Abstract
Epstein-Barr virus (EBV) nuclear antigen 3C (EBNA3C) is essential for primary B-cell transformation. In this report we show that cyclin A, an activator of S phase progression, bound tightly to EBNA3C. EBNA3C interacted with cyclin A in vitro and associated with cyclin A complexes in EBV-transformed lymphoblastoid cell lines. Importantly, EBNA3C stimulated cyclin A-dependent kinase activity and rescued p27-mediated inhibition of cyclin A/Cdk2 kinase activity by decreasing the molecular association between cyclin A and p27 in cells. Additionally, phosphorylation of the retinoblastoma protein, a major regulator of cell cycle progression, was enhanced both in vitro and in vivo in the presence of EBNA3C. Cyclin A interacted with a region of the carboxy terminus of EBNA3C, shown to be important both for stimulation of cyclin A-dependent kinase activity and for cell cycle progression. This provides the first evidence of an essential EBV latent antigen's directly targeting a cell cycle regulatory protein and suggests a novel mechanism by which EBV deregulates the mammalian cell cycle, which is of critical importance in B-cell transformation.
Epstein-Barr virus (EBV) is the etiologic agent of infectious mononucleosis and is associated with numerous human malignancies, including Burkitt's lymphoma, nasopharyngeal carcinoma, posttransplant and AIDS-associated lymphomas, and Hodgkin's disease (5, 40). EBV predominantly infects two human cell types in vivo, establishing lytic infection in the oropharyngeal epithelium and latent infection in B lymphocytes (23, 40). Transformation of B lymphocytes by EBV requires the expression of a number of viral latent genes. A subset of these, including EBV nuclear antigen 3C (EBNA3C), are essential for immortalization in vitro and lymphomagenesis in vivo (1, 3, 15, 24, 40, 53). Indeed, second-site recombination studies demonstrate that replacement of the wild-type EBNA3C gene with a gene encoding a truncated molecule abolishes the transforming potential of EBV (50). These experiments strongly suggest an essential, and to date undefined, role for the carboxy terminus of EBNA3C in B-cell transformation.
Classic work with other DNA tumor viruses has demonstrated that these viruses drive cell proliferation by specifically targeting cell cycle regulatory and checkpoint molecules (10, 17, 20, 25, 32, 51). The simian virus 40 large T antigen, the adenovirus E1A protein, and the papillomavirus E7 protein promote DNA replication, and ultimately cell cycle progression, by inactivating a common target, the retinoblastoma tumor suppressor (Rb) (11, 13, 52). While some studies have shown an association between EBV immediate-early antigens and the Rb and p53 proteins (27, 47, 54), the link between EBV latent antigens and the regulators commonly targeted by tumor viruses has remained unresolved, suggesting that EBV employs unique and complex mechanisms to modulate the cell cycle of infected lymphoid cells.
To date, studies examining the essential EBV nuclear antigen EBNA3C provide perhaps the best link between latent EBV infection and the Rb regulatory pathways, although no direct evidence in human cells has been demonstrated (4, 33, 34). EBNA3C activates the human B-myb promoter in an E2F-dependent manner and induces focus formation similar to papillomavirus E7 in a colony formation assay (33). Also, EBNA3C relieves the block to transformation mediated by the cyclin-dependent kinase inhibitor p16INK4A (33) and drives serum-starved cells through the G1/S restriction point (34). Despite this evidence, a clear molecular link between cell cycle regulatory molecules and EBNA3C has yet to be demonstrated in vivo. Importantly, this study provides the first evidence that EBNA3C directly targets a critical cell cycle regulatory protein in cells, distinctly different from other tumor virus antigens, and describes a potentially fundamental mechanism by which EBV deregulates the mammalian cell cycle.
MATERIALS AND METHODS
Yeast two-hybrid cDNA screen.
An EBV-positive lymphoblastoid cell line (LCL)-derived cDNA library was screened with a yeast two-hybrid system essentially as described previously (8, 16). Transformants were grown on appropriate selective media and screened to identify β-galactosidase-positive colonies. Positive clones were sequenced and identified by Blast search of GenBank.
Plasmids, antibodies, and cell lines.
pA3M-E3C constructs express either full-length EBNA3C or EBNA3C truncations with a C-terminal Myc tag and have been described previously (46). Glutathione S-transferase (GST)-EBNA3C truncation mutants have been described previously (8). The pASIΔEBNA3C construct and pACTcDNA library have been described (8). pCMV-Cdk2, RC-cyclin A, and RC-cyclin E were kindly provided by Philip Hinds (18). The construct expressing GST-cyclin A was provided by Maria Mudryj (12). pCDNA3-p27 was provided by Michele Pagano (31). The construct expressing GST-Rb amino acids 792 to 928 was provided by Ed Harlow (28). Constructs expressing Myc-tagged cyclin A, Rb, and EBNA3C amino acids 1 to 956 were prepared by cloning PCR-amplified cDNAs into the previously described pA3M vector (6).
Rabbit polyclonal antibodies reactive to cyclin A, cyclin E, Cdk2, and p27Kip1 were purchased from Santa Cruz Biotechnology, Inc. A10 monoclonal and rabbit polyclonal antibodies reactive to EBNA3C have been described previously (8). HEK 293 cells are human embryonic kidney cells transformed by adenovirus type 5 DNA; HEK 293T cells stably express the simian virus 40 large T antigen (2). The Burkitt's lymphoma cell line BJAB is derived from an EBV-negative African Burkitt's lymphoma tumor (7). EBNA3C-expressing BJAB cell lines have been described previously (41). U2OS is a human osteosarcoma cell line (36). HEK 293, 293T, U2OS, early-passage Rat-1 fibroblasts, and HeLa cells were grown in Dulbecco's modified Eagle's medium (Gibco-BRL) with 10% fetal bovine serum unless otherwise indicated. 293T cells were transfected by electroporation. BJAB cells were grown in RPMI 1640 (Gibco-BRL) supplemented as for Dulbecco's modified Eagle's medium.
GST pull-down assays.
GST fusion proteins were purified from bulk Escherichia coli cultures following induction with isopropylthiogalactopyranoside (IPTG) as described previously (8). For pull-down assays from cells, lysates were prepared in radioimmunoprecipitation assay buffer (0.5% NP-40, 10 mM Tris, pH 7.5, 2 mM EDTA, 150 mM NaCl, supplemented with protease inhibitors). Lysates were precleared and then rotated with either the GST control or the appropriate GST fusion protein bound to glutathione-Sepharose beads. For in vitro binding experiments, GST fusion proteins were incubated with 35S-labeled in vitro-translated protein in binding buffer (1x phosphate-buffered saline, 0.1% NP-40, 0.5 mM dithiothreitol, 10% glycerol, supplemented with protease inhibitors). In vitro translation was done with the TNT T7 quick coupled transcription-translation system (Promega) according to the manufacturer's instructions.
Immunoprecipitation and immunofluorescence assays.
Viable cells were counted by trypan blue negative staining, and 50 to 100 million cells were collected by centrifugation. Immunoprecipitation was done essentially as described previously (8). Proteins were fractionated by electrophoresis on a sodium dodecyl sulfate-polyacrylamide gel and then transferred to a 0.45-μm nitrocellulose membrane. The membrane was blotted with appropriate rabbit polyclonal antibodies, followed by horseradish peroxidase-linked protein A (Amersham) at a 1:5,000 dilution.
HeLa cells were transfected by Lipofectamine 2000 reagent (Invitrogen) with plasmids expressing either Myc-tagged EBNA3C or Myc-tagged EBNA3C amino acids 366 to 620. Cells were harvested at 48 h, trypsinized, and allowed to adhere to glass slides overnight. Cells were fixed and permeabilized in methanol at −20°C for 10 min, followed by acetone at room temperature for 30 s. Slides were blocked with 5% goat serum and incubated at 4°C with cyclin A and Myc antibodies. Slides were washed with phosphate-buffered saline and then incubated with goat anti-mouse-Texas Red and goat anti-rabbit-fluorescein isothiocyanate antibodies (Jackson Laboratories). Slides were washed and visualized with an Olympus X170 inverted fluorescence microscope. Pictures were captured with a digital PixelFly camera (Cooke, Inc.).
Histone H1 kinase assay.
U2OS cells were seeded into six-well plates and grown to confluence in 0.5% fetal bovine serum for 48 h prior to transfection. Cells were transfected with Lipofectamine 2000 reagent (Invitrogen), harvested after 24 h with a cell scraper, washed with phosphate-buffered saline, and lysed on ice in 500 μl of radioimmunoprecipitation assay buffer (0.5% NP-40, 10 mM Tris, pH 7.5, 2 mM EDTA, 150 mM NaCl, 1 mM EGTA, with protease and phosphatase inhibitors). Lysates were precleared and then rotated with 1 μg of cyclin A antibody overnight at 4°C. Cyclin A complexes were captured by rotating with protein A-Sepharose beads and washed with radioimmunoprecipitation assay buffer. Cyclin A complexes were then washed with histone wash buffer (25 mM Tris, pH 7.5, 70 mM NaCl, 10 mM MgCl2, 1 mM EGTA, 1 mM dithiothreitol, with protease and phosphatase inhibitors). Complexes were incubated in 30 μl of histone wash buffer supplemented with 4 μg of histone H1 (Upstate Biotechnology, Inc.), 10 mM unlabeled ATP, and 0.2 μCi of γ-[32P]ATP per μl for 30 min at 37°C. The reaction was stopped by adding sodium dodecyl sulfate lysis buffer and heating to 95°C for 10 min. Labeled histone H1 was resolved by sodium dodecyl sulfate-12% polyacrylamide gel electrophoresis (SDS-12% PAGE). Quantitation was done with ImageQuant software (Molecular Dynamics). BJAB cells were transfected by electroporation with the Bio-Rad Gene Pulser II. Transfected cells were incubated in RPMI 1640 with 0.5% fetal bovine serum for 24 h and then harvested as for U2OS cells.
Flow cytometry.
Cells were harvested and fixed in 50% ethanol at −20°C. Fixed cells were washed and incubated in phosphate-buffered saline supplemented with 100 μg of RNase A per ml and 50 μg of propidium iodide per ml for 30 min at room temperature and then overnight at 4°C. Samples were then analyzed by flow cytometry with a FACScan instrument (Becton Dickinson). Total populations were gated to remove doublets and small debris.
RESULTS
Search for cellular molecules associated with EBNA3C identified cyclin A as a binding partner.
Genetic studies in EBV demonstrate that a genetic knock-out of the EBNA3C gene neutralizes the transforming potential of EBV in mediating human primary B-cell transformation (50) (Fig. 1A). To identify protein binding partners, a truncated form of EBNA3C corresponding to amino acids 365 to 992 was fused in frame with the GAL4 DNA binding domain and tested against an LCL-derived cDNA library in a yeast two-hybrid screen. Positive clones were sequenced and potential interacting partners were identified by a Blast search of GenBank. Partial sequences from two positive cDNA clones identified coding sequence within the amino-terminal one-third of the human cyclin A gene. Based on this identification, the complete cDNA of cyclin A was obtained for further studies in vitro.
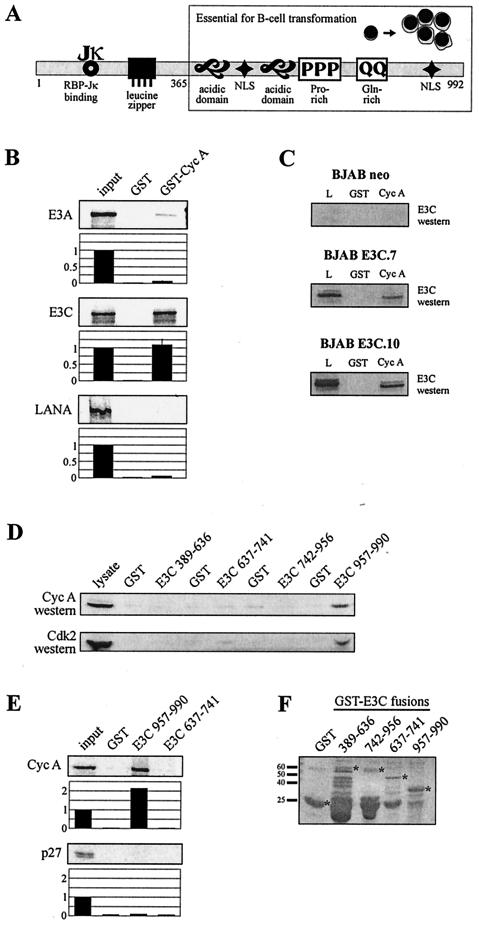
FIG.1.
EBNA3C binds cyclin A. (A) The schematic illustrates structural motifs of EBNA3C that potentially contribute to protein-protein interactions. Introduction of a stop codon at amino acid 365 blocks in vitro transformation by EBV as determined by second-site recombination studies (50). NLS, nuclear localization signal. (B) GST-cyclin A fusion protein was expressed in E. coli and purified with glutathione-Sepharose beads. Full-length EBNA3A, EBNA3C, and Kaposi’s sarcoma-associated herpesvirus LANA were labeled with [35S]methionine by in vitro translation and then incubated with either GST control or GST-cyclin A beads normalized by Coomassie staining. In each case, 5% of in vitro translation input was used for comparison. Precipitated proteins were resolved by SDS-PAGE and bands were visualized and quantified with a phosphorimager screen. (C) Either GST control or GST-cyclin A beads were incubated with lysates from BJAB cells or BJAB cells stably expressing EBNA3C (E3C.7 and E3C.10). EBNA3C was detected by Western blot with the A10 monoclonal antibody. L, 5% BJAB lysate. (D) GST-EBNA3C fusion proteins representing truncation mutants spanning amino acids 389 to 990 were expressed in E. coli and purified with glutathione-Sepharose beads. 293 cells were lysed and incubated with either GST control or GST-EBNA3C beads. Cyclin A and Cdk2 were detected by Western blot; 5% 293 input lysate was loaded as a Western blot control. (E) Human cyclin A and p27 were labeled with [35S]methionine by in vitro transcription-translation and then incubated with either GST control or GST-EBNA3C 957 to 990 beads as described above. Precipitated proteins were resolved by SDS-PAGE and bands were visualized and quantified with a phosphorimager screen. In each case, 5% of in vitro translation input was used for comparison. (F) Coomassie staining of SDS-PAGE-resolved GST-EBNA3C fusion proteins.
Cell cycle progression is partially dependent on the activity of cyclin-dependent kinases (Cdks). These kinases are allosterically activated by binding to cyclins, a family of proteins whose levels oscillate in synchrony with cell cycle progression (29). Cyclin A, synthesized beginning in G1 and maximally at the onset of S phase, is necessary both for progression through S phase and for entry into mitosis (30). Additionally, exogenous expression of cyclin A has been shown to accelerate exit from G1 (38).
Bacterially expressed cyclin A binds EBNA3C.
The results in Saccharomyces cerevisiae suggested that EBNA3C and cyclin A are specific binding partners with potential implications for cell cycle deregulation in human cells. The specificity of this interaction was first determined in the context of bacterially expressed human cyclin A. GST-cyclin A beads precipitated in vitro-translated, 35S-labeled EBNA3C, while negligible signal was observed with GST control beads (Fig. 1B). As negative controls, GST-cyclin A beads demonstrated no specific binding for Kaposi’s sarcoma-associated herpesvirus LANA and very weak binding for EBNA3A (Fig. 1B). The cyclin A-EBNA3C interaction was further assessed in the context of the EBV-negative B-cell line BJAB stably transfected with either an EBNA3C-expressing or a control construct. GST-cyclin A specifically precipitated EBNA3C from these lysates, as determined by Western blot analysis (Fig. 1C).
Carboxy-terminal tail of EBNA3C interacts with cyclin A.
Numerous domains responsible for both transcriptional activity and protein-protein interactions have previously been mapped on EBNA3C (37, 41, 45, 46) (Fig. 1A). To identify the region or regions of EBNA3C responsible for the interaction in S. cerevisiae, we prepared GST fusion proteins corresponding to four EBNA3C truncations spanning amino acids 389 to 990 (Fig. 1D). Beads were normalized by Coomassie staining, incubated with 293 cell lysates, and washed. Western blot analysis demonstrated relatively higher binding activity for both cyclin A and Cdk2 above that of a GST control for the EBNA3C carboxy terminus, corresponding to amino acids 957 to 990. Additionally, amino acids 957 to 990 were able to precipitate in vitro-translated cyclin A but not p27 (Fig. 1E).
EBNA3C forms complexes with cyclin A in EBV-transformed lymphoblastoid cells.
While one study has demonstrated limited binding activity of EBNA3C to the Rb protein in an in vitro assay (33), neither Rb nor any other key cell cycle regulatory molecule has been shown to associate with EBNA3C or other EBV latent antigens in vivo. Therefore, we sought to confirm the presence of EBNA3C-cyclin A complexes in the context of B cells expressing EBNA3C and in EBV-transformed cells. We first immunoprecipitated EBNA3C from BJAB cell lines stably expressing EBNA3C. Western blot analysis of EBNA3C precipitates revealed coimmunoprecipitation of both cyclin A and Cdk2 (Fig. 2A). This result was specific, as cyclin A and Cdk2 were not precipitated by normal rabbit serum and protein A beads. These molecules were also not precipitated from BJAB control cells (Fig. 2A).
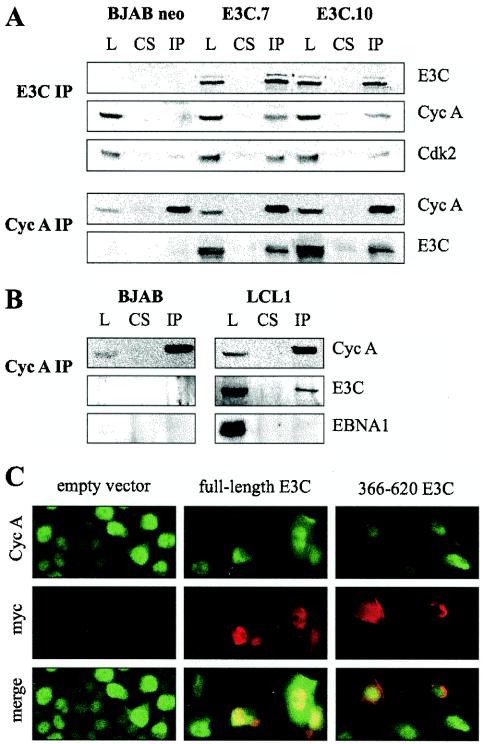
FIG. 2.
EBNA3C and cyclin A form complexes in cells. (A) BJAB cells or BJAB cells stably expressing EBNA3C were lysed (L) and precleared with control rabbit serum (CS). Either EBNA3C (E3C IP) or cyclin A (Cyc A IP) complexes were then captured with the appropriate rabbit polyclonal antibody and protein A beads. In these assays, 5% of the total lysate was used in comparative control lanes. EBNA3C, cyclin A, and Cdk2 were detected by Western blot. (B) BJAB cells and EBV-infected LCLs were lysed and immunoprecipitated for cyclin A as above. EBNA3C was detected by A10 monoclonal antibody. EBNA1 was detected with EBV-reactive human serum. As for panel A, 5% of the total lysate was loaded as a control. (C) HeLa cells were transfected with vector, a plasmid expressing full-length Myc-tagged EBNA3C, or a plasmid expressing Myc-tagged EBNA3C amino acids 366 to 620. Cells were fixed and probed with anti-cyclin A rabbit polyclonal and anti-Myc mouse monoclonal antibodies. Staining was visualized with goat anti-rabbit immunoglobulin-fluorescein isothiocyanate and goat anti-mouse immunoglobulin-Texas Red antibodies.
As further confirmation, cyclin A was immunoprecipitated from EBNA3C-expressing BJAB lysates with a rabbit polyclonal antibody specific to cyclin A. Western blot analysis of cyclin A precipitates revealed that specific EBNA3C binding was not present in normal rabbit serum control precipitates (Fig. 2A). To demonstrate that this complex was also present in the context of EBV-infected cell lines, a similar coimmunoprecipitation analysis was performed on an EBV-immortalized LCL cell line with BJAB as an uninfected control. Again, immunoprecipitation of cyclin A complexes resulted in a reproducible coimmunoprecipitation of EBNA3C but not EBNA1, indicating the existence of this complex in EBV-transformed human B lymphocytes (Fig. 2B). It should be noted that in comparison to BJAB cell lines overexpressing EBNA3C (approximately 10-fold relative to levels in LCLs), cyclin A and EBNA3C coimmunoprecipitated less robustly in LCLs.
EBNA3C and cyclin A colocalize in HeLa cell nuclei.
To determine if cyclin A and EBNA3C exist in similar nuclear compartments, HeLa cells were transfected with a vector expressing either full-length EBNA3C or EBNA3C amino acids 366 to 620, which did not interact with cyclin A in GST binding assays. Full-length EBNA3C staining was tightly nuclear, demonstrating a stippled, punctate pattern with exclusion of nucleoli (Fig. 2C). Cyclin A staining, while concentrated in the nucleus, was more diffuse than for EBNA3C (Fig. 2C). Nevertheless, cyclin A clearly colocalized with EBNA3C complexes in the nucleus, as visualized by yellow fluorescence (Fig. 2C). In contrast, the EBNA3C 366 to 620 mutant localized diffusely throughout the nucleus and nuclear periphery compared to full-length EBNA3C and showed little specific colocalization with cyclin A staining (Fig. 2C). These data corroborate the in vivo association between cyclin A and EBNA3C, as these molecules in part localized to similar compartments in the cell nucleus.
EBNA3C enhances cyclin A-dependent kinase activity in U2OS cells.
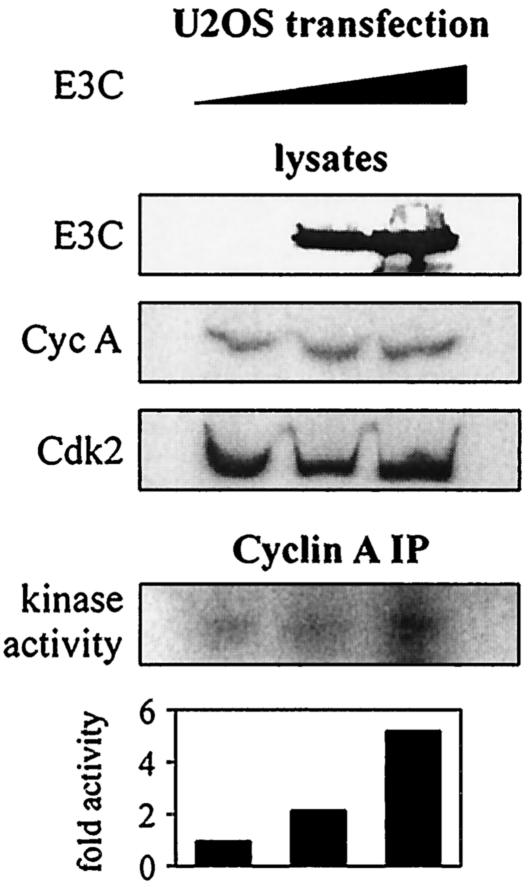
To address the functional consequences of the association of cyclin A and EBNA3C, we tested the activity of cyclin A complexes in phosphorylating a known substrate, histone H1 (43). U2OS cells were transfected with titrated amounts of an EBNA3C expression construct, and cyclin A immunoprecipitate complexes were assayed for in vitro kinase activity as determined by histone H1 phosphorylation. Cyclin A-dependent kinase activity increased in a dose-responsive manner with increased expression of EBNA3C (Fig. 3). Arbitrary counts (with a phosphorimager screen) of labeled histone H1 indicated a change in phosphorylation level up to fivefold (Fig. 3). In repeated experiments, cyclin A protein levels mostly remained constant or occasionally showed a slight increase in the presence of EBNA3C. This suggests that EBNA3C is capable of enhancing the kinase activity of baseline cyclin A complexes.
FIG. 3.
EBNA3C enhances cyclin A-dependent kinase activity. U2OS cells were transfected with 0, 3, or 10 μg of pA3M-EBNA3C. Empty pA3M vector was used to balance total transfected DNA; 24 h later, cyclin A immunoprecipitates were captured and assayed for in vitro kinase activity toward histone H1.
EBNA3C decreases the coimmunoprecipitation of p27 with cyclin A complexes.
As cyclin A-dependent kinase activity is enhanced when cells express EBNA3C, we decided to investigate whether EBNA3C affects the recruitment of cellular molecules that may act as activators or inhibitors of cyclin A-kinase complexes. Potential inhibitors involved in cyclin A-kinase complexes include p21 and p27. In comparison to p21, p27 was significantly more potent in its ability to inhibit cyclin A/Cdk2 kinase activity in our initial experiments, and we therefore chose to focus on this inhibitor in subsequent experiments.
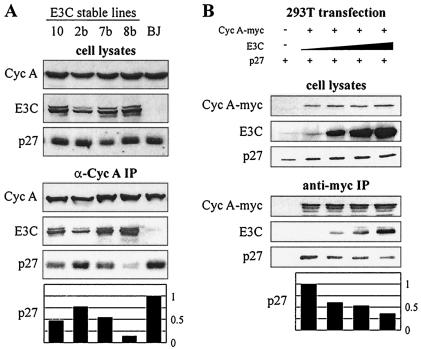
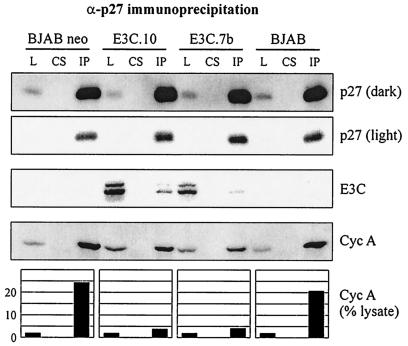
p27Kip1 is a putative tumor suppressor and potent inhibitor of diverse cyclin-Cdk complexes, including cyclin A-Cdk2 (35, 38). To assess the role that p27 might be playing in differential cyclin A-dependent kinase activity, cyclin A complexes were immunoprecipitated from EBNA3C-expressing BJAB cells. While p27 coimmunoprecipitated strongly with cyclin A from the lysates of control BJAB cell lines, stable expression of EBNA3C reproducibly reduced this interaction (Fig. 4A). Interestingly, levels of reduction were typically proportional to the levels of EBNA3C expression (compare E3C and p27 blots in the lower panel of Fig. 4A).
FIG. 4.
EBNA3C disrupts p27 from cyclin A immunoprecipitates. (A) BJAB cells and BJAB cells stably expressing EBNA3C were lysed, precleared with normal rabbit serum and protein A beads, and then immunoprecipitated with antibody reactive to cyclin A. Samples were resolved by SDS-PAGE. Cyclin A, EBNA3C, and p27 were detected by Western blot. (B) 293T cells were transfected by electroporation with constructs expressing Myc-tagged cyclin A, EBNA3C, and p27. Specifically, 10 million 293T cells were transfected with 10 μg of pA3M-cyclin A, 0 to 16 μg of pSG5-EBNA3C, and 5 μg of pCDNA3-p27 as indicated. Cells were harvested at 36 h, lysates were normalized by Bradford assay, and cyclin A complexes were captured by anti-Myc immunoprecipitation.
To confirm this result in a transient system, 293T cells were transfected with expression constructs for Myc-tagged cyclin A, p27, and EBNA3C (Fig. 4B). Overexpression of EBNA3C resulted in a modest but dose-responsive decrease in the amount of p27 coimmunoprecipitating with cyclin A, further suggesting a decreased association between cyclin A and p27 in the context of EBNA3C expression (Fig. 4B).
EBNA3C decreases the coimmunoprecipitation of cyclin A with p27 complexes.
To further address whether p27 and EBNA3C are mutually exclusive members of cyclin A complexes, we immunoprecipitated p27 from the EBNA3C-expressing BJAB lines employed above. As expected, cyclin A associated strongly with p27 precipitated from two independently growing BJAB control cell lines (Fig. 5, cyclin A blot, left and right samples). However, this interaction was significantly reduced with stable expression of EBNA3C in two BJAB isogenic cell backgrounds (Fig. 5, cyclin A blot, middle samples). Interestingly, levels of EBNA3C in p27 immunoprecipitates were, as expected, weakly detectable, which is consistent with a model in which the entry of EBNA3C into cyclin A complexes results in the displacement of p27. Additionally, cellular p27 levels were neither reproducibly increased nor decreased in BJAB lines expressing EBNA3C, suggesting that, at least in this context, EBNA3C is specifically targeting p27 at the level of cyclin A complexes (compare lysate lanes, Fig. 5).
FIG. 5.
EBNA3C disrupts cyclin A from p27 immunoprecipitates. BJAB cells and BJAB cells stably expressing EBNA3C were lysed, precleared with normal rabbit serum and protein A beads, and then immunoprecipitated with antibody reactive to p27. p27 (dark) and p27 (light) represent different exposure times of the same Western blot. L, 2% of total lysate. CS, control serum. IP, immunoprecipitate. Western blot quantification was done with ImageGauge software (FujiFilm).
EBNA3C rescues p27-mediated suppression of cyclin A-dependent kinase activity in U2OS and BJAB cells.
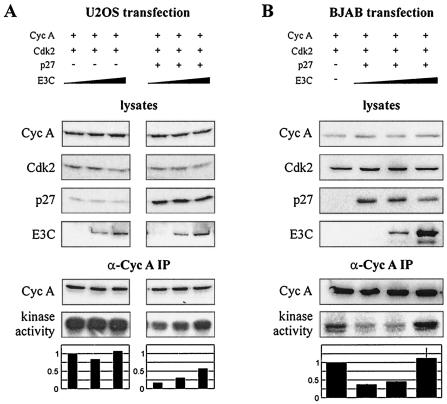
The previous kinase assays did not utilize the exogenous expression of any molecule other than EBNA3C, as we hypothesized that the relative protein levels of cyclin A and other members of the cyclin A complex might be critical to EBNA3C modulation of activity. To more directly assess the role of p27 in EBNA3C activation, cyclin A, Cdk2, and p27 were expressed from heterologous promoters in the context of increased levels of EBNA3C. When cyclin A and Cdk2, without p27, were introduced exogenously, cyclin A-dependent kinase activity was relatively unresponsive to EBNA3C expression (Fig. 6A, left three samples). However, when p27 was exogenously expressed along with cyclin A and Cdk2, increased expression of EBNA3C resulted in a clear stimulation of kinase activity (Fig. 6A, right three samples). Importantly, EBNA3C also rescued p27-mediated suppression of cyclin A/Cdk2 activity in BJAB cells, an EBV-negative B-cell line demonstrating functional relevance in a human B-cell background (Fig. 6B).
FIG. 6.
EBNA3C relieves p27-mediated suppression of cyclin A-dependent kinase activity. (A) U2OS cells were transfected with cyclin A and Cdk2 expression constructs as well as increasing amounts of an EBNA3C expression construct. The right three samples were additionally transfected with a p27 expression plasmid. Specifically, cells were transfected with 2 μg of RC-cyclin A, 1 μg of CMV-Cdk2, 0-1 μg of pCDNA3-p27, and 0 to 10 μg of pA3M-EBNA3C as indicated. After 24 h, cyclin A immunoprecipitates were captured and assayed for in vitro kinase activity toward histone H1. (B) Ten million BJAB cells were transfected with 6 μg of RC-cyclin A, 3 μg of CMV-Cdk2, 0 to 6 μg of pCDNA3-p27, and 0 to 12 μg of pA3M-EBNA3C as indicated. After 24 h, cyclin A immunoprecipitates were assayed for in vitro kinase activity toward histone H1.
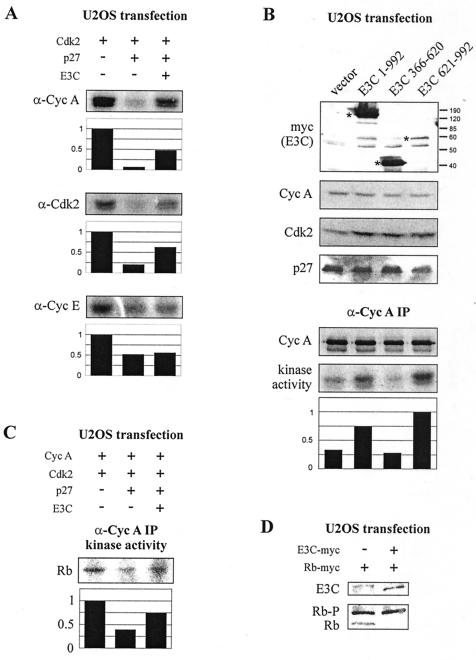
To address the possibility that EBNA3C rescue of p27 suppression might be the result of general cell cycle stimulation unrelated to the association of EBNA3C with cyclin A complexes, U2OS cells were again transfected with Cdk2, p27, EBNA3C, and either cyclin A or cyclin E expression plasmids. Cells were lysed at 24 h and immunoprecipitated with cyclin A-, Cdk2-, or cyclin E-specific antibodies. For both cyclin A and Cdk2 immunoprecipitations, kinase activity toward the histone H1 substrate was significantly suppressed by p27. This suppression, as expected, was rescued with EBNA3C expression (Fig. 7A). However, for cyclin E immunoprecipitation, in repeated attempts, EBNA3C was unable to rescue the p27-mediated suppression of kinase activity, suggesting that EBNA3C is specifically targeting cyclin A/Cdk2-associated activity and is not globally regulating all cyclin-dependent kinase activities in this system (Fig. 7A). Additionally, DNA content determination of U2OS cells harvested as for immunoprecipitation analysis did not demonstrate dramatic differences in the cell cycle profile of either p27- or EBNA3C-expressing cells as would be expected if EBNA3C were globally affecting cell cycle changes under the conditions of these experiments (data not shown).
FIG. 7.
EBNA3C relieves p27-mediated suppression of cyclin A-dependent kinase activity toward the retinoblastoma protein. (A) U2OS cells were transfected with 1 μg of CMV-Cdk2, 0 to 1 μg of pCDNA3-p27, and 0 to 10 μg of pA3M-EBNA3C as indicated. After 24 h, cyclin A, Cdk2, and cyclin E immunoprecipitates were captured and assayed for kinase activity toward histone H1. Cyclin A- and cyclin E-immunoprecipitated samples were additionally transfected with 2 μg of RC-cyclin A and 2 μg of RC-cyclin E, respectively. (B) All samples were transfected with cyclin A, Cdk2, and p27 expression plasmids as for panel A. Samples were additionally transfected with 10 μg of Myc-tagged EBNA3C expression constructs as indicated. Asterisks designate Myc-tagged proteins as detected by Western blot. After 24 h, cyclin A immunoprecipitates were assayed for in vitro kinase activity toward histone H1. (C) U2OS cells were transfected with cyclin A, Cdk2, p27, and EBNA3C expression plasmids as for panel A. After 24 h, cyclin A immunoprecipitates were captured and assayed for in vitro kinase activity toward GST-Rb. (D) U2OS cells were transfected with 5 μg of pA3M-Rb and 0 to 5 μg of pA3M-EBNA3C as indicated. After 24 h, lysates were prepared and resolved by SDS-6% PAGE. EBNA3C, hypophosphorylated Rb, and hyperphosphorylated Rb (Rb-P) were detected by Myc-specific Western blot.
Carboxy terminus of EBNA3C rescues p27-mediated suppression of cyclin A-dependent kinase activity.
To determine whether rescue of cyclin A-dependent kinase activity could be mapped to the same carboxy-terminal region of EBNA3C as in vitro cyclin A binding, U2OS cells were transfected with cyclin A, Cdk2, and p27 expression plasmids along with either full-length EBNA3C or EBNA3C truncation mutants. EBNA3C amino acids 621 to 992 were able to reproducibly rescue p27 suppression of cyclin A-dependent kinase activity similar to, and perhaps more efficiently than, wild-type EBNA3C, a result not seen with nonbinding amino acids 366 to 620 of EBNA3C (Fig. 7B). Interestingly, amino acids 621 to 992 could not be expressed to the levels of wild-type EBNA3C, as determined by Western blotting for the carboxy-terminal Myc tag, possibly due to a cell toxicity effect. Nevertheless, the comparatively low levels of expression that were obtained were even more efficient than the full-length molecule in rescuing the associated kinase activity, suggesting that the carboxy terminus of EBNA3C may be an even more potent mediator of cyclin A-dependent kinase activity than the full-length EBNA3C molecule.
EBNA3C may regulate Rb phosphorylation in cells.
Previously, studies have demonstrated that exogenously expressed cyclin A can overcome an Rb block to cell cycle progression by inducing hyperphosphorylation of the Rb protein (18). Additionally it has been suggested that Rb is a physiologic substrate of cyclin A/Cdk2 complexes and that one role of p27 may be to prevent premature activation of cyclin A-dependent kinase activity (39). To assess whether EBNA3C stimulated the cyclin A-dependent phosphorylation of Rb in vitro, a region of the Rb carboxy terminus with known consensus Cdk phosphorylation sites was fused to GST and purified from bacteria (28). U2OS cells were transfected and cyclin A immunoprecipitation was performed as described for the rescue experiments above; however, this time the Rb carboxy terminus was employed as the kinase substrate (Fig. 7C). As for histone H1, EBNA3C rescued p27 suppression of cyclin A-dependent kinase activity toward the Rb carboxy terminus, suggesting a role for Rb as a relevant substrate phosphorylated by cyclin A/Cdk2 and regulated by EBNA3C (Fig. 7C). When similar amounts of GST protein alone were added as a control, no significant phosphorylation was detected (data not shown). The in vitro phosphorylation of the Rb carboxy terminus is consistent with a possible in vivo role for EBNA3C in driving Rb into its hyperphosphorylated and inactive state.
To determine whether EBNA3C could induce a phosphorylation shift in the Rb protein in cells, U2OS cells were transiently transfected with expression constructs for Myc-tagged Rb and EBNA3C. Cells were harvested at 24 h as for in vitro kinase assay transfections, and Rb phosphorylation status was assessed by Myc-specific Western blot. EBNA3C expression resulted in a virtual elimination of the hypophosphorylated Rb species (Fig. 7D).
EBNA3C expression correlates with accumulation of cells in G2/M.
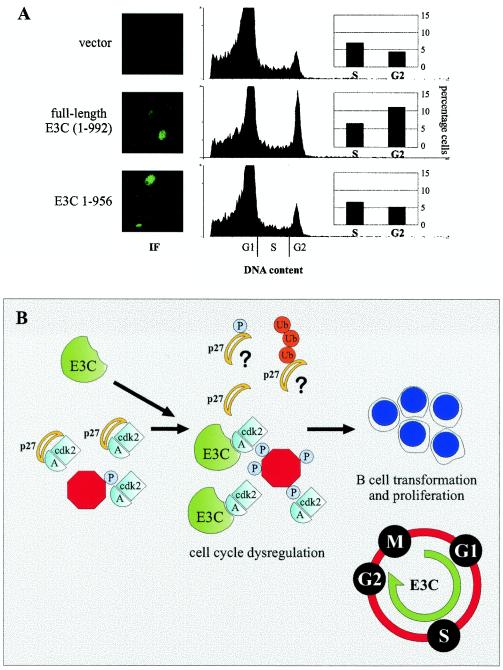
To assess whether the carboxy terminus of EBNA3C contributes to previously described G1 escape and S/G2 accumulation mediated by EBNA3C (34), early-passage Rat-1 fibroblasts were transfected with either cytomegalovirus promoter-driven EBNA3C or EBNA3C 1 to 956 expression constructs. Cells were selected for 2 weeks and macroscopic colonies were pooled. Cells were incubated in 0.1% fetal bovine serum for 72 h to induce G1 arrest (34) and then assayed for DNA content by propidium iodide staining.
Cells transfected with EBNA3C but neither vector nor EBNA3C 1 to 956 showed a reproducible accumulation of cells in G2/M of approximately threefold compared to vector or the carboxy-terminally truncated EBNA3C (Fig. 8A). In contrast, the percentage of cells in S phase was not reproducibly modulated by EBNA3C in this assay in multiple experiments. Immunofluorescence analysis for EBNA3C confirmed the expression of both full-length and carboxy-terminally truncated EBNA3C in the appropriate selected populations. Interestingly, only 5 to 10% of cells expressed levels of EBNA3C that were clearly detectable (by immunofluorescence) above the background in these selected populations, suggesting that the population of cells accumulating in G2/M is likely the same population of cells strongly expressing EBNA3C (Fig. 8A). This cell cycle profile is consistent with constitutive stimulation of cyclin A, as the expression of nondegradable and constitutively active cyclin A has been shown to arrest cells in late G2/early mitosis in numerous cellular contexts (49).
FIG. 8.
EBNA3C expression correlates with accumulation of cells in G2/M. (A) Early-passage Rat-1 fibroblasts were transfected with pA3M (vector), pA3M-E3C, or pA3M-E3C 1 to 956 and selected with G418 for 2 weeks. Pooled colonies were subjected to 0.1% fetal bovine serum for 72 h and then assayed for EBNA3C expression by immunofluorescence and for DNA content by propidium iodide staining. (B) Schematic demonstrating a potential role for EBNA3C in cyclin A complexes, namely, to decrease the association of the cyclin-dependent kinase inhibitor p27 with active kinase complexes. These complexes may phosphorylate cellular substrates with potential relevance for G1/S and S/G2 cell cycle progression.
DISCUSSION
EBV has previously been shown to indirectly affect cell cycle regulatory proteins, with viral immortalization resulting in strong upregulation of cdc2, cyclin E, and cyclin D2 in B cells (19). EBV latent antigens have also been implicated in stimulation of cyclin D2, with independent studies suggesting a role for either LMP1 or EBNA2 and EBNA-LP (19, 44). Interestingly, it has also been suggested, based solely on colocalization assays, that EBNA-LP may complex with the Rb and p53 proteins (48), although subsequent studies questioned the functionality of these strictly in vitro interactions not reproduced in cells (21). More recently, another latent antigen, EBNA3C, has been shown to target the mammalian cell cycle in a manner more reminiscent of the small DNA tumor virus antigens.
EBNA3C has limited binding activity to GST-Rb in vitro in a pocket domain-dependent manner and has some transforming potential in cooperation with the Ras oncoprotein, similar to papillomavirus E7 in colony assays (33). Additionally, expression of EBNA3C increases the DNA content of both NIH 3T3 and U2OS cells under low serum conditions (34). These studies, though clearly suggestive of a role for EBNA3C in regulating the mammalian cell cycle, have not definitively demonstrated an association between EBNA3C and any critical cell cycle regulatory molecule in human cells or EBV-transformed cells. In this report, we show for the first time that complexes form between EBNA3C and cyclin A both in cell lines overexpressing EBNA3C and in EBV-transformed LCLs.
While EBNA3C may yet be shown to directly target Rb in cells, the inability to reproducibly coimmunoprecipitate these molecules from EBNA3C-expressing cells (33) likely suggests that other cell cycle regulatory molecules are involved in EBNA3C-mediated cell cycle deregulation. Indeed, small DNA tumor virus gene products initially shown to target Rb have subsequently been implicated in additional cell cycle regulatory pathways, including the cyclin-dependent kinase inhibitors. Adenovirus E1A interacts with and inactivates p27 in transforming growth factor beta-treated mink lung epithelial cells (26). E1A also blocks induction of p15 and p21 in HaCaT human keratinocyte cells (9). Similarly, papillomavirus E7 has been shown to upregulate Cdk2-associated activity by interacting with and antagonizing the inhibitory function of both p21 and p27 (14, 22, 55).
While our data suggest that EBNA3C may similarly regulate p27 activity, we and others have been unable to demonstrate a significant and direct interaction between EBNA3C and p27 (34). One possibility is that EBNA3C competes with p27 for binding to cyclin A complexes (Fig. 8B). Another possibility is that EBNA3C recruits other factors to cyclin A complexes that may play a role in either p27 phosphorylation or p27 stability (Fig. 8B). Furthermore, as the crystal structure of the cyclin A-Cdk2-p27 complex is available (42), the mapping of cyclin A domains targeted by EBNA3C may provide further insight into how EBNA3C enters and stimulates the activity of these complexes.
While we have not thoroughly ruled out a role for EBNA3C in modulating other cyclin-Cdk complexes, the clear targeting of cyclin A may uniquely distance EBNA3C from small DNA tumor virus antigens. The small DNA tumor viruses drive cells into S phase to promote their own replication; however, EBV is able to efficiently infect and transform nondividing peripheral B lymphocytes, suggesting that EBV may target the cell cycle with a distinctly novel strategy. As was mentioned previously, other EBV latent antigens such as EBNA2, EBNA-LP, and LMP1 may play a role in stimulating cyclin D and E kinase activities by upregulating expression of these proteins (19, 44). EBNA3C may subsequently stimulate cyclin A activity to further promote cell cycle progression (Fig. 8B).
Interestingly, a number of recent studies have suggested that termination of cyclin A-dependent kinase activity is necessary for completion of mitosis and subsequent reentry into G1 (49). To this end, it is possible that EBNA3C overexpression not only results in constitutive cyclin A-dependent kinase activity, but also promotes the accumulation of cells in G2/M. It should, however, be noted that this accumulation of cells would not necessarily be seen in the normal EBV latency growth program, where EBNA3C is expressed at a lower level and other latent antigens also modulate cell cycle progression.
While further experimentation will help to clarify the precise timing of the EBNA3C cell cycle effect, this report for the first time directly links an EBV latent antigen essential for B-cell transformation to cell cycle regulation in human cells and demonstrates a potential role for this interaction in driving EBV-mediated transformation of human primary B lymphocytes.
Acknowledgments
We thank Ed Harlow, Philip Hinds, Elliot Kieff, Maria Mudryj, Michele Pagano, and Martin Rowe for generously providing reagents.
J.S.K. is supported by the Lady Tata Memorial Trust. The project is supported by NIH grants NCI CA72150-07, NCI CA91792-01, and DCR DE14136-01 to E.S.R. E.S.R. is a scholar of the Leukemia and Lymphoma Society of America.
REFERENCES
- 1.Abbot, S. D., M. Rowe, K. Cadwallader, A. Ricksten, J. Gordon, F. Wang, L. Rymo, and A. B. Rickinson. 1990. Epstein-Barr virus nuclear antigen 2 induces expression of the virus-encoded latent membrane protein. J. Virol. 64:2126-2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aiello, L., R. Guilfoyle, K. Huebner, and R. Weinmann. 1979. Adenovirus 5 DNA sequences present and RNA sequences transcribed in transformed human embryo kidney cells (HEK-Ad-5 or 293). Virology 94:460-469. [DOI] [PubMed] [Google Scholar]
- 3.Alfieri, C., M. Birkenbach, and E. Kieff. 1991. Early events in Epstein-Barr virus infection of human B lymphocytes. Virology 181:595-608. (Erratum, 185:946.) [DOI] [PubMed]
- 4.Allday, M. J., and P. J. Farrell. 1994. Epstein-Barr virus nuclear antigen EBNA3C/6 expression maintains the level of latent membrane protein 1 in G1-arrested cells. J. Virol. 68:3491-3498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ambinder, R. F. 1990. Hum. lymphotropic viruses associated with lymphoid malignancy: Epstein-Barr and HTLV-1. Hematol. Oncol. Clin. North Am. 4:821-833. [PubMed] [Google Scholar]
- 6.Aster, J. C., E. S. Robertson, R. P. Hasserjian, J. R. Turner, E. Kieff, and J. Sklar. 1997. Oncogenic forms of NOTCH1 lacking either the primary binding site for RBP-Jkappa or nuclear localization sequences retain the ability to associate with RBP-Jkappa and activate transcription. J. Biol. Chem. 272:11336-11343. [DOI] [PubMed] [Google Scholar]
- 7.Clements, G. B., G. Klein, and S. Povey. 1975. Production by EBV infection of an EBNA-positive subline from an EBNA-negative human lymphoma cell line without detectable EBV DNA. Int. J. Cancer 16:125-133. [DOI] [PubMed] [Google Scholar]
- 8.Cotter, M. A., 2nd, and E. S. Robertson. 2000. Modulation of histone acetyltransferase activity through interaction of Epstein-Barr nuclear antigen 3C with prothymosin alpha. Mol. Cell. Biol. 20:5722-5735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Datto, M. B., P. P. Hu, T. F. Kowalik, J. Yingling, and X. F. Wang. 1997. The viral oncoprotein E1A blocks transforming growth factor beta-mediated induction of p21/WAF1/Cip1 and p15/INK4B. Mol. Cell. Biol. 17:2030-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Debbas, M., and E. White. 1993. Wild-type p53 mediates apoptosis by E1A, which is inhibited by E1B. Genes Dev. 7:546-554. [DOI] [PubMed] [Google Scholar]
- 11.DeCaprio, J. A., J. W. Ludlow, J. Figge, J. Y. Shew, C. M. Huang, W. H. Lee, E. Marsilio, E. Paucha, and D. M. Livingston. 1988. Simian virus 40 large tumor antigen forms a specific complex with the product of the retinoblastoma susceptibility gene. Cell 54:275-283. [DOI] [PubMed] [Google Scholar]
- 12.Devoto, S. H., M. Mudryj, J. Pines, T. Hunter, and J. R. Nevins. 1992. A cyclin A-protein kinase complex possesses sequence-specific DNA binding activity: p33Cdk2 is a component of the E2F-cyclin A complex. Cell 68:167-176. [DOI] [PubMed] [Google Scholar]
- 13.Dyson, N., P. M. Howley, K. Munger, and E. Harlow. 1989. The human papilloma virus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science 243:934-937. [DOI] [PubMed] [Google Scholar]
- 14.Funk, J. O., S. Waga, J. B. Harry, E. Espling, B. Stillman, and D. A. Galloway. 1997. Inhibition of CDK activity and PCNA-dependent DNA replication by p21 is blocked by interaction with the HPV-16 E7 oncoprotein. Genes Dev. 11:2090-2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hammerschmidt, W., and B. Sugden. 1989. Genetic analysis of immortalizing functions of Epstein-Barr virus in human B lymphocytes. Nature 340:393-397. [DOI] [PubMed] [Google Scholar]
- 16.Harper, J. W., G. R. Adami, N. Wei, K. Keyomarsi, and S. J. Elledge. 1993. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell 75:805-816. [DOI] [PubMed] [Google Scholar]
- 17.Hickman, E. S., S. M. Picksley, and K. H. Vousden. 1994. Cells expressing HPV16 E7 continue cell cycle progression following DNA damage induced p53 activation. Oncogene 9:2177-2181. [PubMed] [Google Scholar]
- 18.Hinds, P. W., S. Mittnacht, V. Dulic, A. Arnold, S. I. Reed, and R. A. Weinberg. 1992. Regulation of retinoblastoma protein functions by ectopic expression of human cyclins. Cell 70:993-1006. [DOI] [PubMed] [Google Scholar]
- 19.Hollyoake, M., A. Stuhler, P. Farrell, J. Gordon, and A. Sinclair. 1995. The normal cell cycle activation program is exploited during the infection of quiescent B lymphocytes by Epstein-Barr virus. Cancer Res. 55:4784-4787. [PubMed] [Google Scholar]
- 20.Howes, K. A., N. Ransom, D. S. Papermaster, J. G. Lasudry, D. M. Albert, and J. J. Windle. 1994. Apoptosis or retinoblastoma: alternative fates of photoreceptors expressing the HPV-16 E7 gene in the presence or absence of p53. Genes Dev. 8:1300-1310. [DOI] [PubMed] [Google Scholar]
- 21.Inman, G. J., and P. J. Farrell. 1995. Epstein-Barr virus EBNA-LP and transcription regulation properties of pRB, p107 and p53 in transfection assays. J. Gen. Virol. 76:2141-2149. [DOI] [PubMed] [Google Scholar]
- 22.Jones, D. L., R. M. Alani, and K. Munger. 1997. The human papillomavirus E7 oncoprotein can uncouple cellular differentiation and proliferation in human keratinocytes by abrogating p21Cip1-mediated inhibition of Cdk2. Genes Dev. 11:2101-2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kieff, E., and A. B. Rickinson. 2002. Epstein-Barr virus and its replication, p. 2511-2573. In D. Knipe and P. Howley (ed.), Fields virology, 4th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 24.Longnecker, R., C. L. Miller, B. Tomkinson, X. Q. Miao, and E. Kieff. 1993. Deletion of DNA encoding the first five transmembrane domains of Epstein-Barr virus latent membrane proteins 2A and 2B. J. Virol. 67:5068-5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lowe, S. W., and H. E. Ruley. 1993. Stabilization of the p53 tumor suppressor is induced by adenovirus 5 E1A and accompanies apoptosis. Genes Dev. 7:535-545. [DOI] [PubMed] [Google Scholar]
- 26.Mal, A., R. Y. Poon, P. H. Howe, H. Toyoshima, T. Hunter, and M. L. Harter. 1996. Inactivation of p27Kip1 by the viral E1A oncoprotein in TGFbeta-treated cells. Nature 380:262-265. [DOI] [PubMed] [Google Scholar]
- 27.Mauser, A., S. Saito, E. Appella, C. W. Anderson, W. T. Seaman, and S. Kenney. 2002. The Epstein-Barr virus immediate-early protein BZLF1 regulates p53 function through multiple mechanisms. J. Virol. 76:12503-12512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meyerson, M., and E. Harlow. 1994. Identification of G1 kinase activity for Cdk6, a novel cyclin D partner. Mol. Cell. Biol. 14:2077-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morgan, D. O. 1997. Cyclin-dependent kinases: engines, clocks, and microprocessors. Annu. Rev. Cell Dev. Biol. 13:261-291. [DOI] [PubMed] [Google Scholar]
- 30.Pagano, M., R. Pepperkok, F. Verde, W. Ansorge, and G. Draetta. 1992. Cyclin A is required at two points in the human cell cycle. EMBO J. 11:961-971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pagano, M., S. W. Tam, A. M. Theodoras, P. Beer-Romero, G. Del Sal, V. Chau, P. R. Yew, G. F. Draetta, and M. Rolfe. 1995. Role of the ubiquitin-proteasome pathway in regulating abundance of the cyclin-dependent kinase inhibitor p27. Science 269:682-685. [DOI] [PubMed] [Google Scholar]
- 32.Pan, H., and A. E. Griep. 1994. Altered cell cycle regulation in the lens of HPV-16 E6 or E7 transgenic mice: implications for tumor suppressor gene function in development. Genes Dev. 8:1285-1299. [DOI] [PubMed] [Google Scholar]
- 33.Parker, G. A., T. Crook, M. Bain, E. A. Sara, P. J. Farrell, and M. J. Allday. 1996. Epstein-Barr virus nuclear antigen (EBNA)3C is an immortalizing oncoprotein with similar properties to adenovirus E1A and papillomavirus E7. Oncogene 13:2541-2549. [PubMed] [Google Scholar]
- 34.Parker, G. A., R. Touitou, and M. J. Allday. 2000. Epstein-Barr virus EBNA3C can disrupt multiple cell cycle checkpoints and induce nuclear division divorced from cytokinesis. Oncogene 19:700-709. [DOI] [PubMed] [Google Scholar]
- 35.Polyak, K., M. H. Lee, H. Erdjument-Bromage, A. Koff, J. M. Roberts, P. Tempst, and J. Massague. 1994. Cloning of p27Kip1, a cyclin-dependent kinase inhibitor and a potential mediator of extracellular antimitogenic signals. Cell 78:59-66. [DOI] [PubMed] [Google Scholar]
- 36.Ponten, J., and E. Saksela. 1967. Two established in vitro cell lines from human mesenchymal tumours. Int J. Cancer 2:434-447. [DOI] [PubMed] [Google Scholar]
- 37.Radkov, S. A., R. Touitou, A. Brehm, M. Rowe, M. West, T. Kouzarides, and M. J. Allday. 1999. Epstein-Barr virus nuclear antigen 3C interacts with histone deacetylase to repress transcription. J. Virol. 73:5688-5697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Resnitzky, D., L. Hengst, and S. I. Reed. 1995. Cyclin A-associated kinase activity is rate limiting for entrance into S phase and is negatively regulated in G1 by p27Kip1. Mol. Cell. Biol. 15:4347-4352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Resnitzky, D., L. Hengst, and S. I. Reed. 1995. Cyclin A-associated kinase activity is rate limiting for entrance into S phase and is negatively regulated in G1 by p27Kip1. Mol. Cell. Biol. 15:4347-4352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rickinson, A. B., and E. Kieff. 2002. Epstein-Barr virus, p. 2575-2627. In D. Knipe and P. Howley (ed.), Fields virology, vol. 2. Lippincott Williams & Wilkins, Philadelphia, Pa..
- 41.Robertson, E. S., S. Grossman, E. Johannsen, C. Miller, J. Lin, B. Tomkinson, and E. Kieff. 1995. Epstein-Barr virus nuclear protein 3C modulates transcription through interaction with the sequence-specific DNA-binding protein Jκ. J. Virol. 69:3108-3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Russo, A. A., P. D. Jeffrey, A. K. Patten, J. Massague, and N. P. Pavletich. 1996. Crystal structure of the p27Kip1 cyclin-dependent-kinase inhibitor bound to the cyclin A-Cdk2 complex. Nature 382:325-331. [DOI] [PubMed] [Google Scholar]
- 43.Schulman, B. A., D. L. Lindstrom, and E. Harlow. 1998. Substrate recruitment to cyclin-dependent kinase 2 by a multipurpose docking site on cyclin A. Proc. Natl. Acad. Sci. USA 95:10453-10458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sinclair, A. J., I. Palmero, G. Peters, and P. J. Farrell. 1994. EBNA-2 and EBNA-LP cooperate to cause G0 to G1 transition during immortalization of resting human B lymphocytes by Epstein-Barr virus. EMBO J. 13:3321-3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Subramanian, C., S. Hasan, M. Rowe, M. Hottiger, R. Orre, and E. S. Robertson. 2002. Epstein-Barr virus nuclear antigen 3C and prothymosin alpha interact with the p300 transcriptional coactivator at the CH1 and CH3/HAT domains and cooperate in regulation of transcription and histone acetylation. J. Virol. 76:4699-4708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Subramanian, C., and E. S. Robertson. 2002. The metastatic suppressor Nm23-H1 interacts with EBNA3C at sequences located between the glutamine- and proline-rich domains and can cooperate in activation of transcription. J. Virol. 76:8702-8709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Swenson, J. J., A. E. Mauser, W. K. Kaufmann, and S. C. Kenney. 1999. The Epstein-Barr virus protein BRLF1 activates S phase entry through E2F1 induction. J. Virol. 73:6540-6550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Szekely, L., G. Selivanova, K. P. Magnusson, G. Klein, and K. G. Wiman. 1993. EBNA-5, an Epstein-Barr virus-encoded nuclear antigen, binds to the retinoblastoma and p53 proteins. Proc. Natl. Acad. Sci. USA 90:5455-5459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tin Su, T. 2001. Cell cycle: how, when and why cells get rid of cyclin A. Curr. Biol. 11:R467-R469. [DOI] [PubMed] [Google Scholar]
- 50.Tomkinson, B., E. Robertson, and E. Kieff. 1993. Epstein-Barr virus nuclear proteins EBNA-3A and EBNA-3C are essential for B-lymphocyte growth transformation. J. Virol. 67:2014-2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.White, A. E., E. M. Livanos, and T. D. Tlsty. 1994. Differential disruption of genomic integrity and cell cycle regulation in normal human fibroblasts by the HPV oncoproteins. Genes Dev. 8:666-677. [DOI] [PubMed] [Google Scholar]
- 52.Whyte, P., N. M. Williamson, and E. Harlow. 1989. Cellular targets for transformation by the adenovirus E1A proteins. Cell 56:67-75. [DOI] [PubMed] [Google Scholar]
- 53.Young, L., C. Alfieri, K. Hennessy, H. Evans, C. O'Hara, K. C. Anderson, J. Ritz, R. S. Shapiro, A. Rickinson, E. Kieff, and et al. 1989. Expression of Epstein-Barr virus transformation-associated genes in tissues of patients with EBV lymphoproliferative disease. N. Engl. J. Med. 321:1080-1085. [DOI] [PubMed] [Google Scholar]
- 54.Zacny, V. L., J. Wilson, and J. S. Pagano. 1998. The Epstein-Barr virus immediate-early gene product, BRLF1, interacts with the retinoblastoma protein during the viral lytic cycle. J. Virol. 72:8043-8051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zerfass-Thome, K., W. Zwerschke, B. Mannhardt, R. Tindle, J. W. Botz, and P. Jansen-Durr. 1996. Inactivation of the Cdk inhibitor p27KIP1 by the human papillomavirus type 16 E7 oncoprotein. Oncogene 13:2323-2330. [PubMed] [Google Scholar]