ASK1 and inhibits ASK1 activity and the subsequent ASK1-dependent apoptosis, which demonstrates the effect of antioxidants on cytokine- and stress-induced apoptosis (Saitoh et al., 1998). The JNK cascade is involved in the regulation of many cellular events, including growth control, tumor development, and apoptosis. Despite the many studies on the mechanisms of JNK regulation, the role of JNK activation in apoptosis is highly controversial. Numerous studies have revealed that the JNK pathway possesses both pro-apoptotic and anti-apoptotic functions (Huang et al., 2010). Fig. 2 shows the schematic representation of the three MAPK pathways and where in the pathway the dietary phytochemicals potentially exert their effect.
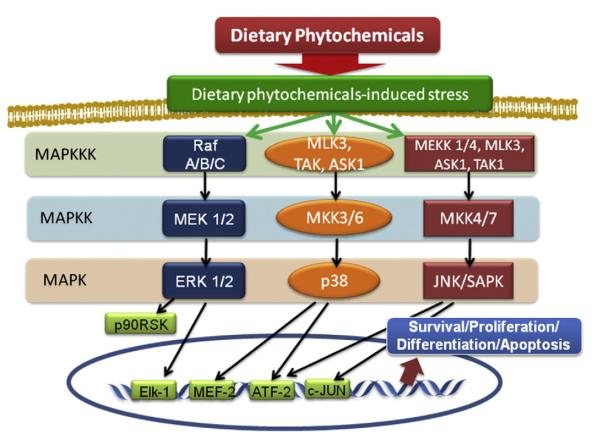
Fig. 2.
MAPK signaling transduction in mammalian cells. Activation of MAPK is regulated by hierarchical cascade known as the MAPK module. Mitogens such as growth factors, cytokines, and environmental stress typically activate MAP3Ks, subsequently resulting in phosphorylation of MAP2Ks. Phosphorylated MAP2Ks activate terminal MAPKs such as Erk1/2, p38, and JNK/SAPK through phosphorylating specific threonine (T) and tyrosine (Y) residues of T-X-Y motif. Several downstream targets of MAPKs including p90RSK and transcription factors such as Elk-1, MEF-2, ATF-2, and c-JUN are specifically activated and resulted in subsequent diverse cellular events, including cell survival, proliferation, differentiation, and apoptosis. The exposure to chemopreventive dietary phytochemicals can activate MAPK pathway, leading to gene expression, apoptotic cell death and differentiation of cancer cells.
Big mitogen-activated protein kinase-1 (BMK-1), also known as Erk5, is a MAPK identified in 1995 (Lee et al., 1995; Zhou et al., 1995). Similar to other three MAPKs, BMK-1 has been shown to be activated various extracellular stimuli such as epidermal growth factor (Kato et al., 1998), IL-6 (Carvajal-Vergara et al., 2005), and hypoxia (Sohn et al., 2002) and regulated by MAPK cascade. As a member of MAPK family, BMK-1 has also been linked to various cellular events including proliferation, migration, and apoptosis (Nithianandarajah-Jones et al., 2012).
2.4. Nuclear factor erythroid-2 (NF-E2)-related factor 2 activity and regulation by kinases
Activation of the ERK2 and JNK1 pathways appears to enhance Nrf2 signaling (Yu et al., 1999; Alam et al., 2000; Yu et al., 2000a; Zipper & Mulcahy, 2000, 2003; Shen et al., 2004; Xu et al., 2006b). In contrast, the activation of the p38 MAPK pathway seems to inhibit Nrf2 signaling (Yu et al., 2000b; Zipper & Mulcahy, 2000; Shen et al., 2004; Keum et al., 2006). It was found that phosphorylation of Nrf2 by p38 promotes the association between the Keap1 and Nrf2 proteins (Keum et al., 2006). This finding suggests that p38 may inhibit Nrf2 signaling by enhancing Keap1 sequestration of Nrf2 in the cytoplasm. In contrast, by using a cell-free system, Huang et al. demonstrated that PKC can directly phosphorylate Nrf2 at Ser40 (Huang et al., 2000), thereby promoting its dissociation from Keap1 (Huang et al., 2002; Bloom & Jaiswal, 2003; Numazawa et al., 2003). Phosphoinositide 3-kinase (PI3K) has also been reported to play a role in Nrf2 activation (Lee et al., 2001; Kang et al., 2002a, 2002b; Nakaso et al., 2003). In response to oxidative stress, PI3K signaling is activated and results in depolymerization of actin microfilaments, which somehow facilitates Nrf2 nuclear translocation (Kang et al., 2002b). PKR-like endoplasmic reticulum kinase (PERK) has been shown to phosphorylate Nrf2 and enhance its nuclear translocation by disrupting Keap1 binding (Cullinan & Diehl, 2004). Transcriptional activation of Nrf2/ARE can be partially modulated by the blockade of the p38 MAPK signaling pathway (Keum et al., 2006). Multiple serine/threonine residues in Nrf2 have been identified as targets of MAPK-mediated phosphorylation. However, it was reported that these modifications only caused a slight reduction in Nrf2 nuclear accumulation, and the protein stability was not altered by Nrf2 phosphorylation in vivo (Sun et al., 2009). Direct phosphorylation of Nrf2 by MAPKs may have limited contributions in the modulation Nrf2 activity, and MAPKs may regulate the Nrf2 signaling pathway through indirect mechanisms (Sun et al., 2009). However, in the context of Nrf2 signaling, the study of the exact biological relevance among different kinases would require genetic knockout or overexpression mouse models. In addition, it has recently been shown that redox stress could enhance the translation of Nrf2, which leads to the current model of Nrf2 activation against stress responses (He et al., 2007; Li & Kong, 2009; Li et al., 2010b).
2.5. Nuclear factor erythroid-2 (NF-E2)-related factor 2 activation by dietary phytochemicals
Sulfur-containing dietary phytochemicals such as phenethyl isothiocyanate (PEITC) and sulforaphane (SFN) are known as potent phase II gene inducers, and these inductions are Nrf2 dependent (Cheung et al., 2009; Cheung & Kong, 2010). PEITC can induce the phosphorylation of ERK and JNK and subsequently phosphorylate Nrf2 and induce its nuclear translocation. Attenuated PEITC-induced ARE activity was observed when ERK and JNK signaling were inhibited (Xu et al., 2006b). Treating HeLa cells with PEITC transiently stimulated ARE-reporter gene expression in a dose-dependent manner. Overexpression of a dominant-negative JNK1 suppressed Nrf2-induced ARE reporter gene expression. PEITC treatment enhanced the expression of the ARE reporter gene in either Nrf2- or JNK1-transfected cells, suggesting that PEITC activates ARE-mediated phase II drug metabolism gene expression via JNK1- and Nrf2-dependent pathways (Keum et al., 2003). SFN attenuates Nrf2 degradation by modifying the Keap1-Nrf2 interaction, which results in the translocation of Nrf2. SFN can react with thiols of Keap1 by forming thionoacyl adducts, thereby releasing Nrf2 from Keap1 binding (Hong et al., 2005). Topical application of SFN decreased the incidence of 7,12-dimethylbenz(a)anthracene (DMBA)/12-O-tetradecanoylphorbol-13-acetate (TPA)-induced skin tumors in Nrf2 (+/+) wildtype (Nrf2 WT) mice. Importantly, there were no chemoprotective effects in sulforaphane-treated Nrf2 KO mice. The data demonstrated that the chemopreventative effect of SFN on DMBA/TPA-induced skin tumors was mediated by Nrf2 (Xu et al., 2006a). SFN treatment of Nrf2 WT but not Nrf2 KO mice restored the number of sunburn cells back to their basal level by 8 days post-UVB irradiation. Inflammatory biomarkers were decreased in SFN-treated Nrf2 WT mice but not KO mice, revealing an SFN-mediated protective effect of Nrf2 against UVB-induced skin inflammation (Saw et al., 2011). Turmeric curcumin and green tea (−)-epigallocatechin-3-gallate (EGCG) have been reported to regulate Nrf2 activity via a similar pathway (Balogun et al., 2003; Na et al., 2008). EGCG treatment increased the nuclear accumulation, ARE binding and transcriptional activity of Nrf2, as well as the expression of Akt and ERK1/2 in human breast epithelial cells, MCF10A. Loss of Nrf2 decreased sensitivity to the EGCG-induced expression of Nrf2-mediated enzymes, such as HO-1, suggesting that Nrf2 mediates EGCG-induced expression of antioxidant enzymes via Akt and ERK1/2 signaling (Na et al., 2008). It was found that oral administration of curcumin induced high levels of HO-1 and reduced hepatic injury in dimethylnitrosamine (DMN)-induced rats. Curcumin administration resulted in enhanced nuclear translocation and ARE-binding of Nrf2, suggesting that curcumin has hepatoprotection potential in DMN-induced hepatotoxicity through Nrf2 activation (Farombi et al., 2008).
Many studies have shown that inhibition of MAPKs by dietary phytochemicals greatly attenuated Nrf2 activity. Procyanidin B2 increased nuclear translocation of Nrf2 and activated ERKs and p38 (Rodriguez-Ramiro et al., 2012). Quercetin, a glycosylated form of flavonoid compounds, potentially inhibits neoplastic transformation by blocking the activation of the MAPK pathway and stimulating cellular protection signaling (Ding et al., 2010). Rune et al. demonstrated dietary bioactive compounds and food-extracts with human HepG2 cells. Interestingly, liver HepG2 cells treated with several dietary plant extracts, including coffee, thyme, broccoli, rosemary, turmeric and red onion, showed stronger EpRE activity than cells treated with pure compounds. This study suggested that there are synergistic effects of combinational compounds in whole foods (Balstad et al., 2011).
2.6. Glycogen synthase kinase-3 signaling cascade and regulation of nuclear factor erythroid-2 (NF-E2)-related factor 2 activity
In addition to MAPK, glycogen synthase kinase-3 (GSK-3) is a conserved kinase that regulates protein turnover in response to cellular signals, which ultimately regulates a variety of cellular processes, including cell proliferation, differentiation, apoptosis, inflammation and tumorigenesis (Mishra, 2010). Various diseases have been linked to this signaling pathway, including mood disorders, Alzheimer’s disease, diabetes, and cancer (Jope et al., 2007). GSK-3-dependent control of β-catenin stability is a central regulatory mechanism of the canonical Wnt signaling pathway, wherein β-catenin phosphorylated by GSK-3β is recognized by β-Trcp, which is rapidly degraded by the 26S proteasome (Hernandez et al., 2010). In addition, GSK-3 has been reported to be necessary for the full transcriptional activity of NF-κB, demonstrating that GSK-3 selectively supports the expression of a subset of genes activated by NF-κB proliferative signals (Hoeflich et al., 2000; Jope et al., 2007).
Interestingly, GSK-3β and Fyn tyrosine kinases have been shown to attenuate the transcriptional activity of Nrf2 (Salazar et al., 2006; Jain & Jaiswal, 2007). Lithium, a GSK-3β inhibitor, promotes Nrf2 transcriptional activity (Rojo et al., 2008; Correa et al., 2011). GSK-3β phosphorylates a group of Ser residues in the Neh6 domain of mouse Nrf2 that overlaps with an SCF/β-TrCP destruction motif (DSGIS, residues 334 to 338) and targets Nrf2 for degradation in a Keap1-independent manner (Rada et al., 2011) or prevents it from entering the nucleus (Salazar et al., 2006). An indirect mechanism may also exist for this regulation. The non-receptor tyrosine kinase Src appears to play a critical role in the nuclear export and degradation of Nrf2, and GSK-3β may act upstream and phosphorylate Src kinase. This may provide a negative feedback mechanism to switch off Nrf2 activation and restore normal cellular homeostasis (Niture et al., 2011). Similarly, GSK-3β phosphorylates Fyn, leading to the nuclear localization of Fyn, which can then phosphorylate Nrf2 on tyrosine 568, resulting in the nuclear export and further degradation of Nrf2 (Jain & Jaiswal, 2007; Kaspar et al., 2009). However, the in vivo and physiological relevance of these signaling mechanisms requires further study in humans.
2.7. Signal transducer and activator of transcription-3 signaling cascade and chemoprevention
The signal transducer and activator of transcription-3 (STAT3) is a transcription factor member of the STAT family (Raptis et al., 2011). Mutational activation of Rac1, RhoA, and Cdc42 regulates STAT3 activation and promotes tumor cell growth and survival, tumor angiogenesis and metastasis (Ram & Iyengar, 2001; Yu & Jove, 2004). High expression of STAT3 is frequently detected in human cancer, and a constitutively active form of STAT3 is able to transform cultured cells, which suggests that STAT3 is an important player in tumor progression (Bromberg et al., 1999; Raptis et al., 2011). Disruption of STAT3 signaling leads to growth inhibition and apoptosis in tumor cell lines and impaired tumor growth in mouse xenograft cancer models (Jing & Tweardy, 2005; Leeman et al., 2006; Germain & Frank, 2007; Zhang et al., 2007b; Johnston & Grandis, 2011). Dietary compounds, including guggulsterone, honokiol, curcumin, resveratrol, flavopiridol, and cucurbitacin, have been shown to possess anti-cancer effects by inhibiting the growth of cancer cell lines and xenograft tumors, and the inhibition of STAT3 activation appears to be involved (Jing & Tweardy, 2005; Leeman et al., 2006; Aggarwal et al., 2009; Leeman-Neill et al., 2010; Zhang et al., 2010a).
3. Dietary phytochemicals activate signaling pathways to suppress cancer progression in initiated tumor cells
3.1. Intrinsic/Extrinsic pathway inducing apoptosis
One of the hallmarks of cancer is a deficiency in apoptosis (Hanahan & Weinberg, 2000). The intrinsic pathway refers to apoptosis induced by the outer-membrane permeabilization of the mitochondria, which releases the pro-apoptotic factors cytochrome c, apoptosis inducing factor (AIF), smac-DIABLO and endonuclease G from the mitochondria into the cytoplasm (Hanahan & Weinberg, 2000; Li et al., 2001; Wang, 2001), thereby promoting caspase activation through the cytochrome c/Apaf-1/caspase-9 cascade (Danial & Korsmeyer, 2004). This intrinsic pathway involves complex interactions between the pro- and anti-apoptotic members of the Bcl-2 (B-cell lymphoma 2) family of proteins. BH3-only proteins, including BID, BAD, BIM, BMF, PUMA, and NOXA, activate the pro-apoptotic multi-BH domain proteins Bax and Bak (Scorrano et al., 2003). When death signals are received by the mitochondria, Bax and Bak form a requisite gateway by integrating into the mitochondrial outer membrane as homo-oligomerized multimers (Suzuki et al., 2000; Letai, 2005; Shu et al., 2010).
For the extrinsic apoptotic cascade, the cell-death pathway is activated through the binding of extracellular ligands of the tumor necrosis factor (TNF) family of proteins to pro-apoptotic death receptors by forming a death-inducing signaling complex that activates caspases 8 and 10, followed by the activation of caspases 3, 6, and/or 7, which is the same caspase machinery utilized by the intrinsic pathway (Ashkenazi & Dixit, 1998; Sprick & Walczak, 2004).
A large spectrum of dietary phytochemicals induces apoptosis by regulating the expression of Bcl-2 family proteins, thereby activating the extrinsic apoptotic pathway. Some phytochemicals, such as PEITC, ganoderic acid T, and polyphyllin D, induce apoptosis via caspases, mitochondrial dysfunction and/or activation of the tumor suppressor gene p53 (Yu et al., 1998; Cheung et al., 2005; Lee et al., 2005; Tang et al., 2006; Ma et al., 2009). PEITC and other structurally related isothiocyanates, except phenyl isothiocyanate (PITC), are capable of inducing apoptosis and Poly (ADP-ribose) polymerase (PARP) cleavage in a dose-dependent manner in HeLa cells through a caspase-3-dependent mechanism (Yu et al., 1998). Polyphyllin D (PD), a saponin found in the Chinese medicinal herb Paris polyphylla, induced the mitochondrial apoptotic pathway by reducing mitochondria membrane potential (Delta psi(m)) and releasing cytochrome c and AIF (Cheung et al., 2005). PD also showed similar effects on the human breast cancer cell lines MCF-7 and MDA-MB-231 and induced apoptosis. PD administration reduced the growth of xenografted MCF-7 cells by 50% (Lee et al., 2005). Ganoderic acid T from the Chinese medicinal herb Ganoderma lucidum markedly inhibited the proliferation of a highly metastatic lung cancer cell line (95-D) by inducing apoptosis and cell cycle arrest at the G(1) phase (Tsang & Kwok, 2010). A large body of research has found that both well-known compounds, such as EGCG, SFN, PEITC, and curcumin, and new dietary phytochemicals, have apoptosis-inducing anti-cancer effects.
3.2. c-JUN NH2-terminal protein kinase regulation and cell apoptosis
As discussed above, the JNK cascade has both pro- and anti-apoptotic characteristics. p53 and c-Myc have been reported to be JNK substrates, and they may play a role in the pro-apoptotic cellular response. JNK is not required for the death receptor signaling mediated by the initiator caspase-8, but it is required for the stress-induced release of mitochondrial cytochrome c, suggesting a role for JNK in the intrinsic apoptosis regulating pathway (Lin, 2003). The studies using mouse embryonic fibroblasts of JNK1 (−/−) JNK2 (−/−) demonstrated that the cells were resistant to apoptosis induced by stress, such as UV irradiation, whereas a fusion expression of JNK1-JNKK2 was sufficient to induce apoptosis (A. Lin, 2003). It has also been reported that JNK could phosphorylate and inactivate Bcl-2 and Bcl-XL, which are both negative regulators of mitochondrial cytochrome C release. An alternate hypothesis proposed by Anning Lin suggested that JNK serves as a modulator rather an intrinsic component of the apoptotic machinery. Activated JNK inactivates suppressors of the apoptotic machinery, which facilitates but does not alone induce apoptosis (Lin, 2003; Liu & Lin, 2005).
In many instances, the environmental stress sufficient for JNK activation does not cause apoptosis. That could be due to JNK-dependent apoptotic signaling pathways that can be blocked by activation of survival signaling pathways, including NF-kB, Akt/PKB and ERK (Wang et al., 2004; Nakano et al., 2006). The apoptotic inducing effects of the JNK cascade are both cell-type and stimulus dependent. The dynamic balance between growth factor-activated ERK and stress-activated JNK-p38 pathways may also be important in determining whether a cell survives or undergoes apoptosis (Lin, 2003; Liu & Lin, 2005; Bode & Dong, 2007).
A large number of dietary phytochemicals activate JNK and induce apoptosis. For instance, PEITC, a natural chemopreventive agent, is capable of inducing JNK activation and apoptotic signaling that is different from DNA-damaging agents. PEITC will not target JNK and JNK upstream kinases directly because it does not induce significant MKK4 or MKK7 activation. JNK dephosphorylation and inactivation rates were decreased in cells exposed to PEITC. PEITC promotes the proteasome-dependent degradation of the JNK-specific phosphatase M3/6 (Yu et al., 1998; Chen et al., 2002).
4. Nuclear factor erythroid-2 (NF-E2)-related factor 2 pharmacogenomics and dietary phytochemicals
Pharmacogenomics is the study of genetic perturbations in the global gene expression profile that are impacted by xenobiotics, pharmaceutical agents or dietary phytochemicals. The development of advanced DNA microarrays coupled with sophisticated bioinformatics technology has made it possible to perform this large-scale research simply on our bench top (Schena et al., 1995; Crettol et al., 2010). Thousands of genes from different tissues can be analyzed and quantified by hybridizing fluorescence-labeled nucleic acid with the DNA microarray platform (Fodor et al., 1993; Gerhold et al., 1999). Due to its broad spectrum of genes, the DNA microarray enables the global assessment of the induction/suppression of genes elicited by pharmacological agents or toxicological drugs (Afshari et al., 1999; Nuwaysir et al., 1999; Kudoh et al., 2000; Voehringer et al., 2000; Rushmore & Kong, 2002). In the context of Nrf2-dependent pharmacogenomics, Nrf2 KO and Nrf2 WT mice are typically used. For instance, using the Affymetrix murine genome U74Av2 oligonucleotide array, SFN treated WT mice showed up-regulation of various Nrf2-mediated genes, such as GST, UGT, and NQO1, in their small intestine compared with the vehicle-treated WT and Nrf2 KO mice (Thimmulappa et al., 2002). SFN treatment did not induce the expression of these genes in the Nrf2 KO mice. This study demonstrated that Nrf2-mediated genes were inducible by SFN and that the lack of Nrf2 severely dampened the induction (Thimmulappa et al., 2002). Similarly, pharmacogenomic studies investigating the role of SFN and PEITC in the modulation of Nrf2-dependent genes in the liver used Nrf2 KO mice and the Affymetrix mouse genome 430 2.0 array chip. We identified a large number of genes associated with the phase I and II xenobiotic-metabolizing enzymes, phase III transporters, antioxidant enzymes, anti-inflammatory, ubiquitin, the cell cycle, cell growth, metabolism, the stress response, kinases, phosphatases, cell adhesion, immune proteins, and transcription factors, among many others (Hu et al., 2006a, 2006b). But there are also differences in Nrf2-dependent gene expression between the two isothiocyanates SFN and PEITC, suggesting that in addition to Nrf2, other indirect interacting pathways could also play a role between the two closely chemically-related analogs.
Further pharmacogenomic studies on other dietary cancer preventive phytochemicals, such as green tea polyphenol EGCG and curcumin, in Nrf2 KO and WT mice were conducted (Shen et al., 2005, 2006). EGCG induced and suppressed the expression of 671 and 228 Nrf2-dependent genes in the liver and small intestine, respectively (Shen et al., 2005). With curcumin, 822 and 222 Nrf2-dependent genes were modulated in the liver and small intestine, respectively (Shen et al., 2006). These genes, in addition to the predicted anti-oxidative stress/phase II detoxifying enzymes/phase III transporters, many other groups of genes including ubiquitination and proteolysis, electron transport, anti-inflammatory, cell growth and apoptosis, cell adhesion, kinases and phosphatases, nuclear co-regulators and transcription factors, among others, are Nrf2-dependent (Shen et al., 2005). Most recently, the effects of Chrysanthemum zawadskii (CZ) tea and licorice Glycyrrhiza uralensis (LE) extracts on the pharmacogenomic regulation of Nrf2-mediated genes in the livers of Nrf2 KO and WT mice have also been studied. DNA microarray results have revealed that the CZ extract exhibited stronger effects on the expression of Nrf2-dependent phase II (detoxification and antioxidant enzymes) and phase III (transporter) genes than LE in the liver (Wu et al., 2011).
In addition to the liver and small intestine, Nrf2-mediated pharmacogenomics studies in the prostates of Nrf2 KO and WT mice were also conducted (Barve et al., 2008). Oral administration of soy isoflavone modulated the expression of a large number of Nrf2-dependent genes, which can be classified into several broad categories: adhesion, apoptosis/autophagy, cell cycle, cell differentiation, DNA associated proteins, mRNA processing, transport, electron transport, lipid metabolism, signaling proteins, ubiquitination, phase II drug metabolizing enzymes, G-protein coupled receptors, cytoskeleton/extracellular matrix/smooth muscle associated, kinases/phosphatases, and protein folding/proteolysis (Barve et al., 2008). For example, genes regulating the cell cycle and cell growth, such as epiregulin, STAT, cyclin dependent kinase (cdk) inhibitor 1A, transforming growth factor (TGF) β, insulin-like growth factor (IGF), and hepatocyte growth factor (HGF), have been identified as Nrf2-dependent genes and were up-regulated by soy isoflavone. In contrast, a number of cell cycle and cell differentiation genes, including cyclin B3, G1 to S phase transition 2, MAD2-like 1, and microtubule associated protein 1b, were found to be down-regulated by soy isoflavone in an Nrf2-dependant fashion (Barve et al., 2008). Pharmacogenomic studies on the combinational treatment of SFN and EGCG in the prostate also showed increased Nrf2-dependent gene expression in the following categories: apoptosis/cell cycle, calcium ion binding, digestion, extracellular space, integral to plasma membrane, intracellular kinases, phosphatases, metal ion binding, transcription factors/interacting partners, transferases, ubiquitination, and other less well-known Nrf2-mediated genes (Nair et al., 2010). Overall, these Nrf2 pharmacogenomic studies suggest the tissue specificity (e.g., differences between the liver and the small intestine), and chemically-related specificity of Nrf2-mediated gene expression (differences between the different phytochemicals), and in general, the liver appears to be more responsive than the small intestine in the context of Nrf2-dependent genes. This poses the questions about the biologic basis and clinical implications of discovering differences between Nrf2-targeted phytochemicals or therapeutic agents in the context of drug discovery and development. Fig. 3 shows a schematic diagram of the impact of dietary phytochemicals on the regulation of Nrf2-dependent pharmacogenomics.
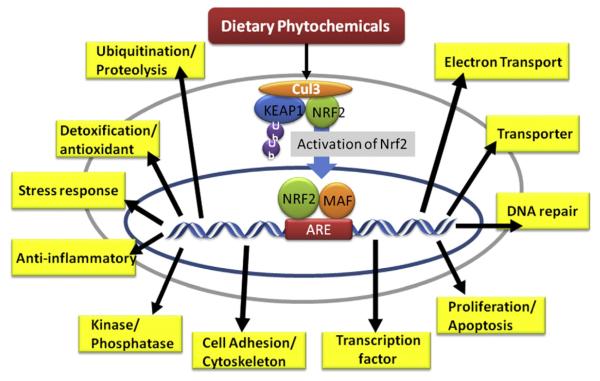
Fig. 3.
The impact of dietary phytochemicals on the regulation of Nrf2-dependent pharmacogenomics. Both Nrf2 KO and WT mice were treated with dietary phytochemicals or with vehicle (control group). DNA was extracted from the organ of interest, such as the liver, prostate, or small intestine, and then hybridized to a DNA microarray. Through a comparison of the compound-treated Nrf2 KO group vs. the treated WT and non-treated Nrf2 KO groups, a large amount of Nrf2-dependant compound-induced genes was found (Thimmulappa et al., 2002; Shen et al., 2005; Hu et al., 2006a, 2006b; Nair et al., 2006; Shen et al., 2006; Barve et al., 2008; Wu et al., 2011), and some of the representative groups of genes are presented.
To investigate the potential regulatory crosstalk between the two important transcription factors Nrf2 and NF-κB1, bioinformatic analyses on the promoter regions of human and murine Nrf2 and NF-κB1 were investigated. NF-κB1 is involved in oxidative stress, inflammation and carcinogenesis (Nair et al., 2008). A canonical regulatory network shows concerted modulation of Nrf2 and NF-κB1 that involves several members of the MAPK family. This elucidation of the relationship between Nrf2 and NF-κB1 will help to better understand the transcriptional regulation and networks of transcription factors associated with the etiopathogenesis of inflammation and cancer (Nair et al., 2008). The regulatory network between Nrf2 and AP-1 in prostate cancer mediated by the dietary phytochemicals EGCG and SFN was also examined (Nair et al., 2010). In silico bioinformatic analysis was utilized to delineate the conserved transcription factor binding sites (TFBS) in the promoter region of Nrf2 and AP-1. A conserved TFBS signature was identified in the promoter regions of Nrf2 and AP-1. Interestingly, NF-κB was also conserved in the human sequence of Nrf2 and AP-1. The in vivo combination studies using EGCG and SFN described above show down-regulation of Nrf2-dependant genes, including several co-factors and co-repressors of Nrf2, Nrf3, and Apc. This study suggests that the in vivo chemopreventive effects of SFN+EGCG in the prostate could be mediated via the relationship between the Nrf2 and AP-1 signaling pathways (Nair et al., 2010).
5. Nuclear factor erythroid-2 (NF-E2)-related factor 2 knockout models
As described above, Nrf2 is one of the key ARE-binding transcription factors that regulate the expression of more than 200 genes, including phase II detoxifying and antioxidant genes (Chen & Kong, 2004, 2005; Yu & Kensler, 2005). Many dietary phytochemicals have been previously shown to exert their cancer chemopreventive effects via the Nrf2 signaling pathway. As a master regulator of stress adaptation and cytoprotection, the role of Nrf2 in the carcinogenesis of various types of cancers has been delineated using Nrf2 deficient mice.
Previously it was reported that Nrf2 KO mice were more susceptible to dextran sulfate sodium (DSS)-induced colitis (Khor et al., 2006). Similarly, Kensler’s laboratory also reported that Nrf2 KO mice have increased colonic inflammatory injury and formation of aberrant crypt foci upon DSS treatment (Osburn et al., 2007). Nrf2 KO mice were also found to be more susceptible to azoxymethane (AOM)-DSS-induced colorectal carcinogenesis (Khor et al., 2008).
In addition to colorectal carcinogenesis, the role of Nrf2 in the carcinogenesis of different cancer types has been previously reported. Nrf2 KO mice were more susceptible to DMBA/TPA-induced skin tumors (Xu et al., 2006a). The incidence of urinary bladder carcinoma by N-nitrosobutyl(4-hydroxybutyl)amine (BBN) was significantly higher in Nrf2-deficient mice (Iida et al., 2004). The multiplicity and incidence of liver tumors in male and female 2-amino-3-methylimidazo [4,5-f] quinoline (IQ)-treated Nrf2-deficient mice were significantly higher compared with the WT counterparts (Kitamura et al., 2007). In addition, Nrf2-deficient mice were also found to have significantly higher burdens of gastric neoplasias after treatment with benzo-[a]pyrene than WT mice (Ramos-Gomez et al., 2001). Although Nrf2 KO mice had no apparent difference in the formation of premalignant lesions, the mammary carcinoma growth rate, size, and weight are dramatically increased in Nrf2 KO mice (Becks et al., 2010).
Many chemopreventive compounds are strong activators of Nrf2. Treating transgenic adenocarcinoma of the mouse prostate (TRAMP) mice with γ-rich tocopherols inhibited prostate carcinogenesis via the restoration of Nrf2 and its target genes, including UGT, GST, Gpx and HO-1 (Barve et al., 2010). Similarly, broccoli sprouts were found to suppress prostate tumorigenesis in TRAMP mice through activation of the Nrf2 pathway (Keum et al., 2009). Using the AOM/DSS colitis-associated colon cancer model, DBM was reported to inhibit colorectal carcinogenesis through the Nrf2 signaling pathway (Cheung et al., 2010). Although most of the dietary phytochemicals are known to target multiple signaling pathways, the cancer chemopreventive effects of some of these phytochemicals have been reported to be Nrf2-dependent. SFN blocked benzo[a]pyrene-induced forestomach tumors in ICR mice by inducing phase II detoxification and antioxidant enzymes. The chemopreventive effect of SFN is Nrf2-dependent because the protective effect was abrogated in Nrf2 KO mice (Fahey et al., 2002). Similarly, we reported that the cancer chemopreventive effect of SFN on DMBA/TPA-induced skin carcinogenesis was also Nrf2-dependent (Xu et al., 2006a). The anti-inflammatory effects of polyunsaturated fatty acids (PUFAs) docosahexaenoic acid (DHA) and eicosapentaenoic acid (EPA) on LPS-induced inflammation in mouse peritoneal macrophages were also found to be mediated via the Nrf2-dependent pathway (Lin et al., 2008; Wang et al., 2010).
6. Nuclear factor erythroid-2 (NF-E2)-related factor 2 mutations in advanced tumor cells
The Neh2 domain of Nrf2 binds to the Kelch/double glycine repeat (DGR) of Keap1, thereby preventing Nrf2 from entering the nucleus and simultaneously targeting Nrf2 for degradation (Li & Kong, 2009). Thus, this interaction is critical for the basal control of Nrf2 transcriptional ability (Li & Kong, 2009). Interestingly, recent reports have demonstrated that in certain types of advanced human cancers, this interaction between Nrf2 and Keap1 is genetically modified via somatic mutations (Shibata et al., 2008; Hayes & McMahon, 2009; Kim et al., 2010; Hu et al., 2012; Shibata et al., 2011). Furthermore, polymorphisms in the promoter region of Nrf2 have been implicated in the predisposition of individuals to lung injury and oxidative stress-related diseases (Yamamoto et al., 2004; Marzec et al., 2007).
Somatic mutations in the ETGE and DLG motifs of Neh2 (the hinge and latch model, respectively) have been found in different types of human cancers, including lung, esophageal, skin, larynx and ovarian cancer. Interestingly, the reported mutations within the ETGE motif mainly correspond with changes, substitutions and duplications of conserved residues for Keap1 recognition (E79→duplication-K-Q-G, T80→I-K-P, G81→D-V, E82→D-L-G-Q-L), whereas mutations in the DLG motif are changes and substitutions (D29→G-H, L30→F, G31→A) (Shibata et al., 2008; Hayes & McMahon, 2009; Kim et al., 2010; Hu et al., 2012; Shibata et al., 2011). Components of the hinge and latch model process, which is required to immobilize the Lys residues between the DLG and ETGE motifs and enable Cul3-Rbx1-mediated ubiquitylation of Nrf2 (Hayes & McMahon, 2009), could be prone to mutation in advanced tumors. Such mutations could potentially result in constitutive over-expression of Nrf2, leading to increased survival of these tumor cells and resistance to chemotherapeutic drugs (Lau et al., 2008). Conversely, several somatic mutations have been reported in the Kelch/DGR domain of KEAP1 that are associated with uncontrolled Nrf2 expression (Padmanabhan et al., 2006). These mutations include G430→C in lung tumors, G364→C in adenocarcinoma and small cell lung cancer cell lines (Padmanabhan et al., 2006), G333→C in a lung tumor-derived cell line, frameshift (stop codon)348 in lung tumor patients, G350→S in a lung tumor-derived cell line, L413→R in a lung tumor-derived cell line, frameshift (stop codon)413 in lung tumor patients, frameshift (stop codon)443 in a lung tumor-derived cell line, frameshift (stop codon)457 in lung tumor patients and 6-aa deletion555-560 in lung tumor patients (Singh et al., 2006). In addition, somatic mutations outside the Kelch/DGR domain (BTB and IVR domains) have also been reported that result in the inactivation of Keap1 and over-expression of Nrf2 (Singh et al., 2006; Zhang et al., 2010b). Interestingly, loss of Keap1 expression has also been found by epigenetic hypermethylation of the Keap1 gene in human lung cancer cell lines and lung cancer tissues (Wang et al., 2008a).
Based on these data, it can be estimated that Keap1 and Nrf2 mutations occur at a frequency of ~25% during promotion and/or progression rather than the initiation stage of lung cancer tumorigenesis (Hayes & McMahon, 2009). This suggests that mutations in the Nrf2-Keap1 system may represent a novel oncogenic mechanism that leads to malignancy and may set a new paradigm for cancer therapy in which the inhibition of Nrf2 could be utilized to enhance the chemotherapeutic sensitivity of these tumor cells (Shibata et al., 2008). Furthermore, the aberrant over-expression of Nrf2 in the more advanced cancer cells could cause drug resistance to chemotherapy. However, in these tumor cells, Nrf2 might already be hyperactivated, and the Nrf2 activating ability of some dietary phytochemicals may not further enhance the activation of Nrf2-mediated drug resistance. Instead, these dietary phytochemicals would intercept the cell growth/survival signaling pathways as discussed above, overriding the Nrf2 survival pathway, resulting in tumor cell apoptosis and cell death. Further studies are necessary to choose the correct method to reduce drug resistance and induce cell killing in advanced cancer cells. Dissecting the Nrf2 activating ability versus the inhibition of cell growth ability in Nrf2 over-expressed tumor cells could shed light on this problem, especially in the context of prevention of cancer initiation versus treatment/killing cancer cells.
7. Epigenetic alteration in cancer
Epigenetics is defined as heritable changes in gene expression without changes in the DNA sequence (Wolffe & Matzke, 1999). DNA methylation, histone modifications, and altered microRNA (miRNA) expression are components of epigenetic alterations and essential mechanisms for mammalian development and the regulation of gene expression (Yoo & Jones, 2006; Winter et al., 2009). Recently, accumulating evidences have shown that epigenetic alterations can largely contribute to the development of cancer. The initiation and progression of cancer is driven not only by acquired genetic alterations but also epigenetic disruptions of gene expression (Esteller, 2008). In cancer cells, hypermethylation on certain promoter regions of tumor suppressor genes causes gene silencing, thereby blocking the expression of these tumor suppressor genes (Bird, 2002). Changes in chromatin structure influence gene expression by either activating or inactivating the transcription of those genes (Rodenhiser & Mann, 2006). Aberrant expression of miRNAs induces the degradation or inhibition of the translation of target messenger RNA (He & Hannon, 2004). It has been suggested that epigenetic mechanisms of gene regulation, including DNA methylation, chromatin modification, and miRNAs, are influenced by various endogenous factors, such as nutrients, infections, physical activity, and other environmental factors (Dolinoy et al., 2007). Recently, dietary chemopreventive phytochemicals, including curcumin, EGCG, resveratrol, SFN, and PEITC have all been shown to alter epigenetic processes, which appear to facilitate the inhibition of tumor growth (Li & Tollefsbol, 2010; Li et al., 2010a). Below, the three epigenetic mechanisms, DNA methylation, histone modification, and miRNA, are reviewed to potentially yield insight into cancer chemoprevention. The impact of dietary chemopreventive phytochemicals in terms of epigenetic modifications of genes will also be discussed below.
7.1. Deoxyribonucleic acid methylation
DNA methylation is a heritable modification of the DNA structure that occurs without sequence alteration and inhibits gene expression directly (Jones & Baylin, 2002). CpG dinucleotides are enriched in short regions known as CpG islands. The distribution of CpG islands is not uniform throughout the whole genome. Methylation of DNA is regulated by DNA methyltransferases (DNMT), including DNMT1, DNMT3a, and DNMT3b, and occurs at the 5′ position of the cytosine (C) residues within the CpG dinucleotides via the addition of a methyl (Me or CH3) group that forms 5-methylcytosine (Bird, 2002; Esteller, 2002). In general, most of the CpG islands throughout the genome are known to be methylated in normal cells. The global hypomethylation of DNA has been reported in variety of malignancies such as prostate and colonic neoplasms (Bedford & van Helden, 1987; Feinberg et al., 1988). The overall level of DNA hypomethylation in malignant tissue appears to be associated with the tumor progression. Most of the metastatic neoplasms had significantly lower 5-methylcytosine contents in their genomic DNA compared to most of the benign neoplasms or normal tissues (Diala et al., 1983; Gama-Sosa et al., 1983). But in contrast, hypermethylation in the promoter region of the CpG islands is usually found in cancer cells (Jones & Baylin, 2002) An imbalance of DNA methylation between widespread hypomethylation and regional hypermethylation may be a key characteristic of human neoplasia (Baylin et al., 1991). It has been reported that chronic methyl-deficient diet causes several cellular alterations such as increased oxidative stress, and hypomethylation of genomic DNA, resulting in development of liver tumors in rodent model (Pogribny et al., 2004, 2012). Hypermethylation of CpG islands is known to cause gene silencing by preventing the recruitment of transcription factors to the promoters of the gene (Bird, 2002; Esteller, 2008). In addition, methylated CpG islands may provide the binding sites for interactions with other methyl-CpG binding domain proteins (MBDs), including MBD1–MBD4 and methyl CpG binding protein 2 (MeCP2) (Lewis et al., 1992; Hendrich & Bird, 1998). Subsequently, these binding proteins further recruit the co-repressor complex, including histone deacetylases (HDACs), which results in transcriptional repression (Nan et al., 1998; Feng & Zhang, 2001). DNA methylation in cancer cells is now believed to be one of the major causes in transcriptional gene silencing in carcinogenesis. Many cancer related genes, such as hMLH1, MGMT (DNA repair), p16INK4a, p15INK4b, p14ARF (cell cycle), death-associated protein kinase (DAPK) (apoptosis), CDH1, CDH13 (cell cadherin), and GSTP1 (detoxification), are inactivated by this epigenetic alteration (Esteller, 2002).
In terms of functional consequences of DNA methylation, the flip side has been reported. Global hypomethylation of DNA has been shown in a variety of malignancies such as prostate and colonic neoplasms (Bedford & van Helden, 1987; Feinberg et al., 1988). The overall level of DNA hypomethylation in malignant tissue appears to be associated with the tumor progression. Most of the metastatic neoplasms had significantly lower 5-methylcytosine contents in their genomic DNA compared to most of the benign neoplasms or normal tissues (Diala et al., 1983; Gama-Sosa et al., 1983). However, in general, hypermethylation in the promoter region of the CpG islands is usually found in cancer cells (Jones & Baylin, 2002). An imbalance of DNA methylation between widespread hypomethylation and regional hypermethylation may be a key characteristic of human neoplasia (Baylin et al., 1991). It has been reported that chronic methyl-deficient diet causes several cellular alterations such as increased oxidative stress, and hypomethylation of genomic DNA, resulting in development of liver tumors in rodent model (Pogribny et al., 2004, 2012).
7.2. Histone modifications
Together with DNA methylation, epigenetic alterations are also attained through histone modifications (Khorasanizadeh, 2004). The nucleosome is the basic subunit of chromatin, which further condenses the chromatin forming the chromosome. A histone octamer consisting of an H3/H4 tetramer and two H2A/H2B dimers and wrapped with 146 bp of DNA forms the nucleosome. Histone plays a key role in maintaining the stability of the highly folded chromatin architecture. Post-translational modifications of histones regulate the chromatin structure, which is closely linked to gene expression (Berlowitz & Pallotta, 1972; Luger et al., 1997; Tremethick, 2007). There are two types of chromatin structures: heterochromatin (condensed) and euchromatin (extended) (Jenuwein & Allis, 2001). In general, heterochromatin is a tightly packed structure and hard for transcription factors to access, which yields repressed gene transcription. However, euchromatin is loosely packed and more accessible to transcriptional factors, which enables active gene expression (Lund & van Lohuizen, 2004). The majority of histone modifications occur at the lysine, arginine, and serine residues of the N-terminal tails extending from the histone core via modifications such as acetylation, methylation, phosphorylation, ubiquitination, and sumoylation (Fig. 4) (Luger et al., 1997; Berger, 2007; Bhaumik et al., 2007; Kouzarides, 2007; Ellis et al., 2009). Histone modifications can either activate or repress the transcriptional status, depending on their location and the type of the modifications involved (Kouzarides, 2007). For instance, euchromatin modifications, such as acetylation of histone 3 and histone 4 (H3 and H4) or di- or trimethylation (me) of H3K4, are associated with active transcription, whereas di- or trimethylation of H3K9 and trimethylation of H3K27 are involved in gene repression (Barski et al., 2007; Li et al., 2007a). Various histone-modifying enzymes are involved in histone modifications. Histone acetyltransferases (HATs) and histone methyltransferases (HMTs) add acetyl groups and methyl groups, respectively, to the histone tails covalently, whereas HDACs and histone demethylases (HDMs) remove acetyl and methyl groups, respectively, from histones (Shi, 2007; Haberland et al., 2009). The global disruption of the trimethylation of H4K20 and monoacetylation of H4K16 is commonly found in cancer cells (Fraga et al., 2005). Overexpression of HDACs such as HDAC1, HDAC2, and HDAC6 has also been observed in cancer cells (Bolden et al., 2006). A large number of dietary phytochemicals have been shown to have modifying effects on various histone-modifying enzymes (Li & Tollefsbol, 2010). Methylation at a cytosine of CpGs can recruit regulatory binding proteins or provide binding sites for these binding proteins. A family of MBDs is thought to lead transcriptional repression through its interaction with HDAC (Jones et al., 1998; Jones & Baylin, 2002). Thus, it has been suggested that histone modification collaborates with DNA methylation. Kikuchi et al. treated gastric cancer cells that had hypermethylated COX-2 promoter regions with both the DNMT inhibitor 5-deoxy-2′-azacytidine and HDAC inhibitor trichostatin A. The combined treatment of DNMT and HDAC inhibitors synergistically restored the expression of the COX-2 gene, while the DNMT inhibitor alone only partially restored gene expression (Kikuchi et al., 2002).
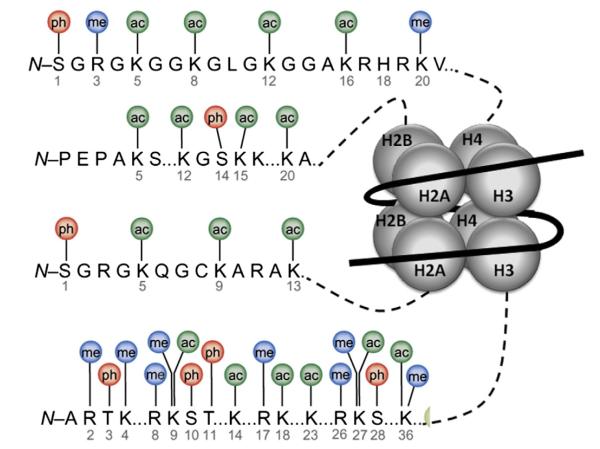
Fig. 4.
Histone tails affecting chromatin modification. Chromatin modifications usually appear at the amino acids in the N-terminal tails of histones (H2A, H2B, H3, H4), which provide the site for a wide range of posttranslational modifications. Various enzymes, such as histone acetyltransferases (HATs), histone methyltransferases (HMTs), histone deacetylases (HDACs), and histone demethylases (HDMs), are involved in these modifications, which cause covalent changes at the marked amino acids. ac: acetyl group, me: methyl group, ph: phosphate group. Modified from (Bhaumik et al., 2007).
7.3. Microribonucleic acids
MicroRNAs (miRNAs) were initially identified by an antisense RNA–RNA interaction in C. elegans and are largely conserved in various organisms (Lee et al., 1993; Pasquinelli et al., 2000). These short non-coding RNAs, which range from 17 to 25 nucleotides in length, are key players in regulating gene expression by inhibiting translation and/or triggering degradation of their target messenger RNAs (mRNAs) (Croce, 2009). miRNAs are also involved in a wide spectrum of biological events, including cell proliferation, apoptosis, tumor development, cell cycle, and immunity (Carleton et al., 2007; Negrini et al., 2007; Baltimore et al., 2008). More than 650 human miRNAs have been identified, and their extensive roles in global gene expression have been investigated. The database of miRNAs and their potential target genes is growing rapidly (Griffiths-Jones et al., 2006; Zhang et al., 2007a). The effect of miRNAs on epigenetic mechanisms and the mutual epigenetic regulations of miRNA expression appear to occur during carcinogenesis (Jones & Baylin, 2002; Esteller, 2007; Kai & Pasquinelli, 2010). In addition, accumulating data have demonstrated that dietary phytochemicals alter miRNA expression in cancer cells. Therefore, targeting specific miRNAs with dietary phytochemicals can be a novel therapeutic approach for chemoprevention (Sun et al., 2008; Izzotti et al., 2010; Li et al., 2010c; Tsang & Kwok, 2010).
7.4. Epigenetic modifications by dietary phytochemicals
Various dietary phytochemicals have been reported to have potential regulatory effects on DNMTs and histone-modifying enzymes, thereby restoring the expression of many tumor suppressor genes (Fang et al., 2003). Treating human prostate cancer LNCaP cells with green tea polyphenols (GTPs) for different days facilitates a concentration- and time-dependent re-expression of GSTP1, which correlates with DNMT1 inhibition. GTP treatment also decreased the mRNA and protein levels of MBD1, MBD4 and MeCP2, and HDAC 1–3 and increased the levels of acetylated histone H3 (LysH9/18) and H4 in a time-dependent manner. This result demonstrated that GTP altered DNA methylation and chromatin modeling (Pandey et al., 2010). Exposing human epidermoid carcinoma A431 cells to EGCG decreased global DNA methylation levels in a dose-dependent manner. EGCG decreased the levels of 5-methylcytosine, activity of DNMT, and mRNA and protein levels of DNMT1, DNMT3a and DNMT3b. EGCG also decreased histone deacetylase activity and increased the levels of acetylated lysine 9 and 14 on histone H3 (H3-Lys9 and 14) and acetylated lysine 5, 12 and 16 on histone H4 but decreased the levels of methylated H3-Lys 9. In addition, EGCG treatment resulted in the re-expression of the mRNA and proteins of silenced tumor suppressor genes, such as p16INK4a and Cip1/p21 (Nandakumar et al., 2011). SFN suppressed the growth of human PC-3 prostate cancer cells by 40% in nude mice via decreasing HDAC activity in the xenografts. SFN also showed a trend towards increased global histone acetylation in prostate cancer PC-3 xenografts (Myzak et al., 2007). In a human study, a single dose of broccoli sprouts significantly inhibited HDAC activity in peripheral blood mononuclear cells (Myzak et al., 2007).
Most recently, the expression of Nrf2 was found to be controlled epigenetically via promoter methylation (Yu et al., 2010). The expression of Nrf2 was suppressed epigenetically by CpG methylation of the promoter region associated with MBD2 and histone modifications in the prostate tumors of TRAMP mice. Treatment of TRAMP C1 cells with the DNMT inhibitor 5-aza-2′-deoxycytidine (5-aza) and HDAC inhibitor Trichostatin A (TSA) restored Nrf2 expression (Yu et al., 2010). Similarly, the treatment of TRAMP C1 cells with curcumin reduced the methylation level of the first five CpGs at the Nrf2 gene and binding of anti-mecyt antibody to the Nrf2 promoter. Demethylation of these CpGs by curcumin contributed to the re-expression of Nrf2 and its downstream gene NQO-1 (Khor et al., 2011). This study demonstrated that Nrf2 expression, a master regulator of the anti-oxidative stress system, appears to be controlled by epigenetic alterations, such as DNA methylation and histone modifications, and that dietary phytochemicals, such as curcumin, can block or reverse these epigenetic alterations, which results in the prevention of carcinogenesis in the TRAMP prostate cancer model. Interestingly, the CpG demethylation effect of curcumin was found not only in TRAMP C1 cells but also in human LNCaP prostate cancer cells (Shu et al., 2011). Neurog1 was found to be highly methylated in various cancer types and has been used as a specific marker for the CpG island methylator phenotype (CIMP) (Ogino et al., 2007; Herbst et al., 2011). We found that the first fourteen CpG sites of Neurog1 were dramatically demethylated by curcumin in LNCaP cells. Curcumin treatment had limited effects on the expression of MBD2, MeCP2, DNMT1, and DNMT3a but dramatically decreased MeCP2 binding to the promoter of Neurog1. Curcumin treatment decreased the total histone deacetylation (HDAC) activity and global enrichment of tri-methylation of histone 3 at lysine 27 (H3K27me3). However, at the individual protein expression level, curcumin increased the expression of HDAC1, 4, 5, and 8 but decreased HDAC3, suggesting a potential role for curcumin on histone modifications (Shu et al., 2011). However, there are also reports showing that DNMT inhibitors 5-azacitidine and its analogs, may induce in vitro malignant transformation and in vivo tumorigenesis (Carr et al., 1984; Rimoldi et al., 1991). Nevertheless, studies thus far suggest that the CpG demethylation ability of curcumin on Nrf2 and Neurog1 genes may lead to the subsequent activation of the expression of these genes and suggest a potential epigenetic modifying role for dietary phytochemicals in human prostate cancer cells.
8. Cancer stem cells
8.1. Cancer stem cell theory and supporting evidence
In recent years, the concept of the cancer stem cells (CSCs) has emerged to account for the heterogeneous cell population within a tumor and perpetuation of the tumor. We will briefly introduce the recent findings in CSC research. In tumors, there is a small subset of cancer cells that possess the constitutive ability to self-renew and differentiate and sustain the growth and spread of tumors (CSCs) (Reya et al., 2001). Accumulating data have demonstrated the existence of cancer stem cells in several human cancers (Lapidot et al., 1994; Bonnet & Dick, 1997; Al-Hajj et al., 2003; Singh et al., 2003; Li et al., 2007b; Schatton et al., 2008). However, the field of cancer stem cell research still has many questions to address, and the mechanisms of CSCs are different from the organ site where they occur. A pioneering study on acute myeloid leukemia (AML) brought much attention to the CSC theory. Particular fractionated human AML cells with the surface marker phenotype CD34+ CD38− regenerated large numbers of progenitors in transplanted severe combined immune-deficient (SCID) mice, whereas cells with a different surface phenotype did not show this property (Lapidot et al., 1994). Additional studies have supported the evidence of CSCs in various solid tumors, such as breast (Al-Hajj et al., 2003), colon (O’Brien et al., 2007), brain (Singh et al., 2003), pancreatic (Li et al., 2007b), liver (Yang et al., 2008) and melanoma (Schatton et al., 2008) cancer. In breast tumorigenesis, CD44+CD24−/lowLineage-cells isolated from patients were able to initiate tumors in mice from only 100 cells, whereas tens of thousands of cells with alternate phenotypes failed to form tumors. During the serial passages of the tumorigenic subpopulation, cells in this population generated tumors containing cells with the CD44+CD24−/lowLineage− surface marker phenotype and diverse phenotypic populations of non-tumorigenic cells present in the initial tumor (Al-Hajj et al., 2003). Similarly, CD133 was used to identify CSCs in brain, pancreatic and colorectal cancers. Several in vitro assays, including tumor-sphere culture, have been used to isolate and identify CSCs. When cancer cells were cultured on nonadherent surfaces with serum-free media, the majority of differentiated cancer cells fail to survive, whereas self-renewing CSCs grew and formed spheroids (Dontu et al., 2003; Hemmati et al., 2003; Charafe-Jauffret et al., 2008).
In normal tissues, stem cells maintain their numbers and self-renewal ability by interacting with adjacent tissue stromal cells (niche) and generate common and more-restricted progenitor cells. Through serial mitotic divisions, these progenitor cells are differentiated into all types of mature cells comprising specific tissues (Reya et al., 2001; Clarke & Fuller, 2006; Visvader & Lindeman, 2008). Through the oxidative stress and/or inflammatory response, the niche can provide mutation signals. Stem cells that accumulate these mutation signals may acquire a tumorigenic capacity, resulting in CSCs. Using similar mutation signals provided by the niche, normal undifferentiated progenitor cells can continue to proliferate and ultimately generate a pool of self-renewing cells, and cancer cells can also be generated during these proliferations (Clarke & Fuller, 2006; Clarke et al., 2006). It has been shown that very few cells leaving the localized tumor can form metastasized tumor in distance site. With the self-renewal capacity and the compatible niche, CSCs are also key players in the metastatic process (Croker & Allan, 2008). Fig. 5 shows a schematic diagram illustrating the potential differences between CSCs, non-tumorigenic cells and tumor cells.
Fig. 5.
Potential differences between cancer stem cells (CSCs) and tumor cells. In normal tissues, stem cells maintain self-renewal ability ( ) by interacting with the tissue stroma (niche = green). Within the normal niche, stem cells generate progenitor cells that differentiate into mature cells. During cell division, the replication potential of daughter cells is decreased at each step. If oxidative stress and inflammation stimulate the niche, it could provide stem cells with mutation signals. In response to the mutation signals, stem cell may acquire a tumorigenic capacity and result in CSCs (star-shaped red). Undifferentiated progenitor cells (blue diamond) may also become CSC-like cells driven by the mutation signals. While CSCs proliferate within tumors, they may give a rise to all types of tumor cells at each stage of tumor progression. Benign and localized tumor cells can be potentially (dashed arrow) generated from CSCs. Advanced metastasized and malignant tumor can be formed driven by further mutational signals including additional epigenetic modifications and genetic mutations. In the context of CSC hypothesis, the subset of tumor cells containing CSCs may grow to tumor in a manner that is hierarchical and heterogeneous. In contrast, non-tumorigenic cancer cell may not form tumor successfully.
) by interacting with the tissue stroma (niche = green). Within the normal niche, stem cells generate progenitor cells that differentiate into mature cells. During cell division, the replication potential of daughter cells is decreased at each step. If oxidative stress and inflammation stimulate the niche, it could provide stem cells with mutation signals. In response to the mutation signals, stem cell may acquire a tumorigenic capacity and result in CSCs (star-shaped red). Undifferentiated progenitor cells (blue diamond) may also become CSC-like cells driven by the mutation signals. While CSCs proliferate within tumors, they may give a rise to all types of tumor cells at each stage of tumor progression. Benign and localized tumor cells can be potentially (dashed arrow) generated from CSCs. Advanced metastasized and malignant tumor can be formed driven by further mutational signals including additional epigenetic modifications and genetic mutations. In the context of CSC hypothesis, the subset of tumor cells containing CSCs may grow to tumor in a manner that is hierarchical and heterogeneous. In contrast, non-tumorigenic cancer cell may not form tumor successfully.
Because it is important to isolate and identify CSCs from the diverse population of cancer cells, CSC research has strongly relied on cell surface markers. To be classified as a CSC, it is essential for the cell to not only express a specific surface marker of CSCs but also to proliferate in a self-renewal assay. There is no surface marker that is only expressed on CSCs, and no universal marker can be used across the different types of tissues and cells (Clarke et al., 2006; Visvader & Lindeman, 2008). A variety of CSC markers have been used to identify the CSC, depending on the types of tumors and cells. For example, as introduced above, CD44+CD24−/lowLineage− expressing cells isolated from breast cancer patients generated tumors in SCID mice (Al-Hajj et al., 2003). A tumorigenic subpopulation of pancreatic cancer cells also expressed cell surface markers CD44 and CD24. Pancreatic cancer cells with the CD44+CD24+ESA+ phenotype had a 100-fold increase in tumorigenic potential (Li et al., 2007b). Surface markers used in both CSC models, CD44 and CD24, do not describe the self-renewal ability per se. The commonly used marker CD44 is mostly involved in the expression of adhesion molecules (Goodison et al., 1999). In addition, the widely used CD133 is a successful surface marker for isolating brain tumor stem cells (Singh et al., 2003), but it is also present in normal brain stem cells (Boivin et al., 2009) and in many non-stem cells (Mizrak et al., 2008).
8.2. Epigenetic events in cancer stem cells
It has been relatively well known that the fate of embryonic stem cells and somatic stem cells is partially governed by proteins from the polycomb group (PcG), which are epigenetic chromatin modifiers (van der Lugt et al., 1994; Valk-Lingbeek et al., 2004). It has been shown that the PcG gene B-cell-specific Moloney murine leukemia virus integration site 1 (Bmi-1) plays a key role in regulating the proliferative activity of normal stem/progenitor cells, and the lack of Bmi-1 in leukemic stem/progenitor cells diminishes the proliferation capacity, indicating the importance of Bmi-1 in the self-renewal of multiple stem cell types (Lessard & Sauvageau, 2003; Park et al., 2003). Overexpression of Bmi-1 promotes cell proliferation and induces leukemia through repression of the Cdkn2a (also known as ink4a/Arf) tumor suppressor (Leung et al., 2004). The enhancer of zeste homolog 2 (Ezh2), the histone methyltransferase of the polycomb repressor complex 2 (PRC2), is upregulated in many cancers, including leukemia, prostate cancer, and breast cancer. High expression of Ezh2 promotes localized, more primitive malignant cell types and is associated with high expression of Bmi-1 (Raaphorst et al., 2001). In addition, Ezh2 catalyzes tri-methylation of histone H3 lysine (H3K27me3). In turn, PRC1 complexes bind to the H3K27me3 and then ubiquitinate lysine 119 of H2A. Bmi-1 plays a key role in this ubiquitination. The action of these cooperating complexes promotes the silencing of tumor suppressor expression by elevating stem cell survival (Fischle et al., 2003; Orlando, 2003). Recent reports suggest that stem cell PcG targets are up to 12-fold more likely to have cancer-specific promoter DNA hypermethylation than non-targets, supporting epigenetic alterations in CSCs (Widschwendter et al., 2007). In the context of CSCs, which are considered more resistant to conventional chemotherapies and targeted therapies, preventive consumption of active reagents that can prevent normal stem/progenitor cells from becoming cancerous could be a promising strategy to overcome cancer development. Scc-13 skin cancer cells treated with EGCG showed reduced cell survival through the decreased expression of Bmi-1 and Ezh2, which correlated with a global reduction of H3K27me3 (Balasubramanian et al., 2010). SFN treatment also caused a concentration-dependent reduction in the Bmi-1 and Ezh2 expression in SCC-13 (Balasubramanian et al., 2011).
9. Conclusions and perspectives
As discussed above, compelling data demonstrating the anti-cancer effects of phytochemicals have been accumulating. Throughout this review, we introduced the great potential of dietary phytochemicals: (1) blocking the initiation of carcinogenesis via the induction of detoxifying/antioxidant enzymes; (2) inhibiting the progression of carcinogenesis via the activation of the apoptotic pathway and cell cycle arrest; (3) the restoration of aberrant epigenetic alterations as an anti-cancer mechanism; and (4) the removal of the self-renewal potential of CSCs. Table 1 describes various phytochemicals that have demonstrated anti-cancer effects using in vitro and in vivo approaches involving these mechanisms. Daily exposure to various toxicants, such as environmental pollutants, carcinogens, dietary mutagens, and solar radiation, is inevitable. Oxidative stress and inflammatory damages caused by these toxins result in the dysregulation of oncogenes and tumor suppressor genes, aberrant epigenetic alterations, and the initiation of CSCs. These stresses and reactive metabolites of carcinogens are strong driving forces behind carcinogenesis. Thus, integrating the Nrf2-Keap1 signaling system in chemoprevention using relatively non-toxic dietary phytochemicals would be a logical approach to block the initiation of carcinogenesis through the inhibition of the oxidative stress/inflammation/reactive metabolites of carcinogens. In addition, at the later stage of carcinogenesis, induction of apoptosis and cell cycle arrest in pre-cancerous cells and carcinoma cells can also be an appealing biological target of dietary phytochemicals to block the progression of these tumor cells. Furthermore, epigenetic modifications and the CSC model are emerging key players in carcinogenesis due to their critical roles in cellular evolution and regulation. A greater understanding of the global pattern of these epigenetic modifications by dietary phytochemicals can yield improved insights into chemopreventive strategies and the potential of dietary phytochemicals to inhibit CSC development.Fig. 6 summarizes our thoughts on the potential cancer chemopreventive strategies discussed above.
Table 1.
Various dietary phytochemicals with anti-cancer effects
| Dietary compound(s) |
Plant | Model | Molecular effect(s) | Treatment(s) | References |
|---|---|---|---|---|---|
| Anacardic acid | Cashew nuts | HeLa; 293 T; SQ20B; SCC35; KBM-5; Jurkat; H1299; A293; Du145; SCC4 |
Inhibited both inducible and constitutive NF-κB activation; down-regulated p300 histone acetyltransferase gene; Inhibited Tip60 HAT |
10–100 μM | Sun et al., 2006, 2008 |
| Benzyl isothiocyanate (BITC) |
Cruciferous vegetables |
RL34 | Induced apoptosis through a mitochondrial redox-sensitive mechanism; Induced GSTP1 mediated redox-regulation |
10–25 μM | Nakamura et al., 2000, 2002 |
| Caffeic acid | Coffee | MCF-7; MDA-MB-231; T-47D | Inhibited DNA methylation catalyzed by DNMT |
3.0 μM | Lee and Zhu, 2006 |
| Chlorogenic acid | Coffee | MCF-7; MDA-MB-231; T-47D; Molt 4; U937; THP-1; REH; KU812; KCL22; K562 (in vivo xenograft) |
Inhibited DNA methylation catalyzed by DNMT; Induced apoptosis of several Bcr-Abl-positive CML via activation of p38; Inhibited growth of xenografted K562 cells |
0.75–25 μM |
Bandyopadhyay et al., 2004; Lee and Zhu, 2006 |
| Curcumin | Curcuma longa | MJ (G11); Hut78; HH; LNCaP; BxPC-3; Tramp C1; Male Albino rats |
Induced apoptosis through the downregulation of STAT3 and NF-κB signaling pathways in CTCL cells; Decreased the total HDAC activity and the enrichment of H3K27me3 at the Neurog1 promoter region; Altered miRNA expression in human pancreatic cells; Induced demethylation ofNrf2; Activated NQO-1 through re-expression ofNrf2; Protected DMN-induced hepatotoxicity through Nrf2 related induction of HO-1 expression |
2.5–20 μM; 100–200 mg/kg (in vivo) |
Farombi et al., 2008; Sun et al.,2008; Zhang et al., 2010a; Khor et al., 2011; Shu et al., 2011 |
| Genistein | Soy | LNCaP; PC-3; DuPro; RWPE-1; COLO-357; L3.6pl; PC-3; H460; BxPC-3; MDA-MB-231; MCF-7; MDA-MB-468; MCF10A |
Activated TSGs by modulating either histone H3-Lysine 9 (H3-K9) methylation or deacetylation at gene promoters; Inhibited the AKT signaling pathway; Increased acetylated histones 3, 4, and H3/ K4 at the p21 and p16 transcription start sites; Inactivated gemcitabine-induced NF-κB in pancreatic tumor cells; Inactivated NF-κB and increased growth inhibition and apoptosis; Demethylated the promoter of the GSTP1 tumor suppressor gene in MDA-MB-468 cells |
10–50 μM | Banerjee et al., 2005; Li et al., 2005; Kikuno et al., 2008; King-Batoon et al., 2008; Majid et al., 2008 |
| ( − )-Epigallocatechin 3 gallate (EGCG) |
Green tea | HT-29; PC-3; KYSE 510; KYSE 150; MCF10A; HepG2 |
Reversed hypermethylation of p16INK4a, RARβ, MGMT, hMLHl in KYSE 510 and KYSE 150 cells; Induced MnSOD and HO-1 via Akt and ERK1/2 signaling; Induced apoptosis and down-regulated Bcl-2 in HepG2 |
5–100 μM; | Fang et al., 2003; Na et al., 2008; Tsang and Kwok, 2010 |
| Indole-3-carbinol (I3C) | Cruciferous vegetable |
PC-3 | Induced cell cycle arrest, inhibition of cell growth, and apoptosis; Up-regulated Bax and down-regulated Bcl-2 via the down-regulation of NF-κB; Induced apoptosis through the inhibition of Akt activation |
30–100 μM | Chinni et al., 2001; Chinni and Sarkar, 2002 |
| 6-Gingerol | Ginger | HaCaT; Hairless mice; Female ICR mice; NF-κB luciferase transgenic mice |
Reduced UVB-induced expression and transactivation of COX-2 and NF-κB translocation in vitro and in vivo; inhibited TPA-induced COX-2 expression in mouse skin in vivo by blocking the p38 MAP kinase- NF-κB signaling pathway |
5–30 μM | Kim et al., 2005, 2007 |
| Lycopene | Tomato | MDA-MB-468; MCF10A; THP-1 | Demethylated the promoter of the GSTP1 tumor suppressor gene in MDA-MB-468 cells; Induced demethylation ofRARb2 and the HIN-1 genes in the noncancerous cells MCF10A; Reduced ROS production and prevented cell cycle arrest in G0/G1 phase induced by oxysterol |
0.5–5 μM | King-Batoon et al., 2008; Palozza et al., 2010 |
| Phenethyl isothiocyanate (PEITC) |
Cruciferous vegetable |
LNCaP; Male C57BL/6 mice | Inhibited the activity and level of HDACs; Demethylated the promoter and restored the unmethylated GSTP1; Activated p21 by inhibition of HDACs; Attenuated cell growth by inhibiting c-myc and HDACs; Induced apoptosis and cell cycle arrest in AOM/DSS-induced mouse colorectal cancer |
0.5–10 μM; 0.05% in diet |
Wang et al., 2007; Wang et al., 2008b; Cheung et al., 2010 |
| Quercetin | Citrus | C6; RAW264.7 | Inhibited LPS/TPA induced transformation and iNOS expression; Reduced intracellular ROS production and mitochondria dysfunction via induction ofHO-1 |
25–100 μM | Chow et al., 2005; Chen et al., 2006 |
| Resveratrol | Grape | PC12; C7H2-2A10; CEM-C7H2; | Attenuated hydrogen peroxide-induced cytotoxicity, DNA fragmentation, and intracellular accumulation of ROS; Induced apoptosis via a mitochondrial pathway mediated by Bcl-2 |
25–200 μM | Jang and Surh, 2001; Tinhofer et al., 2001 |
| Sulforaphane (SFN) | Cruciferous vegetable |
MCF-7; MDA-MB-231; Male athymic nude BALB/c (nu/nu) mice; PC-3; HepG2; Nrf2 KO mice; C57BL/6 mice (Nrf2 WT) |
Repressed hTERT by impacting epigenetic modifiers, such as DNMT1 and DNMT3a; Decreased HDAC activity in PC-3 xenografts; Induced HO-1 modulated by the blockade of p38 MAPK signaling pathway; Induced the protective enzyme HO-1 in DMBA/TPA induced Nrf2 WT mice, not KO mice |
2.5–20 μM 100 nmol (topical application in vivo) |
Keum et al., 2006; Xu, Huang et al., 2006; Myzak et al., 2007; Meeran et al., 2010 |
Fig. 6.
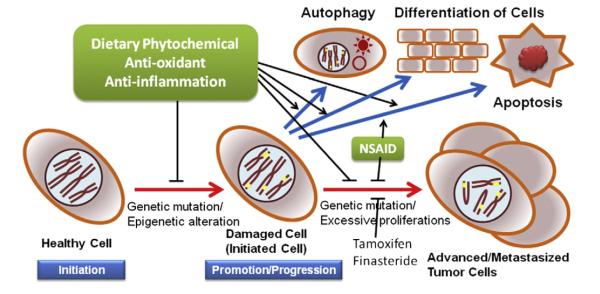
Cancer chemoprevention strategy using dietary phytochemicals and non-toxic therapeutic drugs. Oxidative stress, inflammation and reactive intermediates of carcinogens can cause genetic mutations and epigenetic alterations. Through the promotion/progression stages, initiated cells become advanced/metastatic tumor cells. Applying dietary phytochemicals at the early stage of carcinogenesis may block further development of carcinogenesis. Treatment with dietary phytochemicals and/or relatively non-toxic therapeutic drugs on cancer cells may induce positive results, including autophagy, cell cycle arrest, apoptosis, and differentiation, and may block tumor development.
Although significant progress has been achieved for current cancer imaging and diagnostic tools, the detection of tumorigenesis in its early stages is still not promising because cancers are often asymptomatic, with long latencies, and they continue to threaten human health. Thus, cancer preventative strategies to inhibit the initiation of carcinogenesis using naturally occurring dietary phytochemicals consumed daily are key players in chemoprevention.
In summary, cancer chemoprevention by dietary phytochemicals, either alone and or in combination with relatively non-toxic therapeutic drugs, such as NSAIDs (e.g., aspirin), statins, tamoxifen, or finasteride, offers substantial clinical benefits in the management of human cancers.
Acknowledgments
This work was supported in part by institutional funds and by R01-CA118947, R01-CA152826, R01-CA73674, R01-CA94828 from the National Cancer Institute (NCI), and R01AT007065 from the National Center for Complementary and Alternative Medicines (NCCAM) and the Office of Dietary Supplements (ODS) awarded to Dr. Ah-Ng Tony Kong from the National Institutes of Health (NIH). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NCI, NCCAM, ODS, or the National Institutes of Health.
Abbreviations
- AOM
azoxymethane
- ARE
antioxidant responsive element
- Bcl-2
B-cell lymphoma 2
- BMK-1
big mitogen-activated protein kinase-1
- DGR
double glycine repeat
- DMBA
7,12-dimethylbenz(a)anthracene
- DMN
dimethylnitrosamine
- DNMT
DNA methyltransferases
- DSS
dextran sulfate sodium
- EGCG
(−)-epigallocatechin-3-gallate
- ERK
extracellular signal-regulated kinase
- GSK-3
glycogen synthase kinase-3
- GST
glutathione S-transferase
- GTP
green tea polyphenol
- H3K27me3
tri-methylation of histone 3 at lysine 27
- HAT
histone acetyltranseferase
- HDAC
histone deaccetylase
- HO-1
heme oxygenase-1
- JNK
c-JUN NH2-terminal protein kinase
- Keap1
Kelch-like ECH associated protein 1
- MAP2K
MAPK kinase
- MAPK
mitogen-activated protein kinase
- MAPK3K
MAPKK kinase
- MBD
methyl-CpG binding domain protein
- MeCP2
methyl CpG binding protein 2
- miRNA
microRNA
- mRNA
messenger RNA
- NF-κB
nuclear factor-κB
- NQO
NADP(H):quinine oxidoreductase
- Nrf2 KO
Nrf2 (−/−) knockout
- Nrf2 WT
Nrf2 (+/+) wildtype
- Nrf2
nuclear factor erythroid-2 (NF-E2)-related factor 2
- PEITC
phenethyl isothiocyanate
- RNS
reactive nitrogen species
- ROS
reactive oxygen species
- SFN
sulforaphane
- SOD
superoxide dismutase
- STAT
signal transducer and activator of transcription
- STAT3
signal transducer and activator of transcription-3
- TGF-β
transforming growth factor-β
- TPA
12-O-tetradecanoylphorbol-13-acetate
- UGT
UDP-glucuronosyltransferase
References
- Afshari CA, Nuwaysir EF, Barrett JC. Application of complementary DNA microarray technology to carcinogen identification, toxicology, and drug safety evaluation. Cancer Res. 1999;59:4759–4760. [PubMed] [Google Scholar]
- Aggarwal BB, Kunnumakkara AB, Harikumar KB, Gupta SR, Tharakan ST, Koca C, et al. Signal transducer and activator of transcription-3, inflammation, and cancer: how intimate is the relationship? Ann N Y Acad Sci. 2009;1171:59–76. doi: 10.1111/j.1749-6632.2009.04911.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam J, Wicks C, Stewart D, Gong P, Touchard C, Otterbein S, et al. Mechanism of heme oxygenase-1 gene activation by cadmium in MCF-7 mammary epithelial cells. Role of p38 kinase and Nrf2 transcription factor. J Biol Chem. 2000;275:27694–27702. doi: 10.1074/jbc.M004729200. [DOI] [PubMed] [Google Scholar]
- Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashkenazi A, Dixit VM. Death receptors: signaling and modulation. Science. 1998;281:1305–1308. doi: 10.1126/science.281.5381.1305. [DOI] [PubMed] [Google Scholar]
- Babior BM. Phagocytes and oxidative stress. Am J Med. 2000;109:33–44. doi: 10.1016/s0002-9343(00)00481-2. [DOI] [PubMed] [Google Scholar]
- Balasubramanian S, Adhikary G, Eckert RL. The Bmi-1 polycomb protein antagonizes the (−)-epigallocatechin-3-gallate-dependent suppression of skin cancer cell survival. Carcinogenesis. 2010;31:496–503. doi: 10.1093/carcin/bgp314. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- Balasubramanian S, Chew YC, Eckert RL. Sulforaphane suppresses polycomb group protein level via a proteasome-dependent mechanism in skin cancer cells. Mol Pharmacol. 2011;80:870–878. doi: 10.1124/mol.111.072363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balogun E, Hoque M, Gong P, Killeen E, Green CJ, Foresti R, et al. Curcumin activates the haem oxygenase-1 gene via regulation of Nrf2 and the antioxidant-responsive element. Biochem J. 2003;371:887–895. doi: 10.1042/BJ20021619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balstad TR, Carlsen H, Myhrstad MCW, Kolberg M, Reiersen H, Gilen L, et al. Coffee, broccoli and spices are strong inducers of electrophile response element-dependent transcription in vitro and in vivo—studies in electrophile response element transgenic mice. Mol Nutr Food Res. 2011;55:185–197. doi: 10.1002/mnfr.201000204. [DOI] [PubMed] [Google Scholar]
- Baltimore D, Boldin MP, O’Connell RM, Rao DS, Taganov KD. MicroRNAs: new regulators of immune cell development and function. Nat Immunol. 2008;9:839–845. doi: 10.1038/ni.f.209. [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay G, Biswas T, Roy KC, Mandal S, Mandal C, Pal BC, et al. Chlorogenic acid-inhibits Bcr-Abl tyrosine kinase and triggers p38 mitogen-activated protein kinase-dependent apoptosis in chronic myelogenous leukemic cells. Blood. 2004;104:2514–2522. doi: 10.1182/blood-2003-11-4065. [DOI] [PubMed] [Google Scholar]
- Banerjee S, Zhang YX, Ali S, Bhuiyan M, Wang ZW, Chiao PJ, et al. Molecular evidence for increased antitumor activity of gemcitabine by genistein in vitro and in vivo using an orthotopic model of pancreatic cancer. Cancer Res. 2005;65:9064–9072. doi: 10.1158/0008-5472.CAN-05-1330. [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Barve A, Khor TO, Nair S, Lin W, Yu S, Jain MR, et al. Pharmacogenomic profile of soy isoflavone concentrate in the prostate of Nrf2 deficient and wild-type mice. J Pharm Sci. 2008;97:4528–4545. doi: 10.1002/jps.21311. [DOI] [PubMed] [Google Scholar]
- Barve A, Khor TO, Reuhl K, Reddy B, Newmark H, Kong AN. Mixed tocotrienols inhibit prostate carcinogenesis in TRAMP mice. Nutr Cancer. 2010;62:789–794. doi: 10.1080/01635581003605896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baylin SB, Makos M, Wu JJ, Yen RW, de Bustros A, Vertino P, et al. Abnormal patterns of DNA methylation in human neoplasia: potential consequences for tumor progression. Cancer Cells. 1991;3:383–390. [PubMed] [Google Scholar]
- Beckman KB, Ames BN. The free radical theory of aging matures. Physiol Rev. 1998;78:547–581. doi: 10.1152/physrev.1998.78.2.547. [DOI] [PubMed] [Google Scholar]
- Becks L, Prince M, Burson H, Christophe C, Broadway M, Itoh K, et al. Aggressive mammary carcinoma progression in Nrf2 knockout mice treated with 7,12-dimethylbenz[a]anthracene. BMC Cancer. 2010;10:540. doi: 10.1186/1471-2407-10-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedford MT, van Helden PD. Hypomethylation of DNA in pathological conditions of the human prostate. Cancer Res. 1987;47:5274–5276. [PubMed] [Google Scholar]
- Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- Berlowitz L, Pallotta D. Acetylation of nuclear protein in the heterochromatin and euchromatin of mealy bugs. Exp Cell Res. 1972;71:45–48. doi: 10.1016/0014-4827(72)90261-3. [DOI] [PubMed] [Google Scholar]
- Bhaumik SR, Smith E, Shilatifard A. Covalent modifications of histones during development and disease pathogenesis. Nat Struct Mol Biol. 2007;14:1008–1016. doi: 10.1038/nsmb1337. [DOI] [PubMed] [Google Scholar]
- Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- Bloom DA, Jaiswal AK. Phosphorylation of Nrf2 at Ser40 by protein kinase C in response to antioxidants leads to the release of Nrf2 from INrf2, but is not required for Nrf2 stabilization/accumulation in the nucleus and transcriptional activation of antioxidant response element-mediated NAD(P)H:quinone oxidoreductase-1 gene expression. J Biol Chem. 2003;278:44675–44682. doi: 10.1074/jbc.M307633200. [DOI] [PubMed] [Google Scholar]
- Bode AM, Dong Z. The functional contrariety of JNK. Mol Carcinog. 2007;46:591–598. doi: 10.1002/mc.20348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boivin D, Labbe D, Fontaine N, Lamy S, Beaulieu E, Gingras D, et al. The stem cell marker CD133 (Prominin-1) is phosphorylated on cytoplasmic Tyrosine-828 and Tyrosine-852 by Src and Fyn tyrosine kinases. Biochemistry. 2009;48:3998–4007. doi: 10.1021/bi900159d. [DOI] [PubMed] [Google Scholar]
- Bolden JE, Peart MJ, Johnstone RW. Anticancer activities of histone deacetylase inhibitors. Nat Rev Drug Discov. 2006;5:769–784. doi: 10.1038/nrd2133. [DOI] [PubMed] [Google Scholar]
- Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3:730–737. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- Bromberg JF, Wrzeszczynska MH, Devgan G, Zhao Y, Pestell RG, Albanese C, et al. Stat3 as an oncogene. Cell. 1999;98:295–303. doi: 10.1016/s0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- Burgering BM, Bos JL. Regulation of Ras-mediated signalling: more than one way to skin a cat. Trends Biochem Sci. 1995;20:18–22. doi: 10.1016/s0968-0004(00)88944-6. [DOI] [PubMed] [Google Scholar]
- Busciglio J, Yankner BA. Apoptosis and increased generation of reactive oxygen species in Down’s syndrome neurons in vitro. Nature. 1995;378:776–779. doi: 10.1038/378776a0. [DOI] [PubMed] [Google Scholar]
- Carleton M, Cleary MA, Linsley PS. MicroRNAs and cell cycle regulation. Cell Cycle. 2007;6:2127–2132. doi: 10.4161/cc.6.17.4641. [DOI] [PubMed] [Google Scholar]
- Carr BI, Reilly JG, Smith SS, Winberg C, Riggs A. The tumorigenicity of 5-azacytidine in the male Fischer rat. Carcinogenesis. 1984;5:1583–1590. doi: 10.1093/carcin/5.12.1583. [DOI] [PubMed] [Google Scholar]
- Carvajal-Vergara X, Tabera S, Montero JC, Esparis-Ogando A, Lopez-Perez R, Mateo G, et al. Multifunctional role of Erk5 in multiple myeloma. Blood. 2005;105:4492–4499. doi: 10.1182/blood-2004-08-2985. [DOI] [PubMed] [Google Scholar]
- Chae HZ, Kang SW, Rhee SG. Isoforms of mammalian peroxiredoxin that reduce peroxides in presence of thioredoxin. Methods Enzymol. 1999;300:219–226. doi: 10.1016/s0076-6879(99)00128-7. [DOI] [PubMed] [Google Scholar]
- Chang L, Karin M. Mammalian MAP kinase signalling cascades. Nature. 2001;410:37–40. doi: 10.1038/35065000. [DOI] [PubMed] [Google Scholar]
- Charafe-Jauffret E, Monville F, Ginestier C, Dontu G, Birnbaum D, Wicha MS. Cancer stem cells in breast: current opinion and future challenges. Pathobiology. 2008;75:75–84. doi: 10.1159/000123845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YR, Han J, Kori R, Kong AN, Tan TH. Phenylethyl isothiocyanate induces apoptotic signaling via suppressing phosphatase activity against c-Jun N-terminal kinase. J Biol Chem. 2002;277:39334–39342. doi: 10.1074/jbc.M202070200. [DOI] [PubMed] [Google Scholar]
- Chen TJ, Jeng JY, Lin CW, Wu CY, Chen YC. Quercetin inhibition of ROS-dependent and -independent apoptosis in rat glioma C6 cells. Toxicology. 2006;223:113–126. doi: 10.1016/j.tox.2006.03.007. [DOI] [PubMed] [Google Scholar]
- Chen C, Kong AN. Dietary chemopreventive compounds and ARE/EpRE signaling. Free Radic Biol Med. 2004;36:1505–1516. doi: 10.1016/j.freeradbiomed.2004.03.015. [DOI] [PubMed] [Google Scholar]
- Chen C, Kong AN. Dietary cancer-chemopreventive compounds: from signaling and gene expression to pharmacological effects. Trends Pharmacol Sci. 2005;26:318–326. doi: 10.1016/j.tips.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Cheung KL, Khor TO, Huang MT, Kong AN. Differential in vivo mechanism of chemoprevention of tumor formation in azoxymethane/dextran sodium sulfate mice by PEITC and DBM. Carcinogenesis. 2010;31:880–885. doi: 10.1093/carcin/bgp285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung KL, Khor TO, Kong AN. Synergistic effect of combination of phenethyl isothiocyanate and sulforaphane or curcumin and sulforaphane in the inhibition of inflammation. Pharm Res. 2009;26:224–231. doi: 10.1007/s11095-008-9734-9. [DOI] [PubMed] [Google Scholar]
- Cheung K, Kong A-N. Molecular targets of dietary phenethyl isothiocyanate and sulforaphane for cancer chemoprevention. AAPS J. 2010;12:87–97. doi: 10.1208/s12248-009-9162-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung JY, Ong RC, Suen YK, Ooi V, Wong HN, Mak TC, et al. Polyphyllin D is a potent apoptosis inducer in drug-resistant HepG2 cells. Cancer Lett. 2005;217:203–211. doi: 10.1016/j.canlet.2004.06.042. [DOI] [PubMed] [Google Scholar]
- Chinni SR, Li YW, Upadhyay S, Koppolu PK, Sarkar FH. Indole3-carbinol (I3C) induced cell growth inhibition, G1 cell cycle arrest and apoptosis in prostate cancer cells. Oncogene. 2001;20:2927–2936. doi: 10.1038/sj.onc.1204365. [DOI] [PubMed] [Google Scholar]
- Chinni SR, Sarkar FH. Akt inactivation is a key event in indole-3-carbinol-induced apoptosis in PC-3 cells. Clin Cancer Res. 2002;8:1228–1236. [PubMed] [Google Scholar]
- Chow JM, Shen SC, Huan SK, Lin HY, Chen YC. Quercetin, but not rutin and quercitrin, prevention of H2O2-induced apoptosis via anti-oxidant activity and heme oxygenase 1 gene expression in macrophages. Biochem Pharmacol. 2005;69:1839–1851. doi: 10.1016/j.bcp.2005.03.017. [DOI] [PubMed] [Google Scholar]
- Clarke MF, Dick JE, Dirks PB, Eaves CJ, Jamieson CH, Jones DL, et al. Cancer stem cells—perspectives on current status and future directions: AACR Workshop on cancer stem cells. Cancer Res. 2006;66:9339–9344. doi: 10.1158/0008-5472.CAN-06-3126. [DOI] [PubMed] [Google Scholar]
- Clarke MF, Fuller M. Stem cells and cancer: two faces of eve. Cell. 2006;124:1111–1115. doi: 10.1016/j.cell.2006.03.011. [DOI] [PubMed] [Google Scholar]
- Cobb MH, Goldsmith EJ. How MAP kinases are regulated. J Biol Chem. 1995;270:14843–14846. doi: 10.1074/jbc.270.25.14843. [DOI] [PubMed] [Google Scholar]
- Correa F, Mallard C, Nilsson M, Sandberg M. Activated microglia decrease histone acetylation and Nrf2-inducible anti-oxidant defence in astrocytes: restoring effects of inhibitors of HDACs, p38 MAPK and GSK3beta. Neurobiol Dis. 2011;44:142–151. doi: 10.1016/j.nbd.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420:860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crettol S, Petrovic N, Murray M. Pharmacogenetics of phase I and phase II drug metabolism. Curr Pharm Des. 2010;16:204–219. doi: 10.2174/138161210790112674. [DOI] [PubMed] [Google Scholar]
- Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croker AK, Allan AL. Cancer stem cells: implications for the progression and treatment of metastatic disease. J Cell Mol Med. 2008;12:374–390. doi: 10.1111/j.1582-4934.2007.00211.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullinan SB, Diehl JA. PERK-dependent activation of Nrf2 contributes to redox homeostasis and cell survival following endoplasmic reticulum stress. J Biol Chem. 2004;279:20108–20117. doi: 10.1074/jbc.M314219200. [DOI] [PubMed] [Google Scholar]
- Danial NN, Korsmeyer SJ. Cell death: critical control points. Cell. 2004;116:205–219. doi: 10.1016/s0092-8674(04)00046-7. [DOI] [PubMed] [Google Scholar]
- Diala ES, Cheah MS, Rowitch D, Hoffman RM. Extent of DNA methylation in human tumor cells. J Natl Cancer Inst. 1983;71:755–764. [PubMed] [Google Scholar]
- Ding M, Zhao J, Bowman L, Lu Y, Shi X. Inhibition of AP-1 and MAPK signaling and activation of Nrf2/ARE pathway by quercitrin. Int J Oncol. 2010;36:59–67. [PubMed] [Google Scholar]
- Dolinoy DC, Weidman JR, Jirtle RL. Epigenetic gene regulation: linking early developmental environment to adult disease. Reprod Toxicol. 2007;23:297–307. doi: 10.1016/j.reprotox.2006.08.012. [DOI] [PubMed] [Google Scholar]
- Dontu G, Abdallah WM, Foley JM, Jackson KW, Clarke MF, Kawamura MJ, et al. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003;17:1253–1270. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan SE, Giddings BW, Brooks MW, Buday L, Sizeland AM, Weinberg RA. Association of Sos Ras exchange protein with Grb2 is implicated in tyrosine kinase signal transduction and transformation. Nature. 1993;363:45–51. doi: 10.1038/363045a0. [DOI] [PubMed] [Google Scholar]
- Ellis L, Atadja PW, Johnstone RW. Epigenetics in cancer: targeting chromatin modifications. Mol Cancer Ther. 2009;8:1409–1420. doi: 10.1158/1535-7163.MCT-08-0860. [DOI] [PubMed] [Google Scholar]
- Esteller M. CpG island hypermethylation and tumor suppressor genes: a booming present, a brighter future. Oncogene. 2002;21:5427–5440. doi: 10.1038/sj.onc.1205600. [DOI] [PubMed] [Google Scholar]
- Esteller M. Epigenetic gene silencing in cancer: the DNA hypermethylome. Hum Mol Genet. 2007;16(Spec No 1):R50–R59. doi: 10.1093/hmg/ddm018. [DOI] [PubMed] [Google Scholar]
- Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358:1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- Fahey JW, Haristoy X, Dolan PM, Kensler TW, Scholtus I, Stephenson KK, et al. Sulforaphane inhibits extracellular, intracellular, and antibiotic-resistant strains of Helicobacter pylori and prevents benzo[a]pyrene-induced stomach tumors. Proc Natl Acad Sci U S A. 2002;99:7610–7615. doi: 10.1073/pnas.112203099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang MZ, Wang Y, Ai N, Hou Z, Sun Y, Lu H, et al. Tea polyphenol (−)-epigallocatechin-3-gallate inhibits DNA methyltransferase and reactivates methylation-silenced genes in cancer cell lines. Cancer Res. 2003;63:7563–7570. [PubMed] [Google Scholar]
- Farombi EO, Shrotriya S, Na H-K, Kim S-H, Surh Y-J. Curcumin attenuates dimethylnitrosamine-induced liver injury in rats through Nrf2-mediated induction of heme oxygenase-1. Food Chem Toxicol. 2008;46:1279–1287. doi: 10.1016/j.fct.2007.09.095. [DOI] [PubMed] [Google Scholar]
- Feinberg AP, Gehrke CW, Kuo KC, Ehrlich M. Reduced genomic 5-methylcytosine content in human colonic neoplasia. Cancer Res. 1988;48:1159–1161. [PubMed] [Google Scholar]
- Feinberg AP, Ohlsson R, Henikoff S. The epigenetic progenitor origin of human cancer. Nat Rev Genet. 2006;7:21–33. doi: 10.1038/nrg1748. [DOI] [PubMed] [Google Scholar]
- Feng Q, Zhang Y. The MeCP1 complex represses transcription through preferential binding, remodeling, and deacetylating methylated nucleosomes. Genes Dev. 2001;15:827–832. doi: 10.1101/gad.876201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley JW, Kong A-N, Hintze KJ, Jeffery EH, Ji LL, Lei XG. Antioxidants in foods: state of the science important to the food industry. J Agric Food Chem. 2011;59:6837–6846. doi: 10.1021/jf2013875. [DOI] [PubMed] [Google Scholar]
- Fischle W, Wang Y, Jacobs SA, Kim Y, Allis CD, Khorasanizadeh S. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev. 2003;17:1870–1881. doi: 10.1101/gad.1110503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fodor SP, Rava RP, Huang XC, Pease AC, Holmes CP, Adams CL. Multiplexed biochemical assays with biological chips. Nature. 1993;364:555–556. doi: 10.1038/364555a0. [DOI] [PubMed] [Google Scholar]
- Fraga MF, Ballestar E, Villar-Garea A, Boix-Chornet M, Espada J, Schotta G, et al. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat Genet. 2005;37:391–400. doi: 10.1038/ng1531. [DOI] [PubMed] [Google Scholar]
- Gama-Sosa MA, Slagel VA, Trewyn RW, Oxenhandler R, Kuo KC, Gehrke CW, et al. The 5-methylcytosine content of DNA from human tumors. Nucleic Acids Res. 1983;11:6883–6894. doi: 10.1093/nar/11.19.6883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhold D, Rushmore T, Caskey CT. DNA chips: promising toys have become powerful tools. Trends Biochem Sci. 1999;24:168–173. doi: 10.1016/s0968-0004(99)01382-1. [DOI] [PubMed] [Google Scholar]
- Germain D, Frank DA. Targeting the cytoplasmic and nuclear functions of signal transducers and activators of transcription 3 for cancer therapy. Clin Cancer Res. 2007;13:5665–5669. doi: 10.1158/1078-0432.CCR-06-2491. [DOI] [PubMed] [Google Scholar]
- Goodison S, Urquidi V, Tarin D. CD44 cell adhesion molecules. Mol Pathol. 1999;52:189–196. doi: 10.1136/mp.52.4.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberland M, Montgomery RL, Olson EN. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat Rev Genet. 2009;10:32–42. doi: 10.1038/nrg2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliwell B, Gutteridge J. Free Radicals in Biology and Medicine. Oxford University Press; Oxford, UK: 1999. [Google Scholar]
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Harman D. Aging: a theory based on free radical and radiation chemistry. J Gerontol. 1956;11:298–300. doi: 10.1093/geronj/11.3.298. [DOI] [PubMed] [Google Scholar]
- Hayes JD, McMahon M. NRF2 and KEAP1 mutations: permanent activation of an adaptive response in cancer. Trends Biochem Sci. 2009;34:176–188. doi: 10.1016/j.tibs.2008.12.008. [DOI] [PubMed] [Google Scholar]
- He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- He X, Lin GX, Chen MG, Zhang JX, Ma Q. Protection against chromium (VI)-induced oxidative stress and apoptosis by Nrf2. Recruiting Nrf2 into the nucleus and disrupting the nuclear Nrf2/Keap1 association. Toxicol Sci. 2007;98:298–309. doi: 10.1093/toxsci/kfm081. [DOI] [PubMed] [Google Scholar]
- Hemmati HD, Nakano I, Lazareff JA, Masterman-Smith M, Geschwind DH, Bronner-Fraser M, et al. Cancerous stem cells can arise from pediatric brain tumors. Proc Natl Acad Sci U S A. 2003;100:15178–15183. doi: 10.1073/pnas.2036535100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrich B, Bird A. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol Cell Biol. 1998;18:6538–6547. doi: 10.1128/mcb.18.11.6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbst A, Rahmig K, Stieber P, Philipp A, Jung A, Ofner A, et al. Methylation of NEUROG1 in serum is a sensitive marker for the detection of early colorectal cancer. Am J Gastroenterol. 2011;106:1110–1118. doi: 10.1038/ajg.2011.6. [DOI] [PubMed] [Google Scholar]
- Hernandez F, Gomez de Barreda E, Fuster-Matanzo A, Lucas JJ, Avila J. GSK3: a possible link between beta amyloid peptide and tau protein. Exp Neurol. 2010;223:322–325. doi: 10.1016/j.expneurol.2009.09.011. [DOI] [PubMed] [Google Scholar]
- Hibi M, Lin A, Smeal T, Minden A, Karin M. Identification of an oncoprotein- and UV-responsive protein kinase that binds and potentiates the c-Jun activation domain. Genes Dev. 1993;7:2135–2148. doi: 10.1101/gad.7.11.2135. [DOI] [PubMed] [Google Scholar]
- Hoeflich KP, Luo J, Rubie EA, Tsao MS, Jin O, Woodgett JR. Requirement for glycogen synthase kinase-3beta in cell survival and NF-kappaB activation. Nature. 2000;406:86–90. doi: 10.1038/35017574. [DOI] [PubMed] [Google Scholar]
- Hong F, Freeman ML, Liebler DC. Identification of sensor cysteines in human Keap1 modified by the cancer chemopreventive agent sulforaphane. Chem Res Toxicol. 2005;18:1917–1926. doi: 10.1021/tx0502138. [DOI] [PubMed] [Google Scholar]
- Hu Y, Ju Y, Lin D, Wang Z, Huang Y, Zhang S, et al. Mutation of the Nrf2 gene in non-small cell lung cancer. Mol Biol Rep. 2012;39(4):4743–4747. doi: 10.1007/s11033-011-1266-4. (Electronic publication ahead of print) [DOI] [PubMed] [Google Scholar]
- Hu R, Xu C, Shen G, Jain MR, Khor TO, Gopalkrishnan A, et al. Gene expression profiles induced by cancer chemopreventive isothiocyanate sulforaphane in the liver of C57BL/6J mice and C57BL/6J/Nrf2 (−/−) mice. Cancer Lett. 2006a;243:170–192. doi: 10.1016/j.canlet.2005.11.050. [DOI] [PubMed] [Google Scholar]
- Hu R, Xu C, Shen G, Jain MR, Khor TO, Gopalkrishnan A, et al. Identification of Nrf2-regulated genes induced by chemopreventive isothiocyanate PEITC by oligonucleotide microarray. Life Sci. 2006b;79:1944–1955. doi: 10.1016/j.lfs.2006.06.019. [DOI] [PubMed] [Google Scholar]
- Huang P, Han J, Hui L. MAPK signaling in inflammation-associated cancer development. Protein Cell. 2010;1:218–226. doi: 10.1007/s13238-010-0019-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang HC, Nguyen T, Pickett CB. Regulation of the antioxidant response element by protein kinase C-mediated phosphorylation of NF-E2-related factor 2. Proc Natl Acad Sci U S A. 2000;97:12475–12480. doi: 10.1073/pnas.220418997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang HC, Nguyen T, Pickett CB. Phosphorylation of Nrf2 at Ser-40 by protein kinase C regulates antioxidant response element-mediated transcription. J Biol Chem. 2002;277:42769–42774. doi: 10.1074/jbc.M206911200. [DOI] [PubMed] [Google Scholar]
- Hussain SP, Harris CC. Inflammation and cancer: an ancient link with novel potentials. Int J Cancer. 2007;121:2373–2380. doi: 10.1002/ijc.23173. [DOI] [PubMed] [Google Scholar]
- Iida K, Itoh K, Kumagai Y, Oyasu R, Hattori K, Kawai K, et al. Nrf2 is essential for the chemopreventive efficacy of oltipraz against urinary bladder carcinogenesis. Cancer Res. 2004;64:6424–6431. doi: 10.1158/0008-5472.CAN-04-1906. [DOI] [PubMed] [Google Scholar]
- Itoh K, Chiba T, Takahashi S, Ishii T, Igarashi K, Katoh Y, et al. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem Biophys Res Commun. 1997;236:313–322. doi: 10.1006/bbrc.1997.6943. [DOI] [PubMed] [Google Scholar]
- Itoh K, Wakabayashi N, Katoh Y, Ishii T, Igarashi K, Engel JD, et al. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 1999;13:76–86. doi: 10.1101/gad.13.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izzotti A, Calin GA, Steele VE, Cartiglia C, Longobardi M, Croce CM, et al. Chemoprevention of cigarette smoke-induced alterations of MicroRNA expression in rat lungs. Cancer Prev Res (Phila) 2010;3:62–72. doi: 10.1158/1940-6207.CAPR-09-0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks T, Weinberg RA. Taking the study of cancer cell survival to a new dimension. Cell. 2002;111:923–925. doi: 10.1016/s0092-8674(02)01229-1. [DOI] [PubMed] [Google Scholar]
- Jain AK, Jaiswal AK. GSK-3beta acts upstream of Fyn kinase in regulation of nuclear export and degradation of NF-E2 related factor 2. J Biol Chem. 2007;282:16502–16510. doi: 10.1074/jbc.M611336200. [DOI] [PubMed] [Google Scholar]
- Jang JH, Surh YJ. Protective effects of resveratrol on hydrogen peroxide-induced apoptosis in rat pheochromocytoma (PC12) cells. Mutat Res Genet Toxicol Environ Mutagen. 2001;496:181–190. doi: 10.1016/s1383-5718(01)00233-9. [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Jeong WS, Jun M, Kong AN. Nrf2: a potential molecular target for cancer chemoprevention by natural compounds. Antioxid Redox Signal. 2006;8:99–106. doi: 10.1089/ars.2006.8.99. [DOI] [PubMed] [Google Scholar]
- Jeong WS, Keum YS, Chen C, Jain MR, Shen G, Kim JH, et al. Differential expression and stability of endogenous nuclear factor E2-related factor 2 (Nrf2) by natural chemopreventive compounds in HepG2 human hepatoma cells. J Biochem Mol Biol. 2005;38:167–176. doi: 10.5483/bmbrep.2005.38.2.167. [DOI] [PubMed] [Google Scholar]
- Jing N, Tweardy DJ. Targeting Stat3 in cancer therapy. Anticancer Drugs. 2005;16:601–607. doi: 10.1097/00001813-200507000-00002. [DOI] [PubMed] [Google Scholar]
- Johnston PA, Grandis JR. STAT3 signaling: anticancer strategies and challenges. Mol Interv. 2011;11:18–26. doi: 10.1124/mi.11.1.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- Jones PL, Veenstra GJC, Wade PA, Vermaak D, Kass SU, Landsberger N, et al. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- Jope RS, Yuskaitis CJ, Beurel E. Glycogen synthase kinase-3 (GSK3): inflammation, diseases, and therapeutics. Neurochem Res. 2007;32:577–595. doi: 10.1007/s11064-006-9128-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kai ZS, Pasquinelli AE. MicroRNA assassins: factors that regulate the disappearance of miRNAs. Nat Struct Mol Biol. 2010;17:5–10. doi: 10.1038/nsmb.1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang KW, Choi SH, Kim SG. Peroxynitrite activates NF-E2-related factor 2/antioxidant response element through the pathway of phosphatidylinositol 3-kinase: the role of nitric oxide synthase in rat glutathione S-transferase A2 induction. Nitric Oxide. 2002;7:244–253. doi: 10.1016/s1089-8603(02)00117-9. [DOI] [PubMed] [Google Scholar]
- Kang KW, Lee SJ, Park JW, Kim SG. Phosphatidylinositol 3-kinase regulates nuclear translocation of NF-E2-related factor 2 through actin rearrangement in response to oxidative stress. Mol Pharmacol. 2002;62:1001–1010. doi: 10.1124/mol.62.5.1001. [DOI] [PubMed] [Google Scholar]
- Kaspar JW, Niture SK, Jaiswal AK. Nrf2:INrf2 (Keap1) signaling in oxidative stress. Free Radic Biol Med. 2009;47:1304–1309. doi: 10.1016/j.freeradbiomed.2009.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato Y, Tapping RI, Huang S, Watson MH, Ulevitch RJ, Lee JD. Bmk1/Erk5 is required for cell proliferation induced by epidermal growth factor. Nature. 1998;395:713–716. doi: 10.1038/27234. [DOI] [PubMed] [Google Scholar]
- Kensler TW, Curphey TJ, Maxiutenko Y, Roebuck BD. Chemoprotection by organosulfur inducers of phase 2 enzymes: dithiolethiones and dithiins. Drug Metabol Drug Interact. 2000;17:3–22. doi: 10.1515/dmdi.2000.17.1-4.3. [DOI] [PubMed] [Google Scholar]
- Keum YS, Jeong WS, Kong AN. Chemoprevention by isothiocyanates and their underlying molecular signaling mechanisms. Mutat Res. 2004;555:191–202. doi: 10.1016/j.mrfmmm.2004.05.024. [DOI] [PubMed] [Google Scholar]
- Keum YS, Khor TO, Lin W, Shen G, Kwon KH, Barve A, et al. Pharmacokinetics and pharmacodynamics of broccoli sprouts on the suppression of prostate cancer in transgenic adenocarcinoma of mouse prostate (TRAMP) mice: implication of induction of Nrf2, HO-1 and apoptosis and the suppression of Akt-dependent kinase pathway. Pharm Res. 2009;26:2324–2331. doi: 10.1007/s11095-009-9948-5. [DOI] [PubMed] [Google Scholar]
- Keum Y-S, Owuor ED, Kim B-R, Hu R, Kong ANT. Involvement of Nrf2 and JNK1 in the activation of antioxidant responsive element (ARE) by chemopreventive agent phenethyl isothiocyanate (PEITC) Pharm Res. 2003;20:1351–1356. doi: 10.1023/a:1025737622815. [DOI] [PubMed] [Google Scholar]
- Keum YS, Yu S, Chang PP, Yuan X, Kim JH, Xu C, et al. Mechanism of action of sulforaphane: inhibition of p38 mitogen-activated protein kinase isoforms contributing to the induction of antioxidant response element-mediated heme oxygenase-1 in human hepatoma HepG2 Cells. Cancer Res. 2006;66:8804–8813. doi: 10.1158/0008-5472.CAN-05-3513. [DOI] [PubMed] [Google Scholar]
- Khor TO, Huang MT, Kwon KH, Chan JY, Reddy BS, Kong AN. Nrf2-deficient mice have an increased susceptibility to dextran sulfate sodium-induced colitis. Cancer Res. 2006;66:11580–11584. doi: 10.1158/0008-5472.CAN-06-3562. [DOI] [PubMed] [Google Scholar]
- Khor TO, Huang MT, Prawan A, Liu Y, Hao X, Yu S, et al. Increased susceptibility of Nrf2 knockout mice to colitis-associated colorectal cancer. Cancer Prev Res (Phila) 2008;1:187–191. doi: 10.1158/1940-6207.CAPR-08-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khor TO, Huang Y, Wu T-Y, Shu L, Lee J, Kong A-NT. Pharmacodynamics of curcumin as DNA hypomethylation agent in restoring the expression of Nrf2 via promoter CpGs demethylation. Biochem Pharmacol. 2011;82:1073–1078. doi: 10.1016/j.bcp.2011.07.065. [DOI] [PubMed] [Google Scholar]
- Khorasanizadeh S. The nucleosome: from genomic organization to genomic regulation. Cell. 2004;116:259–272. doi: 10.1016/s0092-8674(04)00044-3. [DOI] [PubMed] [Google Scholar]
- Kikuchi T, Itoh F, Toyota M, Suzuki H, Yamamoto H, Fujita M, et al. Aberrant methylation and histone deacetylation of cyclooxygenase 2 in gastric cancer. Int J Cancer. 2002;97:272–277. doi: 10.1002/ijc.1612. [DOI] [PubMed] [Google Scholar]
- Kikuno N, Shiina H, Urakami S, Kawamoto K, Hirata H, Tanaka Y, et al. Genistein mediated histone acetylation and demethylation activates tumor suppressor genes in prostate cancer cells. Int J Cancer. 2008;123:552–560. doi: 10.1002/ijc.23590. [DOI] [PubMed] [Google Scholar]
- Kim JK, Kim Y, Na KM, Surh YJ, Kim TY. 6 -Gingerol prevents UVB-induced ROS production and COX-2 expression in vitro and in vivo. Free Radic Res. 2007;41:603–614. doi: 10.1080/10715760701209896. [DOI] [PubMed] [Google Scholar]
- Kim SO, Kundu JK, Shin YK, Park JH, Cho MH, Kim TY, et al. 6 -Gingerol inhibits COX-2 expression by blocking the activation of p38 MAP kinase and NF-kappa B in phorbol ester-stimulated mouse skin. Oncogene. 2005;24:2558–2567. doi: 10.1038/sj.onc.1208446. [DOI] [PubMed] [Google Scholar]
- Kim YR, Oh JE, Kim MS, Kang MR, Park SW, Han JY, et al. Oncogenic NRF2 mutations in squamous cell carcinomas of oesophagus and skin. J Pathol. 2010;220:446–451. doi: 10.1002/path.2653. [DOI] [PubMed] [Google Scholar]
- King-Batoon A, Leszczynska JM, Klein CB. Modulation of gene methylation by genistein or lycopene in breast cancer cells. Environ Mol Mutagen. 2008;49:36–45. doi: 10.1002/em.20363. [DOI] [PubMed] [Google Scholar]
- Kitamura Y, Umemura T, Kanki K, Kodama Y, Kitamoto S, Saito K, et al. Increased susceptibility to hepatocarcinogenicity of Nrf2-deficient mice exposed to 2-amino-3-methylimidazo[4,5-f]quinoline. Cancer Sci. 2007;98:19–24. doi: 10.1111/j.1349-7006.2006.00352.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong AN, Mandlekar S, Yu R, Lei W, Fasanmande A. Pharmacodynamics and toxicodynamics of drug action: signaling in cell survival and cell death. Pharm Res. 1999;16:790–798. doi: 10.1023/a:1011953431486. [DOI] [PubMed] [Google Scholar]
- Kong AN, Owuor E, Yu R, Hebbar V, Chen C, Hu R, et al. Induction of xenobiotic enzymes by the MAP kinase pathway and the antioxidant or electrophile response element (ARE/EpRE) Drug Metab Rev. 2001;33:255–271. doi: 10.1081/dmr-120000652. [DOI] [PubMed] [Google Scholar]
- Kong AN, Yu R, Hebbar V, Chen C, Owuor E, Hu R, et al. Signal transduction events elicited by cancer prevention compounds. Mutat Res. 2001;480–481:231–241. doi: 10.1016/s0027-5107(01)00182-8. [DOI] [PubMed] [Google Scholar]
- Kong AN, Yu R, Lei W, Mandlekar S, Tan TH, Ucker DS. Differential activation of MAPK and ICE/Ced-3 protease in chemical-induced apoptosis. The role of oxidative stress in the regulation of mitogen-activated protein kinases(MAPKs) leading to gene expression and survival or activation of caspases leading to apoptosis. Restor Neurol Neurosci. 1998;12:63–70. [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Kudoh K, Ramanna M, Ravatn R, Elkahloun AG, Bittner ML, Meltzer PS, et al. Monitoring the expression profiles of doxorubicin-induced and doxorubicin-resistant cancer cells by cDNA microarray. Cancer Res. 2000;60:4161–4166. [PubMed] [Google Scholar]
- Kwak MK, Egner PA, Dolan PM, Ramos-Gomez M, Groopman JD, Itoh K, et al. Role of phase 2 enzyme induction in chemoprotection by dithiolethiones. Mutat Res. 2001;480–481:305–315. doi: 10.1016/s0027-5107(01)00190-7. [DOI] [PubMed] [Google Scholar]
- Kyriakis JM, Banerjee P, Nikolakaki E, Dai T, Rubie EA, Ahmad MF, et al. The stress-activated protein kinase subfamily of c-Jun kinases. Nature. 1994;369:156–160. doi: 10.1038/369156a0. [DOI] [PubMed] [Google Scholar]
- Lapidot T, Sirard C, Vormoor J, Murdoch B, Hoang T, Caceres-Cortes J, et al. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367:645–648. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- Lau A, Villeneuve NF, Sun Z, Wong PK, Zhang DD. Dual roles of Nrf2 in cancer. Pharmacol Res. 2008;58:262–270. doi: 10.1016/j.phrs.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Lee JM, Hanson JM, Chu WA, Johnson JA. Phosphatidylinositol 3-kinase, not extracellular signal-regulated kinase, regulates activation of the antioxidant-responsive element in IMR-32 human neuroblastoma cells. J Biol Chem. 2001;276:20011–20016. doi: 10.1074/jbc.M100734200. [DOI] [PubMed] [Google Scholar]
- Lee JC, Laydon JT, McDonnell PC, Gallagher TF, Kumar S, Green D, et al. A protein kinase involved in the regulation of inflammatory cytokine biosynthesis. Nature. 1994;372:739–746. doi: 10.1038/372739a0. [DOI] [PubMed] [Google Scholar]
- Lee JD, Ulevitch RJ, Han J. Primary structure of BMK1: a new mammalian map kinase. Biochem Biophys Res Commun. 1995;213:715–724. doi: 10.1006/bbrc.1995.2189. [DOI] [PubMed] [Google Scholar]
- Lee MS, Yuet-Wa JC, Kong SK, Yu B, Eng-Choon VO, Nai-Ching HW, et al. Effects of polyphyllin D, a steroidal saponin in Paris polyphylla, in growth inhibition of human breast cancer cells and in xenograft. Cancer Biol Ther. 2005;4:1248–1254. doi: 10.4161/cbt.4.11.2136. [DOI] [PubMed] [Google Scholar]
- Lee WJ, Zhu BT. Inhibition of DNA methylation by caffeic acid and chlorogenic acid, two common catechol-containing coffee polyphenols. Carcinogenesis. 2006;27:269–277. doi: 10.1093/carcin/bgi206. [DOI] [PubMed] [Google Scholar]
- Leeman RJ, Lui VW, Grandis JR. STAT3 as a therapeutic target in head and neck cancer. Expert Opin Biol Ther. 2006;6:231–241. doi: 10.1517/14712598.6.3.231. [DOI] [PubMed] [Google Scholar]
- Leeman-Neill RJ, Cai Q, Joyce SC, Thomas SM, Bhola NE, Neill DB, et al. Honokiol inhibits epidermal growth factor receptor signaling and enhances the antitumor effects of epidermal growth factor receptor inhibitors. Clin Cancer Res. 2010;16:2571–2579. doi: 10.1158/1078-0432.CCR-10-0333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessard J, Sauvageau G. Bmi-1 determines the proliferative capacity of normal and leukaemic stem cells. Nature. 2003;423:255–260. doi: 10.1038/nature01572. [DOI] [PubMed] [Google Scholar]
- Letai A. Pharmacological manipulation of Bcl-2 family members to control cell death. J Clin Invest. 2005;115:2648–2655. doi: 10.1172/JCI26250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung C, Lingbeek M, Shakhova O, Liu J, Tanger E, Saremaslani P, et al. Bmi1 is essential for cerebellar development and is overexpressed in human medulloblastomas. Nature. 2004;428:337–341. doi: 10.1038/nature02385. [DOI] [PubMed] [Google Scholar]
- Lewis JD, Meehan RR, Henzel WJ, Maurer-Fogy I, Jeppesen P, Klein F, et al. Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell. 1992;69:905–914. doi: 10.1016/0092-8674(92)90610-o. [DOI] [PubMed] [Google Scholar]
- Li YW, Ahmed F, Ali S, Philip PA, Kucuk O, Sarkar FH. Inactivation of nuclear factor kappa B by soy isoflavone genistein contributes to increased apoptosis mduced by chemotherapeutic agents in human cancer cells. Cancer Res. 2005;65:6934–6942. doi: 10.1158/0008-5472.CAN-04-4604. [DOI] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Li C, Heidt DG, Dalerba P, Burant CF, Zhang L, Adsay V, et al. Identification of pancreatic cancer stem cells. Cancer Res. 2007;67:1030–1037. doi: 10.1158/0008-5472.CAN-06-2030. [DOI] [PubMed] [Google Scholar]
- Li W, Kong AN. Molecular mechanisms of Nrf2-mediated antioxidant response. Mol Carcinog. 2009;48:91–104. doi: 10.1002/mc.20465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Kong D, Wang Z, Sarkar FH. Regulation of microRNAs by natural agents: an emerging field in chemoprevention and chemotherapy research. Pharm Res. 2010a;27:1027–1041. doi: 10.1007/s11095-010-0105-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li LY, Luo X, Wang X. Endonuclease G is an apoptotic DNase when released from mitochondria. Nature. 2001;412:95–99. doi: 10.1038/35083620. [DOI] [PubMed] [Google Scholar]
- Li W, Thakor N, Xu EY, Huang Y, Chen C, Yu R, et al. An internal ribosomal entry site mediates redox-sensitive translation of Nrf2. Nucleic Acids Res. 2010b;38:778–788. doi: 10.1093/nar/gkp1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Tollefsbol TO. Impact on DNA methylation in cancer prevention and therapy by bioactive dietary components. Curr Med Chem. 2010;17:2141–2151. doi: 10.2174/092986710791299966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Vandenboom TG, II, Wang Z, Kong D, Ali S, Philip PA, et al. miR-146a suppresses invasion of pancreatic cancer cells. Cancer Res. 2010c;70:1486–1495. doi: 10.1158/0008-5472.CAN-09-2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin A. Activation of the JNK signaling pathway: breaking the brake on apoptosis. Bioessays. 2003;25:17–24. doi: 10.1002/bies.10204. [DOI] [PubMed] [Google Scholar]
- Lin W, Wu RT, Wu T, Khor TO, Wang H, Kong AN. Sulforaphane suppressed LPS-induced inflammation in mouse peritoneal macrophages through Nrf2 dependent pathway. Biochem Pharmacol. 2008;76:967–973. doi: 10.1016/j.bcp.2008.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Lin A. Role of JNK activation in apoptosis: a double-edged sword. Cell Res. 2005;15:36–42. doi: 10.1038/sj.cr.7290262. [DOI] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Lund AH, van Lohuizen M. Epigenetics and cancer. Genes Dev. 2004;18:2315–2335. doi: 10.1101/gad.1232504. [DOI] [PubMed] [Google Scholar]
- Ma Q. Transcriptional responses to oxidative stress: pathological and toxicological implications. Pharmacol Ther. 2010;125:376–393. doi: 10.1016/j.pharmthera.2009.11.004. [DOI] [PubMed] [Google Scholar]
- Ma DD, Lu HX, Xu LS, Xiao W. Polyphyllin D exerts potent anti-tumour effects on Lewis cancer cells under hypoxic conditions. J Int Med Res. 2009;37:631–640. doi: 10.1177/147323000903700305. [DOI] [PubMed] [Google Scholar]
- Maheo K, Morel F, Langouet S, Kramer H, Le Ferrec E, Ketterer B, et al. Inhibition of cytochromes P-450 and induction of glutathione S-transferases by sulforaphane in primary human and rat hepatocytes. Cancer Res. 1997;57:3649–3652. [PubMed] [Google Scholar]
- Majid S, Kikuno N, Nelles J, Noonan E, Tanaka Y, Kawamoto K, et al. Genistein induces the p21WAF1/CIP1 and p16INK4a tumor suppressor genes in prostate cancer cells by epigenetic mechanisms involving active chromatin modification. Cancer Res. 2008;68:2736–2744. doi: 10.1158/0008-5472.CAN-07-2290. [DOI] [PubMed] [Google Scholar]
- Marshall CJ. MAP kinase kinase kinase, MAP kinase kinase and MAP kinase. Curr Opin Genet Dev. 1994;4:82–89. doi: 10.1016/0959-437x(94)90095-7. [DOI] [PubMed] [Google Scholar]
- Marzec JM, Christie JD, Reddy SP, Jedlicka AE, Vuong H, Lanken PN, et al. Functional polymorphisms in the transcription factor NRF2 in humans increase the risk of acute lung injury. FASEB J. 2007;21:2237–2246. doi: 10.1096/fj.06-7759com. [DOI] [PubMed] [Google Scholar]
- Meeran SM, Patel SN, Tollefsbol TO. Sulforaphane causes epigenetic repression of hTERT expression in human breast cancer cell lines. PLoS One. 2010;5:e11457. doi: 10.1371/journal.pone.0011457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra R. Glycogen synthase kinase 3 beta: can it be a target for oral cancer. Mol Cancer. 2010;9:144. doi: 10.1186/1476-4598-9-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizrak D, Brittan M, Alison MR. CD133: molecule of the moment. J Pathol. 2008;214:3–9. doi: 10.1002/path.2283. [DOI] [PubMed] [Google Scholar]
- Motohashi H, Yamamoto M. Nrf2–Keap1 defines a physiologically important stress response mechanism. Trends Mol Med. 2004;10:549–557. doi: 10.1016/j.molmed.2004.09.003. [DOI] [PubMed] [Google Scholar]
- Myzak MC, Tong P, Dashwood WM, Dashwood RH, Ho E. Sulforaphane retards the growth of human PC-3 xenografts and inhibits HDAC activity in human subjects. Exp Biol Med (Maywood) 2007;232:227–234. [PMC free article] [PubMed] [Google Scholar]
- Na HK, Kim EH, Jung JH, Lee HH, Hyun JW, Surh YJ. (−)-Epigallocatechin gallate induces Nrf2-mediated antioxidant enzyme expression via activation of PI3K and ERK in human mammary epithelial cells. Arch Biochem Biophys. 2008;476:171–177. doi: 10.1016/j.abb.2008.04.003. [DOI] [PubMed] [Google Scholar]
- Nair S, Barve A, Khor TO, Shen GX, Lin W, Chan JY, et al. Regulation of Nrf2- and AP-1-mediated gene expression by epigallocatechin-3-gallate and sulforaphane in prostate of Nrf2-knockout or C57BL/6J mice and PC-3 AP-1 human prostate cancer cells. Acta Pharmacol Sin. 2010;31:1223–1240. doi: 10.1038/aps.2010.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair S, Doh ST, Chan JY, Kong AN, Cai L. Regulatory potential for concerted modulation of Nrf2- and Nfkb1-mediated gene expression in inflammation and carcinogenesis. Br J Cancer. 2008;99:2070–2082. doi: 10.1038/sj.bjc.6604703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair S, Xu C, Shen G, Hebbar V, Gopalakrishnan A, Hu R, et al. Pharmacogenomics of phenolic antioxidant butylated hydroxyanisole (BHA) in the small intestine and liver of Nrf2 knockout and C57BL/6J mice. Pharm Res. 2006;23:2621–2637. doi: 10.1007/s11095-006-9099-x. [DOI] [PubMed] [Google Scholar]
- Nakahara S, Yone K, Sakou T, Wada S, Nagamine T, Niiyama T, et al. Induction of apoptosis signal regulating kinase 1 (ASK1) after spinal cord injury in rats: possible involvement of ASK1-JNK and -p38 pathways in neuronal apoptosis. J Neuropathol Exp Neurol. 1999;58:442–450. doi: 10.1097/00005072-199905000-00003. [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Kawakami M, Yoshihiro A, Miyoshi N, Ohigashi H, Kawai K, et al. Involvement of the mitochondrial death pathway in chemopreventive benzyl isothiocyanate-induced apoptosis. J Biol Chem. 2002;277:8492–8499. doi: 10.1074/jbc.M109760200. [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Ohigashi H, Masuda S, Murakami A, Morimitsu Y, Kawamoto Y, et al. Redox regulation of glutathione S-transferase induction by benzyl isothiocyanate: correlation of enzyme induction with the formation of reactive oxygen intermediates. Cancer Res. 2000;60:219–225. [PubMed] [Google Scholar]
- Nakano H, Nakajima A, Sakon-Komazawa S, Piao JH, Xue X, Okumura K. Reactive oxygen species mediate crosstalk between NF-kappaB and JNK. Cell Death Differ. 2006;13:730–737. doi: 10.1038/sj.cdd.4401830. [DOI] [PubMed] [Google Scholar]
- Nakaso K, Yano H, Fukuhara Y, Takeshima T, Wada-Isoe K, Nakashima K. PI3K is a key molecule in the Nrf2-mediated regulation of antioxidative proteins by hemin in human neuroblastoma cells. FEBS Lett. 2003;546:181–184. doi: 10.1016/s0014-5793(03)00517-9. [DOI] [PubMed] [Google Scholar]
- Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, et al. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- Nandakumar V, Vaid M, Katiyar SK. (−)-Epigallocatechin-3-gallate reactivates silenced tumor suppressor genes, Cip1/p21 and p16INK4a, by reducing DNA methylation and increasing histones acetylation in human skin cancer cells. Carcinogenesis. 2011;32:537–544. doi: 10.1093/carcin/bgq285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathan C, Ding A. SnapShot: reactive oxygen intermediates (ROI) Cell. 2010;140(951–951):e952. doi: 10.1016/j.cell.2010.03.008. [DOI] [PubMed] [Google Scholar]
- Negrini M, Ferracin M, Sabbioni S, Croce CM. MicroRNAs in human cancer: from research to therapy. J Cell Sci. 2007;120:1833–1840. doi: 10.1242/jcs.03450. [DOI] [PubMed] [Google Scholar]
- Nithianandarajah-Jones GN, Wilm B, Goldring CE, Muller J, Cross MJ. ERK5: structure, regulation and function. Cell Signal. 2012;24:2187–2196. doi: 10.1016/j.cellsig.2012.07.007. [DOI] [PubMed] [Google Scholar]
- Niture SK, Jain AK, Shelton PM, Jaiswal AK. Src subfamily kinases regulate nuclear export and degradation of transcription factor Nrf2 to switch off Nrf2-mediated antioxidant activation of cytoprotective gene expression. J Biol Chem. 2011;286:28821–28832. doi: 10.1074/jbc.M111.255042. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Numazawa S, Ishikawa M, Yoshida A, Tanaka S, Yoshida T. Atypical protein kinase C mediates activation of NF-E2-related factor 2 in response to oxidative stress. Am J Physiol Cell Physiol. 2003;285:C334–C342. doi: 10.1152/ajpcell.00043.2003. [DOI] [PubMed] [Google Scholar]
- Nuwaysir EF, Bittner M, Trent J, Barrett JC, Afshari CA. Microarrays and toxicology: the advent of toxicogenomics. Mol Carcinog. 1999;24:153–159. doi: 10.1002/(sici)1098-2744(199903)24:3<153::aid-mc1>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- O’Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445:106–110. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- Ogino S, Kawasaki T, Kirkner GJ, Kraft P, Loda M, Fuchs CS. Evaluation of markers for CpG island methylator phenotype (CIMP) in colorectal cancer by a large population-based sample. J Mol Diagn. 2007;9:305–314. doi: 10.2353/jmoldx.2007.060170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlando V. Polycomb, epigenomes, and control of cell identity. Cell. 2003;112:599–606. doi: 10.1016/s0092-8674(03)00157-0. [DOI] [PubMed] [Google Scholar]
- Osburn WO, Karim B, Dolan PM, Liu G, Yamamoto M, Huso DL, et al. Increased colonic inflammatory injury and formation of aberrant crypt foci in Nrf2-deficient mice upon dextran sulfate treatment. Int J Cancer. 2007;121:1883–1891. doi: 10.1002/ijc.22943. [DOI] [PubMed] [Google Scholar]
- Padmanabhan B, Tong KI, Ohta T, Nakamura Y, Scharlock M, Ohtsuji M, et al. Structural basis for defects of Keap1 activity provoked by its point mutations in lung cancer. Mol Cell. 2006;21:689–700. doi: 10.1016/j.molcel.2006.01.013. [DOI] [PubMed] [Google Scholar]
- Palozza P, Simone R, Catalano A, Boninsegna A, Bohm V, Frohlich K, et al. Lycopene prevents 7-ketocholesterol-induced oxidative stress, cell cycle arrest and apoptosis in human macrophages. J Nutr Biochem. 2010;21:34–46. doi: 10.1016/j.jnutbio.2008.10.002. [DOI] [PubMed] [Google Scholar]
- Pandey M, Shukla S, Gupta S. Promoter demethylation and chromatin remodeling by green tea polyphenols leads to re-expression of GSTP1 in human prostate cancer cells. Int J Cancer. 2010;126:2520–2533. doi: 10.1002/ijc.24988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park IK, Qian D, Kiel M, Becker MW, Pihalja M, Weissman IL, et al. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature. 2003;423:302–305. doi: 10.1038/nature01587. [DOI] [PubMed] [Google Scholar]
- Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- Paul G, Bataille F, Obermeier F, Bock J, Klebl F, Strauch U, et al. Analysis of intestinal haem-oxygenase-1 (HO-1) in clinical and experimental colitis. Clin Exp Immunol. 2005;140:547–555. doi: 10.1111/j.1365-2249.2005.02775.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogribny IP, James SJ, Beland FA. Molecular alterations in hepatocarcinogenesis induced by dietary methyl deficiency. Mol Nutr Food Res. 2012;56:116–125. doi: 10.1002/mnfr.201100524. [DOI] [PubMed] [Google Scholar]
- Pogribny IP, James SJ, Jernigan S, Pogribna M. Genomic hypomethylation is specific for preneoplastic liver in folate/methyl deficient rats and does not occur in non-target tissues. Mutat Res Fundam Mol Mech Mutagen. 2004;548:53–59. doi: 10.1016/j.mrfmmm.2003.12.014. [DOI] [PubMed] [Google Scholar]
- Pool-Zobel B, Veeriah S, Bohmer FD. Modulation of xenobiotic metabolising enzymes by anticarcinogens—focus on glutathione S-transferases and their role as targets of dietary chemoprevention in colorectal carcinogenesis. Mutat Res. 2005;591:74–92. doi: 10.1016/j.mrfmmm.2005.04.020. [DOI] [PubMed] [Google Scholar]
- Raaphorst FM, Otte AP, Meijer CJ. Polycomb-group genes as regulators of mammalian lymphopoiesis. Trends Immunol. 2001;22:682–690. doi: 10.1016/s1471-4906(01)02082-8. [DOI] [PubMed] [Google Scholar]
- Rada P, Rojo AI, Chowdhry S, McMahon M, Hayes JD, Cuadrado A. SCF/{beta}-TrCP promotes glycogen synthase kinase 3-dependent degradation of the Nrf2 transcription factor in a Keap1-independent manner. Mol Cell Biol. 2011;31:1121–1133. doi: 10.1128/MCB.01204-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ram PT, Iyengar R. G protein coupled receptor signaling through the Src and Stat3 pathway: role in proliferation and transformation. Oncogene. 2001;20:1601–1606. doi: 10.1038/sj.onc.1204186. [DOI] [PubMed] [Google Scholar]
- Raman M, Chen W, Cobb MH. Differential regulation and properties of MAPKs. Oncogene. 2007;26:3100–3112. doi: 10.1038/sj.onc.1210392. [DOI] [PubMed] [Google Scholar]
- Ramos-Gomez M, Kwak MK, Dolan PM, Itoh K, Yamamoto M, Talalay P, et al. Sensitivity to carcinogenesis is increased and chemoprotective efficacy of enzyme inducers is lost in Nrf2 transcription factor-deficient mice. Proc Natl Acad Sci U S A. 2001;98:3410–3415. doi: 10.1073/pnas.051618798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raptis L, Arulanandam R, Geletu M, Turkson J. The R(h)oads to Stat3: Stat3 activation by the Rho GTPases. Exp Cell Res. 2011;317:1787–1795. doi: 10.1016/j.yexcr.2011.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- Rimoldi D, Srikantan V, Wilson VL, Bassin RH, Samid D. Increased sensitivity of nontumorigenic fibroblasts expressing ras or myc oncogenes to malignant transformation induced by 5-aza-2′-deoxycytidine. Cancer Res. 1991;51:324–330. [PubMed] [Google Scholar]
- Rodenhiser D, Mann M. Epigenetics and human disease: translating basic biology into clinical applications. CMAJ. 2006;174:341–348. doi: 10.1503/cmaj.050774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Ramiro I, Ramos S, Bravo L, Goya L, Martin MA. Procyanidin B2 induces Nrf2 translocation and glutathione S-transferase P1 expression via ERKs and p38-MAPK pathways and protect human colonic cells against oxidative stress. Eur J Nutr. 2012;51(7):881–892. doi: 10.1007/s00394-011-0269-1. (Electronic publication ahead of print) [DOI] [PubMed] [Google Scholar]
- Rojo AI, Rada P, Egea J, Rosa AO, Lopez MG, Cuadrado A. Functional interference between glycogen synthase kinase-3 beta and the transcription factor Nrf2 in protection against kainate-induced hippocampal cell death. Mol Cell Neurosci. 2008;39:125–132. doi: 10.1016/j.mcn.2008.06.007. [DOI] [PubMed] [Google Scholar]
- Rushmore TH, Kong AN. Pharmacogenomics, regulation and signaling pathways of phase I and II drug metabolizing enzymes. Curr Drug Metab. 2002;3:481–490. doi: 10.2174/1389200023337171. [DOI] [PubMed] [Google Scholar]
- Saitoh M, Nishitoh H, Fujii M, Takeda K, Tobiume K, Sawada Y, et al. Mammalian thioredoxin is a direct inhibitor of apoptosis signal-regulating kinase (ASK) 1. EMBO J. 1998;17:2596–2606. doi: 10.1093/emboj/17.9.2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar M, Rojo AI, Velasco D, de Sagarra RM, Cuadrado A. Glycogen synthase kinase-3beta inhibits the xenobiotic and antioxidant cell response by direct phosphorylation and nuclear exclusion of the transcription factor Nrf2. J Biol Chem. 2006;281:14841–14851. doi: 10.1074/jbc.M513737200. [DOI] [PubMed] [Google Scholar]
- Sarti P, Avigliano L, Gorlach A, Brune B. Superoxide and nitric oxide-participation in cell communication. Cell Death Differ. 2002;9:1160–1162. doi: 10.1038/sj.cdd.4401099. [DOI] [PubMed] [Google Scholar]
- Saw CL, Huang MT, Liu Y, Khor TO, Conney AH, Kong AN. Impact of Nrf2 on UVB-induced skin inflammation/photoprotection and photoprotective effect of sulforaphane. Mol Carcinog. 2011;50:479–486. doi: 10.1002/mc.20725. [DOI] [PubMed] [Google Scholar]
- Schatton T, Murphy GF, Frank NY, Yamaura K, Waaga-Gasser AM, Gasser M, et al. Identification of cells initiating human melanomas. Nature. 2008;451:345–349. doi: 10.1038/nature06489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- Schetter AJ, Heegaard NH, Harris CC. Inflammation and cancer: interweaving microRNA, free radical, cytokine and p53 pathways. Carcinogenesis. 2010;31:37–49. doi: 10.1093/carcin/bgp272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scorrano L, Oakes SA, Opferman JT, Cheng EH, Sorcinelli MD, Pozzan T, et al. BAX and BAK regulation of endoplasmic reticulum Ca2+: a control point for apoptosis. Science. 2003;300:135–139. doi: 10.1126/science.1081208. [DOI] [PubMed] [Google Scholar]
- Shen G, Hebbar V, Nair S, Xu C, Li W, Lin W, et al. Regulation of Nrf2 transactivation domain activity. The differential effects of mitogen-activated protein kinase cascades and synergistic stimulatory effect of Raf and CREB-binding protein. J Biol Chem. 2004;279:23052–23060. doi: 10.1074/jbc.M401368200. [DOI] [PubMed] [Google Scholar]
- Shen G, Xu C, Hu R, Jain MR, Gopalkrishnan A, Nair S, et al. Modulation of nuclear factor E2-related factor 2-mediated gene expression in mice liver and small intestine by cancer chemopreventive agent curcumin. Mol Cancer Ther. 2006;5:39–51. doi: 10.1158/1535-7163.MCT-05-0293. [DOI] [PubMed] [Google Scholar]
- Shen G, Xu C, Hu R, Jain MR, Nair S, Lin W, et al. Comparison of (−)-epigallocatechin-3-gallate elicited liver and small intestine gene expression profiles between C57BL/6J mice and C57BL/6J/Nrf2 (−/−) mice. Pharm Res. 2005;22:1805–1820. doi: 10.1007/s11095-005-7546-8. [DOI] [PubMed] [Google Scholar]
- Shi Y. Histone lysine demethylases: emerging roles in development, physiology and disease. Nat Rev Genet. 2007;8:829–833. doi: 10.1038/nrg2218. [DOI] [PubMed] [Google Scholar]
- Shibata T, Kokubu A, Saito S, Narisawa-Saito M, Sasaki H, Aoyagi K, et al. NRF2 mutation confers malignant potential and resistance to chemoradiation therapy in advanced esophageal squamous cancer. Neoplasia. 2011;13:864–873. doi: 10.1593/neo.11750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata T, Ohta T, Tong KI, Kokubu A, Odogawa R, Tsuta K, et al. Cancer related mutations in NRF2 impair its recognition by Keap1-Cul3 E3 ligase and promote malignancy. Proc Natl Acad Sci U S A. 2008;105:13568–13573. doi: 10.1073/pnas.0806268105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu L, Cheung KL, Khor TO, Chen C, Kong AN. Phytochemicals: cancer chemoprevention and suppression of tumor onset and metastasis. Cancer Metastasis Rev. 2010;29:483–502. doi: 10.1007/s10555-010-9239-y. [DOI] [PubMed] [Google Scholar]
- Shu L, Khor TO, Lee JH, Boyanapalli SS, Huang Y, Wu TY, et al. Epigenetic CpG demethylation of the promoter and reactivation of the expression of Neurog1 by curcumin in prostate LNCaP cells. AAPS J. 2011;13:606–614. doi: 10.1208/s12248-011-9300-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SK, Clarke ID, Terasaki M, Bonn VE, Hawkins C, Squire J, et al. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63:5821–5828. [PubMed] [Google Scholar]
- Singh A, Misra V, Thimmulappa RK, Lee H, Ames S, Hoque MO, et al. Dysfunctional KEAP1–NRF2 interaction in non-small-cell lung cancer. PLoS Med. 2006;3:e420. doi: 10.1371/journal.pmed.0030420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn SJ, Sarvis BK, Cado D, Winoto A. ERK5 MAPK regulates embryonic angiogenesis and acts as a hypoxia-sensitive repressor of vascular endothelial growth factor expression. J Biol Chem. 2002;277:43344–43351. doi: 10.1074/jbc.M207573200. [DOI] [PubMed] [Google Scholar]
- Sprick MR, Walczak H. The interplay between the Bcl-2 family and death receptor-mediated apoptosis. Biochim Biophys Acta. 2004;1644:125–132. doi: 10.1016/j.bbamcr.2003.11.002. [DOI] [PubMed] [Google Scholar]
- Stokoe D, Macdonald SG, Cadwallader K, Symons M, Hancock JF. Activation of Raf as a result of recruitment to the plasma membrane. Science. 1994;264:1463–1467. doi: 10.1126/science.7811320. [DOI] [PubMed] [Google Scholar]
- Sun M, Estrov Z, Ji Y, Coombes KR, Harris DH, Kurzrock R. Curcumin (diferuloylmethane) alters the expression profiles of microRNAs in human pancreatic cancer cells. Mol Cancer Ther. 2008;7:464–473. doi: 10.1158/1535-7163.MCT-07-2272. [DOI] [PubMed] [Google Scholar]
- Sun Z, Huang Z, Zhang DD. Phosphorylation of Nrf2 at multiple sites by MAP kinases has a limited contribution in modulating the Nrf2-dependent antioxidant response. PLoS One. 2009;4:e6588. doi: 10.1371/journal.pone.0006588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun YL, Jiang XF, Chen SJ, Price BD. Inhibition of histone acetyltransferase activity by anacardic acid sensitizes tumor cells to ionizing radiation. FEBS Lett. 2006;580:4353–4356. doi: 10.1016/j.febslet.2006.06.092. [DOI] [PubMed] [Google Scholar]
- Sung B, Pandey MK, Ahn KS, Yi TF, Chaturvedi MM, Liu MY, et al. Anacardic acid (6-nonadecyl salicylic acid), an inhibitor of histone acetyltransferase, suppresses expression of nuclear factor-kappa B-regulated gene products involved in cell survival, proliferation, invasion, and inflammation through inhibition of the inhibitory subunit of nuclear factor-kappa B alpha kinase, leading to potentiation of apoptosis. Blood. 2008;111:4880–4891. doi: 10.1182/blood-2007-10-117994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Youle RJ, Tjandra N. Structure of Bax: coregulation of dimer formation and intracellular localization. Cell. 2000;103:645–654. doi: 10.1016/s0092-8674(00)00167-7. [DOI] [PubMed] [Google Scholar]
- Talalay P. Chemoprotection against cancer by induction of phase 2 enzymes. Biofactors. 2000;12:5–11. doi: 10.1002/biof.5520120102. [DOI] [PubMed] [Google Scholar]
- Tang W, Liu JW, Zhao WM, Wei DZ, Zhong JJ. Ganoderic acid T from Ganoderma lucidum mycelia induces mitochondria mediated apoptosis in lung cancer cells. Life Sci. 2006;80:205–211. doi: 10.1016/j.lfs.2006.09.001. [DOI] [PubMed] [Google Scholar]
- Thimmulappa RK, Mai KH, Srisuma S, Kensler TW, Yamamoto M, Biswal S. Identification of Nrf2-regulated genes induced by the chemopreventive agent sulforaphane by oligonucleotide microarray. Cancer Res. 2002;62:5196–5203. [PubMed] [Google Scholar]
- Tinhofer I, Bernhard D, Senfter M, Anether G, Loeffler M, Kroemer G, et al. Resveratrol, a tumor-suppressive compound from grapes, induces apoptosis via a novel mitochondrial pathway controlled by Bcl-2. FASEB J. 2001;15:1613–1615. doi: 10.1096/fj.00-0675fje. [DOI] [PubMed] [Google Scholar]
- Tremethick DJ. Higher-order structures of chromatin: the elusive 30 nm fiber. Cell. 2007;128:651–654. doi: 10.1016/j.cell.2007.02.008. [DOI] [PubMed] [Google Scholar]
- Tsang WP, Kwok TT. Epigallocatechin gallate up-regulation of miR-16 and induction of apoptosis in human cancer cells. J Nutr Biochem. 2010;21:140–146. doi: 10.1016/j.jnutbio.2008.12.003. [DOI] [PubMed] [Google Scholar]
- Valk-Lingbeek ME, Bruggeman SW, van Lohuizen M. Stem cells and cancer; the polycomb connection. Cell. 2004;118:409–418. doi: 10.1016/j.cell.2004.08.005. [DOI] [PubMed] [Google Scholar]
- van der Lugt NM, Domen J, Linders K, van Roon M, Robanus-Maandag E, te Riele H, et al. Posterior transformation, neurological abnormalities, and severe hematopoietic defects in mice with a targeted deletion of the bmi-1 protooncogene. Genes Dev. 1994;8:757–769. doi: 10.1101/gad.8.7.757. [DOI] [PubMed] [Google Scholar]
- Visvader JE, Lindeman GJ. Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat Rev Cancer. 2008;8:755–768. doi: 10.1038/nrc2499. [DOI] [PubMed] [Google Scholar]
- Voehringer DW, Hirschberg DL, Xiao J, Lu Q, Roederer M, Lock CB, et al. Gene microarray identification of redox and mitochondrial elements that control resistance or sensitivity to apoptosis. Proc Natl Acad Sci U S A. 2000;97:2680–2685. doi: 10.1073/pnas.97.6.2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakabayashi N, Dinkova-Kostova AT, Holtzclaw WD, Kang MI, Kobayashi A, Yamamoto M, et al. Protection against electrophile and oxidant stressby induction of the phase 2 response: fate of cysteines of the Keap1 sensor modified by inducers. Proc Natl Acad Sci U S A. 2004;101:2040–2045. doi: 10.1073/pnas.0307301101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. The expanding role of mitochondria in apoptosis. Genes Dev. 2001;15:2922–2933. [PubMed] [Google Scholar]
- Wang R, An J, Ji F, Jiao H, Sun H, Zhou D. Hypermethylation of the Keap1 gene in human lung cancer cell lines and lung cancer tissues. Biochem Biophys Res Commun. 2008;373:151–154. doi: 10.1016/j.bbrc.2008.06.004. [DOI] [PubMed] [Google Scholar]
- Wang LG, Beklemisheva A, Liu XM, Ferrari AC, Feng J, Chiao JW. Dual action on promoter demethylation and chromatin by an isothiocyanate restored GSTP1 silenced in prostate cancer. Mol Carcinog. 2007;46:24–31. doi: 10.1002/mc.20258. [DOI] [PubMed] [Google Scholar]
- Wang H, Khor TO, Saw CL, Lin W, Wu T, Huang Y, et al. Role of Nrf2 in suppressing LPS-induced inflammation in mouse peritoneal macrophages by polyunsaturated fatty acids docosahexaenoic acid and eicosapentaenoic acid. Mol Pharm. 2010;7:2185–2193. doi: 10.1021/mp100199m. [DOI] [PubMed] [Google Scholar]
- Wang LG, Liu XM, Fang Y, Dai W, Chiao FB, Puccio GM, et al. De-repression of the p21 promoter in prostate cancer cells by an isothiocyanate via inhibition of HDACs and c-Myc. Int J Oncol. 2008;33:375–380. [PubMed] [Google Scholar]
- Wang JH, Yao MZ, Zhang ZL, Zhang YH, Wang YG, Liu XY. HSF1 blockade-induced tumor thermotolerance abolishment is mediated by JNK-dependent caspase-3 activation. Biochem Biophys Res Commun. 2004;321:736–745. doi: 10.1016/j.bbrc.2004.07.018. [DOI] [PubMed] [Google Scholar]
- Wattenberg LW. Chemoprophylaxis of carcinogenesis: a review. Cancer Res. 1966;26:1520–1526. [PubMed] [Google Scholar]
- Widschwendter M, Fiegl H, Egle D, Mueller-Holzner E, Spizzo G, Marth C, et al. Epigenetic stem cell signature in cancer. Nat Genet. 2007;39:157–158. doi: 10.1038/ng1941. [DOI] [PubMed] [Google Scholar]
- Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11:228–234. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- Wolffe AP, Matzke MA. Epigenetics: regulation through repression. Science. 1999;286:481–486. doi: 10.1126/science.286.5439.481. [DOI] [PubMed] [Google Scholar]
- Wu TY, Khor TO, Saw CL, Loh SC, Chen AI, Lim SS, et al. Anti-inflammatory/Anti-oxidative stress activities and differential regulation of Nrf2-mediated genes by non-polar fractions of tea Chrysanthemum zawadskii and licorice Glycyrrhiza uralensis. AAPS J. 2011;13:1–13. doi: 10.1208/s12248-010-9239-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Huang MT, Shen G, Yuan X, Lin W, Khor TO, et al. Inhibition of 7,12-dimethylbenz(a)anthracene-induced skin tumorigenesis in C57BL/6 mice by sulforaphane is mediated by nuclear factor E2-related factor 2. Cancer Res. 2006;66:8293–8296. doi: 10.1158/0008-5472.CAN-06-0300. [DOI] [PubMed] [Google Scholar]
- Xu C, Yuan X, Pan Z, Shen G, Kim JH, Yu S, et al. Mechanism of action of isothiocyanates: the induction of ARE-regulated genes is associated with activation of ERK and JNK and the phosphorylation and nuclear translocation of Nrf2. Mol Cancer Ther. 2006;5:1918–1926. doi: 10.1158/1535-7163.MCT-05-0497. [DOI] [PubMed] [Google Scholar]
- Yamamoto T, Yoh K, Kobayashi A, Ishii Y, Kure S, Koyama A, et al. Identification of polymorphisms in the promoter region of the human NRF2 gene. Biochem Biophys Res Commun. 2004;321:72–79. doi: 10.1016/j.bbrc.2004.06.112. [DOI] [PubMed] [Google Scholar]
- Yang ZF, Ho DW, Ng MN, Lau CK, Yu WC, Ngai P, et al. Significance of CD90+ cancer stem cells in human liver cancer. Cancer Cell. 2008;13:153–166. doi: 10.1016/j.ccr.2008.01.013. [DOI] [PubMed] [Google Scholar]
- Yoo CB, Jones PA. Epigenetic therapy of cancer: past, present and future. Nat Rev Drug Discov. 2006;5:37–50. doi: 10.1038/nrd1930. [DOI] [PubMed] [Google Scholar]
- Yu BP. Cellular defenses against damage from reactive oxygen species. Physiol Rev. 1994;74:139–162. doi: 10.1152/physrev.1994.74.1.139. [DOI] [PubMed] [Google Scholar]
- Yu R, Chen C, Mo YY, Hebbar V, Owuor ED, Tan TH, et al. Activation of mitogen-activated protein kinase pathways induces antioxidant response element-mediated gene expression via a Nrf2-dependent mechanism. J Biol Chem. 2000;275:39907–39913. doi: 10.1074/jbc.M004037200. [DOI] [PubMed] [Google Scholar]
- Yu H, Jove R. The STATs of cancer—new molecular targets come of age. Nat Rev Cancer. 2004;4:97–105. doi: 10.1038/nrc1275. [DOI] [PubMed] [Google Scholar]
- Yu X, Kensler T. Nrf2 as a target for cancer chemoprevention. Mutat Res. 2005;591:93–102. doi: 10.1016/j.mrfmmm.2005.04.017. [DOI] [PubMed] [Google Scholar]
- Yu SW, Khor TO, Cheung KL, Li WG, Wu TY, Huang Y, et al. Nrf2 expression is regulated by epigenetic mechanisms in prostate cancer of TRAMP mice. PLoS One. 2010:5. doi: 10.1371/journal.pone.0008579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu R, Lei W, Mandlekar S, Weber MJ, Der CJ, Wu J, et al. Role of a mitogen-activated protein kinase pathway in the induction of phase II detoxifying enzymes by chemicals. J Biol Chem. 1999;274:27545–27552. doi: 10.1074/jbc.274.39.27545. [DOI] [PubMed] [Google Scholar]
- Yu R, Mandlekar S, Harvey KJ, Ucker DS, Kong AN. Chemopreventive isothiocyanates induce apoptosis and caspase-3-like protease activity. Cancer Res. 1998;58:402–408. [PubMed] [Google Scholar]
- Yu R, Mandlekar S, Lei W, Fahl WE, Tan TH, Kong AN. p38 mitogen-activated protein kinase negatively regulates the induction of phase II drug-metabolizing enzymes that detoxify carcinogens. J Biol Chem. 2000;275:2322–2327. doi: 10.1074/jbc.275.4.2322. [DOI] [PubMed] [Google Scholar]
- Zhang C, Li B, Zhang X, Hazarika P, Aggarwal BB, Duvic M. Curcumin selectively induces apoptosis in cutaneous T-cell lymphoma cell lines and patients’ PBMCs: potential role for STAT-3 and NF-kappaB signaling. J Invest Dermatol. 2010;130:2110–2119. doi: 10.1038/jid.2010.86. [DOI] [PubMed] [Google Scholar]
- Zhang B, Pan X, Cobb GP, Anderson TA. MicroRNAs as oncogenes and tumor suppressors. Dev Biol. 2007;302:1–12. doi: 10.1016/j.ydbio.2006.08.028. [DOI] [PubMed] [Google Scholar]
- Zhang P, Singh A, Yegnasubramanian S, Esopi D, Kombairaju P, Bodas M, et al. Loss of Kelch-like ECH-associated protein 1 function in prostate cancer cells causes chemoresistance and radioresistance and promotes tumor growth. Mol Cancer Ther. 2010;9:336–346. doi: 10.1158/1535-7163.MCT-09-0589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zhang J, Wang L, Wei H, Tian Z. Therapeutic effects of STAT3 decoy oligodeoxynucleotide on human lung cancer in xenograft mice. BMC Cancer. 2007;7:149. doi: 10.1186/1471-2407-7-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Xue Y, Oberley TD, Kiningham KK, Lin SM, Yen HC, et al. Overexpression of manganese superoxide dismutase suppresses tumor formation by modulation of activator protein-1 signaling in a multistage skin carcinogenesis model. Cancer Res. 2001;61:6082–6088. [PubMed] [Google Scholar]
- Zhou G, Bao ZQ, Dixon JE. Components of a new human protein kinase signal transduction pathway. J Biol Chem. 1995;270:12665–12669. doi: 10.1074/jbc.270.21.12665. [DOI] [PubMed] [Google Scholar]
- Zipper LM, Mulcahy RT. Inhibition of ERK and p38 MAP kinases inhibits binding of Nrf2 and induction of GCS genes. Biochem Biophys Res Commun. 2000;278:484–492. doi: 10.1006/bbrc.2000.3830. [DOI] [PubMed] [Google Scholar]
- Zipper LM, Mulcahy RT. Erk activation is required for Nrf2 nuclear localization during pyrrolidine dithiocarbamate induction of glutamate cysteine ligase modulatory gene expression in HepG2 cells. Toxicol Sci. 2003;73:124–134. doi: 10.1093/toxsci/kfg083. [DOI] [PubMed] [Google Scholar]