Abstract
Malignant peripheral nerve sheath tumors (MPNSTs) are sarcomas of Schwann cell-lineage origin that occur sporadically or in association with the inherited syndrome, Neurofibromatosis Type 1. To identify genetic drivers of MPNST development, we utilized the Sleeping Beauty (SB) transposon-based somatic mutagenesis system in mice with somatic loss of tumor protein p53 (Trp53) function and/or overexpression of epidermal growth factor receptor (EGFR). Common insertion site (CIS) analysis of 269 neurofibromas and 106 MPNSTs identified 695 and 87 sites with a statistically significant number of recurrent transposon insertions, respectively. Comparison to human data sets revealed novel and known driver genes for MPNST formation at these sites. Pairwise co-occurrence analysis of CIS-associated genes identified many cooperating mutations that are enriched for in Wnt/CTNNB1, PI3K/Akt/mTor, and growth factor receptor signaling pathways. Lastly, we identified several novel proto-oncogenes including forkhead box R2 (Foxr2), which we functionally validated as a proto-oncogene involved in MPNST maintenance.
Malignant peripheral nerve sheath tumors (MPNSTs) are aggressive, metastatic, nerve-associated tumors, which occur sporadically (~50% cases) or in association with the inherited syndrome Neurofibromatosis Type 1 (NF1)1. The lifetime risk of developing a sporadic MPNST is 0.001%, compared to 5–13% for NF1 patients (1:3,000 people) where MPNSTs often arise from pre-existing plexiform neurofibromas1,2. Despite advances in understanding MPNST biology, the primary treatment of MPNSTs, regardless of origin, is tumor resection followed by radiation and non-specific chemotherapeutic agents resulting in 5-year survival rates of less than 25% in patients with metastatic disease3–6. The most commonly altered gene known to cause benign neurofibroma formation and further progression into MPNST is NF1, which encodes Neurofibromin 1, a RAS-GTPase activating protein that causes NF1 syndrome when mutated7–10. NF1 mutation causes increased and aberrant signaling through pro-growth and pro-proliferation signaling pathways (RAS/MAPK/ERK and PI3K/AKT/mTOR) in human neurofibromas and MPNST-derived cell lines11–13. Overexpression of growth factor receptors and ligands like EGFR, NRG, PDGF, HGF, SCF, and TGFβ1 are also observed in neurofibromas and MPNSTs with NF1 mutation14–19. Besides NF1 mutations, genomic aberrations have not been identified in neurofibromas. However, genomic aberrations including deletions and/or mutations of cell cycle regulators (TP53, RB1, and CDKN2A), gene amplification of growth factor receptors (ERBB2, EGFR, KIT, MET, and PDGFR), and the presence of hyperdiploid or near-triploid genomes commonly occur in human MPNSTs14–33. These observations suggest that progression to malignancy requires many cooperating genomic alterations. High levels of genomic complexity make the identification of human MPNST genetic drivers difficult and leave the following questions unanswered: 1) What gene(s) cooperate with NF1 loss for MPNST formation? 2) What additional gene modules cooperate for MPNST formation? 3) What are the epigenetically altered drivers of MPNST formation? 4) What is the signature/identity of MPNST maintenance genes? Though some genes and genetic pathways are implicated in MPNST development, there are still many left to uncover to create effective therapies for human MPNST treatment.
Recently, the Sleeping Beauty (SB) transposon system was utilized, in a tissue-specific manner in mice, to identify genetic drivers of numerous solid and blood cancers34–37. To identify MPNST drivers, we targeted SB transposon mutagenesis to Schwann cells and their precursors using the Cnp-Cre transgene and a conditional SB mutagenesis system35,38. Since mutations/deletions in TP53 and/or amplification or elevated expression of EGFR are commonly associated with human MPNSTs (24–75% and 25–70%, respectively), a conditional dominant-negative Trp53R270H allele and a Cnp-EGFR transgene were included23,39–42. Analysis of 375 SB-accelerated PNSTs uncovered 745 common-insertion site (CIS) associated genes (known and novel). We also identified genes and signaling pathways that cooperate for MPNST formation. We identified several novel proto-oncogenes, including Foxr2 and described new functions for FOXR2 in human MPNST formation and maintenance. Thus, utilizing the SB mutagenesis system, we identified genes and genetic pathways that may provide new therapeutic targets for MPNST treatment.
RESULTS
SB mutagenesis accelerated neurofibroma development and progression to MPNST
Four experimental mouse cohorts underwent SB mutagenesis on wildtype or tumor pre-disposing backgrounds following Cnp-Cre induction in Schwann cells and their precursors (Supplementary Fig. 1a)35,38,41,42. Mice lacking full components for SB mutagenesis served as controls. Predominantly, the Cnp-EGFR (hereafter called EGFR-overexpressing) and Cnp-Cre; Cnp-EGFR; Trp53R270H (hereafter called EGFR-overexpressing and p53-mutant) mice with or without SB mutagenesis developed nerve-associated tumors throughout the body (Supplementary Table 1, Supplementary Fig. 1b). Nerve-associated tumors possessed histological features of Schwann cell tumor stages: Schwann cell hyperplasia, benign grade 1 PNSTs (neurofibromas), and aggressive grade 3 PNSTs (MPNST in humans) (Supplmentary Fig. 1c–d). Mouse grade 3 PNSTs developed in anatomical regions observed in human MPNSTs (Supplementary Fig. 2a–b)6,43,44. Moreover, some control and SB-derived grade 3 PNSTs contained regions of divergent cellular histological subtypes and differentiation events characteristic of some human MPNSTs (Supplementary Fig. 3a–b)45–47. Neurofibromas displayed no divergent differentiation. Collectively, our mouse model data indicate that the common genetic changes in human MPNST produce a phenotype in mice that resembles the human disease.
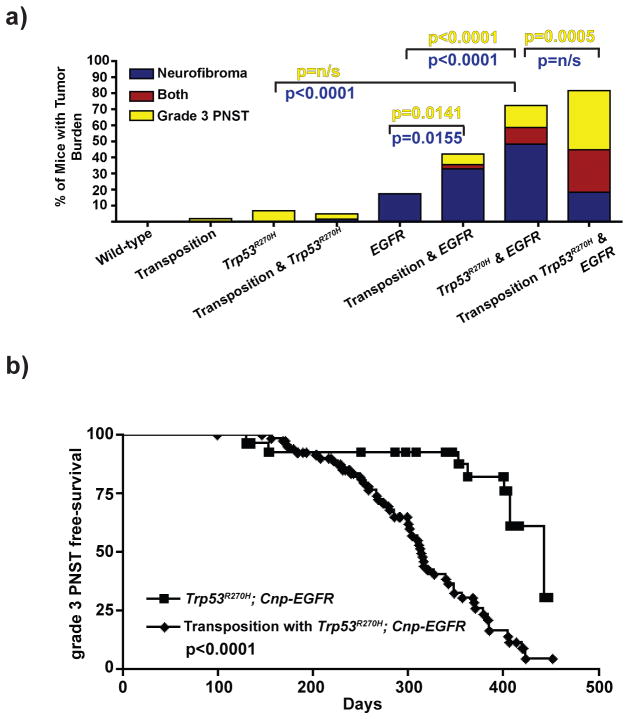
To determine if SB mutagenesis accelerated tumor formation, SB expression and activity in neurofibromas and grade 3 PNSTs were first confirmed by immunohistochemistry and by PCR-excision assay (Supplementary Fig. 1e)48. Wildtype or p53-mutant mice undergoing transposition developed few Schwann cell tumors (~2–7%) (Figure 1a). As previously reported, EGFR-overexpressing mice developed nerve hyperplasia with low incidence of neurofibroma formation (~17%) and no grade 3 PNST formation (Figure 1a)41. In contrast, SB mutagenesis significantly enhanced neurofibroma formation (~35%, p=0.0155) and induced grade 3 PNST formation in EGFR-overexpressing mice (p=0.0141). EGFR-overexpresing and p53-mutant transgenes cooperated to significantly increase neurofibroma and grade 3 PNST formation (p<0.0001, Figure 1a). SB mutagenesis on both pre-disposed alleles led to neurofibroma development, significantly increased grade 3 PNST development (p=0.0005), and significantly reduced grade 3 PNST free-survival compared to both pre-disposed alleles alone (p<0.0001, Figure 1b). Tumor penetrance for each genotype is summarized in Supplementary Table 1. Overall, SB mutagenesis enhanced grade 3 PNST incidence compared to controls.
Figure 1. SB mutagenesis induced and accelerated grade 3 PNST formation.
(a) Bar graph depicting the percentage of mice that developed each tumor type at the time of necropsy based on genotype. p-values reflect FET. (b) Survival curve depicting incidence of grade 3 tumor formation in Trp53R270H; Cnp-EGFR control mice compared to mice undergoing transposition with Trp53R270H; Cnp-EGFR background. Median age of tumor-free survival was 313 days (n=87) with transposition compared to 443 days (n=29) in the control. Log rank test: p<0.0001.
Identification of Schwann cell tumor driver-mutation genes
T2/Onc insertion sites from 269 SB-derived neurofibromas and 106 SB-derived grade 3 PNSTs from 100 mice were analyzed to identify driver-mutations (CISs). We utilized two statistical methods to identify CISs: TAPDANCE CIS (td-CIS) and gene centric CIS (gCIS) analyses (See methods for details, Supplementary Data 1–2, Table 1, Supplementary Table 2)49,50. td-CIS analysis identified 511 neurofibroma and 34 grade 3 PNST driver-mutations with 21 genes overlapping. gCIS analysis identified 535 neurofibroma and 75 grade 3 PNST driver-mutations with 32 genes overlapping. Accounting for coinciding CISs from each analysis, there were 87 grade 3 PNST (25.3% overlap) and 695 neurofibroma driver-mutations (50.1% overlap) with 37 genes common to both tumor types.
Table 1. List of grade 3 PNST CISs.
This table is a combined list of CISs identified by the tdCIS method and the gCIS method. Twenty-two CIS were identified by both statistical analyses independently. p-values and q-values were generated as described by Sarver et al. 2012 and Brett et al. 2011. Prediction on gene function indicates if the T2/Onc insertion would cause transcriptional activation of a gene (“Drive” and “Drive N-terminal Truncation) or disrupt gene transcription (“Disrupt”) based on position and orientation of T2/Onc insertion to gene transcription.
| CIS-Associated Gene | % of Tumors with tdCIS | tdCIS P-Value library # | p-value for grade 3 PNST enrichment | % of Tumors with gCIS | gCIS q-Value library # | Prediction on gene function | Neurofibroma CIS | Human Homolog |
|---|---|---|---|---|---|---|---|---|
| Nf1 | 34.9 | 5.03 x 10−25 | 8.04 x 10−06 | 36.8 | 2.61 x 10−62 | Disrupt | X | NF1 |
| Pten | 30.2 | 3.21 x 10−41 | 3.85 x 10−06 | 31.1 | 0 | Disrupt | X | PTEN |
| Stag2 | 19.8 | 6.72 x 10−12 | - | 19.8 | 7.33 x 10−26 | Disrupt | X | STAG2 |
| Taok1 | 19.8 | 1.32 x 10−18 | 0.003062 | 19.8 | 6.41 x 10−49 | Disrupt | X | TAOK1 |
| Srgap2 | 16.0 | 2.90 x 10−05 | 0.001182 | 17.9 | 3.40 x 10−11 | Disrupt | X | SRGAP2 |
| Faf1 | 14.2 | 0.047642812 | 0.03222 | 17.9 | 3.05 x 10−06 | Disrupt | X | FAF1 |
| Crebbp | 16.0 | 1.20 x 10−07 | 0.002837 | 16.0 | 3.49 x 10−21 | Disrupt | X | CREBBP |
| Dyrk1a | 11.3 | 1.44 x 10−06 | 0.009831 | 11.3 | 1.48 x 10−09 | Disrupt | X | DYRK1A |
| Spag9 | 11.3 | 0.046952457 | - | 11.3 | 1.22 x 10−07 | Disrupt | X | SPAG9 |
| Ppp6r3 | 10.4 | 0.047173607 | - | 10.4 | 7.46 x 10−09 | Disrupt | X | PPP6R3 |
| Bmpr2 | 9.4 | 0.005118412 | - | 10.4 | 4.70 x 10−07 | Drive N-term truncation | - | BMPR2 |
| Cdk13 | 9.4 | 0.005118412 | 0.01161 | 10.4 | 2.57 x 10−08 | Disrupt | X | CDK13 |
| Ccny | 4.7 | 0.042667588 | - | 10.4 | 4.25 x 10−06 | Drive N-term truncation | X | CCNY |
| Copg2 | 8.5 | 0.048819435 | - | 8.5 | 7.08 x 10−05 | Disrupt | X | COPG2 |
| Eras | 8.5 | 1.46 x 10−08 | - | 8.5 | 1.20 x 10−74 | Drive N-term truncation | - | ERAS |
| gm9766 | 5.7 | 0.043383381 | - | 8.5 | 0.000234 | Disrupt | X | C6orf204 |
| Jak2 | 5.7 | 0.00127532 | - | 8.5 | 9.07 x 10−09 | Drive N-term truncation | X | JAK2 |
| Sec63 | 7.5 | 0.047985716 | - | 7.5 | 1.28 x 10−05 | Disrupt | - | SEC63 |
| Foxr2 | 6.6 | 3.27 x 10−05 | - | 7.5 | 7.96 x 10−22 | Drive | - | FOXR2 |
| Cnot1 | 5.7 | 0.043383381 | - | 7.5 | 7.14 x 10−05 | Disrupt | - | CNOT1 |
| 2610044O15Rik | 4.7 | 0.042667588 | - | 5.7 | 5.60 x 10−11 | Drive N-term truncation | - | - |
| Baz1b | 4.7 | 0.042667588 | 0.0309 | 5.7 | 0.000157 | Drive N-term truncation | - | BAZ1B |
| Cdc27 | 15.1 | 0.001665472 | - | - | - | Disrupt | - | CDC27 |
| Pcdh10 | 11.3 | 0.046952457 | - | - | - | Disrupt | - | PCDH10 |
| Top2b | 11.3 | 0.046952457 | - | - | - | Disrupt | X | TOP2B |
| Plaa | 8.5 | 0.048819435 | - | - | - | Disrupt | - | PLAA |
| Zfp521 | 8.5 | 0.048819435 | - | - | - | Drive N-term truncation | - | ZNF521 |
| Gdi2 | 7.5 | 0.047985716 | - | - | - | Disrupt | X | GDI2 |
| Gmds | 7.5 | 0.047985716 | - | - | - | Disrupt | - | GMDS |
| Gsk3b | 7.5 | 0.047985716 | - | - | - | Disrupt | X | GSK3B |
| Picalm | 6.6 | 0.049919619 | - | - | - | Disrupt | X | PICALM |
| Gria3 | 5.7 | 0.00127532 | - | - | - | Disrupt | - | GRIA3 |
| Jmy | 5.7 | 0.043383381 | - | - | - | Drive N-term truncation | - | JMY |
| Nf2 | 5.7 | 0.00127532 | - | - | - | Disrupt | X | NF2 |
| Dip2c | - | - | - | 17.9 | 9.86 x 10−05 | Disrupt | X | DIP2C |
| Dmd | - | - | - | 16.0 | 1.61 x 10−05 | Disrupt | X | DMD |
| Zswim6 | - | - | - | 11.3 | 1.40 x 10−06 | Disrupt | X | ZSWIM6 |
| Kdm6a | - | - | - | 10.4 | 6.11 x 10−05 | Disrupt | X | KDM6A |
| Eml4 | - | - | - | 9.4 | 9.21 x 10−05 | Disrupt | - | EML4 |
| Trip12 | - | - | - | 9.4 | 9.41 x 10−05 | Disrupt | X | TRIP12 |
| Npepps | - | - | - | 7.5 | 3.95 x 10−06 | Drive | - | NPEPPS |
| Ptpn14 | - | - | - | 7.5 | 0.000211 | Disrupt | - | PTPN14 |
| Setd5 | - | - | - | 7.5 | 0.000103 | Disrupt | X | SETD5 |
| Smc1a | - | - | - | 7.5 | 6.93 x 10−11 | Disrupt | - | SMC1A |
| Strn3 | - | - | - | 7.5 | 0.000448 | Disrupt | X | STRN3 |
| 5830433M19Rik | - | - | - | 6.6 | 0.000103 | Disrupt | - | C9orf82 |
| Cpne3 | - | - | - | 6.6 | 5.75 x 10−06 | Disrupt | - | CPNE3 |
| Eif4enif1 | - | - | - | 6.6 | 1.24 x 10−08 | Disrupt | X | EIF4ENIF1 |
| Map3k4 | - | - | - | 6.6 | 7.51 x 10−05 | Disrupt | - | MAP3K4 |
| Ppp2r2a | - | - | - | 6.6 | 1.18 x 10−05 | Disrupt | X | PPP2R2A |
| Ptch1 | - | - | - | 6.6 | 1.28 x 10−09 | Disrupt | X | PTCH1 |
| Rab2a | - | - | - | 6.6 | 0.000438 | Disrupt | X | RAB2A |
| Sec24b | - | - | - | 6.6 | 0.000117 | Disrupt | - | SEC24B |
| Sppl3 | - | - | - | 6.6 | 0.000178 | Disrupt | - | SPPL3 |
| Atl2 | - | - | - | 5.7 | 9.72 x 10−05 | Disrupt | - | ATL2 |
| Fam168b | - | - | - | 5.7 | 5.81 x 10−10 | Drive N-term truncation | X | FAM168B |
| Maea | - | - | - | 5.7 | 4.65 x 10−08 | Disrupt | X | MAEA |
| Mark2 | - | - | - | 5.7 | 4.48 x 10−05 | Disrupt | X | MARK2 |
| Shfm1 | - | - | - | 5.7 | 1.17 x 10−08 | Drive N-term truncation | - | SHFM1 |
| Txndc11 | - | - | - | 5.7 | 0.000367 | Disrupt | X | TXNDC11 |
| Ube2l3 | - | - | - | 5.7 | 4.32 x 10−06 | Disrupt | X | UBE2L3 |
| Ccm2 | - | - | - | 4.7 | 0.000296 | Disrupt | - | CCM2 |
| Cntfr | - | - | - | 4.7 | 3.66 x 10−09 | Disrupt | - | CNTFR |
| Gosr1 | - | - | - | 4.7 | 8.84 x 10−05 | Disrupt | - | GOSR1 |
| Klf13 | - | - | - | 4.7 | 7.38 x 10−05 | Disrupt | - | KLF13 |
| Myst2 | - | - | - | 4.7 | 0.000131 | Disrupt | - | KAT7 |
| Nr1d2 | - | - | - | 4.7 | 0.000131 | Disrupt | X | NR1D2 |
| Plekhb2 | - | - | - | 4.7 | 1.55 x 10−06 | Disrupt | - | PLEKHB2 |
| Rab12 | - | - | - | 4.7 | 6.99 x 10−08 | Disrupt | - | RAB12 |
| Zbtb10 | - | - | - | 4.7 | 0.000292 | Disrupt | - | ZBTB10 |
| Zfp217 | - | - | - | 4.7 | 7.38 x 10−06 | Drive N-term truncation | - | ZNF217 |
| 2310044G17Rik | - | - | - | 3.8 | 4.37 x 10−06 | Drive | - | KIAA1737 |
| Cdkn2a | - | - | - | 3.8 | 0.000128 | Disrupt | - | CDKN2A |
| Ddx3x | - | - | - | 3.8 | 3.07 x 10−05 | Disrupt | - | DDX3X |
| Manbal | - | - | - | 3.8 | 0.000157 | Drive | - | MANBAL |
| Mycn | - | - | - | 3.8 | 8.71 x 10−12 | Disrupt | - | MYCN |
| Pabpc1l | - | - | - | 3.8 | 5.76 x 10−05 | Drive | - | PABPC1L |
| Prdx1 | - | - | - | 3.8 | 2.27 x 10−05 | Disrupt | - | PRDX1 |
| Psmc1 | - | - | - | 3.8 | 1.66 x 10−08 | Disrupt | - | PSMC1 |
| Sntn | - | - | - | 3.8 | 4.48 x 10−05 | Disrupt | - | SNTN |
| 2610507I01Rik | - | - | - | 2.8 | 1.41 x 10−06 | Drive | - | - |
| Ifng | - | - | - | 2.8 | 0.000154 | Drive | - | IFNG |
| Igsf8 | - | - | - | 2.8 | 1.14 x 10−07 | Drive | - | IGSF8 |
| Mir1b | - | - | - | 2.8 | 0.000125 | Disrupt | - | MIR206 |
| Mir99a | - | - | - | 2.8 | 1.61 x 10−05 | Drive | - | MIR99A |
| P4hb | - | - | - | 2.8 | 0.000167 | Disrupt | - | P4HB |
| Trip10 | - | - | - | 2.8 | 0.000104 | Disrupt | - | TRIP10 |
Analysis of td-CIS association with tumor type identified significant driver-mutations in neurofibromas (n=47) and grade 3 PNSTs (n=10) (Table 1, Supplementary Table 2, Supplementary Fig. 4a–b). Known Schwann cell tumor suppressor genes (TSG), Nf1 and Pten, were significantly associated with grade 3 PNSTs and identified in 34.9% (p=8.04E-06) and 30.2% (p=3.85E-06) of tumors, respectively13,51. In neurofibromas, Nf1 and Pten insertions contributed to 16.4% and 14.1% of tumors, respectively. The most commonly mutated neurofibroma CISs were Csmd1 (46.8%, p=1.12E-05), Utrn (30.5%, p=0.02189) and Zbtb20 (29.4%, p=1.15E-05). Genes previously implicated in human MPNST formation were also identified: Bmpr2 (9.4%, p=0.005), Jak2 (5.7%, p=0.0013), and Nf2 (5.7%, p=0.0013).
The position/orientation of the T2/Onc murine stem cell virus (MSCV) promoter relative to the direction of gene transcription can be used to predict whether T2/Onc is likely to drive or disrupt gene transcription52. Transcriptional activation may occur if the majority of transposon insertions are orientated upstream of a gene or translational start site with MSCV promoters in the same direction as gene transcription; the gene would be a putative proto-oncogene (e.g. Eras, Foxr2, and Zfp521) (Supplementary Fig. 5a, Supplementary Table 3). Disruption of transcription may occur if the transposons land within a gene with no MSCV promoter orientation or insertion site bias within the locus; the gene would be a putative TSG (e.g. Nf1 and Pten) (Supplementary Fig. 5b, Supplementary Table 3). Twenty putative proto-oncogenes and sixty-seven putative TSGs were identified in grade 3 PNST CISs (Table 1, Supplementary Table 3).
Comparative analysis of grade 3 PNST CISs identified novel genes involved in MPNSTs
Relevance of the CIS-associated genes to human MPNST formation was evaluated by cross-species comparative analysis of the grade 3 PNST CISs to previously generated human array comparative genomic hybridization (aCGH), single nucleotide polymorphism (SNP) arrays, human microarray expression, and methylome data from normal Schwann cells, neurofibromas, and MPNSTs (Figure 2)5,53,54. Utilizing human MPNST aCGH data, we identified CISs with a tendency toward CNA-gains (n=31, ZBTB10, CDK13, RAB2A, SHFM1, CPNE3) and CNA-losses (n= 34, CDKN2A, JAK2, NF1, NF2, FAF1, PLAA, PICLAM) (Supplementary Table 4, Figure 2, Supplementary Fig. 6–7)54. Comparably, whole genome SNP analysis on additional MPNSTs (n=14) identified several CISs in chromosomal regions gained (BMPR2, CPNE3, EML4) and lost (CDKN2A, JAK2, PLAA) in >35% of the samples (Supplementary Table 5). However, many recurrently gained/lost genes were found in regions of large chromosomal rearrangements making it difficult to discern whether they are driver or passenger mutations. To address this concern, methylome and microarray expression data from human Schwann cell tumors were analyzed.
Figure 2. Comparative analysis of grade 3 PNST CISs to human MPNSTs.
CNA, methylome, and microarray expression data from human MPNST data (Supplementary Fig. 6–8, Supplementary Tables 3,5) were combined into a “bubble plot”. The y-axis displays the methylation state (negative number indicating hypomethylation and positive number indicating hypermethylation) of each gene’s CpG-IS nearest to the transcriptional start site (TSS) in MPNSTs versus NHSCs (methylation analysis described in detail in online methods and Supp Table 5). The x-axis depicts the CNA observed in MPNSTs versus NHSCs (determination of CNA is described in online methods and Supp Table 3). Numbers reflect the percentage of the 51 patient samples that have a CNA. Negative numbers indicated CNA loss while positive numbers indicate CNA gains. The yellow shaded area indicates the 95th percentile for significant recurrent CNAs. Microarray expression was represented by size from 1X-10X (determination of expression changes are described in online methods and Supp Fig 8) and color to depict genes that are upregulated in red (a), downregulated in blue (b), or have no change or have no probes on the microarray in gray (c) in gene expression comparing MPNSTs to NHSCs. For recurrent CNAs, we considered a gene that shows the gain (or loss) pattern if the ratio of gain (or loss) out of all 51 patients is more than 1.5X that of the ratio of loss (or gain). Significant recurrent CNAs must be observed in 23/51 patient samples (95th percentile).
Promoter CpG Island Shore (CpG-IS) methylation status is predictive of gene expression: hypermethylation is associated with gene silencing and hypomethylation is associated with gene expression53. We used MPNST whole methylome data to analyze the methylation patterns of the promoter CpG-IS regions53. Fifty-five CIS genes showed significant differential hypo- or hyper-methylation patterns in their CpG-IS regions (see Online Methods, Supplementary Table 6, Figure 2). Six CISs displayed hypomethylation in >10% of tumors (BMPR2, PRDX1, PTPN14, BAZ1B, JMY, MAEA), while only KLF13 was hypermethylated in >10% of MPNSTs.
To assess the impact of CNAs and methylation patterns on gene expression, we utilized microarray expression data. Seventy-seven CISs had a human homolog on the microarray (Supplementary Fig. 8)5. Microarray expression data predict CISs either upregulated (EIF4ENIF1, EML4, PTCH1, TRIP10, ZBTB10, ZNF217, ZNF521) or downregulated (TAOK1, PICALM, NF1, NF2, SPAG9, SRGAP2, PTEN) in disease progression to MPNSTs (Supplementary Fig. 8, Figure 2). Overall, genes we predicted to be oncogenes and TSGs from the SB screen displayed the expected biases in overexpression/CNA-gains/hypomethylation and reductions in gene expression/CNA-losses/hypermethylation (Figure 2). These genes include known drivers of MPNST formation (CDKN2A, PTEN, NF1) and novel candidate drivers (BAZ1B, FOXR2, ERAS, PLAA, DIP2C, PICALM).
Lastly, we utilized the catalog of somatic mutations in cancer (COSMIC-v63) database to assess alterations of the CISs in other human cancers (Supplementary Table 7)55. The grade 3 PNST CIS list is significantly enriched for in the Cancer Gene Census (12/488 genes, FET p=2.99x10−06, hypergeometric test p=3.206496x10−06) indicating we identified driver-mutations common to many cancer types.
MPNST candidate genes are enriched in specific signaling pathways and processes
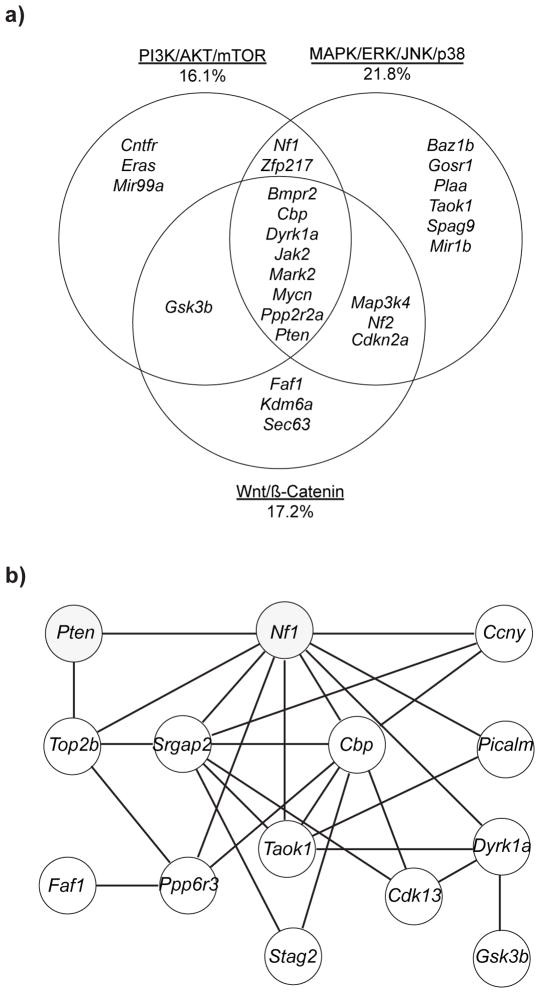
Ingenuity Pathway Analysis (IPA) and the Database for Annotations, Visualization and Integrated Discover (DAVID) were used to identify significantly altered signaling pathways in grade 3 PNST CISs (Supplementary Tables 8–9). IPA identified significant enrichment in cancer-associated signaling pathways (p=8.69x10−09–3.89x10−03, n=25 genes) including PI3K/AKT (p=1.55x10−03), ERK/MAPK (p=3.79x10−02), Wnt/CTNNB1 (p=5.20x10−04) and BMP/TGF-β (p=4.15x10−02). Cell cycle regulation was the most significantly altered pathway (n=21 genes, p=9.87x10−07-3.89x10−03 and n=6 genes, FET p=2.85x10−05, respectively). Additional cancer-associated signaling pathways included Jak-STAT (FET p=5.90x10−03) and TGF-beta (FET p=8.40x10−03). Combining data from DAVID analysis, IPA, and literature reviews, we classified the CISs into three major signal transduction pathways implicated in MPNST development: PI3K/AKT/mTOR (16.1%), MAPK/ERK/JNK/p38 (21.8%), and Wnt/CTNNB1 (17.2%) (Figure 3a)51,56–58. Seven CISs (Bmpr2, Cbp, Dyrk1a, Mark2, Mycn, Ppp2r2a, Pten) are involved in signaling through all three pathways as canonical members and/or effectors.
Figure 3. CIS analysis for cooperating genes and pathways for grade 3 PNST formation.
(a) Venn diagram depicting the clustering of the 87 grade 3 PNST CISs into three cancer-associated signaling pathways: PI3K/AKT/mTOR, MAPK/ERK/p38, Wnt/CTNNB1. Values indicate the percentage of the 87 CISs contributing to each pathway. CISs were classified into each pathway based on IPA, DAVID, GENECARD, and PUBMED analysis and literature review to identify canonical members and pathway effectors. (b) This module of genes depicts co-occurrence analysis of grade 3 PNST td-CISs. p-values for CIS interactions are listed in Supplementary Table 10.
Co-occurring CISs in grade 3 PNSTs
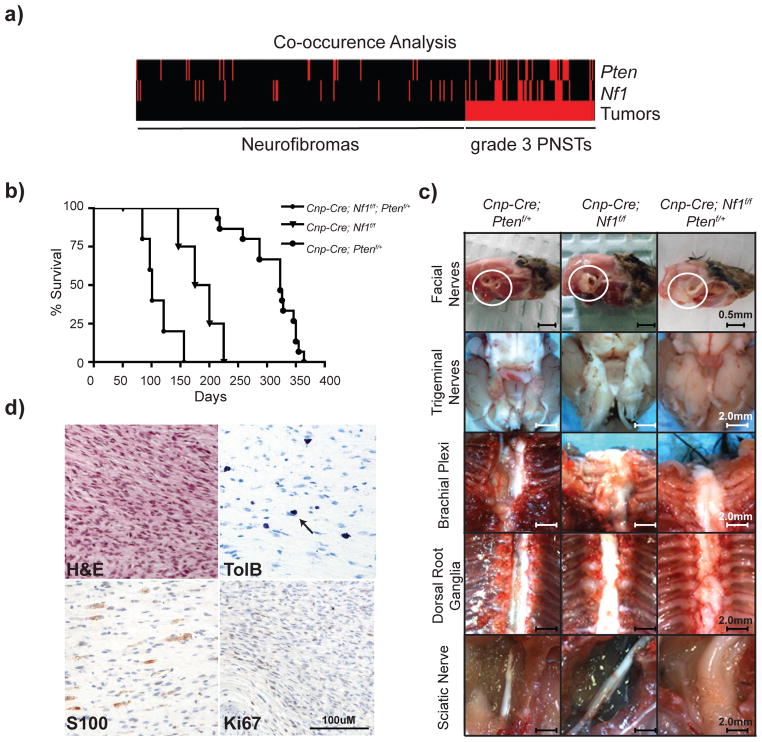
Co-occurrence analysis was performed to identify genes frequently mutated together in grade 3 PNSTs at a higher frequency than expected by chance49. Co-occurrence analysis of the 34 td-CISs identified 99 pairs of co-occurring CISs (co-CIS) (Supplementary Table 10). Several genes were identified as co-CISs with the known MPNST driver Nf1: Pten, Dyrk1a, Ppp6r3, Taok1, Picalm, Top2b, Srgap2, and Ccny (Supplementary Table 10, Figure 3b). Nf1 and Pten were present in the highest percentage of tumors as single CISs (34.9% and 30.2% respectively, Table 1) and co-occurred in 13/106 grade 3 PNSTs (FET p=7.05x10−05) compared to 1/269 neurofibromas (Figure 4a). Human microarray expression data demonstrated a reduction in NF1 and PTEN with disease progression to MPNSTs. (Supplementary Fig. 8). These data suggest that co-occurring mutations in Nf1 and Pten cooperate to form grade 3 PNSTs and may cooperate in human MPNST progression.
Figure 4. Loss of Nf1 and Pten cooperate to form high-grade PNSTs.
(a) Co-occurrence analysis heat map depicting each mouse tumor (neurofibromas left, grade 3 PNST right) and the presence of an insertion into either the Pten or the Nf1 locus (red bars). Tumors from 13/62 mice (106 grade 3 PNSTS) contained insertions in both Nf1 and Pten, which is statistically significantly different from the neurofibroma profile of 1/55 mice (269 neurofibromas) (FET p<7.94x10−05). (b) Survival curve of three genetic cohorts: Cnp-Cre; Nf1f/f; Ptenf/+(n=5), Cnp-Cre; Nf1f/f (n=5), Cnp-Cre; Ptenf/+(n=15). Statistical analysis of curves: Cnp-Cre; Nf1f/f; Ptenf/+ vs Cnp-Cre; Nf1f/f p<0.05; Cnp-Cre; Nf1f/f; Ptenf/+ vs Cnp-Cre; Ptenf/+p<0.0001; Cnp-Cre; Nf1f/f vs Cnp-Cre; Ptenf/+p=0.0018. p-values reflect Logrank test. (c) Necropsy images from a 275 day old Cnp-Cre; Ptenf/+mouse, a 175 day old Cnp-Cre; Nf1f/f mouse, and a 120 day old Cnp-Cre; Nf1f/f; Ptenf/+mouse. (d) Histological analysis of sciatic nerve tumor in (c). H&E staining depicts high cellularity with few mitotic figures corroborated with Ki67 IHC. Toluidine blue stain depicts presence of Mast cells indicative of nerve origin. S100 positive staining depicts presence of Schwann cells. This tumor contained regions of high-grade PNST formation.
To address whether Nf1 and Pten mutations cooperate to form Schwann cell tumors in vivo, we generated transgenic mice with biallelic inactivation of Nf1 and heterozygous loss of Pten in Cnp-Cre expressing cells. Cnp-Cre; Nf1f/f; Ptenf/+ mice had a significantly decreased median survival of 101 days compared to control Cnp-Cre; Nf1f/f (185 days, Logrank p=0.017) and Cnp-Cre; Ptenf/+ (323 days, Logrank p<0.0001) mice (Figure 4b). Cnp-Cre; Nf1f/f; Ptenf/+ mice (n=5) succumbed to paralysis-related deaths with peripheral nerve hyperplasia and tumors, the majority of which were grade 1 neurofibromas (Figure 4c). Importantly, grade 1 neurofibromas in the extremities (sciatic nerve, brachial plexus) contained regions of high-grade PNST histology (Figure 4d). Cnp-Cre; Nf1f/f mice (n=4) developed nerve hyperplasia and neurofibromas but no high-grade PNSTs. Cnp-Cre; Ptenf/+ mice (n=15) developed hematopoietic malignancies rather than Schwann cell-related phenotypes.
These data are consistent with our previous experiments in which Dhh-Cre; Nf1f/f; Ptenf/+mice only developed neurofibromas, while Cnp-Cre; Nf1f/f; Ptenf/+ mice had shorter median survival and possessed regions of high-grade PNST development59. Both mouse models demonstrate the cooperativity of Nf1 and Pten mutations for accelerating Schwann cell tumorigenesis. Additional cooperating mutations identified in the co-occurrence analysis may provide insight into cooperating signaling pathways that promote MPNST formation in humans.
The novel proto-oncogene FOXR2 has a role in human MPNST maintenance
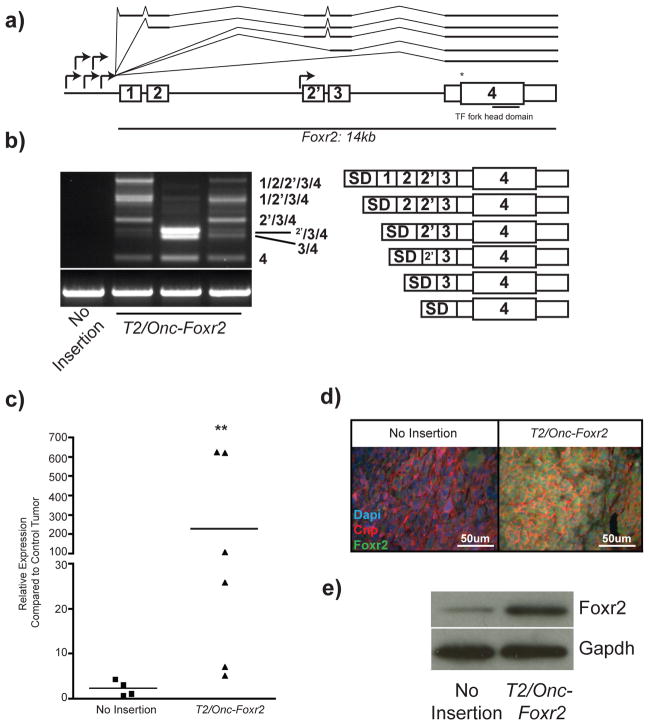
Foxr2 is a grade 3 PNST CIS predicted to be a proto-oncogene due to the orientation and position of T2/Onc insertions (Figure 5a). mRNA fusion transcripts between the T2/Onc MSCV promoter/SD and Foxr2 were identified in tumors with T2/Onc insertions in the Foxr2 locus (Figure 5b). The sequenced products demonstrated splicing from the T2/Onc splice donor into each exon of the Foxr2 gene, including a cryptic exon not previously annotated in intron 2 (labeled 2′) (Figure 5a–b). T2/Onc fusion transcripts led to increased Foxr2 mRNA and protein expression in tumors and tumor-derived cell lines, suggesting that Foxr2 functions as a proto-oncogene (Figures 5c–e).
Figure 5. T2/Onc insertions in the Foxr2 locus cause overexpression of Foxr2 in SB-derived grade 3 PNSTs.
(a) Schematic depicting the mouse Foxr2 locus. The MSCV promoter of T2/Onc for all transposon insertions faces the same orientation as Foxr2 transcription (arrows). Annotated exons are marked as 1, 2, 3, 4. * represents the translational start site. The exon 2′ is a putative unannotated exon. (b) RT-PCR on cDNA from SB-derived grade 3 PNSTs. Bands indicate mRNA fusions between T2/Onc and Foxr2. Lane 1 is an SB-derived grade 3 PNST that did not contain a Foxr2 insertion. Lanes 2 and 4 are T2/Onc insertions upstream of exon 1. Lane 3 is the T2/Onc insertion immediately upstream of exon 2′. The schematics are the sequenced splicing events from each of the excised products. The superscript 2′ represents splicing into the putative 2′ exon. (c) Quantitative PCR for Foxr2 expression in SB-derived grade 3 PNSTs with and without a T2/Onc insertion in Foxr2. ** p<0.001 based on two-tailed student t-test. (d) Immunofluorescent staining for Foxr2 (green), Cnp (red), and Dapi (blue) on tumor sections containing (right) and not containing (left) a T2/Onc insertion in Foxr2. (e) Western blot analysis for Foxr2 expression from two SB-derived tumor cell lines containing (right) and not containing (left) a T2/Onc insertion into Foxr2.
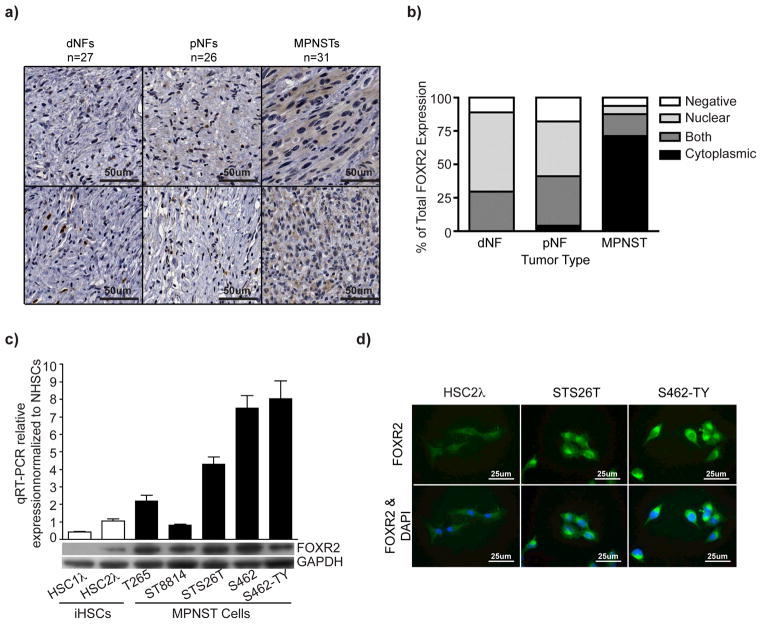
Immunohistochemical analysis of a human tissue microarray demonstrated increased cytoplasmic FOXR2 expression in MPNSTs compared to neurofibromas (Figure 6a–b). FOXR2 mRNA and protein levels were increased in MPNST cell lines compared to normal Schwann cells and CDK4/TERT immortalized human Schwann cells (iHSCs) (Figure 6c)59. MPNST cell lines also demonstrated cytoplasmic FOXR2 expression (Figure 6d).
Figure 6. Increased FOXR2 expression is associated with human MPNSTs.
(a) Immunohistochemical staining for FOXR2 of a tissue microarray (TMA) containing 27 dermal neurofibroma samples (dNF), 26 plexiform neurofibroma samples (pNF), and 31 MPNST samples. Representative images for FOXR2 staining for each tumor type are shown. (b) Bar graph depicts percentages for either staining localization for each tumor type (nuclear, cytoplasmic, both, or negative). (c) The bar graph depicts quantitative PCR analysis for FOXR2 expression in iHSCs (HSC1λ and HSC2λ) and MPNST (T265, ST8814, STS26T, S462, S462-TY) cell lines. Data are normalized to purified normal human Schwann cells (NHSCs). The lower bands indicate western blot analysis for FOXR2 for the same cell lines. (d) Immunofluorescent imaging of FOXR2 expression in HSC2λ, STS26T, and S462-TY. Staining is predominantly cytoplasmic as observed in the MPNST samples in (a).
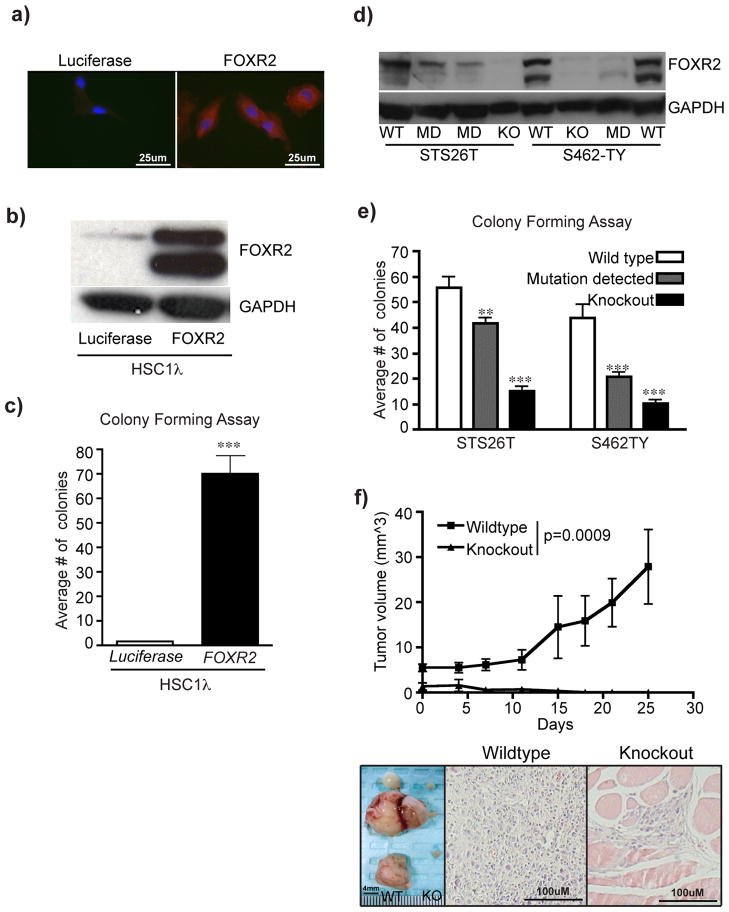
FOXR2 is a novel gene with unknown function. To explore the role of FOXR2 in MPNST formation, we developed constructs to overexpress the full-length cDNA or knock out the gene using TAL-effector nucleases (TALENs) (Supplementary Fig. 9a–b) in iHSCs and MPNST cell lines5,59. FOXR2 overexpression in the iHSC clone HSC1λ increased total FOXR2 protein and cytoplasmic staining, minimally increased in vitro proliferation, and significantly increased soft-agar colony formation compared to luciferase control (two-tailed t-test p<0.0001) (Figure 7a–c, Supplementary Fig. 10a). Xenograft experiments demonstrated no tumor formation in the control HSC1λ cells, but 3/5 mice harboring FOXR2 overexpressing cells developed a tumor (Supplementary Fig. 11a–b). This phenotype was confirmed by TALEN-mediated targeted knockout of FOXR2 in S462-TY and STS26T human MPNST cell lines (Supplementary Fig. 9b). TALENs that generated knockout or heterogeneous mutations (called mutation detected) of FOXR2 caused loss of protein by western blot analysis (Supplementary Fig. 9c, Figure 7d). FOXR2 knockout and mutation detected cell lines had reduced proliferation and significant reductions in colony formation ability compared to HPRT knockout control cells (t-test p<0.0001) (Supplementary Fig. 10b–c, Figure 7e). Additionally, FOXR2 loss inhibited the ability of STS26T cells to form tumors in xenograft experiments compared to HRPT knockout controls (Figure 7f). shRNA-mediated FOXR2 knockdown in STS26T and S462-TY cell lines confirmed the FOXR2 TALEN results in vitro (Supplementary Fig. 10d–e). Collectively, FOXR2 has minor effects on proliferation in vitro, whereas effects on anchorage independent growth and tumorigenicity are more profound.
Figure 7. Modulating FOXR2 expression significantly alters MPNST tumorigenic properties.
(a) Immunofluorescent imaging of FOXR2 expression in HSC1λ targeted with either a Luciferase expression construct (left) or a FOXR2 expression construct (right). (b) FOXR2 western blot on cells from (a). (c) Bar graph depicts results from a soft-agar colony-forming assay performed in triplicate. Statistical analysis was done using a two-tailed student t-test, *** p<0.0001 (d) Western blot analysis of FOXR2 expression on STS26T and S462-TY cell lines targeted with FOXR2 TALENs. WT = wildtype, KO = knockout, MD = mutation detected. (e) Bar graph depicts results from a soft agar colony-forming assay performed in triplicate with biological replicate cell lines. STS26T wildtype (n= 3), STS26T mutation detected (n=4), STS26T knockout (n=4), S462-TY wildtype (n=2), S462-TY mutation detected (n=4 ), and S462-TY knockout (n=4). Statistics were done using a two-tailed student t-test comparing to the respective wildtype control. **p-=0.0032, ***p<0.0001. (f) One million STS26T wildtype (n=8, left flank) and STS26T FOXR2 KO (n=8, right flank) cells were injected into Nu/Nu mice. Tumors were measured over a 1 month period. Wildtype STS26T tumors grew significantly larger than paired KO (two-tailed student t-test ***p=0.0009). Images were captured at time of necropsy of the tumors from WT (n=5, left) or regions were injections occurred (n=5, right). H&E staining of tissue sections from the masses indicate the wildtype masses were tumors while the masses from the KO cells were predominantly fat pad tissue that contained the injected cells.
DISCUSSION
We performed a large, forward genetic screen in Schwann cells utilizing SB somatic cell mutagenesis. We identified hundreds of genes that cooperated with EGFR overexpression and p53-loss of function to form neurofibromas and grade 3 PNSTs. Many co-CISs were identified, suggesting that specific, cooperating genetic mutations are required for tumor development. Also, CISs were identified in known signaling pathways altered in human MPNSTs: Wnt/CTNNB1, PI3K/Akt/mTor, and growth factor signaling11–13,58. Lastly, we identified several novel proto-oncogenes, including Foxr2, and elucidated a role for FOXR2 in human MPNST maintenance.
SB mutagenesis alone or on the Trp53R270H background was insufficient for Schwann cell tumorigenesis compared to previous reports in which SB alone caused tumors35,60. This may be explained by differences in the number of cells susceptible to SB mutagenesis. Compared to colonic epithelial cells, which have a high turnover rate, that of Schwann cells is near zero unless injury occurs60,61. Moreover, SB mutagenesis is a random event, and mutations that confer selective advantage for clonal outgrowth are rare events. Taken together, targeting a small population of cells with a low turnover rate may produce fewer SB-induced tumors unless a pre-existing mutation(s), such as EGFR-overexpression, is present to expand target cell populations41.
CIS overlap between two methods for defining CISs in grade 3 PNSTs and neurofibromas was 65% and 68%, respectively (Table 1, Supplementary Table 2). The discordance in CIS calling is based predominantly on methods used to identify a CIS49,50. gCIS analysis is based on window sizes corresponding to annotated genes and 15,000bp upstream to account for the promoter region50. Therefore, the number of insertions required to be deemed a gCIS depends on gene size (i.e., large genes require many insertions while smaller genes require fewer insertions). TAPDANCE analysis generates random window sizes throughout the genome and does not require gene annotation to identify CISs, as 4 ‘gene barren’ regions were identified in the neurofibroma CIS list (Supplementary Table 2)49. These regions could contain important enhancer elements, unannotated genes, non-coding RNAs, or other gene regulatory elements. The two methods are complimentary and enhance CIS identification.
Based on the number of neurofibromas (n=269) and grade 3 PNSTs (n=106) sequenced, there is a disproportionate number of CISs between neurofibromas (695) and grade 3 PNSTs (87). Analysis of unique transposon insertion site non-redundant (nr) regions identified 33,022 and 139,543 nr-regions for grade 3 PNSTs and neurofibromas, respectively. This discrepancy can be reconciled by the genetic heterogeneity of the SB-induced tumors. The fewer grade 3 PNSTs nr-regions were sequenced at a higher frequency compared to the neurofibroma nr-regions, suggesting that few cells acquired mutations in strong genetic drivers that conferred a selective advantage for clonal outgrowth. These clonal insertions were sequenced thousands of times more than in the more genetically heterogeneous neurofibromas. Additionally, it is possible only a few, strong driver mutations in Schwann cells are required for progression to grade 3 PNSTs, while there are many alterations that are sufficient to cause neurofibroma formation. For example, cells may achieve many mutations but only the right combination of two or more genes may drive high-grade tumor formation, as we observed in the Nf1/Pten model. Lastly, we can reliably identify low-penetrant CISs with higher statistical power given the larger neurofibroma sample size compared to the grade 3 PNSTs.
CIS analysis identified a disproportionate number of putative TSGs compared to proto-oncogenes (Table 1, Supplementary Table 2), which also occurred in other solid tumor SB screens34,60,62 and likely has several explanations: 1) The strength of the T2/Onc MSCV promoter is not strong enough to sufficiently transcriptionally activate proto-oncogenes. A body-wide SB screen using the CAGGs promoter instead of MSCV in T2/Onc identified a larger spectrum of tumors and CISs35. 2) T2/Onc can insert in both orientations with less insertion position bias within or near a gene to cause inactivation meaning a greater chance of genes being inactivated. Alternatively, for driving oncogenes we observed a clustering of T2/Onc insertions in the promoter region or 5′UTR that predominantly orient to drive transcription.
Comparative genomic analysis reliably identified genes previously implicated in human MPNST formation (i.e., CDKN2A, NF1, PTEN)27,63. We also identified novel genes recurrently overexpressed/CNA-gains/hypomethylated (BAZ1B, FOXR2, ERAS) and underexpressed/biallelically inactivated/hypermethylated (DIP2C, PLAA, PICALM) in human MPNSTs (Figure 2, Supplementary Figs. 6–8, Supplementary Tables 4–6)63,64. However, we identified genes (e.g., ZBTB10 and EML4) with CNA gains in human MPNSTs predicted to be disrupted by SB mutagenesis. This discrepancy may reflect differences in mouse versus human MPNST formation and/or CNAs observed in human MPNSTs may mask the function of the gene in tumor progression. Additionally, many of the CISs could be categorized into signaling pathways (PI3K/AKT/mTOR, MAPK/ERK/JNK/p38, and Wnt/CTNNB1) (Figure 3a, Supplementary Tables 8–9). Collectively, these data provide novel gene drivers/signaling pathways that potentially promote human MPNST formation. Further experimental evidence is required to determine the contribution any candidate driver gene(s)/pathways may have on MPNST development.
Nf1 and Pten were most frequently mutated in our screen, and Pten was significantly co-mutated with Nf1 in grade 3 PNSTs. Loss of NF1 alone is not sufficient for MPNST formation in genetically engineered mouse models of Nf1 loss65–68. Gregorian et. al demonstrated that monoallelic inactivation of Pten in Schwann cells with activated KRasG12D overexpression caused grade 3 PNST formation51. Neither mutation alone was sufficient to generate grade 3 PNSTs. These data suggest that, like Nf1, Pten mutations alone are insufficient for MPNST formation. Our mouse model of biallelic Nf1 inactivation and Pten heterozygosity demonstrated cooperation for high-grade PNST formation compared to loss of either allele alone (Figure 3a–d). We attempted to generate biallelic loss of Nf1 and Pten with the Cnp-cre model, but no live mouse originated from the crosses with the expected Mendelian ratio of 1:8. Cnp-Cre; Nf1f/f; Ptenf/f may be embryonic lethal. However, Nf1 loss in the context of Pten heterozygosity produced tumors with regions containing high-grade PNSTs, which is consistent with the observation that PTEN expression is not completely lost in human MPNSTs. The previously reported Dhh-Cre; Nf1f/f; Ptenf/+ mouse model presented with neurofibromas and required biallelic inactivation of the second Pten allele for high-grade PNST formation59. Therefore, it is important to consider gene dosage, co-mutations, and timing of mutation induction to determine effective mouse models for developmental and therapeutic studies.
FOXR2 is a forkhead-box (FOX) transcription factor family member identified in silico on chromosome Xp11.21 in humans. FOXR2 is an ortholog of FOXR1 that shares 57.7% amino acid identity69. FOXR1 is a transcriptional inhibitor of FOX family target genes such as p27Kip170. Similar to FOXR1, FOXR2 is expressed in mouse embryonic development (E9.5) with reduced expression in adult tissues69. Human Protein Atlas and ONCOMINE indicate FOXR2 expression is low and variegated in normal adult human tissues, but expression is increased at the genomic (CNA gains) and protein levels in several cancers71–73. COSMIC-v63 reports 18 point mutations within FOXR2 from 4,862 samples55. We utilized the MutationAssessor program to determine the functional impact of the 18 point mutations on cancer, which ranged from neutral to medium with 4 point-mutations identified in the Fork-head DNA binding domain (Supplementary Table 12). This may indicate that FOXR2 mutations are not as pivotal to cancer formation as increased expression. Our comparative genomic analysis and functional data predict that FOXR2 is a proto-oncogene in human MPNST maintenance. FOX superfamily members have been implicated in human cancers through mechanisms like gene amplifications and translocations74. Further studies are warranted to determine the extent of FOXR2 involvement in tumor development and its function in normal somatic cells.
Interestingly, in the grade 3 PNSTs containing T2/Onc-Foxr2 fusion transcripts we observed splicing into the first annotated exon of Foxr2 where splice acceptor elements were not previously described (Figure 5). We identified RNA splicing elements (branch point ‘A’, polyprimidine tract, and ‘AG’ boundary) within 35 nucleotides of the first annotated Foxr2 exon indicating the potential for splicing. Furthermore, FOXR2 full-length cDNA unexpectedly encodes two unique protein isoforms (Figure 7b,d), and we observed a bias in isoform loss following TALEN knockout (Figure 7d). The FOXR2 locus contains five in-frame methionines of which four have optimal Kozak sequences (amino acid positions 1, 25, 53, 58, and 81) with the following predicted molecular weights: 35.93kDa, 32.94 kDa, 29.76kDa, 29.17kDa, and 26.81kDas. Further studies are necessary to define the Foxr2 locus in mouse and human and to determine the implications of the use of the alternative start codons in tumorigenesis.
Overall, using a SB forward genetic screen we identified hundreds of candidate cancer driver genes that promote Schwann cell tumorigenesis. Moreover, we determined a new functional role for the novel proto-oncogene FOXR2 in human MPNST maintenance. Further functional testing of additional CISs may reveal new genetic pathways to target for treatment of human MPNSTs.
URLs
Ingenuity Pathway Analysis, http://www.ingenuity.com; Catalogue of Somatic Mutations in Cancer - COSMIC, http://www.sanger.ac.uk/genetics/CGP/cosmic/; Oncomine, http://www.oncomine.org/; Database for Annotation, Visualization and Integrated Discovery – DAVID, http://david.abcc.ncifcrf.gov; gene centric CIS analysis, http://ias.eng.uiowa.edu/uploader/; UCSC Genome Browser, http://genome.ucsc.edu; Mutation Assessor program, http://mutationassessor.org/, TALE-NT (https://boglab.plp.iastate.edu/node/add/talen)
ONLINE METHODS
Transgenic animals and tumor isolation
Three transgenes were used to induce SB mutagenesis in Schwann cells: a conditionally expressed SB transposase enzyme (R26-lsl-SB11)35, a Schwann cell specific Cre-recombinase controlled by the CNP promoter (Cnp-Cre)38, and a concatomer of oncogenic transposons (T2/Onc15)48. Triple transgenic mice (Cnp-Cre; R26-lsl-SB11; T2/Onc15) undergo insertional mutagenesis in Schwann cells. MPNST predisposing alleles, Cnp-EGFR and conditional Trp53R270H alleles, were also included41,42. Mouse cohorts are shown in Figure 1a. Genotyping PCR was performed on phenol-chloroform extracted mouse-tail DNA as previously described34,38,41,42.
Peripheral nerves and tumors were resected, assessed, and histologically evaluated as previously described59. Each sample was pathologically graded using established criteria for tumors generated in genetically engineered mouse models (GEMM)75. Grade 1 PNSTs are called neurofibromas. Tumors with features of human MPNSTs (GEMM grade 3 PNSTs) are called grade 3 PNSTs.
Transposon Insertion Site Analysis
DNA isolated from SB-derived neurofibromas and grade 3 PNSTs underwent linker-mediated PCR to amplify transposon-genomic DNA junctions and were sequenced as previously described34. Identification of TAPDANCE CISs and gene centric CISs from sequenced transposon-genomic DNA PCR products was performed as previously described49,50. The local-hopping phenomenon of T2/Onc was accounted for by excluding chromosome 15 (T2/Onc donor locus) from the analysis. Known CIS artifacts originating from the T2/Onc sequence elements (En2, Foxf2) and amplified regions of the mouse genome (Serinc3, Sfi) were also excluded48,49. Thirty-three histologically normal nerves from SB mutagenized mice were sequenced to assess potential transposon integration hotspots; no CISs were identified.
Gene expression data analysis
Published data (GEO accession #:GSE14038, Affymetrix GeneChip HU133 Plus 2.0) were used for gene expression pattern analysis. For gene annotation, custom CDF (custom GeneChip library file) based on RefSeq target definitions (Hs133P REFSEQ Version 8) was downloaded and used to provide accurate interpretation of GeneChip data76.
Statistical comparisons were done using R/Bioconductor packages and GeneSpring GX v7.3.1 (Agilent Technologies). Differentially expressed (DE)-genes were defined as genes with expression levels at least three-fold higher or lower in target groups (MPNST) compared to normal human Schwann cells (NHSCs) after applying Benjamini and Hochberg false discovery rate correction (FDR/BH p ≤ 0.05)77.
MPNST whole-methylome data analysis
Feber et al. published unbiased whole-methylome data of NHSCs, neurofibroma, and MPNST genomes (GEO accession #: GSE21714)53. We adopted the Feber et al. method for detecting differentially methylated regions (DMR) in MPNST compared to NHSC53,78. Briefly, Batman methylation scores per 100bp were averaged for each 1Kb window. A conservative threshold for DMR calling was used based on the 95th percentile of the difference in methylation score. DMR regions were mapped to human genome hg18 version (Build NCBI-36).
The nearest CpG island shores (CpG-IS) to the transcription start sites (TSS) of each gene/miRNA were scanned53,78. We defined CpG-IS as areas up to 2Kb distant from CpG islands. We considered the nearest upstream CpG-IS from the transcription start site, within 5K bp ranges from each TSS. The genomic coordinates of miRNAs, genes, and CpG islands (NCBI36/hg18) were extracted from corresponding tracks of UCSC Genome Browser (http://genome.ucsc.edu). For intragenic miRNAs, we assumed their expression is influenced by the nearest CpG-IS to the TSS of their host gene.
Copy number alteration (CNA) data analysis
CNA data on 51 primary MPNSTs were from the published GSE3388 data set (Agilent Human Genome CGH Microarray kit (4x44K)54,79. A circular binary segmentation (CBS) algorithm was applied to the log2 ratios of intensity values from tumor and normal to reduce local noise effects. CBS calculates a likelihood-ratio statistic for each array probe by permutation to locate change-points80. After segmentation step, CGHcall algorithm was used to assign each segment an aberration label: gain, loss, or normal81. A visualization program was written in R to present overall gain/loss patterns of all 51 MPNSTs. We considered a given genomic region as significantly recurrent CNA region if the number of CNA labels (gain or loss) given to them exceeded a threshold of statistical significance that we estimated using a permutation test. Briefly, CNA labels, three state calls (gain/loss/normal) by CGHcall algorithm, of the targets were permuted 10,000 times to get distribution, and sums of gain or loss labels were computed for each target. The 95th percentile values for both sums (23 patients out of 51) were chosen as thresholds of statistically significant recurrent CNA. Genomic coordinates used in plots were based on hg19/GRCh37.p8 (R/Bioconductor biomaRt package)82.
SNP-Array Analysis
SNP-array analysis was performed on 14 MPNST primary tumors using the Illumina Infinium technology (Illumina, San Diego, CA). Tumors were analyzed using the Illumina Human660W-Quad beadchip. The data were processed with GenoCNA to segment the genome and determine copy number status82.
Tissue Microarray
Representative areas of disease were identified on hematoxylin and eosin-stained sections for 30 dermal neurofibromas, 31 plexiform neurofibromas, and 32 MPNSTs. Blocks consisting of duplicate 1.0 mm core samples were constructed with a manual tissue arrayer (MTA-1, Beecher Inc, WI) with 64 cores per recipient block. Immunohistochemistry (IHC) for FOXR2 (1:100, SIGMA) was performed utilizing an immunohistochemical staining platform (Nemesis 7200, Biocare) following standard IHC protocols83. Digital images of IHC stained TMA slides were obtained as previously described by Rizzardi et al. 201284.
Cell Culture/Assays
Immortalized human Schwann cells (iHSCs) were acquired from the laboratory of Dr. Margaret Wallace. iHSCs and human MPNST cell lines (ST88-14, STS26T, S462, S462-TY, and T265) were cultured in complete media (1xDMEM, 10% fetal bovine serum, 1x penicillin/streptomycin) and grown at 37°C in 5% CO2. shRNA GIPZ lentiviral constructs were purchased from Openbiosystems. Proliferation assays were setup in a 96-well plate format with 500 cells per well in full DMEM media containing 1ug/ml of puromycin (Life Technologies). Proliferation was assessed every 24 hours for 5 days using the MTS assay (Promega) following the manufacturer’s protocols. Experiments were performed in triplicate. Soft agar anchorage independent colony formation assays were carried out as previously described85. After 2 weeks of growth, cells were fixed in 10% formalin containing 0.005% Crystal Violet for 1 hour at room temperature. Formalin was removed and colonies were imaged on a Leica S8 AP0 microscope. Twelve images per cell line were taken and automated colony counts were done using ImageJ software.
FOXR2 overexpression construct
FOXR2 cDNA (Open Biosystems) was cloned into the Gateway Vector System (Invitrogen) and subcloned into a piggyBac (PB) transposon vector. The PB control vector contains the Luciferase and Gfp reporter genes. Cells were transfected with 2ug of FOXR2 or Luciferase PB transposon (Supplementary Fig. 8a) and 500ng of PB7 transposase plasmid using the NEON transfection system following manufacturers protocols (Invitrogen). Successfully transfected cells were enriched with 1ug/ml puromycin.
Generation of TALEN Knockout Cell Lines
TALENs targeting FOXR2 were designed using TALE-NT and constructed as previously described86–88. An HPRT TALEN pair served as a control86. TALENs were assembled using Golden Gate cloning with the truncated Δ152+63 TALEN backbone86,89. Assembled TALENs were tested by transient transfection into K562 cells using the NEON electroporation system (Invitrogen) and subsequent CEL-I assay90.
Knockout cell lines were generated using the PB Co-transposition method (Moriarity et al., unpublished data). Briefly, cells were electroporated with plasmids encoding the left and right TALENs, the hyperactive PB7 transposase, and Puromycin encoding transposon vectors. TALEN-treated cells were incubated at 37°C, 5% CO2 for 1 day followed by two days of ‘cold shock’ at 30°C to increase nuclease activity91. Cells were seeded at 50 cells per well in 96 well plates with Puromycin containing DMEM. Single cell clones were expanded, DNA was extracted, and CEL-I PCR was performed with amplicons sequenced via standard single pass Sanger sequencing (ACGT, Inc). Individual clone sequence data were analyzed for insertion/deletions (indels) formed by non-homologous end joining at the TALEN cut site and were classified as wildtype (WT), knockout (KO), or mutation detected (MD). Mutation detected clones were further analyzed by standard TOPO cloning and sequencing to determine the indels at each allele.
Quantitative-PCR analysis
1ug of total RNA was DNase (Life Technologies) treated and used for reverse transcription with the Super Script III first strand synthesis kit (Life Technologies) following manufacturer’s protocol. qPCR reactions utilized the LightCycler 480 SYBR I Green (Roche) and were analyzed on an Eppendorf Mastercycler ep gradient S. Primers are listed in Supplementary Table 12. Data were analyzed using the RealPlex software, calibrated to ACTB levels, and normalized to respective controls.
Immunoblotting and immunofluorescence
FOXR2 protein was detected in whole cell lysates harvested with modified RIPA buffer (0.5% (vol/vol) NP-40, 50 mM Tris-HCl pH 7.4, 150 mM NaCl, 1 mM EDTA) containing phosphatase inhibitors (SIGMA) and a complete mini protease inhibitor pellet (Roche). Lysates were separated on a 10% resolving gel, transferred to PVDF membrane, and probed with antibodies against FOXR2 (SIGMA) and GAPDH (Cell Signaling Tech.) following manufacturers’ protocols. Immunofluorescence on tissue sections and cell lines with antibodies probing FOXR2 (SIGMA) and CNPase (Santa Cruz) were carried out following manufacturers’ protocols followed by incubation with fluorescently labeled secondary antibodies (Life Technologies). Slides were counter stained and mounted with anti-FADE Gold (Life Technologies) containing DAPI. Images were acquired using AxioVision software.
Supplementary Material
Acknowledgments
We would like to thank the Biomedical Genomics Center at the University of Minnesota for performing the illumina deep sequencing. We would like to thank the Biological Materials Procurement Network (BioNet) specifically Dr. Stephen Schmechel, Anthony Rizzardi, Colleen Forster, and Sarah Bowell for construction, immunohistochemical staining, and scanning of the tissue microarray. We also acknowledge the following shared resources of the Masonic Cancer Center at the University of Minnesota: The Mouse Genetics Laboratory, Biostatistics and Bioinformatics, Comparative Pathology, and the Tissue Procurement Facility. We thank the Minnesota Supercomputing Institute for computational resources. We thank the Research Animal Resources at the University of Minnesota specifically Alwan Aliye for his technical support in mouse maintenance. This work received funding from the NIH-NINDS-P50 N5057531, the Margaret Harvey Schering Trust, The Zachary Neurofibromatosis Research Fund and The Jacqueline Dunlap Neurofibromatosis Research Fund. Work performed by A.L. Watson was supported by the Children’s Tumor Foundation Young Investigators Award #2011-01-018.
Footnotes
DATABASE ACCESSION NUMBERS. Microarray analysis GEO accession #:GSE14038, Affymetrix GeneChip HU133 Plus 2.076. MPNST whole genome methylation data (GEO accession #: GSE21714)53. Copy number alteration data GEO accession #GSE338854.
AUTHOR CONTRIBUTIONS
E.P.R., A.L.W., B.S.M., D.A.B., and N.W. performed laboratory experiments and/or analyzed the data. K.C. performed bioinformatic data analysis of microarray expression data, methylome data, and CNA data. A.S. analyzed deep sequencing data for CIS analysis. M.H.C. assessed histology and graded mouse tumors. C.L.M. provided MRIs of human MPNSTs and data analysis. M.R.W. generated the immortalized human Schwann cells. B.G. and E.S. generated and analyzed SNP array data. N.R. and D.A.L. supervised laboratory experiments and assisted in writing the manuscript. E.P.R. wrote the manuscript.
COMPETING FINANCIAL INTERESTS
D.A. Largaespada has ownership interest (including patents) in Discovery Genomics, Inc. and NeoClone Biotechnologies International. He is also a consultant/Advisory Board member of Discovery Genomics, Inc. and NeoClone Biotechnologies International.
References
- 1.Ducatman BS, Scheithauer BW, Piepgras DG, Reiman HM, Ilstrup DM. Malignant peripheral nerve sheath tumors. A clinicopathologic study of 120 cases. Cancer. 1986;57:2006–21. doi: 10.1002/1097-0142(19860515)57:10<2006::aid-cncr2820571022>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 2.Evans DG, et al. Malignant peripheral nerve sheath tumours in neurofibromatosis 1. Journal of medical genetics. 2002;39:311–4. doi: 10.1136/jmg.39.5.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Watson MA, et al. Gene Expression Profiling Reveals Unique Molecular Subtypes of Neurofibromatosis Type I-associated and Sporadic Malignant Peripheral Nerve Sheath Tumors. Brain pathology. 2004;14:297–303. doi: 10.1111/j.1750-3639.2004.tb00067.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Holtkamp N, et al. Subclassification of nerve sheath tumors by gene expression profiling. Brain pathology. 2004;14:258–264. doi: 10.1111/j.1750-3639.2004.tb00062.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miller SJ, et al. Large-scale molecular comparison of human schwann cells to malignant peripheral nerve sheath tumor cell lines and tissues. Cancer Res. 2006;66:2584. doi: 10.1158/0008-5472.CAN-05-3330. [DOI] [PubMed] [Google Scholar]
- 6.Zou C, et al. Clinical, pathological, and molecular variables predictive of malignant peripheral nerve sheath tumor outcome. Annals of surgery. 2009;249:1014–22. doi: 10.1097/SLA.0b013e3181a77e9a. [DOI] [PubMed] [Google Scholar]
- 7.Messiaen LM, et al. Exhaustive mutation analysis of the NF1 gene allows identification of 95% of mutations and reveals a high frequency of unusual splicing defects. Hum Mutat. 2000;15:541–555. doi: 10.1002/1098-1004(200006)15:6<541::AID-HUMU6>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 8.Leppig KA, et al. Familial neurofibromatosis 1 microdeletions: cosegregation with distinct facial phenotype and early onset of cutaneous neurofibromata. Am J Med Genet. 1998;73:197–204. doi: 10.1002/(sici)1096-8628(1997)73:2<197::aid-ajmg17>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 9.De Raedt T, et al. Elevated Risk for MPNST in< i> NF1 Microdeletion Patients. The American Journal of Human Genetics. 2003;72:1288–1292. doi: 10.1086/374821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ballester R, et al. The NF1 locus encodes a protein functionally related to mammalian GAP and yeast IRA proteins. Cell. 1990;63:851–9. doi: 10.1016/0092-8674(90)90151-4. [DOI] [PubMed] [Google Scholar]
- 11.Sherman LS, Atit R, Rosenbaum T, Cox AD, Ratner N. Single cell Ras-GTP analysis reveals altered Ras activity in a subpopulation of neurofibroma Schwann cells but not fibroblasts. J Biol Chem. 2000;275:30740. doi: 10.1074/jbc.M001702200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Basu TN, et al. Aberrant Regulation Of Ras Proteins In Malignant-tumor Cells From Type-1 Neurofibromatosis Patients. Nature. 1992;356:713–715. doi: 10.1038/356713a0. [DOI] [PubMed] [Google Scholar]
- 13.Cichowski K, Jacks T. NF1 tumor suppressor gene function: narrowing the GAP. Cell. 2001;104:593–604. doi: 10.1016/s0092-8674(01)00245-8. [DOI] [PubMed] [Google Scholar]
- 14.Forus A, et al. Comparative genomic hybridization analysis of human sarcomas: I. Occurrence of genomic imbalances and identification of a novel major amplicon at 1q21-q22 in soft tissue sarcomas. Genes, chromosomes & cancer. 1995;14:8–14. doi: 10.1002/gcc.2870140103. [DOI] [PubMed] [Google Scholar]
- 15.Lothe RA, et al. Gain of 17q24-qter detected by comparative genomic hybridization in malignant tumors from patients with von Recklinghausen’s neurofibromatosis. Cancer research. 1996;56:4778–81. [PubMed] [Google Scholar]
- 16.Mechtersheimer G, et al. Analysis of chromosomal imbalances in sporadic and NF1-associated peripheral nerve sheath tumors by comparative genomic hybridization. Genes, chromosomes & cancer. 1999;25:362–9. [PubMed] [Google Scholar]
- 17.Mertens F, et al. Cytogenetic findings in malignant peripheral nerve sheath tumors. International journal of cancer. 1995;61:793–8. doi: 10.1002/ijc.2910610609. [DOI] [PubMed] [Google Scholar]
- 18.Plaat BE, et al. Computer-assisted cytogenetic analysis of 51 malignant peripheral-nerve-sheath tumors: sporadic vs. neurofibromatosis-type-1-associated malignant schwannomas. International journal of cancer. 1999;83:171–8. doi: 10.1002/(sici)1097-0215(19991008)83:2<171::aid-ijc5>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 19.Schmidt H, et al. Genomic imbalances of 7p and 17q in malignant peripheral nerve sheath tumors are clinically relevant. Genes, chromosomes & cancer. 1999;25:205–11. [PubMed] [Google Scholar]
- 20.Schmidt H, et al. Gains in chromosomes 7, 8q, 15q and 17q are characteristic changes in malignant but not in benign peripheral nerve sheath tumors from patients with Recklinghausen’s disease. Cancer letters. 2000;155:181–90. doi: 10.1016/s0304-3835(00)00426-2. [DOI] [PubMed] [Google Scholar]
- 21.Schmidt H, et al. Cytogenetic characterization of six malignant peripheral nerve sheath tumors: comparison of karyotyping and comparative genomic hybridization. Cancer genetics and cytogenetics. 2001;128:14–23. doi: 10.1016/s0165-4608(01)00393-4. [DOI] [PubMed] [Google Scholar]
- 22.Birindelli S, et al. Rb and TP53 pathway alterations in sporadic and NF1-related malignant peripheral nerve sheath tumors. Laboratory investigation; a journal of technical methods and pathology. 2001;81:833–44. doi: 10.1038/labinvest.3780293. [DOI] [PubMed] [Google Scholar]
- 23.Legius E, et al. TP53 mutations are frequent in malignant NFI tumors. Genes, Chromosomes and Cancer. 2006;10:250–255. doi: 10.1002/gcc.2870100405. [DOI] [PubMed] [Google Scholar]
- 24.Perry A, et al. Differential NF1, p16, and EGFR patterns by interphase cytogenetics (FISH) in malignant peripheral nerve sheath tumor (MPNST) and morphologically similar spindle cell neoplasms. Journal of neuropathology and experimental neurology. 2002;61:702–9. doi: 10.1093/jnen/61.8.702. [DOI] [PubMed] [Google Scholar]
- 25.Menon A, et al. Chromosome 17p deletions and p53 gene mutations associated with the formation of malignant neurofibrosarcomas in von Recklinghausen neurofibromatosis. Proc Natl Acad Sci U S A. 1990;87:5435. doi: 10.1073/pnas.87.14.5435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kourea HP, Orlow I, Scheithauer BW, Cordon-Cardo C, Woodruff JM. Deletions of the INK4A gene occur in malignant peripheral nerve sheath tumors but not in neurofibromas. Am J Pathol. 1999;155:1855. doi: 10.1016/S0002-9440(10)65504-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nielsen GP, et al. Malignant transformation of neurofibromas in neurofibromatosis 1 is associated with CDKN2A/p16 inactivation. Am J Pathol. 1999;155:1879. doi: 10.1016/S0002-9440(10)65507-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mantripragada KK, et al. High-resolution DNA copy number profiling of malignant peripheral nerve sheath tumors using targeted microarray-based comparative genomic hybridization. Clinical Cancer Research. 2008;14:1015. doi: 10.1158/1078-0432.CCR-07-1305. [DOI] [PubMed] [Google Scholar]
- 29.Mawrin C, et al. Immunohistochemical and molecular analysis of p53, RB, and PTEN in malignant peripheral nerve sheath tumors. Virchows Archiv. 2002;440:610–615. doi: 10.1007/s00428-001-0550-4. [DOI] [PubMed] [Google Scholar]
- 30.Holtkamp N, et al. Mutation and expression of PDGFRA and KIT in malignant peripheral nerve sheath tumors, and its implications for imatinib sensitivity. Carcinogenesis. 2006;27:664. doi: 10.1093/carcin/bgi273. [DOI] [PubMed] [Google Scholar]
- 31.Storlazzi C, et al. Identification of a novel amplicon at distal 17q containing the BIRC5/SURVIVIN gene in malignant peripheral nerve sheath tumours. J Pathol. 2006;209:492–500. doi: 10.1002/path.1998. [DOI] [PubMed] [Google Scholar]
- 32.Badache A, De Vries GH. Neurofibrosarcoma-derived Schwann cells overexpress platelet-derived growth factor (PDGF) receptors and are induced to proliferate by PDGF BB. J Cell Physiol. 1998;177:334–342. doi: 10.1002/(SICI)1097-4652(199811)177:2<334::AID-JCP15>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 33.Badache A, Muja N, De Vries GH. Expression of Kit in neurofibromin-deficient human Schwann cells: role in Schwann cell hyperplasia associated with type 1 neurofibromatosis. Oncogene. 1998;17:795–800. doi: 10.1038/sj.onc.1201978. [DOI] [PubMed] [Google Scholar]
- 34.Keng VW, et al. A conditional transposon-based insertional mutagenesis screen for genes associated with mouse hepatocellular carcinoma. Nature biotechnology. 2009;27:264–74. doi: 10.1038/nbt.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dupuy AJ, et al. A modified sleeping beauty transposon system that can be used to model a wide variety of human cancers in mice. Cancer research. 2009;69:8150–6. doi: 10.1158/0008-5472.CAN-09-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu X, et al. Clonal selection drives genetic divergence of metastatic medulloblastoma. Nature. 2012;482:529–533. doi: 10.1038/nature10825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Quintana RM, et al. A Transposon-Based Analysis of Gene Mutations Related to Skin Cancer Development. J Invest Dermatol. 2012 doi: 10.1038/jid.2012.245. [DOI] [PubMed] [Google Scholar]
- 38.Lappe-Siefke C, et al. Disruption of Cnp1 uncouples oligodendroglial functions in axonal support and myelination. Nature genetics. 2003;33:366–74. doi: 10.1038/ng1095. [DOI] [PubMed] [Google Scholar]
- 39.Tabone-Eglinger S, et al. Frequent EGFR Positivity and Overexpression in High-Grade Areas of Human MPNSTs. Sarcoma. 2008;2008:849156. doi: 10.1155/2008/849156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Holtkamp N, et al. MMP-13 and p53 in the progression of malignant peripheral nerve sheath tumors. Neoplasia (New York, N Y. 2007;9:671–7. doi: 10.1593/neo.07304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ling BC, et al. Role for the epidermal growth factor receptor in neurofibromatosis-related peripheral nerve tumorigenesis. Cancer cell. 2005;7:65–75. doi: 10.1016/j.ccr.2004.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de Vries A, et al. Targeted point mutations of p53 lead to dominant-negative inhibition of wild-type p53 function. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:2948–53. doi: 10.1073/pnas.052713099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carli M, et al. Pediatric malignant peripheral nerve sheath tumor: the Italian and German soft tissue sarcoma cooperative group. Journal of Clinical Oncology. 2005;23:8422–8430. doi: 10.1200/JCO.2005.01.4886. [DOI] [PubMed] [Google Scholar]
- 44.Stucky CCH, et al. Malignant peripheral nerve sheath tumors (MPNST): the Mayo Clinic experience. Annals of surgical oncology. 2012;19:878–885. doi: 10.1245/s10434-011-1978-7. [DOI] [PubMed] [Google Scholar]
- 45.Pytel P, Taxy JB, Krausz T. Divergent differentiation in malignant soft tissue neoplasms: the paradigm of liposarcoma and malignant peripheral nerve sheath tumor. International journal of surgical pathology. 2005;13:19–28. doi: 10.1177/106689690501300103. [DOI] [PubMed] [Google Scholar]
- 46.Magro G, et al. Multinucleated floret-like giant cells in sporadic and NF1-associated neurofibromas: a clinicopathologic study of 94 cases. Virchows Arch. 456:71–6. doi: 10.1007/s00428-009-0859-y. [DOI] [PubMed] [Google Scholar]
- 47.Rodriguez FJ, Folpe AL, Giannini C, Perry A. Pathology of peripheral nerve sheath tumors: diagnostic overview and update on selected diagnostic problems. Acta Neuropathol. 2012:1–25. doi: 10.1007/s00401-012-0954-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Collier LS, Carlson CM, Ravimohan S, Dupuy AJ, Largaespada DA. Cancer gene discovery in solid tumours using transposon-based somatic mutagenesis in the mouse. Nature. 2005;436:272–6. doi: 10.1038/nature03681. [DOI] [PubMed] [Google Scholar]
- 49.Sarver A, Erdman J, Starr T, Largaespada D, Silverstein KA. TAPDANCE: An Automated tool to identify and annotate Transposon insertion CISs and associations between CISs from next generation sequence data. BMC Bioinformatics. 2012;13:154. doi: 10.1186/1471-2105-13-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brett BT, et al. Novel molecular and computational methods improve the accuracy of insertion site analysis in Sleeping Beauty-induced tumors. PloS one. 2011;6:e24668. doi: 10.1371/journal.pone.0024668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gregorian C, et al. PTEN dosage is essential for neurofibroma development and malignant transformation. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:19479–84. doi: 10.1073/pnas.0910398106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Largaespada DA, Collier LS. Transposon-mediated mutagenesis in somatic cells: identification of transposon-genomic DNA junctions. Methods in molecular biology (Clifton, N J. 2008;435:95–108. doi: 10.1007/978-1-59745-232-8_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Feber A, et al. Comparative methylome analysis of benign and malignant peripheral nerve sheath tumors. Genome Res. 2011;21:515–524. doi: 10.1101/gr.109678.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang J, et al. Genomic and molecular characterization of malignant peripheral nerve sheath tumor identifies the IGF1R pathway as a primary target for treatment. Clinical Cancer Research. 2011;17:7563–7573. doi: 10.1158/1078-0432.CCR-11-1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Forbes SA, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011;39:D945–D950. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ghadimi MP, et al. Targeting the PI3K/mTOR Axis, Alone and in Combination with Autophagy Blockade, for the Treatment of Malignant Peripheral Nerve Sheath Tumors. Molecular Cancer Therapeutics. 2012;11:1758–1769. doi: 10.1158/1535-7163.MCT-12-0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Saito T, et al. Nuclear< i> β-catenin correlates with cyclin D1 expression in spindle and pleomorphic sarcomas but not in synovial sarcoma. Hum Pathol. 2006;37:689–697. doi: 10.1016/j.humpath.2006.01.017. [DOI] [PubMed] [Google Scholar]
- 58.Mo W, et al. CXCR4/CXCL12 Mediate Autocrine Cell-Cycle Progression in NF1-Associated Malignant Peripheral Nerve Sheath Tumors. Cell. 2013 doi: 10.1016/j.cell.2013.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Keng VW, et al. PTEN and NF1 Inactivation in Schwann Cells Produces a Severe Phenotype in the Peripheral Nervous System That Promotes the Development and Malignant Progression of Peripheral Nerve Sheath Tumors. Cancer Res. 2012;72:3405–3413. doi: 10.1158/0008-5472.CAN-11-4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Starr TK, et al. A transposon-based genetic screen in mice identifies genes altered in colorectal cancer. Science (New York, N Y. 2009;323:1747–50. doi: 10.1126/science.1163040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jessen KR. Glial cells. Int J Biochem Cell Biol. 2004;36:1861–1867. doi: 10.1016/j.biocel.2004.02.023. [DOI] [PubMed] [Google Scholar]
- 62.Pérez-Mancera PA, et al. The deubiquitinase USP9X suppresses pancreatic ductal adenocarcinoma. Nature. 2012;486:266–270. doi: 10.1038/nature11114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Subramanian S, et al. Genome-wide transcriptome analyses reveal p53 inactivation mediated loss of miR-34a expression in malignant peripheral nerve sheath tumours. J Pathol. 2010;220:58–70. doi: 10.1002/path.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Je EM, An CH, Yoo NJ, Lee SH. Mutational analysis of PIK3CA, JAK2, BRAF, FOXL2, IDH1, AKT1 and EZH2 oncogenes in sarcomas. APMIS. 2012 doi: 10.1111/j.1600-0463.2012.02878.x. [DOI] [PubMed] [Google Scholar]
- 65.Jacks T, et al. Tumour predisposition in mice heterozygous for a targeted mutation in Nf1. Nat Genet. 1994;7:353–361. doi: 10.1038/ng0794-353. [DOI] [PubMed] [Google Scholar]
- 66.Cichowski K, et al. Mouse models of tumor development in neurofibromatosis type 1. Science (New York, N Y. 1999;286:2172–6. doi: 10.1126/science.286.5447.2172. [DOI] [PubMed] [Google Scholar]
- 67.Zhu Y, Ghosh P, Charnay P, Burns DK, Parada LF. Neurofibromas in NF1: Schwann cell origin and role of tumor environment. Science (New York, N Y. 2002;296:920–2. doi: 10.1126/science.1068452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wu J, et al. Plexiform and dermal neurofibromas and pigmentation are caused by Nf1 loss in desert hedgehog-expressing cells. Cancer Cell. 2008;13:105–116. doi: 10.1016/j.ccr.2007.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Katoh M. Identification and characterization of human FOXN6, mouse Foxn6, and rat Foxn6 genes in silico. Int J Oncol. 2004;25:219. [PubMed] [Google Scholar]
- 70.Santo E, et al. Oncogenic activation of FOXR1 by 11q23 intrachromosomal deletion-fusions in neuroblastoma. Oncogene. 2011;31:1571–1581. doi: 10.1038/onc.2011.344. [DOI] [PubMed] [Google Scholar]
- 71.Uhlen M, et al. Towards a knowledge-based human protein atlas. Nat Biotechnol. 2010;28:1248–1250. doi: 10.1038/nbt1210-1248. [DOI] [PubMed] [Google Scholar]
- 72.Rhodes DR, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia (New York, NY) 2004;6:1. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rhodes DR, et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia (New York, NY) 2007;9:166. doi: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Myatt SS, Lam EWF. The emerging roles of forkhead box (Fox) proteins in cancer. Nature Reviews Cancer. 2007;7:847–859. doi: 10.1038/nrc2223. [DOI] [PubMed] [Google Scholar]
- 75.Stemmer-Rachamimov AO, et al. Comparative pathology of nerve sheath tumors in mouse models and humans. Cancer Res. 2004;64:3718–3724. doi: 10.1158/0008-5472.CAN-03-4079. [DOI] [PubMed] [Google Scholar]
- 76.Dai M, et al. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 2005;33:e175–e175. doi: 10.1093/nar/gni179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B (Methodological) 1995:289–300. [Google Scholar]
- 78.Irizarry RA, et al. The human colon cancer methylome shows similar hypo-and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41:178–186. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yang J, et al. Deletion of the< i> WWOX gene and frequent loss of its protein expression in human osteosarcoma. Cancer Lett. 2010;291:31–38. doi: 10.1016/j.canlet.2009.09.018. [DOI] [PubMed] [Google Scholar]
- 80.Olshen AB, Venkatraman E, Lucito R, Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5:557–572. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- 81.Van De Wiel MA, et al. CGHcall: calling aberrations for array CGH tumor profiles. Bioinformatics. 2007;23:892–894. doi: 10.1093/bioinformatics/btm030. [DOI] [PubMed] [Google Scholar]
- 82.Sun W, et al. Integrated study of copy number states and genotype calls using high-density SNP arrays. Nucleic Acids Res. 2009;37:5365–5377. doi: 10.1093/nar/gkp493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.DeRycke MS, et al. S100A1 expression in ovarian and endometrial endometrioid carcinomas is a prognostic indicator of relapse-free survival. Am J Clin Pathol. 2009;132:846–856. doi: 10.1309/AJCPTK87EMMIKPFS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rizzardi A, et al. Quantitative comparison of immunohistochemical staining measured by digital image analysis versus pathologist visual scoring. Diagnostic Pathology. 2012;7:42. doi: 10.1186/1746-1596-7-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Daniel AR, Faivre EJ, Lange CA. Phosphorylation-dependent antagonism of sumoylation derepresses progesterone receptor action in breast cancer cells. Molecular Endocrinology. 2007;21:2890–2906. doi: 10.1210/me.2007-0248. [DOI] [PubMed] [Google Scholar]
- 86.Cermak T, et al. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011;39:e82–e82. doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wood AJ, et al. Targeted genome editing across species using ZFNs and TALENs. Science. 2011;333:307–307. doi: 10.1126/science.1207773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mussolino C, et al. A novel TALE nuclease scaffold enables high genome editing activity in combination with low toxicity. Nucleic Acids Res. 2011;39:9283–9293. doi: 10.1093/nar/gkr597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Miller JC, et al. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol. 2010;29:143–148. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- 90.Guschin DY, et al. A rapid and general assay for monitoring endogenous gene modification. Methods Mol Biol. 2010;649:247–256. doi: 10.1007/978-1-60761-753-2_15. [DOI] [PubMed] [Google Scholar]
- 91.Doyon Y, et al. Transient cold shock enhances zinc-finger nuclease–mediated gene disruption. Nature methods. 2010;7:459–460. doi: 10.1038/nmeth.1456. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.