Abstract
The gastrointestinal (GI) microbiome contributes significantly to host nutrition and health. However, relationships involving GI microbes, their hosts and host macrohabitats remain to be established. Here, we define clear patterns of variation in the GI microbiomes of six groups of Mexican black howler monkeys (Alouatta pigra) occupying a gradation of habitats including a continuous evergreen rainforest, an evergreen rainforest fragment, a continuous semi-deciduous forest and captivity. High throughput microbial 16S ribosomal RNA gene sequencing indicated that diversity, richness and composition of howler GI microbiomes varied with host habitat in relation to diet. Howlers occupying suboptimal habitats consumed less diverse diets and correspondingly had less diverse gut microbiomes. Quantitative real-time PCR also revealed a reduction in the number of genes related to butyrate production and hydrogen metabolism in the microbiomes of howlers occupying suboptimal habitats, which may impact host health.
Keywords: Alouatta, gut microbiome, habitat, primate
Introduction
The mammalian gastrointestinal (GI) microbiome contributes significantly to host digestive efficiency (Bauchop, 1971; Lambert, 1998; Flint and Bayer, 2008), while diet strongly influences the microbiome's composition (Ley et al., 2006, 2008; Turnbaugh et al., 2009; Yildirim et al., 2010; Wu et al., 2011). Across species, host diet and GI microbiome composition are correlated (Ley et al., 2008), while within species, dietary shifts alter GI microbiome composition (Ley et al., 2006; Turnbaugh et al., 2009). For most wild, mammalian populations, dietary shifts are common across seasons and habitats. However, while experimental studies imply that shifts in host diet in wild, mammalian populations should impact GI microbiome composition, the relationship between host habitat, diet and GI microbiome composition remains to be examined.
Understanding the interaction between host habitat and microbiome composition may also be useful for conservation efforts because changes in the GI microbiome have been shown to affect host nutrition and health (Dethlefsen et al., 2007; Sekirov et al., 2010; Flint et al., 2011; Hooper et al., 2012). Dysbioses may manifest themselves in a number of different ways, but within the GI-tract they are most often seen as losses of microbial species diversity. Reductions in GI microbiome diversity may reduce microbial functional groups, making the microbiome less efficient, less resistant to disturbance and more susceptible to pathogenic invasion (Rosenfeld, 2002; Costello et al., 2012). Hosts with low GI microbiome diversity also exhibit increased stress responses (higher glucocorticoid levels) and reduced immune function (fewer cells that secrete local, strain-specific immunoglobulin A) (Macpherson et al., 2008; Neish, 2009; Messaoudi et al., 2010; Hooper et al., 2012).
Here, we explore cross-habitat patterns in the GI microbiome richness, diversity and composition of an endangered Mesoamerican primate, the black howler monkey (A. pigra; IUCN, 2012) using 16S ribosomal RNA gene sequencing to survey colonizing bacteria. The howler monkey (Alouatta sp.) provides an exceptional model for understanding how variation in host habitat and diet impacts GI microbiome structure, and ultimately, host nutrition and health. Howlers can consume a leaf-heavy diet (Milton, 1980; Lambert, 1998; Estrada and Coates-Estrada, 2005; Di Fiore and Campbell, 2007), permitting them to occupy areas with limited resource availability and to persist longer in degraded habitats than more frugivorous primate taxa (Estrada and Coates-Estrada, 1996; Zunino et al., 2007). As many leaves contain high levels of resistant fiber and toxins, GI microbiome contributions to daily energy flows through metabolism of xenobiotics are critical to howler nutrition (Milton, 1979; Lambert, 1998; Lambert and Fellner, 2012). Fibrolytic processes of the GI microbiome contribute up to 31% of a howler's required daily energy via the formation of short-chain fatty acids (93.7% acetate, 5.9% propionate, 0.4% butyrate in A. palliata; Milton and McBee, 1983). Therefore, if shifts in howler diet change GI microbiome composition or reduce microbiome richness and diversity, there may be consequences for howler nutrition and health.
In this study, we test the hypothesis that howlers occupying habitats of different qualities develop different GI microbiomes. Animals in disturbed habitats generally consume different types of food resources than their counterparts in undisturbed habitats (Kamler et al., 2007; Flaherty et al., 2010; Abbas et al., 2011; Chaves et al., 2012). As a result, their diets typically deviate from the host species archetypical dietary regime and are likely to be associated with shifts in GI microbiome composition. Specifically, because habitat degradation leads to differences in available plant species and reductions in plant diversity (Benitez-Malvido and Martinez-Ramos, 2003; Vellend, 2003; Lindborg and Eriksson, 2004), we predict that howlers inhabiting disturbed habitats will exhibit signs of dysbiosis, such as lower microbial richness and diversity. If this is the case, then host nutrition and health may be negatively affected.
To explore the relationship between host habitat, diet and GI microbiome composition, we analyzed fecal samples from six howler groups in four distinct habitats along a gradient of disturbance (detailed in Supplementary Information) in southeastern Mexico: a continuous, evergreen rainforest (CR), an evergreen rainforest fragment 4.5 km from the CR (FR), a continuous, semi-deciduous forest (SD), and a wildlife center (captive). Analyses utilize important perspectives from non-human primate ecology, biological anthropology, community ecology and microbial ecology (Benezra et al., 2012; Costello et al., 2012), and illustrate a ‘co-degradation' of howler habitat, diet and GI microbiomes that presents new concerns for conservation biology.
Materials and methods
Fecal sample collection
Samples were collected across an 8-week period (May–June 2009) from two neighboring groups of black howlers inhabiting a CR in Palenque National Park, Mexico (Balam Group, n=6 and Motiepa Group, n=8). Additional samples were obtained from two groups of howlers in a continuous, but SD forest ∼200 km from the national park (Tormento Group 1, n=3, and Tormento Group 2, n=4), a FR located 7 km outside of the national park near human dwellings (Cuartel Group, n=4), and a rehabilitation center located 5 km outside of the national park, which housed locally rescued monkeys on an atypical diet (AcaJungla, n=7; Supplementary Figure S1; additional information about the collection sites is provided in Supplementary Information). No continuous forest exists between the sample sites, and they are separated by distances far exceeding the territories occupied by A. pigra groups, precluding migration of groups among habitats.
Fecal samples were collected immediately upon defecation, and then stored in 96% ethanol at 4 °C until transport to the University of Illinois at Champaign-Urbana where they were stored at −20 °C until processing. A total of 128 samples were collected. Individuals in the CR were sampled weekly when possible for the duration of the study.
Behavioral data collection
Feeding data for the CR and SD groups were obtained from ∼225 h of focal individual sampling with binoculars. Twenty-minute focal samples of behavior were collected between 0600 and 1600 hours each day, with activity recorded instantaneously every 2 min. During a feeding bout (active consumption of food resources), the food type (e.g. young leaves, mature fruit, flowers) and plant species were recorded. Qualitative dietary data were collected for rainforest fragments during fieldwork. General diet data for the captive group were obtained from veterinary records at the rehabilitation center.
As quantitative dietary data were expressed in time spent consuming resources, comparisons were standardized by body weight depending on individual age and sex (Kelaita et al., 2011). The time each individual spent feeding on food types and plant species was analyzed as proportion data after arcsine transformation. Non-parametric Mantel tests comparing diet and fecal microbiomes were performed using PRIMER 6 for Windows v 6.1.10 (PRIMER-E, Plymouth, UK).
DNA analyses
Microbial DNA was extracted from fecal samples using the MoBio UltraClean Soil Kit (Mo Bio Laboratories Inc., Carlsbad, CA, USA). The V1–V3 region of the 16S ribosomal RNA gene was amplified by polymerase chain reaction (20 cycles of 94 °C (30 s), 48 °C (30 s), 72 °C (2 min)) using primers 27f 5′-(CGTATCGCCTCCCTCGCGCCATCAG-3′, 5′-AGAGTTTGATYMTGGCTCAG-3′, corresponding to nucleotides 8–27 of the Escherichia coli 16s ribosomal RNA gene) and 534r 5′-(CTATGCGCCTTGCCAGCCCGCTCAG-3′ (MID tag 1–50) 5′-ATTACCGCGGCTGCTGGCA)-3′. The amplicons were pyrosequenced using 454 FLX-Titanium technology at the J. Craig Venter Institute (Rockville, MD, USA).
One-hundred and twelve samples were successfully sequenced, and the resulting sequences were processed using mothur (Schloss et al., 2009) and Tornado (Sipos et al., 2010). After removing low quality sequences (<Q30) and sequences shorter than 250 nt, with homopolymers longer than six nucleotides, and containing ambiguous base calls or incorrect primer sequences, the average amplicon length was 456.9 nt, which we trimmed to the common 385 nt (E. coli positions: 534−>149) shared by all sequences. Sequences were aligned against the silva (Pruesse et al., 2007) and infernal (Nawrocki et al., 2009) databases and merged to give an optimal alignment using Tornado (Sipos et al., 2010). Potentially chimeric sequences were detected using chimera slayer (<6% http://www.mothur.org/wiki/chimera.slayer/) and removed. The remaining reads were pre-clustered (http://www.mothur.org/wiki/Pre.cluster) and then clustered using mothur's average algorithm (Schloss et al., 2009). Operational taxonomic units were defined as sharing >97% sequence identity. Operational taxonomic units detected fewer than twice across the entire data set were removed as probable artifacts.
Rarefaction data, Simpson, Shannon-Weaver and Chao1 indices were created using mothur (Schloss et al., 2009). Taxonomic profiles were generated using the Ribosomal Database Project MultiClassifier (Wang et al., 2007). Dendrograms were constructed and visualized in PRIMER 6 for Windows v 6.1.10 (PRIMER-E, Plymouth, UK) using Bray-Curtis similarity indexes (Clark and Gorley, 2006). Sample clustering patterns were tested using analysis of similarity (ANOSIM) in PRIMER 6 (Clark and Gorley, 2006). T-tests were used to compare the relative abundances of bacterial taxa across habitats.
Initial sequence processing demonstrated that samples collected weekly from CR howlers clustered by individual (ANOSIM R=0.43, P=0.001) and weekly variation explained only 3.5% of the total variation in microbial community composition. Therefore, weekly samples were pooled by individual for sequence analysis to represent overall microbiome structure without excluding low-read samples and inflating of degrees of freedom in statistical analyses. Analyses of each individual sample from CR animals returned results consistent with pooled samples (Supplementary Figure S2).
Quantitative real-time PCR (qRT–PCR) was performed on a single sample from each individual using the SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA, USA) as described previously (Nava et al., 2012). Primers ACSF1/ACSR1 (Gagen et al., 2010) and FTHFSf/FTHFSr (Leaphart and Lovell, 2001) targeting functional genes acs (acetyl-CoA synthase involved in the acetyl-CoA pathway) of acetogens were used. Primers ME1/ME2 (Hales et al., 1996) and DSR1fdeg/DSR4rdeg (Leloup et al., 2007) were used for mcrA (coding for the methyl-coenzyme M reductase catalyzing methane formation) and dsrA (dissimilatory (bi)sulfite reductase catalyzing the anaerobic respiration of sulfite or sulfate) genes. Primers BcoAf/BcoAr (Louis et al., 2010) were used to quantify the butyryl-CoA:acetate CoA-transferase gene, involved in the microbial production of butyrate. Primers NiFe-uniF/NiFe-uniR (Schmidt et al., 2011) were used to quantify the (Ni-Fe)-hydrogenase ((NF)hyd) gene, involved in the microbial production of hydrogen. Samples were run on a 7900HT Fast Real-Time PCR System (Applied Biosystems) using a dissociation curve. Standard curves were determined simultaneously using plasmids containing dsrA, mcrA, (NF)hyd and BcoA or diluted PCR products from reference strains for acs. T-tests were used to compare the relative abundance of each gene across habitats.
Results
Habitat-microbiome clusters
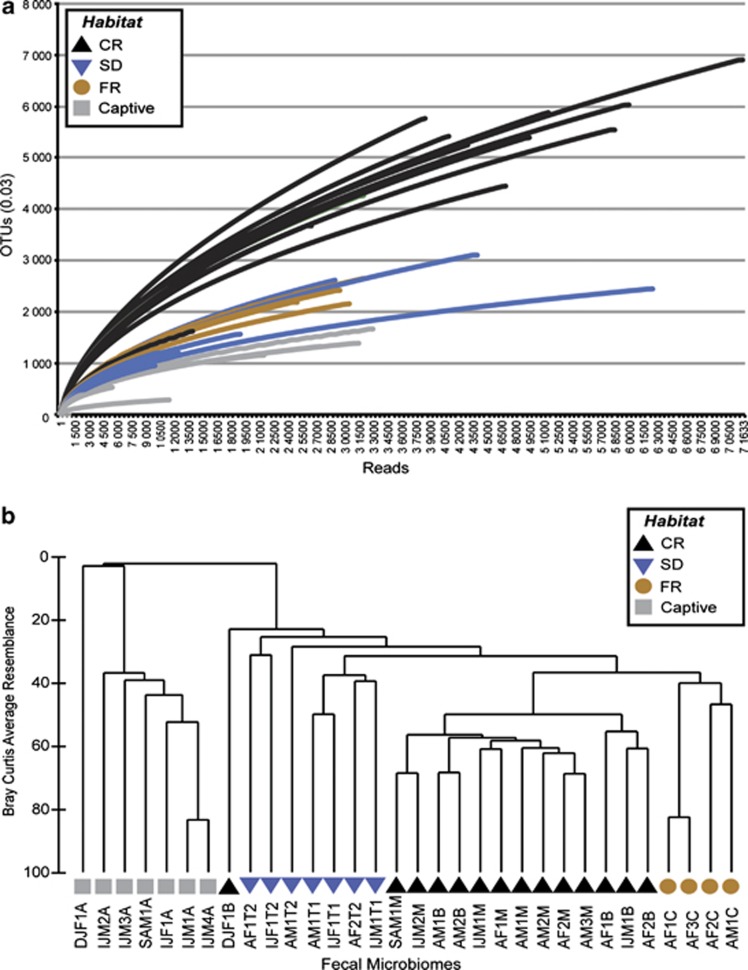
Microbial species richness and diversity differed strikingly among howler monkey groups occupying distinct habitats (Figure 1a). Chao1 richness estimates (Chao, 1987) clustered by habitat, revealing that howlers inhabiting the CR harbored as many as six times the number of microbial operational taxonomic units as captives, and up to twice as many operational taxonomic units as the howlers inhabiting the continuous, SD forest and the FR (Figure 1a, Supplementary Tables S1, S2). This pattern was maintained when unpooled data from a given week were used to represent CR howlers (Supplementary Figure S2). CR howler GI microbiome diversity varied little from week to week (Supplementary Table S3) and exceeded diversity in all other groups (Supplementary Tables S1, S2). With regard to GI microbiome composition, Firmicutes and Bacteroidetes phyla predominated, and were detected in similar proportions across all habitats (Supplementary Figure S3; 72±0.06% and 19±0.05%, respectively). However, both ANOSIM's and dendrograms based on Bray-Curtis similarity matrices indicate that abundance-weighted phylogenetic composition of individual howler microbiomes varied significantly by habitat (Figure 1b, Table 1). The most extreme distinction was observed among the microbiomes of captive and wild groups (ANOSIM R>0.85, P<0.003), but all samples exhibited habitat-specific clusters of microbial taxa (Figure 1b). For example, the microbiomes of captive howlers were enriched with Proteobacteria (4.8% vs 1.1±0.001% in other groups), with a 36% increase in the genus Succinivibrio (Table 2). On the other hand, FR howlers were enriched with Verrucomicrobia (3% vs 0.32±0.002%), especially genera of the class Opitutae (a 53% increase relative to other habitats, Table 2).
Figure 1.
Howler microbiomes clustered by host habitat. (a) Rarefaction curves show the number of unique Operational taxonomic units (sharing⩾97% sequence identity) per total reads for each sample. Analysis of variance of the log-linear regression coefficients for each curve confirmed habitat differences (F3, 31=28.91, P<0.0001). (b) Dendrogram compares sample pairs using Bray-Curtis similarity indices. Individual identities and groups are listed in Table 1. ANOSIM tests habitat clustering using permutations (ANOSIM R=0.743, P=0.001). The CR outlier is the same individual in both panels.
Table 1. Inter-group ANOSIM results for gut microbiome composition.
| CR | SD | FR | |
|---|---|---|---|
| SD | 0.663*** | ||
| RF | 0.721*** | 0.429* | |
| Captive | 0.954*** | 0.912*** | 0.852** |
Abbreviations: ANOSIM, analysis of similarity; CR, continuous evergreen rainforest; SD, continuous semi-deciduous forest; FR, fragmented evergreen rainforest.
R-statistic with P-value: *<0.025, **<0.005, ***<0.001.
ANOSIM of inter-habitat differences based on the abundance-weighted phylogenetic composition of individual howler samples.
Table 2. Relative abundance (± s.d.) of selected microbial genera in each howler group binned using the Ribosomal Database Project (RDP) multiclassifier with bootstrapping >70% across howler habitats and groups.
| Genus | CR–Motiepa | CR–Balam | FR | SD–T2 | SD–T1 | Captive | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| |
Mean (%) |
s.d. |
Mean (%) |
s.d. |
Mean (%) |
s.d. |
Mean (%) |
s.d. |
Mean (%) |
s.d. |
Mean (%) |
s.d. |
| Bacteroides | 0.003 | 0.002 | 0.002 | 0.002 | 0.014 | 0.015 | 0.000 | 0.000 | 0.003 | 0.006 | 0.186* | 0.143 |
| Blautia | 1.009 | 0.180 | 0.875 | 0.144 | 0.374* | 0.204 | 1.830 | 0.558 | 1.500 | 0.558 | 2.860 | 1.834 |
| Butyricicoccus | 0.269 | 0.097 | 0.268 | 0.232 | 0.156* | 0.149 | 0.106 | 0.055 | 0.231 | 0.155 | 0.036* | 0.044 |
| Butyricimonas | 0.000 | 0.000 | 0.000 | 0.000 | 0.000* | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.022* | 0.013 |
| Butyrivibrio | 0.013 | 0.003 | 0.017 | 0.012 | 0.008 | 0.003 | 0.010 | 0.006 | 0.008 | 0.013 | 0.001* | 0.002 |
| Desulfovibrio | 0.002 | 0.003 | 0.000 | 0.000 | 0.006* | 0.003 | 0.002 | 0.002 | 0.000 | 0.002 | 0.000+ | 0.000 |
| Lactobacillus | 0.015 | 0.035 | 0.005 | 0.005 | 0.029+ | 0.018 | 0.001* | 0.001 | 0.005* | 0.004 | 0.282# | 0.333 |
| Marvinbryantia | 0.127 | 0.037 | 0.108 | 0.045 | 0.083+ | 0.028 | 0.053* | 0.012 | 0.038* | 0.012 | 0.000# | 0.000 |
| Opitutus | 0.007 | 0.003 | 0.003 | 0.002 | 0.026+ | 0.013 | 0.000* | 0.000 | 0.000* | 0.000 | 0.001* | 0.002 |
| Prevotella | 0.052 | 0.027 | 0.037 | 0.011 | 0.007 | 0.008 | 0.003 | 0.003 | 0.016 | 0.014 | 9.177* | 5.153 |
| Pseudobutyrivibrio | 0.000 | 0.000 | 0.000 | 0.000 | 0.000* | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.006* | 0.007 |
| Ruminococcus | 0.510 | 0.234 | 0.384 | 0.178 | 0.808 | 0.406 | 0.302 | 0.350 | 0.565 | 0.504 | 0.231 | 0.351 |
| Succinivibrio | 0.000 | 0.000 | 0.000 | 0.000 | 0.001 | 0.001 | 0.000 | 0.000 | 0.000 | 0.000 | 2.255* | 2.742 |
Abbreviations: CR, a continuous evergreen rainforest; FR, evergreen rainforest fragment; SD, semi-deciduous forest.
Symbols indicate significantly different abundances (t-tests, α=0.05). Abundances with matching symbols were not significantly different from each other.
Dietary impacts
As the captive howlers exhibited similar GI microbiomes but came from distinct geographic regions of Mexico and possessed no known kin relationships, the habitat-associated patterns in microbiome composition are most likely an effect of environmental influences, such as diet, rather than genetic influence. Detailed observations of monkey diets collected during the study period revealed nearly identical diets in the two CR groups, which consumed mature fruits 53.0% of their total feeding time (Supplementary Table S4). In contrast, the two SD groups consumed mostly young leaves (52.3% Supplementary Table S4), essentially substituting leaves for fruits when compared with the CR howlers. The CR howlers also consumed a more diverse diet than SD howlers (15 vs 9 tree species), and the tree species exploited by each individual differed significantly between the forest habitats (ANOSIM R=0.637, P=0.001; Supplementary Table S5). Non-parametric Mantel tests comparing Bray-Curtis similarity matrices of diet and microbiome composition for all CR and SD individuals (n=22 howlers) revealed a correlation between diet and microbiome composition (plant part: Spearman's ρ=0.406, P=0.08; plant species: Spearman's ρ=0.820, P=0.01). Howlers feeding on different plant species also exhibited distinct profiles of GI microbiome richness and diversity (Supplementary Table S1, S5, Figure 1).
Qualitative dietary information obtained for the FR and captive groups illustrated similar patterns. Both FR and captive howlers consumed a diet with severely reduced diversity. Rainforest fragment howlers used a subset of tree species found in the CR, while captives were fed an atypical diet, including fruit, fiber-rich cereal and Leaf-Eater Primate Diet feed (Mazuri, Purina Mills, St Louis, MO, USA). The reduced number of plant species utilized by the FR and captive howlers was also associated with compositional changes and reductions in richness and diversity of their fecal microbiomes (Supplementary Table S1, Figure 1).
Taxonomic shifts in microbiome composition corresponded to diet variation. Prevotella, a common bacterial genus capable of degrading hemicellulose, pectin and simpler carbohydrates, such as those expected in fruits and low-complexity fibrous resources (Russell and Baldwin, 1979; Salyers, 1979), was found in greater abundance in the captive howlers than in any other individuals (Table 2), and all howler groups exhibited more Prevotella than Bacteroides, except the FR howlers (Table 2). Differences in relative abundance of Prevotella and Bacteroides have been associated with diet-related gut microbial enterotypes in humans (Wu et al., 2011). Additionally, all howlers, except the SD howlers, possessed lactobacilli (Table 2). Lactobacilli abundance has been found to be favored by dietary calcium (Bovee-Oudenhoven et al., 1999), and Ficus fruits, which are present in every wild habitat except the SD, are higher in calcium content compared with other fruit species (O'Brien et al., 1998). The highest levels of lactobacilli were observed in captive howlers (Table 2) and may be a result of high calcium supplementation in the Leaf-Eater Primate Diet feed and occasional probiotic administration.
Potential health consequences
Habitat-associated reductions in GI microbiome richness and diversity appear to influence the stability of the gut community and may affect howler health. Microbiome composition varied least among individual CR howlers, while SD and captive howlers showed significant inter-individual variation (Tables 2 and 3). This may be indicative of increased stochasticity in GI microbiome colonization (Costello et al., 2012), which may evoke a reduced resilience to disturbance or invasion by pathogens. However, obvious pathogenic bacteria were rare (except in DJF1A who was experiencing severe diarrhea and harbored high numbers of potentially pathogenic E. coli).
Table 3. Average inter- and intra-group Bray-Curtis similarity indices.
| CR | SD | FR | Captive | |
|---|---|---|---|---|
| Inter-group | 20.12 | 20.72 | 24.34 | 2.16 |
| Intra-group | 51.20 | 30.54 | 48.25 | 32.27 |
Abbreviations: CR, a continuous evergreen rainforest; FR, evergreen rainforest fragment; SD, semi-deciduous forest.
The table shows the average Bray-Curtis similarity measures among howler groups and among individual howlers within groups.
Although potentially pathogenic sequences were not related to gut microbiome diversity, all captives died within 6 months following the sampling period, and DJF1B (CR infant outlier) also died (predation cannot be excluded). None of these individuals was considered a mature adult, but all but two individuals (DJF1B and DJF1A) were old enough to forage independently on an adult diet, and age-matched wild animals had GI microbiomes that were compositionally consistent with adults. Indeed, there were no GI microbiome differences according to age or sex in either the CR (ANOSIM R=0.20, P=0.12) or the SD (ANOSIM R=−0.13, P=0.74) howlers. These patterns warrant investigation in future studies to determine whether or not reductions in GI microbiome richness may have a role in disease susceptibility and host mortality.
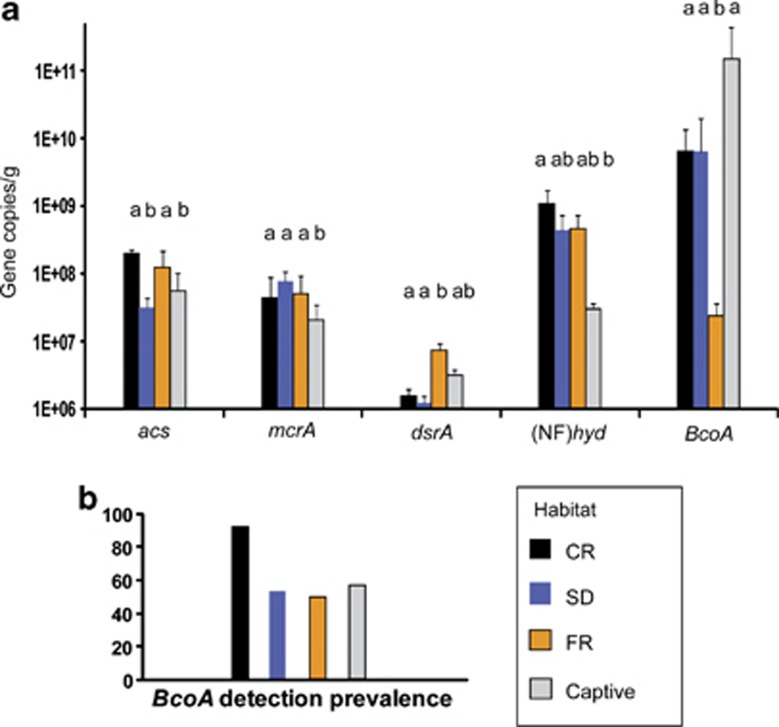
Shifts in microbial taxa and metabolic functional genes across habitats also demonstrate effects of GI microbiome compositional changes on howler health. Butyrate is the primary energy source for the colonic epithelium and is recognized for numerous health-promoting benefits (Donohoe et al., 2011; Grootaert et al., 2011; Hu et al., 2010; Rawluszko et al., 2011; Sharma and Singh, 2011). Butyrate-producing taxa, such as Butyrivibrio spp. were most abundant in howlers occupying the CR and were detected at very low levels in all but one of the seven captives (Table 2). Similarly, Butyricicoccus spp., were found in all non-captive individuals but in only five of the captives (at significantly lower levels than the CR individuals; Table 2). qRT–PCR of the butyryl-CoA:acetate CoA-transferase gene, involved in the microbial production of butyrate, indicated that this pathway was most prevalent in the CR individuals compared with other sites (Figure 2b). Although the captive howlers exhibited the highest average abundance of BcoA, this pattern was driven by one individual, and overall prevalence of BcoA was low (detected in only four of seven individuals). Additionally, in captives, known butyrate producers (for example, Pseudobutyrivibrio spp. only seen in captives) may have been further offset by butyrate oxidizing genera (for example, Butyricimonas spp. also only observed in the captives).
Figure 2.
(a) Abundance (±s.d.) of metabolic functional genes in the microbiomes of howlers across habitats as determined by qRT–PCR. Genes targeted included acs for acetogens, mcrA for methanogens, dsrA for sulfate reducers, (NF)hyd for hydrogen producers, and BcoA for butyrate producers. (b) Prevalence of the butyryl-CoA:acetate CoA-transferase gene for butyrate producers across habitats. Letters indicate significantly different abundances among habitats (t-tests, P<0.05). Prevalence of all genes except those involved in butyrate production was 100% for all habitats. Patterns are similar when groups are taken into account within habitats.
Clear cross-habitat distinctions also occurred in other microbial functional groups with known impacts on host health. Hydrogen-production provides a proximal measure of the fermentative activity in the microbiome, and qRT–PCR detected more (Ni-Fe)-hydrogenase ((NF)hyd) genes in the CR than any other habitat (Figure 2a). Similarly, the relative abundance of different hydrogenotrophs varied with habitat. Acetogens can convert dihydrogen and carbon dioxide to one of the primary short-chain fatty acids, acetate, which can then be absorbed and used by host epithelia (Rey et al., 2010; Carbonero et al., 2012). Acetogenic Marvinbryantia, were observed in all non-captive howlers and were most abundant in the CR (Table 2). This genus was not detected in any of the captives. Although captives hosted another common acetogenic genus, Blautia (Table 2), qRT–PCR revealed a significantly greater number of acetyl-CoA synthase genes in the CR howlers (Figure 2a). Methanogens, which convert hydrogen and carbon dioxide to methane, a product of no nutritive value (Carbonero et al., 2012), are not detected using the 534r primer used in our pyrosequencing evaluation of 16S ribosomal RNA genes. However, qRT–PCR of the methyl-coenzyme M reductase gene revealed methanogenesis to also be least abundant in captives (Figure 2a). Instead, hydrogenotrophy in captives appeared to be facilitated by sulfate-reducing bacteria, which dissipate dihydrogen through the production of hydrogen sulfide (H2S). Hydrogen sulfide is a toxic gas that affects smooth muscle and has been linked to colonic nociception, inflammatory bowel disease and colorectal cancer (Medani et al., 2011; Carbonero et al., 2012). Desulfovibrionales, the prominent intestinal sulfate-reducing bacteria order, were most abundant in the FR and captive groups and rare in the CR and SD (Table 2). qRT–PCR of the dissimilatory (bi)sulfite reductase gene confirmed that sulfate-reducing genes were more abundant in the FR and captive groups of howlers (Figure 2).
Discussion
Our results demonstrate that non-human primate GI microbiome richness, diversity and composition vary strongly with habitat. Although howler diets likely shift seasonally within habitats, this study suggests that dietary shifts associated with habitat disturbance influence non-human primate GI microbiomes. The howler diet in minimally disturbed and relatively pristine macrohabitats appears to promote the acquisition and maintenance of a diverse set of GI-tract microbes, while diets in suboptimal habitats lead to greater variability in microbiome composition, as well as reduced diversity.
While results from this study suggest that the identity and diversity of plants in the diet are driving patterns in microbiome richness and diversity, further research is necessary to determine if dietary diversity or certain plant metabolites that vary between plant species promote a stable GI microbiome composition with high alpha diversity. A diverse host diet, such as that of the CR howlers, delivers a large array of nutrients and carbohydrate substrates to the gut thereby providing a variety of feeding niches to support a high diversity of microbial taxa or functional groups (Louis et al., 2007). Therefore, less diverse diets typical of degraded or suboptimal habitats may sustain lower GI microbiome richness and diversity regardless of the plant species being utilized. In contrast, a specific food item or plant species may strongly influence the composition of the gut microbiome by determining the amount of one or two key nutrients in the gut. Understanding the importance of each of these mechanisms is critical to improving our knowledge of host–microbe interactions in wild primate populations.
As an observational field study of endangered primates, the present study cannot establish causality experimentally. However, our results provide some evidence that habitat degradation may affect host health negatively via diet-associated shifts in the gut microbiome. In general, efficient fermentation is inferred by the microbial activity of butyrate- and hydrogen-producing fermenters and hydrogen-utilizing acetogens and methanogens. Butyrogenic and acetogenic metabolism are particularly beneficial as they allow optimal energy gain, and butyrate promotes intestinal development and health (Donohoe et al., 2011; Grootaert et al., 2011). In contrast, sulfate-reducing bacteria represent a less desirable group of hydrogen-utilizers, whose end-product H2S can provoke carcinogenesis and inflammation (Medani et al., 2011; Carbonero et al., 2012). Thus, high butyrogenic and acetogenic metabolism indicate potentially healthier microbiomes in CR while both captive and FR howlers present less fermentative potential along with high numbers of potentially harmful sulfate-reducing bacteria.
Future studies should examine the relationship between howler GI microbiome composition and health directly by measuring health factors that might be influenced by the microbiome such as glucocorticoid levels, parasite abundance and diversity and immunoglobulin A levels (Macpherson et al., 2008; Neish, 2009; Messaoudi et al., 2010). Black howler monkeys living in small, highly disturbed forest fragments are reported to exhibit higher fecal glucocorticoid levels than those inhabiting less-disturbed forest (Martinez-Mota et al., 2007), and studies of several howler species, including black howlers in Palenque, have reported higher GI parasite diversity and abundance in primates inhabiting degraded areas compared with those in relatively undisturbed habitats (Eckert et al., 2006; Stoner and Gonzalez Di Pierro, 2006; Trejo-Macias et al., 2007; Vitazkova and Wade, 2007; Trejo-Macias and Estrada, 2012). The depletion of the GI microbiome in degraded environments may explain these patterns, at least in part.
This study indicates that primates in degraded habitats risk ‘double jeopardy' from both external factors (reduced resource availability) and internal factors (diminished microbiomes). These findings likely apply more broadly to other primate taxa and to mammals in general. Further research should prospectively investigate the mechanisms by which diet impacts the microbiome in wild mammals and by which degraded host microbiomes negatively affect host health. Using this information as a basis for developing new techniques and tools, conservation efforts can begin to better assess the impacts of human activity on animal health and the challenges faced by animal populations forced into suboptimal habitats.
Acknowledgments
We would like to thank David Muñoz for his assistance with forest fragment sample collection, Jenna Leinberger and Maeve Leurck for their assistance with qRT–PCR, and Dr Salomon Gonzalez for his permission to collect captive howler samples at Acajungla/Aluxes. Thanks are also due to Dr Roderick Mackie for use of lab supplies and space, to Dr Paul Garber as Ph.D. advisor to KRA, and to two anonymous reviewers for their comments. Research was carried out under permits from the Mexican environmental agencies, the Secretaria del Medio Ambiente y Recursos Naturales (SEMARNAT) and the Comision Nacional de Areas Naturales Protegidas (CONANP), granted to AE. We appreciate the logistical support provided by Universidad Nacional Autonoma de Mexico (UNAM) through AE. INIFAP and Ing. Antonio Sanchez Martinez granted permission to NR and AE to work at El Tormento. This project was funded by the NSF grant #0935347 (Human Origins Moving in New Directions (HOMINID). Fieldwork and preliminary lab work were funded by grants from the University of Illinois (Beckman, Tinker, and the Program in Ecology, Evolution and Conservation Biology) to KRA. KRA is supported by an NSF Graduate Research Fellowship. 16S sequence data are available from the authors upon request.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Abbas F, Morellet N, Hewison AJM, Merlet J, Cargnelutti B, Lourtet B, et al. Landscape fragmentation generates spatial variation of diet composition and quality in a generalist herbivore. Oecologia. 2011;167:401–411. doi: 10.1007/s00442-011-1994-0. [DOI] [PubMed] [Google Scholar]
- Bauchop T. Stomach microbiology of primates. Annu Rev Microbiol. 1971;25:429–436. doi: 10.1146/annurev.mi.25.100171.002241. [DOI] [PubMed] [Google Scholar]
- Benezra A, DeStefano J, Gordon JI. Anthropology of microbes. PNAS. 2012;109:6378–6381. doi: 10.1073/pnas.1200515109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benitez-Malvido J, Martinez-Ramos M. Impact of forest fragmentation on understory plant species richness in Amazonia. Conserv Biol. 2003;17:389–400. [Google Scholar]
- Bovee-Oudenhoven IM, Wissink ML, Wouters JT, Van der Meer R. Dietary calcium phosphate stimulates intestinal Lactobacilli and decreases the severity of a Salmonella infection in rats. J Nutr. 1999;129:607–612. doi: 10.1093/jn/129.3.607. [DOI] [PubMed] [Google Scholar]
- Carbonero F, Benefiel AC, Gaskins HR. Contributions of the microbial hydrogen economy on colonic homeostasis. Nat Rev Gastroenterol Hepatol. 2012;9:504–518. doi: 10.1038/nrgastro.2012.85. [DOI] [PubMed] [Google Scholar]
- Chao A. Estimating the population size for capture-recapture data with unequal catchability. Biometrics. 1987;11:265–270. [PubMed] [Google Scholar]
- Chaves OM, Stoner KE, Arroyo-Rodriguez V. Differences in diet between spider monkey groups living in forest fragments and continuous forest in Mexico. Biotropica. 2012;44:105–113. [Google Scholar]
- Clark KR, Gorley RN. PRIMER v6: User Manual/Tutorial. PRIMER-E, Plymouth 2006.
- Costello EK, Stagaman K, Dethlefsen L, Bohannan BJ, Relman DA. An application of ecological theory toward an understanding of the human microbiome. Science. 2012;336:1255–1262. doi: 10.1126/science.1224203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dethlefsen L, McFall-Ngai M, Relman DA. An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature. 2007;449:811–818. doi: 10.1038/nature06245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Fiore A, Campbell C.2007The Atelines: variations in ecology, behavior, and social organizationIn: Campbell C, Fuentes A, MacKinnon KC, Panger M, Bearder SK, (eds)Primates in Perspective Oxford University Press: New York; 155–186. [Google Scholar]
- Donohoe DR, Garge N, Zhang X, Sun W, O'Connell TM, Bunger MK, et al. The microbiome and butyrate regulate energy metabolism and autophagy in the mammalian colon. Cell Metab. 2011;13:517–526. doi: 10.1016/j.cmet.2011.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert KA, Hahn NE, Genz AK, Kitchen DM, Stuart MD, Averbeck GA, Stromberg BE, et al. Coprological surveys of Alouatta pigra at two sites in Belize. Int J Primatol. 2006;27:227–238. [Google Scholar]
- Estrada AE, Coates-Estrada R. Tropical rain forest fragmentation and wild populations of primates at Los Tuxtlas. Int J Primatol. 1996;5:759–783. [Google Scholar]
- Estrada AE, Coates-Estrada R. Use of leaf resources by howling monkeys (Alouatta palliata) and leaf-cutting ants (Atta cephalotes) in the tropical rain forest of Los Tuxtlas, Mexico. Am J Primatol. 2005;10:51–66. doi: 10.1002/ajp.1350100106. [DOI] [PubMed] [Google Scholar]
- Flaherty EA, Ben-David M, Smith WP. Diet and food availability: Implications for foraging and dispersal of prince of wales northern flying squirrels across managed landscapes. J Mammal. 2010;91:79–91. [Google Scholar]
- Flint HJ, Bayer EA. Plant cell wall breakdown by anaerobic microorganisms from the mammalian digestive tract. Ann NY Acad Sci. 2008;1125:280–288. doi: 10.1196/annals.1419.022. [DOI] [PubMed] [Google Scholar]
- Flint HJ, Duncan SH, Louis P.2011Impact of intestinal microbial communities upon healthIn: Rosenberg E, Gophna U, (eds)Beneficial Microorganisms in Multicellular Life Forms Springer: Berlin; 243–252. [Google Scholar]
- Gagen EJ, Denman SE, Padmanabha J, Zadbuke S, Al Jassim R, Morrison M, et al. Functional gene analysis suggests different acetogen populations in the bovine rumen and tammar wallaby forestomach. Appl Environ Microbiol. 2010;76:7785–7795. doi: 10.1128/AEM.01679-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grootaert C, Van de Wiele T, Van Roosbroeck I, Possemiers S, Vercoutter-Edouart AS, Verstraete W, et al. Bacterial monocultures, propionate, butyrate and H2O2 modulate the expression, secretion and structure of the fasting-induced adipose factor in gut epithelial cell lines. Environ Microbiol. 2011;13:1778–1789. doi: 10.1111/j.1462-2920.2011.02482.x. [DOI] [PubMed] [Google Scholar]
- Hales BA, Edwards C, Ritchie DA, Hall G, Pickup RW, Saunders JR. Isolation and identification of methanogen-specific DNA from blanket bog peat by PCR amplification of sequence analysis. Appl Environ Microbiol. 1996;62:668–675. doi: 10.1128/aem.62.2.668-675.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336:1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Xu C, Zhou X, He B, Wu L, Cui B, et al. 2010Sodium butyrate protects against myocardial ischemia and reperfusion injury by inhibiting high mobility group box 1 protein in rats Biomed Pharmacothere-pub ahead of print 25 Sept 2010; doi: 10.1016/j.biopha2010.09.005 [DOI] [PubMed]
- IUCN 2012. IUCN Red List of Threatened Species. Version 2012.2 www.iucnredlist.org Downloaded on 14 November 2012.
- Kamler JF, Ballard WB, Wallace MC, Gipson PS. Diets of swift foxes (Vulpes velox) in continuous and fragmented prairie in Northwestern Texas. Southwest Nat. 2007;52:504–510. [Google Scholar]
- Kelaita M, Dias PAD, Aguilar Cucurachi MS, Canales-Espinosa D, Cortes-Ortiz L. Impact of intrasexual selection on sexual dimorphism and testes size in the Mexican howler monkeys Alouatta palliata and A. pigra. Am J Phys Anthr. 2011;146:179–187. doi: 10.1002/ajpa.21559. [DOI] [PubMed] [Google Scholar]
- Lambert JE. Primate digestion: interactions among anatomy, physiology, and feeding ecology. Evol Anthr. 1998;7:8–20. [Google Scholar]
- Lambert JE, Fellner V. In vitro fermentation of dietary carbohydrate consumed by african apes and monkeys: preliminary results for interpreting microbial and digestive strategy. Int J Primatol. 2012;33:263–281. [Google Scholar]
- Leaphart AB, Lovell CR. Recovery and analysis of formyltetrahydrofolate synthetase gene sequences from natural populations of acetogenic bacteria. Appl Environ Microbiol. 2001;67:1392–1395. doi: 10.1128/AEM.67.3.1392-1395.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leloup J, Loy A, Knab NJ, Borowski C, Wagner M, Jorgensen BB. Diversity and abundance of sulfate-reducing microorganisms in the sulfate and methane zones of a marine sediment, Black Sea. Environ Microbiol. 2007;9:131–142. doi: 10.1111/j.1462-2920.2006.01122.x. [DOI] [PubMed] [Google Scholar]
- Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS, et al. Evolution of mammals and their gut microbes. Science. 2008;320:1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- Lindborg R, Eriksson O. Historical landscape connectivity affects present plant species diversity. Ecology. 2004;85:1840–1845. [Google Scholar]
- Louis P, Scott KP, Duncan P, Flint HJ. Understanding the effects of diet on bacterial metabolism in the large intestine. J Appl Microbiol. 2007;102:1197–1208. doi: 10.1111/j.1365-2672.2007.03322.x. [DOI] [PubMed] [Google Scholar]
- Louis P, Young P, Holtrop G, Flint HJ. Diversity of human colonic butyrate-producing bacteria revealed by analysis of the butyryl-CoA: acetate coA-transferase gene. Environ Microbiol. 2010;12:304–314. doi: 10.1111/j.1462-2920.2009.02066.x. [DOI] [PubMed] [Google Scholar]
- Macpherson AJ, McCoy KD, Johansen FE, Brandtzaeg P. The immune geography of IgA induction and function. Nat Rev. 2008;1:11–22. doi: 10.1038/mi.2007.6. [DOI] [PubMed] [Google Scholar]
- Martinez-Mota R, Valdespino C, Sanchez-Ramos MA, Serio-Silva JC. Effects of forest fragmention on the physiological stress of black howler monkeys. Anim Cons. 2007;10:374–379. [Google Scholar]
- Medani M, Collins D, Docherty NG, Baird AW, O'Connell PR, Winter DC. Emerging role of hydrogen sulfide in colonic physiology and pathophysiology. Inflammatory Bowel Dis. 2011;17:1620–1625. doi: 10.1002/ibd.21528. [DOI] [PubMed] [Google Scholar]
- Messaoudi M, Lalonde R, Violle N, Javelot H, Desor D, Nejdi A, et al. Assessment of psychotropic-like properties of a probiotic fomulation (Lactobacillus helveticus R0052 and Bifidobacterium longum R0175) in rats and human subjects. Br J Nutr. 2010;105:755–764. doi: 10.1017/S0007114510004319. [DOI] [PubMed] [Google Scholar]
- Milton K. Factors influencing leaf choice by howler monkeys: a test of some hypotheses of food selection by generalist herbivores. Am Nat. 1979;114:362–378. [Google Scholar]
- Milton K. The Foraging Strategy of Howler Monkeys. Columbia University Press: New York; 1980. [Google Scholar]
- Milton K, McBee RH. Rates of fermentative digestion in the howler monkey, Alouatta palliata (Primates: Ceboidea) Comp Biochem Physiol. 1983;74A:29–31. doi: 10.1016/0300-9629(83)90706-5. [DOI] [PubMed] [Google Scholar]
- Nava GM, Carbonero F, Croix JA, Greenberg E, Gaskins HR. Abundance and diversity of mucosa-associated hydrogenotrophic microbes in the healthy human colon. ISME J. 2012;6:57–70. doi: 10.1038/ismej.2011.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrocki EP, Kolbe DL, Eddy SR. Infernal 1.0: Inference of RNA alignments. Bioinforma. 2009;25:1335–1337. doi: 10.1093/bioinformatics/btp157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neish AS. Microbes in gastrointestinal health and disease. Gastroenterol. 2009;136:65–80. doi: 10.1053/j.gastro.2008.10.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien TG, Kinnaird M, Dierenfeld ES, Conklin-Brittain NL, Wrangham RW, Silver SC. What's so special about figs. Nature. 1998;392:668. [Google Scholar]
- Pruesse EC, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, et al. SILVA: A comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawluszko AA, Krokowicz P, Jagodzinski PP. Butyrate induces expression of 17B-hydroxysteroid dehydrogenase type 1 in HT29 and SW707 colorectal cancer cells. DNA Cell Biol. 2011;30:661–669. doi: 10.1089/dna.2010.1192. [DOI] [PubMed] [Google Scholar]
- Rey FE, Faith JJ, Bain J, Muehlbauer MJ, Stevens RD, Newgard CB, et al. Dissecting the in vivo metabolic potential of two human gut acetogens. J Biol Chem. 2010;285:22082–22090. doi: 10.1074/jbc.M110.117713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld JS. Functional redundancy in ecology and conservation. Oikos. 2002;98:156. [Google Scholar]
- Russell JB, Baldwin RL. Comparison of maintenance energy expenditures and growth yields among several rumen bacteria grown on continuous culture. Appl Environ Microbiol. 1979;37:537–543. doi: 10.1128/aem.37.3.537-543.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salyers AA. Energy sources of major intestinal fermentative anaerobes. Am J Clin Nutr. 1979;32:158–163. doi: 10.1093/ajcn/32.1.158. [DOI] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt O, Wust PK, Hellmuth S, Borst K, Horn MA, Drake HL. Novel [NiFe]- and [FeFe]-hydrogenase gene transcripts indicative of active facultative aerobes and obligate anaerobes in earthworm gut contents. Appl Environ Microbiol. 2011;77:5842–5850. doi: 10.1128/AEM.05432-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekirov I, Russel SI, Antunes CM, Finlay BB. Gut microbiota in health and disease. Physiol Rev. 2010;90:859–904. doi: 10.1152/physrev.00045.2009. [DOI] [PubMed] [Google Scholar]
- Sharma B, Singh N. Attenuation of vascular dementia by sodium butyrate in streptozotocin diabetic rats. Psychopharm. 2011;215:677–687. doi: 10.1007/s00213-011-2164-0. [DOI] [PubMed] [Google Scholar]
- Sipos M, Jeraldo P, Chia N, Qu A, Dhillon AS, Konkel ME, et al. Robust computational analysis of rRNA hypervariable tag datasets. PLoS One. 2010;5:e15220. doi: 10.1371/journal.pone.0015220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoner KE, Gonzalez Di Pierro A.2006Intestinal parasitic infections in Alouatta pigra in tropical rainforest in Lacandona, Chiapas, Mexico: implications for behavioral ecology and conservationIn Estrada AE, Garber PA, Pavelka MS, Luecke L, (eds)New perspectives in the study of Mesoamerican primates: Distribution, ecology, and behavior Springer: New York; 215–240. [Google Scholar]
- Trejo-Macias G, Estrada AE. Risk factors connected to gastrointestinal parasites in mantled Alouatta palliata mexicana and black howler monkeys Alouatta pigra living in continuous and in fragmented rainforests in Mexico. Curr Zoo. 2012;58:375–383. [Google Scholar]
- Trejo-Macias G, Estrada AE, Mosqueda Cabrera MA. Survey of helminth parasites in populations of Alouatta palliata mexicana and A. pigra in continuous and in fragmented habitat in Southern Mexico. Int J Primatol. 2007;28:931–945. [Google Scholar]
- Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon HA. The effect of diet on the human gut microbiome: A metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med. 2009;1:6ra14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vellend M. Habitat loss inhibits recovery of plant diversity as forests regrow. Ecol. 2003;84:1158–1164. [Google Scholar]
- Vitazkova SK, Wade SE. Effects of ecology on the gastrointestinal parasites of Alouatta pigra. Int J Primatol. 2007;28:1327–1343. [Google Scholar]
- Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildirim S, Yeoman CJ, Sipos M, Torralba M, Wilson BA, Goldberg TL, et al. Characterization of the fecal microbiome from non-human wild primates reveals species specific microbial communities. PLoS One. 2010;5:e13963. doi: 10.1371/journal.pone.0013963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zunino GE, Kowalewski MM, Oklander LI, Gonzalez V. Habitat fragmentation and population size of the black and gold howler monkey (Alouatta caraya) in a semideciduous forest in Northern Argentina. Am J Primatol. 2007;69:966–975. doi: 10.1002/ajp.20389. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.