Abstract
The adenovirus major late transcription unit (MLTU) encodes multiple proteins from five regions, L1 to L5, through differential splicing and polyadenylation. MLTU expression is temporally regulated; only a single product from L1 (52/55K) is expressed prior to replication, but a subsequent switch, the mechanism of which has not been defined, leads to full expression that encompasses L1 IIIa and all L2 to L5 products. Transfection of a plasmid containing the complete MLTU gave a full array of proteins in proportions similar to those in a late infection, and in a time course, the temporal pattern of expression in a natural infection was reproduced. However, a plasmid truncated after the L3 poly(A) site exclusively expressed the L1 52/55K protein and was defective in the switch to full gene expression from L1 to L3. The L4 33K protein, supplied in trans, was sufficient to upregulate cytoplasmic mRNA for MLTU products characteristic of the late pattern of expression to levels comparable to those produced by the full-length MLTU. There was a corresponding increase in expression of the L1 IIIa, L2, and L3 proteins, except hexon. Hexon protein expression additionally required both the L4 100K protein in trans and sequences downstream of the L3 poly(A) site in cis. These results indicate that induction of L4 protein expression is a key event in the early-late switch in MLTU expression, which we propose is precipitated by small amounts of L4 expression in a feed-forward activation mechanism.
Human adenoviruses, especially group C viruses such as serotype 5 (Ad5), have been studied extensively; their tropism, gene expression, host cell interactions, and transforming capabilities are well characterized. Despite this knowledge, however, there remain important aspects of adenovirus biology that are not understood. One of these is the control of protein expression from the major late transcription unit (MLTU) during the infectious cycle.
Upon Ad5 infection, a temporally phased program of gene expression occurs. The E1A gene products are produced immediately and upregulate transcription from all of the early regions: E1A, E1B, E2, E3, and E4. The MLTU, which encodes the majority of the virus structural proteins within five subregions, L1 to L5, is also active at a low level during this early phase, but transcription proceeds only as far as the L3 region and mRNA production is focused on the L1 52/55K reading frame (33). After replication, the major late promoter (MLP) becomes fully activated and transcription is allowed to proceed through the full length of the MLTU to supply the proteins for the packaging of newly replicated genomes into virions. The MLP is activated by the E1A 289R protein (32) and also by the intermediate-late protein IVa2 (28). However, unreplicated genomes from virus allowed to superinfect cells already in the late phase of infection, and hence containing these activators, continue to display the early pattern of MLTU expression until they themselves have begun to replicate (44). This indicates a role for a cis-acting element(s) that must be activated before trans factors are able to exert their influence on the MLP.
Multiple mRNAs are produced from the MLTU primary transcript via differential usage of 3′ splice and poly(A) sites. All contain the tripartite leader (TPL) sequence (leaders 1, 2, and 3) to which 3′ acceptor sites in the main MLTU body are spliced. The TPL is important for nuclear export, translation, and stability of late mRNAs (19, 27). An optional fourth exon, called the i-leader, is inserted between leaders 2 and 3 in the majority of L1 52/55K mRNA produced at early times, whereas it is mostly excluded from this and all other MLTU mRNAs at late times (8). The inclusion or exclusion of the i-leader is regulated by E4 Orf3 and E4 Orf6, respectively (35, 36), and its presence destabilizes the mRNA (42).
The relative efficiencies of individual 3′ splice and poly(A) sites within the body of the MLTU change as infection proceeds, leading to an altered pattern of mRNA production. In L1, the distal IIIa 3′ splice site is inactive until the late phase of infection, due in part to the inhibitory action of SR proteins on a cis-acting element identified upstream of the branch site (22). This inhibition is relieved by dephosphorylation of SR proteins, dependent on the effect of E4 Orf4 on protein phosphatase 2A (23, 34). In addition, there is specific activation of the IIIa splice site in the late phase via its atypical branch site, or polypyrimidine tract, that responds to a factor present only in late infected cells (31). The balance of use of the L1 and L2 to L5 poly(A) sites changes as infection progresses, due in part to changes in the availability of the polyadenylation factor CStF coupled with differences in the intrinsic affinity of each site for this factor (29). In addition, cis-acting sequences have been described adjacent to the core elements of the L1 poly(A) site that either control site switching or destabilize nuclear RNA in which processing at the L1 site has not occurred (10, 39).
The L4 region encodes two of the three nonstructural late viral proteins, 100K and 33K, the other being L1 52/55K. Immediately following the onset of replication, L4 poly(A) site usage becomes as prominent as that of L1, significantly before L2, L3, and L5 poly(A) sites are used (24). This timing of L4 expression would fit with one or more of its products having a role in enhancing expression from the other MLTU subregions. In this paper, we provide evidence that both the 100K and 33K L4 proteins, and a cis-acting element(s) within the MLTU body, are necessary for progression from the early to the late pattern of MLTU protein expression.
MATERIALS AND METHODS
Plasmid cloning and PCR amplification.
The MLTU was cloned from the Ad5 genome in two parts, which were then combined. To obtain the right-hand part, Ad5 bp 27082 to 35938 (with a deletion in the E3 region between 28592 and 30470) was taken from pFGdXI (kindly donated by F. L. Graham) by a BamHI/SpeI digest and cloned into pBR322SpeI, a derivative of pBR322 with an SpeI linker at the PvuII site, to give pAdJY1. A PacI site was then engineered into pAdJY1 at Ad5 32884 by site-directed mutagenesis to give pAdJPacI. A full-length MLTU was cloned by constructing a plasmid containing the left end of the MLTU that overlapped with the MLTU sequence in pAdJPacI. To obtain the left-hand part, a PacI site was first engineered in pBR322 at the NdeI site (2297) to give pBRPacI. The Bst1107I fragment from Ad5 genomic DNA (5764 to 29010) was then ligated into the Bst1107I site (2245) in pBR322PacI, and the desired orientation was selected as pBRZ17I. The full-length clone, pAdMLTU, containing Ad5 sequence from 5764 to 32884 flanked by PacI sites, was obtained by transferring the right-end sequences from pAdJPacI as a SpeI/ClaI fragment into pBRZ17I. Finally, the complete MLTU under MLP control was transferred as a PacI fragment into a derivative of the Epstein-Barr virus episomal plasmid pMEP4 (Invitrogen) that contained a unique PacI site and had its hMTIIa promoter removed to give pMLPL1-5.
The full-length MLTU under the control of a tetracycline-regulated (Tet) promoter (pBiL1-5) was obtained as follows. The MLP was removed from a clone of the Ad5 fragment 6009 to 10589 in pBRPacI by inserting PacI linkers at the PvuII site within the first leader (6081), digestion with PacI, and recircularization. The Ad5 sequence in this clone was then extended by inserting a genomic XbaI fragment, 10589 to 28592, that contained a frame-shifting XhoI linker insertion (23912) in E2A. The resulting plasmid was then extended to the right end of the MLTU sequence, using the SpeI/ClaI fragment from pAdMLTU, to give pAdMLTUΔP. Finally, the promoterless MLTU was transferred as a PacI fragment into pBiEGFPPacI (formed from pBiEGFP [Clontech] with a PacI linker at the PvuII site), placing it under the bidirectional Tet promoter.
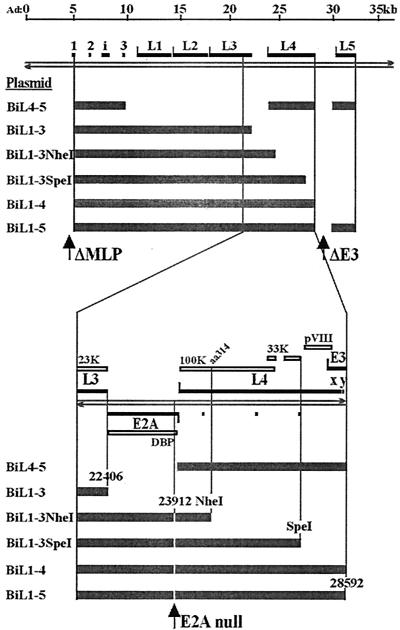
Deletion variants of pBiL1-5 were constructed as follows (their MLTU contents are summarized in Fig. 1). pBiL1-3 was cloned by ligation of the PacI-BamHI (6081 to 21562) sequence from pBiL1-5 and a BamHI (21562)-XbaI (22406; virus E2X) (5) fragment into PacI/NheI-cut pBiEGFPPacI. To obtain pBiL4-5, sequences 9461 to 27331 (encompassing the third leader to L4) were removed from pAdMLTUΔP and replaced with SalI (24056; virus E2S2) (5)-EcoRI (27331) comprising the L4 region alone. The third leader was then reinserted as a SalI fragment (9461 to 9841). Finally, the truncated MLTU was moved, as a PacI fragment, into PacI-linearized pBiEGFPPacI. pBiL1-3NheI was constructed like pBiL1-3 but using the BamHI-NheI fragment (21562 to 24999) from pAdMLTUΔP. Similarly, pBiL1-3SpeI and pBiL1-4 were cloned using the BamHI-SpeI (21562 to 27082) and BamHI-XbaI (21562 to 28592) fragments from pAdMLTUΔP, respectively.
FIG. 1.
(Top) Ad5 MLTU and cloned fragments. The Ad5 genome is represented as paired left- and right-pointing arrows with a line scale above in kilobase pairs. The solid blocks represent the principal exons of the MLTU, which spans ∼29 kbp. L1 to L5 indicate the five MLTU regions, each having its own poly(A) site. The thick shaded bars indicate the Ad5 DNA contents of plasmids used in this study, placed under the control of a TET(OFF)-regulated promoter. (Bottom) The MLTU region from the 3′ end of L3 to the 5′ end of E3 is expanded to show the positions of clone ends and the E2A null point mutation with respect to the significant sequence features of the region. Rightward transcription-translation products are shown above the genome, and leftward products are shown below. The open blocks represent protein coding regions. For further details, see the text.
Plasmids expressing specific L4 proteins carrying a C-terminal FLAG epitope tag were obtained by PCR amplification from L4 template DNA using specific primers and cloning the products, as EcoRI-PacI fragments, into pCMV-FLAG. The 33K reading frame was amplified using a 5′ primer annealed to Ad5 26194 to 26222 with a 5′ GGGGAATTCTCTA overhang and a 3′ primer annealed to Ad5 27057 to 27083 with a 5′ CCCTTAATTAA overhang. The 100K reading frame was amplified using a 5′ primer annealed to Ad5 24061 to 24087 with a 5′ GGGGAATTCTCTAG overhang and a 3′ primer annealed to Ad5 26456 to 26481 with a 5′ CCCTTAATTAA overhang. pCMVFLAG was constructed from pEGFP-N1 (Clontech) by replacing the EGFP gene as a BamHI-NotI fragment with a double-stranded oligonucleotide encoding the FLAG epitope and having suitable overhangs for ligation.
Cells, transfection, and virus infection.
293TET cells (Clontech) were maintained in Dulbecco modified Eagle medium plus 10% Tet-approved fetal calf serum (Clontech) supplemented with 100 μg of Geneticin/ml (G418 sulfate; Gibco). Tissue culture plates were pretreated with poly-d-lysine (Sigma; 1 μg/ml in phosphate-buffered saline [PBS]) at 4°C overnight. Transient-expression assays were performed by transfecting ∼9 × 106 cells with 10 μg of pBiL1-5 (a 30-kbp plasmid [see below]) and 5 μg of salmon sperm DNA by calcium phosphate coprecipitation. The amounts of other plasmid DNAs transfected were adjusted to be equimolar with this amount of pBiL1-5 and thus varied according to the plasmid size. Unrelated plasmid DNA was added to single-plasmid transfections to normalize the total plasmid DNA input for comparison with cotransfections. Transfected cell extracts were prepared directly in sodium dodecyl sulfate (SDS) gel sample buffer 48 h posttransfection except where indicated. Infections with Ad5 wt300 (21) were carried out at a multiplicity of infection of 10 PFU/cell, and infected cells were assayed for gene expression 24 h postinfection.
Western blotting.
Extract from ∼2 × 106 transfected cells or 5 × 104 infected cells was loaded onto SDS-10% polyacrylamide gels, and the proteins were separated by electrophoresis. Western blotting and detection were carried out as previously described (26) using the following antisera: anti-DBP rabbit polyclonal serum raised against the bacterially expressed E2-72K protein (E. Harfst and K.N. Leppard, unpublished data) used at 1:1,000; anti-100K mouse monoclonal antibody against the L4-100K protein (W. C. Russell, University of St. Andrews) used at 1:1,000; anti-52/55K rabbit polyclonal serum raised against the bacterially expressed L1 52/55K protein (2) used at 1:2,000; and AbJLB1 rabbit polyclonal serum against viral particle proteins used at 1:5,000. To generate AbJLB1, purified Ad5 wild-type particles in PBS were dialyzed against 8 M urea for 4 h at 4°C and then against PBS to provide denatured immunogen. A rabbit was immunized with 100 μg of protein in nonulcerative Freund's complete adjuvant and boosted with the same amount of antigen in incomplete adjuvant after 4 weeks (4). When characterized against other available sera, it had strong activity against a variety of capsid and core proteins but did not react with fiber. Presumably, the urea denaturation prevented the production of antibodies capable of recognizing fiber epitopes displayed in a Western blot assay. FLAG-tagged proteins were detected with M2, a mouse monoclonal antibody against the FLAG epitope (Sigma), used at 1:2,000. Secondary antibodies were donkey anti-rabbit immunoglobulin and goat anti-mouse immunoglobulin, both horseradish peroxidase conjugated and used at 1:2,000. Detection of secondary antibodies was done by enhanced chemiluminescence (NEN).
Detection of mRNA by reverse transcription (RT)-PCR.
Total cytoplasmic RNA was extracted from infected or transfected cells in isotonic buffer containing Nonidet P-40 and purified as previously described (25). For each sample, 200 U of Superscript Reverse II (Invitrogen) was used to generate cDNA from 4 μg of total cytoplasmic RNA in a 50-μl volume at 42°C for 50 min with 0.2 μM 3′ primer and recommended amounts of other reaction constituents. Five-microliter aliquots of serially diluted cDNA templates were removed into a PCR mixture (0.8 mM dNTP, 3 mM MgCl2, 2 U of Taq polymerase [Fermentas] under the manufacturer's buffer conditions; final volume, 50 μl) containing the appropriate primer pairs at a 0.3 μM final concentration. The primers used were complementary to Ad sequences (5′-3′ basepair positions) as follows: second leader (to detect i-leader inclusion), 7113 to 7138, or third leader, 9684 to 9710, paired with either L1 52/55K, 11945 to 11918; L1 IIIa, 12815 to 12789; L2 penton, 14889 to 14863; or L3 hexon, 19254 to 19228. The reaction mixtures were incubated at 94°C for 1 min, 65°C for 30 s, and 72°C for 1 min 40 s for 40 cycles; 50% of each sample was analyzed on 0.7% (wt/vol) agarose gels.
Nuclear RNA was prepared from transfected cells using GenElute (Sigma) and treated with DNase I. Equivalent amounts of RNA were subjected to RT-PCR as described above, except that 10-μl aliquots of diluted cDNA were used. Control reactions omitted reverse transcriptase. Primers were complementary to the Ad sequences 22034 to 22061 and 22749 to 22722, detecting RNA unprocessed at the L3 poly(A) site.
RESULTS
Analysis of late gene expression from MLTU plasmids.
As part of attempts to establish cell lines expressing Ad5 late proteins, a plasmid (pBiL1-5) was constructed containing the MLTU under the control of a tetracycline-regulated promoter in pBiEGFP (Fig. 1). A frameshift mutation was included in E2 72K DNA-binding protein (DBP), since its expression is toxic to cells and would preclude the isolation of cell lines. Also, part of the E3 region was removed, and sequences downstream of the L5 poly(A) site were excluded. Related plasmids described in the remainder of this paper were constructed to maintain these modifications where appropriate. All of these plasmids contained genomic DNA encoding the tripartite leader sequence, from the PvuII site in leader 1, so that late RNA processing occurred in the correct context. The bidirectional promoter in this plasmid allowed the coexpression of enhanced green fluorescent protein (EGFP), which enabled the assessment of transfection efficiency independent of any MLTU expression (data not shown).
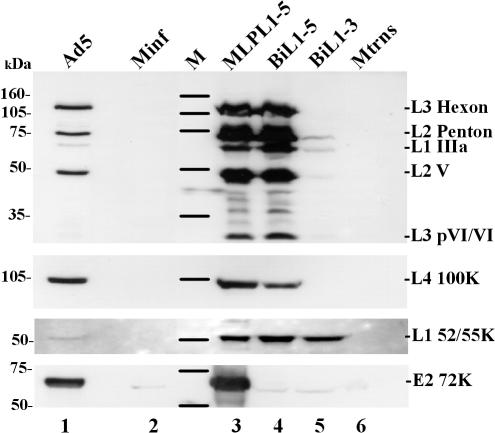
When transfected into 293TET(OFF) cells, pBiL1-5 expressed a full range of late proteins from across the MLTU (Fig. 2 lane 4) in a pattern similar to that seen in the late phase of Ad5 infection (lane 1). This pattern and, more significantly, the amounts of protein present were also similar to those seen when the same MLTU sequence (but without the mutation in DBP) was transfected under the control of the MLP (lane 3). Thus, replacement of the MLP and mutation of E2A do not affect expression from the cloned MLTU in this context. A second plasmid, pBiL1-3, which was derived from pBiL1-5 and contained regions L1 to L3 only (Fig. 1), expressed L1 52/55K at levels similar to those of pBiL1-5, whereas it expressed considerably lower levels of L1 IIIa, L2 penton, and L2 V and negligible amounts of pVI and hexon proteins from L3 (Fig. 2, lane 5). Thus, whereas pBiL1-5 expression included the whole MLTU with a balance of products similar to that from a late infection, pBiL1-3 expression appeared very reminiscent of the early expression pattern from the MLTU, with little expression of proteins other than L1 52/55K.
FIG. 2.
Late protein expression from a cloned MLTU. Proteins from 293TET(OFF) cells, transfected or infected with plasmid or virus as indicated, were separated on an SDS-10% polyacrylamide gel and transferred to nitrocellulose, and Ad5 proteins were detected by incubation with specific antisera followed by chemiluminescent detection. (Top) Polyclonal anti-denatured-particle protein serum. (Bottom) Monospecific antisera as indicated (see Materials and Methods). M, molecular mass markers (masses are indicated on the left); Minf, mock infected; Mtrns, mock transfected.
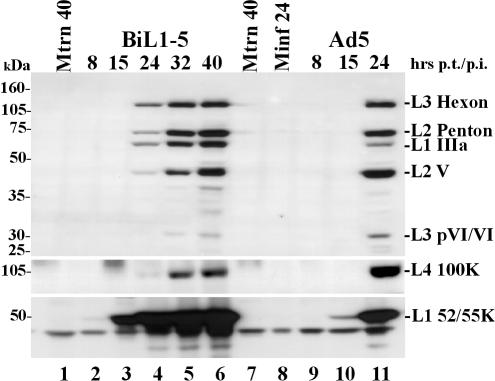
To determine if the full late protein expression profile observed for pBiL1-5 had developed via an early phase, the production of late proteins was monitored over a 40-h period posttransfection (Fig. 3). Two phases of expression were apparent: first, the expression to significant levels of L1 52/55K protein (lane 3), leading to a second phase, commencing about 10 h later, when all the late proteins were expressed (lanes 4 to 6). Expression seen from pBiL1-5 during the first phase mimicked that seen at all times posttransfection from pBiL1-3. Thus, temporal regulation of pBiL1-5 expression resembles that seen during viral infection (lanes 9 to 11), indicating that the expression from pBiL1-5 follows principles of regulation and processing similar to those of the MLTU in vivo, while pBiL1-3 appears unable to carry out the early-late switch and expression from its L1 IIIa, L2, and L3 sequences remains in the early pattern.
FIG. 3.
Time course of expression of late proteins from virus or cloned MLTU. Proteins from 293TET(OFF) cells transfected with pBiL1-5 or infected with Ad5 wt300 and harvested at the times indicated were analyzed and detected as for Fig. 2. Minf, mock infected; Mtrns, mock transfected. The positions of molecular mass markers are indicated on the left.
To test whether these protein data were mirrored at the RNA level, the levels of specific transcripts in total cytoplasmic RNA were analyzed by semiquantitative RT-PCR. Primers were designed for detection of mRNAs from L1 (52/55K and IIIa), L2 (penton), and L3 (hexon). Except for L1 52/55K, the 5′ primer was designed to anneal to the third leader sequence, and the appropriate late region-specific 3′ primers (also used for the RT step) were designed so that PCR bands could be easily distinguished by size where multiple products were predicted. The 5′ primer for the L1 52/55K PCR was designed to anneal to the second leader sequence so that the amount of i-leader inclusion in this mRNA could be assessed. If the pBiL1-3 expression pattern truly resembled that seen from the MLTU early in infection, then a higher proportion of L1 52/55K mRNA from this plasmid than from pBiL1-5 should include the i-leader at 40 h posttransfection.
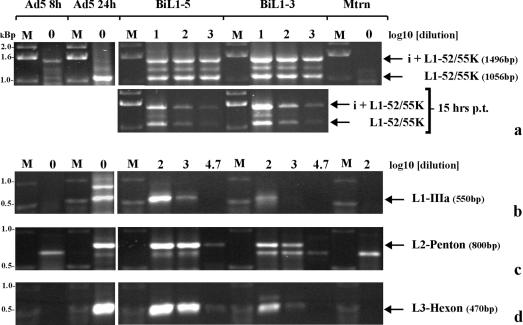
The L1 52/55K mRNA yielded RT-PCR products at ∼1.0 (i-leader excluded) and ∼1.5 (i-leader included) kbp. At both 15 and 40 h posttransfection, taking these two products together, the levels of total L1 52/55K mRNA for pBiL1-5 and pBiL1-3 were similar (Fig. 4a), in agreement with the amounts of 52/55K protein expressed by these plasmids. Both plasmids also showed the same small excess of i-leader-containing product at 15 h, and for pBiL1-3, this remained true at 40 h. However, the positions were reversed for pBiL1-5, with a modest excess of i-leader-excluded product at this time. These data fit with the idea that MLTU expression from the L1 to L5 plasmid progresses to a late pattern while that from pBiL1-3 does not. The change in balance between the two forms of L1 mRNA from pBiL1-5 was not as dramatic as was seen during the progression of an infection (Fig. 4a). This is likely to be due in part to the persistence at 40 h posttransfection of i-leader-included mRNA made earlier. The absence of any replicative increase in the template DNA copy number, a factor which amplifies the difference between the amounts of late-expressed and early-expressed L1 52/55K mRNA during infection, will make this persistence appear more significant. In addition, all of these experiments were conducted without proteins from the E4 gene; the relative levels of expression of E4 Orf3 and Orf6 have been shown to affect i-leader inclusion (35, 36).
FIG. 4.
Cytoplasmic mRNA levels expressed from full-length or truncated MLTU sequences. Total cytoplasmic RNA was prepared from 293TET(OFF) cells either 15 or 40 h posttransfection (p.t.) with pBiL1-5 or pBiL1-3 or mock transfection or 8 or 24 h postinfection with Ad5 wt300. Four micrograms of RNA was reverse transcribed under standard conditions, and then serial dilutions of this cDNA template (the dilution factor is indicated above each lane) were amplified by PCR, using primer pairs specific for L1 52/55K mRNA (a), L1 IIIa mRNA (b), L2 penton mRNA (c), or L3 hexon mRNA (d). The PCR products were separated by electrophoresis on agarose gels and detected by ethidium bromide staining. M, DNA size markers as indicated.
The level of L3 hexon mRNA in cytoplasmic RNA from pBiL1-3-transfected cells was at least 50-fold less than in RNA from pBiL1-5-transfected cells (Fig. 4d), in agreement with the very low levels of hexon protein expressed by pBiL1-3 (Fig. 2). The levels of L1 IIIa and L2 penton mRNA produced from pBiL1-3 were also greatly reduced, by 10- and 50-fold, respectively, compared to pBiL1-5 (Fig. 4b and c), again in agreement with protein levels from the two plasmids. Taking the mRNA and protein data together, it is clear that the limited pattern of expression from pBiL1-3 closely resembled that seen from the MLTU during the early phase of infection, while expression from pBiL1-5 was similar to that seen during the late phase of an infection.
L4 proteins are implicated in the early-to-late switch.
To investigate the minimum MLTU content required to observe a late-like pattern of expression, further plasmids were constructed that had extensions beyond the L3 poly(A) site into the L4 region. pBiL1-4, containing all of L4 (Fig. 1), generated a pattern of expression almost identical to that of pBiL1-5; IIIa, penton, V, and hexon proteins were expressed at high levels (Fig. 5A, lane 2). Thus, L4, but not L5, is required for the early-late switch. Since pBiL1-4 was produced by extending the sequence present in pBiL1-3, this result also shows that the L2 and L3 sequences in pBiL1-3 were capable of expressing proteins, discounting trivial explanations for the absence of expression from this plasmid.
FIG. 5.
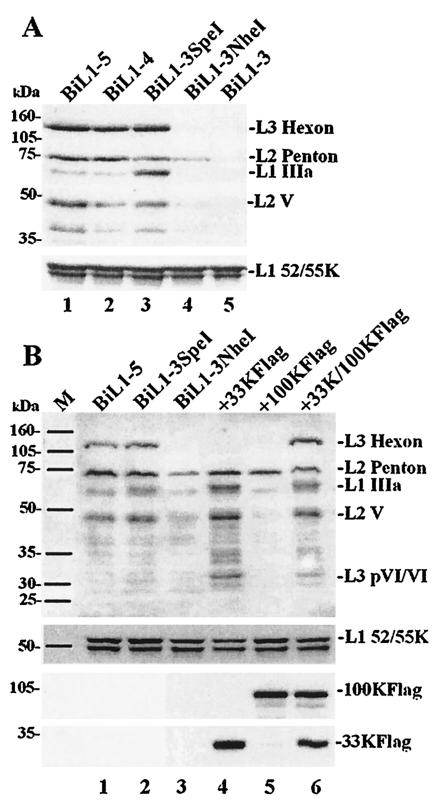
Complementation of late protein expression from a truncated MLTU by L4 proteins. Proteins from transfected 293TET(OFF) cells were analyzed and detected as for Fig. 2. (A) Single transfections with increasingly truncated MLTU plasmids; top, polyclonal anti-denatured-particle protein serum; bottom, monospecific anti-L1 52/55K serum. (B) Single transfections with MLTU plasmids as indicated (lanes 1 to 3) or cotransfections of pBiL1-3NheI with L4 expression plasmid(s) as indicated (lanes 4 to 6); top, polyclonal anti-denatured-particle protein serum; middle, monospecific anti-L1 52/55K serum; bottom, anti-FLAG epitope antibody M2. The positions of molecular mass markers are indicated on the left.
Two further plasmids that contained only part of L4 were tested. The pBiL1-3SpeI MLTU is able to encode the L4 100K and 33K proteins, but not pVIII. This truncated MLTU also excludes both the E2-E and E3 promoters, which are present in pBiL1-4. The pBiL1-3NheI MLTU extends to a site in the L4 100K reading frame; it is unable to express L4 33K or pVIII and has the potential only to produce an N-terminal fragment of 100K; the E2-L promoter is also absent from this clone. Upon transfection, pBiL1-3SpeI was able to express its MLTU in the late-like manner (Fig. 5A, lane 3), similar to the L1 to L4 and L1 to L5 plasmids. However, L1 IIIa protein levels were higher than for these longer plasmids, indicating some aberration in expression control. In contrast, MLTU expression from pBiL1-3NheI (lane 4) appeared to be very similar to the early-like expression pattern observed for pBiL1-3 (lane 5), although L2 penton levels were possibly greater. All the plasmids expressed L1 52/55K to the same level, discounting the possibility that the other differences observed were due to variable transfection efficiency. Taken together, these data indicate that L4 sequences in the region coding for 100K and 33K are required for a full late pattern of expression from cloned L1, L2, and L3 MLTU sequences.
To test directly whether the 100K and/or 33K protein was required for full expression from L1-3, expression clones for each protein were cotransfected with pBiL1-3NheI. To create these clones, genomic DNA fragments carrying each of these reading frames (for 33K this included an intron) were cloned fused to a C-terminal FLAG epitope tag under the control of the cytomegalovirus promoter. The two plasmids expressed FLAG-tagged proteins of the expected sizes in immunoblots (Fig. 5B, bottom). The expression of 100K alone with pBiL1-3NheI had little effect on MLTU expression, although L2 penton was slightly up-regulated (Fig. 5B, lane 5). In contrast, expression of 33K alone was able to drive expression of L1 to L3 proteins from pBiL1-3NheI (except hexon) into the ‘late’-like pattern, with up-regulation of IIIa, penton, V, and pVI (lane 4). Combining 33K and 100K gave MLTU expression from pBiL1-3NheI that was very similar to that observed from pBiL1-3SpeI and longer constructs (lane 6); in particular, hexon protein was now detected at high levels. Thus, 33K is a key trans-acting factor in the early-to-late switch, but 100K is also required for hexon protein expression.
L4 33K acts at the RNA level.
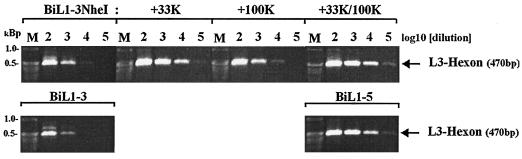
To determine whether 33K was exerting its effect at the RNA or protein level, the amounts of specific cytoplasmic mRNAs in cells transfected with pBiL1-3NheI alone or together with the L4 clones were assessed (Fig. 6). Hexon mRNA from pBiL1-3NheI in the absence of L4 proteins was at a low level compared to pBiL1-5, similar to pBiL1-3. Cotransfection with 33K alone was sufficient to increase hexon mRNA between 10- and 100-fold, while 100K alone had only a marginal effect. When 100K was supplied with 33K, there was a small additive effect, but the principal factor in the increase of hexon mRNA was L4 33K. Levels of hexon mRNA from pBiL1-3NheI in the presence of both L4 proteins were indistinguishable from those produced by the full-length clone pBiL1-5. Since addition of 100K in the presence of 33K had little effect on hexon mRNA levels but a major effect on hexon protein (Fig. 5B), the latter effect must be at the level of translation or protein stability.
FIG. 6.
Cytoplasmic L3 hexon mRNA levels expressed from truncated MLTU clone pBiL1-3NheI in the presence of L4 proteins. 293TET(OFF) cells were transfected with pBiL1-3NheI alone or with expression plasmids for one or both of the L4 100K and 33K proteins. L3 hexon mRNA was detected as in Fig. 4. Analyses of RNA from pBiL1-3 and pBiL1-5 transfections carried out in parallel are shown for comparison. M, DNA size markers as indicated.
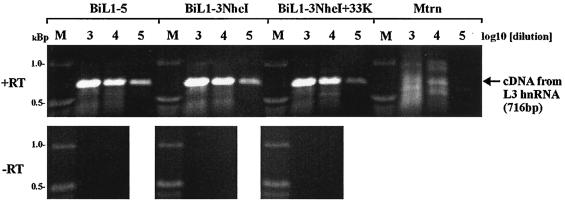
The increase in amounts of cytoplasmic L1 IIIa, L2 penton, and L3 hexon mRNAs produced by pBiL1-3 or pBiL1-3NheI in the presence of L4 33K represented a net increase in MLTU-derived cytoplasmic mRNA, since L1 52/55K mRNA was not diminished (Fig. 4 and 6 and data not shown). To determine if this increase was a reflection of increased levels of MLTU transcript in the nucleus, RT-PCR was carried out on DNase I-treated nuclear RNA using primers that detected unprocessed RNA from the 3′ end of L3. As shown in Fig. 7, nuclei from cells transfected with pBiL1-3NheI contained amounts of this unprocessed RNA similar to those from pBiL1-5, and cotransfection of pBiL1-3NheI with L4 33K had no effect on the amount of this RNA. In each case, controls omitting the reverse transcriptase step gave no product, confirming that the results were not due to plasmid DNA contaminating the samples. These data suggest that L4 33K acts posttranscriptionally to increase cytoplasmic L1 IIIa, L2, and L3 mRNAs, affecting either RNA processing or RNA export from the nucleus.
FIG. 7.
Nuclear L3 RNA levels expressed from truncated MLTU clone pBiL1-3NheI. DNase I-treated nuclear RNA harvested from 293(TET) cells 40 h posttransfection with the plasmids indicated was reverse transcribed (+RT) or mock reverse transcribed (−RT), and then serial dilutions of these cDNA templates (the dilution factor is indicated above each lane) were amplified by PCR, using primers specific for the 3′ end of L3. The 716-bp product derives from sequences unprocessed at the L3 poly(A) site. M, DNA size markers as indicated.
Activation of hexon protein expression by L4 100K requires downstream sequences.
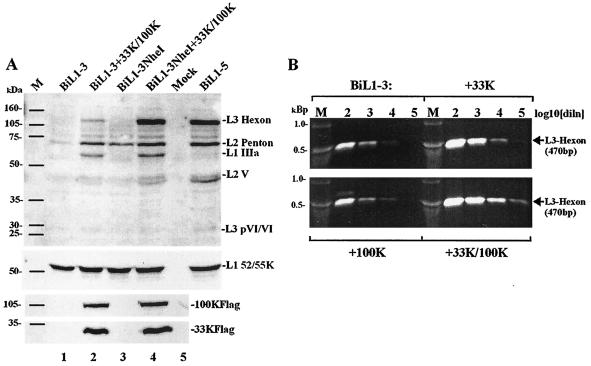
The two plasmids pBiL1-3 and pBiLi1-3NheI showed similarly restricted patterns of MLTU expression in the absence of L4 proteins (Fig. 5A). However, the two plasmids were distinguishable by their responses to L4 proteins in trans. Cotransfection of pBiL1-3 with L4 33K produced results very similar to those observed for pBiL1-3NheI, with induction of late proteins except hexon (data not shown) and induction of hexon mRNA (Fig. 8B). However a combination of 33K and 100K, which was sufficient to give high levels of hexon protein from pBiL1-3NheI (Fig. 8A, lanes 3 and 4), gave only low-level induction of hexon protein in similar cotransfections with pBiL1-3 (Fig. 8A, lanes 1 and 2), even though hexon cytoplasmic mRNA was induced to levels comparable to those seen from pBiL1-3NheI (Fig. 6 and 8B). This result indicated that sequences lying downstream of the L3 poly(A) site that were present in the L1-3NheI MLTU but not in the L1-3 MLTU were required for 100K to be able to mediate its effect on hexon protein accumulation. Since these sequences are not present in mature hexon mRNA, we considered whether their effects might be indirect, via an influence on i-leader inclusion in hexon mRNA. However, there was no difference between the amounts of hexon mRNA that included i-leader in pBiL1-3 and pBiL1-3NheI; both levels were low relative to standard hexon mRNA (data not shown).
FIG. 8.
Induction of hexon expression by 100K requires sequences downstream from L3. (A) Proteins from 293TET(OFF) cells transfected with plasmid(s) as indicated were analyzed and detected as in Fig. 2. Top, polyclonal anti-denatured-particle protein serum; middle, monospecific anti-L1 52/55K serum; bottom, anti-FLAG epitope antibody M2. M, molecular mass markers (masses are indicated on the left). (B) 293TET(OFF) cells were transfected with pBiL1-3NheI alone or with expression plasmids for L4 100K and/or 33K. L3 hexon mRNA was detected as in Fig. 4. M, DNA size markers as indicated. diln, dilution.
DISCUSSION
During the late phase of infection, the Ad5 MLTU encodes multiple protein products from mRNAs produced from each of its five segments, whereas earlier in the infection, its activity is limited to L1 52/55K expression. The data presented here show that this temporal progression to a full late pattern of MLTU expression is recapitulated by a full-length MLTU clone, driven by a heterologous promoter in a transient transfection. In contrast, plasmids truncated to lack a functional L4 region remained in the early-phase expression pattern throughout. The difference in MLTU expression observed between the L1-L5 and L1-L3 constructs initially suggested to us that transcriptional regulatory elements embedded within the distal MLTU might be required for the early-late switch in MLTU activity. For example, protein recruitment at such a site might alter the conformation of the viral chromatin to facilitate full MLTU expression. However, two of the obvious candidate sequences, the E3 and E2-E promoters, could be deleted (as in pBiL1-3SpeI) without impairing the switch. A third candidate, E2-L, lay within the region that was additionally deleted in pBiL1-3NheI. This plasmid failed to switch normally, potentially implicating E2-L or other elements in this 2.0-kb region. However, the fact that pBiL1-3NheI could be induced to switch by provision of L4 proteins in trans showed conclusively that no elements in the L4 region between the NheI and SpeI sites were needed in cis.
The L4 33K protein increased levels of specific late cytoplasmic mRNAs by up to 50-fold, whereas the 100K protein alone had little effect on MLTU expression but was specifically required, with 33K, for hexon protein production. This effect of L4 100K in trans required the presence in cis of sequences within a 2.5-kb region downstream of the L3 poly(A) site. In preliminary experiments, complementation of expression from pBiL1-3 by proteins from an L4 to L5 plasmid, pBiL4-5 (Fig. 1), was also tested. Induced L2-L3 protein expression was weaker than that achieved by the 33K and 100K expression constructs, but there was also a reduction in L1 52/55K expression (data not shown). Specific down-regulation of L1 52/55K expression by a product of L4-L5 could be discounted because there was no such down-regulation by the L4 expression constructs and no difference in L1 52/55K expression between L1 to L4 and L1 to L5 plasmids (Fig. 5A). Instead, we believe this result was due to promoter competition between pBiL1-3 and pBiL4-5 for a limiting protein factor, possibly the TET transactivator protein itself.
There have been only a few previous studies of L4 33K. The protein was described in early work as a major phosphoprotein in the infected cell that localized to the nucleus (3, 15, 37, 41). More recently, a viable mutant strain of Ad5, v33K.1, was constructed that had a premature stop codon created at position 20 of the 33K reading frame (13). This virus showed reduced production of progeny in a growth curve leading ultimately to a fivefold reduction in final yield. Both DNA replication and late protein production were normal, but assembly of particles was impaired. A second group attempted to isolate virus carrying a mutation that would remove 47 residues from the C terminus of 33K but were unable to recover virus from transfections of full-length cloned mutant genomes (14). Viral DNA replication could be detected, but onset posttransfection was delayed compared to the wild type. Expression of hexon and 100K proteins was detected by immunofluorescence, but no empty or full virions could be purified from the cells.
The difference in severity of phenotype between the 33K mutations studied by Finnen and by Fessler may be because the v33K.1 mutation is leaky, perhaps due to internal initiation of translation downstream from the premature stop codon. Alternatively, Finnen et al. suggested that their truncation may have created a dominant-negative form of the protein (14). However, both studies point to a role for 33K in the assembly of particles, whereas data presented here show that 33K is required to turn on full late gene expression. There are two potential explanations for this difference. First, 33K may be a multifunctional protein, with roles both as a regulator of MLTU expression and in assembly. Of the three studies, only the present one unambiguously examined the effect of a total lack of 33K; the other two retained substantial parts of the 33K reading frame. According to this reasoning, the central part of 33K, deleted here but retained and potentially expressed in the studies of Fessler and Young and of Finnen et al. (13, 14), would be necessary and sufficient for regulating the MLTU switch. Alternatively, the primary function of 33K may be in activating late gene expression, leading secondarily to defective assembly. Although in the study of v33K.1, late protein synthesis as assessed by pulse radiolabeling was judged to be normal (13), minor or unapparent differences from the wild type may have been sufficient to lead to reduced steady-state levels of late proteins and hence to reduced particle yield. In the study of the lethal C-terminal truncation of 33K (14), although both 100K and hexon were detected by fluorescence, it is hard to make a quantitative assessment of MLTU expression from these data, which may have been impaired. Finally, it may be that, in the context of virus infection, there are redundant mechanisms for activating late gene expression so that lack of the 33K activity is compensated for by other factors.
The mechanism by which L4 33K upregulates MLTU cytoplasmic mRNA levels is unclear. The fact that similar amounts of unprocessed nuclear RNA from the 3′ end of L3 were detected in the presence and absence of 33K suggests that it does not act on transcription initiation or elongation. An effect of 33K on initiation might also be considered unlikely because the MLTU plasmids employed all used a heterologous promoter, although the downstream elements important for MLP regulation were retained (20). Instead, it may be inferred that L4 33K either increased RNA processing at distal sites within the MLTU transcript or increased the export of these distal mRNAs from the nucleus. Further experiments are required to distinguish between these possibilities. Although no cis-acting element that responds to 33K has been defined in this study, one or more such elements must exist, either binding 33K or binding a factor whose activity is regulated by 33K. Since the action of 33K appears to be posttranscriptional, these elements are most likely to be present in the RNA transcript and to function as RNA.
There are more data available on the L4 100K protein, which is clearly multifunctional. This protein is initially cytoplasmic but accumulates in the nucleus as infection proceeds (15). First, 100K chaperones the assembly of hexon polypeptide into trimers and their nuclear accumulation (7). 100K binds specifically to nascent or newly synthesized hexon polypeptide (16), and cells infected by conditional 100K mutants show cytoplasmic hexon monomers but no nuclear hexon trimers by immunofluorescence with conformation-specific antibodies (6, 38). Second, 100K has a role in particle assembly (30). Third, 100K is tightly bound with messenger ribonucleoprotein (mRNP) complexes from infected cells (43) and binds to RNA directly (1), though without apparent sequence specificity (40). Finally, 100K regulates the production of late proteins through an effect on translation initiation. A 10-fold reduction in late-protein synthesis was seen during infection by the 100K mutant ts1 (17). It is now clear that this was due to a loss of inhibition of host mRNA translation (18). 100K, but not ts1 100K, displaces a kinase from the cap initiation complex, resulting in reduced phosphorylation of an initiation factor and hence reduced stability of the translation initiation complex (9). This activity favors late viral mRNA translation because sequences in the TPL permit translation of MLTU mRNAs by an alternative mechanism, now called ribosome shunting, when normal initiation is impaired (12).
We considered whether the observed effect of 100K on hexon protein synthesis was related to its established function in chaperoning hexon trimer assembly. Three lines of argument suggest that these functions are distinct. First, the loss of hexon expression in the absence of 100K was absolute, whereas considerable amounts of hexon polypeptide were observed in cells blocked in trimerization due to a 100K temperature-sensitive lesion (6). Second, the ability of 100K to rescue hexon polypeptide expression in our experiments depended on the presence in cis of sequences lying outside the hexon coding region and so could not be due to direct interaction between the proteins. Finally, the hexon protein detected here by Western blotting was, by definition, denatured and monomeric; hexon proteins produced in the presence and absence of 100K should therefore be detected equally. Although some renaturation may have occurred after transfer, it is implausible that the protein could be “remembering” its 100K-chaperoning experience in the cell so as to be detectable on the filter.
The cloned MLTU in pBiL1-5 and related plasmids lacked the 5′ 33 nucleotides of the tripartite leader. The unstructured conformation of this region is crucial to effective translation in the face of eIF4E inactivation by 100K (11). However, since translation from pBiL1-5 was qualitatively and quantitatively similar to that from a clone with an intact leader 1, the heterologous sequences forming the 5′ end of our cloned MLTU must be able to function in the same way. The same substitution of sequence in leader 1 of the TPL also removed one of the three regions of complementarity to 18S rRNA. Although these are important for efficient ribosome shunting, the first is completely dispensable while the others remain intact (46). Thus, it can be concluded that the cis-acting elements known to be required for translation by a ribosome-shunting mechanism are present in pBiL1-5 and its deletion variants. In transfections where 100K is not expressed, translation will proceed by a combination of scanning and shunting mechanisms, whereas when 100K is present, only the shunting mechanism will operate but competition from cell mRNA is reduced (45). The overall translation efficiencies in these two situations are expected to be similar. It seems very unlikely that inhibition of normal translation initiation by 100K could account for its specific upregulation of hexon synthesis. First, a lack of functional 100K has been shown to affect all late protein expression similarly (17), and second, the action of 100K on the efficiency of late protein translation is secondary to its effect on host translation; such a mechanism cannot generate an effect that is specific to just one out of the family of TPL-containing MLTU mRNAs.
The sequences that are required in cis for 100K to upregulate hexon translation lie outside the region contained in fully processed hexon mRNA. Therefore, we propose that these sequences must act prior to or during RNA processing to alter the structure or composition of the mRNP particle that is the actual substrate for translation. Either the secondary structure of the mRNA might be altered by the different availabilities of base-pairing interactions during maturation or the constellation of proteins loaded onto the RNA might be altered. Further experiments will be needed to resolve this question.
No discernible difference in expression was observed between the nonreplicating pBiL1-5 plasmid and the potentially replicating Epstein-Barr virus replicon plasmid pMLPL1-5, and both resembled a late viral infection. This is in contrast to an infected cell, where the early-late switch depends in some way on viral-genome replication (44). The absence of this requirement in our system may reflect the fact that DNA is delivered naked into the nucleus or that transfection causes it to localize in the nucleus differently from infecting genomes. In the context of infection, the trans-acting requirement for L4 proteins in the early-late switch detailed here would be expected to occur after whatever cis-acting effect is imparted by replication.
In conclusion, our data show a previously unrecognized effect of the L4 33K protein on the production of cytoplasmic mRNA from the Ad5 MLTU. The characteristics of this effect suggest that it is a key component of the early-to-late switch in Ad5 late gene expression. We propose that small amounts of L4 expression precipitate this switch in a feed-forward activation process. The timing of L4 expression during infection, early in the onset of full MLTU activity, supports this idea (24). Further work is now required to determine the detailed molecular mechanism involved.
Acknowledgments
D.C.F. and J.L.B. were supported by studentships from the Biotechnology and Biological Sciences Research Council, Swindon, United Kingdom.
REFERENCES
- 1.Adam, S. A., and G. Dreyfuss. 1987. Adenovirus proteins associated with mRNA and hnRNA in infected Hela cells. J. Virol. 61:3276-3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arslanoglu, A. 1999. Studies of the adenovirus L1 gene aimed at L1 gene deficiencies for use in gene therapy vectors. Ph.D. thesis. University of Warwick, Coventry, United Kingdom.
- 3.Axelrod, N. 1978. Phosphoproteins of adenovirus 2. Virology 87:366-383. [DOI] [PubMed] [Google Scholar]
- 4.Brown, J. L. 2001. Expression systems for adenovirus proteins. Ph.D. thesis. University of Warwick, Coventry, United Kingdom.
- 5.Caravokyri, C., and K. N. Leppard. 1996. Human adenovirus type-5 variants with sequence alterations flanking the E2a gene—effects on E2 expression and DNA replication. Virus Genes 12:65-75. [DOI] [PubMed] [Google Scholar]
- 6.Cepko, C. L., and P. A. Sharp. 1983. Analysis of Ad5 hexon and 100K ts mutants using conformation-specific monoclonal antibodies. Virology 129:137-154. [DOI] [PubMed] [Google Scholar]
- 7.Cepko, C. L., and P. A. Sharp. 1982. Assembly of adenovirus major capsid protein is mediated by a nonvirion protein. Cell 31:407-415. [DOI] [PubMed] [Google Scholar]
- 8.Chow, L. T., T. R. Broker, and J. B. Lewis. 1978. Complex splicing patterns of RNAs from the early regions of adenovirus-2. J. Mol. Biol. 134:265-303. [DOI] [PubMed] [Google Scholar]
- 9.Cuesta, R., Q. Xi, and R. J. Schneider. 2000. Adenovirus-specific translation by displacement of kinase Mnk1 from cap-initiation complex eIF4F. EMBO J. 19:3465-3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeZazzo, J. D., E. Falck-Pedersen, and M. J. Imperiale. 1991. Sequences regulating temporal poly(A) site switching in the adenovirus major late transcription unit. Mol. Cell. Biol. 11:5977-5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dolph, P. J., J. Huang, and R. J. Schneider. 1990. Translation by the adenovirus tripartite leader: elements which determine independence from cap-binding protein complex. J. Virol. 64:2669-2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dolph, P. J., V. Racaniello, A. Villamarin, F. Palladino, and R. J. Schneider. 1988. The adenovirus tripartite leader may eliminate the requirement for cap-binding protein complex during translation initiation. J. Virol. 62:2059-2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fessler, S. P., and C. S. H. Young. 1999. The role of the L4 33K gene in adenovirus infection. Virology 263:507-516. [DOI] [PubMed] [Google Scholar]
- 14.Finnen, R. L., J. F. Biddle, and J. Flint. 2001. Truncation of the human adenovirus type 5 L4 33 kDa protein: evidence for an essential role of the carboxy terminus in the viral replication cycle. Virology 289:388-399. [DOI] [PubMed] [Google Scholar]
- 15.Gambke, C., and W. Deppert. 1981. Late nonstructural 100,000- and 33,000-dalton proteins of adenovirus type 2. I. Subcellular localization during the course of infection. J. Virol. 40:585-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gambke, C., and W. Deppert. 1983. Specific complex of the late nonstructural 100,000-dalton protein with newly synthesized hexon in adenovirus type 2-infected cells. Virology 124:1-12. [DOI] [PubMed] [Google Scholar]
- 17.Hayes, B. W., G. C. Telling, M. M. Myat, J. F. Williams, and S. J. Flint. 1990. The adenovirus L4 100-kilodalton protein is necessary for efficient translation of viral late mRNA species. J. Virol. 64:2732-2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang, J., and R. J. Schneider. 1991. Adenovirus inhibition of cellular protein synthesis involves inactivation of cap-binding protein. Cell 65:271-280. [DOI] [PubMed] [Google Scholar]
- 19.Huang, W., and S. J. Flint. 1998. The tripartite leader sequence of subgroup C adenovirus major late mRNAs can increase the efficiency of mRNA export. J. Virol. 72:225-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jansen-Durr, P., H. Boeuf, and C. Kedinger. 1988. Replication induced stimulation of the major late promoter of adenovirus is correlated to the binding of a factor to sequences in the first intron. Nucleic Acids Res. 16:3771-3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones, N. C., and T. Shenk. 1978. Isolation of deletion and substitution mutants of adenovirus type 5. Cell 13:181-186. [DOI] [PubMed] [Google Scholar]
- 22.Kanopka, A., O. Muhlemann, and G. Akusjarvi. 1996. Inhibition by SR proteins of splicing of a regulated adenovirus pre-mRNA. Nature 381:535-538. [DOI] [PubMed] [Google Scholar]
- 23.Kanopka, A., O. Muhlemann, S. Petersen-Mahrt, C. Estmer, C. Ohrmalm, and G. Akusjarvi. 1998. Regulation of adenovirus alternative RNA splicing by dephosphorylation of SR proteins. Nature 393:185-187. [DOI] [PubMed] [Google Scholar]
- 24.Larsson, S., C. Svensson, and G. Akusjarvi. 1992. Control of adenovirus major late gene-expression at multiple levels. J. Mol. Biol. 225:287-298. [DOI] [PubMed] [Google Scholar]
- 25.Leppard, K. N., and T. Shenk. 1989. The adenovirus E1b-55 kD protein influences mRNA transport via an intranuclear effect on RNA metabolism. EMBO J. 8:2329-2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lethbridge, K. J., G. E. Scott, and K. N. Leppard. 2003. Nuclear matrix localization and SUMO-1 modification of adenovirus type 5 E1b 55K protein are controlled by E4 Orf6 protein. J. Gen. Virol. 84:259-268. [DOI] [PubMed] [Google Scholar]
- 27.Logan, J., and T. Shenk. 1984. Adenovirus tripartite leader sequence enhances translation of mRNAs late after infection. Proc. Natl. Acad. Sci. USA 81:3655-3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lutz, P., and C. Kedinger. 1996. Properties of the adenovirus IVa2 gene product, an effector of late phase-dependent activation of the major late promoter. J. Virol. 70:1396-1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mann, K. P., E. A. Weiss, and J. R. Nevins. 1993. Alternative poly(A) site utilization during adenovirus infection coincides with a decrease in the activity of a poly(A) site processing factor. Mol. Cell. Biol. 13:2411-2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morin, N., and P. Boulanger. 1986. Hexon trimerization occurring in an assembly-defective, 100K temperature-sensitive mutant of adenovirus 5. Virology 152:11-31. [DOI] [PubMed] [Google Scholar]
- 31.Muhlemann, O., B.-G. Yue, S. Petersen-Mahrt, and G. Akusjarvi. 2000. A novel type of splicing enhancer regulating adenovirus pre-mRNA splicing. Mol. Cell. Biol. 20:2317-2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nevins, J. R. 1981. Mechanism of activation of early viral transcription by the adenovirus E1a gene-product. Cell 26:213-220. [DOI] [PubMed] [Google Scholar]
- 33.Nevins, J. R., and M. C. Wilson. 1981. Regulation of adenovirus-2 gene-expression at the level of transcriptional termination and RNA processing. Nature 290:113-118. [DOI] [PubMed] [Google Scholar]
- 34.Nilsson, C. E., S. Petersen-Mahrt, C. Durot, R. Shtrichman, A. R. Krainer, T. Kleinberger, and G. Akusjarvi. 2001. The adenovirus E4-ORF4 splicing enhancer protein interacts with a subset of phosphorylated SR proteins. EMBO J. 20:864-871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nordqvist, K., K. Ohman, and G. Akusjarvi. 1994. Human adenovirus encodes 2 proteins which have opposite effects on accumulation of alternatively spliced mRNAs. Mol. Cell. Biol. 14:437-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ohman, K., K. Nordqvist, and G. Akusjarvi. 1993. Two adenovirus proteins with redundant activities in virus growth facilitate tripartite leader mRNA accumulation. Virology 194:50-58. [DOI] [PubMed] [Google Scholar]
- 37.Oosterom-Dragon, E. A., and C. W. Anderson. 1983. Polypeptide structure and encoding location of the adenovirus serotype-2 late, non-structural 33K protein. J. Virol. 45:251-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oosterom-Dragon, E. A., and H. S. Ginsberg. 1981. Characterization of two temperature-sensitive mutants of type-5 adenovirus with mutations in the 100,000-dalton protein gene. J. Virol. 40:491-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prescott, J. C., L. Liu, and E. Falck-Pedersen. 1997. Sequence-mediated regulation of adenovirus gene expression by repression of mRNA accumulation. Mol. Cell. Biol. 17:2207-2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Riley, D., and S. J. Flint. 1993. RNA-binding properties of a translational activator, the adenovirus L4 100-kilodalton protein. J. Virol. 67:3586-3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Russell, W. C., and G. E. Blair. 1977. Polypeptide phosphorylation in adenovirus-infected cells. J. Gen. Virol. 34:19-35. [DOI] [PubMed] [Google Scholar]
- 42.Soloway, P. D., and T. Shenk. 1990. The adenovirus type 5 i-leader open reading frame functions in cis to reduce the half-life of L1 mRNAs. J. Virol. 64:551-558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tasseron-DeJong, J. G., J. Brouwer, K. Rietveld, C. E. M. Zoetemelk, and L. Bosch. 1979. Messenger ribonucleoprotein complexes in human KB cells infected with adenovirus type 5 contain tightly bound viral-coded ‘100K’ proteins. Eur. J. Biochem. 100:271-283. [DOI] [PubMed] [Google Scholar]
- 44.Thomas, G. P., and M. B. Mathews. 1980. DNA replication and the early to late transition in adenovirus infection. Cell 22:523-533. [DOI] [PubMed] [Google Scholar]
- 45.Yueh, A., and R. J. Schneider. 1996. Selective translation initiation by ribosome jumping in adenovirus-infected and heat-shocked cells. Genes Dev. 10:1557-1567. [DOI] [PubMed] [Google Scholar]
- 46.Yueh, A., and R. J. Schneider. 2000. Translation by ribosome shunting on adenovirus and hsp70 mRNAs facilitated by complementarity to 18S rRNA. Genes Dev. 14:414-421. [PMC free article] [PubMed] [Google Scholar]