Abstract
Coral reefs provide habitat for an array of marine invertebrates that host symbiotic microbiomes. Photosynthetic symbionts including Symbiodinium dinoflagellates and diatoms potentially influence the diversity of their host-associated microbiomes by releasing carbon-containing photosynthates and other organic compounds that fuel microbial metabolism. Here we used 16S ribosomal RNA (rRNA) gene amplicon pyrosequencing to characterise the microbiomes of 11 common Great Barrier Reef marine invertebrate species that host photosynthetic symbionts and five taxa in which they are absent. The presence of photosynthetic symbionts influenced the composition but not the species richness, evenness and phylogenetic diversity of invertebrate-associated microbiomes. Invertebrates without photosynthetic symbionts were dominated by Alphaproteobacteria, whereas those hosting photosynthetic symbionts were dominated by Gammaproteobacteria. Interestingly, many microbial species from photosymbiont-bearing invertebrates, including Oceanospirillales spp., Alteromonas spp., Pseudomonas spp., Halomonas spp., are implicated in the metabolism of dimethylsulfoniopropionate (DMSP). DMSP is produced in high concentrations by photosynthetic dinoflagellates and is involved in climate regulation by facilitating cloud formation. Microbiomes correlated with host taxa and replicate individuals from most sampled species grouped in distance-based redundancy analysis of retrieved 16S rRNA gene sequences. This study highlights the complex nature of invertebrate holobionts and confirms the importance of photosynthetic symbionts in structuring marine invertebrate bacterial communities.
Keywords: marine invertebrate, microbial diversity, 16S rRNA gene, pyrosequencing, coral reefs
Coral reefs harbour abundant and diverse marine invertebrates that perform important ecosystem functions such as: calcification, bioerosion, consolidation and benthic-pelagic coupling (Glynn and Enochs, 2011). Animal–plant/microbe symbioses are vital to these ecosystems as they facilitate photosynthetic productivity, mineral recycling, nutrient provision to the host and secondary metabolite production (Smith and Douglas, 1987). Although patterns of microbial diversity and putative symbiotic functions have been well explored in corals and sponges (Sunagawa et al., 2009; Mouchka et al., 2010; Webster and Taylor, 2012; Bourne and Webster, 2013), there is a lack of data on microbial associations in other reef taxa including Bivalves, Foraminifera and Ascidians.
The diversity of microbial communities associated with corals and sponges is known to be influenced by host interactions (Wegley et al., 2007; Kimes et al., 2010; Raina et al., 2010; Fan et al., 2012), the production of antimicrobial compounds (Ritchie, 2006; Shnit-Orland and Kushmaro, 2009) and environmental conditions (Hong et al., 2009; Ceh et al., 2011). Recent studies, however, indicate that other members of the coral holobiont (in particular Symbiodinium dinoflagellates) also influence microbial community structure through release of complex carbon-containing exudates including dimethylsulfoniopropionate (DMSP; Ikeda and Miyachi, 1995; Raina et al., 2009, 2010). DMSP can be degraded to dimethylysulphide, a central molecule in the global sulphur cycle, which diffuses from the ocean into the atmosphere where it influences cloud formation, with consequences for atmospheric chemistry, local climate and water temperature (Ayers and Gras, 1991; Andreae and Crutzen, 1997). A complex array of other organic exudates including amino acids and polysaccharides can also influence invertebrate-associated microbiomes, which may affect holobiont fitness. For example, Symbiodinium spp. have been shown to influence the response of bacterial communities to thermal stress, which affects the susceptibility of the holobiont to bleaching (van Oppen et al., 2009; Stat et al., 2012), disease (Stat et al., 2008) and colonisation by opportunistic potential pathogens (Littman et al., 2010).
In this study, we used 16S ribosomal RNA (rRNA) gene amplicon pyrosequencing (Supplementary Methods) to characterise the microbiomes of 16 common Great Barrier Reef marine invertebrate species representing five invertebrate families (Table 1). These families included 11 species that host photosynthetic symbionts (Symbiodinium and diatoms) and five species that do not host these symbionts. The microbiomes for three replicate samples from each invertebrate species and seawater controls were characterised. Briefly, 16S rRNA gene amplicons generated using primers 63F and 533R (Engelbrektson et al., 2010) were subjected to 454 pyrosequencing. Sequences were checked for chimeras using UCHIME ver. 3.0.617 (Edgar et al., 2011), denoised using Acacia (Bragg et al., 2012) and then parsed using the QIIME pipeline with default settings (Caporaso et al., 2010). We tested the hypotheses that: (1) the presence of photosynthetic symbionts influences the diversity of marine invertebrate-associated microbiomes, and (2) that the diversity of marine invertebrate-associated microbiomes differs between host species.
Table 1. List of samples, phylogentic classification, associated pyrosequence reads and symbiont type.
| Sample/species name | Taxa/group | No. of raw reads | No. of cleaned reads | % Removed | Symbiont | Symbiont type |
|---|---|---|---|---|---|---|
| Acropora millipora #1 | Scleractinea | 9016 | 8043 | 10.8 | Yes | Symbiodinium |
| Acropora millipora #2 | Scleractinea | 6618 | 6221 | 6.0 | Yes | Symbiodinium |
| Acropora millipora #3 | Scleractinea | 15 986 | 13 909 | 13.0 | Yes | Symbiodinium |
| Pocillopora damicornis #1 | Scleractinea | 10 168 | 9254 | 9.0 | Yes | Symbiodinium |
| Pocillopora damicornis #2 | Scleractinea | 14 733 | 13 636 | 7.4 | Yes | Symbiodinium |
| Pocillopora damicornis #3 | Scleractinea | 12 701 | 11 380 | 10.4 | Yes | Symbiodinium |
| Seriatopora hystrix #1 | Scleractinea | 12 302 | 11 758 | 4.4 | Yes | Symbiodinium |
| Seriatopora hystrix #2 | Scleractinea | 12 839 | 11 857 | 7.6 | Yes | Symbiodinium |
| Seriatopora hystrix #3 | Scleractinea | 11 151 | 10 500 | 5.8 | Yes | Symbiodinium |
| Nephtea sp. #3 | Octocorallia | 10 596 | 9505 | 10.3 | Yes | Symbiodinium |
| Sarcophyton sp. #1 | Octocorallia | 18 070 | 16 505 | 8.7 | Yes | Symbiodinium |
| Sarcophyton sp. #2 | Octocorallia | 10 750 | 9680 | 10.0 | Yes | Symbiodinium |
| Sarcophyton sp. #3 | Octocorallia | 14 179 | 11 761 | 17.1 | Yes | Symbiodinium |
| Sinularia flexibilis #1 | Octocorallia | 12 068 | 11 595 | 3.9 | Yes | Symbiodinium |
| Sinularia flexibilis #2 | Octocorallia | 12 231 | 10 947 | 10.5 | Yes | Symbiodinium |
| Sinularia flexibilis #3 | Octocorallia | 11 358 | 10 522 | 7.4 | Yes | Symbiodinium |
| Tridacna cf. crocea #1 | Bivalvia | 12 910 | 11 523 | 10.7 | Yes | Symbiodinium |
| Tridacna cf. crocea #2 | Bivalvia | 4516 | 4015 | 11.1 | Yes | Symbiodinium |
| Tridacna cf. crocea #3 | Bivalvia | 3931 | 3495 | 11.1 | Yes | Symbiodinium |
| Tridacna cf. maxima #1 | Bivalvia | 13 246 | 10 453 | 21.1 | Yes | Symbiodinium |
| Tridacna cf. maxima #2 | Bivalvia | 12 597 | 10 579 | 16.0 | Yes | Symbiodinium |
| Tridacna cf. maxima #3 | Bivalvia | 8403 | 6945 | 17.4 | Yes | Symbiodinium |
| Heterostegina depressa #1 | Foraminifera | 13 868 | 12 056 | 13.1 | Yes | Diatom |
| Heterostegina depressa #2 | Foraminifera | 14 328 | 12 860 | 10.2 | Yes | Diatom |
| Heterostegina depressa #3 | Foraminifera | 15 993 | 14 126 | 11.7 | Yes | Diatom |
| Marginopora vertebralis #1 | Foraminifera | 10 917 | 9626 | 11.8 | Yes | Symbiodinium |
| Marginopora vertebralis #2 | Foraminifera | 16 713 | 15 239 | 8.8 | Yes | Symbiodinium |
| Marginopora vertebralis #3 | Foraminifera | 28 568 | 25 942 | 9.2 | Yes | Symbiodinium |
| Sorites sp. #1 | Foraminifera | 5467 | 4855 | 11.2 | Yes | Symbiodinium |
| Sorites sp. #2 | Foraminifera | 7361 | 6439 | 12.5 | Yes | Symbiodinium |
| Sorites sp. #3 | Foraminifera | 15 979 | 14 292 | 10.6 | Yes | Symbiodinium |
| Bryozoan sp. #1 | Bryozoa | 14 520 | 12 493 | 14.0 | No | NA |
| Bryozoan sp. #2 | Bryozoa | 11 004 | 9488 | 13.8 | No | NA |
| Bryozoan sp. #3 | Bryozoa | 16 807 | 13 562 | 19.3 | No | NA |
| Diademnum molle #1 | Ascidiacaea | 24 826 | 23 150 | 6.8 | No | NA |
| Diademnum molle #2 | Ascidiacaea | 12 348 | 8590 | 30.4 | No | NA |
| Diademnum molle #3 | Ascidiacaea | 14 772 | 11 266 | 23.7 | No | NA |
| Lissoclinum patella #1 | Ascidiacaea | 13 130 | 11 493 | 12.5 | No | NA |
| Lissoclinum patella #2 | Ascidiacaea | 15 511 | 13 994 | 9.8 | No | NA |
| Lissoclinum patella #3 | Ascidiacaea | 13 700 | 12 621 | 7.9 | No | NA |
| Polycarpa aurata #1 | Ascidiacaea | 15 176 | 14 203 | 6.4 | No | NA |
| Polycarpa aurata #2 | Ascidiacaea | 13 526 | 12 576 | 7.0 | No | NA |
| Polycarpa aurata #3 | Ascidiacaea | 10 968 | 9925 | 9.5 | No | NA |
| Rhopaloiedes odorabile | Porifera | 16 382 | 11 999 | 26.8 | No | NA |
| Seawater #1 | Seawater | 56 633 | 45 068 | 20.4 | No | NA |
| Seawater #2 | Seawater | 19 560 | 15 614 | 20.2 | No | NA |
Abbreviation: NA, not applicable.
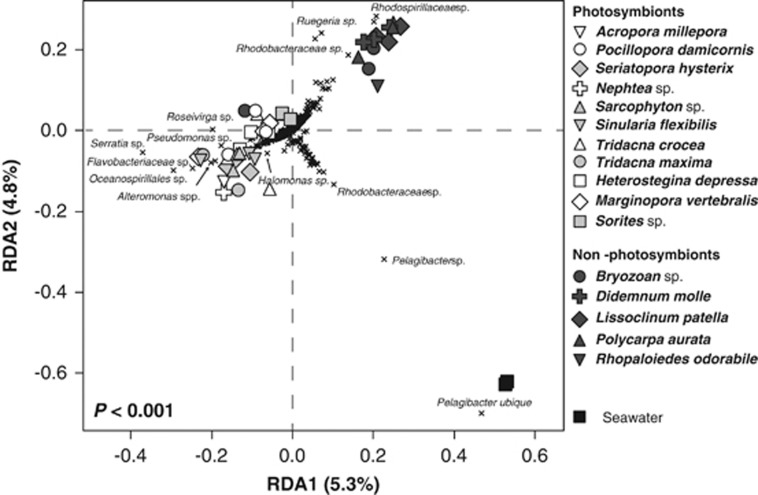
The presence of photosynthetic symbionts influenced the composition (Figure 1), but not the species richness, evenness and phylogenetic diversity (P>0.05, linear regression; Supplementary Table S1) of invertebrate-associated microbiomes. At the class level, the presence of photosynthetic symbionts explained 21% of variation in the composition of microbial communities between samples (PERMANOVA, F1,44=11.37, P<0.001). At the level of operational taxonomic units (OTUs), defined as groups of sequences that shared 97% nucleotide sequence similarity (‘species' level), the presence of photosynthetic symbionts explained a significant, albeit smaller (6%) proportion of variation in the composition of microbial communities between samples (PERMANOVA, F1,44=2.66, P<0.001). Unifrac analysis based on OTUs also confirmed that the presence of photosynthetic symbionts influenced microbial community composition (unweighted P=0.002, weighted P<0.001).
Figure 1.
Redundancy analysis (RDA) summarising variation in the composition of marine invertebrate-associated microbial communities that was attributable to the presence–absence of photosymbionts. The filled shapes represent individual samples collected from each invertebrate species. The black crosses represent bacterial OTUs. For clarity, taxonomic affiliations are shown for the most discriminating OTUs only. The distance of an object (sample or OTU) from the origin is proportional to its variance along an axis and its angle relative to the axes reflects its correlation with those axes. Full sample collection and processing details can be found in the Supplementary Methods and the sequence data set deposited in the NCBI Sequence Read Archive (SRA) database with the accession number SRA4494953.
Alphaproteobacteria and Gammaproteobacteria were the dominant classes of bacteria associated with reef invertebrates. Invertebrates without photosynthetic symbionts (with the exception of one replicate Bryozoan sp.) were associated with a larger abundance of Alphaproteobacteria, whereas those with photosynthetic symbionts generally hosted a higher relative abundance of Gammaproteobacteria (Supplementary Figure S1). The only exceptions were Seriatopora hysterix and two of the Sinularia sp. samples in which Flavobacteria were particularly abundant. Other bacterial classes including the Deltaproteobacteria, Sphingobacteria and Cyanobacteria differed between invertebrate species but were not influenced by the presence of photosymbionts (Supplementary Figure S1). Although present in all samples, the Cyanobacteria were particularly abundant (8–24% relative abundance) in Heterostegina depressa and in one Marginopora vertebralis sample (17% relative abundance). The composition of microbial communities associated with the sponge Rhopaloeides odorabile was different to those associated with other invertebrates, although this community pattern is consistent with a previous investigation (Webster et al., 2010).
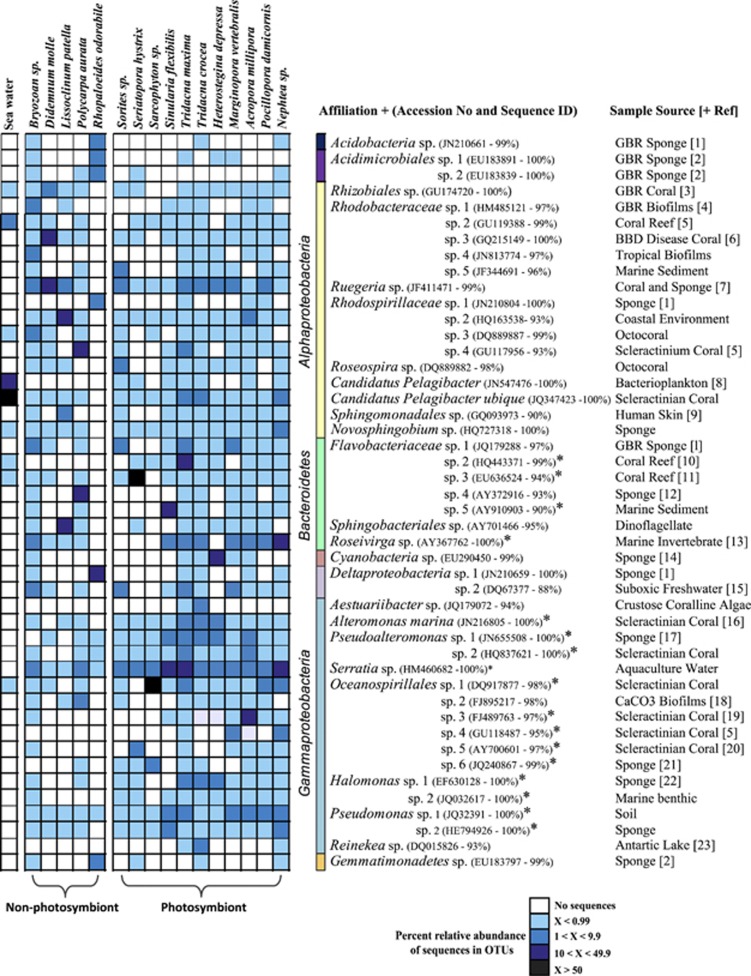
Most dominant OTUs (that is, >5% relative abundance) were affiliated with bacterial populations previously retrieved from marine environments including corals and sponges (Figure 2). Invertebrates that host photosynthetic symbionts were positively correlated with OTUs related to Oceanospirillales spp., a Roseivirga sp., an Alteromonas sp., Pseudoalteromonas spp., Halomonas spp., Pseudomonas spp. and Flavobacteriacae spp. (Figure 1). Indicator species analysis (Dufrene and Legendre, 1997) confirmed these OTUs were significantly correlated with the presence of photosynthetic symbionts by having high relative abundance and frequency of occurrence (Figure 2). These OTUs are all affiliated with species implicated in the metabolism of complex organic molecules such as DMSP and dimethylysulphide. For example, previous studies have identified abundant bacteria within the Oceanospirillales that are able to metabolise DMSP in the coral Acropora millepora (Raina et al., 2009). Halomonas spp. have been shown to be capable of metabolism of DMSP and its breakdown product acrylic acid (Todd et al., 2010), while members of the Flavobacteriacae respond rapidly to high DMSP concentrations in phytoplankton blooms, although the genetic pathways for metabolism of this compound in this group of bacteria is unknown (Howard et al., 2011). In the marine environment, DMSP has been the focus of considerable attention because of its fundamental role as carbon and sulphur sources for bacteria (Sievert et al., 2007). Coral reefs are one of the largest producers of DMSP with the source thought to be derived from marine invertebrates harbouring symbiotic dinoflagellates (Broadbent et al., 2002; Van Alstyne et al., 2006). In fact, the concentrations of DMSP and its breakdown products dimethylysulphide and acrylate in reef-building corals are the highest recorded in the marine environment (Broadbent and Jones, 2004). These results further support the concept that sulphur-based organic compounds derived from photosymbionts influence the microbial communities of marine invertebrates by providing nutrient sources readily available for metabolism by associated microbiomes. Although compounds such as DMSP appear to have a role in structuring microbial communities associated with the host organism, there are likely to be many other organic exudates derived from photosymbionts that also influence microbial associations. In addition, host animal factors can have an important role in structuring microbial communities. Results from this study and other recent reports highlight that members of the Oceanospirillales, specifically, Endozoicomonas spp. are commonly found in marine invertebrates with and without photosymbionts and potentially have important functional roles within their host species (Yang et al., 2010; Nishijima et al., 2012; Speck and Donachie, 2012).
Figure 2.
Heatmap of OTUs (97% sequence identity, averaged within each invertebrate species) that represent >5% of sequence tags within a particular sample type or represented an OTU with a significant indicator value (representative of high relative abundance and high relative frequency of occurrence in photosymbiont-bearing invertebrates). OTUs with highest indicator values are represented by an asterisk (*). The closest sequence match determined in a BLAST database query (Altschul et al., 1997) and its corresponding accession number and derived sample source are also represented. If these affiliated sequences are represented in a published study the associated reference can be found in Supplementary References.
Indicator species analysis demonstrated that no OTUs were significantly correlated with invertebrates that do not host photosymbionts, although Rugeria-, Rhodobacteraceae- and Rhodospirillaceae-related sequences were more commonly retrieved in these samples as observed in the redundancy analysis (Figure 1). Microbial communities associated with the seawater samples were distinct from those associated with both photosymbiont and non-photosymbiont-bearing invertebrates (P=0.002, PERMANOVA). This difference was related to a larger abundance of the ubiquitous bacterioplankton Candidatus pelagibacter (SAR11) comprising ∼66% of sequence reads from this control group (Figure 1).
The composition of microbial communities also differed between invertebrate groups (Foramaninfera, Scleractinia, Octocorallia, Bivalvia, Bryozoa and Ascidiacaea; P=0.001, see Supplementary Figure S2) and this was further supported by both unweighted and weighted unifrac distances (P<0.001, redundancy analysis). Many replicate microbiomes from the same invertebrate species grouped well at both class and OTU taxonomic assignment levels and was reflected by the composition of microbial communities being different between invertebrate species (P<0.001, PERMANOVA). Most within-species variability existed for the samples derived from A. millepora, Sinularia flexibilis, Tridacna spp. and Bryozoan sp. (Supplementary Figure S2). Although the redundancy analysis and Heatmap/Cluster analysis generally group individual specimens from one species closely together and showed significant relationships at the taxa level, there is no apparent higher phylogenetic grouping. However, both analyses clearly separate samples by presence or absence of photosymbionts. Further studies comparing microbiomes among taxa with and without photosymbionts will be useful in further clarifying the strength of these relationships and the role photosymbionts have in driving microbial associations.
Rarefaction analysis demonstrated that all three Foraminifera species hosted the largest bacterial diversity among the invertebrate samples (Supplementary Figure S3), which may reflect their lifestyle closely associated with reef rubble, filamentous algae and reef sediment. Heterostegina depressa was the only diatom-bearing invertebrate species and richness of these samples (1123 OTUs) exceeded that of all other invertebrate taxa. The two Octocoral species, Sarcophyton sp. and Sinularia flexibilis hosted the lowest microbial richness (80–180 OTUs, respectively), which may relate to the high antimicrobial activity previously identified in these species (Kim, 1994; Jensen et al., 1996; Harder et al., 2003). Many invertebrate species including all Foraminifera, Bivalvia, Bryozoa and the Scleractinian corals A. millepora and Pocillopora damicornis hosted higher bacterial richness than the surrounding seawater (Supplementary Figure S3). Rarefaction analysis of the Scleractinian corals in this study is consistent with earlier estimates of coral microbial diversity (Sunagawa et al., 2010).
The documented decline in coral reef ecosystems worldwide (Wilkinson, 2008) has prompted research into understanding how changing environmental conditions affect the close symbiotic associations of marine invertebrates (Webster et al., 2001; Bourne et al., 2008; Vega Thurber et al., 2008; Webster et al., 2008; Littman et al., 2010, 2011; Webster et al., 2011). Only by studying marine invertebrates as holobionts (the host and all associated microbial communities) and better characterising the forces that structure their microbial associations will we be able to fully assess their capacity to adapt or acclimatise to environmental stress. From this study, 16S rRNA gene amplicon pyrosequencing revealed high diversity of bacterial symbionts within 16 common Great Barrier Reef species. Importantly, although microbial composition was related to host species, a significant amount of the variation in community composition was attributed to the presence or absence of photosymbionts. These results highlight the importance of photosymbionts in structuring reef bacterial symbioses.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreae MO, Crutzen PJ. Atmospheric aerosols: biogeochemical sources and role in atmospheric chemistry. Science. 1997;276:1052–1058. [Google Scholar]
- Ayers GP, Gras JL. Seasonal relationship between cloud condensation nuclei and aerosol methanesulphonate in marine air. Nature. 1991;353:834–835. [Google Scholar]
- Bourne D, Lida Y, Uthicke S, Smith-Keune C. Changes in coral-associated microbial communities during a bleaching event. ISME J. 2008;2:350–363. doi: 10.1038/ismej.2007.112. [DOI] [PubMed] [Google Scholar]
- Bourne DG, Webster NS.2013Coral reef microbial communitiesIn: Rosenberg E, Stackebrandt E, DeLong EF, Thompson FL, Lory S (eds)The Prokaryotes Springer: NewYork, USA [Google Scholar]
- Bragg L, Stone G, Imelfort M, Hugenholtz P, Tyson GW. Fast, accurate error-correction of amplicon pyrosequences using Acacia. Nat Methods. 2012;9:425–426. doi: 10.1038/nmeth.1990. [DOI] [PubMed] [Google Scholar]
- Broadbent AD, Jones GB. DMS and DMSP in mucus ropes, coral mucus, surface films and sediment pore waters from coral reefs in the Great Barrier Reef. Mar Freshwater Res. 2004;55:849–855. [Google Scholar]
- Broadbent AD, Jones GB, Jones RJ. DMSP in corals and benthic algae from the Great Barrier Reef. Estuar Coast Shelf Sci. 2002;55:547–555. [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Costello EK, Fierer N, Peña AG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceh J, van Keulen M, Bourne DG. Coral-associated bacterial communities on Ningaloo Reef, Western Australia. FEMS Microbiol Ecol. 2011;75:134–144. doi: 10.1111/j.1574-6941.2010.00986.x. [DOI] [PubMed] [Google Scholar]
- Dufrene M, Legendre P. Species assemblages and indicator species: the need for a flexible asymmetrical approach. Ecol Monogr. 1997;67:345–366. [Google Scholar]
- Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelbrektson A, Kunin V, Wrighton KC, Zvenigorodsky N, Chen F, Ochman H, et al. Experimental factors affecting PCR-based estimates of microbial species richness and evenness. ISME J. 2010;4:642–647. doi: 10.1038/ismej.2009.153. [DOI] [PubMed] [Google Scholar]
- Fan L, Reynolds D, Liu M, Stark M, Kjelleberg S, Webster NS, et al. Functional equivalence and evolutionary convergence in complex communities of sponge symbionts. Proc Natll Acad Sci. 2012;109:E1878–E1887. doi: 10.1073/pnas.1203287109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glynn PW, Enochs IC.2011Invertebrates and their roles in coral reef ecosystemsIn: Dubinsky Z, Stambler N (eds)Coral Reefs: An Ecosystem in Transition Springer: London; 273–326. [Google Scholar]
- Harder T, Lau SCK, Dobretsov S, Fang TK, Qian P-Y. A distinctive epibiotic bacterial community on the soft coral Dendronephthya sp. and antibacterial activity of coral tissue extracts suggest a chemical mechanism against bacterial epibiosis. FEMS Microbiol Ecol. 2003;43:337–347. doi: 10.1111/j.1574-6941.2003.tb01074.x. [DOI] [PubMed] [Google Scholar]
- Hong M-J, Yu Y-T, Chen CA, Chiang P-W, Tang S-L. Influence of species specificity and other factors on bacteria associated with the coral Stylophora pistillata in Taiwan. Appl Environ Microbiol. 2009;75:7797–7806. doi: 10.1128/AEM.01418-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard EC, Sun S, Reisch CR, del Valle DA, Bürgmann H, Kiene RP, et al. Changes in dimethylsulfoniopropionate demethylase gene assemblages in response to an induced phytoplankton bloom. Appl Environ Microbiol. 2011;77:534–531. doi: 10.1128/AEM.01457-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda Y, Miyachi S. Carbon dioxide fixation by photosynthesis and calcification for a solitary coral, Fungia sp. Bulletin de l'Institut Oceanographique. 1995;14:61–67. [Google Scholar]
- Jensen PR, Harvell CD, Wirtz K, Fenical W. Antimicrobial activity of extracts of Caribbean gorgonian corals. Marine Biology. 1996;125:411–419. [Google Scholar]
- Kim K. Antimicrobial activity in Gorgonian corals (Coelenterata, Octocorallia) Coral Reefs. 1994;13:75–80. [Google Scholar]
- Kimes NE, Van Nostrand JD, Weil E, Zhou J, Morris PJ. Microbial functional structure of Montastraea faveolata, an important Caribbean reef-building coral, differs between healthy and yellow-band diseased colonies. Environ Microbiol. 2010;12:541–556. doi: 10.1111/j.1462-2920.2009.02113.x. [DOI] [PubMed] [Google Scholar]
- Littman R, Willis BL, Bourne DG. Metagenomic analysis of the coral holobiont during a natural bleaching event on the Great Barrier Reef. Environ Microbiol Rep. 2011. [DOI] [PubMed]
- Littman RA, Bourne DG, Willis BL. Responses of coral-associated bacterial communities to heat stress differ with Symbiodinium type on the same coral host. Mol Ecol. 2010;19:1978–1990. doi: 10.1111/j.1365-294X.2010.04620.x. [DOI] [PubMed] [Google Scholar]
- Mouchka ME, Hewson I, Harvell DC. Coral-associated bacterial assemblages: current knowledge and the potential for climate-driven impacts. Intergr Comp Biol. 2010;50:662–674. doi: 10.1093/icb/icq061. [DOI] [PubMed] [Google Scholar]
- Nishijima M, Adachi K, Katsuta A, Shizuri Y, Yamasato K.2012Endozoicomonas numazuensis sp. nov., a gamaproteobacterium isolated from marine sponges, and emended description of the genus Endozoicomonas Kurahashi and Yokota 2007 Int J Syst Evol Microbiol;doi: 10.1099/ijs.0.042077-0 [DOI] [PubMed]
- Raina J-B, Dinsdale E, Willis BL, Bourne DG. Do organic sulphur compounds DMSP and DMS drive coral microbial associations. Trends Microbiol. 2010;18:101–108. doi: 10.1016/j.tim.2009.12.002. [DOI] [PubMed] [Google Scholar]
- Raina J-B, Tapiolas D, Willis BL, Bourne DG. Coral-associated bacteria and their role in the biogeochemical cycling of sulfur. Appl Environ Microbiol. 2009;75:3492–3501. doi: 10.1128/AEM.02567-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie KB. Regulation of microbial populations by coral surface mucus and mucus-associated bacteria. Mar Ecol Prog Ser. 2006;322:1–14. [Google Scholar]
- Shnit-Orland M, Kushmaro A. Coral mucus-associated bacteria: a possible first line of defense. FEMS Microbiol Ecol. 2009;67:371–380. doi: 10.1111/j.1574-6941.2008.00644.x. [DOI] [PubMed] [Google Scholar]
- Sievert SM, Kiene RP, Schulz-Vogt HN. The sulfur cycle. Oceanography. 2007;20:117–123. [Google Scholar]
- Smith DC, Douglas AE. The Biology of Symbiosis. Edward Arnold Ltd: London; 1987. [Google Scholar]
- Speck MD, Donachie SP.2012Widespread oceanospirillaceae bacteria in Porites spp J Mar Biol;doi: 10.1155/2012/746720 [DOI]
- Stat M, Baker AC, Bourne DG, Correa AM, Forsman Z, Huggett MJ, et al. Molecular delineation of species in the coral holobiont. Adv Mar Biol. 2012;63:1–65. doi: 10.1016/B978-0-12-394282-1.00001-6. [DOI] [PubMed] [Google Scholar]
- Stat M, Morris E, Gates RD. Functional diversity in coraldinoflagellate symbiosis. Proc Natl Acad Sci USA. 2008;105:9256–9261. doi: 10.1073/pnas.0801328105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunagawa S, DeSantis TZ, Piceno YM, Brodie EL, DeSalvo MK, Voolstra CR, et al. Bacterial diversity and white plague disease-associated community changes in the Caribbean coral Montastraea faveolata. ISME J. 2009;3:512–521. doi: 10.1038/ismej.2008.131. [DOI] [PubMed] [Google Scholar]
- Sunagawa S, Woodley CM, Medina M. Threatened corals provide underexplored microbial habitats. PLoS One. 2010;5:e9554. doi: 10.1371/journal.pone.0009554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd JD, Curson AR, Nikolaidou-Katsaraidou N, Brearley CA, Watmough NJ, Chan Y, et al. Molecular dissection of bacterial acrylate catabolism--unexpected links with dimethylsulfoniopropionate catabolism and dimethyl sulfide production. Environ Microbiol. 2010;12:327–343. doi: 10.1111/j.1462-2920.2009.02071.x. [DOI] [PubMed] [Google Scholar]
- Van Alstyne K, Schupp P, Slattery M. The distribution of dimethylsulfoniopropionate in tropical Pacific coral reef invertebrates. Coral Reefs. 2006;25:321–327. [Google Scholar]
- van Oppen MJH, Baker AC, Coffroth MA, Willis BL.2009Bleaching resistance and the role of algal endosymbiontsIn: van Oppen, MJH, Lough, JM (eds).Coral Bleaching Patterns, Processes, Causes and Consequences Springer Ecological Series: Berlin [Google Scholar]
- Vega Thurber RLV, Barott KL, Hall D, Desnues C, Edwards RA, Haynes M. Metagenomic analysis indicates that stressors induce production of herpeslike viruses in the coral Porites compressa. Proc Natl Acad Sci USA. 2008;105:18413–18418. doi: 10.1073/pnas.0808985105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster NS, Cobb RE, Negri AP. Temperature thresholds for bacterial symbiosis with a sponge. ISME J. 2008;2:830–842. doi: 10.1038/ismej.2008.42. [DOI] [PubMed] [Google Scholar]
- Webster NS, Soo R, Cobb R, Negri AP. Elevated seawater temperature causes a microbial shift on crustose coralline algae with implications for the recruitment of coral larvae. ISME J. 2011;5:759–770. doi: 10.1038/ismej.2010.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster NS, Taylor MW. Marine sponges and their microbial symbionts: love and other relationships. Environ Microbiol. 2012;14:335–346. doi: 10.1111/j.1462-2920.2011.02460.x. [DOI] [PubMed] [Google Scholar]
- Webster NS, Taylor MW, Behnam F, Lücker S, Rattei T, Whalan S, et al. Deep sequencing reveals exceptional diversity and modes of transmission for bacterial sponge symbionts. Environ Microbiol. 2010;12:2070–2082. doi: 10.1111/j.1462-2920.2009.02065.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster NS, Webb RI, Ridd MJ, Hill RT, Negri AP. The effects of copper on the microbial community of a coral reef sponge. Environ Microbiol. 2001;3:19–31. doi: 10.1046/j.1462-2920.2001.00155.x. [DOI] [PubMed] [Google Scholar]
- Wegley L, Edwards RA, Rodriguez-Brito B, Liu H, Rohwer F. Metagenomic analysis of the microbial community associated with the coral Porites astreoides. Environ Microbiol. 2007;9:2707–2719. doi: 10.1111/j.1462-2920.2007.01383.x. [DOI] [PubMed] [Google Scholar]
- Wilkinson CR.2008Status of Coral Reefs of the World.Global Coral Reef Monitoring Network and Reef and Rainforest Research Center: Townsville, p296
- Yang CS, Chen MH, Arun AB, Chen CA, Wang JT, Chen WM. Endozoicomonas montiporae sp. nov., isolated from encrusting pore coral Montipora aequituberculata. Int J Syst Evol Microbiol. 2010;60:1158–1162. doi: 10.1099/ijs.0.014357-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.