Abstract
Envelope protein precursors of many viruses are processed by a basic endopeptidase to generate two molecules, one for receptor binding and the other for membrane fusion. Such a cleavage event has not been demonstrated for the hepatitis B virus family. Two binding partners for duck hepatitis B virus (DHBV) pre-S envelope protein have been identified. Duck carboxypeptidase D (DCPD) interacts with the full-length pre-S protein and is the DHBV docking receptor, while duck glycine decarboxylase (DGD) has the potential to bind several deletion constructs of the pre-S protein in vitro. Interestingly, DGD but not DCPD expression was diminished following prolonged culture of primary duck hepatocytes (PDH), which impaired productive DHBV infection. Introduction of exogenous DGD promoted formation of protein-free viral genome, suggesting restoration of several early events in viral life cycle. Conversely, blocking DGD expression in fresh PDH by antisense RNA abolished DHBV infection. Moreover, addition of DGD antibodies soon after virus binding reduced endogenous DGD protein levels and impaired production of covalently closed circular DNA, the template for DHBV gene expression and genome replication. Our findings implicate this second pre-S binding protein as a critical cellular factor for productive DHBV infection. We hypothesize that DCPD, a molecule cycling between the cell surface and the trans-Golgi network, targets DHBV particles to the secretary pathway for proteolytic cleavage of viral envelope protein. DGD represents the functional equivalent of other virus receptors in its interaction with processed viral particles.
The early events in the virus infectious cycle may serve as critical targets for therapeutic intervention. These events are difficult to study due to the transient and dynamic nature of the entry process, the low signal intensity, and the lack of a systemic approach to identify the cellular components required for individual steps. Despite such difficulties, receptors for many important viral pathogens have been identified and general themes in viral attachment and entry are beginning to emerge. For instance, the envelope proteins of orthomyxoviruses, paramyxoviruses, and retroviruses are translated as single polypeptides that are subsequently processed by a basic endopeptidase to generate two polypeptides, one for receptor binding and the other for membrane fusion (13, 22). The fusion activity is not triggered until following receptor-ligand interaction and may occur at the site of virus binding (retroviruses) or in the acidic environment of the endosome following virus endocytosis (influenza virus). Furin and PC7 are the known endopeptidases capable of processing gp160 of human immunodeficiency virus (HIV) into the N-terminal gp120 (surface protein) and C-terminal gp41 (transmembrane protein) in the secretory pathway (9). The gp120 remains associated with virus particles via its noncovalent interaction with gp41. Binding of gp120 to CD4, the docking receptor, is accompanied by gp41 interaction with CXCR4 or CCR5, the viral coreceptor, thus leading to fusion of the viral membrane with the plasma membrane of the target cell (1, 6).
Such a cleavage event has not been documented for hepatitis B virus (HBV) or its related animal viruses, a group of enveloped hepatotropic DNA viruses (Hepadnaviridae) that, similar to retroviruses, requires reverse transcription for genome replication (7). Instead, HBV expresses three coterminal envelope proteins via alternative translational initiation. Thus, the abundant small envelope protein contains the S domain only, while the large and middle envelope proteins contain additional pre-S1/pre-S2 domains and pre-S2 domain, respectively. The cellular receptor for HBV remains enigmatic following decades of extensive search, as no binding protein for the pre-S1 domain, the candidate site for receptor engagement, turned out to confer HBV susceptibility in reconstitution experiments. Due to the difficulty in obtaining primary human hepatocytes, which may have variable susceptibility to HBV infection, it is not clear whether antibodies against some of the pre-S1 binding partners could block HBV infection.
In this regard, duck hepatitis B virus (DHBV) is an ideal model to study the early events in the hepadnaviral life cycle. DHBV can infect ducklings in vivo and primary duck hepatocytes (PDH) in vitro, which remain susceptible to DHBV infection for 2 weeks if maintained in serum-free medium supplemented with dimethyl sulfoxide (DMSO) (25). DHBV expresses two envelope proteins, with a 161-amino-acid pre-S domain unique to the large envelope protein. At least two cellular proteins have been found to bind to the pre-S domain: duck carboxypeptidase D (DCPD) and duck glycine decarboxylase (DGD) (11, 14, 15, 18, 19, 31). The DCPD is believed to be the DHBV docking receptor, since it confers DHBV binding to transfected cells and its antibody has been found to block DHBV infection of PDH (32, 34). The DCPD may also determine host specificity of DHBV infection, as carboxypeptidase D molecules from other species fail to interact with DHBV pre-S domain (15, 18, 27). On the other hand, restricted expression of DGD in the liver, kidney, and pancreas coincides with DHBV tissue tropism (18, 19). Interestingly, DCPD and DGD have overlapping and possibly competing binding sites in the pre-S domain (18, 31), although DCPD prefers full-length pre-S polypeptide whereas DGD binds much more efficiently to a few truncated versions of the pre-S domain such as 98-161 and 1-102. The sequence 97REAFRRY103 conforms to the PC7 recognition site (35), thus raising the intriguing possibility of DHBV-DGD interaction upon proteolytic cleavage of the large envelope protein.
Before such a hypothesis can be tested in earnest, it will be essential to confirm that DGD is indeed an essential component for DHBV infection. Here we addressed this question by applying DGD-specific antisense RNA and antibodies to freshly prepared PDH culture. We also monitored DGD protein expression over the course of in vitro culture of duck hepatocytes, which is characterized by declined cellular susceptibility to DHBV infection (25). The overall results suggest that DGD is a cellular factor critical for an early step in productive DHBV infection. A model to account for the requirement of both DCPD and DGD in DHBV infectivity is proposed.
MATERIALS AND METHODS
PDH culture.
Three-day-old DHBV-free ducklings were perfused as initially described by Tuttleman et al. (33), with sequential infusion of 0.5 mM EGTA and 0.5 mg of collagenase/ml into the portal vein (31). Hepatocytes were seeded in 60-mm-diameter petri dishes (2 × 106/well), 6-well plates (7 × 105/well), or 12-well plates (3 × 105/well) overnight with L15 medium supplemented with 5% fetal bovine serum and were kept in a 37°C humidified incubator. Cells were maintained in serum-free medium containing 1% DMSO, with medium change every 2 or 3 days.
DGD expression and DHBV susceptibility following in vitro culture of duck hepatocytes.
PDH cultured in 6-well plates were harvested at different time points using cell scrapers and collected by low-speed centrifugation. The cell pellet was lysed on ice for 10 min with 100 μl of lysis buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1% Triton X-100, 1% sodium deoxycholate). Nuclei were removed by centrifugation, and a 5-μl aliquot of lysate was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) using 0.1% SDS-8% polyacrylamide gels. Proteins were transferred to polyvinylidene difluoride membranes, which were treated at room temperature for 3 h with 3% bovine serum albumin-phosphate-buffered saline containing 0.05% Tween 20 (PBST) and incubated at 4°C overnight with rabbit polyclonal antibodies against DCPD (32) or DGD (19) diluted at 1:1,000 in PBST. Blots were washed and incubated for 2 to 3 h with 125I-labeled protein A (low specific activity; Perkin-Elmer Life Sciences, Inc.) diluted 1:1,000 with PBST. Following extensive wash, bound 125I signals were revealed by autoradiography. To measure DHBV binding and entry, PDH cultured in 6-well plates were incubated overnight (around 16 h) with 12 μl of viremic duck serum diluted in 1.5 ml of medium (a single batch of a highly viremic duck serum was used throughout this study). Cells were washed, harvested with a cell scraper, and divided into two equal parts before being spun down. One cell pellet was treated with 500 μl of 0.25% trypsin-10 mM EDTA at 37°C for 5 min to remove cell surface-bound virus particles (due to some cell damage caused by mechanical force, the amount of endocytosed virions may be underestimated). The reaction was stopped by addition of complete medium, and cells were spun down again. Both cell pellets were resuspended in TEN buffer and digested at 37°C for several hours with 0.5 mg of proteinase K/ml in the presence of 0.5% SDS. Following phenol extraction and ethanol precipitation, DNA was separated by using 1.5% agarose gel lacking ethidium bromide. DHBV DNA was detected by Southern blot analysis using a random primed DHBV DNA probe. The DHBV DNA was obtained through several rounds of PCR amplification and was thus of high purity. To study productive DHBV infection, triplicates of PDH maintained in 60-mm-diameter dishes for different durations were incubated overnight with 3 ml of medium supplemented with 20 μl of viremic duck serum. The three dishes were harvested immediately after binding and 2 and 6 days later, respectively. Levels of total DHBV DNA were measured.
Effect of exogenous DGD on DHBV infection of cultured PDH.
PDH cultured in 60-mm-diameter petri dishes for 17 days were cotransfected with 1 μg of a green fluorescent protein (GFP) reporter plasmid together with 2 μg of pcDNA vector or the same plasmid expressing DGD (AUU346AUG) (19) or full-length DCPD (32) by using Lipofectamine PLUS reagent (GIBCO/BRL). Two days later cells were infected overnight with 20 μl of viremic duck serum diluted in 3 ml of medium. Cells were harvested 6 days postinfection into two equal parts, one for total viral DNA extraction and the other for protein-free DNA extraction according to the protocol of Yu and Summers (37). Basically, cells were incubated with 0.5 ml of 10 mM Tris-HCl [pH 7.5]-10 mM EDTA-1% SDS at 37°C for 5 min, supplemented with 125 μl of 2.5 M KCl, and chilled on ice for 5 min. The detergent-protein complexes were removed by centrifugation, and the supernatant was extracted with phenol. Nucleic acids were precipitated with ethanol, separated in 1.5% agarose gel without ethidium bromide, transferred to nylon membranes, and hybridized with a randomly primed DHBV probe.
In additional experiments, DGD and DCPD were delivered to cultured hepatocytes by adenovirus vectors. Recombinant adenoviruses were generated using the system of He et al. (10). The full-length DGD (AUU346AUG) (19) and DCPD cDNAs were subcloned into the BglII-XhoI sites and NotI-XbaI sites of the pAdTrackCMV shuttle vector, respectively, and subsequently linearized with PmeI to generate recombinants with coelectroporated adenovirus backbone pAdEasy-1 in Escherichia coli strain BJ5183. The recombinant DNA was amplified in DH10B cells, purified by CsCl gradient centrifugation, and transfected into human embryonic kidney cell line 293 following linearization with PacI. Adenoviral stocks were amplified through several rounds of infection of 293 cells and concentrated by ultracentrifugation through cesium chloride gradient. Viral titer was estimated from the optical density at 260 nm (assuming 1012 viral particles in 1 A260 unit). PDH grown in 60-mm-diameter dishes for 3 weeks were incubated at 37°C overnight with 5 μl of adenoviruses corresponding to a multiplicity of infection (MOI) of circa 100. After medium change, cells were infected for 6 h with 20 μl of DHBV viremic serum diluted in 3 ml of medium. Cells were harvested 4 and 9 days later and lysed with 200 μl of lysis buffer. Viral DNA was analyzed from 100 μl of lysate as described above. For core protein detection, rabbit polyclonal anti-core antibody was raised against core protein expressed as glutathione S-transferase (GST) fusion protein. The GST tag was removed by thrombin cleavage. An aliquot of lysate (2 μl) was run through 0.1% SDS-12% PAGE. The blot was blocked with 3% bovine serum albumin in PBST for 4 h and incubated at 4°C overnight with the rabbit anti-core antibody diluted 1:5,000 in PBST. Following wash and incubation at room temperature for 1 h with a 1:10,000 dilution of goat anti-rabbit antibody conjugated to horseradish peroxidase, signals were revealed by enhanced chemiluminescence. Use of the Kodak Image Station 440 CF and serial exposures helped avoid signal saturation.
Cell viability assay.
PDH grown in 12-well plates were incubated overnight with different adenoviral constructs at MOIs of 100 and 500. The adenoviruses were removed and cell viability was measured 24, 48, and 72 h later by the WST-8 assay (a modified MTT assay from Dojindo Laboratory, Kumamoto, Japan). Duplicate cell samples were incubated with 20 μl of tetrazolium in 200 μl of medium for 2 h, and absorbance at 490 nm was measured with a reference wavelength at 620 nm. Similar assays were performed on PDH incubated with DGD antiserum as described below.
Inhibition of DHBV infectivity by DGD antisense RNAs or antibodies.
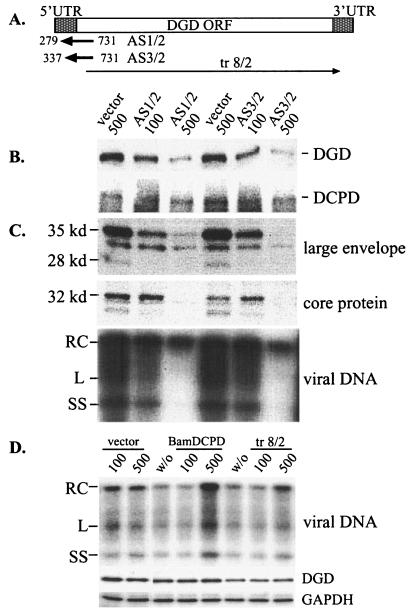
DGD cDNA fragments 279-731 and 337-731, encompassing the translation initiation codon (see Fig. 3A), were cloned into pAdTrackCMV vector in the reverse orientation. Construct tr8/2, which lacks the coding sequence for the first 94 residues of DGD, was cloned into the KpnI-XhoI sites in the sense orientation. An antisense construct against DCPD, BamDCPD, was generated by cloning the 1.8-kb NotI-BamHI fragment into NotI-BglII sites of pAdTrackCMV. Adenoviruses were generated. PDH in 6-well plates cultured for 2 to 3 days were infected overnight with adenoviruses at MOIs of 100 and 500. After removal of adenoviruses, cells were incubated with 12 μl of DHBV viremic serum diluted in 1.5 ml of medium for 6 h. Cells were harvested 7 days later. For experiments involving DGD antibodies, PDH cultured in 12-well plates for 1 or 2 days were infected with 5 μl of viremic duck serum diluted in 600 μl for 6 h, followed by washing. Cells were incubated for various times with different dilutions of a rabbit DGD antiserum (19) prior to, during, or following DHBV infection. The PDH were harvested at day 7 postinfection, and viral large envelope protein and core protein as well as total viral DNA were analyzed.
FIG. 3.
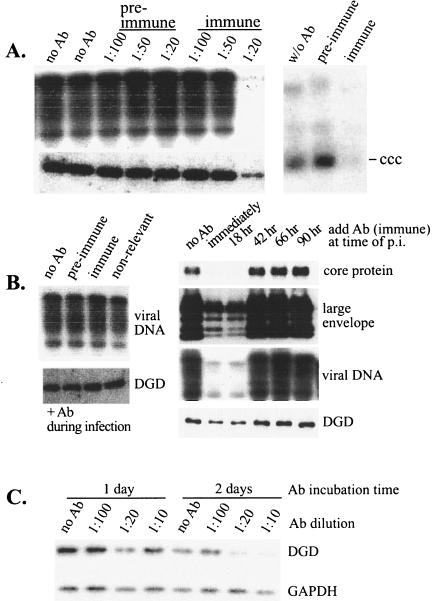
Inhibition of DGD expression and DHBV infectivity by adenovirus-mediated antisense constructs. (A) Schematic representation of two DGD antisense constructs (AS1/2 and AS3/2) and one sense construct lacking the N-terminal 94 residues (tr8/2). The DGD open reading frame (ORF) and untranslated regions (UTR) are shown at the top. (B) Effect of DGD antisense constructs on endogenous DGD and DCPD expression. Freshly cultured PDH were infected overnight with adenovirus vector or the two antisense constructs at an MOI of 100 or 500 as indicated. Cells were harvested 48 h postinfection, and both DGD and DCPD proteins were analyzed in Western blots. (C) Effect of antisense constructs on DHBV infectivity. Cells were infected with adenovirus for 2 days, incubated with viremic duck serum for 6 h, and harvested 7 days later. Viral large envelope protein and core protein, as well as viral DNA, were analyzed. (D) Effects of a DCPD antisense construct (BamDCPD) and DGD sense construct (tr8/2) on DHBV infectivity and DGD expression. Experimental conditions were the same as described for panel C, and DHBV DNA at day 7 postinfection is shown. Western blot analysis of GAPDH expression serves as a loading control.
Influence of DGD antibodies on DGD-pre-S interaction.
Twenty milligrams of duck liver proteins was incubated with 10 μg of pre-S peptide 80-102 fused to GST and immobilized on beads (18) at 4°C for 1.5 h in the presence of DGD or DCPD antibodies. The beads were washed extensively, and bound DGD protein was revealed by Western blot.
Binding of DGD antibodies to PDH culture and depletion of DGD.
Duplicates of PDH maintained in 12-well plates were incubated at 37°C for 1, 2, or 3 days with 30 μl of DGD preimmune or immune serum diluted 1:20 in L15 medium. Cells were washed with medium and harvested, one well with and the other without trypsin treatment. Retained antibodies present in cell lysate were detected by Western blot analysis with horseradish peroxidase-conjugated anti-rabbit antibodies. To specifically monitor the retention of DGD antibodies, PDH grown in 60-mm-diameter dishes were metabolically labeled with [35S]methionine-cysteine for 4 h (31), followed by further incubation in the presence or absence of a 1:20 dilution of DGD antiserum (75 μl of antiserum in 1.5 ml medium) for 6, 24, and 48 h. Cells were lysed with HEPES lysis buffer (100 mM NaCl, 10 mM HEPES [pH 7.5], 1 mM EDTA, 1% NP-40) containing a 1:100 dilution of protease inhibitor cocktail (Sigma). The cell lysate was incubated with 50 μl of protein A beads (50% slurry) at 4°C overnight. Retained DGD was analyzed in 0.1% SDS-8% PAGE and revealed by fluorography. To determine the effect of DGD antibodies on steady-state DGD protein level, PDH cultured in 12-well plates were incubated with preimmune or immune serum (from 1:10 to 1:100 dilution) for 1 and 2 days, respectively, and endogenous DGD was detected by Western blot. As a control, a nonrelevant cellular protein GAPDH (glyceraldehyde-3-phosphate dehydrogenase) was monitored in parallel by Western blot using a 1:5,000 dilution of polyclonal antibody (Trevigen) followed by enhanced chemiluminescence.
RESULTS
Diminished PDH susceptibility to DHBV infection correlates with declined DGD protein expression.
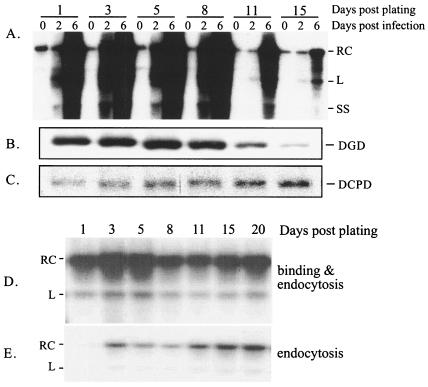
The ability of duck hepatocytes to support productive DHBV infection, as defined by amplification of viral proteins and genomes, is gradually impaired over the course of in vitro culture of the PDH. Thus, cells cultured for 3 weeks are usually no longer infectible with DHBV (25). In the present study, triplicates of PDH maintained in DMSO medium for 1 to 15 days were infected with DHBV and harvested immediately or 2 or 6 days later. The amplification of viral DNA signals over the three time points was robust when PDH were infected within the first 8 days of plating (Fig. 1A). Its magnitude declined with delayed infection at day 11 postplating and was minimal with infection at day 15. The reduced DHBV infectivity was not caused by impairment in viral attachment or endocytosis (Fig. 1D and E). In agreement with this finding, sustained DCPD expression was observed during the entire study period (Fig. 1C). The lack of DCPD down-regulation is consistent with its broad tissue distribution (11, 31) and suggests the lack of liver specificity of DCPD promoter. On the other hand, expression of DGD declined over time, which correlated nicely with the reduction in cellular susceptibility to DHBV infection (Fig. 1B). Similar results were obtained in PDH maintained in a medium lacking DMSO but containing 5% fetal bovine serum, which prompted a more rapid loss of cellular susceptibility to DHBV infection (25) and a steep decline of DGD expression (data not shown).
FIG. 1.
Concordant decline of DHBV infectivity and DGD expression in cultured duck hepatocytes. (A) DHBV DNA replication. PDH were infected overnight with DHBV at different days after plating as indicated. Cells were harvested immediately (day 0) or 2 or 6 days later. DHBV DNA was detected by Southern blot. RC, linear (L), and single-stranded (SS) viral DNA forms are indicated. The signal at day 6 relative to that at day 0 indicates the degree of productive infection. (B and C) Western blot analysis of the expression of DGD (B) and DCPD (C) from the same batch of uninfected PDH at the same time points postplating. Bound antibodies were revealed by 125I-labeled protein A. (D and E) DHBV binding (D) and endocytosis (E). Following overnight infection of PDH at the same time points as shown in panels A to C, cells were harvested immediately by scraping and divided into two parts. One part was treated with trypsin to remove cell surface-bound virus (E). Cell-associated DHBV DNA was analyzed by Southern blotting. The weak DHBV signal shown in lane 1 of panel E could reflect enhanced permeability of the cells to trypsin, since the experiment was performed immediately following attachment of collagenase-treated hepatocytes.
Introduction of DGD into old PDH cultures restored an early step in viral life cycle.
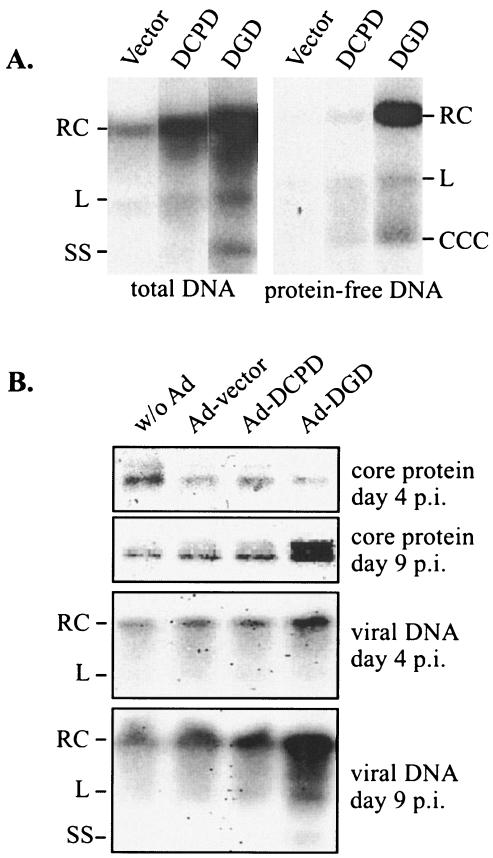
A concomitant decline in DGD expression and cellular susceptibility to DHBV infection is still insufficient to establish a direct role for this protein, because expression of numerous liver-specific proteins may have tapered off. To directly address the role of DGD, PDH cultured in DMSO medium for 17 days were transiently transfected with pcDNA vector, pcDNA-DGD, or pcDNA-DCPD. Cells were subsequently infected with DHBV and harvested 6 days later. Cells receiving DCPD, and especially DGD, displayed increased viral replication signals compared with cells transfected with empty vector (Fig. 2A, left panel).
FIG. 2.
Effect of exogenous DGD on DHBV infection in aged PDH. (A) PDH cultured for 17 days were transfected with pcDNA vector or pcDNA-based DCPD or DGD construct. Cells were infected with DHBV 2 days later and harvested 6 days postinfection for analysis of total viral DNA (left panel) and protein-free DNA (right panel) forms. Transfection efficiency as measured by cotransfected GFP was found to be approximately 10%. RC, RC DNA; L, linear DNA; SS, single-stranded DNA; CCC, cccDNA. (B) PDH cultured for 3 weeks were infected with empty adenovirus or adenovirus expressing DCPD or DGD. Cells were incubated with viremic duck serum and harvested 4 and 9 days later. Both viral core protein and total viral DNA were analyzed.
During productive DHBV infection, the incoming relaxed circular (RC) virion DNA is converted to covalently closed circular DNA (cccDNA), which serves as the template for viral DNA replication (28). Whereas both virion-associated DNA and newly replicated viral DNA inside core particles are physically attached to viral polymerase, the cccDNA is not. Thus, phenol extraction without a prior proteinase K digestion step enriches the cccDNA, which runs at a position (2 kb) distinct from RC DNA (4 kb) and linear DNA (3.2 kb) (37). Through this procedure, we found that the cccDNA was moderately increased in DGD-transfected cells (Fig. 2A, right panel). Moreover, a marked increase in the protein-free RC DNA form was also found (Fig. 2A), suggesting that DGD is required for the path leading to removal of polymerase from RC DNA, a step likely to precede cccDNA formation. In a separate experiment, we delivered DCPD and DGD cDNAs by adenoviral vectors into PDH cultured for a longer time (3 weeks). Nine days postinfection, cells transduced with DGD displayed stronger signals of DHBV core protein and viral DNA compared with cells receiving DCPD or empty adenoviral vector (Fig. 2B).
Reduction of endogenous DGD levels by antisense RNAs abolished DHBV infectivity.
In the reverse experiment, we applied antisense RNA to specifically block DGD expression in freshly cultured PDH. The two antisense RNA constructs delivered by adenoviral vector covered the translation initiation codon of DGD (Fig. 3A). A sense DGD construct missing the N-terminal 94 residues of DGD protein, including the signal peptide (tr8/2), and an antisense construct of DCPD that had no effect on DCPD expression (BamDCPD) were applied in parallel as negative controls. The endogenous DGD, but not DCPD, levels were greatly reduced 2 days later in cells infected with both DGD antisense constructs at an MOI of 500 (Fig. 3B). Such a dose of adenovirus was needed to infect most cells in the culture (>90% as indicated by GFP expression) but did not cause cell toxicity (Fig. 4A and data not shown). No alteration in DGD levels was found in cells receiving the same dose of adenovirus vector. Importantly, DHBV infectivity was severely impaired by such a dose of antisense constructs, as revealed by reduced levels of viral large envelope protein, core protein, and viral DNA (Fig. 3C). No inhibitory effect of DHBV infection or DGD expression was seen with the BamDCPD or tr8/2 construct (Fig. 3D).
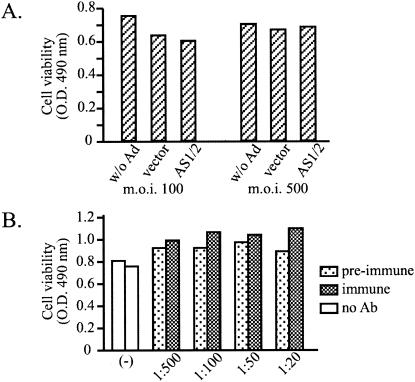
FIG. 4.
Effects of adenovirus infection (A) and DGD antiserum (B) on PDH cell viability. (A) Cells were infected with adenoviruses (Ad) overnight at an MOI of 100 or 500. Cell viability was analyzed at days 1, 2, and 3 postinfection using a modified MTT (WST-8) assay. Shown are data from day 2, but similar results were obtained from cells at day 1 and day 3. (B) PDH were incubated with various dilutions of preimmune or immune serum for 48 h, and cell viability was measured by WST-8 reagent 2, 3, and 5 days later. Shown are data from cells at day 2 following antibody (Ab) incubation. O.D., optical density.
DGD antibodies blocked viral infectivity at a postbinding step.
DGD binding to truncated pre-S peptide 80-102 could be inhibited quantitatively by the rabbit polyclonal antibodies (Fig. 5A), which failed to interfere with DCPD binding to full-length pre-S domain (data not shown). This observation prompted us to investigate the impact of DGD antibodies on productive DHBV infection in PDH. Indeed, incubation of cells with DGD antibodies for 48 h immediately following removal of virus inoculum totally blocked DHBV replication, but only at a 1:20 dilution (Fig. 6A, upper left panel). No such inhibition was achieved with more diluted immune serum or the same dilution of the preimmune rabbit serum (Fig. 6A, upper left panel). The 1:20 diluted DGD antibodies were not toxic to cells (Fig. 4B). Analysis of cccDNA immediately following antibody treatment revealed its reduction by the DGD antibodies (Fig. 6A, right panel), thus confirming the intervention of DHBV infection at an early step.
FIG. 5.
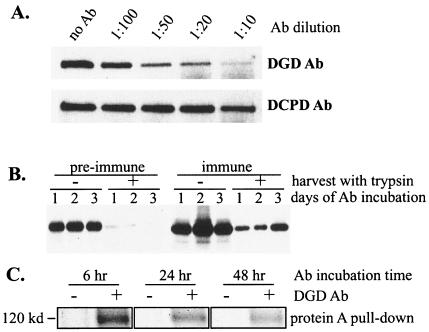
(A) Effect of DGD and DCPD antibodies on DGD binding to a truncated pre-S peptide. Duck liver lysate was incubated with pre-S peptide 80-102 immobilized on beads via the GST tag. DGD or DCPD antibodies (Ab) in various dilutions were present during the incubation, and DGD retained on the beads was detected by Western blot. (B) Antibody binding to duck hepatocytes and internalization. PDH were incubated with a 1:20 dilution of preimmune or immune serum for 1, 2, or 3 days, washed, and harvested with (+) or without (−) trypsin treatment. Cell-associated immunoglobulin was detected. (C) Specificity of antibodies attached to PDH. PDH were metabolically labeled and subsequently incubated for 6, 24, or 48 h with or without a 1:20 dilution of DGD antiserum. Cells were harvested, and the cell lysate was incubated with 30 μl of protein A beads. Bound proteins were revealed by SDS-PAGE and fluorography. The 120-kDa DGD protein was pulled down only in cells preincubated with DGD immune serum.
FIG. 6.
Reduction of DGD protein levels and impairment of DHBV infection by a 1:20 dilution of DGD antibodies added soon after virus binding. (A) Effect of rabbit serum added immediately following DHBV attachment. Left panels, PDH were incubated with a low dose of viremic duck serum for 6 h, washed, and immediately incubated with DGD preimmune or immune serum at various dilutions for 48 h. Cells were harvested 1 week postinfection. Viral DNA replication (upper left panel) and DGD levels (lower left panel) were measured. Right panel, cells infected with DHBV for 6 h were incubated with a 1:20 dilution of preimmune or immune serum for 24 h and harvested immediately. The protein-free DNA forms were analyzed. (B) Temporal effect of DGD antibodies. PDH were infected with viremic duck serum for 6 h and harvested 1 week postinfection. Rabbit serum (preimmune, immune, or nonrelevant) at a 1:20 dilution was present during the 6-h DHBV infection (left panels) or at different points following infection for 48 h (right panels). DGD protein levels at the time of harvest are shown in the lower panels, while markers for productive infection, such as viral DNA and core and envelope proteins, are shown in the upper panels. (C) Effect of DGD antibodies on DGD protein levels in DHBV-free cells. PDH were incubated with various dilutions of DGD immune serum for 1 or 2 days and harvested immediately for analysis of DGD levels. The GAPDH levels were measured to control for protein loading.
To determine the effective window of infection inhibition by DGD antibodies, we added a 1:20 dilution of DGD antibodies either during the short period (6 h) of DHBV incubation or for 48 h at various times following virus incubation. Productive DHBV infection as measured by core protein, large envelope protein, and viral DNA was inhibited only by antibodies added either immediately postinfection or 18 h later (Fig. 6B, upper panels). Interestingly, inhibition of DHBV infection was closely associated with down-regulation of DGD protein levels. For example, the endogenous DGD level was markedly reduced in cells treated with a 1:20 dilution, but not higher dilutions, of DGD antibodies (Fig. 6A, lower left panel). A 1:20 dilution of DGD antibodies added 42 h following virus infection, which failed to block DHBV replication, was also incapable of depleting endogenous DGD (Fig. 6B, lower right panel). In uninfected PDH, such a concentration of DGD antibodies could significantly reduce DGD levels following 2 days of incubation (Fig. 6C). These findings suggest that DGD degradation was mediated by its complex formation with corresponding antibodies, which became less efficient following DHBV infection (possibly through DGD sequestering by viral large envelope protein).
To directly measure the attachment and internalization of DGD antibodies, fresh PDH culture was incubated with a 1:20 dilution of preimmune or immune serum for 1, 2, or 3 days. Cells were harvested by scraping or trypsin treatment, which would remove cell surface antibodies. Western blot analysis revealed binding of antibodies from immune serum to cells, a fraction of which became resistant to trypsin treatment and was hence internalized (Fig. 5B, right panel). The DGD specificity of the bound antibodies was demonstrated by the ability of protein A beads alone to pull down endogenous DGD from such samples (Fig. 5C). Although a small amount of immunoglobulins from preimmune serum also bound to PDH, there was little internalization (Fig. 5B, left panel).
DISCUSSION
DGD was discovered serendipitously during the mapping of the DCPD binding site on the pre-S domain (18). It was found to interact strongly with several truncated pre-S polypeptides, such as 92-161, 98-161, and 1-102, but only barely with slightly extended versions, such as 87-161 and 1-104. Residues 98 to 102 constitute the DGD contact site, with a critical contribution provided by the C-terminal three residues, especially Arg102 (its conservative change to lysine was sufficient to abolish the DGD interaction) (18). This unique binding pattern suggests that the DGD contact site is masked by the folding of the full-length large envelope protein but can be activated by a cleavage event close to either side of this binding motif. Indeed, the sequence 97REAFRRY103 conforms to the PC7 recognition site (35). Circumstantial evidence supports a role for DGD in DHBV infection. First, the DGD binding site is highly conserved among the diverse DHBV strains and corresponds to a known neutralizing epitope (5). Second, DGD expression is restricted to DHBV-infectible duck tissues such as the liver, pancreas, and kidney (18, 19), although liver-specific expression of transcription factors required for production of viral pregenomic RNA could also restrict hepadnaviral infection to the liver (30). Third, double amino acid substitutions at positions 100 to 102 reduced DHBV infectivity (18).
Extending the observed tissue-specific expression of DGD, the present study documents a decline in DGD expression following prolonged culture of PDH. Under these experimental conditions, hepatocytes undergo de-differentiation, possibly through the methylation of liver-specific genes (26). The reduction in DGD expression correlated nicely with a gradual drop of cellular susceptibility to DHBV infection (Fig. 1). In contrast, neither expression of DCPD, the viral receptor, nor binding of DHBV to hepatocytes was impaired following prolonged culture of duck hepatocytes (Fig. 1). Introduction of DGD but not DCPD cDNA into 17-day-old PDH cultures enhanced formation of cccDNA; even more pronounced was the detection of protein-free RC DNA (Fig. 2A), a DNA form that is not abundantly present for DHBV infection established at an early time point. The accumulation of this DNA species suggests that additional factors required for productive DHBV infection are depleted in old culture of PDH. In another experiment, DGD reconstitution via adenovirus vector was associated with higher levels of core protein and viral DNA than that seen with DCPD- or vector-transduced cells (Fig. 2B). The less robust effect of DGD in this experiment (Fig. 2B) compared with that in the transfection experiment (Fig. 2A) may be related to the fact that the cells were older at the time of adenovirus infection.
Besides the reconstitution experiments performed in aged duck hepatocytes, we also carried out depletion experiments in freshly prepared cells. Blocking DGD expression via antisense RNA or depleting its protein pool by antibodies prevented DHBV infection (Fig. 3 and 6). Data obtained from the antibody studies are more extensive and informative. The DGD antibodies apparently inhibited an early step in viral life cycle, as indicated by reduction in the cccDNA levels (Fig. 6A, right panel). The antibodies present at the time of DHBV binding alone for 6 h failed to inhibit productive DHBV infection, as did antibodies supplied 42 h postbinding. Although the effectiveness of antibodies added 18 h postinfection is somewhat surprising, with only 6 h of incubation with diluted virus inoculum (5 μl per well of 12-well plates) very little cccDNA was formed during the first 24 h postinfection (our unpublished observations). Considering the time needed for antibodies to be internalized and to deplete intracellular DGD (Fig. 6C), DGD may be required in the viral life cycle between 1 to 2 days postbinding to DCPD. Whether that reflects a block in cccDNA formation from incoming RC virion DNA or prevention of cccDNA amplification from newly generated core particles or both remains an open question. The finding that DGD antibodies are effective following DHBV binding is consistent with the observations from prolonged culture of PDH, where DHBV binding per se was not affected by reduced DGD expression (Fig. 1D) while generation of protein-free RC DNA could be partially restored by exogenous DGD (Fig. 2). Once the cccDNA pool is established, DGD is not required for viral transcription or replication. We found that DHBV DNA replication continued for more than 1 month, when DGD expression was no longer present (J. Li et al., unpublished data).
The exact mechanism of DGD interaction with DHBV during natural infection remains to be determined. Despite DGD cell surface availability (18, 19), its pre-S binding domain appears inaccessible on the exterior of the plasma membrane (unpublished observations). This phenomenon may explain why anti-DGD antibodies block DHBV infectivity only at high concentrations (1:10 or 1:20 dilution), despite the fact that they inhibited the interaction between DGD and truncated pre-S peptide in cell lysate in a dose-dependent manner (Fig. 5A). The high concentrations of antibodies were needed to deplete the cellular pool of DGD (Fig. 6C), apparently through binding to cell surface DGD followed by intracellular degradation of the antigen-antibody complex (Fig. 5 and 6C) as reported previously (20). We have never encountered a situation where DGD expression was reduced by antibodies yet productive DHBV infection continued to proceed.
This study suggests that DHBV requires a second pre-S binding protein for initiation of infection. We envision this requirement to be related to the need for proteolytic cleavage of viral envelope proteins, which is known to occur for many other enveloped viruses inside the lumen of the secretory pathway. Indeed, HIV particles released to the circulation contain heterodimers of gp120 (the N-terminal part of gp160) and gp41 (the membrane-bound C-terminal part), with gp120 mediating HIV attachment to CD4-positive cells. In contrast, circulating virus particles of the hepadnavirus family do not display processed envelope proteins (at least at position 102), possibly owing to their second role as matrix proteins during virion formation. The 3′ one-third of both the HBV and DHBV pre-S domain has been implicated in envelopment of core particles (3, 17), and the pre-S domain of both HBV and DHBV faces cytosol rather than lumen during viral morphogenesis (4, 8, 23, 24, 29). This orientation would likely preclude processing by the protease inside the lumen. Only at a certain stage of virion formation would half of the large envelope protein undergo a dramatic conformational change to be exposed on the virion surface (4, 8, 23, 24, 29). We suggest that DCPD serves to retarget DHBV particles to the secretory pathway, where viral large envelope protein may be processed. The cleaved viral particles subsequently interact with DGD, the functional equivalent of other viral receptors, at an unknown subcellular location. This hypothesis could explain, in part, the long incubation period required to initiate DHBV replication and is consistent with the following experimental observations. First, pre-S peptide 1-102 corresponds to the N-terminal region of DHBV large envelope protein and binds strongly to DGD. Second, the shared pre-S binding sites of DCPD and DGD may allow a switch of DHBV binding from DCPD to DGD following cleavage. Third, DCPD is known to transit between the cell surface and the trans-Golgi apparatus (2, 36), a venue for proteolytic cleavage of viral envelope proteins. Fourth, furin is the major protease involved in processing viral envelope proteins and it also translocates between the cell surface and the trans-Golgi apparatus (12, 21). Indeed, furin was found to process DCPD and enhance DHBV retention (S. Tong et al., unpublished data).
Alternatively, DGD may work at the stage of cccDNA amplification without the need for a proteolytic cleavage. Although the secondary structure of the intact pre-S domain apparently makes it poorly recognizable by DGD (which can be overcome by cleavage at either side of the DGD binding site), the DGD binding motif might be available in the nascent large envelope protein, when it is bound to molecular chaperons. In this regard, cytosolic heat shock protein Hsc70 has been found to bind to the pre-S domain of HBV large envelope protein, which facilitates its interaction with core particles leading to virion formation (16). Since the DGD binding site on the pre-S domain (98-102) is adjacent to the pre-S domain required for envelopment (3, 17), binding of DGD to the cytosolic pre-S domain may prevent core-envelope interaction and inhibit virion formation. At the early stage of infection, delay of virion formation may recruit the core particles to the nucleus for cccDNA amplification, which is essential for efficient viral replication. A third possibility is that DGD interaction, at the site of cell-cell junction, facilitates spread of DHBV infection into neighboring cells, rather than retaining the virus at the original cells of attachment. Further studies will be needed to sort through these various possibilities of DGD action. At any rate, establishment of the critical role of DGD in the life cycle of a hepadnavirus may provide a potential molecular target for inhibition of productive viral replication in the liver.
Acknowledgments
We are grateful to T. He and B. Vogelstein from Johns Hopkins University for adenoviral vectors.
J. Li was a Liver Scholar of the American Liver Foundation. This work was supported by NIH grants CA095490, CA-35711, and P20RR15578, and Lifespan Research Funds.
REFERENCES
- 1.Bour, S., R. Geleziunas, and M. Wainberg. 1995. The human immunodeficiency virus type 1 (HIV-1) CD4 and its central role in promotion of HIV-1 infection. Microbiol. Rev. 59:63-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Breiner, K., S. Urban, and H. Schaller. 1998. Carboxypeptidase D (gp180), a Golgi-resident protein, functions in the attachment and entry of avian hepatitis B viruses. J. Virol. 72:8098-8104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bruss, V. 1997. A short linear sequence in the pre-S domain of the large hepatitis B virus envelope protein required for virion formation. J. Virol. 71:9350-9357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bruss, V., X. Lu, R. Thomssen, and W. Gerlich. 1994. Post-translational alterations in transmembrane topology of the hepatitis B virus large envelope protein. EMBO J. 13:2273-2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chassot, S., V. Lambert, A. Kay, C. Godinot, B. Roux, C. Trepo, and L. Cova. 1993. Fine mapping of neutralization epitopes on duck hepatitis B virus (DHBV) pre-S protein using monoclonal antibodies and overlapping peptides. Virology 192:217-223. [DOI] [PubMed] [Google Scholar]
- 6.Clapham, P. R., and A. McKnight. 2002. Cell surface receptors, virus entry and tropism of primate lentiviruses. J. Gen. Virol. 83:1809-1829. [DOI] [PubMed] [Google Scholar]
- 7.Ganem, D., and R. Schneider. 2001. Hepadnaviridae: the viruses and their replication, p. 2923-2970. In D. Knipe and P. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 8.Guo, J., and J. Pugh. 1997. Topology of the large envelope protein of duck hepatitis B virus suggests a mechanism for membrane translocation during particle morphogenesis. J. Virol. 71:1107-1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hallenberger, S., M. Moulard, M. Sordel, H. D. Klenk, and W. Garten. 1997. The role of eukaryotic subtilisin-like endoproteases for the activation of human immunodeficiency virus glycoproteins in natural host cells. J. Virol. 71:1036-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He, T., S. Zhou, L. da Costa, J. Yu, K. Kinzler, and B. Vogelstein. 1998. A simplified system for generating recombinant adenoviruses. Proc. Natl. Acad. Sci. USA 95:2509-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ishikawa, T., K. Kuroki, R. Lenhoff, J. Summers, and D. Ganem. 1994. Analysis of the binding of a host cell surface glycoprotein to the pre-S protein of duck hepatitis B virus. Virology 202:1061-1064. [DOI] [PubMed] [Google Scholar]
- 12.Jones, B., L. Thomas, S. Molloy, C. Thulin, M. Fry, K. Walsh, and G. Thomas. 1995. Intracellular trafficking of furin is modulated by the phosphorylation state of a casein kinase II site in its cytoplasmic tail. EMBO J. 14:5869-5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klenk, H., and W. Garten. 1994. Activation cleavage of viral spike proteins by host proteases, p. 241-280. In E. Wimmer (ed.), Cellular receptors for animal viruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 14.Kuroki, K., R. Cheung, P. Marion, and D. Ganem. 1994. A cell surface protein that binds avian hepatitis B virus particles. J. Virol. 68:2091-2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuroki, K., F. Eng, T. Ishikawa, C. Turck, F. Harada, and D. Ganem. 1995. gp180, a host cell glycoprotein that binds duck hepatitis B virus particles, is encoded by a member of the carboxypeptidase gene family. J. Biol. Chem. 270:15022-15028. [DOI] [PubMed] [Google Scholar]
- 16.Lambert, C., and R. Prange. 2003. Chaperone action in the posttranslational topological reorientation of the hepatitis B virus large envelope protein: implications for translocational regulation. Proc. Natl. Acad. Sci. USA 100:5199-5204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lenhoff, R. J., and J. Summers. 1994. Coordinate regulation of replication and virus assembly by the large envelope protein of an avian hepadnavirus. J. Virol. 68:4565-4571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li, J., S. Tong, and J. Wands. 1996. Characterization of a 120-kilodalton pre-S-binding protein as a candidate duck hepatitis B virus receptor. J. Virol. 70:6029-6035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li. J., S., Tong, and J. Wands. 1999. Identification and expression of glycine decarboxylase (p120) as a duck hepatitis B virus pre-S envelope-binding protein. J. Biol. Chem. 274:27658-27665. [DOI] [PubMed] [Google Scholar]
- 20.Mohr, L., J. I. Schauer, R. H. Boutin, D. Moradpour, and J. Wands. 1999. Targeted gene transfer to hepatocellular carcinoma cells in vitro using a novel monoclonal antibody-based gene delivery system. Hepatology 29:82-89. [DOI] [PubMed] [Google Scholar]
- 21.Molloy, S., L. Thomas, J. VanSlyke, P. Stenberg, and G. Thomas. 1994. Intracellular trafficking and activation of the furin proprotein convertase: localization to the TGN and recycling from the cell surface. EMBO J. 13:18-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakayama, K. 1997. Furin: a mammalian subtilisin/Kex2p-like endoprotease involved in processing of a wide variety of precursor proteins. Biochem. J. 327:625-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ostapchuck, P., P. Hearing, and D. Ganem. 1994. A dramatic shift in the transmembrane topology of a viral envelope glycoprotein accompanies hepatitis B viral morphogenesis. EMBO J. 13:1048-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prange, P., and R. Streeck. 1995. Novel transmembrane topology of the hepatitis B virus envelope proteins. EMBO J. 14:247-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pugh, J., and J. Summers. 1989. Infection and uptake of duck hepatitis B virus by duck hepatocytes maintained in the presence of dimethyl sulfoxide. Virology 172:564-572. [DOI] [PubMed] [Google Scholar]
- 26.Pugh, J., and H. Simmons. 1994. Duck hepatitis B virus infection of Muscovy duck hepatocytes and nature of virus resistance in vivo. J. Virol. 68:2487-2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spangenberg, H., H. Lee, J. Li, F. Tan, R. Skidgel, J. Wands, and S. Tong. 2001. A short sequence within domain C of duck carboxypeptidase D is critical for duck hepatitis B virus binding and determines host specificity. J. Virol. 75:10630-10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Summers, J., and W. S. Mason. 1982. Replication of the genome of a hepatitis B-like virus by reverse transcription of an RNA intermediate. Cell 29:403-415. [DOI] [PubMed] [Google Scholar]
- 29.Swameye, I., and H. Schaller. 1997. Dual topology of the large envelope protein of duck hepatitis B virus: determinants preventing pre-S translocation and glycosylation. J. Virol. 71:9434-9441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tang, H., and A. McLachlan. 2001. Transcriptional regulation of hepatitis B virus by nuclear hormone receptors is a critical determinant of viral tropism. Proc. Natl. Acad. Sci. USA 98:1841-1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tong, S., J. Li, and J. Wands. 1995. Interaction between duck hepatitis B virus and a 170-kilodalton cellular protein is mediated through a neutralizing epitope of the pre-S region and occurs during viral infection. J. Virol. 69:7106-7112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tong, S., J. Li, and J. Wands. 1999. Carboxypeptidase D is an avian hepatitis B virus receptor. J. Virol. 73:8696-8702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tuttleman, J., J. Pugh, and J. Summers. 1986. In vitro experimental infection of primary duck hepatocyte cultures with duck hepatitis B virus. J. Virol. 58:17-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Urban, S., C. Schwarz, U. Marx, H. Zentgraf, H. Schaller, and G. Multhaup. 2000. Receptor recognition by a hepatitis B virus reveals a novel mode of high affinity virus-receptor interaction. EMBO J. 19:1217-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van de Loo, J., J. Creemers, N. Bright, B. Young, A. Roebroek, and W. Van de Ven. 1997. Biosynthesis, distinct post-translational modifications, and functional characterization of lymphoma proprotein convertase. J. Biol. Chem. 272:27116-27123. [DOI] [PubMed] [Google Scholar]
- 36.Varlamov, O., and L. Fricker. 1998. Intracellular trafficking of metallocarboxypeptidase D in AtT-20 cells: localization to the trans-Golgi network and recycling from the cell surface. J. Cell Sci. 111:877-885. [DOI] [PubMed] [Google Scholar]
- 37.Yu, M., and J. Summers. 1994. Multiple functions of capsid protein phosphorylation in duck hepatitis B virus replication. J. Virol. 68:4341-4348. [DOI] [PMC free article] [PubMed] [Google Scholar]