Abstract
Previously we have described a stepwise, energy-dependent pathway for human immunodeficiency virus type 1 (HIV-1) capsid assembly in a cell-free system. In this pathway, Gag polypeptides utilize the cellular factor HP68 and assemble into immature capsids by way of assembly intermediates that have defined biochemical characteristics. Here we address whether this pathway is universally conserved among primate lentiviruses and can be observed in mammalian cells. We demonstrate that HIV-2 Gag associates with human HP68 in a cell-free system and that Gag proteins of HIV-2, simian immunodeficiency virus SIVmac239, and SIVagm associate with endogenous HP68 in primate cells, as is seen for HIV-1. Analysis of primate cells expressing lentivirus Gag proteins revealed Gag-containing complexes with the same sedimentation values as seen for previously described HIV-1 assembly intermediates in the cell-free system (10S, 80-150S, and 500S). These complexes fit criteria for assembly intermediates as judged by energy sensitivity, pattern of HP68 association, and the failure of specific complexes to be formed by assembly-incompetent Gag mutants. We also demonstrate that virus-like particles released from cells do not appear to contain HP68, suggesting that HP68 is released from Gag upon completion of capsid assembly in cells, as was observed previously in the cell-free system. Together these findings support a model in which all primate lentivirus capsids assemble by a conserved pathway of HP68-containing, energy-dependent assembly intermediates that have specific biochemical features.
Human immunodeficiency virus type 1 (HIV-1) particle production can be viewed as a series of distinct biochemical events: first, translation and targeting of newly synthesized Gag polypeptides; next, the posttranslational events of capsid assembly and genome encapsidation; and finally, budding, release, and protease-mediated maturation. With new advances, it is becoming increasingly clear that each of these steps requires complex virus-host interactions (19). This is best exemplified by recent studies showing that budding requires a number of cellular factors, including Tsg101 and the ESCRT proteins (17, 41, 42, 55; reviewed in reference 16).
The steps that precede budding are the posttranslational events of immature HIV-1 capsid assembly. The exact mechanism by which Gag assembles posttranslationally in cells remains poorly understood. Capsids of some viruses, such as tobacco mosaic virus (reviewed in reference 32), have been shown to self-assemble, suggesting that the property of capsid protein multimerization is intrinsic to the capsid proteins themselves and is not dependent on other proteins. In the case of retroviruses, a self-assembly model for capsid formation has been proposed based on studies in which purified HIV-1 or Rous sarcoma virus Gag polypeptides and fragments produce spherical structures when incubated with nucleic acid (6, 7, 12, 20-22, 56, 61) and inositol phosphate (5). However, these self-assembly studies were performed with high concentrations (typically 1 mg/ml) of polypeptides encoding Gag mutants that were produced recombinantly and purified. While demonstrating that Gag polypeptides self-associate in isolation, these studies leave unanswered the question of how wild-type (WT) Gag polypeptides assemble into immature capsids within the unique and crowded environment provided by the cytoplasm. In the cytosol, newly translated WT Gag is present at relatively low concentrations amid cellular protein levels of as high as 100 to 300 mg/ml (13), in marked contrast to self-assembly systems, which contain high concentrations of Gag with no significant cellular protein. A precedent for a difference in protein behavior when proteins are studied in isolation versus under cytosolic conditions is seen with protein folding: many proteins fold spontaneously when they are purified and incubated at high concentrations without other proteins but require molecular chaperones for proper folding when they are newly translated in a cellular environment (reviewed in reference 27).
Thus, systems that examine capsid proteins in isolation may not provide the best model for understanding the mechanism by which viral capsids form within the protein-rich environment of cells. However, cellular systems can be problematic for studying this question because posttranslational events of capsid assembly occur rapidly in cells, making biochemical detection and analysis of this process difficult. Historically, cellular mechanisms have been dissected successfully by utilizing cell-free systems, which reconstitute complex cellular events using cellular extracts (reviewed in reference 43). Cell-free systems, like cells, link protein synthesis to posttranslational events, contain cellular factors at quasiphysiological concentrations, and typically reproduce translational and posttranslational events with great fidelity but in a less rapid manner than in cells (2, 23, 46).
For these reasons, eukaryotic cell-free systems have been used to reconstitute and dissect cellular mechanisms involved in immature HIV-1 capsid formation. In these systems, Gag polypeptides are translated de novo and then assemble into capsids that closely resemble immature HIV-1 capsids produced in cells (37, 50). Phenotypes of 16 known assembly-defective and assembly-competent mutations in Gag, which were initially characterized in cells, are faithfully reproduced in a cell-free assembly system (37, 49). Dissection of this cell-free system for immature HIV-1 capsid assembly demonstrated that capsid formation occurs in a stepwise manner, requiring membranes, energy, and host factors (37). Pulse-chase experiments revealed that Gag chains progress sequentially through discrete posttranslational assembly intermediates with sedimentation values of approximately 10S, 80-150S, and 500S, culminating in production of fully assembled immature capsids of 750S. Cell-free studies also showed that ATP depletion prevents assembly of completed immature capsids, indicating that posttranslational events in capsid assembly are energy dependent (37). In addition, a cellular factor, termed HP68 (for host protein of 68 kDa), was found to play a critical role in immature HIV-1 capsid formation (62). In this cell-free system, HP68 associates with Gag polypeptides in the 80-150S and 500S assembly intermediates. Depletion-reconstitution in the cell-free system and dominant-negative experiments in primate cells demonstrated that HP68 is essential for posttranslational events in immature HIV-1 capsid formation (62). While it remains unclear how HP68 functions, HP68 contains ATP-binding motifs and data suggest that it promotes a conformational change in the structure of the nascent capsid (62). HP68 was previously described as an RNase L inhibitor that prevents host RNase L from degrading RNA (1a, 39, 40). It remains to be determined whether the ability of HP68 to protect viral RNA is linked to its role in promoting assembly.
Based on these studies, we have proposed a model in which HP68 plays an essential role in promoting energy-dependent, posttranslational events required for immature HIV-1 capsid formation in cells (62). In contrast to the self-assembly model, the stepwise pathway model implicates specific virus-host interactions as critical for proper immature capsid formation and is therefore important to verify further. The first question we examined here is whether Gag polypeptides from other primate lentiviruses associate with HP68 in primate cells. The Gag proteins studied here represent members of three major primate lentivirus lineages (24) and share only approximately 60% amino acid conservation between lineages. An interesting precedent exists for lack of conservation of a host factor interaction with Gag proteins of different primate lentivirus lineages. Binding of Gag by the host factor cyclophilin A, which is required for HIV-1 infectivity (4, 15, 38, 51), is restricted to members of the HIV-1 primate lentivirus lineage (3, 59). In contrast, the cellular machinery involved in budding, which is recruited by different retroviral Gag proteins and governed by specific late-domain motifs, appears to be well conserved (see e.g., reference 42). These opposing precedents underscore the significance of determining whether the HP68-Gag association is conserved for other primate lentiviruses.
Second, we address the prediction that capsid assembly intermediates corresponding to those found in the cell-free system can be identified for HIV-1 and other primate lentiviruses in primate cells. Specific HIV-1 Gag-containing complexes in the cell-free system were initially shown to fit the definition of assembly intermediates by the demonstration that a radiolabeled cohort of Gag polypeptides progressed sequentially through these complexes and into completely assembled capsids over time (37). Once identified as assembly intermediates by their precursor-product relationship to completed capsids, these complexes were further defined by specific biochemical characteristics in the cell-free system (37, 62). While high-molecular-weight complexes containing Gag have been observed in cellular systems (10, 11, 25, 34, 35, 37, 48, 53, 54, 60), these complexes have not been analyzed in detail to determine if in fact they are assembly intermediates. This characterization is particularly important to address in cellular systems that are likely to harbor a variety of other Gag-containing complexes, including those destined for degradation. Here, we identify Gag-containing complexes from primate cells expressing a variety of primate lentivirus Gag polypeptides and demonstrate by three criteria that they closely resemble assembly intermediates that were originally defined in the HIV-1 cell-free assembly system. Finally, we show that the critical, energy-dependent 80-150S assembly intermediate found in the cell-free system is highly transient and therefore difficult to detect in cells. We demonstrate that, as predicted from cell-free studies, energy depletion allows this previously unidentified complex to be trapped in cells. Together our findings support the idea that primate lentiviruses assemble their capsids in cells via a common posttranslational mechanism that is energy dependent and involves the host factor HP68.
MATERIALS AND METHODS
Constructs for Gag expression.
Cell-free plasmid vectors were derived from the SP64 vector (Promega) into which the 5′ untranslated region (UTR) of Xenopus laevis globin had been inserted at the HindIII site (44). The Gag-encoding open reading frame from the molecular clone HIV-2 304 (obtained from Shiu-Lok Hu) was amplified by PCR (forward primer, 5′-ATCATCTCTAGATCTACCATGGGCGCGAGAGGCTCCGTC-3′; reverse primer, 5′-GATGATGAATTCTCACTACTGGTCTTCTCCAAAGAG-3′) and introduced downstream of the SP6 promoter and the globin UTR by using primer-encoded BglII and EcoRI restriction sites, similar to the approach used previously for the construction of the pSP HIV-1 SF2 Gag plasmid (37). Plasmid pSP HIV-1 Gag Tr, used in cell-free experiments, was constructed by inserting into the modified SP64 vector a PCR amplicon made from HIV-1 SF2 Gag by using primers encoding a double stop codon after bp 1170 (37, 49). The Flag-HuHP68 plasmid was constructed from a pcDNA 3.1 (Invitrogen) construct encoding WT human HP68 (HuHP68) (62) by inserting complementary oligonucleotides encoding a Kozak's consensus, the initiating methionine, and the eight-amino-acid Flag tag into HindIII and HpaI restriction sites (forward primer, 5′-AGCTTACCATGGATTACAAAGACGACGACGACAAAGCAGACAAGTT-3′; reverse primer, 5′-AACTTGTCTGCTTTGTCGTCGTCGTCTTTGTAATCCATGGTA-3′) and then subcloned into the modified SP64 vector by using NcoI and EcoRI.
Plasmids for cellular studies included HIV-1 pBru ΔEnv, which carries the full-length HIV-1 genome with a deletion in envelope and was previously described by Kimpton and Emerman (31), who referred to it as pME337. HIV-1 pBru ΔEnv Gag Tr was constructed by site-directed insertion of a double stop codon after bp 1425 in pBru as described previously (37), resulting in truncation of Gag after residue 361. To construct plasmids for production of immature non-HIV-1 primate lentivirus particles by mammalian cells, relevant Gag open reading frames were amplified by PCR from the following sources: molecular clone HIV-2 506 (generated by Elisabeth Beattie and provided by Shiu-Lok Hu), simian immunodeficiency virus SIVmac239 (generated by Elizabeth Beattie and provided by Shiu-Lok Hu), and pSAB-1 (obtained from the AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases [NIAID], National Institutes of Health [NIH]) (29). Coding regions for WT and truncated Gag proteins were introduced into the NheI and EcoRI sites in pcDNA 3.1 (Invitrogen). Truncation mutations (Tr) were constructed by using primers encoding double stop codons after the last arginine in the capsid domain of Gag from HIV-2 506 (forward primer, 5′-ATCATCGCTAGCACCATGGGCGCGAGAGGCTCCGTC-3′; reverse primer, 5′-GATGATGAATTCATCACCTTGCCTTCTGCCCTGGGCCTCC-3′), SIVmac239 (forward primer, 5′-ATCATCGCTAGCACCATGGGCGTGAGAAACTCCGTCTTGTC-3′; reverse primer, 5′-GATGATGAATTCATCACCTTGCCTTCTGCCCTGGGCCTCC-3′), and SIVagmSab (forward primer, 5′-ATCATCAAGCTTACCATGGGTGCGAGTAACTCAGTCTTAAGTGG-3′; reverse primer, 5′-GATGATGAATTCTCATCAACGTGCCTTATGTTGTGCTCCTCCAATGC-3′), which correspond to amino acid 361 in HIV-1 Bru Gag. Gag truncation constructs lack most or all of nucleocapsid (NC) and all of p6, and the position of the final amino acid encoded by each truncated construct is as follows: HIV-1 Bru Gag Tr, 361; HIV-2 506 Gag Tr, 361; SIVmac239 Gag, 362; and SIVagmSab Gag Tr, 375). All plasmid coding regions were sequenced.
For generation of H9 cells stably expressing HIV-1, 293T cells were transfected with a plasmid encoding the vesicular stomatitis virus G envelope and a pBru ΔEnv plasmid (obtained from Michael Emerman) encoding a deletion in envelope from bp 6636 to 7216, a frameshift in Vpr created by addition of four nucleotides at the unique NcoI site at bp 5260, and a deletion in Vif between bp 4707 and 4989 (all described previously [36]) and a deletion of Nef from bp 8390 to 8661, into which a polylinker was added followed by insertion of a puromycin-coding region into the XbaI and XhoI sites in the polylinker. Vesicular stomatitis virus G-pseudotyped virions produced from these cells were purified and used to infect H9 cells, which were selected with puromycin.
cDNA amplification by RT-PCR.
For reverse transcription-PCR (RT-PCR) of the African green monkey (AGM) HP68 and macaque HP68 orthologues, mRNAs were prepared (Micro-Fast Track kit; Invitrogen) from Cos-1 cells (Cercopithecus aethiops) (American Type Culture Collection) and from Macaca fascicularis peripheral blood mononuclear cells (obtained from Nancy Haigwood). RT was performed with oligo(dT) primers (cDNA cycle kit; Invitrogen). AGM and macaque HP68 cDNAs were first amplified with primers corresponding to the 5′ and 3′ ends of the HuHP68-coding region. The coding sequence of macaque and AGM HP68 from bp 19 to 1781 was obtained by using primers corresponding to the HuHP68-coding region (GenBank accession number X76388, HuHP68 mRNA [also called RNase L inhibitor or 2-5A binding protein]) (forward primer, 5′-ATGGCAGACAAGTTAACG-3′ [specific to HuHP68 bp 1 to 18]; reverse primer, 5′-CTAATCATCCAAGAAAAAG-3′ [specific to bp 1782 to 1800]). To obtain 5′ and 3′ ends of AGM HP68, amplification was performed with primers specific to the 5′ and 3′ HuHP68 UTRs anchored by primers specific to the internal coding regions. The oligonucleotides for obtaining 5′ and 3′ ends of AGM HP68 were as follows: forward primer 5′-TGTGGCTGAAAAGTGAAGGC-3′ (specific to 5′ HuHP68 UTR, bp −99 to −79) with reverse primer 5′-CCTGAAAGATCTTCAACATTTCG-3′ (bp 634 to 656) and forward primer 5′-GTTGAAGATCTTTCAGGAGGAGAG-3′ (bp 640 to 659) with reverse primer 5′-CAAGTTCCAGAGTATGTGGG-3′ (specific to 3′ HuHP68 UTR, bp 1900 to 1919). Two independent RTs and amplifications were performed to confirm sequences of coding regions. The sequence obtained for AGM HP68 corresponds to the complete protein (amino acids 1 to 599), while the sequence obtained for macaque HP68 corresponds to amino acids 7 to 592.
Transfections and cell harvests.
Cos-1 (C. aethiops; AGM kidney) cells in 60-mm-diameter dishes were transfected by using 24 μl of Lipofectamine reagent (Invitrogen) and 4 to 8 μg of plasmids as indicated. For immunoprecipitation (IP) and gradient analysis, approximately 3 × 106 cells were washed in phosphate-buffered saline (PBS) and harvested on ice approximately 36 h posttransfection in 200 μl (for gradient experiments) or 300 μl (for IP experiments) of harvest buffer containing 0.625 to 1.0% NP-40, 10 mM Tris acetate (TrisAc) (pH 7.4), 50 mM KAc, 100 mM NaCl, and either 4 mM MgAc (for Fig. 4 and 6A) or 10 mM EDTA (for Fig. 2 and 6C), in addition to a 1 mM concentration of the protease inhibitor phenylmethylsulfonyl fluoride. In certain cases (for Fig. 4, 5, and 6B), MgAc-containing harvest buffer also contained 1 U of apyrase (Sigma) per μl, as indicated. Approximately 50 × 106 H9 cells stably expressing HIV-1 pBru ΔEnv (see Fig. 6C) were collected and harvested in 500 μl of harvest buffer containing EDTA. Lysates were sheared by 25 passes through a 20-gauge needle and clarified by centrifugation at 365 × g for 5 min at 4°C in a GH-3.8 rotor (Beckman Coulter Allegra 6R centrifuge) and at 18,000 × g for 20 s in a conventional microcentrifuge.
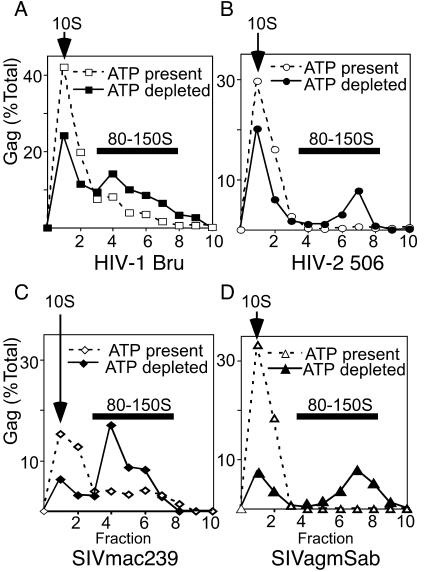
FIG. 4.
Effect of ATP depletion on the 80-150S complex in primate cells expressing primate lentivirus Gag polypeptides. Cos-1 cells were transfected with expression plasmids for the WT primate lentivirus Gag constructs HIV-1 pBru ΔEnv (A), HIV-2 506 Gag (B), SIV mac239 Gag (C), and SIVagmSab Gag (D). Lysates were harvested in harvest buffer in the presence of ATP (dashed lines and open symbols) or the ATP-hydrolyzing enzyme apyrase (solid lines and closed symbols) and analyzed by velocity sedimentation on step gradients, SDS-PAGE, immunoblotting, and densitometry to generate profiles showing S values of Gag-containing complexes. Arrows indicate positions of 10S complexes; dark bars indicate positions of 80-150S complexes. The experiment was repeated three independent times, with the results of one representative experiment shown.
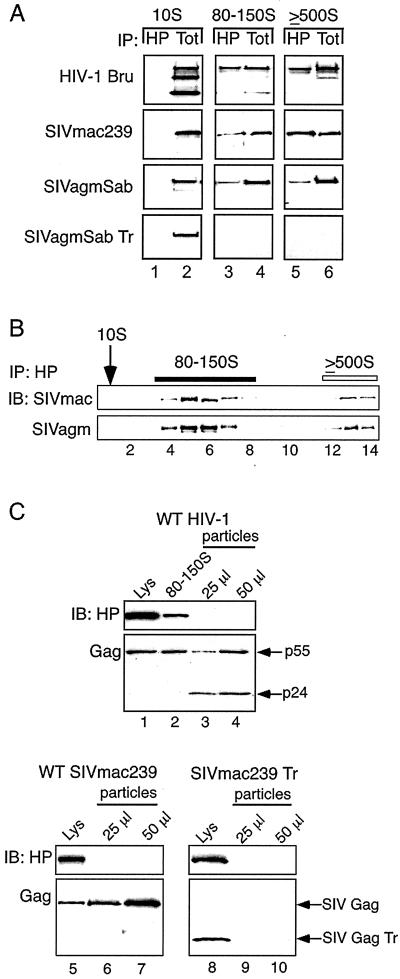
FIG. 6.
Association of HP68 with Gag in assembly intermediates and absence of HP68 in VLPs. (A) Cos-1 cells were transfected to express HIV-1 pBruΔEnv, SIVmac239 Gag, SIVagmSab Gag, or SIVagmSab GagTr. Lysates were harvested in harvest buffer and subjected to velocity sedimentation with step gradients. Gradient fractions corresponding to 10S, 80-150S, and ≥500S were pooled and subjected to IP with α-HuHP68 (HP) (lanes 1, 3, and 5). IP reaction mixtures and aliquots of co-IP input (Tot) (lanes 2, 4, and 6) were analyzed by SDS-PAGE and immunoblotting with antibody for relevant Gag. Tot represents 2.5 to 5% of co-IP input, except in the case of the SIVagmSab 80-150S and SIVagmSab Tr 10S, in which Tot represents 1 and 10% of input, respectively. (B) Cos-1 cells transfected to express SIVmac239 Gag or SIVagmSab Gag were harvested and analyzed as for panel A. Individual gradient fractions were subjected to IP with α-HuHP68 (HP) followed by SDS-PAGE and immunoblotting (IB) for relevant Gag. Positions of 10S complexes (arrow), 80-150S complexes (dark bar), and ≥500S complexes (open bar) are indicated. (C) Levels of HP68 and Gag in total cell lysate (lanes 1, 5, and 8), in 80-150S complexes from cells (lane 2), and in VLPs (lanes 3, 4, 6, 7, 9, 10) released from H9 cells expressing HIV-1 pBru ΔEnv (WT HIV-1) or Cos-1 cells expressing either SIVmac239 Gag or SIVmac239 Gag Tr were determined by immunoblotting. Aliquots of total lysate and 80-150S complexes shown represent 0.5% of H9 lysate, 2% of 80-150S fractions, and 0.25% of Cos-1 lysate expressing either SIVmac239 Gag or SIVmac239Gag Tr, harvested as described in Materials and Methods. The experiment was repeated three independent times, with the results of one representative experiment shown.
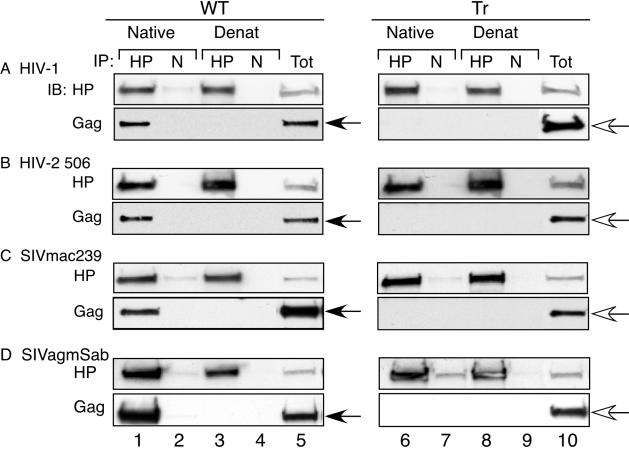
FIG. 2.
Association of genetically divergent primate lentivirus Gag polypeptides with primate HP68 in Cos-1 cells. Lysates from cells expressing WT (left panels) or truncated (Tr) (right panels) Gag proteins were harvested in nonionic detergent, and IPs were performed with α-HuHP68 antibody (lanes 1, 3, 6, and 8) or nonimmune antibody (N) (lanes 2, 4, 7, and 9) under native conditions (lanes 1, 2, 6, and 7) or following denaturation of lysate (Denat) (lanes 3, 4, 8, and 9), as described in Materials and Methods. Aliquots representing 5% of the IP input cell lysate are shown (Tot) (lanes 5 and 10). IPs were analyzed by SDS-PAGE followed by immunoblotting (IB) for HP68 (HP) or the relevant Gag. Closed arrows indicate WT Gag, and open arrows indicate truncated Gag. The experiment was repeated three independent times, with the results of one representative experiment shown. Gag truncation constructs lack most or all of NC and all of p6, as described in Materials and Methods.
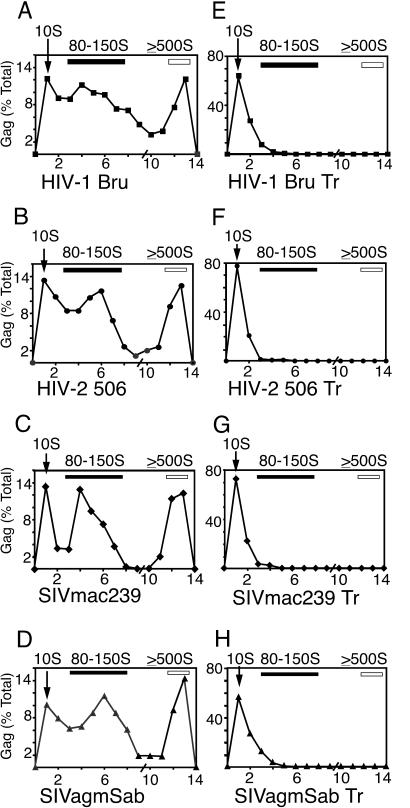
FIG. 5.
Assembly intermediates of WT and truncated primate lentivirus Gag polypeptides. Cos-1 cells were transfected with expression plasmids for the WT or truncated primate lentivirus Gag constructs HIV-1 pBru ΔEnv (A), HIV-2 506 Gag (B), SIVmac239 Gag (C), SIVagmSab Gag (D), HIV-1 pBru ΔEnv Gag Tr (E), HIV-2 506 Gag Tr (F), SIVmac239 Gag Tr (G), and SIVagmSab Gag Tr (H). Cells were harvested in harvest buffer containing apyrase to deplete ATP. Lysates were analyzed by velocity sedimentation on step gradients, followed by SDS-PAGE, immunoblotting, and densitometry to generate profiles showing Gag-containing complexes of different S values. Positions of 10S complexes (arrows), 80-150S complexes (dark bars), and ≥500S complexes (open bars) are indicated. The slash on the x axis indicates the splice point of the two different gradients used. The experiment was repeated three independent times, with the results of one representative experiment shown.
In vitro transcription, cell-free translation, and assembly.
In vitro transcription was performed with SP6 polymerase by using the cell-free expression plasmids described above and was followed by in vitro translation and assembly for 180 min at 26°C with wheat germ extract and [35S]methionine (ICN Biochemicals), as described previously (14, 37). For Fig. 1B, cotranslation of Flag-HuHP68 and Gag was performed by programming the cell-free reaction with an input transcription mixture consisting of 40% Flag-HuHP68 transcript and 60% Gag transcript. Where indicated, cell-free reaction mixtures were treated with the translation inhibitor emetine (0.2 mM final concentration) and either 20 mM TrisAc buffer (pH 7.5) or apyrase (Sigma) (final concentration of 0.5 U/μl).
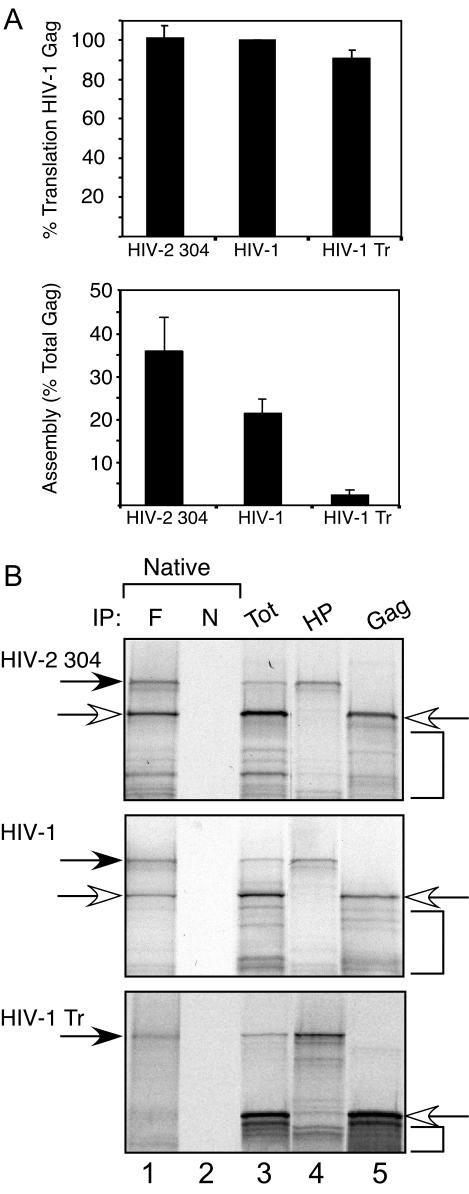
FIG. 1.
Assembly of HIV-2 Gag and association with HP68 in the cell-free system. (A) Radiolabeled cell-free reaction mixtures were programmed with transcripts for HIV-2 304 Gag, WT HIV-1 Gag, or HIV-1 Gag Tr. Reactions were analyzed by velocity sedimentation with step gradients and SDS-PAGE followed by autoradiography. Top panel, amount of Gag translated in equivalent aliquots of each reaction mixture as percentage of WT HIV-1 Gag translated, quantitated by densitometry. Bottom panel, amount of Gag assembly (percentage of total Gag) assessed by densitometry of assembly profiles as described in Materials and Methods. Error bars show standard errors of the means from three independent experiments. (B) Lanes 1 to 3, in separate parallel reactions, Flag-tagged HuHP68 was cotranslated with HIV-2 304 Gag, WT HIV-1 Gag, or HIV-1 Gag Tr Gag transcripts in the presence of [35S]methionine. Aliquots of cell-free reaction mixtures were subjected to IP with anti-Flag (F) (lane 1) or nonimmune antibody (N) (lane 2) under native conditions. Lane 3 shows 10% of the IP input total (Tot). Lane 4, translation of Flag-HuHP68 alone (HP) to indicate migration of Flag-HuHP68 (dark arrow). Lane 5, translation of each relevant primate lentivirus Gag alone to indicate their migrations (open arrows). Brackets indicate positions of early termination and late initiation products that are typically seen in cell-free translations. High background is seen in immune lanes due to use of commercially made polyacrylamide gels lacking SDS with SDS-containing sample buffer and running buffer, as discussed in Materials and Methods. Coomassie blue staining revealed equivalent amount of immune and nonimmune antibodies eluted from IP. The experiment was repeated three independent times, with the results of one representative experiment shown.
Gradient analysis of cell-free reactions and cellular lysates.
Calibration of gradients to determine S value positions has been described previously (37). Either 200 μl of cell lysate or 10 μl of cell-free assembly reaction mixtures diluted in 90 μl of harvest buffer was layered onto gradients containing sucrose prepared in harvest buffer. Modified velocity sedimentation gradient analysis used the following 2-ml sucrose step gradients: for resolution of 10S and 80-150S complexes (see Fig. 3 to 6), gradients consisted of 500 μl each of 5%, 10%, 30%, and 40% sucrose, and for resolution of ≥500S complexes (see Fig. 1A, 3, 5, and 6A and B), gradients consisted of 500 μl of 20% sucrose, 900 μl of 40% sucrose, and 600 μl of 66% sucrose. These gradients maximize separation of the 10S, 80-150S, and 500S peaks, at the expense of resolving the 500S and 750S peaks. Step gradients can make heterogeneous peaks appear somewhat more homogeneous, but they are used here because they improve resolution of peaks that have been characterized previously by linear gradient analysis (37, 49, 62). Centrifugation of all gradients was performed at 135,000 × g for 45 min at 4°C in a TLS55 rotor (Beckman Coulter Optima Max-E centrifuge), and 200-μl fractions were collected serially from the top. Equivalent aliquots of each gradient fraction were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by autoradiography or immunoblotting. For Fig. 1A (bottom panel), cell-free assembly was determined by quantitating the amount of Gag present in fractions 9 and 10 as the percent Gag present in all gradient fractions. We have previously demonstrated that these fractions largely correspond to completely assembled capsids by assessing immature HIV-1 particles released from cells (37). For other quantitation methods, see below.
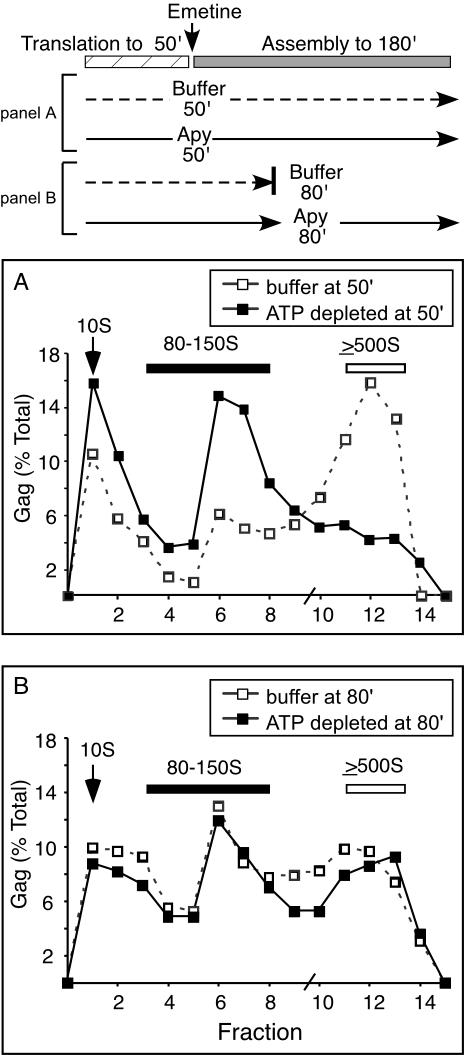
FIG. 3.
Effect of ATP depletion on HIV-1 capsid assembly intermediates in the cell-free system. Parallel radiolabeled cell-free reaction mixtures were programmed with transcript for HIV-1 Gag and were treated as shown in the schematic at the top. (A) Reaction mixtures were treated with emetine, an inhibitor of translation elongation, at 50 min to create an isolated translation phase (hatched bar) and posttranslational assembly phase (50 to 180 min) (gray bar). Immediately after emetine treatment, the reaction mixtures were treated either with buffer (dashed line in schematic and open squares in graph) or with the ATP-hydrolyzing enzyme apyrase (Apy) (solid line in schematic and closed squares in graph), and the reactions were allowed to proceed until 180 min and then analyzed by velocity sedimentation on step gradients, SDS-PAGE, autoradiography, and densitometry to generate profiles showing HIV-1 Gag assembly intermediates. (B) To assess the effect of ATP depletion on the stability of higher-molecular-weight assembly intermediates (500S) during the posttranslational phase, a separate reaction mixture was treated with apyrase at 80 min and incubated until 180 min (solid line in schematic and closed squares in graph), while a control reaction mixture was put on ice to halt assembly at 80 min (dashed line in schematic and open squares in graph). Positions of 10S complexes (arrows), 80-150S complexes (dark bars), and ≥500S complexes (open bars) are indicated. The slash mark on the x axis indicates the splice point of the two different velocity sedimentation step gradients used. The experiment was repeated three independent times, with the results of one representative experiment shown.
Co-IPs and immunoblotting.
IPs of 35S-labeled cell-free reaction mixtures (see Fig. 1B) were performed at 120 min by incubating 2 μl of cell-free reaction mixtures in harvest buffer containing MgAc (described above), along with 6 μl of either mouse anti-Flag antibody coupled to agarose beads (Sigma) or mouse immunoglobulin G (IgG) (Sigma) and protein G beads (Pierce). For IP of pooled gradient fractions (see Fig. 6A), fractions 1 and 2 from gradient 1 (10S complexes), fractions 3 to 7 from gradient 1 (80-150S complexes), and fractions 7, 8, and 9 from gradient 2 (≥500S complexes) were combined. IP of individual gradient fractions (see Fig. 6B) was performed on fractions 1 to 9 of gradient 1 (lanes 1 to 9) and fractions 6 to 10 of gradient 2 (lanes 10 to 14). IP of cell lysates was performed by incubating 100 μl of cell lysate (see Fig. 2) or aliquots of pooled or unpooled 150 μl gradient fractions (see Fig. 6A and B, respectively) with either affinity-purified anti-HuHP68 antibody or nonimmune antibody coupled to protein A-conjugated agarose beads (Bio-Rad). For IP of denatured lysates, the input was boiled for 3 min in the presence of 1.0% SDS, after which denatured aggregates were removed by clarification at 18,000 × g for 45 s in a conventional microcentrifuge. The supernatant was diluted with harvest buffer to reduce the SDS concentration to below 0.1%, and antibody was then added.
Affinity purification and coupling of HuHP68 antiserum were performed according to standard protocols (26) and have been described previously (62). IP reaction mixtures were incubated by rotation for 2 to 4 h at 4°C and washed twice with detergent wash buffer containing 0.1 M TrisAc (pH 8.0), 1.0% NP-40, 50 mM KAc, 100 mM NaCl, and either 1 mM EDTA (for Fig. 2) or 4 mM MgAc (for Fig. 1B, 6A and B) and twice with the corresponding wash buffer without NP-40. The immunoprecipitated product was eluted in SDS-PAGE loading buffer and subjected to SDS-PAGE. IP reaction products were analyzed by autoradiography (cell-free reactions) or immunoblotting (cell lysates). Note that for SDS-PAGE analysis of IP samples, commercial gels that do not contain SDS were used with SDS-containing running and loading buffers. This leads to high-background lanes with large amounts of radiolabeled protein (such as immune lanes and total lanes), due to slight alterations in migration of SDS-coated proteins upon migration through the SDS-free gel. Also, to confirm that the same amounts of immune and nonimmune antibodies were eluted from co-IPs, IgG in each lane of the SDS-polyacrylamide gels was examined by Coomassie blue staining before autoradiography was performed.
Immunoblotting was performed with anti-HIV-1 Gag p24 (Dako) at 1:600 or monoclonal antibody 183-H12-5C (Bruce Chesebro, NIH AIDS Research and Reference Reagent Program) (8, 52, 57) at 1:250 to 1:500 as primary antibodies for HIV-1 Gag and non-HIV-1 Gag, respectively. Anti-IgG or anti-IgG1 coupled to horseradish peroxidase (Santa Cruz) was used as the secondary antibody, and enhanced chemiluminescence (Pierce) was performed. Immunoblotting for HP68 was performed with crude anti-HuHP68 antiserum at 1:500 (62) followed by anti-protein A coupled to horseradish peroxidase (Pierce).
Preparation of VLPs.
For preparation of virus-like particles (VLPs), medium from 10 × 106 H9 cells (4.5 × 105 cells/ml) stably expressing HIV-1 pBru ΔEnv or from 9 × 106 Cos-1 cells transiently transfected to express SIVmac239 Gag or SIVmac239 Gag Tr was clarified by centrifugation at 650 × g for 10 min at 4°C in a GH-3.8 rotor (Beckman Coulter Allegra 6R centrifuge), followed by centrifugation at 18,000 × g for 2 min in a conventional microcentrifuge. Each sucrose cushion was prepared by applying 3.3 ml of clarified medium to 1.5 ml of 20% sucrose in PBS layered over 50 μl of 60% sucrose in PBS and centrifuged at 162,000 × g for 45 min at 4°C as previously described (45) with an MLS50 rotor (Beckman-Coulter Optima Max-E centrifuge), and the resulting VLP preparation was resuspended in 0.625% NP-40 buffer containing 1 mM EDTA and 1 mM phenylmethylsulfonyl fluoride. For H9 cells expressing HIV-1, 26 ml of medium was concentrated into 850 μl, and for Cos-1 cells expressing SIVmac239 Gag or SIVmac239 Gag Tr, 10 ml of medium was concentrated into 110 μl. VLP inputs for immunoblotting were normalized to contain comparable amounts of WT Gag as cell lysate inputs in adjacent immunoblot lanes (see Fig. 6C).
Quantitation.
Autoradiographs and Western blots were digitized by using an AGFA Duoscan T1200 scanner and Photoshop 5.5 software (Adobe Systems Incorporated). Mean band densities were determined and adjusted for band size and background. For quantitation of autoradiographs and immunoblots, dilution standards were performed to ensure that bands analyzed were within the linear range for signal intensity. Assembly profiles were generated by plotting the amount of total Gag present in each fraction as a percentage of the total Gag in all gradient fractions (see Fig. 3 to 5). Gradient profiles and autoradiographs examining all three major peaks (10S, 80-150S, and ≥500S) (see Fig. 3, 5, and 6B) were generated by splicing fractions 1 to 9 of gradient 1 (which resolves 10S and 80-150S complexes) to fraction 6 or 7 to 10 of gradient 2 (which resolves ≥500S complexes) (see Fig. 3 and 5).
Nucleotide sequence accession numbers.
GenBank accession numbers are AY450957 (for macaque HP68) and AY450958 (for AGM HP68).
RESULTS
Divergence of primate lentivirus Gag sequences and conservation of primate HP68 sequences.
The first goal of this study was to determine whether Gag polypeptides of different primate lentiviruses associate with the host factor HP68, as does HIV-1 Gag (62). We first assessed the degree of conservation of Gag protein sequences from three different primate lentivirus lineages (reviewed in reference 24) and the HP68 orthologues present in the natural or experimental hosts that they infect. Comparison of sequences of HIV-2 304 and 506 (obtained from S. L. Hu) as well as SIVmac239 and SIVagmSab (obtained from GenBank) revealed that the sequences of any two Gag proteins from different primate lentivirus lineages are not well conserved, sharing only approximately 55% identity and 65% conservation (data not shown). This is consistent with the significant degree of genetic divergence that distinguishes different primate lentivirus lineages (24). To assess the sequence homology of HP68 orthologues from natural or experimental hosts of these primate lentiviruses, HP68 cDNA was amplified by RT-PCR from mRNAs of macaque (M. fascicularis) peripheral blood mononuclear cells and AGM kidney cells (Cos-1). These cells were chosen because SIVmac (47) as well as the two HIV-2 molecular clones studied here (1) have been shown to cause AIDS in macaques, and SIVagm infects AGMs (47). The deduced amino acid sequences revealed that these simian HP68 orthologues are over 97% identical to HuHP68 (data not shown). The only amino acid sequence differences found relative to HuHP68 were Q44ΔR, V242ΔE, G367ΔR, G382ΔE, and G473ΔA in macaque HP68 and V109ΔA, I99ΔV, V242ΔE, and G473ΔA in AGM HP68 (data not shown) (see sequences AY450957 and AY450958 deposited in GenBank).
HIV-2 Gag assembles in the cell-free system and associates with HuHP68.
To determine whether genetically divergent primate lentivirus Gag polypeptides associate with the highly conserved HP68 protein, we first examined the assembly of HIV-2 Gag and its association with HuHP68 in a cell-free system similar to the one that was used initially to identify the interaction between HP68 and HIV-1 Gag (62). We have demonstrated previously that HIV-1 Gag, when translated de novo in wheat germ extract, assembles into particles that very closely resemble completely assembled authentic HIV-1 capsids from cells, as indicated by the criteria of velocity sedimentation, buoyant density, and electron microscopic appearance (37). Here we examined whether HIV-2 Gag also assembles into capsids as judged by velocity sedimentation on step gradients. As a control, we used HIV-1 Gag Tr, an assembly-incompetent Gag mutant (18, 28, 30, 37, 49) that lacks the NC and p6 domains. When cell-free reaction mixtures were programmed with transcripts encoding WT HIV-2 304 Gag, WT HIV-1 Gag, and the truncation mutant HIV-1 Gag Tr, equivalent amounts of translated product were produced (Fig. 1A, top panel). Velocity sedimentation analysis using step gradients revealed that WT HIV-2 304 Gag assembled as well as or better than WT HIV-1 Gag, while HIV-1 Gag Tr was assembly incompetent, as expected (Fig. 1A, bottom panel).
To examine the association of HIV-2 304 Gag with HuHP68, 35S-labeled cell-free translations were programmed with transcripts for HIV-2 304 Gag and for HuHP68 encoding a Flag tag at the N terminus (Flag-HuHP68), both together (Fig. 1B, lanes 1 to 3) and separately (Fig. 1B, lanes 4 and 5). IPs performed on cotranslations revealed that anti-Flag antibody immunoprecipitated Flag-HuHP68, as expected, and coimmunoprecipitated radiolabeled HIV-2 304 Gag under native conditions (Fig. 1B, top panel, lane 1) but not after denaturation of the cell-free reaction input (data not shown), which disrupts native protein-protein interactions. Nonimmune antibody failed to immunoprecipitate Flag-HuHP68 or HIV-2 304 Gag (Fig. 1B, top panel, lane 2), showing the specificity of the interaction. Similar results were obtained when WT HIV-1 Gag was cotranslated with Flag-HuHP68 and analyzed by IP (Fig. 1B, middle panel, lanes 1 and 2). In contrast, when the assembly-incompetent HIV-1 Gag Tr mutant was cotranslated with Flag-HuHP68, anti-Flag antibody immunoprecipitated Flag-HuHP68 but not HIV-1 Gag Tr (Fig. 1B, lower panel, lanes 1 and 2). This is consistent with previous results indicating that HIV-1 Gag Tr does not associate with HP68 (62). Thus, in the cell-free system, WT HIV-2 304 Gag, like HIV-1, appears to assemble into immature capsids and to associate with HuHP68.
Gags from three different primate lentivirus lineages associate with HP68 in primate cells.
Previously, we demonstrated by co-IP that WT HIV-1 Gag (expressed alone with HIV-1 Rev or expressed in the context of the full-length HIV-1 genome by transfection or infection) associates with endogenous HP68 in various human as well as nonhuman primate cells (62). To determine whether HIV-2 Gag associates with endogenous HP68 in primate cells, Cos-1 cells, which are of AGM origin, were transfected to express either the nearly complete HIV-1 Bru genome (HIV-1 Bru ΔEnv) or HIV-2 506 Gag. Cell lysates were subjected to IP with an affinity-purified antibody directed against the C terminus of HuHP68 (α-HuHP68), which recognizes endogenous HP68 in human and nonhuman primate cells (62). Immunoblotting of IP products with antibody to Gag revealed that HIV-1 Bru Gag and HIV-2 506 Gag were coimmunoprecipitated to equal extents under native conditions by α-HuHP68 but were not immunoprecipitated by nonimmune serum (Fig. 2A and B, Gag panels, compare lanes 1 and 2). Similar results were obtained when WT SIVmac239 and SIVagmSab (from the second and third major primate lentivirus lineages, respectively) were expressed in Cos-1 cells (Fig. 2C and D, Gag panels, lanes 1 and 2). WT Gag proteins were not coimmunoprecipitated by α-HuHP68 or nonimmune serum after denaturation (Fig. 2, Gag panels, lanes 3 and 4). In contrast, corresponding Gag mutants that are truncated proximal to NC were not coimmunoprecipitated by α-HuHP68 under native conditions or after denaturation (Fig. 2, Gag panels, lanes 6 to 9). In each instance truncation mutants were expressed as well or better than their WT counterparts (data not shown). Reprobing of the same blots with α-HuHP68 revealed that HP68 was immunoprecipitated by α-HuHP68 but not by nonimmune antiserum under native conditions and following denaturation of the input lysate, for all samples, as expected (Fig. 2, HP panels, lanes 1, 3, 6, and 8; compare to nonimmune controls in lanes 2, 4, 7, and 9). Together, these data indicate that WT Gag polypeptides from three of the major primate lentivirus lineages associate with endogenous HP68 in primate cells, while the corresponding mutants truncated proximal to NC do not. Similar results were obtained upon expression of these Gag constructs in 293T cells, which are of human origin (data not shown).
ATP depletion results in accumulation of the 80-150S assembly intermediate.
Previous studies have revealed that HIV-1 Gag assembles into immature capsids via an energy-dependent pathway of discrete posttranslational assembly intermediates in the cell-free system (37, 49). While some data from cellular studies support the existence of Gag-containing complexes in primate cells (10, 11, 25, 34, 35, 37, 48, 53, 54, 60), little has been done to establish whether Gag-containing complexes found in cells are in fact assembly intermediates. To determine this, we first examined whether complexes with the expected sedimentation values could be identified in Cos-1 cells expressing primate lentivirus Gag proteins. Cell lysates were prepared with harvest buffer (containing the nonionic detergent NP-40 and physiological salts) and analyzed by using modified velocity sedimentation gradients. Gag polypeptides from HIV-1 Bru, HIV-2 506, SIVmac239, and SIVagmSab were present primarily in the 10S fraction, with a small amount in the ≥500S region (data not shown; also see Fig. 4). When the harvest temperature was carefully maintained at 4°C, a small peak could be seen in the 80-150S fractions (data not shown), suggesting the presence of a complex that might correspond to the 80-150S assembly intermediate. (It should be noted that two closely migrating complexes at approximately 80S and 150S are not resolved completely by gradients used in this study and for this reason are referred to here as the 80-150S complex.) The failure to see a significant peak in the 80-150S region raised the possibility that the 80-150S complex is highly transient in cells.
Our earlier studies with the cell-free system had suggested that posttranslational ATP depletion blocks assembly, with accumulation of Gag polypeptides in the 80-150S complexes (37). These studies raised the possibility that the 80-150S assembly intermediate in cells could be trapped and revealed by ATP depletion. However, these earlier studies in the cell-free system did not determine whether ATP depletion actually traps the 80-150S intermediate by preventing progression of Gag polypeptides out of this complex or simply disrupts higher-molecular-weight Gag-containing complexes as they form posttranslationally, causing them to disassemble into 80-150S complexes. To distinguish between these possibilities, we performed a set of experiments in the cell-free system. First, we programmed cell-free reaction mixtures with HIV-1 Gag and treated them with the translation inhibitor emetine at 50 min into the reaction, creating an isolated posttranslational phase (50 to 180 min) (Fig. 3, schematic). We have demonstrated previously that this treatment divides the cell-free reaction into two phases: in the first 50 min, translation but not assembly of HIV-1 Gag occurs; in the second phase, further translation of Gag is prevented by emetine, and Gag that is already synthesized proceeds to assemble (37). At 50 min, parallel cell-free reaction mixtures were treated with either buffer or the ATP-hydrolyzing enzyme apyrase (33). In both cases, the reactions were allowed to proceed for an additional 130 min and then analyzed by velocity sedimentation with step gradients. Examination of translation totals revealed no difference in the total amount of Gag present in the two different reaction mixtures (data not shown). As expected from our previous results (37), Gag polypeptides assembled into complexes of ≥500S (Fig. 3A) in the reaction mixture treated with emetine and buffer at 50 min. In contrast, Gag polypeptides in the reaction mixture treated with emetine and apyrase at 50 min accumulated 10S and 80-150S complexes, with dramatically less Gag present in complexes of ≥500S (Fig. 3A), demonstrating the importance of ATP for posttranslational events in assembly, as shown previously (37). We also examined whether accumulation of Gag polypeptides in the 80-150S complex upon ATP depletion was due to inhibition of Gag polypeptide progression from the 80-150S complex or to disruption of Gag chains which had already formed ≥500S complexes. Cell-free reaction mixtures were incubated for a longer time prior to apyrase treatment (80 min rather than 50 min) to allow significant accumulation of ≥500S assembly intermediates. These reaction mixtures were then treated with buffer and the reactions were halted on ice (Fig. 3B), or the reaction mixtures were treated with apyrase and allowed to incubate for another 100 min (Fig. 3B). The control reaction mixture treated with buffer at 80 min reveals the presence of ≥500S complexes (Fig. 3B). Treatment with apyrase at 80 min followed by incubation for another 100 min did not alter the amount of ≥500S Gag-containing complexes (Fig. 3B). Similar results were obtained with apyrase treatment at later times (data not shown). Together these findings indicate that in the cell-free system, ATP depletion at the beginning of the posttranslational phase of assembly (50 min) traps the 80-150S complex by preventing progression of Gag polypeptides from the 80-150S complex and does not cause disruption of higher-molecular-weight complexes. The possibility that the 80-150S complexes represent monosomes and disomes was ruled out by demonstrating that EDTA treatment, which disrupts ribosomes (9), does not disrupt posttranslational, Gag-containing 80-150S complexes (data not shown). Similar results have been obtained upon EDTA treatment of complexes obtained from cells (J. E. Dooher and J. R. Lingappa, unpublished data).
To determine whether the 80-150S complex in primate cells can be trapped and revealed in a similar manner by ATP depletion, Cos-1 cells expressing primate lentivirus Gag proteins were harvested in NP-40-containing harvest buffer, either under standard conditions (normal levels of cellular ATP present) or in the presence of apyrase (ATP depleted) (Fig. 4). ATP depletion resulted in a significant accumulation of the 80-150S complex for all primate lentivirus Gag polypeptides, as seen by using the modified velocity sedimentation gradients that separate the 10S complex from the 80-150S complex (Fig. 4), with no change seen in the amount of Gag present (data not shown). Thus, energy depletion at the time of harvest allows the transient 80-150S complex to be trapped and revealed in cells expressing Gag proteins from different primate lentivirus lineages.
Assembly-incompetent primate lentivirus Gag mutants fail to form high-molecular-weight complexes in cells.
If the 80-150S complex seen with apyrase treatment in cells is an assembly intermediate, we would predict, based on our studies of HIV-1 in the cell-free system (37, 49), that this complex would not be formed by assembly-incompetent Gag mutants. Cos-1 cells expressing WT or truncated primate lentivirus Gag proteins were harvested using apyrase, and subjected to velocity sedimentation on step gradients that resolve the 10S and 80-150S complexes, as well as complexes of ≥500S. Gradient profiles demonstrated that all four WT Gag polypeptides form complexes of 10S, 80-150S, and ≥500S in cells (Fig. 5A to D). It should be noted that unlike the cell-free experiment in Fig. 3, which was synchronized by the use of a translation inhibitor to allow one cohort of chains to be examined, the experiment using apyrase in cells (Fig. 5) was performed under steady-state, unsynchronized conditions, explaining why complexes of ≥500S are seen. Furthermore, while the assembly intermediate profiles seen for the different WT primate lentivirus Gag polypeptides in cells are remarkably similar overall, some subtle differences do exist. In particular, in the case of HIV-2 506 and SIVagmSab (Fig. 4B and D and 5B and D), the peak of the 80-150S complex is shifted to the right relative to the peak of the corresponding complex for HIV-1 Bru and SIVmac239 (Fig. 4A and C and 5A and C). The reason for this difference is not known, but it could reflect a difference in composition of this heterogeneous complex for certain primate lentiviruses, or subtle differences in the dynamics of assembly.
In contrast to profiles obtained with WT constructs, none of the assembly-incompetent Gag mutants formed complexes of >10S in primate cells in the presence (Fig. 5E to H) or absence (data not shown) of apyrase. In each instance truncation mutants were expressed as well as or better than their WT counterparts (data not shown), indicating that their failure to form 80-150S complexes was not due to reduced expression levels. Thus, these assembly-incompetent primate lentivirus Gag mutants fail to form the 80-150S and 500S complexes in primate cells, as was seen for HIV-1 in the cell-free system.
Only high-molecular-weight assembly intermediates are associated with HP68 in cells.
The data described above indicate that Gag-containing complexes from cells have the same biochemical features as the assembly intermediates in the cell-free system that were defined by pulse-chase studies. These characteristics include energy sensitivity and the failure of specific complex formation by assembly-incompetent Gag mutants. To further verify that the complexes found in cells are assembly intermediates, we examined a third criterion, namely, the pattern of Gag association with HP68. In the cell-free system, only high-molecular-weight assembly intermediates (80-150S and 500S) contained HP68 in association with Gag polypeptides (62). Cos-1 cells expressing primate lentivirus Gag proteins from three different lineages were subjected to velocity sedimentation with step gradients. IPs were performed with α-HuHP68 on pooled adjacent fractions containing the putative assembly intermediates (Fig. 6A), as well as on individual fractions across the entire gradient (Fig. 6B). WT Gag polypeptides were coimmunoprecipitated by α-HuHP68 in the 80-150S and 500S fractions (Fig. 6A, lanes 3 and 5, and B) but not in the 10S fraction (Fig. 6A, lane 1, and B), even though Gag was present in the 10S fraction as indicated by an immunoblot of the input (Fig. 6A, lane 2). As expected, assembly-incompetent Gag truncation mutants failed to form high molecular-weight assembly intermediates or to be immunoprecipitated by α-HuHP68 in any fraction (Fig. 6A, SIVagmSab Tr panel, lanes 4 and 6, and data not shown). The finding that Gag-containing complexes from cells have the same pattern of HP68 association seen for assembly intermediates in the cell-free system supports the model that these complexes in cells represent true capsid assembly intermediates.
The association of HP68 with assembly intermediates in primate cells raised the question of whether Gag remains associated with HP68 when capsid assembly is completed. To address this, we examined whether HP68 is present in VLPs that have undergone assembly and release from cells. The amounts of HP68 in samples from cells and VLPs that contained equivalent amounts of Gag were compared. Cellular lysates, pooled 80-150S complexes, and VLP preparations were immunoblotted for Gag and HP68. Cell lysates and 80-150S fractions from H9 cells expressing HIV-1 Bru ΔEnv (Fig. 6C, lanes 1 and 2) and Cos-1 cells expressing WT SIVmac239 Gag (Fig. 6C, lane 5) contained both Gag and HP68, as expected (Fig. 2 and 6A and B). In contrast, HP68 was not detected in VLP samples containing levels of Gag comparable to that in cell lysate samples, in the case of both HIV-1 (Fig. 6C, lanes 3 and 4) and SIVmac239 (Fig. 6C, lanes 6 and 7). Controls were prepared in parallel from the assembly-incompetent SIV mac239 Gag Tr mutant, which fails to form VLPs. The SIV mac239 Gag Tr cell lysate contained both HP68 and SIV mac239 Gag Tr (Fig. 6C, lane 8), while equivalent amounts of VLP preparations from these cells contained neither Gag nor HP68 (Fig. 6C, lanes 9 and 10), as expected given the failure of this mutant to form VLPs. Thus, while significant amounts of HP68 are present and associated with WT Gag in total cell lysates and specific cellular fractions, HP68 does not appear to be present in significant quantities in VLPs formed by different WT primate lentivirus Gag proteins. Even if very small amounts of HP68 (below the level of detection by immunoblotting) are present in VLPs, the dramatic change seen in the ratio of HP68 to Gag supports the model that HP68 is released from Gag upon the completion of capsid assembly in cells. This is consistent with results obtained previously from the cell-free system (62).
DISCUSSION
We have previously demonstrated that the host factor HP68, also called RNase L inhibitor, is critical for posttranslational events in immature HIV-1 capsid assembly (62). The findings presented here reveal that, despite their significant sequence divergence, Gag polypeptides from primate lentiviruses of three different lineages associate in primate cells with endogenous HP68. These primate lentivirus Gag proteins form complexes that correspond to capsid assembly intermediates by a variety of criteria. These findings support a model in which primate lentivirus Gag polypeptides assemble into immature capsids in primate cells via an energy-dependent, stepwise pathway of assembly intermediates.
The finding that HP68 associates with a broad spectrum of primate lentivirus Gags raises the possibility that this virus-host interaction may exist for other retroviruses and possibly for viruses outside the retroviral lineage. Previously, we have found that the capsid protein of the hepadnavirus hepatitis B virus does not appear to associate with HP68 in a cell-free system (62). It remains to be determined whether other retroviral Gag proteins, including those of Alpharetroviridae, Betaretroviridae, and Gammaretroviridae (formerly called avian type C, type B and D, and nonavian type C retroviruses, respectively), associate with HP68. In addition, exactly what governs the association of primate lentivirus Gag proteins with HP68 remains to be defined. Additional data suggest that specific residues in NC are critical for association of HIV-1 Gag with HP68 (P. Kiser and J. Lingappa, unpublished observations), in agreement with the finding shown here that truncation of various primate lentiviruses proximal to the NC domain inhibits association with HP68. Interestingly, while the overall Gag sequence is not well conserved between different primate lentivirus lineages, selected regions of Gag are relatively more conserved. One of these more conserved areas is the part of NC that encompasses the two Cys-His boxes (i.e., corresponding to amino acids 392 to 426 in HIV-1 Bru Gag). When we examined this region from Gags of the different primate lentivirus lineages studied here, we found 67% sequence identity and 80% sequence similarity on average, which is higher than those seen for Gag overall (55 and 65%, respectively). Thus, our finding that the HP68-Gag interaction is observed for diverse primate lentiviruses is consistent with the apparent requirement of NC for HP68-Gag association and the higher degree of sequence conservation within NC.
Previously, we have demonstrated that HP68 associates with Gag in high-molecular-weight complexes that behave as intermediates in the assembly of immature HIV-1 capsids in a cell-free system. Our finding that HP68 is associated with Gag polypeptides of other primate lentiviruses raises the possibility that all primate lentiviruses assemble by way of a stepwise pathway of discrete, HP68-containing assembly intermediates. Consistent with this, we find that complexes resembling the previously defined 10S, 80-150S, and 500S assembly intermediates can be found in lysates of primate cells expressing assembly-competent primate lentivirus Gag polypeptides. By three criteria, these complexes fit the characteristics expected of assembly intermediates that were defined by pulse-chase studies with the cell-free system. These criteria include the failure of assembly-incompetent Gag mutants to form high-molecular-weight assembly intermediates and the association of specific assembly intermediates with the cellular factor HP68. Third, we demonstrate that the 80-150S complex is extremely transient and energy sensitive in mammalian cells. This is in agreement with the observation that posttranslational ATP depletion resulted in blockade of the assembly pathway with accumulation of the 80-150S assembly intermediate in the cell-free system (37) (Fig. 3). The finding that HIV-1 capsid assembly is energy dependent in the cell-free system was subsequently confirmed in cellular systems expressing HIV-1 (54) and other retroviruses (58). However, the exact step during capsid assembly in cells at which this energy dependence occurs has not been defined. The results shown here demonstrate the presence of a primate lentivirus capsid assembly intermediate in cells (80-150S complex) that is likely to be wholly or partially responsible for the energy dependence of posttranslational events in assembly. The finding that the 80-150S complex is very difficult to detect in lysates of mammalian cells expressing HIV-1 Gag or other primate lentivirus Gag proteins prepared under standard conditions, but is present in significant quantities when the harvest is performed under ATP-depleted conditions, explains why this complex has not been observed previously in cellular systems.
The exact composition and function of the highly transient and dynamic assembly intermediates described here remain to be determined. Previously we have demonstrated that the viral proteins Vif and Gag-Pol are present along with HP68 and Gag in these assembly intermediates (62; K. Klein and J. Lingappa, unpublished observations). Preliminary data suggest an approximate Gag-to-HP68 stoichiometry of 20:1 when assembly intermediates of ≥80S are examined together (Dooher and Lingappa, unpublished observations). Our findings in the cell-free system and in cells suggest a model for primate lentivirus capsid assembly in which newly synthesized Gag first appears in a 10S complex, which then associates with cellular factors and other viral proteins to form the energy-sensitive 80-150S assembly intermediate(s). In the presence of ATP, Gag polypeptides progress from the HP68-containing 80-150S assembly intermediate into the 500S HP68-containing intermediate and then into the completed 750S immature capsid. Upon progression into the completely assembled 750S immature capsid, HP68 is released from Gag, as indicated by our finding that VLPs from cells (Fig. 6C) and completed capsids in the cell-free system (62) do not contain significant quantities of HP68 compared to assembly intermediates from cells. Exactly how the ATP-binding protein HP68 functions in this assembly process remains unclear. HP68 may promote a conformational change that is important for targeting or for capsid formation. Alternatively, HP68, which is known to protect viral RNA from degradation (1a, 39), may promote or regulate encapsidation of genomic RNA into assembly intermediates. Isolation of these critical capsid assembly intermediates from cells will allow these possibilities for HP68 function to be tested.
While the studies presented here support the existence of HP68-containing assembly intermediates in cells expressing primate lentiviruses, they do not address whether HP68 is required for formation of primate lentivirus capsid assembly intermediates or completed immature capsids in cellular systems. To address this question, cell lines stably expressing a small interfering RNA oligonucleotide that knocks down HP68 levels are now being prepared. Nevertheless, the results presented here have important implications for understanding the mechanism of primate lentivirus capsid assembly. We demonstrate that complexes with the same biochemical features as capsid assembly intermediates defined in the cell-free system are found in cells expressing primate lentivirus Gag polypeptides. Two of these assembly intermediates contain the host factor HP68, both in cells and in the cell-free system, and one of the HP68-containing complexes is highly transient and energy sensitive in cells, raising the possibility that this complex may be an important point of regulation during the assembly process. Together, our findings suggest the existence of a universal mechanism of immature capsid formation for primate lentiviruses that is energy dependent and involves the cellular factor HP68, and they establish new approaches for further testing this model.
Acknowledgments
pSAB-1 was obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH, from Mojun Jin and Beatrice Hahn. Monoclonal antibody 183-H12-5C was obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH, from Bruce Chesebro and Kathy Wehrly. The HIV-expressing H9 cell line was engineered by Kevin Klein with the help of Lily Yu and Michael Emerman. HIV-2 clones were generated by E. Beattie and provided by S.-L. Hu. We acknowledge Jonathan Allen, Michael Emerman, Nancy Haigwood, Shiu-Lok Hu, Kevin Klein, Vishwanath Lingappa, Maxine Linial, Paul Luciw, Julie Overbaugh, Lorne Walker, and Lily Yu for reagents and/or advice and Mona Dellos and Dara Lehman for technical assistance.
These studies were funded by NIH grant R29 AI41881 and the NIH-funded University of Washington CFAR grant AI27757 to J.R.L. and by NSF Graduate Fellowship DGE9616736 to J.E.D.
J.R.L. is a cofounder of Prosetta Corporation.
REFERENCES
- 1.Beattie, E., C. Sherbert, J. McClure, L. Misher, N. L. Haigwood, L. Kuller, A. Schmidt, D. A. Anderson, and S. L. Hu. Pathogenicity and sequence analysis of molecular clones derived from HIV-2/287, an isolate highly pathogenic in pig-tailed macaques. J. Med. Primatol., in press.
- 1a.Bisbal, C., C. Martinand, M. Silhol, B. Lebleu, and T. Salehzada. 1995. Cloning and characterization of a RNAse L inhibitor. A new component of the interferon-regulated 2-5A pathway. J. Biol. Chem. 270:13308-13317. [DOI] [PubMed] [Google Scholar]
- 2.Blobel, G. 1995. Unidirectional and bidirectional protein traffic across membranes. Cold Spring Harbor Symp. Quant. Biol. 60:1-10. [DOI] [PubMed] [Google Scholar]
- 3.Braaten, D., E. K. Franke, and J. Luban. 1996. Cyclophilin A is required for the replication of group M human immunodeficiency virus type 1 (HIV-1) and simian immunodeficiency virus SIV(CPZ)GAB but not group O HIV-1 or other primate immunodeficiency viruses. J. Virol. 70:4220-4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Braaten, D., and J. Luban. 2001. Cyclophilin A regulates HIV-1 infectivity, as demonstrated by gene targeting in human T cells. EMBO J. 20:1300-1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campbell, S., R. J. Fisher, E. M. Towler, S. Fox, H. J. Issaq, T. Wolfe, L. R. Phillips, and A. Rein. 2001. Modulation of HIV-like particle assembly in vitro by inositol phosphates. Proc. Natl. Acad. Sci. USA 98:10875-10879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campbell, S., and A. Rein. 1999. In vitro assembly properties of human immunodeficiency virus type 1 Gag protein lacking the p6 domain. J. Virol. 73:2270-2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campbell, S., and V. M. Vogt. 1997. In vitro assembly of virus-like particles with Rous sarcoma virus Gag deletion mutants: identification of the p10 domain as a morphological determinant in the formation of spherical particles. J. Virol. 71:4425-4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chesebro, B., K. Wehrly, J. Nishio, and S. Perryman. 1992. Macrophage-tropic human immunodeficiency virus isolates from different patients exhibit unusual V3 envelope sequence homogeneity in comparison with T-cell-tropic isolates: definition of critical amino acids involved in cell tropism. J. Virol. 66:6547-6554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Connolly, T., and R. Gilmore. 1986. Formation of a functional ribosome-membrane junction during translocation requires the participation of a GTP-binding protein. J. Cell Biol. 103:2253-2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dawson, L., and X. F. Yu. 1998. The role of nucleocapsid of HIV-1 in virus assembly. Virology 251:141-157. [DOI] [PubMed] [Google Scholar]
- 11.Ding, L., A. Derdowski, J. J. Wang, and P. Spearman. 2003. Independent segregation of human immunodeficiency virus type 1 Gag protein complexes and lipid rafts. J. Virol. 77:1916-1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ehrlich, L. S., B. E. Agresta, and C. A. Carter. 1992. Assembly of recombinant human immunodeficiency virus type 1 capsid protein in vitro. J. Virol. 66:4874-4883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ellis, R. J. 2001. Macromolecular crowding: an important but neglected aspect of the intracellular environment. Curr. Opin. Struct. Biol. 11:114-119. [DOI] [PubMed] [Google Scholar]
- 14.Erickson, A. H., and G. Blobel. 1983. Cell-free translation of messenger RNA in a wheat germ system. Methods Enzymol. 96:38-50. [DOI] [PubMed] [Google Scholar]
- 15.Franke, E. K., H. E. Yuan, and J. Luban. 1994. Specific incorporation of cyclophilin A into HIV-1 virions. Nature 372:359-362. [DOI] [PubMed] [Google Scholar]
- 16.Freed, E. O. 2003. The HIV-TSG101 interface: recent advances in a budding field. Trends Microbiol. 11:56-59. [DOI] [PubMed] [Google Scholar]
- 17.Garrus, J. E., U. K. von Schwedler, O. W. Pornillos, S. G. Morham, K. H. Zavitz, H. E. Wang, D. A. Wettstein, K. M. Stray, M. Cote, R. L. Rich, D. G. Myszka, and W. I. Sundquist. 2001. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 107:55-65. [DOI] [PubMed] [Google Scholar]
- 18.Gheysen, D., E. Jacobs, F. de Foresta, C. Thiriart, M. Francotte, D. Thines, and M. De Wilde. 1989. Assembly and release of HIV-1 precursor Pr55gag virus-like particles from recombinant baculovirus-infected insect cells. Cell 59:103-112. [DOI] [PubMed] [Google Scholar]
- 19.Greene, W. C., and B. M. Peterlin. 2002. Charting HIV's remarkable voyage through the cell: basic science as a passport to future therapy. Nat. Med. 8:673-680. [DOI] [PubMed] [Google Scholar]
- 20.Gross, I., H. Hohenberg, C. Huckhagel, and H. G. Krausslich. 1998. N-terminal extension of human immunodeficiency virus capsid protein converts the in vitro assembly phenotype from tubular to spherical particles. J. Virol. 72:4798-4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gross, I., H. Hohenberg, and H. G. Krausslich. 1997. In vitro assembly properties of purified bacterially expressed capsid proteins of human immunodeficiency virus. Eur. J. Biochem. 249:592-600. [DOI] [PubMed] [Google Scholar]
- 22.Gross, I., H. Hohenberg, T. Wilk, K. Wiegers, M. Grattinger, B. Muller, S. Fuller, and H. G. Krausslich. 2000. A conformational switch controlling HIV-1 morphogenesis. EMBO J. 19:103-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gruenberg, J., and K. E. Howell. 1989. Membrane traffic in endocytosis: insights from cell-free assays. Annu. Rev. Cell Biol. 5:453-481. [DOI] [PubMed] [Google Scholar]
- 24.Hahn, B. H., G. M. Shaw, K. M. De Cock, and P. M. Sharp. 2000. AIDS as a zoonosis: scientific and public health implications. Science 287:607-614. [DOI] [PubMed] [Google Scholar]
- 25.Halwani, R., A. Khorchid, S. Cen, and L. Kleiman. 2003. Rapid localization of Gag/GagPol complexes to detergent-resistant membrane during the assembly of human immunodeficiency virus type 1. J. Virol. 77:3973-3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harlow, E., and D. Lane. 1999. Using antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 27.Hartl, F. U. 1996. Molecular chaperones in cellular protein folding. Nature 381:571-579. [DOI] [PubMed] [Google Scholar]
- 28.Hockley, D. J., M. V. Nermut, C. Grief, J. B. Jowett, and I. M. Jones. 1994. Comparative morphology of Gag protein structures produced by mutants of the gag gene of human immunodeficiency virus type 1. J. Gen. Virol. 75:2985-2997. [DOI] [PubMed] [Google Scholar]
- 29.Jin, M. J., H. Hui, D. L. Robertson, M. C. Muller, F. Barre-Sinoussi, V. M. Hirsch, J. S. Allan, G. M. Shaw, P. M. Sharp, and B. H. Hahn. 1994. Mosaic genome structure of simian immunodeficiency virus from west African green monkeys. EMBO J. 13:2935-2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jowett, J. B., D. J. Hockley, M. V. Nermut, and I. M. Jones. 1992. Distinct signals in human immunodeficiency virus type 1 Pr55 necessary for RNA binding and particle formation. J. Gen. Virol. 73:3079-3086. [DOI] [PubMed] [Google Scholar]
- 31.Kimpton, J., and M. Emerman. 1992. Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated beta-galactosidase gene. J. Virol. 66:2232-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klug, A. 1999. The tobacco mosaic virus particle: structure and assembly. Philos. Trans. R. Soc. Lond. B 354:531-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Komoszynski, M., and A. Wojtczak. 1996. Apyrases (ATP diphosphohydrolases, EC 3.6.1.5): function and relationship to ATPases. Biochim. Biophys. Acta 1310:233-241. [DOI] [PubMed] [Google Scholar]
- 34.Lee, Y. M., B. Liu, and X. F. Yu. 1999. Formation of virus assembly intermediate complexes in the cytoplasm by wild-type and assembly-defective mutant human immunodeficiency virus type 1 and their association with membranes. J. Virol. 73:5654-5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee, Y. M., and X. F. Yu. 1998. Identification and characterization of virus assembly intermediate complexes in HIV-1-infected CD4+ T cells. Virology 243:78-93. [DOI] [PubMed] [Google Scholar]
- 36.Lewis, P., M. Hensel, and M. Emerman. 1992. Human immunodeficiency virus infection of cells arrested in the cell cycle. EMBO J. 11:3053-3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lingappa, J. R., R. L. Hill, M. L. Wong, and R. S. Hegde. 1997. A multistep, ATP-dependent pathway for assembly of human immunodeficiency virus capsids in a cell-free system. J. Cell Biol. 136:567-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luban, J., K. L. Bossolt, E. K. Franke, G. V. Kalpana, and S. P. Goff. 1993. Human immunodeficiency virus type 1 Gag protein binds to cyclophilins A and B. Cell 73:1067-1078. [DOI] [PubMed] [Google Scholar]
- 39.Martinand, C., C. Montavon, T. Salehzada, M. Silhol, B. Lebleu, and C. Bisbal. 1999. RNase L inhibitor is induced during human immunodeficiency virus type 1 infection and down regulates the 2-5A/RNase L pathway in human T cells. J. Virol. 73:290-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martinand, C., T. Salehzada, M. Silhol, B. Lebleu, and C. Bisbal. 1998. RNase L inhibitor (RLI) antisense constructions block partially the down regulation of the 2-5A/RNase L pathway in encephalomyocarditis-virus-(EMCV)-infected cells. Eur. J. Biochem. 254:248-255. [DOI] [PubMed] [Google Scholar]
- 41.Martin-Serrano, J., T. Zang, and P. D. Bieniasz. 2001. HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat. Med. 7:1313-1319. [DOI] [PubMed] [Google Scholar]
- 42.Martin-Serrano, J., T. Zang, and P. D. Bieniasz. 2003. Role of ESCRT-I in retroviral budding. J. Virol. 77:4794-4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mellman, I., and G. Warren. 2000. The road taken: past and future foundations of membrane traffic. Cell 100:99-112. [DOI] [PubMed] [Google Scholar]
- 44.Melton, D. A., P. A. Krieg, M. R. Rebagliati, T. Maniatis, K. Zinn, and M. R. Green. 1984. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 12:7035-7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mergener, K., M. Facke, R. Welker, V. Brinkmann, H. R. Gelderblom, and H. G. Krausslich. 1992. Analysis of HIV particle formation using transient expression of subviral constructs in mammalian cells. Virology 186:25-39. [DOI] [PubMed] [Google Scholar]
- 46.Rothman, J. E., and L. Orci. 1992. Molecular dissection of the secretory pathway. Nature 355:409-415. [DOI] [PubMed] [Google Scholar]
- 47.Sharp, P. M., E. Bailes, F. Gao, B. E. Beer, V. M. Hirsch, and B. H. Hahn. 2000. Origins and evolution of AIDS viruses: estimating the time-scale. Biochem. Soc. Trans. 28:275-282. [DOI] [PubMed] [Google Scholar]
- 48.Simon, J. H. M., E. A. Carpenter, R. A. M. Fouchier, and M. H. Malim. 1999. Vif and the p55Gag polyprotein of human immunodeficiency virus type 1 are present in colocalizing membrane-free cytoplasmic complexes. J. Virol. 73:2667-2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Singh, A. R., R. L. Hill, and J. R. Lingappa. 2001. Effect of mutations in Gag on assembly of immature human immunodeficiency virus type 1 capsids in a cell-free system. Virology 279:257-270. [DOI] [PubMed] [Google Scholar]
- 50.Spearman, P., and L. Ratner. 1996. Human immunodeficiency virus type 1 capsid formation in reticulocyte lysates. J. Virol. 70:8187-8194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thali, M., A. Bukovsky, E. Kondo, B. Rosenwirth, C. T. Walsh, J. Sodroski, and H. G. Gottlinger. 1994. Functional association of cyclophilin A with HIV-1 virions. Nature 372:363-365. [DOI] [PubMed] [Google Scholar]
- 52.Toohey, K., K. Wehrly, J. Nishio, S. Perryman, and B. Chesebro. 1995. Human immunodeficiency virus envelope V1 and V2 regions influence replication efficiency in macrophages by affecting virus spread. Virology 213:70-79. [DOI] [PubMed] [Google Scholar]
- 53.Tritel, M., and M. D. Resh. 2000. Kinetic analysis of human immunodeficiency virus type 1 assembly reveals the presence of sequential intermediates. J. Virol. 74:5845-5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tritel, M., and M. D. Resh. 2001. The late stage of human immunodeficiency virus type 1 assembly is an energy-dependent process. J. Virol. 75:5473-5481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.VerPlank, L., F. Bouamr, T. J. LaGrassa, B. Agresta, A. Kikonyogo, J. Leis, and C. A. Carter. 2001. Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55(Gag). Proc. Natl. Acad. Sci. USA 98:7724-7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.von Schwedler, U. K., T. L. Stemmler, V. Y. Klishko, S. Li, K. H. Albertine, D. R. Davis, and W. I. Sundquist. 1998. Proteolytic refolding of the HIV-1 capsid protein amino-terminus facilitates viral core assembly. EMBO J. 17:1555-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wehrly, K., and B. Chesebro. 1997. p24 antigen capture assay for quantification of human immunodeficiency virus using readily available inexpensive reagents. Methods 12:288-293. [DOI] [PubMed] [Google Scholar]
- 58.Weldon, R. A., Jr., W. B. Parker, M. Sakalian, and E. Hunter. 1998. Type D retrovirus capsid assembly and release are active events requiring ATP. J. Virol. 72:3098-3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wiegers, K., and H. G. Krausslich. 2002. Differential dependence of the infectivity of HIV-1 group O isolates on the cellular protein cyclophilin A. Virology 294:289-295. [DOI] [PubMed] [Google Scholar]
- 60.Yang, L., and L. Ratner. 2002. Interaction of HIV-1 gag and membranes in a cell-free system. Virology 302:164-173. [DOI] [PubMed] [Google Scholar]
- 61.Yu, F., S. M. Joshi, Y. M. Ma, R. L. Kingston, M. N. Simon, and V. M. Vogt. 2001. Characterization of Rous sarcoma virus Gag particles assembled in vitro. J. Virol. 75:2753-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zimmerman, C., K. C. Klein, P. K. Kiser, A. R. S. Singh, B. L. Firestein, S. C. Riba, and J. R. Lingappa. 2002. Identification of a host protein essential for assembly of immature HIV-1 capsids. Nature 415:88-92. [DOI] [PubMed] [Google Scholar]