Abstract
Although the left and right human cerebral hemispheres differ both functionally and anatomically, the mechanisms that underlie the establishment of these hemispheric specializations, as well as their physiological and behavioral implications, remain largely unknown. Since cerebral asymmetry is strongly correlated with handedness, and handedness is assumed to be influenced by a number of genetic and environmental factors, we performed an association study of LRRTM1 rs6733871 and a number of polymorphisms in PCSK6 and different aspects of handedness assessed with the Edinburgh handedness inventory in a sample of unrelated healthy adults (n = 1113). An intronic 33bp variable-number tandem repeat (VNTR) polymorphism in PCSK6 (rs10523972) shows a significant association (significance threshold: p<0.0025, adjusted for multiple comparisons) with a handedness category comparison (P = 0.0005) and degree of handedness (P = 0.001). These results provide further evidence for the role of PCSK6 as candidate for involvement in the biological mechanisms that underlie the establishment of normal brain lateralization and thus handedness and support the assumption that the degree of handedness, instead the direction, may be the more appropriate indicator of cerebral organization.
Introduction
Handedness is the most obvious manifestation of cerebral lateralization in humans, with more than 90% preferring to use their right hand [1]. In addition to the usually measured direction of this lateralization (left versus right), handedness is also characterized by its degree (consistent vs. inconsistent) [2], [3]. Handedness is a heritable trait, and many studies are consistent in assuming a polygenic model of handedness which means that a large number of genetic factors with a small additive effect may contribute to the trait variance [4]. Additive genetic effects were shown to account for about a quarter of the variance with the remainder accounted for by non-shared environmental influences [5]–[7]. Linkage analyses have identified a number of loci of interest including chromosome regions 2p12-q11, 10q26 and 12q21–23 [8]–[11]. Yet, to date, only few specific genes have been suggested as candidates influencing handedness: Francks et al. (2007) reported evidence for the involvement of the imprinted gene leucine-rich repeat transmembrane neuronal 1 (LRRTM1) on chromosome 2p12 in the development of schizophrenia/schizoaffective disorder and human handedness [12]. LRRTMs bind neurexins and act as control factors of presynaptic and postsynaptic glutamatergic differentiation [13]. In detail, Francks et al. (2007) report the association of a three-marker haplotype within an intron of CTNNA2, 137kb upstream of LRRTM1, with schizophrenia when inherited paternally [12]. Furthermore, this haplotype was also found to be paternally associated with handedness in a sample of reading-disabled sibships, implying the existence of specific imprinting effects on human brain [12], [14], [15]. The imprinting effects within LRRTM1 and their association with schizophrenia could be supported by a study analysing a cohort of German patients. In addition to the haplotype specific markers, imprinting effects were also observed for the non-synonymous SNP rs6733871 that leads to an amino acid change (N330S) in LRRTM1. Furthermore, using the quantitative transmission disequilibrium test (QTDT), association between variation in LRRTM1 rs6733871 and handedness in a well-characterized dyslexia sample of German descent was observed [16].
Recently, a genome-wide association study for a quantitative measure of relative handskill in individuals with reading disability (RD) revealed association with SNPs in PCSK6 (proprotein convertase subtilisin/kexin type 6, also known as PACE4) which is known to have regulatory function during anterior CNS patterning and left-right axis formation [17], [18]. The strongest association with the quantitative measure of relative hand skill (peg-board task) was found for two intronic PCSK6 SNPs, rs9806256 and rs11855415. RD individuals carrying the minor allele showed significantly greater relative right-hand skill as compared with the frequent wild-type allele. Replication in independent cohorts of individuals with RD revealed the same effect for the minor rs11855415 allele, while the same allele showed a significant trend towards reduced hand skill in the general population [17].
Here we report an association study of LRRTM1 rs6733871 and a number of polymorphisms in PCSK6 and different aspects of handedness assessed with the Edinburgh handedness inventory in a sample of 1113 unrelated healthy Caucasian adults. The results show a significant association with an intronic 33bp variable-number tandem repeat (VNTR) polymorphism in PCSK6 (rs10523972) and handedness.
Results
By using data from 1113 unrelated individuals, we tested the LRRTM1 SNP rs6733871 and a number of SNPs in PCSK6 for association with different aspects of handedness (Table 1). Each individual reported their hand preference (i.e., right, left or ambidextrous) in response to 10 questions (e.g., which hand do you prefer to use when: writing? throwing? using scissors?). All responses were converted to a laterality quotient (LQ) using the formula (R − L)/(R+L)×100. LQ scores thus ranged from −100 for strong left-handedness to +100 for strong right-handedness. Individuals were assigned to one of six categories of handedness: 1) consistent left (LQ = −100, 1.9%), 2) inconsistent left (LQ>−100 and LQ ≤ −50, 4.1%), 3) ambidextrous with a tendency towards left (LQ>−50 and LQ >0, 2.0%), 4) ambidextrous with a tendency towards right (LQ = 0 and LQ<+50, 4.0%),5) inconsistent right (LQ ≥ +50 and LQ<+100, 40.5%), or 6) consistent right (LQ = +100, 47.5%).
Table 1. p-values for the different association tests (the LQ has been analyzed using an ANOVA, while handedness groups, handedness direction and degree of handedness have been analyzed using Kruskal-Wallis-tests).
| Gene | SNP | LQ | Handedness groups | Handedness direction | Degree of handedness |
| LRRTM1 | rs6733871 | 0.35 | 0.34 | 0.54 | 0.41 |
| PCSK6 | rs11855415 | 0.33 | 0.84 | 0.13 | 0.88 |
| rs9806256 | 0.89 | 0.77 | 0.69 | 0.43 | |
| rs1947942 | 0.37 | 0.21 | 0.41 | 0.33 | |
| rs10523972 | 0.02 | 0.0005* | 0.07 | 0.001* |
Effects significant at the p<0.0025 level (the adjusted p value for multiple comparision) are given in bold.
Note: * = p<0.0025 (Bonferroni corrected p value).
The p-values for all genotype group comparisons for LQ, handedness groups (consistent left-handers, inconsistent left-handers, ambidexter with a tendency towards left-handedness, ambidexter with a tendency towards right-handedness, inconsistent right-handers, consistent right-handers), handedness direction (left, right) and degree of handedness (consistent, inconsistent).
When testing LQ, handedness direction (left, right), degree of handedness (consistent vs. inconsistent) and affiliation to one of the six categories of handedness for association with the LRRTM1 and PCSK6 one SNP rendered p-values below p<0.0025, the adjusted significance level for multiple comparisons (Table 1). The analyzed SNPs in PCSK6 lie in a 12-kb region spanning introns 14–18 and are in low to high linkage disequilibrium (LD, D’ = 0.07–0.87) with each other. Rs10523972 is an intronic 33bp variable-number tandem repeat (VNTR) polymorphism in PCSK6 with eight different alleles (4–11 copies); of which 6 and 9 copies were most frequently observed (20.4% and 70.2%). Dichotomizing these alleles into short (≤6 repeats) and long (≥9 repeats) alleles reveals significant associations for the handedness category comparison (Kruskal-Wallis test: χ2 = 15.07; p = 0.0005) and the degree of handedness (Kruskal-Wallis test: χ2 = 13.32; p = 0.001).
Moreover, the results show that among individuals heterozygous for the long and short PCSK6 VNTR alleles the number of consistent right-handers was much lower (39.03%) than in individuals homozygous for the long allele (50.52%). The findings for individuals homozygous for the short allele should be interpreted carefully, since this group is very small (n = 55, Table 2).
Table 2. Observed frequencies of handedness in relation to the PCSK6 rs10523972 alleles.
| PCSK6 VNTR alleles (n) | CLH | ILH | ALH | ARH | IRH | CRH |
| long/long (685) | 13 (1.93%) | 23 (3.42%) | 11 (1.63%) | 25 (3.71%) | 261 (38.78%) | 340 (50.52%) |
| long/short (339) | 7 (1.99%) | 21 (5.98%) | 9 (2.56%) | 17 (4.84%) | 160 (45.58%) | 137 (39.03%) |
| short/short (55) | 1 (1.82%) | 0 (0%) | 1 (1.82%) | 2 (3.64%) | 21 (38.18%) | 30 (54.55%) |
Numbers and percentages of consistent left-handers (CLH), inconsistent left-handers (ILH), ambidexter with a tendency towards left-handedness (ALH), ambidexter with a tendency towards right-handedness (ARH), inconsistent right-handers (IRH) and consistent right-handers (CRH) for the combined PCSK6 rs10523972 alleles.
The significant degree of handedness effect (Kruskal-Wallis test: χ2 = 13.32; p = 0.001) indicates a difference in the number of participants with consistent and non-consistent handedness. To further investigate this effect, we compared the distribution of consistent and non-consistent handedness for the different allele combinations against a uniform 50/50 distribution, resembling the overall distribution of consistent or inconsistent handed participants (consistent: 527 individuals, inconsistent: 552 individuals). This analysis revealed that heterozygous participants were more prone to inconsistent hand preference (consistent: 140; inconsistent: 199; χ2 = 10.27; P = 0.001, Table 1). Yet, the effect failed to reach significance for both homozygous allele combinations (long/long, consistent: 357; inconsistent: 328; χ2 = 1.23; p = 0.27 and short/short, consistent: 30; inconsistent: 25; χ2 = 0.46; p = 0.5).
Scerri et al. (2011) report a significant association with the minor allele of rs11855415 and increased relative right-hand skill in individuals with RD. Yet, the minor allele showed a significant trend towards reduced laterality of hand skill in the general population [17]. In order to compare the results we also z-standardized the LQ scores (mean = 0, SD = 1) and plotted their distribution for both rs118555415 and rs10523972 (see Fig. S1 and Fig. S2). The distribution of LQ scores for the minor alleles (rs118555415 AA and rs10523972 short/short), appear more tightly clustered around the mean than the frequent alleles, suggesting that carriers of the minor alleles show reduced variability in relative hand skill as compared to carriers of the frequent alleles. Further comparing the absolute value of the z-standardized LQ as a quantitative trait between these groups (see Fig. S3 and Fig. S4) indeed revealed that the homozygous carriers of the minor alleles for rs118555415 and rs10523972 showed the lowest absolute z-standardized scores (0.52 and 0.52) as compared with the homozygous carriers of the frequent alleles and the heterozygotes (0.60 and 0.61; 0.69 and 0.68). Yet, the effect for both SNPs failed to reach nominal significance (p = 0.14 and p = 0.22).
Discussion
Our data show a statistical trend supporting earlier findings for an association of PCSK6 with handedness. No supportive evidence was obtained for a possible association of the non-synonymous LRTTM1 SNP rs6733871. However, neither the three-marker haplotype nor parental effects were analyzed. The finding that tandem repeat variation in PCSK6 (rs10523972) shows a significant association with the degree of handedness could be explained by altered physiological and biological changes associated with this polymorphism. For example, VNTRs in regulating sequences can affect gene expression by altering the number of transcription factor binding sites or introducing changes in spacing between critical promoter elements [19]. Yet, gene expression can also be affected by modulating the activity of RNA-binding proteins or mediating effects on chromatin structure [19]. According to the Encyclopedia of DNA Elements (ENCODE) dataset, available for visualization and download via the UCSC Genome Browser (http://genome.ucsc.edu/), the region containing the PCSK6 VNTR sequence clusters multiple transcription factor binding sites and is enriched for histone H3 lysine 27 acetylation (H3K27ac), a reliable chromatin marker of active (Fig. 1) [20], [21]. Additionally, the UCSC genome browser lists different spliced expressed sequence tags (ESTs) that emanate from the VNTR region, suggestive of a bidirectional promoter that can drive the expression of further truncated PCSK6 mRNA variants that differ in their C- or N-terminal segments [22]. Thus, variation in its repeat number might well have considerable physical impact on the DNA, which can influence in a tissue-dependent manner gene regulation, DNA organization and many other mechanisms [23]. As already reviewed and discussed in detail in Scerri et al. (2011), PCSK6 plays an important role in the pathway for left-right asymmetry and has also been described to be a direct target of the transcription factor FOXP2, whose gene was the first one known to be involved in the development of speech and language [17], [24].
Figure 1. Graphical representation of the analyzed single-nucleotide polymorphisms in relation to the exon-intron structure at the 3’ end of PCSK6.
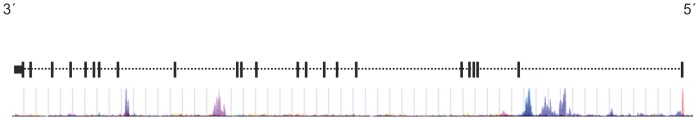
H3K7Ac ChIP-Seq data from the ENCODE project, presented beneath, were obtained from the UCSC genome browser.
The lack of replication in our sample of the reported association with LRRTM1 might be explained by different study designs, including the participant populations and definition of handedness. Individual differences in handedness were often reduced on the direction of hand preference, left versus right-handers. Yet, evidence has accumulated that the degree (consistent versus inconsistent) of hand preference probably reflects a more critical dimension on which the handedness groups differ [25], [26].
In summary, the results of our study provide evidence for an association of PCSK6 variation and the degree of handedness in the general population. Together with previously published data on a PCSK6 association and reduced variability in relative hand skill a healthy cohort [17] these results further support PCSK6s likely role as candidate for involvement in the biological mechanisms that underlie the establishment of normal brain lateralization and thus handedness. However, independent replication in larger cohorts is needed in order to further validate the present results.
Furthermore, our data support the assumption that the degree of handedness, i.e. how consistently a person prefers to use one versus the other hand over a wide variety of tasks may be the more appropriate indicator of cerebral organization. Future association studies should therefore take into consideration the different measures to define handedness. Understanding the complex genetic contribution to handedness could be crucial in order to advance our knowledge of normal brain functioning, but also the understanding of neurodevelopmental and psychiatric diseases.
Methods
Ethics Statement
The study was approved by the ethics committee, Ruhr-University Bochum. All participants gave written informed consent and were treated in accordance with the declaration of Helsinki.
Study Participants
In total, 1113 genetically unrelated, healthy adult participants of Caucasian descent for at least two generations participated in the present study (655 women and 458 men). The participants had a mean age of 32.97 (SEM = 0.55) and none of them had a history of any neurological or psychiatric diseases. 895 of the participants were young adults (mostly university students), that were recruited for the present study, while the remaining 218 participants came from a sample of healthy elderly volunteers. All participants were native German speakers. No participants are included in the cohort that had been forced to write with the right hand in school, although they actually would have preferred to use the left.
Handedness Assessment
Handedness was assessed using the Edinburgh handedness inventory [27]. In this questionnaire, participants have to indicate whether they prefer to use the left or right hand for ten different activities. Based on the participants answers, an individual laterality quotient (LQ) can be calculated using the formula LQ = [(R−L)/(R+L)]×100 (R = number of right sided preferences; L = number of left-sided preferences). The LQ has a range between +100 and −100. Positive values indicate right handedness and negative values left handedness, while higher absolute values indicate more consistent handedness and lower absolute values more inconsistent handedness or ambidexterity. Based on each participant’s individual LQ, three different measures were calculated in order to separately investigate different aspects of handedness that may be associated with different genetic variations. In order to gain a measure of handedness direction, participants were grouped into right-handers (RH; LQ between 0 and 100) and left-handers (LH; LQ between−100 and 0), an approach, that is commonly used in large-scale studies [6]. Additionally, we applied a more complex grouping method, classifying participants into six different handedness categories: consistent left handers (LQ = -100), inconsistent left-handers (LQ = -99 to −50), ambidexter with a tendency towards left-handedness (LQ = −49 to 0), ambidexter with a tendency towards right-handedness (LQ = 0 to 49), inconsistent right-handers (50 to 99) and consistent right handers (LQ = 100). Also, in order to gain a dichotomous measure of the degree of handedness independent of the individuals preferred hand, participants were grouped into individuals with consistent (LQ either 100 or −100) or individuals with inconsistent (all other LQ’s) handedness.
Genotyping
For non-invasive sampling, exfoliated cells were brushed from the oral mucosa of the participants. DNA isolation was performed with QIAamp DNA mini Kit (50) (Qiagen GmbH, Hilden, Germany). SNPs were chosen for genotyping by selecting SNPs from the literature (LRRTM1 rs6733871, PCSK6 rs11855415, rs9806256) or from the UCSC genome browser, with particular attention to SNPs in conserved sequences (rs1947942) or SNPs that disrupt putative transcription factor binding sites (rs10523972). Genotyping of four SNPs (rs6733871, rs11855415, rs9806256 and rs1947942) was conducted by polymerase chain reaction (PCR) and differential enzymatic analysis with the PCR restriction fragment length polymorphism method. Genotyping of the PCSK6 VNTR, rs10523972, was performed on the Beckman Coulter CEQ8000 8-capillary system using ’Fragment Analysis Module’ software (Beckman Coulter, Inc., Fullerton, USA). The genotypes were confirmed by sequence analysis. Oligonucleotides were designed using Primer Express 2.0 Software (Applied Biosystems). Further details of methodology and primer sequences are available upon request.
Statistical Analysis
The statistical analyses were performed assuming a co-dominant effect for each polymorphism. Thus, all three genotypes for each polymorphism were analyzed separately. Since the LQ is an interval-scaled variable, it was analyzed parametrically using univariate analyses of variance (ANOVAs). Greenhouse-Geisser correction was applied when appropriate. Nominal-scaled variables (handedness categories, handedness direction and degree of handedness) were analyzed non-parametrically using the Kruskal-Wallis Test. In order to account for multiple comparisons, an effect was considered significant if the p-value was smaller than 0.05/20 (the overall number comparisons) = 0.0025 (Bonferroni correction), a highly conservative significance threshold. All statistics were calculated using IBM SPSS Statistics 20.
Supporting Information
Plot of z-standardized LQ score (mean = 0, SD = 1) distribution (y-axis) for each genotype of rs11855415 (x-axis) in 1113 genetically unrelated, healthy adult participants of Caucasian descent.
(JPG)
Plot of z-standardized LQ score (mean = 0, SD = 1) distribution (y-axis) for the dichotomized genotypes of rs10523972 (x-axis) in 1113 genetically unrelated, healthy adult participants of Caucasian descent.
(JPG)
Plot of the absolute value of the LQ score (y-axis) for each genotype of rs11855415 (x-axis) in 1113 genetically unrelated, healthy adult participants of Caucasian descent.
(JPG)
Plot of the absolute value of the LQ score (y-axis) for the dichotomized genotypes of rs10523972 (x-axis) in 1113 genetically unrelated, healthy adult participants of Caucasian descent.
(JPG)
Funding Statement
This work was supported by a DFG Emmy-Noether grant [BE4045/10-1 to C.B.] and a Cutting-Edge Grant from the Ruhr-University of Bochum. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Sun T, Walsh CA (2006) Molecular approaches to brain asymmetry and handedness. Nat Rev Neurosci 7: 655–662. [DOI] [PubMed] [Google Scholar]
- 2. Lyle KB, Hanaver-Torrez SD, Hackländer RP, Edlin JM (2012) Consistency of handedness, regardless of direction, predicts baseline memory accuracy and potential for memory enhancement. J Exp Psychol Learn Mem Cogn 38: 187–193. [DOI] [PubMed] [Google Scholar]
- 3. Kempe V, Brooks PJ, Christman SD (2009) Inconsistent handedness is linked to more successful foreign language vocabulary learning. Psychon Bull Rev 16: 480–485. [DOI] [PubMed] [Google Scholar]
- 4.McManus IC, Davison A, Armour JAL (2013) Multi-locus genetic models of handedness closely resemble single-locus models in explaining family data and are compatible with genome-wide association studies. Annals of the New York Academy of Sciences, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Geschwind DH, Miller BL, DeCarli C, Carmelli D (2002) Heritability of lobar brain volumes in twins supports genetic models of cerebral laterality and handedness. Proc Natl Acad Sci U S A, 99, 3176–3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Medland SE, Duffy DL, Wright MJ, Geffen GM, Martin NG (2006) Handedness in twins: joint analysis of data from 35 samples. Twin Res Hum Genet 9: 46–53. [DOI] [PubMed] [Google Scholar]
- 7. Medland SE, Duffy DL, Wright MJ, Geffen GM, Hay DA, et al. (2009) Genetic influences on handedness: data from 25,732 Australian and Dutch twin families. Neuropsychologia 47: 330–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Francks C, Fisher SE, MacPhie IL, Richardson AJ, Marlow AJ, et al. (2002) A genomewide linkage screen for relative handskill in siblingpairs. Am J Hum Genet 70: 800–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Francks C, DeLisi LE, Fisher SE, Laval SH, Rue JE, et al. (2003) Confirmatory evidence for linkage of relative hand skill to 2p12-q11. Am J Hum Genet 72: 499–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Van Agtmael T, Forrest SM, Williamson R (2002) Parametric and non-parametric linkage analysis of several candidate regions for genes for human handedness. Eur J Hum Genet 10: 623–630. [DOI] [PubMed] [Google Scholar]
- 11. Warren DM, Stern M, Duggirala R, Dyer TD, Almasy L (2006) Heritability and linkage analysis of hand, foot, and eye preference in Mexican Americans. Laterality 11: 508–524. [DOI] [PubMed] [Google Scholar]
- 12. Francks C, Maegawa S, Laurén J, Abrahams BS, Velayos-Baeza A, et al. (2007) LRRTM1 on chromosome 2p12 is a maternally suppressed gene that is associated paternally with handedness and schizophrenia. Mol Psychiatry 12: 1129–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Siddiqui TJ, Pancaroglu R, Kang Y, Rooyakkers A, Craig AM (2010) LRRTMs and neuroligins bind neurexins with a differential code to cooperate in glutamate synapse development. J Neurosci 30: 7495–7506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Crow TJ, Close JP, Dagnall AM, Priddle TH (2009) Where and what is the right shift factor or cerebral dominance gene? A critique of Francks et al. (2007). Laterality 14: 3–10. [DOI] [PubMed] [Google Scholar]
- 15. McManus C, Nicholls M, Vallortigara G (2009) Editorial commentary: is LRRTM1 the gene for handedness? Laterality 14: 1–2. [DOI] [PubMed] [Google Scholar]
- 16. Ludwig KU, Mattheisen M, Mühleisen TW, Roeske D, Schmäl C, et al. (2009) Supporting evidence for LRRTM1 imprinting effects in schizophrenia. Mol Psychiatry 14: 743–745. [DOI] [PubMed] [Google Scholar]
- 17. Scerri TS, Brandler WM, Paracchini S, Morris AP, Ring SM, et al. (2011) PCSK6 is associated with handedness in individuals with dyslexia. Hum Mol Genet 20: 608–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Constam DB, Robertson EJ (2000) SPC4/PACE4 regulates a TGFbeta signaling network during axis formation. Genes Dev 14: 1146–1155. [PMC free article] [PubMed] [Google Scholar]
- 19. Gemayel R, Cho J, Boeynaems S, Verstrepen KJ (2012) Beyond Junk-Variable Tandem Repeats as Facilitators of Rapid Evolution of Regulatory and Coding Sequences. Genes 3: 461–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rosenbloom KR, Dreszer TR, Long JC, Malladi VS, Sloan CA, et al. (2012) ENCODE whole-genome data in the UCSC Genome Browser: update 2012. Nucleic Acids Res 40: D912–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, et al. (2010) Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci USA 107: 21931–21936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhong M, Benjannet S, Lazure C, Munzer S, Seidah NG (1996) Functional analysis of human PACE4-A and PACE4-C isoforms: identification of a new PACE4-CS isoform. FEBS Lett 396: 31–36. [DOI] [PubMed] [Google Scholar]
- 23. Gemayel R, Vinces MD, Legendre M, Verstrepen KJ (2010) Variable tandem repeats accelerate evolution of coding and regulatory sequences. Annu Rev Genet 44: 445–477. [DOI] [PubMed] [Google Scholar]
- 24. Enard W (2011) FOXP2 and the role of cortico-basal ganglia circuits in speech and language evolution. Curr Opin Neurobiol 21: 415–424. [DOI] [PubMed] [Google Scholar]
- 25. Prichard E, Propper RE, Christman SD (2013) Degree of Handedness, but not Direction, is a Systematic Predictor of Cognitive Performance. Front Psychol 4: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lyle KB, Orsborn AE (2011) Inconsistent handedness and saccade execution benefit face memory without affecting interhemispheric interaction. Memory 19: 613–624. [DOI] [PubMed] [Google Scholar]
- 27. Oldfield RC (1971) The assessment and analysis of handedness: the Edinburgh inventory. Neuropsychologia 9: 97–113. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Plot of z-standardized LQ score (mean = 0, SD = 1) distribution (y-axis) for each genotype of rs11855415 (x-axis) in 1113 genetically unrelated, healthy adult participants of Caucasian descent.
(JPG)
Plot of z-standardized LQ score (mean = 0, SD = 1) distribution (y-axis) for the dichotomized genotypes of rs10523972 (x-axis) in 1113 genetically unrelated, healthy adult participants of Caucasian descent.
(JPG)
Plot of the absolute value of the LQ score (y-axis) for each genotype of rs11855415 (x-axis) in 1113 genetically unrelated, healthy adult participants of Caucasian descent.
(JPG)
Plot of the absolute value of the LQ score (y-axis) for the dichotomized genotypes of rs10523972 (x-axis) in 1113 genetically unrelated, healthy adult participants of Caucasian descent.
(JPG)


