Abstract
Homocysteine (Hcy) is a thiol-containing amino acid formed during methionine metabolism. Elevated level of Hcy is known as hyperhomocysteinemia (HHcy). HHcy is an independent risk factor for cerebrovascular diseases such as stroke, dementia, Alzheimer’s disease, etc. Stroke, which is caused by interruption of blood supply to the brain, is one of the leading causes of death and disability in a number of people worldwide. The HHcy causes an increased carotid artery plaque that may lead to ischemic stroke but the mechanism is currently not well understood. Though mutations or polymorphisms in the key genes of Hcy metabolism pathway have been well elucidated in stroke, emerging evidences suggested epigenetic mechanisms equally play an important role in stroke development such as DNA methylation, chromatin remodeling, RNA editing, noncoding RNAs (ncRNAs), and microRNAs (miRNAs). However, there is no review available yet that describes the role of genetics and epigenetics during HHcy in stroke. The current review highlights the role of genetics and epigenetics in stroke during HHcy and the role of epigenetics in its therapeutics. The review also highlights possible epigenetic mechanisms, potential therapeutic molecules, putative challenges, and approaches to deal with stroke during HHcy.
Keywords: Brain, CBS, Cerebrovascular diseases, Chromatin remodeling, DNA methylation, Epigenomics, MTHFR, Noncoding RNA, Vascular diseases
Introduction
Stroke is a medical emergency caused by interrupted blood supply to the brain that further leads to rapid loss of brain functions. It is the second leading cause of death worldwide and associated with long-term disability [1]. Stroke can be classified into ischemic (blockage of blood supply) and hemorrhagic (rupture of blood vessel). Thrombolytic agents along with antiplatelet drugs have been used for stroke treatment; however, some cases benefited with neurosurgery. Besides other factors (high blood pressure, diabetes, previous stroke or transient ischemic attack (TIA), high cholesterol, tobacco smoking, and arterial fibrillation), homocysteine (Hcy) is also now largely accepted as a risk factor for stroke [2–7]. Increase in the levels of Hcy, a sulfur-containing amino acid derived from methionine, is known as hyperhomocysteinemia (HHcy) that is found to be associated with more than fivefold increased risk of stroke [3, 8]. HHcy is most commonly associated with large-artery atherosclerosis and venous thrombosis, but it also contributes to cerebral ischemia stroke by other mechanisms as, for example, involvement of genetic and epigenetic factors [9–13]. The genetic regulation of enzymes involved in Hcy metabolism and the levels of vitamin cofactors (folate, B6, and B12) determine the level of Hcy [14]. Evidences suggest that genetic variations in these pathway genes such as methylenetetrahydrofolate reductase (MTHFR), cystathionine-β-synthase (CBS), nicotinamide N-methyl-transferase (NNMT), and DNA methyltransferase (DNMT) increase the risk of stroke during HHcy [11, 15–17]. Clinical trials for lowering Hcy levels by vitamin interventions such as folate, pyridoxine (vitamin B6), and cobalamin (vitamin B12) have failed to reduce the risk of recurrent stroke. However, there are ongoing clinical trials evaluating the potential benefits of vitamin supplementation. Apart from genetic mechanisms, there are epigenetic mechanisms that regulate stroke and could be further helpful in HHcy-induced stroke [12, 18, 19]. Epigenetic mechanisms refer to molecular processes such as DNA methylation, histone modification, nucleosome remodeling, and small noncoding RNAs (ncRNAs) (e.g., miRNAs) that modulate gene expression and function within the cell of the body. One example is miR-140 that helps in tissue repair after cerebral ischemia [34]. Understanding genetic and epigenetic mechanisms may be helpful in discovering sensitive biomarkers and determining therapeutic approaches in HHcy-induced stroke. Therapeutic approaches may include inhibition of DNMTs and histone deacetylase (HDAC) activities with potential epigenetic agents (drugs, dietary components, and miRNAs) that selectively activate or inactivate gene expression. Though the therapeutic approaches are still in their infancy, preliminary results suggest that targeting DNMTs and histone-modifying agents can promote brain repair and functional reorganization in stroke [20]. This review, for the first time, suggests the role of epigenetics, microRNA (miRNA), and ncRNA in modulating gene expression in stroke and possible utilization of epigenetic mechanisms in HHcy-induced stroke and its therapeutics.
Factors Involved in Hyperhomocysteinemia During Stroke
Genetic Factors
Earlier studies have indicated that the metabolism of Hcy is regulated by several genes and mutations in these genes affect Hcy level that is the risk factor for stroke. Hence, we have included polymorphism studies to take into consideration genetic factors in stroke. Individual polymorphisms need not be targeted and clinical trials are still in their preliminary phase. Hcy is metabolized by two pathways: (1) remethylation and (2) transsulfuration (Fig. 1). Protein-bound Hcy and mixed Hcy disulfides are collectively referred as total Hcy (tHcy). They are spontaneously formed in the plasma and are measured in most assays [21]. Many studies have evaluated the role of Hcy in atherosclerosis through endothelial injury, platelet activation, smooth muscle proliferation, oxidative modification of low density lipoproteins, and endothelial leukocyte interactions [13]. However, there are few studies that elucidate genetic mechanisms behind stroke. Mutations in the genes of Hcy metabolic pathway may lead to elevated Hcy levels that result in increased risk for stroke. Some of the important molecules have been discussed here.
Fig. 1.
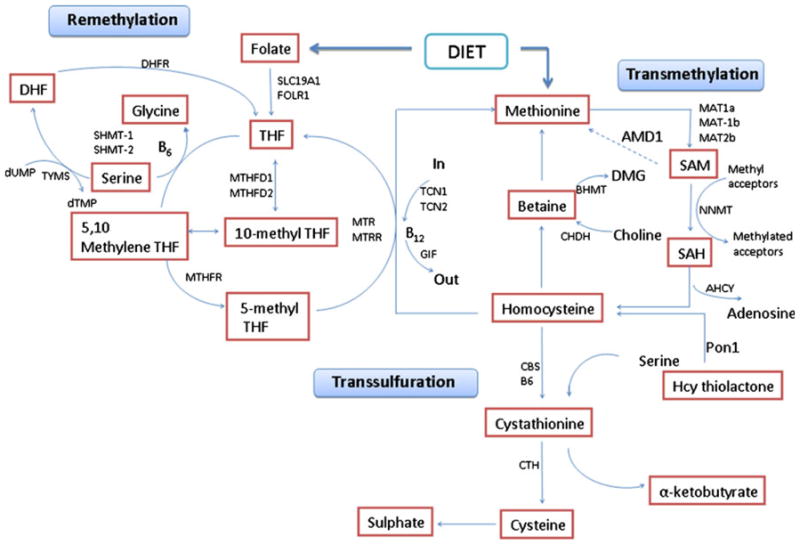
Genes, pathways, and the mechanisms of homocysteine metabolism
Methylenetetrahydrofolate Reductase
MTHFR is a key enzyme in the synthesis of 5-methyl tetrafolate that serves as a cofactor in the synthesis of methionine from Hcy by methionine synthase (Fig. 1). The human MTHFR gene is located on chromosome 1p36.3. A single base pair substitution (677 C→T) results in the conversion of alanine to valine at amino acid 222. This variation results in reduced enzyme activity and elevation of Hcy in the homozygous state TT. Many studies have reported that 677C>T MTHFR is associated with elevated plasma Hcy levels and increased stroke risk [15, 22]. A study by Zhang et al. [23] demonstrates that high Hcy levels are associated with risk of stroke recurrence and cause mortality in stroke patients, and MTHFR C677T was not associated with stroke [23]. However, a meta-analysis has also confirmed the association of plasma Hcy levels (1.93 μmol/L) with increased risk of stroke (odds ratio 1.26 for TT vs. CC) [10].
Cystathionine-β-Synthase
CBS enzyme is involved in the conversion of Hcy to cystathionine in the presence of vitamin B6. The CBS gene is located on chromosome 21q22.3. More than 100 mutations have been identified in this gene and the most frequently detected is 833C→T that results in amino acid change of isoleucine to threonine at 278 positions. Another mutation is 919G→A substitution that results in amino acid change of glycine to serine at amino acid 307. Both these mutations are present in exon 8. There is a 68-bp insertion in exon 8 at position 844. There is also a 31-bp variable number tandem repeat (VNTR) that spans the exon 13–intron 13 boundary of the CBS gene and may consist of 16, 17, 20, and 19 or 21 repeat units. The increase in repeat units results in progressively higher plasma Hcy levels. Several studies have correlated these polymorphisms with stroke [9, 11].
Methoinine Synthase
Methoinine synthase (MS) along with MTHFR sequentially catalyzes the remethylation of Hcy to methionine and deficiency in its activity results in HHcy. The gene is mapped to chromosome 1q43. The polymorphism 2756A→G results in a change of aspartic acid to glycine D919G and has been reported as a genetic risk factor for thromboembolic events [24]. Mutation in this gene results in increased enzyme activity and trapping of methyl-THF.
Other Genes
The first genomewide linkage analysis performed in Spanish population showed that the nicotinamide N-methyltransferase (NNMT) gene is the most significant genetic determinant of plasma Hcy [17]. Low et al. [14] have reported that Hcy pathway genes such as 5-methyltetrahydrofolate-Hcy methyltransferase reductase (MTRR), serine hydroxymethyltransferase 1 (SHMT1), and transcobalamin II (TCN2) are associated with stroke risk (P< 0.05). MTRR and SHMT1 are involved in regulating folate cycle and TCN2 is involved in transportation of vitamin B12 [14]. The cellular availability of folate and vitamin B12 is important in determining stroke risk. The Hcy levels in plasma are inversely related to plasma concentrations of vitamin B12 as well as the intake of these vitamins [25]. The deficiency of both folate and vitamin B12 levels either caused by low dietary intake or a combination of genetic variants of SHMT1, MTRR, and TCN2 may increase stroke risk.
Overall, there are very few publications that evaluate the role of Hcy metabolic genes in stroke, and further studies are needed with other important genes of Hcy metabolism as shown in Table 1. The genetic polymorphism studies reported in case of stroke during HHcy is also summarized in Table 2.
Table 1.
Genes involved in homocysteine metabolism pathway
| Gene symbols | Names |
|---|---|
| AHCY | S-Adenosylhomocysteine hydrolase |
| AMD1 | S-Adenosylmethionine decarboxylase 1 |
| BHMT | Betaine-homocysteine methyltransferase |
| CBS | Cystathionine-beta-synthase |
| CHDH | Choline dehydrogenase |
| CTH | Cystathionase |
| DHFR | Dihydrofolate reductase |
| FOLR1 | Folate receptor 1 |
| GIF | Gastric intrinsic factor |
| MAT1a | Methionine adenosyltransferase I, alpha |
| MAT2a | Methionine adenosyltransferase I, alpha |
| MAT2b | Methionine adenosyltransferase II, beta |
| MTHFD1 | Methylenetetrahydrofolate dehydrogenase 1 |
| MTHFD2 | Methylenetetrahydrofolate dehydrogenase 2 |
| MTHFR | 5,10-Methylenetetrahydrofolate dehydrogenase |
| MTR | 5-Methyltetrahydrofolate-homocysteine methyltransferase |
| MTRR | 5-Methyltetrahydrofolate-homocysteine methyltransferase reductase |
| NNMT | Nicotinamide N-methyltransferase |
| PON1 | Paraoxonase 1 |
| SHMT1 | Serine hydroxymethyltransferase 1 |
| SHMT2 | Serine hydroxymethyltransferase 2 |
| SLC19A1 | Solute carrier family 19 member 1 |
| TCN1 | Transcobalamin I |
| TCN2 | Transcobalamin II |
| TYMS | Thymidylate synthase |
Table 2.
Genetic polymorphism studies involved in stroke related to homocysteine metabolism pathway
| S. no. | Gene | Mutation | Significance in stroke | Odds ratio (95 % CI) | Reference |
|---|---|---|---|---|---|
| 1 | MTHFR (1p36.3) | 677 C→T | Associated with increased Hcy levels and stroke risk | 1.26 (1.14–1.40) | [10, 15, 22] |
| 2 | CBS (21q22.3) | 833 C→T 919G→A |
C833T associated with higher plasma Hcy levels and correlated with stroke | 1.57 (1.02–2.41) | [9, 11] |
| 3 | MS (1q43) | 2,756 A→G | Associated with HHcy and risk factor for thromboembolic events | 1.58 | [24] |
| 4 | MTRR (5p15.31) | rs16879248 (T/C) | Associated with Hcy metabolism and ischemia stroke risk | 0.75 (0.61–0.91) | [14] |
| 5 | SHMT1 (17p11) | rs11868708 (T/C) | Associated with Hcy metabolism and ischemia stroke risk | 1.24 (1.05–1.46) | [14] |
| 6 | TCN2 (22q12) | rs11703570 (C/T) | Associated with Hcy metabolism and ischemia stroke risk | 1.28 (1.04–1.57) | [14] |
| 7 | NNMT (11q23) | rs694539 (A/G) | Influences plasma Hcy levels: the GAIT project | – | [17] |
Epigenetic Mechanisms in Stroke During Hyperhomocysteinemia
Epigenetics mechanisms regulate gene expression and functional networks and also encompass gene environment interactions [26]. Epigenetic mechanisms include DNA methylation, histone code modifications, nucleosome remodeling, higher order chromatin formation, ncRNA including miRNA, and RNA editing. These changes help in maintaining cellular homeostasis, controlling normal development, and mediating response to external stimuli [27]. In earlier literature, Qureshi and Mehler [28] have described epigenetic mechanisms in stroke [28]. The emerging evidences show that several epigenetic mechanisms are involved in stroke pathogenesis and suggest that understanding these processes may be critical for early diagnosis, risk assessment, and promotion of tissue repair and functional reorganization. Epigenetics therapeutic agents could be applied in stroke therapy and also provide a platform for targeting activation of neural stem cells in the brain that promote regenerative processes for restoring brain functions.
Role of DNA Methylation
DNA methylation plays a vital role in epigenetic mechanisms by regulating gene expression profiles. Abnormal DNA methylations have been reported to be associated with stroke as well as other disorders such as atherosclerosis, obesity, kidney disease, cancer, insulin resistance, and auto-immunity [16, 29]. DNA methylation inhibits the process of transcription by binding and recruiting epigenetic factors to the methylated sites and mediating reversible gene silencing. DNA methylation is the transfer of methyl group from an activated donor such as S-adenosylmethionine (SAM) to cytosine residues in specific regions of the gene by DNMTs. It is a gene family and consists of members DNMT 3a and DNMT 3b (de novo methylation), and DNMT 1 (maintains methylation). Hcy metabolism plays an important role in DNA methylation and HHcy has been reported to be associated with stroke [3, 16]. High levels of Hcy lead to increase in the levels of SAM and higher activity of DNMTs. This leads to hypermethylation of DNA and eventually, gene silencing. Many studies have shown that low folic acid and vitamin levels (B6 and B12) in daily diet cause an elevation in Hcy level that may lead to diseases like stroke [25]. Collectively, these studies suggest that altered DNA methylation levels due to Hcy or key enzymes of Hcy metabolism lead to vulnerability of the brain to ischemic injury. The fact that increase in DNA methylation levels leads to cerebral ischemic stroke and may be responsible for promoting cell death is explained by middle cerebral artery occlusion (MCAO) experiments [30–33]. Following MCAO in mice, DNA methylation levels are increased in ischemic brain tissue that might be responsible for promoting cell death. One of these studies demonstrates that animals heterozygous for the conditional allele (Dnmt11lox/+) have significantly smaller infarcts following 1-h MCAO/reperfusion compared to their wild type litters. Mice with a deletion of Dnmt1 in postmitotic neurons (Dnmt11lox/c) were not protected. Thus, reduced levels of Dnmt1, but not its absence, in postmitotic neurons protect from ischemic brain injury [31]. Also, in MCAO mice, treatment with methylation inhibitor reduces extent of ischemic injury [34]. Besides, there are other factors that cause DNA methylation such as X chromosome inactivation and genomic imprinting that include Fabry disease, Prader–Willi syndrome, and imprinted GNAS genomic locus [35–39].
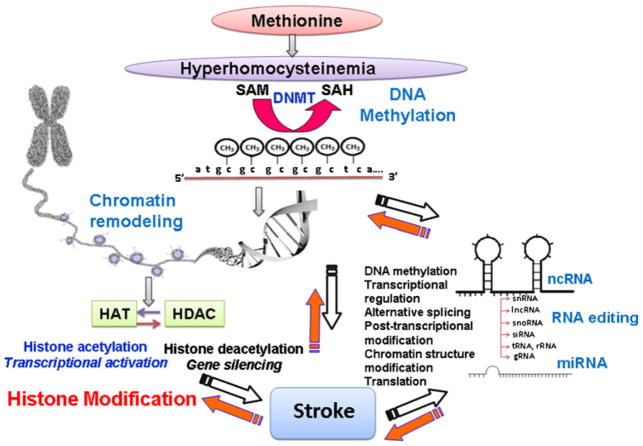
DNA methylation that is mediated by many factors further leads to regulation of specific genes and plays an important role in pathophysiology of stroke. As an example, DNA methylation influences the expression of thrombospondin 1 (THBS1), an important gene that plays a role in response to cerebral ischemia and intracerebral hemorrhage [18, 19]. In murine cerebral endothelial cells, deprivation of oxygen and glucose in vitro is associated with decrease in THBS1 expression due to promoter methylation. This review selectively highlighted the role of Hcy in DNA methylation and stroke pathophysiology although DNA methylation has also been implicated in modulation of homeostasis, cell cycle dynamics, cell viability, and vascular and endothelia stress responses that are related to stroke (Fig. 2).
Fig. 2.
Epigenetic mechanisms that could be involved in hyperhomocysteinemia-induced stroke. These mechanisms also control genes of homocysteine pathway that leads to hyperhomocysteinemia. SAM S-adenosylmethionine, SAH S-adenosylhomocysteine, ncRNA noncoding RNA, HAT histone acetyltransferase, HDAC histone deacetylase, miRNA microRNA, DNMT DNA methyltransferase, snRNA small nuclear RNA, lncRNA long noncoding RNA, snoRNA small nucleolar RNA, siRNA small interfering RNA, tRNA transfer RNA, rRNA ribosomal RNA, gRNA guide RNA
Histone Modification and Chromatin Remodeling
Abnormal chromatin is representative of apoptotic and necrotic cell death that are associated with neural injury in stroke. The roles of chromatin remodeling are not well characterized in cerebral ischemia, though increasing evidences suggest that these functions are extremely important. Also, the chromatin-modifying agents {histone acetyltransferase (HAT) and HDAC and their inhibitors (sirtuins (SIRTs), histone methyltransferase trichostatin A, sodium butyrate, and sodium-4 phenyl butyrate)} may be neuroprotective [20]. Histone modifications such as acetylation, methylation, phosphorylation, ubiquitination, and adenosine diphosphate ribosylation are responsible for modulating chromosome structure and function at many genomic regions [40–42]. Histone modifications are catalyzed by specific enzymes such as HAT and HDAC. Histone modification is a major event that leads to repression or activation of transcription machinery and chromatin remodeling [41]. Classical histone proteins (H2A, H2B, H3, and H4), linker histones [H1], and variant histones (H2A.Z) are wrapped around DNA to form a histone core called nucleosome, which is a basic unit of chromatin. Histone-modifying enzymes modify the conformation of DNA that leads to repositioning of nucleosomes. This alters the binding of transcription factors and other regulatory proteins to genomic regions and modulates expression profiles that lead to chromatin remodeling [40]. Chromatin is further characterized into euchromatin, which is open and functionally active, and heterochromatin, which is more densely packed and inactive [42].
Histone nucleosome and chromatin modifications comprise epigenetic mechanisms and affect genome structure and functions that are critical in understanding the molecular pathophysiology of stroke. Atherosclerosis, which is a major factor in determining stroke risk, is affected by modulation of cholesterol biosynthesis and transport pathways by chromatin-modifying enzymes [43–45]. SIRTs are HDAC enzymes and play an important role in stroke by mediating diverse functions such as cellular stress resistance, energy metabolism, and genomic stability [46]. Many studies have shown that inhibition of HDAC enzymes in stroke models decreases neuronal injury and improves functional outcomes [47]. Histone acetylation has a role in protecting neurons against oxidative damage by enhancing neuroprotective antioxidant enzymes, e.g., peroxiredoxins [48]. In view of the above points, it is evident that histone modifications play a significant role in stroke pathophysiology and understanding epigenetic mechanisms will be helpful in targeting epigenetic events that could further be used in HHcy-induced stroke therapeutics (Fig. 2).
MicroRNA in Stroke: A Class of Noncoding RNAs
ncRNAs are derived from nonprotein coding sequences that were earlier thought to be junk DNA and constitute more than 98 % of the genome [49]. These ncRNAs have been shown to have diverse functions in DNA methylation, transcriptional regulation, alternative splicing, post-transcriptional modification, chromatin structure modification, and translation [50]. miRNA is the subclass of ncRNAs that regulate the expression of genes involved in development and homeostatic processes. These processes are implicated in neural differentiation, maintenance, and plasticity [51]. The miRNAs are 20–22 nucleotides long, processed from stem loop pre-miRNAs to mature miRNAs, and regulate a large number of target genes by binding to the 3′ region of their corresponding messenger RNA (mRNA). The role of miRNAs have been studied in animal models of cerebral ischemia and found to be associated with postischemic cerebral regeneration [12, 52]. These miRNAs bind to their corresponding target mRNAs and modulate transcription, stress responses, ionic flux, and inflammation. In MCAO mice, for example, miRNA-140 is one of the miRNAs that are upregulated in the brain after 3 h of MCAO. The miRNA-140 regulates stromal cell-derived factor 1, which mediates tissue repair after cerebral ischemia by vascular progenitor cell proliferation and migration [34]. The studies suggest that miRNA-140 may be responsible for regeneration responses in postischemic brain. Some miRNAs are significantly detected in peripheral blood such as: rno-miR-19b, rno-miR-290, and rno-miR-292-5p, after 24 h of MCAO and rno-miR-352, rno-miR-26b, rno-miR-26a, rno-miR-20a, rno-miR-17, rno-miR-140, rno-miR-92, rno-miR-214, rno-miR-15b, and rno-miR-328 after 48 h of MCAO, and suggest that they may serve as future biomarkers for ischemic stroke [12] in addition to regulating systemic responses to cerebral ischemia.
Apart from miRNAs, long ncRNAs (lncRNA) that are a subclass of ncRNAs play an important role in stroke by regulating transcriptional modification and chromatin remodeling [53]. One of the lncRNAs binds to the cyclin D1 gene that is a critical mediator of ischemia neuronal cell death [54]. The translocated in liposarcoma (TLS) RNA-binding protein then binds to cyclin D1 gene to repress the transcription of the gene [55]. Another lncRNA, ANRIL (NCBI ID 100048912), is associated with the development of atherosclerosis through smooth muscle proliferation and migration [56–62].
In mouse brain, some repeated mobile genetic elements (e.g., retroposons) are increased in cerebral ischemia that represents another class of ncRNA apart from miRNA and lncRNA [63]. These ncRNA transcripts resemble viruslike 30 families of interspersed genetic elements and are induced by cerebral ischemia. The above points conclude that ncRNAs that also include miRNA and lncRNAs are important in pathogenesis of stroke and represent an area to be explored further in HHcy-induced stroke (Fig. 2).
RNA Editing in Stroke
RNA editing is the process in which nucleotides in the RNA molecule are changed or edited that results in the generation of transcript diversity [27]. Many studies have associated RNA editing abnormalities with stroke and neurodegenerative diseases [27]. The miRNAs that bind to target mRNAs and regulate their function are themselves altered by RNA editing that changes their processing stability and dynamics [64, 65]. Hence, RNA editing in miRNAs regulates their function that may play a critical role in stroke. For example, the immature form of miRNA-151 is subject to RNA editing and it affects processing of primary miRNA into mature miRNA in the brain [66]. The miRNA-151 is found in neurons and is upregulated following MCAO; hence, it is associated with stroke [66]. The miRNA-151 is predicted to bind to target cell cycle regulators and tyrosine kinase 2 protein (focal adhesive kinase). It is a nonreceptor tyrosine kinase and plays a role in integrin and growth factor signaling pathways that are differentially regulated in MCAO. Hence, these miRNAs are important in modulating neurite outgrowth and restoring neural network integrity postcerebral ischemia [67–69]. These findings are suggestive of miRNA regulatory mechanisms through RNA editing influencing important mechanisms in stroke and could further be checked in HHcy-induced stroke (Fig. 2).
Epigenetic Regulators and MicroRNAs: An Era of Epigenetic Therapy for HHcy-Induced Stroke
For the treatment of stroke, the US Food and Drug Administration has approved some drugs that target DNA methylation and histone modification molecules, which are promising for stroke in HHcy as well as in hematological, immunological, and neuropsychiatric diseases. Epigenetic agents (DNMT and HDAC modulators), which may be beneficial in stroke, are still in their preliminary stages and are being evaluated and summarized in Table 3. The commonly used Aza drugs, 5-azacytidine, 5-aza-2-deoxycytidine (decetabine), and zebularine, are potent DNMT inhibitors by acting as analogs of cytosine. Adipose stromal cells are induced by factors like 5-azacytidine to differentiate into neuronlike cells and are termed as epigenetically engineered cells. These are transplanted into an animal model of stroke and have led to improved neurological outcomes and are used in cellular therapies [70, 71]. In other studies, animals with MCAO have shown improved neurological consequences and reduction in ischemic injury upon treatment with 5-aza-deoxycytidine. In comparison to the transplantation studies, the in vivo drug treatments have shown better results, suggesting it to be a better strategy than transplantation of exogenous cells [30]. However, these inhibitors are nonspecific for DNMT and cause toxicity; therefore, second-generation agents that specifically inhibit DNMT are under development [72]. As an example, MG98 specifically targets DNMT1 mRNA and inhibits its translation as it is an antisense oligonucleotide [72]. Many other drugs such as valproic acid, hydralazine, and procainamide exhibit a mode of action that affects DNA methylation [73]. One of these drugs, valproic acid (valproate), is a known drug and believed to act on GABAergic neurons. Since valproate action does not require cell division, it potentially affects the epigenomic status of genes in mature postmitotic cells such as central nervous system (CNS) neurons. It has been shown that it acts on epigenetic reprogramming that involves demethylation of specific genes [74] and HDAC inhibition that can alter chromatin structure by increasing histone acetylation [75]. There is an adverse condition called systemic lupus erythematous that is also found to be associated with global DNA hypomethylation and cerebrovascular complications [76] and is provoked by some epigenetic agents (for instance, procainamide and hydralazines) that cause DNA hypomethylation.
Table 3.
Epigenetic agents in stroke therapeutics
| S. No. | Mechanism | Significance in stroke | Epigenetic agents | Structure | Brand name | Reference |
|---|---|---|---|---|---|---|
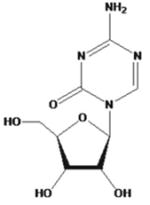
| 1 | DNMT inhibition, acting as analog of cytosine | DNMT inhibition reduces DNA methylation reported in ischemia stroke brain injury after MCAO, improved neurological functions | 5-azacytidine |
|
Vidaza | 34, 42, 53, 61, 79, 10 |
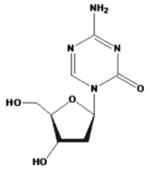
| 5-aza-2-deoxycytidine (5-Aza-2dc or decitabine) |
|
Dacogen | ||||
| zebularine |
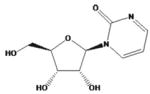
|
Not assigned | ||||
| hydralazine |
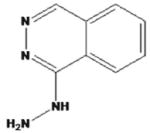
|
Nepresol Zinepress | ||||
| procainamide |
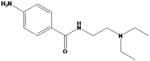
|
Procanbid Pronestyl Procan | ||||
| 2. | HDAC inhibition, modify histones | known to decrease neuronal injury and improve regeneration, provide neuroprotection in cerebral ischemia stroke | trichostatin A |
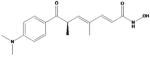
|
Trichostatin A TSA | 46, 59, 53, 61, 10, 41, 46, 59 |
| suberoylanilide hydroxamic acid |
|
Vorinostat Zolinza | ||||
| sodium butyrate |
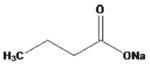
|
CM3000 | ||||
| sodium 4-phenylbutyrate |
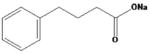
|
Buphenyl | ||||
| valproic acid |
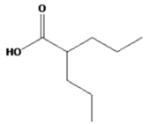
|
Depakene Depakote Depakote ER | ||||
| curcumin |
|
Cavamax® W8 Curcumin | ||||
| resveratrol |
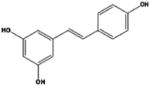
|
Longevinex |
In addition to DNA methylation, drugs that affect histone modifications are also used such as HDAC inhibitors (trichostatin A, sodium butyrate, sodium-4 phenyl butyrate, and suberoylanilide hydroxamic acid). These drugs are known to decrease neuronal injury and improve regeneration [20]. The activities of these inhibitors vary according to different classes of HDAC. There are some dietary components, such as curcumin (turmeric) and resveratrol (grapes, berries, and peanuts), which are found to be having HDAC inhibitory activity and have been reported to provide neuroprotection in cerebral ischemia stroke [77, 78]. The beneficial effects induced by inhibiting HDAC include better stress response (nitric oxide synthase, heat shock protein 70, and TNFα), growth, and viability [20]. Apart from DNA methylation and HDAC, RNA-based therapies are also promising that use miRNA, lncRNA, small interfering RNA (siRNA), e.g., Bcl-2 and 19-kDa interacting protein 3 in a rat model of stroke along with cultured primary neurons exposed to hypoxia [79], and carboxyl terminal modulator protein in Sprague–Dawley rat ischemia stroke models [80]. The above studies proved siRNA and miRNA importance in brain dysfunction (ischemic stroke and dementia). However, these approaches are yet in their preliminary stage and yet to be validated and evaluated. Two important epigenetic factors repressive element-1 silencing transcription factor (REST) and corepressor for element-1 silencing transcription factor (CoREST) are involved in gene expression, gene activation, and silencing of a large number of genes implicated in neuronal development and regeneration [81] (Fig. 3). These factors regulate the expression of many ncRNAs that include miRNA and lncRNA, e.g., repression of GluR2 subunit and μ-opioid receptor 1 [82, 83]. Such agents that specifically bind to miRNA and regulate their activity in stroke are in their early stage of development [84]. These agents bind to complexes that comprise miRNAs, ncRNAs, and transcription factors and may be termed as RNA operons and the mechanisms regulating RNA operons may be termed regulons. Epigenetic mechanisms also include transport of RNA between adjacent nerve cells, which represents novel targets for therapeutic approaches [85, 86]. However, these therapeutic approaches could also be checked in HHcy-induced stroke as depicted in Fig. 3.
Fig. 3.
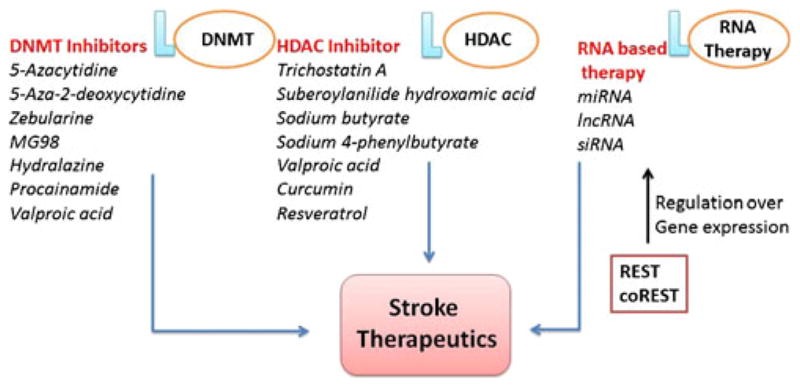
Epigenetic potential agents involved in stroke therapeutics. DNMT DNA methyltransferase, HDAC histone deacetylase, REST repressive element-1 silencing transcription factor, CoREST corepressor for element-1 silencing transcription factor, miRNA microRNA, siRNA small interfering RNA, lncRNA long noncoding RNA
Future Perspectives, Challenges, and Conclusion
The future of epigenomics is fascinating, since it could lead to uncovering of different ways to treat brain disorders including HHcy-induced stroke. The epigenetic studies could also direct the individuals to determine their “at risk” status for different brain diseases and in that regard, the science could also encourage lifestyle changes that can further prevent the wrong genes from switching on or off. Not only in therapeutics, epigenetic approaches can also be looked in order to determine sensitive and specific biomarker for stroke in HHcy. The epigenetic changes could be first determined/screened in the test samples using epigenetic profiling such as DNA methylation profiling, histone modification profiling, and miRNA profiling. The screened molecules could then be validated in terms of specificity and sensitivity using molecular techniques (for example, sequencing, quantitative real-time PCR (qRT-PCR), chromatin immunoprecipitation quantitative PCR (ChIP-qPCR), and miRNA arrays) and can give birth to a new biomarker by correlating their use with clinical outcomes.
The genetic and epigenetic processes are intricately connected in driving the development of brain dysfunction including stroke during HHcy. Though the current approaches using genetic and epigenetic processes are promising, there are several limitations that need attention and exploration in HHcy-induced stroke. The major concerns are relating polymorphisms in Hcy pathway genes with epigenetic mechanisms and identifying of HHcy-related epigenomic patterns that feature transcriptional output and consequences into stroke. There are several challenges in determining specific epigenetic patterns in HHcy-induced stroke and their use as prognostic factors
Another important limitation in HHcy-induced stroke is with the therapeutic approaches with epigenetic-modulating agents, for example, their use, delivery issues, concern with maintenance of a pharmacodynamic response, and achievement of a therapeutic index. A further limitation is associated with determining the role of drug in epigenetic mechanisms (methylation or histone modification), issues concerned with specificity and sensitivity, and their role in establishing prediction of response to the treatment. There are several other questions related to epigenetic agents as well, for instance, with demethylating agents that selectively target replicating cells and clinical outcome of treatment with demethylating agents may be dependent on the specific profile of methylated genes. The toxicity of epigenetic modulators is one more issue that limits their efficacy.
A new group of molecules called “exosomes” are microvesicles that carry miRNA, ncRNA, and proteins and are secreted in local or peripheral circulations [87]. Exosomes also carry regulatory proteins on their surface and signaling molecules for recognition and uptake, and mediate cell to cell transfer of RNA molecules. Exosomes can be manipulated for carrying target RNAs (siRNA/miRNA), expressing signaling molecules on their surface, or carrying drug molecules inside them for the treatment of neurodegeneration, HHcy-induced stroke therapeutics, neuronal plasticity, and expressing as biomarkers in HHcy-induced stroke. Exosomes are potential promising agents in order to determine the therapeutic potential of RNA drugs and for efficient, tissue-specific, and nonimmunogenic delivery of the epigenetic drugs and siRNA/miRNA. A recent report confirmed a similar approach for delivering siRNA to the mouse brain by systemic injection of targeted exosomes [88]. In another study, exosomes have been used to deliver therapeutic mRNA/protein for treatment of cancer [89]. However, there are very few reports and this area still needs to be explored. Hence, taken together, studying epigenetic mechanisms reveals epigenetic agents that can be employed for modulating DNA methylation, histone modification, and regulating RNA molecules to favor neuronal mechanisms in stroke during HHcy.
In the present review, we discussed the genetic and epigenetic role in stroke during HHcy. The high Hcy levels lead to alteration of the activity of DNMTs that consequently causes hypermethylation of the genes and gene silencing. This further affects the genes that play an important role in pathophysiology of stroke. The process also affects the expression of histone modification genes and ncRNA including miRNA. Hence, we highlight the role of Hcy-mediated epigenetic changes and possible mechanisms in stroke since they are poorly explored. Hcy levels that are regulated by many genes may lead to stroke causing epigenetic modifications and potential epigenetic agents/drugs or RNAs that modulate epigenetic changes can be used as therapeutics. In our opinion, epigenetic approaches in HHcy-induced stroke are very promising. Studies are required in this area to explore epigenetic regulatory pathways and mechanisms that could improve the disease and also help in stroke therapeutics.
Acknowledgments
This work was supported by NIH grant HL-107640 to NT and NS-51568 to SCT.
References
- 1.Mathers CD, Boerma T, Ma Fat D. Global and regional causes of death. Br Med Bull. 2009;92:7–32. doi: 10.1093/bmb/ldp028. [DOI] [PubMed] [Google Scholar]
- 2.Donnan GA, Fisher M, Macleod M, Davis SM. Stroke. Lancet. 2008;371:1612–1623. doi: 10.1016/S0140-6736(08)60694-7. [DOI] [PubMed] [Google Scholar]
- 3.Hankey GJ, Eikelboom JW. Homocysteine and stroke. Lancet. 2005;365:194–196. doi: 10.1016/S0140-6736(05)17751-4. [DOI] [PubMed] [Google Scholar]
- 4.Harker LA, Ross R, Slichter SJ, Scott CR. Homocysteine induced arteriosclerosis: the role of endothelial cell injury and platelet response in its genesis. J Clin Invest. 1976;58:731–741. doi: 10.1172/JCI108520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nishinaga M, Ozawa T, Shimada K. Homocysteine, a thrombogenic agent, suppresses anticoagulant heparin sulfate expression in cultured porcine aortic endothelial cells. J Clin Invest. 1993;92:1381–1386. doi: 10.1172/JCI116712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ratnoff OD. Activation of Hageman factor by L-homocystine. Science. 1968;162:1007–1009. doi: 10.1126/science.162.3857.1007-a. [DOI] [PubMed] [Google Scholar]
- 7.Wald DS, Law M, Morris JK. Homocysteine and cardiovascular disease: evidence on causality from a meta-analysis. Br Med J. 2002;325:1202–1206. doi: 10.1136/bmj.325.7374.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McIlroy SP, Dynan KB, Lawson JT, Patterson CC, Passmore AP. Moderately high homocysteine tied to stroke, Alzheimer’s risk. Science news. 2002 Oct 4; doi: 10.1161/01.str.0000032550.90046.38. [DOI] [PubMed] [Google Scholar]
- 9.Biswas A, Ranjan R, Meena A, Akhter MS, Yadav BK, et al. Homocystine levels, polymorphisms and the risk of ischemic stroke in young Asian Indians. J Stroke Cerebrovasc Dis. 2009;18:103–110. doi: 10.1016/j.jstrokecerebrovasdis.2008.09.014. [DOI] [PubMed] [Google Scholar]
- 10.Casas JP, Bautista LE, Smeeth L, Sharma P, Hingorani AD. Homocysteine and stroke: evidence on a causal link from Mendelian randomisation. Lancet. 2005;365:224–232. doi: 10.1016/S0140-6736(05)17742-3. [DOI] [PubMed] [Google Scholar]
- 11.Fang L, Wu W, Wu YQ. Relationship between polymorphisms of cystathionine beta-synthase gene and stroke. Zhongguo Wei Zhong Bing Ji Jiu Yi Xue. 2004;16:161–164. [PubMed] [Google Scholar]
- 12.Jeyaseelan K, Lim KY, Armugam A. MicroRNA expression in the blood and brain of rats subjected to transient focal ischemia by middle cerebral artery occlusion. Stroke. 2008;39:959–966. doi: 10.1161/STROKEAHA.107.500736. [DOI] [PubMed] [Google Scholar]
- 13.Thambyrajah J, Townend JN. Homocysteine and atherothrombosis: mechanisms for injury. Eur Heart J. 2000;21:967–974. doi: 10.1053/euhj.1999.1914. [DOI] [PubMed] [Google Scholar]
- 14.Low H-Q, Chen Christopher PLH, Kasiman K, Thalamathu A, Ng S-S, et al. A comprehensive association analysis of homocysteine metabolic pathway genes in Singaporean Chinese with ischemic stroke. PLoS One. 2011;6:24757. doi: 10.1371/journal.pone.0024757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frosst P, Blom HJ, Milos R, Goyette P, Sheppard CA, et al. A candidate genetic risk factor for vascular disease: a common mutation in methylenetetrahydrofolate reductase. Nat Genet. 1995;10:111–113. doi: 10.1038/ng0595-111. [DOI] [PubMed] [Google Scholar]
- 16.Robertson KD. DNA methylation and human disease. Nat Rev Genet. 2005;6(597):610. doi: 10.1038/nrg1655. [DOI] [PubMed] [Google Scholar]
- 17.Souto JC, Blanco-Vaca F, Soria JM, et al. A genome wide exploration suggests a new candidate gene at chromosome 11q23 as the major determinant of plasma homocysteine levels: results from the GAIT project. Am J Hum Genet. 2005;76:925–933. doi: 10.1086/430409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu CJ, Chen SD, Yang DI, et al. Promoter region methylation and reduced expression of thrombospondin-1 after oxygen-glucose deprivation in murine cerebral endothelial cells. J Cereb Blood Flow Metab. 2006;26:1519–1526. doi: 10.1038/sj.jcbfm.9600304. [DOI] [PubMed] [Google Scholar]
- 19.Zhou HJ, Zhang HN, Tang T, et al. Alteration of thrombospondin-1 and -2 in rat brains following experimental intracerebral hemorrhage. J Neurosurg. 2010;113:820–825. doi: 10.3171/2010.1.JNS09637. [DOI] [PubMed] [Google Scholar]
- 20.Langley B, Brochier C, Rivieccio MA. Targeting histone deacetylases as a multifaceted approach to treat the diverse outcomes of stroke. Stroke. 2009;40:2899–2905. doi: 10.1161/STROKEAHA.108.540229. [DOI] [PubMed] [Google Scholar]
- 21.Ubbink JB. Assay methods for the measurement of total homocyst(e)ine in plasma. Semin Thromb Hemost. 2000;26:233–241. doi: 10.1055/s-2000-8468. [DOI] [PubMed] [Google Scholar]
- 22.Kang SS, Zhou J, Wong PW, Kowalisyn J, Strokosch G. Intermediate homocysteinemia: a thermolabile variant of methylenetetrahydrofolate reductase. Am J Hum Genet. 1988;43:414–421. [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang W, Sun K, Chen J, Liao Y, Qin Q, et al. High plasma homocysteine levels contribute to the risk of stroke recurrence and all-cause mortality in a large prospective stroke population. Clin Sci. 2010;118:187–194. doi: 10.1042/CS20090142. [DOI] [PubMed] [Google Scholar]
- 24.Morita H, Kurihara H, Sugiyama T, Hamada C, Kurihara Y, et al. Polymorphism of the methionine synthase gene: association with homocysteine metabolism and late-onset vascular diseases in the Japanese population. Arterioscler Thromb Vasc Biol. 1999;19:298–302. doi: 10.1161/01.atv.19.2.298. [DOI] [PubMed] [Google Scholar]
- 25.Selhub J, Jacques PF, Wilson PW, Rush D, Rosenberg IH. Vitamin status and intake as primary determinants of homocysteinemia in an elderly population. JAMA. 1993;270:2693–2698. doi: 10.1001/jama.1993.03510220049033. [DOI] [PubMed] [Google Scholar]
- 26.Mehler MF. Epigenetic principles and mechanisms underlying nervous system functions in health and disease. Prog Neurobiol. 2008;86:305–341. doi: 10.1016/j.pneurobio.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mehler MF, Mattick JS. Noncoding RNAs and RNA editing in brain development, functional diversification, and neurological disease. Physiol Rev. 2007;87:799–823. doi: 10.1152/physrev.00036.2006. [DOI] [PubMed] [Google Scholar]
- 28.Qureshi IA, Mehler MF. Emerging role of epigenetics in stroke: part 1: DNA methylation and chromatin modifications. Arch Neurol. 2010;67:1316–1322. doi: 10.1001/archneurol.2010.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gluckman PD, Hanson MA, Buklijas T, Low FM, Beedle AS. Epigenetic mechanisms that underpin metabolic and cardiovascular diseases. Nat Rev Endocrinol. 2009;5:401–408. doi: 10.1038/nrendo.2009.102. [DOI] [PubMed] [Google Scholar]
- 30.Endres M, Meisel A, Biniszkiewicz D, et al. DNA methyl-transferase contributes to delayed ischemic brain injury. J Neurosci. 2000;20:3175–3181. doi: 10.1523/JNEUROSCI.20-09-03175.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Endres M, Fan G, Meisel A, Dirnagl U, Jaenisch R. Effects of cerebral ischemia in mice lacking DNA methyltransferase 1 in post-mitotic neurons. Neuroreport. 2001;12:3763–3766. doi: 10.1097/00001756-200112040-00032. [DOI] [PubMed] [Google Scholar]
- 32.Sunday L, Osuna C, Krause DN, Duckles SP. Age alters cerebrovascular inflammation and effects of estrogen. Am J Physiol Heart Circ Physiol. 2007;292:2333–2340. doi: 10.1152/ajpheart.01057.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilson ME, Westberry JM. Regulation of oestrogen receptor gene expression: new insights and novel mechanisms. J Neuroendocrinol. 2009;21:238–242. doi: 10.1111/j.1365-2826.2009.01830.x. [DOI] [PubMed] [Google Scholar]
- 34.Nicolas FE, Pais H, Schwach F, et al. Experimental identification of microRNA-140 targets by silencing and overexpressing miR-140. RNA. 2008;14:2513–2520. doi: 10.1261/rna.1221108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen M, Gavrilova O, Liu J. Alternative Gnas gene products have opposite effects on glucose and lipid metabolism. Proc Natl Acad Sci U S A. 2005;102:7386–7391. doi: 10.1073/pnas.0408268102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dobrovolny R, Dvorakova L, Ledvinova J, et al. Relationship between X-inactivation and clinical involvement in Fabry heterozygotes: eleven novel mutations in the alphagalactosidase A gene in the Czech and Slovak population. J Mol Med. 2005;83:647–654. doi: 10.1007/s00109-005-0656-2. [DOI] [PubMed] [Google Scholar]
- 37.Freson K, Izzi B, Labarque V, et al. GNAS defects identified by stimulatory G protein alpha-subunit signalling studies in platelets. J Clin Endocrinol Metab. 2008;93:4851–4859. doi: 10.1210/jc.2008-0883. [DOI] [PubMed] [Google Scholar]
- 38.Giacomini PS, Shannon PT, Clarke JT, Jaigobin C. Fabry’s disease presenting as stroke in a young female. Can J Neurol Sci. 2004;31:112–114. doi: 10.1017/s0317167100002936. [DOI] [PubMed] [Google Scholar]
- 39.Kusuhara T, Ayabe M, Hino H, Shoji H, Neshige R. A case of Prader–Willi syndrome with bilateral middle cerebral artery occlusion and moyamoya phenomenon. Rinsho Shinkeigaku. 1996;36:770–773. [PubMed] [Google Scholar]
- 40.Cairns BR. The logic of chromatin architecture and remodeling at promoters. Nature. 2009;461:193–198. doi: 10.1038/nature08450. [DOI] [PubMed] [Google Scholar]
- 41.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293(5532):1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 42.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 43.Fang S, Miao J, Xiang L, Ponugoti B, Treuter E, Kemper JK. Coordinated recruitment of histone methyltransferase G9a and other chromatin-modifying enzymes in SHP-mediated regulation of hepatic bile acid metabolism. Mol Cell Biol. 2007;27:1407–1424. doi: 10.1128/MCB.00944-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gilardi F, Mitro N, Godio C, et al. The pharmacological exploitation of cholesterol 7 alpha-hydroxylase, the key enzyme in bile acid synthesis: from binding resins to chromatin remodelling to reduce plasma cholesterol. Pharmacol Ther. 2007;116:449–472. doi: 10.1016/j.pharmthera.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 45.Shafaati M, O’Driscoll R, Björkhem I, Meaney S. Transcriptional regulation of cholesterol 24-hydroxylase by histone deacetylase inhibitors. Biochem Biophys Res Commun. 2009;378:689–694. doi: 10.1016/j.bbrc.2008.11.103. [DOI] [PubMed] [Google Scholar]
- 46.Finkel T, Deng CX, Mostoslavsky R. Recent progress in the biology and physiology of sirtuins. Nature. 2009;460:587–591. doi: 10.1038/nature08197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Faraco G, Pancani T, Formentini L, et al. Pharmacological inhibition of histone deacetylases by suberoylanilide hydroxamic acid specifically alters gene expression and reduces ischemic injury in the mouse brain. Mol Pharmacol. 2006;70:1876–1884. doi: 10.1124/mol.106.027912. [DOI] [PubMed] [Google Scholar]
- 48.Soriano FX, Papadia S, Bell KF, Hardingham GE. Role of histone acetylation in the activity-dependent regulation of sulfiredoxin and sestrin 2. Epigenetics. 2009;4:152–158. doi: 10.4161/epi.4.3.8753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Birney E, Stamatoyannopoulos JA, Dutta A, et al. Identification and analysis of functional elements in 1 % of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mattick JS, Amaral PP, Dinger ME, Mercer TR, Mehler MF. RNA regulation of epigenetic processes. Bioessays. 2009;31:51–59. doi: 10.1002/bies.080099. [DOI] [PubMed] [Google Scholar]
- 51.Schratt G. Fine-tuning neural gene expression with microRNAs. Curr Opin Neurobiol. 2009;19:213–219. doi: 10.1016/j.conb.2009.05.015. [DOI] [PubMed] [Google Scholar]
- 52.Dharap A, Bowen K, Place R, Li LC, Vemuganti R. Transient focal ischemia induces extensive temporal changes in rat cerebral microRNAome. J Cereb Blood Flow Metab. 2009;29:675–687. doi: 10.1038/jcbfm.2008.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 54.Rashidian J, Iyirhiaro G, Aleyasin H, et al. Multiple cyclin-dependent kinases signals are critical mediators of ischemia/hypoxic neuronal death in vitro and in vivo. Proc Natl Acad Sci U S A. 2005;102:14080–14085. doi: 10.1073/pnas.0500099102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang X, Arai S, Song X, et al. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature. 2008;454:126–130. doi: 10.1038/nature06992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Broadbent HM, Peden JF, Lorkowski S, et al. Susceptibility to coronary artery disease and diabetes is encoded by distinct, tightly linked SNPs in the ANRIL locus on chromosome 9p. Hum Mol Genet. 2008;17:806–814. doi: 10.1093/hmg/ddm352. [DOI] [PubMed] [Google Scholar]
- 57.Holdt LM, Beutner F, Scholz M, et al. ANRIL expression is associated with atherosclerosis risk at chromosome 9p21. Arterioscler Thromb Vasc Biol. 2010;30:620–627. doi: 10.1161/ATVBAHA.109.196832. [DOI] [PubMed] [Google Scholar]
- 58.Jarinova O, Stewart AF, Roberts R, et al. Functional analysis of the chromosome 9p21.3 coronary artery disease risk locus. Arterioscler Thromb Vasc Biol. 2009;29:1671–1677. doi: 10.1161/ATVBAHA.109.189522. [DOI] [PubMed] [Google Scholar]
- 59.Liu Y, Sanoff HK, Cho H, et al. INK4/ARF transcript expression is associated with chromosome 9p21 variants linked to atherosclerosis. PLoS One. 2009;4:5027. doi: 10.1371/journal.pone.0005027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pasmant E, Laurendeau I, He’ron D, Vidaud M, Vidaud D, Bièche I. Characterization of a germ-line deletion, including the entire INK4/ARF locus, in amelanomaneural system tumor family. Cancer Res. 2007;67:3963–3969. doi: 10.1158/0008-5472.CAN-06-2004. [DOI] [PubMed] [Google Scholar]
- 61.Schaefer AS, Richter GM, Groessner-Schreiber B, et al. Identification of a shared genetic susceptibility locus for coronary heart disease and periodontitis. PLoS Genet. 2009;5:1000378. doi: 10.1371/journal.pgen.1000378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Visel A, Zhu Y, May D, et al. Targeted deletion of the 9p21 non-coding coronary artery disease risk interval in mice. Nature. 2010;464:409–412. doi: 10.1038/nature08801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Costain WJ, Rasquinha I, Graber T, et al. Cerebral ischemia induces neuronal expression of novel VL30 mouse retrotransposons bound to polyribosomes. Brain Res. 2006;1094:24–37. doi: 10.1016/j.brainres.2006.03.120. [DOI] [PubMed] [Google Scholar]
- 64.Kawahara Y, Megraw M, Kreider E, et al. Frequency and fate of microRNA editing in human brain. Nucleic Acids Res. 2008;36:5270–5280. doi: 10.1093/nar/gkn479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yang W, Chendrimada TP, Wang Q, et al. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat Struct Mol Biol. 2006;13:13–21. doi: 10.1038/nsmb1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kawahara Y, Zinshteyn B, Chendrimada TP, Shiekhattar R, Nishikura K. RNA editing of the microRNA-151 precursor blocks cleavage by the Dicer–TRBP complex. EMBO Rep. 2007;8:763–769. doi: 10.1038/sj.embor.7401011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim JB, Piao CS, Lee KW, et al. Delayed genomic responses to transient middle cerebral artery occlusion in the rat. J Neurochem. 2004;89:1271–1282. doi: 10.1111/j.1471-4159.2004.02429.x. [DOI] [PubMed] [Google Scholar]
- 68.Shani V, Bromberg Y, Sperling O, Zoref-Shani E. Involvement of Src tyrosine kinases (SFKs) and of focal adhesion kinase (FAK) in the injurious mechanism in rat primary neuronal cultures exposed to chemical ischemia. J Mol Neurosci. 2009;37:50–59. doi: 10.1007/s12031-008-9113-3. [DOI] [PubMed] [Google Scholar]
- 69.Shimamura N, Matchett G, Yatsushige H, Calvert JW, Ohkuma H, Zhang J. Inhibition of integrin αvβ3 ameliorates focal cerebral ischemic damage in the rat middle cerebral artery occlusion model. Stroke. 2006;37:1902–1909. doi: 10.1161/01.STR.0000226991.27540.f2. [DOI] [PubMed] [Google Scholar]
- 70.Kang SK, Lee DH, Bae YC, Kim HK, Baik SY, Jung JS. Improvement of neurological deficits by intracerebral transplantation of human adipose tissue-derived stromal cells after cerebral ischemia in rats. Exp Neurol. 2003;183:355–366. doi: 10.1016/s0014-4886(03)00089-x. [DOI] [PubMed] [Google Scholar]
- 71.Lee TH, Yoon JG. Intracerebral transplantation of human adipose tissue stromal cells after middle cerebral artery occlusion in rats. J Clin Neurosci. 2008;15:907–912. doi: 10.1016/j.jocn.2007.03.016. [DOI] [PubMed] [Google Scholar]
- 72.Plummer R, Vidal L, Griffin M, et al. Phase I study of MG98, an oligonucleotide antisense inhibitor of human DNA methyltransferase 1, given as a 7-day infusion in patients with advanced solid tumors. Clin Cancer Res. 2009;5:3177–3183. doi: 10.1158/1078-0432.CCR-08-2859. [DOI] [PubMed] [Google Scholar]
- 73.Csoka AB, Szyf M. Epigenetic side-effects of common pharmaceuticals: a potential new field in medicine and pharmacology. Med Hypotheses. 2009;73:770–780. doi: 10.1016/j.mehy.2008.10.039. [DOI] [PubMed] [Google Scholar]
- 74.Milutinovic S, D’Alessio AC, Detich N, Szyf M. Valproate induces widespread epigenetic reprogramming which involves demethylation of specific genes. Carcinogenesis. 2007;28:560–571. doi: 10.1093/carcin/bgl167. [DOI] [PubMed] [Google Scholar]
- 75.Phiel CJ, Zhang F, Huang EY, et al. Histone deacetylase is a direct target of valproic acid, a potent anticonvulsant, mood stabilizer, and teratogen. J Biol Chem. 2001;276:36734–36741. doi: 10.1074/jbc.M101287200. [DOI] [PubMed] [Google Scholar]
- 76.Szyf M. Epigenetic therapeutics in autoimmune disease. Clin Rev Allergy Immunol. 2010;39:62–77. doi: 10.1007/s12016-009-8172-8. [DOI] [PubMed] [Google Scholar]
- 77.Lu KT, Robin YY, Chen LG, et al. Neuroprotective effects of resveratrol on cerebral ischemia-induced neuron loss mediated by free radical scavenging and cerebral blood flow elevation. J Agric Food Chem. 2006;54:3126–3131. doi: 10.1021/jf053011q. [DOI] [PubMed] [Google Scholar]
- 78.Ovbiagele B. Potential role of curcumin in stroke prevention. Expert Rev Neurother. 2008;8:1175–1176. doi: 10.1586/14737175.8.8.1175. [DOI] [PubMed] [Google Scholar]
- 79.Zhang Z, Yang X, Zhang S, Ma X, Kong J. BNIP3 upregulation and EndoG translocation in delayed neuronal death in stroke and in hypoxia. Stroke. 2007;38:1606–1613. doi: 10.1161/STROKEAHA.106.475129. [DOI] [PubMed] [Google Scholar]
- 80.Miyawaki T, Ofengeim D, Noh KM, et al. The endogenous inhibitor of Akt, CTMP, is critical to ischemia-induced neuronal death. Nat Neurosci. 2009;12:618–626. doi: 10.1038/nn.2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Qureshi IA, Mehler MF. Regulation of non-coding RNA networks in the nervous system: what’s the REST of the story? Neurosci Lett. 2009;466:73–80. doi: 10.1016/j.neulet.2009.07.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Calderone A, Jover T, Noh KM, et al. Ischemic insults derepress the gene silencer REST in neurons destined to die. J Neurosci. 2003;23:2112–2121. doi: 10.1523/JNEUROSCI.23-06-02112.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Formisano L, Noh KM, Miyawaki T, Mashiko T, Bennett MV, Zukin RS. Ischemic insults promote epigenetic reprogramming of μ opioid receptor expression in hippocampal neurons. Proc Natl Acad Sci U S A. 2007;104:4170–4175. doi: 10.1073/pnas.0611704104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li JB, Levanon EY, Yoon JK, et al. Genome-wide identification of human RNA editing sites by parallel DNA capturing and sequencing. Science. 2009;324:1210–1213. doi: 10.1126/science.1170995. [DOI] [PubMed] [Google Scholar]
- 85.Feinberg EH, Hunter CP. Transport of dsRNA into cells by the transmembrane protein SID-1. Science. 2003;301:1545–1547. doi: 10.1126/science.1087117. [DOI] [PubMed] [Google Scholar]
- 86.Sproule DM, Kaufmann P. Mitochondrial encephalopathy, lactic acidosis, and stroke like episodes. Ann N Y Acad Sci. 2008;1142:133–158. doi: 10.1196/annals.1444.011. [DOI] [PubMed] [Google Scholar]
- 87.Smalheiser NR. Exosomal transfer of proteins and RNAs at synapses in the nervous system. Biol Direct. 2007;2:35. doi: 10.1186/1745-6150-2-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Alvarez-Erviti L, Seow Y, Yin H, Betts C, Lakhal S, Wood MJ. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat Biotechnol. 2011;29:341–345. doi: 10.1038/nbt.1807. [DOI] [PubMed] [Google Scholar]
- 89.Mizrak A, Bolukbasi MF, Ozdener GB, et al. Genetically engineered microvesicles carrying suicide mRNA/protein inhibit schwannoma tumor growth. Mol Ther. 2013;21:101–108. doi: 10.1038/mt.2012.161. [DOI] [PMC free article] [PubMed] [Google Scholar]