Abstract
We analyzed the functions of the influenza B virus nonstructural NS1-B protein, both by utilizing a constructed mutant virus (ΔNS1-B) lacking the NS1 gene and by testing the activities of the protein when expressed in cells. The mutant virus replicated to intermediate levels in 6-day-old embryonated chicken eggs that contain an immature interferon (IFN) system, whereas older eggs did not support viral propagation to a significant extent. The ΔNS1-B virus was a substantially stronger inducer of beta IFN (IFN-β) transcripts in human lung epithelial cells than the wild type, and furthermore, transiently expressed NS1-B protein efficiently inhibited virus-dependent activation of the IFN-β promoter. Interestingly, replication of the ΔNS1-B knockout virus was attenuated by more than 4 orders of magnitude in tissue culture cells containing or lacking functional IFN-α/β genes. These findings show that the NS1-B protein functions as a viral IFN antagonist and indicate a further requirement of this protein for efficient viral replication that is unrelated to blocking IFN effects.
Influenza is a severe acute respiratory disease that claims the lives of an estimated 20,000 people on average per year in the United States alone (60). Both influenza A and B viruses have in the past been responsible for such widespread epidemics in humans. The viruses belong to the Orthomyxoviridae family and are characterized by segmented negative-strand RNA genomes that consist of eight viral gene segments adding up to total sizes of 13.6 and 14.6 kb, respectively (36). Most of the 11 known proteins expressed by each virus type are believed to serve analogous functions. However, the proapoptotic PB1-F2 protein is uniquely found in the majority of influenza A virus strains (10), whereas only influenza B viruses express the NB protein that contributes to viral virulence (24, 59). There are further minor differences between influenza A and B viruses in the expression strategies of gene products encoded by the viral NA and M gene segments (35). Significant biological and epidemiological differences are indicated by the almost exclusive confinement of influenza B viruses to humans, whereas type A influenza viruses have a broad host reservoir in many avian and several other mammalian species (76).
A decisive factor for the efficient replication of influenza and several other viruses is the ability to inhibit in their hosts the expression of the antiviral cytokines alpha interferon (IFN-α) and IFN-β (for a review, see references 20 and 38). IFN-α/β gene induction appears to be a biphasic process whereby an immediate-early expression of the single IFN-β gene facilitates a secondary delayed activation of several IFN-α genes through a positive feedback loop (46, 56, 78). The activation of the IFN-β promoter is most likely triggered by virus-derived double-stranded RNA (dsRNA) molecules that are recognized by unidentified molecular sensors that in turn signal for the activation of transcription factors belonging to the NF-κB, IRF-3/-7, and ATF-2/c-Jun families (30, 31, 39, 44, 73, 77). Secreted IFN-α/β bind to a common IFN-α/β receptor and thereby activate the JAK/STAT signaling pathway, which leads to the nuclear formation of the heterotrimeric transcription factor ISGF-3 (62). ISGF-3 mediates the expression of more than 100 IFN-dependent genes including the dsRNA-activated protein kinase R (PKR), the Mx proteins, and the 2′-5′ oligo(A) synthetases, the expression of which creates an intracellular milieu that is unfavorable for viral propagation (13, 55). Moreover, the IFN-α/β sensitize cells for induction of apoptosis, which is thought to further contain viral spread in the infected organism (2, 4, 63, 66). Since IFN-α/β induces expression of the major histocompatibility complex class I genes and stimulate natural killer and dendritic cells, it also potently contributes to the development of adaptive immunity to invading viruses (for a review, see references 6 and 37).
Given the pleiotropic antiviral activities of IFN-α/β, it is not surprising that viruses have evolved a variety of IFN antagonistic proteins that tackle this cellular defense at distinct levels. Hence, viral gene products have been shown (i) to repress transcriptional activation of IFN genes, (ii) to compete for binding of secreted IFNs to their cognate receptors, (iii) to interfere with IFN signaling, or (iv) directly inhibit IFN-controlled antiviral gene products (reviewed in references 16 and 20). Importantly, genetic abolition of IFN antagonists, for instance, in Sendai virus, respiratory syncytial, virus or vaccinia virus, leads to strong attenuation in IFN-competent hosts (8, 32, 68). These findings highlight that countermeasures against the IFN defense system are pivotal chain links in the pathogenic processes that determine viral virulence.
Recent analyses have firmly established that the nonstructural NS1 protein of influenza A virus (termed NS1-A) is a major force that antagonizes activation of PKR and the expression of early defense genes, including IFN-β (5, 17, 19, 22, 41-43, 51, 64). NS1-A is a multifunctional 26-kDa protein that has been reported to bind to single- and double-stranded RNAs, to inhibit the polyadenylation and splicing of cellular pre-mRNAs, and to enhance translation (1, 11, 12, 15, 21, 23, 40, 41, 50, 54, 75). Notably, either the ability to sequester virus-derived dsRNAs and thereby to reduce the signaling for IFN gene activation or the blockade of early antiviral defense transcripts on a posttranscriptional level have been suggested to explain the IFN-antagonistic function of the NS1-A protein (34, 43, 51, 64, 71).
Comparatively little is known about the mechanism(s) used by influenza B viruses to circumvent antiviral responses. Influenza B virus expresses from an unspliced transcript of the viral NS segment a 281-amino-acid nonstructural protein termed NS1-B (9) that shares with its NS1-A counterpart the ability to bind to the same RNA targets and to inhibit activation of PKR in vitro (70). Accordingly, mutational analyses have identified RNA-binding domains in the N-terminal regions of the NS1-A and NS1-B proteins at amino acids 1 to 73 and 1 to 93, respectively (53, 70, 79). However, unique biological activities of the NS1-B protein are indicated by its deficiency to inhibit pre-mRNA processing (70), by its distinctive ability to bind to the antiviral response gene product ISG-15 and to inhibit its conjugation to cellular proteins (80), and by <20% sequence identity to the NS1-A protein. In fact, these differences have recently been considered to explain the observation that some early response genes, including ISG56, 2′-5′ oligo(A) synthetases, and ISG-15, were selectively induced in cells infected with influenza B virus but not in cells infected with influenza A virus (34, 51). Whether these findings indicate that the NS1-A and NS1-B proteins serve overlapping or different functions during infection is not known.
We have taken a reverse genetic approach to define the role(s) of the NS1-B protein during the life cycle of influenza B virus. To do this, we established an eight-plasmid system for the generation of influenza B virus from cloned cDNA, which is similar to the ones described recently by other groups (24, 26, 29), and succeeded to eliminate the NS1-B gene from the viral genome. Replication of a ΔNS1-B knockout virus was attenuated in embryonated chicken eggs and was even more reduced in tissue culture cells, suggesting that the NS1-B protein is important for efficient viral growth. We demonstrate that the ΔNS1-B virus is a considerably stronger inducer of IFN-β transcripts than the wild type and, furthermore, that ectopic expression of the NS1-B protein efficiently inhibited virus-induced IFN-β gene activation. Hence, these results indicate that the NS1-B protein serves to boost viral growth and that the subversion of the host's IFN response is an important part of its activities.
MATERIALS AND METHODS
Cells and viruses.
293T, Vero, and A549 cells were grown in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum, 2 mM l-glutamine, and antibiotics. Madin-Darby canine kidney (MDCK) type II cells were grown in minimal essential medium (MEM) supplemented with 10% fetal calf serum. The MDCK-C3 cell line that contains a stable integrate of a firefly luciferase gene controlled by the human IFN-β promoter was generated by transfection of MDCK cells with plasmid p125-Luc-neoR and selection of a resulting G418 sulfate-resistant cell clone showing strong inducible reporter gene activity after treatment with the dsRNA analogue poly(I)-poly(C). All cells were maintained at 37°C and 5% CO2. Stocks of the influenza B virus strains B/Lee/40 and B/Md/59 (obtained from P. Palese, Mt. Sinai School of Medicine, New York, N.Y.) and influenza A/PR/8/34 virus were grown in the allantoic cavities of 11-day-old embryonated chicken eggs for 3 days at 33°C (type B) or 2 days at 37°C (type A). The influenza B virus mutant ΔNS1-B and the influenza A/delNS1 virus (18) were amplified in 6-day-old chicken eggs. The ΔNS1-B virus was purified and further concentrated for metabolic labeling experiments and reporter gene assays by sedimentation through a 30% sucrose cushion during centrifugation in a SW28 rotor (Beckman) for 90 min at 25,000 rpm and 4°C. To analyze viral replication, confluent MDCK and Vero cell cultures were infected at the indicated multiplicity of infection (MOI) and incubated for 3 days at 33°C in MEM containing 0.2% bovine albumin (MEM-BA) and 1 μg of trypsin/ml. Virus titers were determined on MDCK cells by plaque assay or by counting the numbers of fluorescent cells after infection and indirect immunofluorescence staining with the NP-specific monoclonal antibody BM3149 (DBC Biermann, Bad Nauheim, Germany). Titers were correspondingly expressed as PFU or fluorescence-forming units/ml.
RT-PCR and construction of plasmids.
The RNeasy kit (Qiagen) was used according to the manufacturer's protocol to extract viral RNA from a stock of influenza B/Lee/40 virus. The eight reverse genetic plasmids pHW-Lee-PB2, pHW-Lee-PB1, pHW-Lee-PA, pHW-Lee-HA, pHW-Lee-NP, pHW-Lee-NA, pHW-Lee-M, and pHW-Lee-NS were constructed by reverse transcriptase PCR (RT-PCR) amplification of single viral RNA segments and cloning of the resulting cDNAs into the vector pHW2000 (27). In brief, the viral RNAs were first reverse transcribed with Moloney murine leukemia virus reverse transcriptase (Promega) by using a universal nine-nucleotide primer (UNI-9) that is complementary to the conserved 3′ ends of all eight viral RNA segments. The RT reaction was performed for 60 min at 37°C, followed by 15 min at 70°C. Subsequently, single gene segments were amplified by PCR by using the Pfu Turbo Polymerase (Roche Diagnostics) and segment-specific primers carrying BsmBI (PB1, PB2, PA, NA, M, and NS), BspMI (NP), or AarI (HA) restriction site sequences at their 5′ ends. Primer sequences can be provided on request. The PCR was performed for 35 cycles under the following conditions: 94°C for 1 min, 52°C for 30 s, and 72°C for 3 min (NS, M, NA) or 6 min (NP, HA, PA, PB1, PB2) followed by one cycle at 72°C for 5 min. Human IFN-β and actin transcripts were detected in 1 μg of total RNA (RNeasy) extracted from A549 cells by using gene-specific primers in the OneStep RT-PCR kit (Qiagen).
pHW-Lee-NS-XhoI is a derivative of pHW-Lee-NS that was constructed with the QuikChange mutagenesis kit (Stratagene) by introducing a novel XhoI recognition site at nucleotides 262 to 267 of the viral NS segment. For generation of the pHW-Lee-ΔNS1-B plasmid, polyadenylated RNA was extracted from influenza B/Lee/40 virus-infected MDCK cells by using the Oligotex direct mRNA Midi/Maxi Kit (Qiagen) and NS segment-specific primers were used for RT-PCR amplification. The amplified NEP/NS2 fragment was purified with the QiaEx II gel extraction kit (Qiagen), digested with BsmBI, and cloned into pHW2000. To construct plasmid pcDNA-NS1-B, the NS1 cDNA was PCR amplified with pHW-Lee-NS as a template and cloned between the HindIII/XhoI sites of pcDNA3 (Invitrogen). p125-Luc-neoR was constructed by transferring the SacII/XhoI fragment of p125-Luc (78) containing the IFN-β promoter luciferase reporter gene into pcDNAI-neo (Invitrogen). The integrities of the constructs were confirmed by DNA cycle sequencing by using an ABI Prism 3100 genetic analyzer (Applied Biosystems).
Transfection-mediated recovery of recombinant influenza B virus.
To generate the recombinant influenza B wild-type virus the eight plasmids pHW-Lee-PB2, pHW-Lee-PB1, pHW-Lee-PA, pHW-Lee-HA, pHW-Lee-NP, pHW-Lee-NA, pHW-Lee-M, and pHW-Lee-NS-XhoI (0.5 μg each) were transfected into 106 293T cells in suspension with the Lipofectamine 2000 reagent (Invitrogen). At 72 h after transfection, the supernatant of transfected cells was inoculated into the allantoic cavities of 11-day-old chicken eggs to grow stocks of recombinant virus. The ΔNS1-B virus was rescued by essentially the same procedure, except that pHW-Lee-NS-XhoI was replaced by pHW-Lee-ΔNS1-B and 0.5 μg of pcDNA-NS1 (74) was added to the transfection mix. Supernatants of transfected cells were passaged into 6-day-old chicken eggs. The recovery of recombinant influenza B viruses was verified by gel electrophoretic analyses of RT-PCR products representing the viral NS segments. Sequence analysis indicated that the triplet encoding tyrosine 71 of the NEP/NS2 protein in the parental plasmid pHW-Lee-delNS1 had been changed from TAT to CAT in the recovered ΔNS1-B virus. This alteration was not further investigated.
Metabolic labeling and immunoblot analysis.
Confluent 5 × 105 MDCK cells seeded in 22 mm dishes were either mock-treated or infected with rec. Lee wild-type or ΔNS1-B virus at an MOI of 10. Cells were incubated in MEM-BA at 33°C for various time points and were subsequently labeled for 1 h with 25 μCi of [35S]methionine (Amersham, Braunschweig, Germany) in Met− MEM. The cells were washed with ice-cold phosphate-buffered saline and lysed in 100 μl of radioimmunoprecipitation assay buffer. The extracts were subjected to 12.5% sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and the radiolabeled proteins were visualized by autoradiography. The viral NP and NS1 proteins were detected in lysates of infected MDCK cells by immunoblot analysis with a monoclonal NP-specific mouse antibody (BM3149; DBC Biermann) or rabbit anti-NS1-B serum (a gift of A. García-Sastre, Mt. Sinai School of Medicine, New York, N.Y.).
Analysis of IFN-β promoter activity in infected cells.
MDCK-C3 cells that contain a stably integrated IFN-β promoter luciferase reporter gene (see above) were either mock treated or infected with influenza virus at an MOI of 1 to analyze virus-induced IFN-β promoter activation. Cells were incubated for 8 h at 33° (type B viruses) or 37°C (type A viruses) and were extracted in 100 μl of reporter lysis buffer (Promega). Luciferase activity in lysate aliquots was determined by using Promega's luciferase assay system in a model LB96V luminometer (EG+G Berthold, Bad Wildbad, Germany). The activities were normalized for equal protein amounts, and reporter gene activation in infected cells was expressed in comparison to values obtained from mock-infected cells. For assaying the effects of the NS1-A and NS1-B proteins on viral IFN-β promoter activation normal MDCK cells where transfected by using Lipofectamine 2000 (Invitrogen). Each transfection of 5 × 105 cells contained 50 ng of p125-Luc reporter plasmid, 5 ng of pRL-TK-luc and 1 μg of either pcDNA3, pcDNA-NS1-A, or pcDNA-NS1-B expression vector. At 24 h posttransfection cells were mock treated or infected with influenza ΔNS1-B or A/delNS1 virus at an MOI of 1 and incubated as described above. At 8 h postinfection cells were lysed in 100 μl of passive lysis buffer (Promega), and the activities of the Renilla and the firefly luciferases were determined by using Promega's dual luciferase assay system.
RESULTS
Generation of recombinant influenza B virus from cloned cDNA.
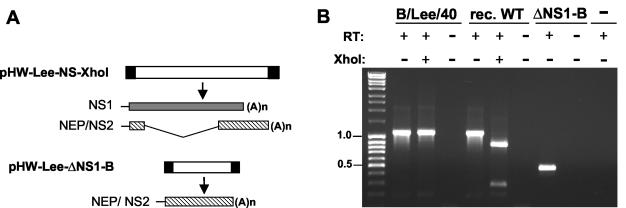
To analyze the roles of the NS1-B protein during the viral life cycle, a purely plasmid-based system for the generation of recombinant influenza B virus was established. Complete cDNA of each of the eight viral RNA gene segments of the influenza B/Lee/40 virus was synthesized and integrated into the plasmid pHW2000 (27). These plasmids facilitate bidirectional transcription of negative-sense viral RNAs and positive-sense mRNAs since the cloned viral cDNAs are flanked upstream by a human RNA polymerase I promoter and downstream by an RNA polymerase II-specific promoter. Transfection of the eight plasmids into 293T cells resulted after 72 h in the generation of virus that was amplified in a subsequent passage in embryonated chicken eggs to high titers. The NS segment of the transfectant virus carried an engineered genetic tag site such that the corresponding cDNA was susceptible to cleavage by the restriction endonuclease XhoI, thereby verifying the recombinant nature of the isolate (Fig. 1B). The natural and the recombinant influenza B viruses grew to basically identical titers in MDCK cells and embryonated chicken eggs (see below), indicating that there were no further differences in their genomes.
FIG. 1.
Generation of recombinant influenza B/Lee wild-type virus and NS1 deletion mutant (ΔNS1-B). (A) Structure of the NS segments of the wild-type and ΔNS1-B viruses. The plasmid pHW-Lee-NS-XhoI expresses the NS wild-type segment that encodes both the NS1 and, from a spliced transcript, the NEP/NS2 proteins. This construct was used with the other seven plasmids encoding the residual viral gene segments for recovery of the recombinant B/Lee wild-type virus in transfected 293T cells. In the plasmid pHW-Lee-ΔNS1-B, the coding region for NS1 was deleted. (B) RT-PCR analysis of viral NS segments. After the extraction of viral RNAs from the natural influenza B/Lee/40, the recombinant wild-type and ΔNS1-B viruses the viral NS segments were reverse transcribed (lanes “+”) and amplified by PCR. The resulting products were separated on a 2% agarose gel and stained with ethidium bromide. DNA size marker fragments were run in parallel on the very left lane. In control reactions, the RT step or an RNA template was omitted (lanes “RT:−” and “−”). The NS segment of the recombinant wild-type (rec. WT) was distinguished from the B/Lee/40 segment by digestion at the introduced XhoI site. Deletion of the NS1 coding region reduced the length of the NS segment from 1,096 to 441 nucleotides. The positions of the 1.0- and 0.5b-kb size marker fragments are indicated to the left.
Rescue of an influenza B mutant virus lacking the NS1 gene.
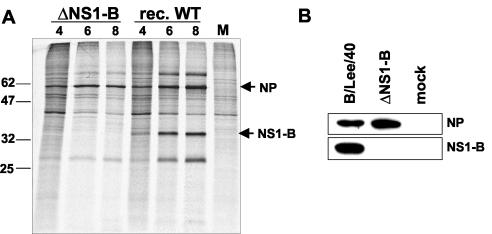
The coding region of the influenza B virus NS1 protein overlaps in part with the reading frame for the NEP/NS2 protein that is expressed from a spliced transcript of the viral NS gene segment (9) (Fig. 1A). For the generation of NS1-deficient influenza B virus, we prepared a derivative of the NS reverse genetic plasmid termed pHW-Lee-ΔNS1-B, in which the sequences specifying the NS1 protein were deleted, while all NEP/NS2 coding sequences and the terminal noncoding regions were maintained (Fig. 1A). This construct yielded recombinant virus in the rescue system when supernatant of transfected 293T cells was passaged in 6-day-old embryonated chicken eggs, which are characterized by an immature IFN system (28, 48). No virus was recovered when supernatants of transfected cells were passaged in MDCK cells or 11-day-old eggs (data not shown). RT-PCR and sequencing analyses confirmed the presence of the shortened ΔNS1-B segment in the obtained virus preparation (Fig. 1B and data not shown). Metabolic labeling of virus-infected MDCK cells with [35S]methionine demonstrated that protein synthesis of the wild-type and ΔNS1-B viruses showed essentially identical kinetics (Fig. 2A). However, extracts of ΔNS1-B virus-infected cells clearly lacked a protein corresponding to NS1-B. This finding was confirmed by immunoblot analysis of extracts from MDCK cells infected with the ΔNS1-B or the B/Lee/40 virus (Fig. 2B). No NS1-B protein was detected in cells infected with the mutant virus, although they expressed substantial amounts of the viral nucleoprotein. These findings established the generation of influenza B virus lacking the NS1-B gene.
FIG. 2.
Lack of NS1 expression in ΔNS1-B virus-infected cells. (A) Metabolic labeling of proteins in virus-infected MDCK cells. Cells were mock treated (lane “M”) or infected with the recombinant wild-type or ΔNS1-B virus at an MOI of 10 and incubated at 33°C for various time points. At 3, 5, or 7 h postinfection, the cultures were metabolically labeled with [35S]methionine for 1 h, and cell extracts were prepared at 4, 6, and 8 h postinfection, followed by analysis by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and autoradiography. The positions of molecular mass markers and of the viral NP and NS1-B proteins are indicated on the left and right sides, respectively. (B) Immunoblot analysis of virus-infected cells. MDCK cells were either mock treated or infected for 8 h with influenza B/Lee/40 or ΔNS1-B virus. Lysates were prepared and analyzed by immunoblotting with NP- and NS1-specific antibodies.
Deletion of the NS1 gene severely attenuates influenza B virus for replication.
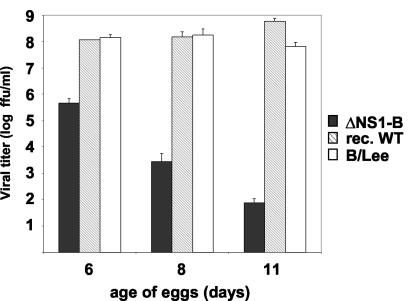
To characterize the impact of the NS1 gene deletion on the replicative properties of influenza B virus, we first compared the growth of the wild-type and ΔNS1-B viruses on embryonated chicken eggs of 6, 8, and 11 days of age (Fig. 3). It has previously been shown that the IFN inducibility and responsiveness in chicken embryo cells increases with age and this maturation process correlates with a higher restriction of influenza virus replication in older eggs (28, 48, 58, 65). Thus, we rationalized that a comparison of the replicative properties in embryonated eggs could identify a putative IFN sensitivity of the ΔNS1-B virus. The natural and the recombinant wild-type viruses grew to high titers in the range of 108 infectious units/ml in embryonated eggs within 3 days regardless of their age at infection (Fig. 3). In striking contrast, the growth capability of the ΔNS1-B virus gradually declined when eggs of increasing age were used for infection. Thus, we determined averages of 5 × 105, 3 × 103, and less than 1 × 102 fluorescence-forming units/ml in 6-, 8-, and 11-day-old eggs, respectively (Fig. 3). These findings indicated that the ΔNS1-B virus is sensitive to IFN to some extent and suggested that the NS1-B protein may have IFN-antagonistic properties.
FIG. 3.
Replication of the ΔNS1-B virus in embryonated chicken eggs. Six-, eight-, and eleven-day-old embryonated chicken eggs were inoculated with 100 infectious units of either influenza B/Lee/40, recombinant wild-type (rec. WT), or ΔNS1B virus and then incubated for 72 h at 33°C. Virus titers were determined as described in Materials and Methods. The indicated values represent the average of at least three independent experiments. Error bars indicate the standard deviations.
We next examined multicyclic replication on MDCK and Vero tissue culture cells that promote high-level growth of influenza B virus wild-type strains (Table 1). In comparison, the ΔNS1-B virus was attenuated for replication by about 6 logs on MDCK cells that can produce and respond to IFN-α/β. Unexpectedly, the ΔNS1-B virus exhibited also a severe growth phenotype on Vero cells that have a chromosomal deletion of the IFN-α and IFN-β genes (14). This was a surprising observation because Vero cells are known to support the replication of several mutant viruses with inactive IFN-antagonistic proteins much better than cell lines in which these antiviral cytokines can be expressed, as was observed with respiratory syncytial virus, human parainfluenza virus type 2, and influenza A virus (18, 33, 57). In parallel infections it was confirmed that the corresponding influenza A/delNS1 mutant virus replicated to a titer of ca. 5 × 106 PFU/ml on the Vero cells used (data not shown). However, the missing antiviral IFN function did not rescue the growth of the ΔNS1-B mutant in these cells, indicating that the NS1-B protein has a crucial role in promoting high-level replication of influenza B virus both in IFN-competent and -defective host cells.
TABLE 1.
Replication of the ΔNS1-B virus is attenuated in MDCK and Vero cellsa
| Cell type and expt no. (MOI) | Virus titer
|
|
|---|---|---|
| Recombinant wild type | ΔNS1-B | |
| MDCK cells | ||
| 1 (0.1) | 3.1 × 108 | 3.2 × 102 |
| 2 (0.1) | 3.1 × 108 | 1.7 × 102 |
| 3 (0.1) | 1.4 × 107 | 6.8 × 101 |
| 4 (0.1) | 3.1 × 107 | 6.8 × 101 |
| 5 (0.01) | 2.7 × 108 | <1.7 × 101 |
| 6 (0.01) | 3.3 × 108 | <1.7 × 101 |
| Vero cells | ||
| 1 (0.1) | 5.9 × 106 | 2.5 × 102 |
| 2 (0.1) | 6.6 × 106 | 1.7 × 102 |
| 3 (0.1) | 1.2 × 105 | 2.2 × 102 |
| 4 (0.1) | 2.0 × 105 | 1.7 × 102 |
| 5 (0.01) | 3.5 × 106 | <1.7 × 101 |
| 6 (0.01) | 4.4 × 106 | <1.7 × 101 |
Confluent monolayer cultures were infected at the indicated MOIs and incubated for 3 days at 33°C. Virus titers were determined as described in Materials and Methods.
The NS1 protein suppresses activation of the IFN-β promoter by influenza B virus.
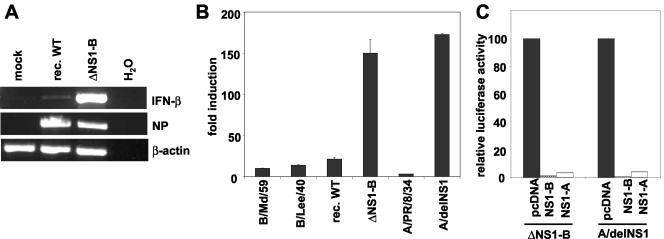
The growth characteristics of the ΔNS1-B virus in the embryonated eggs suggested that it was either a strong inducer of IFN-α/β and/or particularly vulnerable to the antiviral activities of these cytokines. To examine the former possibility, we analyzed the accumulation of IFN-β transcripts in human A549 lung epithelial cells and canine MDCK cells. We demonstrated that the ΔNS1-B mutant virus induced a prominent synthesis of IFN-β transcripts, whereas only low levels were detectable in wild-type-infected cells (Fig. 4A and data not shown). This finding indicated that the NS1-B protein inhibits virus-induced expression of the IFN-β gene.
FIG. 4.
The NS1-B protein antagonizes viral activation of the IFN-β gene. (A) IFN-β gene expression is strongly upregulated in ΔNS1-B virus-infected lung epithelial cells. Human A549 cells were infected with the recombinant B/Lee wild-type virus or the ΔNS1-B mutant at an MOI of 1 or were mock infected. Total RNA was extracted from the cells 10 h postinfection, and equal amounts were analyzed by RT-PCR with DNA oligonucleotide primers specific for IFN-β mRNA (upper panel), the viral NP segment (middle panel), or human β-actin transcripts (lower panel). The amplified products were separated by agarose gel electrophoresis and stained with ethidium bromide. In control reactions, no RNA template was added (lanes H2O). (B) Influenza A and B viruses lacking the NS1 proteins are strong activators of the IFN-β promoter. MDCK-C3 cells that contain a stable firefly luciferase reporter gene under the control of the IFN-β promoter were infected with influenza B virus strain Md/59 or Lee/40, the recombinant Lee wild-type virus (rec. WT), or ΔNS1-B virus at an MOI of 1. For a comparison, the influenza A/PR/8/34 strain and the isogenic A/delNS1 mutant virus were used in parallel infections. Luciferase activity was determined 8 h postinfection and is presented as the fold induction compared to mock-infected cells after normalization of extracts for equal amounts of protein. The graph shows average values of a typical experiment conducted in duplicate that was repeated three times. Error bars indicate the standard deviations. (C) Normal MDCK cells were transfected with the IFN-β promoter luciferase reporter plasmid p125-Luc and expression plasmids for the NS1-B or NS1-A protein or empty vector (pcDNA). In all assays, the plasmid pRL-TK-Luc encoding a Renilla luciferase under the control of a constitutive promoter was cotransfected and used as an internal control to normalize the results. One day posttransfection cells were stimulated by infection with the ΔNS1-B or the A/delNS1A virus at an MOI of 1 or were mock treated. Luciferase activities in cell extracts were determined 8 h postinfection. The value for a virus-induced increase in vector-transfected cells was arbitrarily set to 100% and compared to the induction observed in NS1-expressing cells.
For a more quantitative analysis, we compared the induction of an IFN-β promoter reporter gene by the ΔNS1-B and the recombinant Lee wild-type virus, as well as that of the two natural influenza B/Lee/40 and B/Md/59 strains. Furthermore, we also tested the influenza A/PR/8/34 virus expressing or lacking the corresponding NS1-A gene in parallel to detect possible differences between influenza A and B viruses. For these experiments we constructed and used the MDCK-C3 cell line that contains a stably integrated luciferase reporter gene under transcriptional control of the IFN-β promoter (see Materials and Methods). These assays demonstrated that all influenza B virus wild-type strains moderately activated the IFN promoter by ∼10-fold, which was only slightly higher as was found in parallel for the influenza A/PR/8/34 virus (Fig. 4B). In contrast, the ΔNS1-B virus greatly stimulated expression from the IFN-β promoter by at least 150-fold. Thus, the different levels of IFN-β transcripts in cells infected with the wild-type or the ΔNS1-B virus (Fig. 4A) appear to result from a stronger transcriptional activation of the IFN-β promoter in the absence of the NS1-B protein. A comparable strong induction was also observed for the influenza A/delNS1 virus that lacks the corresponding NS1-A gene and had previously been characterized as a strong IFN inducer (18, 43, 71).
The former experiments showed that the deletion of the NS1 gene dramatically increased the interferonogenicity of influenza B virus and suggested that the NS1-B protein antagonizes IFN-β induction as it is known for its NS1-A counterpart. We therefore examined whether or not the expression of the NS1-B or NS1-A proteins was sufficient to prevent activation of the IFN-β promoter by either the ΔNS1-B or the A/delNS1 virus. Cells were first cotransfected with an IFN-β reporter plasmid and vectors expressing the NS1-A or NS1-B protein. Subsequently, the cells were stimulated by infection with one of the two types of NS1-deficient influenza viruses to activate the IFN-β promoter. In these assays it was demonstrated that both the NS1-B and the NS1-A protein rendered the IFN-β reporter almost completely unresponsive to either virus challenge (Fig. 4C). Taken together, these findings strongly suggest that a prominent function of the influenza B virus NS1 protein is to tame the cellular IFN response and to enable the virus to fully exploit the host's capacity for virus propagation.
DISCUSSION
Although it has been known for a long time that influenza B viruses are sensitive to IFN-α/β (61), there is still surprisingly little knowledge of the viral gene product(s) that suppresses this antiviral defense in infected cells. Based on three key observations, the present study establishes that the NS1-B protein functions as an IFN antagonist of influenza B viruses and can be added to the growing list of viral proteins known to tackle the cellular IFN system (16).
First is the genetic deletion of the NS1-B gene restricted virus replication in 11-day-old eggs with a more mature IFN system much stronger than in younger eggs, in which IFN induction is low in general (28, 48, 58). Thus, the growth capabilities of the constructed ΔNS1-B knockout virus in embryonated chicken eggs were inversely correlated with the IFN inducibility in these hosts. Second, the ΔNS1-B virus proved to be a potent inducer of the IFN-β promoter and correspondingly of IFN-β transcripts, whereas wild-type viruses expressing the NS1-B protein were comparatively moderate stimulators. Interestingly, the differences between the wild-type and the ΔNS1-B mutant virus closely resembled the variations observed for influenza A viruses expressing or lacking the antagonistic NS1-A gene. Strikingly, the ectopic expression of NS1-B was sufficient to silence the activation of the IFN-β promoter by infection with either the ΔNS1-B or the heterotypic A/delNS1 mutant virus. Moreover, in a reversed experimental setup, the NS1-A protein also prevented activation of the IFN-β promoter by either type of NS1 knockout virus. These findings indicate that the major nonstructural proteins of the two influenza virus types exhibit an overlapping or identical activity that blocks activation of the IFN cascade by virus infection at the early stage of IFN gene expression.
At present, we can only speculate on the mechanism(s) by which the NS1-B protein antagonizes IFN-α/β. Previously, it had been shown that the NS1-A and NS1-B proteins share two important biological activities, namely, the binding to the same dsRNA molecules and the ability to block the activation of PKR in vitro (70). There is abundant evidence that the NS1-A protein inhibits in infected cells the activation of dsRNA-responsive transcription factors, including ATF-2/c-Jun, IRF-3, and NF-κB, and thereby reduces the activation of dsRNA-controlled response genes to a tolerable level (43, 64, 71). Given the exchangeability of the two NS1 proteins in blocking IFN-β promoter activation, we suggest that the NS1-B protein neutralizes virus-induced dsRNA signals that trigger antiviral defense genes in influenza B virus-infected cells. This activity could mechanistically be mediated either by dsRNA sequestration or by targeting of the cellular factors that recognize the molecular dsRNA patterns.
Although the similar antagonistic activities apparently argue for a similar function for the nonstructural proteins of the two influenza virus types, we also observed significant differences in terms of the requirements for efficient virus growth. In light of the deleterious effects of IFN-α/β on virus infection, we had anticipated that the deletion of the NS1-B gene would severely restrict multicyclic replication of the ΔNS1-B mutant virus on MDCK cells that can secrete and respond to IFN-α/β. However, it was surprising that growth of the mutant was also strongly reduced on Vero cells by at least 4 logs. The Vero cell line is deficient in IFN expression (14) and has been shown to support quite well the replication of other mutant viruses with defective IFN antagonists, including the influenza A/delNS1 virus (18, 25, 33, 57). Specifically, the A/delNS1 virus replicated on Vero cells to titers of ca. 5 × 106 PFU/ml, which was only 10-fold lower than the isogenic wild-type virus (the present study and reference 18). These findings indicate a much higher dependency of influenza B virus on the functions of its major nonstructural protein compared to influenza A virus.
We suggest two nonexclusive explanations for the strong general impact of the NS1-B gene deletion on viral replication. First, the NS1-B protein may play a further important role in counteracting the activity of an antiviral gene product(s) that is expressed in Vero cells in an IFN-independent manner. For instance, it is known that a subset of IFN-inducible genes, including ISG15, ISG54, and ISG56, can be directly upregulated by virus infection in the absence of IFN, most likely mediated through activated IRF-3 (3, 7, 49, 52, 72). In fact, the NS1-B protein has been reported to bind to and prevent the conjugation of the ubiquitin-like ISG15 protein to cellular target proteins, which may additionally benefit virus replication by weakening the intracellular JAK/STAT signaling pathway (45, 80). Enhanced ISGylation may consequently contribute to the restriction of the ΔNS1-B virus in cells in general. However, there is currently no information available on whether the gene of the ISG15-activating enzyme UBE1L is present in Vero cells or deleted, as was reported for several tumor cell lines (47). A second possible explanation for restriction of the ΔNS1-B virus in Vero cells might be a critical contribution of the NS1-B protein to the principal viral replication process rather than an effect on the host cell. For instance, an influenza B/Lee/40 reassortant virus expressing a truncated NS1-B protein of only 127 amino acids exhibited a deregulated pattern of viral RNA synthesis that was associated with a small plaque phenotype (67). Thus, the complete absence of the NS1-B protein could significantly impart viral RNA synthesis and result in considerable attenuation of viral growth even in the absence of antiviral responses.
We are aware that our model to explain the IFN-antagonistic function of the NS1-B protein is tentatively not in agreement with a previous report, in which influenza B/Lee/40 virus infection activated IRF-3 (34). This outcome would not be expected if all dsRNA-dependent responses were blocked by the NS1-B protein. However, we noticed in the course of our study a strong influence of the multiplicity of infection on the activation of the IFN-β promoter by wild-type influenza B viruses. A moderate activation was determined when a multiplicity of ≤1 was used (see Fig. 4), whereas the promoter was induced five times more strongly when the cells were infected at an MOI of 10 (B. Dauber and T. Wolff, unpublished observations). Thus, infection of a cell with many virus particles may generate an overwhelming quantity of intracellular dsRNAs that simply run down the inhibitory capacities of the nonstructural NS1-B protein, resulting in activation of dsRNA-dependent reactions. Moreover, the second possibility remains that virus-specific components other than dsRNA contribute to the activation of IRF-3 by influenza B virus, as has been reported for the measles virus nucleocapsid protein (69). In any case, our data show that the presence of the NS1-B protein profoundly reduces the activation of such antiviral reactions.
In summary, the establishment and use of a reverse genetic procedure for influenza B virus identified the NS1-B protein as a viral factor that antagonizes IFN-β induction and boosts viral replication. This reverse genetic system will hopefully allow us in future studies to explore the functions of single NS1-B domains during the replication cycle and to assess their contributions to the pathogenesis and virulence of influenza B virus.
Acknowledgments
We thank A. Egorov (Institute of Applied Microbiology, Vienna, Austria) for the generous gift of the influenza A/delNS1 virus, T. Fujita (The Tokyo Metropolitan Institute of Medical Science, Tokyo, Japan) and R. Webster (St. Jude Children's Research Hospital, Memphis, Tenn.) for providing plasmids, Khanh Le (Robert Koch-Institute, Berlin, Germany) for constructing plasmid p125-Luc neo, Adolfo García-Sastre (Mt. Sinai School of Medicine, New York, N.Y.) for anti-NS1-B serum, and Stephan Ludwig (University of Duesseldorf, Duesseldorf, Germany) and Hartmut Hengel (RKI, Berlin, Germany) for critical comments on the manuscript.
B.D. acknowledges financial support from the FAZIT Foundation (Frankfurt, Germany).
REFERENCES
- 1.Aragon, T., S. de la Luna, I. Novoa, L. Carrasco, J. Ortin, and A. Nieto. 2000. Eukaryotic translation initiation factor 4GI is a cellular target for NS1 protein, a translational activator of influenza virus. Mol. Cell. Biol. 20:6259-6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balachandran, S., P. C. Roberts, T. Kipperman, K. N. Bhalla, R. W. Compans, D. R. Archer, and G. N. Barber. 2000. Alpha/beta interferons potentiate virus-induced apoptosis through activation of the FADD/caspase-8 death signaling pathway. J. Virol. 74:1513-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bandyopadhyay, S. K., G. T. Leonard, Jr., T. Bandyopadhyay, G. R. Stark, and G. C. Sen. 1995. Transcriptional induction by double-stranded RNA is mediated by interferon-stimulated response elements without activation of interferon-stimulated gene factor 3. J. Biol. Chem. 270:19624-19629. [DOI] [PubMed] [Google Scholar]
- 4.Barber, G. N. 2000. The interferons and cell death: guardians of the cell or accomplices of apoptosis? Semin. Cancer Biol. 10:103-111. [DOI] [PubMed] [Google Scholar]
- 5.Bergmann, M., A. García-Sastre, E. Carnero, H. Pehamberger, K. Wolff, P. Palese, and T. Muster. 2000. Influenza virus NS1 protein counteracts PKR-mediated inhibition of replication. J. Virol. 74:6203-6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Biron, C. A., and G. C. Sen. 2001. Interferons and other cytokines, p. 321-351. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott/The Williams & Wilkins Co., Philadelphia, Pa.
- 7.Boyle, K. A., R. L. Pietropaolo, and T. Compton. 1999. Engagement of the cellular receptor for glycoprotein B of human cytomegalovirus activates the interferon-responsive pathway. Mol. Cell. Biol. 19:3607-3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brandt, T. A., and B. L. Jacobs. 2001. Both carboxy- and amino-terminal domains of the vaccinia virus interferon resistance gene, E3L, are required for pathogenesis in a mouse model. J. Virol. 75:850-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Briedis, D. J., and R. A. Lamb. 1982. Influenza B virus genome: sequences and structural organization of RNA segment 8 and the mRNAs coding for the NS1 and NS2 proteins. J. Virol. 42:186-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, W., P. A. Calvo, D. Malide, J. Gibbs, U. Schubert, I. Bacik, S. Basta, R. O'Neill, J. Schickli, P. Palese, P. Henklein, J. R. Bennink, and J. W. Yewdell. 2001. A novel influenza A virus mitochondrial protein that induces cell death. Nat. Med. 7:1306-1312. [DOI] [PubMed] [Google Scholar]
- 11.Chen, Z., Y. Li, and R. M. Krug. 1999. Influenza A virus NS1 protein targets poly(A)-binding protein II of the cellular 3′-end processing machinery. EMBO J. 18:2273-2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de la Luna, S., P. Fortes, A. Beloso, and J. Ortin. 1995. Influenza virus NS1 protein enhances the rate of translation initiation of viral mRNAs. J. Virol. 69:2427-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Der, S. D., A. Zhou, B. R. Williams, and R. H. Silverman. 1998. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc. Natl. Acad. Sci. USA 95:15623-15628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diaz, M. O., S. Ziemin, M. M. Le Beau, P. Pitha, S. D. Smith, R. R. Chilcote, and J. D. Rowley. 1988. Homozygous deletion of the alpha- and beta 1-interferon genes in human leukemia and derived cell lines. Proc. Natl. Acad. Sci. USA 85:5259-5263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fortes, P., A. Beloso, and J. Ortin. 1994. Influenza virus NS1 protein inhibits pre-mRNA splicing and blocks mRNA nucleocytoplasmic transport. EMBO J. 13:704-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garcia-Sastre, A. 2002. Mechanisms of inhibition of the host interferon alpha/beta-mediated antiviral responses by viruses. Microbes Infect. 4:647-655. [DOI] [PubMed] [Google Scholar]
- 17.Garcia-Sastre, A., R. K. Durbin, H. Zheng, P. Palese, R. Gertner, D. E. Levy, and J. E. Durbin. 1998. The role of interferon in influenza virus tissue tropism. J. Virol. 72:8550-8558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garcia-Sastre, A., A. Egorov, D. Matassov, S. Brandt, D. E. Levy, J. E. Durbin, P. Palese, and T. Muster. 1998. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology 252:324-330. [DOI] [PubMed] [Google Scholar]
- 19.Geiss, G. K., M. Salvatore, T. M. Tumpey, V. S. Carter, X. Wang, C. F. Basler, J. K. Taubenberger, R. E. Bumgarner, P. Palese, M. G. Katze, and A. Garcia-Sastre. 2002. Cellular transcriptional profiling in influenza A virus-infected lung epithelial cells: the role of the nonstructural NS1 protein in the evasion of the host innate defense and its potential contribution to pandemic influenza. Proc. Natl. Acad. Sci. USA 99:10736-10741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goodbourn, S., L. Didcock, and R. E. Randall. 2000. Interferons: cell signalling, immune modulation, antiviral response and virus countermeasures. J. Gen. Virol. 81:2341-2364. [DOI] [PubMed] [Google Scholar]
- 21.Hatada, E., and R. Fukuda. 1992. Binding of influenza A virus NS1 protein to dsRNA in vitro. J. Gen. Virol. 73:3325-3329. [DOI] [PubMed] [Google Scholar]
- 22.Hatada, E., S. Saito, and R. Fukuda. 1999. Mutant influenza viruses with a defective NS1 protein cannot block the activation of PKR in infected cells. J. Virol. 73:2425-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hatada, E., T. Takizawa, and R. Fukuda. 1992. Specific binding of influenza A virus NS1 protein to the virus minus-sense RNA in vitro. J. Gen. Virol. 73:17-25. [DOI] [PubMed] [Google Scholar]
- 24.Hatta, M., and Y. Kawaoka. 2003. The NB protein of influenza B virus is not necessary for virus replication in vitro. J. Virol. 77:6050-6054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He, B., R. G. Paterson, N. Stock, J. E. Durbin, R. K. Durbin, S. Goodbourn, R. E. Randall, and R. A. Lamb. 2002. Recovery of paramyxovirus simian virus 5 with a V protein lacking the conserved cysteine-rich domain: the multifunctional V protein blocks both interferon-beta induction and interferon signaling. Virology 303:15-32. [DOI] [PubMed] [Google Scholar]
- 26.Hoffmann, E., K. Mahmood, C. F. Yang, R. G. Webster, H. B. Greenberg, and G. Kemble. 2002. Rescue of influenza B virus from eight plasmids. Proc. Natl. Acad. Sci. USA 99:11411-11416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoffmann, E., G. Neumann, Y. Kawaoka, G. Hobom, and R. G. Webster. 2000. A DNA transfection system for generation of influenza A virus from eight plasmids. Proc. Natl. Acad. Sci. USA 97:6108-6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Isaacs, A., and S. Baron. 1960. Antiviral action of interferon in embryonic cells. Lancet ii:946-947. [DOI] [PubMed] [Google Scholar]
- 29.Jackson, D., A. Cadman, T. Zurcher, and W. S. Barclay. 2002. A reverse genetics approach for recovery of recombinant influenza B viruses entirely from cDNA. J. Virol. 76:11744-11747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jacobs, B. L., and J. O. Langland. 1996. When two strands are better than one: the mediators and modulators of the cellular responses to double-stranded RNA. Virology 219:339-349. [DOI] [PubMed] [Google Scholar]
- 31.Juang, Y. T., W. Lowther, M. Kellum, W. C. Au, R. Lin, J. Hiscott, and P. M. Pitha. 1998. Primary activation of interferon A and interferon B gene transcription by interferon regulatory factor 3. Proc. Natl. Acad. Sci. USA 95:9837-9842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kato, A., K. Kiyotani, Y. Sakai, T. Yoshida, and Y. Nagai. 1997. The paramyxovirus, Sendai virus, V protein encodes a luxury function required for viral pathogenesis. EMBO J. 16:578-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kawano, M., M. Kaito, Y. Kozuka, H. Komada, N. Noda, K. Nanba, M. Tsurudome, M. Ito, M. Nishio, and Y. Ito. 2001. Recovery of infectious human parainfluenza type 2 virus from cDNA clones and properties of the defective virus without V-specific cysteine-rich domain. Virology 284:99-112. [DOI] [PubMed] [Google Scholar]
- 34.Kim, M. J., A. G. Latham, and R. M. Krug. 2002. Human influenza viruses activate an interferon-independent transcription of cellular antiviral genes: outcome with influenza A virus is unique. Proc. Natl. Acad. Sci. USA 99:10096-10101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lamb, R. A., and C. M. Horvath. 1991. Diversity of coding strategies in influenza viruses. Trends Genet. 7:261-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lamb, R. A., and R. M. Krug. 2001. Orthomyxoviridae: the viruses and their replication, p. 1487-1531. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott/The Williams & Wilkins Co., Philadelphia, Pa.
- 37.Le Bon, A., and D. F. Tough. 2002. Links between innate and adaptive immunity via type I interferon. Curr. Opin. Immunol. 14:432-436. [DOI] [PubMed] [Google Scholar]
- 38.Levy, D. E., and A. Garcia-Sastre. 2001. The virus battles: IFN induction of the antiviral state and mechanisms of viral evasion. Cytokine Growth Factor Rev. 12:143-156. [DOI] [PubMed] [Google Scholar]
- 39.Lin, R., C. Heylbroeck, P. M. Pitha, and J. Hiscott. 1998. Virus-dependent phosphorylation of the IRF-3 transcription factor regulates nuclear translocation, transactivation potential, and proteasome-mediated degradation. Mol. Cell. Biol. 18:2986-2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu, Y., X. Y. Qian, and R. M. Krug. 1994. The influenza virus NS1 protein: a novel inhibitor of pre-mRNA splicing. Genes Dev. 8:1817-1828. [DOI] [PubMed] [Google Scholar]
- 41.Lu, Y., M. Wambach, M. G. Katze, and R. M. Krug. 1995. Binding of the influenza virus NS1 protein to double-stranded RNA inhibits the activation of the protein kinase that phosphorylates the eIF-2 translation initiation factor. Virology 214:222-228. [DOI] [PubMed] [Google Scholar]
- 42.Ludwig, S., O. Planz, S. Pleschka, and T. Wolff. 2003. Influenza-virus-induced signaling cascades: targets for antiviral therapy? Trends Mol. Med. 9:46-52. [DOI] [PubMed] [Google Scholar]
- 43.Ludwig, S., X. Wang, C. Ehrhardt, H. Zheng, N. Donelan, O. Planz, S. Pleschka, A. García-Sastre, G. Heins, and T. Wolff. 2002. The influenza A virus NS1 protein inhibits activation of Jun N-terminal kinase and AP-1 transcription factors. J. Virol. 76:11166-11171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Majde, J. A. 2000. Viral double-stranded RNA, cytokines, and the flu. J. Interferon Cytokine Res. 20:259-272. [DOI] [PubMed] [Google Scholar]
- 45.Malakhova, O. A., M. Yan, M. P. Malakhov, Y. Yuan, K. J. Ritchie, K. I. Kim, L. F. Peterson, K. Shuai, and D. E. Zhang. 2003. Protein ISGylation modulates the JAK-STAT signaling pathway. Genes Dev. 17:455-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marie, I., J. E. Durbin, and D. E. Levy. 1998. Differential viral induction of distinct interferon-alpha genes by positive feedback through interferon regulatory factor-7. EMBO J. 17:6660-6669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McLaughlin, P. M., W. Helfrich, K. Kok, M. Mulder, S. W. Hu, M. G. Brinker, M. H. Ruiters, L. F. de Leij, and C. H. Buys. 2000. The ubiquitin-activating enzyme E1-like protein in lung cancer cell lines. Int. J. Cancer 85:871-876. [DOI] [PubMed] [Google Scholar]
- 48.Morahan, P. S., and S. E. Grossberg. 1970. Age-related cellular resistance of the chicken embryo to viral infections. I. Interferon and natural resistance to myxoviruses and vesicular stomatitis virus. J. Infect. Dis. 121:615-623. [DOI] [PubMed] [Google Scholar]
- 49.Mossman, K. L., P. F. Macgregor, J. J. Rozmus, A. B. Goryachev, A. M. Edwards, and J. R. Smiley. 2001. Herpes simplex virus triggers and then disarms a host antiviral response. J. Virol. 75:750-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nemeroff, M. E., S. M. Barabino, Y. Li, W. Keller, and R. M. Krug. 1998. Influenza virus NS1 protein interacts with the cellular 30-kDa subunit of CPSF and inhibits 3′ end formation of cellular pre-mRNAs. Mol. Cell 1:991-1000. [DOI] [PubMed] [Google Scholar]
- 51.Noah, D. L., K. Y. Twu, and R. M. Krug. 2003. Cellular antiviral responses against influenza A virus are countered at the posttranscriptional level by the viral NS1A protein via its binding to a cellular protein required for the 3′ end processing of cellular pre-mRNAS. Virology 307:386-395. [DOI] [PubMed] [Google Scholar]
- 52.Preston, C. M., A. N. Harman, and M. J. Nicholl. 2001. Activation of interferon response factor-3 in human cells infected with herpes simplex virus type 1 or human cytomegalovirus. J. Virol. 75:8909-8916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Qian, X. Y., C. Y. Chien, Y. Lu, G. T. Montelione, and R. M. Krug. 1995. An amino-terminal polypeptide fragment of the influenza virus NS1 protein possesses specific RNA-binding activity and largely helical backbone structure. RNA 1:948-956. [PMC free article] [PubMed] [Google Scholar]
- 54.Salvatore, M., C. F. Basler, J. P. Parisien, C. M. Horvath, S. Bourmakina, H. Zheng, T. Muster, P. Palese, and A. García-Sastre. 2002. Effects of influenza A virus NS1 protein on protein expression: the NS1 protein enhances translation and is not required for shutoff of host protein synthesis. J. Virol. 76:1206-1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Samuel, C. E. 2001. Antiviral actions of interferons. Clin. Microbiol. Rev. 14:778-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sato, M., N. Hata, M. Asagiri, T. Nakaya, T. Taniguchi, and N. Tanaka. 1998. Positive feedback regulation of type I IFN genes by the IFN-inducible transcription factor IRF-7. FEBS Lett. 441:106-110. [DOI] [PubMed] [Google Scholar]
- 57.Schlender, J., B. Bossert, U. Buchholz, and K. K. Conzelmann. 2000. Bovine respiratory syncytial virus nonstructural proteins NS1 and NS2 cooperatively antagonize alpha/beta interferon-induced antiviral response. J. Virol. 74:8234-8242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sekellick, M. J., and P. I. Marcus. 1985. Interferon induction by viruses. XIV. Development of interferon inducibility and its inhibition in chick embryo cells “aged” in vitro. J. Interferon Res. 5:651-667. [DOI] [PubMed] [Google Scholar]
- 59.Shaw, M. W., P. W. Choppin, and R. A. Lamb. 1983. A previously unrecognized influenza B virus glycoprotein from a bicistronic mRNA that also encodes the viral neuraminidase. Proc. Natl. Acad. Sci. USA 80:4879-4883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Simonsen, L., M. J. Clarke, G. D. Williamson, D. F. Stroup, N. H. Arden, and L. B. Schonberger. 1997. The impact of influenza epidemics on mortality: introducing a severity index. Am. J. Public Health 87:1944-1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Smart, K. M., and E. D. Kilbourne. 1966. The influence of cortisone on experimental viral infection. VII. Kinetics of interferon formation and its inhibition with hydrocortisone in relation to viral strain and virulence. J. Exp. Med. 123:309-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stark, G. R., I. M. Kerr, B. R. Williams, R. H. Silverman, and R. D. Schreiber. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67:227-264. [DOI] [PubMed] [Google Scholar]
- 63.Takaoka, A., S. Hayakawa, H. Yanai, D. Stoiber, H. Negishi, H. Kikuchi, S. Sasaki, K. Imai, T. Shibue, K. Honda, and T. Taniguchi. 2003. Integration of interferon-alpha/beta signaling to p53 responses in tumour suppression and antiviral defense. Nature 424:516-523. [DOI] [PubMed] [Google Scholar]
- 64.Talon, J., C. M. Horvath, R. Polley, C. F. Basler, T. Muster, P. Palese, and A. García-Sastre. 2000. Activation of interferon regulatory factor 3 is inhibited by the influenza A virus NS1 protein. J. Virol. 74:7989-7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Talon, J., M. Salvatore, R. E. O'Neill, Y. Nakaya, H. Zheng, T. Muster, A. Garcia-Sastre, and P. Palese. 2000. Influenza A and B viruses expressing altered NS1 proteins: a vaccine approach. Proc. Natl. Acad. Sci. USA 97:4309-4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tanaka, N., M. Sato, M. S. Lamphier, H. Nozawa, E. Oda, S. Noguchi, R. D. Schreiber, Y. Tsujimoto, and T. Taniguchi. 1998. Type I interferons are essential mediators of apoptotic death in virally infected cells. Genes Cells 3:29-37. [DOI] [PubMed] [Google Scholar]
- 67.Tanaka, T., T. Odagiri, and K. Tobita. 1988. Biological functions of the NS1 protein of an influenza B virus mutant which has a long carboxyl terminal deletion. Arch. Virol. 102:173-185. [DOI] [PubMed] [Google Scholar]
- 68.Teng, M. N., S. S. Whitehead, and P. L. Collins. 2001. Contribution of the respiratory syncytial virus G glycoprotein and its secreted and membrane-bound forms to virus replication in vitro and in vivo. Virology 289:283-296. [DOI] [PubMed] [Google Scholar]
- 69.tenOever, B. R., M. J. Servant, N. Grandvaux, R. Lin, and J. Hiscott. 2002. Recognition of the measles virus nucleocapsid as a mechanism of IRF-3 activation. J. Virol. 76:3659-3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang, W., and R. M. Krug. 1996. The RNA-binding and effector domains of the viral NS1 protein are conserved to different extents among influenza A and B viruses. Virology 223:41-50. [DOI] [PubMed] [Google Scholar]
- 71.Wang, X., M. Li, H. Zheng, T. Muster, P. Palese, A. A. Beg, and A. García-Sastre. 2000. Influenza A virus NS1 protein prevents activation of NF-κB and induction of alpha/beta interferon. J. Virol. 74:11566-11573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wathelet, M. G., P. M. Berr, and G. A. Huez. 1992. Regulation of gene expression by cytokines and virus in human cells lacking the type-I interferon locus. Eur. J. Biochem. 206:901-910. [DOI] [PubMed] [Google Scholar]
- 73.Wathelet, M. G., C. H. Lin, B. S. Parekh, L. V. Ronco, P. M. Howley, and T. Maniatis. 1998. Virus infection induces the assembly of coordinately activated transcription factors on the IFN-β enhancer in vivo. Mol. Cell 1:507-518. [DOI] [PubMed] [Google Scholar]
- 74.Wolff, T., R. E. O'Neill, and P. Palese. 1996. Interaction cloning of NS1-I, a human protein that binds to the nonstructural NS1 proteins of influenza A and B viruses. J. Virol. 70:5363-5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wolff, T., R. E. O'Neill, and P. Palese. 1998. NS1-Binding protein (NS1-BP): a novel human protein that interacts with the influenza A virus nonstructural NS1 protein is relocalized in the nuclei of infected cells. J. Virol. 72:7170-7180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wright, P. F., and R. G. Webster. 2001. Orthomyxoviruses, p. 1533-1579. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott/The Williams & Wilkins Co., Philadelphia, Pa.
- 77.Yoneyama, M., W. Suhara, Y. Fukuhara, M. Fukuda, E. Nishida, and T. Fujita. 1998. Direct triggering of the type I interferon system by virus infection: activation of a transcription factor complex containing IRF-3 and CBP/p300. EMBO J. 17:1087-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yoneyama, M., W. Suhara, Y. Fukuhara, M. Sato, K. Ozato, and T. Fujita. 1996. Autocrine amplification of type I interferon gene expression mediated by interferon-stimulated gene factor 3 (ISGF3). J. Biochem. 120:160-169. [DOI] [PubMed] [Google Scholar]
- 79.Yuan, W., J. M. Aramini, G. T. Montelione, and R. M. Krug. 2002. Structural basis for ubiquitin-like ISG 15 protein binding to the NS1 protein of influenza B virus: a protein-protein interaction function that is not shared by the corresponding N-terminal domain of the NS1 protein of influenza A virus. Virology 304:291-301. [DOI] [PubMed] [Google Scholar]
- 80.Yuan, W., and R. M. Krug. 2001. Influenza B virus NS1 protein inhibits conjugation of the interferon (IFN)-induced ubiquitin-like ISG15 protein. EMBO J. 20:362-371. [DOI] [PMC free article] [PubMed] [Google Scholar]