Abstract
Few studies have directly measured sulfate reduction at hydrothermal vents, and relatively little is known about how environmental or ecological factors influence rates of sulfate reduction in vent environments. A better understanding of microbially mediated sulfate reduction in hydrothermal vent ecosystems may be achieved by integrating ecological and geochemical data with metabolic rate measurements. Here we present rates of microbially mediated sulfate reduction from three distinct hydrothermal vents in the Middle Valley vent field along the Juan de Fuca Ridge, as well as assessments of bacterial and archaeal diversity, estimates of total biomass and the abundance of functional genes related to sulfate reduction, and in situ geochemistry. Maximum rates of sulfate reduction occurred at 90 °C in all three deposits. Pyrosequencing and functional gene abundance data revealed differences in both biomass and community composition among sites, including differences in the abundance of known sulfate-reducing bacteria. The abundance of sequences for Thermodesulfovibro-like organisms and higher sulfate reduction rates at elevated temperatures suggests that Thermodesulfovibro-like organisms may have a role in sulfate reduction in warmer environments. The rates of sulfate reduction presented here suggest that—within anaerobic niches of hydrothermal deposits—heterotrophic sulfate reduction may be quite common and might contribute substantially to secondary productivity, underscoring the potential role of this process in both sulfur and carbon cycling at vents.
Keywords: hydrothermal vent, microbial ecology, primary productivity, sulfate reduction
Introduction
Deep-sea hydrothermal vent ecosystems are complex dynamic habitats characterized by steep gradients in temperature and geochemistry (Jannasch and Mottl, 1985). In these habitats, as hot hydrothermal fluid mixes with cold seawater, the precipitation of minerals creates large and complex hydrothermal chimney deposits. Within these permeable mineral structures, the continued mixing of chemically reduced, vent-derived fluids and oxidized seawater provides favorable conditions that support the growth of endolithic microbial communities (Schrenk et al., 2003).
Sulfide oxidation is considered to be one of the most important microbial chemosynthetic pathways at ridge ecosystems, as evidenced by the ubiquity of sulfide-oxidizing Epsilon- and Gammaproteobacteria at ridge environments (Nakagawa et al., 2004, 2005; Campbell et al., 2006; Huber et al., 2007; Nakagawa and Takai, 2008). To date, significantly less attention has been paid to the distribution and magnitude of sulfate reduction at vents, though sulfate-reducing bacteria and archaea have frequently been isolated from deep-sea hydrothermal environments (Jannasch et al., 1988; Blöchl et al., 1997; Alazard et al., 2003; Audiffrin et al., 2003; Houghton et al., 2007). Moreover, analyses of functional genes that express key proteins required for sulfate reduction suggest there is a high diversity of sulfate-reducing organisms at vents, higher than predicted via 16S ribosomal RNA (rRNA) gene analyses alone (Nakagawa et al., 2004; Nercessian et al., 2005).
From a biogeochemical and bioenergetic perspective, both sulfide oxidation and sulfate reduction would be favored at hydrothermal vents, although to varying degrees as a function of environmental chemistry. Sulfide oxidation is most favorable when coupled to oxygen or nitrate as an electron acceptor (Amend and Shock, 2001). Around vents, sulfide is typically in micromolar to millimolar concentrations (Butterfield et al., 1994a, 1994b), while oxygen and nitrate are around 110 and 40 μℳ respectively (Johnson et al., 1986). In contrast, sulfate reduction is highly favored in anoxic niches at vents, as it is in other marine anaerobic environments (Muyzer and Stams, 2008). Here, as in most marine systems, sulfate is abundant at 28 mℳ (Dittmar, 1884), two to three orders of magnitude higher than oxygen. At vents, sulfate reduction would occur in regimes where seawater-derived sulfate is still present but oxygen is absent as, for example, within hydrothermal vent deposits. Sulfate-reducing micro-organisms commonly use hydrogen and/or dissolved organic matter as electron donors, both of which are found within hydrothermal fluids (Cruse and Seewald, 2006; Lang et al., 2006). Sulfate reduction—as a function of its extent and magnitude—could readily influence the cycling of sulfur and sulfur isotopes, as well as carbon, within hydrothermal environments.
To date, studies have quantified rates of sulfate reduction in hydrothermal-influenced sediments (Jorgensen et al., 1992; Elsgaard et al., 1994a, 1994b, 1995; Weber and Jorgensen, 2002; Kallmeyer and Boetius, 2004) and isolated vent micro-organisms (Hoek et al., 2003). In contrast to the numerous studies of sulfate reduction in marine sediments (Canfield, 1989), studies of sulfate reduction in hydrothermal deposits are few (Bonch-Osmolovskaya et al., 2011), due in part to the challenges associated with sampling and studying the heterogeneous and consolidated sulfide deposits typical of hydrothermal vent chimneys.
Here we present rates of microbially-mediated sulfate reduction from three distinct hydrothermal mineral deposits from active hydrothermal ‘chimneys' found in the Middle Valley field along the Juan de Fuca Ridge, as well as assessments of bacterial and archaeal diversity, estimates of total biomass and the abundance of functional genes related to sulfate reduction, and in situ geochemistry. These analyses further our understanding of sulfate reduction (including rates, diversity, and distribution of known sulfate-reducing microbes) in vent ecosystems. Moreover, they underscore the potential role of heterotrophic sulfate reduction in hydrothermal systems and constrain its potential influence on both sulfur and carbon cycling.
Materials and methods
Geologic setting and sampling of hydrothermal deposits
Middle Valley (48°27′N, 128°59′ W) is an intermediate spreading, axial rift valley located along the Endeavour Segment of the Juan de Fuca Ridge in the Northwest Pacific ocean. Layers of continentally-derived sediments cover Middle Valley, although active hydrothermal vents remain prominent above the sediments. Hydrothermal deposits were collected from three active hydrothermal spires during dive 4625 with the HOV Alvin (R/V Atlantis expedition AT15-67, July 2010) and brought to the surface in a sealed, temperature-insulated polyethylene box. Samples were recovered from actively venting sulfide deposits at Needles (48.45778, −128.709, 2412.212 m, Tmax=123 °C), Dead Dog (48.45603,−128.71, 2405.268 m, Tmax=261 °C), and Chowder Hill (48.455543, −128.709, 2398.257 m, Tmax =261 °C) vents. Once on board ship, samples were directly transferred to sterile anaerobic seawater and handled/processed using appropriate sterile microbiological techniques. Subsamples were immediately transferred to gastight jars (Freund Container Inc., Lisle, IL, USA), filled with sterile anaerobic seawater containing 2 mℳ sodium sulfide at pH 6, and stored at 4 °C. On return to the laboratory, all samples were maintained in anaerobic seawater (0.2 μm filter-sterilized before use) supplemented with 2 mℳ ΣH2S (defined as the sum of H2S, HS− and S2−) and adjusted to pH 6. The vent-like media was replenished every 8 to 12 weeks, and all samples were kept in the dark at 4 °C before incubation.
Vent fluid volatile geochemistry via in situ mass spectrometry
In situ concentrations of dissolved volatiles (e.g., H2S, H2, CO2, O2 and others) were measured at each site with an in situ mass spectrometer as previously described (Wankel et al., 2011). Briefly, dissolved volatiles were quantified in situ by sampling vent effluent for up to 10 min, until partial pressures reached steady state (data were monitored in real-time within the submersible). Concentrations were determined from empirically derived calibrations and validated by comparison with discrete samples collected using titanium gastight samplers.
Measuring sulfate reduction rates (SRRs)
Hydrothermal deposits were homogenized in a commercial blender (Xtreme blender, Waring Inc., Toringtor, CT, USA) under a nitrogen atmosphere. Anaerobic homogenization was designed to minimize fine-scale geochemical and microbial heterogeneity and facilitate more accurate experimental replication. Hydrothermal homogenate (made up of both mineral deposit and interstitial fluid) was aliquoted volumetrically (7.5 mL, ca 29 g wet weight and ca 20 g dry weight) into Balch tubes in an anaerobic chamber. The tubes were supplemented with 15 mL of sterile artificial vent fluid media designed to mimic the geochemical composition of fluids within the pores of a sulfide deposit (pH 6, 14 mℳ SO42−, 2.3 mℳ NaHCO3, 1 mℳ H2S, and 10 μℳ each of pyruvate, citrate, formate, acetate, and lactate). Organic acid concentrations are comparable to those measured in situ along the Juan de Fuca Ridge (Lang et al., 2006). Sufficient 35SO42− was added to achieve 555 kBq (15 μCi) of activity. Owing to technical difficulties with post processing methodology, shipboard incubations using fresh material were not successful. The data presented here were generated using samples that had been maintained in sulfidic vent-like effluent (as described above) for 1 year. Samples were incubated anaerobically for 7 days at 4 °C, 30 °C, 40 °C, 50 °C, 60 °C, 80 °C, and 90 °C. Controls for sulfate reduction consisted of samples amended with 28 mℳ molybdate, a competitive inhibitor of sulfate reduction (Saleh et al., 1964; Newport and Nedwell, 1988). Six biological replicates were run for each treatment, along with three biological replicates for each control. Reactions were quenched with the injection of 5 mL 25% zinc acetate (which is ∼20-fold more zinc than the maximum sulfide concentration), and all samples were frozen at −20 °C for further analysis.
To determine SRRs, samples were thawed and the supernatant was removed and filtered through a 0.2 μm syringe filter. The crushed deposits that remained in the tube were washed three times with deionized water to remove any remaining sulfate. One gram (wet weight) of crushed deposit was analyzed via chromium distillation (see Supplementary Methods), and SRRs were calculated as in Fossing and Jorgensen (1989) using the following calculation:
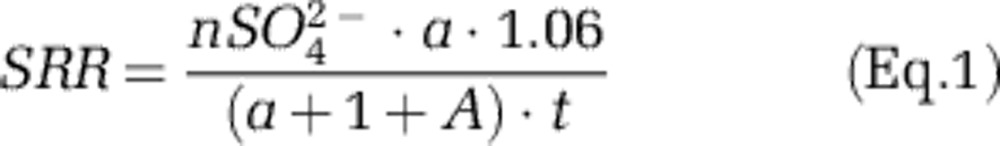 |
Where nSO42− is the quantity (in moles) of sulfate added to each incubation (14 mℳ × 15 mL=210 μmol), a is the activity (d.p.m.) of the trapped sulfide, 1.06 is the fractionation factor between the sulfide and sulfate pools, A is the activity of the sulfate pool at the completion of the incubation, and t is the incubation time (days). The rates are presented in units of nmol S g−1 day−1.
DNA extraction
Immediately before conducting the rate experiments, a subsample of the homogenized hydrothermal deposit was removed and frozen at −80 °C for molecular analysis. This approach ensures that the resulting sequences best represent those communities responsible for the observed activity. DNA was extracted from this crushed deposit sample with a protocol modified from (Santelli et al., 2008). Subsamples were washed with 0.1 𝒩 HCl, followed by two rinses with a sterile solution containing 10 mℳ Tris (pH 8.0) and 50 mℳ EDTA. A known mass of material was added to PowerSoil beadbeating tubes (MoBio Laboratories, Carlsbad, CA, USA), incubated at 70 °C for 10 min, and then amended with 200 ng of poly-A. Subsamples were subjected to beadbeating, followed by three freeze-thaw cycles to further lyse cells. Nucleic acids were extracted using hot phenol (60 °C for 3 min), followed by two chloroform:isoamyl alcohol phase extractions and a final precipitation in ethanol. DNA was resuspended in TE (pH 8.0) and quantified using the Qubit fluorometer (Life Technologies, Grand Island, NY, USA).
Enumeration of gene abundance via quantitative PCR
Quantitative PCR (qPCR) was used to determine the abundance of bacterial and archaeal 16S rRNA genes. To provide additional constraints on the abundance of sulfate-reducing microbes, qPCR was also used to enumerate (A) the adenosine 5′-phosphosulfate reductase (aprA) gene using primers that target both sulfate-reducing bacteria and archaea (Christophersen et al., 2011); (B) bacterial dissimilatory sulfite reductase (dsrA) using primers that target bacteria (Kondo et al., 2004); and (C) Deltaproteobacteria via primers that target their 16S rRNA genes (Stults et al., 2001). Quantification was performed in triplicate with the Stratagene MX3005p qPCR System (Agilent Technologies, Santa Clara, CA, USA) using the Perfecta SYBR FastMix with low ROX (20 μl reactions, Quanta Biosciences, Gaitherburg, MD, USA), specific primers and annealing temperatures (Table 1), and 10 ng of template genomic DNA. The thermal cycling conditions for all assays was 94 °C for 10 min, 35 cycles of 94 °C for 1 min, the annealing temperature for 1 min (Table 1), extension at 72 °C for 30 s, and fluorescence read after 10 s at 80 °C. Following amplification, dissociation curves were determined across a temperature range of 55–95 °C. Ct values for each well were calculated using the manufacturer's software. Plasmids containing bacterial and archaeal 16S rRNA and functional gene inserts (amplified from Arcobacter nitrofigulis (ATCC 33309), Methanosarcina acidovorans, and Desulfovibrio vulgaris Hildenborough (ATCC 29579/NCIMB 8303/ AE017285, respectively) were used as standards for calibration (see Supplementary Methods for more detail).
Table 1. Primers used for the enumeration of 16S rRNA and sulfate reduction functional genes.
| Target gene | Forward primer | Conc (nℳ) | T (°C)a | Reverse primer | Conc (nℳ | T (°C)a | Reference |
|---|---|---|---|---|---|---|---|
| Bacterial 16S | Bact1369F- 5′-CGGTGAATACGTTCYCGG-3′ | 1000 | 59 | Prok1541R- 5′-AAGGAGGTGATCC RGCCGCA-3′ | 1000 | 59 | Suzuki et al. (2001) |
| Bacterial 16Sb | Gray28F -5′-GAGTTTGATCNTGGCTCAG-3′ | 500 | 54 | Gray519R- 5′-GTNTTACNGCGGCKGCTG-3′ | 500 | 54 | Modified from Frias-lopez et al. (2002); modified from Manefield et al. (2002) |
| Archaeal 16S | Arch1-1369F- 5′-CGGTGAATACGTCCCTGC-3′ Arch2- 1369F-5′-CGGTGAATATGCCCCTGC-3′ | 500 (1:1 Mix) | 59 | Prok1541R- 5′-AAGGAGGTGATCC RGCCGCA-3′ | 1000 | 59 | Suzuki et al. (2001) |
| Archaeal 16Sb | Arch349F- 5′-GYGCASCAGKCGMGAAW-3′ | 500 | 54 | Arch806R- 5′-GGACTACVSGGGTATCTAAT | 500 | 54 | Takai and Horikoshi (2000) |
| δ-Proteobacteriac | Delta361GF- 5′-AAGCCTGACGCASCAA-3′ | 600 | 55 | Delta685R- ATCTACGGATTTCACTCCTACA-3′ | 600 | 55 | Stults et al. (2001) |
| Disimmilatoryd sulfite reductase | DSR1-F+-5′-ATCGGNCARGCNTTYCCNTT-3′ | 400 | 58 | DSR-R- 5′-GTGGMRCCGTGCAKRTTGG-3′ | 600 | 58 | Kondo et al. (2004) |
| Disimmilatory sulfite reductasee | DSR2060F 5′-CAACATCGTYCATACMCAGGG-3′ | 500 | 50 | DSR4R- 5′-GTGTAGCAGTTACCGCA-3′ | 500 | 50 | Wagner et al. (1998); Oakley et al. (2011) |
| Adenosine 5′-phosphosulfate reductase | aps3F 5′-TGGCAGATCATGWTYAAYGG-3′ | 400 | 55 | aps2R- 5′-GCGCCGTAACCRTCYTTRAA-3′ | 400 | 55 | Christophersen et al. (2011) |
Annealing temperature.
Primers used for 454-pytrotag sequencing, all other primers used for qPCR.
Primers Delta361F and Delta685R allowed for quantification of iron- and sulfate-reducing genera within the δ-Proteobacteria including Geobacter, Pelobacter (including fermentative species), Desulfovibrio, Desulfomicrobium, Desulfuromusa, and Desulfuromonas (including dissimilatory S reducers) (Stults et al., 2001).
Although these primers amplify bacterial dsrA gene sequences, they may not detect some gram-positive species and cannot detect select thermophilic bacterial and archaeal sulfate-reducing lineages (Kondo et al., 2004).
Targets both bacterial and archaeal aprA gene (Christophersen et al., 2011).
Sequencing and phylogenetic analysis via 454 pyrosequencing
DNA samples were sequenced using 454 pyrotag methods similar to those described previously (Dowd et al., 2008). All samples were sequenced at the Research and Testing Laboratory (Lubbock, TX, USA) using a 454FLX instrument (Roche Inc., Indianapolis, IN, USA) with Titanium reagents. The resulting bacterial and archaeal 16S rRNA (bacterial V1–V3 and archaeal V3–V4 of the 16S rRNA genes; primers are shown in Table 1), as well as drsB sequences, were analyzed via Mothur (Schloss et al., 2009). Sequences were trimmed, quality checked, aligned to the SILVA-compatible alignment database reference alignment (dsrB gene data sets were aligned to a dsrB gene database generated from the Ribosomal Database Project (RDP)), analyzed for chimeras, classified against the Greengenes99 database, and clustered into operational taxonomic units (OTUs; see Supplementary Methods for more detail). Rarefaction curves were used to examine the number of OTUs as a function of sampling depth. Alpha diversity was assessed by generating values from the Chao1 richness estimator and the inverse Simpson diversity index.
Sequence accession numbers
The 16S rRNA and dsrB gene sequences reported in this study have been submitted to the Sequence Read Archive under the accession numbers SRX154520 through SRX154528.
Results
Physical and geochemical characteristics of the study sites
The hydrothermal deposits sampled from Middle Valley were all relatively friable and were composed predominantly of anhydrite (CaSO4, M Tivey, personal communication). Chowder Hill and Dead Dog had the highest observed venting fluid temperatures (measured in situ at 261 °C), followed by Needles (123 °C). In situ measurements of dissolved hydrogen sulfide (H2S) revealed significant differences in hydrothermal fluid composition among hydrothermal deposits. Unfortunately the inline pH probe with the in situ mass spectrometer malfunctioned during the dive. Using previously reported pH values (Butterfield et al., 1994a, 1994b), Chowder Hill would have the highest in situ measurement of total sulfide (3.9 mℳ), followed by Dead Dog (2.2 mℳ) and Needles (0.59 mℳ) (Table 2). These concentrations are within the same magnitude of previously reported H2S in focused vent fluids at Middle Valley (Butterfield et al., 1994a). Chowder Hill did exhibit the highest in situ concentration of hydrogen (1.86 mℳ), followed by Dead Dog (1.66 mℳ) and Needles (∼1.42 mℳ). These values are also consistent with previous studies (Cruse and Seewald, 2006), as well as gastight samples collected and analyzed shipboard (M Lilley, personal communication).
Table 2. In situ ∑H2S measurements compensated for a range of hydrothermally relevant pH.
| pH | Chowder Hill H2S (mℳ) | Dead Dog H2S (mℳ) | Needles H2S (mℳ) |
|---|---|---|---|
| Not pH compensateda | 3.91 | 2.10 | 0.660 |
| 3.5 | 3.9 | 2.1 | 0.55 |
| 4.0 | 3.9 | 2.1 | 0.55 |
| 4.5 | 3.9 | 2.1 | 0.55 |
| 5.0 | 3.9b | 2.1 | 0.56 |
| 5.5 | 4.0 | 2.2b | 0.56b |
| 6.0 | 4.2 | 2.3 | 0.59 |
| 6.5 | 4.8 | 2.6 | 0.68 |
| 7.0 | 6.8 | 3.7 | 0.96 |
| 7.5 | 13 | 7.1 | 1.9 |
| 8.0 | 33 | 18 | 4.7 |
| 8.2 | 50 | 27 | 7.1 |
Values represent the median values of the 10 highest sampling points.
The first row of values refers only to (H2S) and not ∑H2S.
Total sulfide calculation at the most environmentally relevant pH (Butterfield et al., 1994).
Sulfate reduction rates
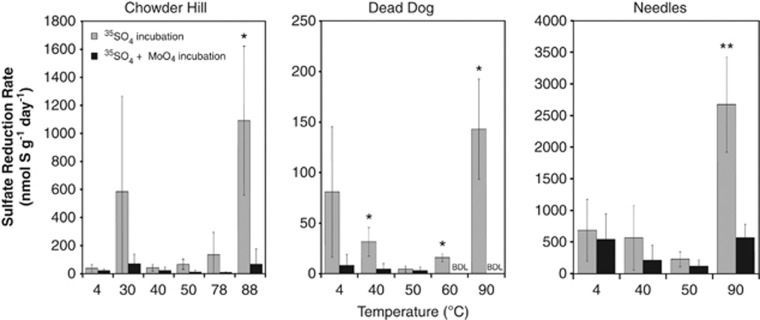
Sulfate reduction was observed in all samples at temperatures ranging from 4 °C to 90 °C (Figure 1). Maximal rates of sulfate reduction were observed between 88 °C and 90 °C (2670 nmol g−1 day−1 at Needles, 1090 nmol g−1 day−1 at Chowder Hill and 142 nmol g−1 day−1 at Dead Dog; Figure 1). Notably, the highest SRRs were observed in Needles samples, which were ∼20-fold higher than those observed at Dead Dog, and ∼2-fold greater than at Chowder Hill. Many of the rates exhibit large deviations because of the high variability among the biological replicates, most likely due to persistent mineralogical and microbiological heterogeneity across incubations, even after homogenization. Sulfate reduction was also observed in molybdate amended experiments, although we suspect that molybdate was scavenged by minerals that attenuated the effect of the inhibitor as has been previously observed in metal-rich environments (Bostick et al., 2003; Xu et al., 2006).
Figure 1.
Temperature-dependent sulfate reduction rates in three hydrothermal deposits (Chowder Hill, Dead Dog, and Needles) recovered from Middle Valley. Slurried samples were incubated for 7 days with ( ) or without (
) or without ( ) the addition of 28 mℳ molybdate (a competitive inhibitor for sulfate reduction). Error bars represent 1 s.d. from the average. Statistical significance from the controls (Wilcox–Mann–Whitney) are shown as *P<0.1, **P<0.05. Scintillation measurements of 0 c.p.m. for radioactive sulfide (Zn35S) were considered below the detection limit (BDL).
) the addition of 28 mℳ molybdate (a competitive inhibitor for sulfate reduction). Error bars represent 1 s.d. from the average. Statistical significance from the controls (Wilcox–Mann–Whitney) are shown as *P<0.1, **P<0.05. Scintillation measurements of 0 c.p.m. for radioactive sulfide (Zn35S) were considered below the detection limit (BDL).
Quantification of taxonomic and functional genes
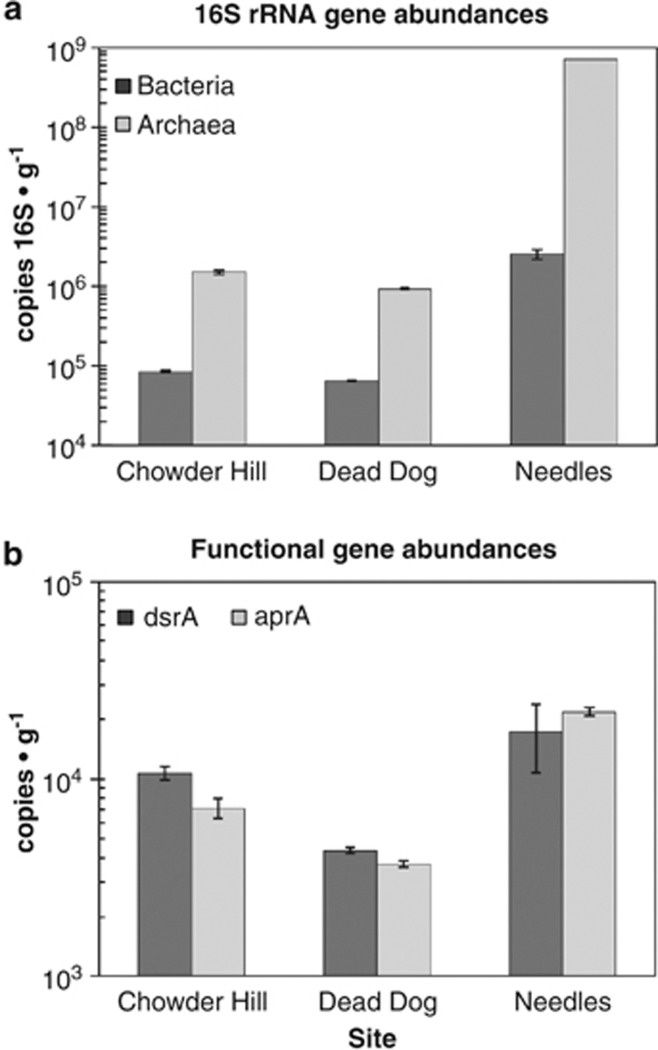
Microbial density (as estimated by 16S rRNA gene copies g−1 mineral) was greatest at Needles and lowest at Dead Dog. Microbial communities at each site were dominated by archaea (Figure 2a), with Needles showing the highest ratio of archaea to bacteria (227:1 as compared with 14:1 at Dead Dog or 17.5:1 at Chowder Hill). Assuming an average of 4.19 copies of 16S rRNA gene per bacterium and 1.71 copies of 16S rRNA gene per archaeon genome (Klappenbach et al., 2001; Lee et al., 2009), Needles hosts a microbial community of 4.12 × 108 cells g−1 sample, three orders of magnitude higher than Chowder Hill (8.96 × 105 cells g−1 sample) and Dead Dog (5.65 × 105 cells g−1 sample).
Figure 2.
Abundance of (a) 16S rRNA genes and (b) functional gene markers for sulfate reduction across three massive sulfide deposits. Samples of hydrothermal sulfide material were collected from each of the three vent sites, frozen on return to the surface, and DNA was extracted using described protocols. Bacterial and archaeal 16S rRNA, dsrA, and aprA genes were enumerated by qPCR using published primer sets and normalized to grams of extracted sulfide. Error bars represent 1 s.d.
16S rRNA gene primers specifically targeting ribotypes allied to Desulfovibrio, Desulfomicrobium, Desulfuromusa, and Desulfuromonas were used to enumerate Deltaproteobacteria known to mediate sulfate reduction in many marine systems (Stults et al., 2001). However, given the difficulty in amplifying 16S rRNA genes from deep-sea thermophiles with typical primer sets—because of mismatches with limited sequence representation in GenBank—it is probable that these assays also underestimate abundances in these environments (Teske and Sorensen, 2008). Deltaproteobacterial abundance at Needles was approximately 4.48 × 105 copies g−1 sample (approximately 26% of the entire bacterial population), although none were detected at Chowder Hill or Dead Dog (data not shown). The abundance of both functional genes for sulfate reduction, dsrA and aprA, was greatest at Needles and lowest at Dead Dog (a pattern similar to that seen in the 16S rRNA gene abundance estimates; Figure 2b). If we assume an average of one dsrA gene copy per genome (Klein et al., 2001; Kondo et al., 2004), the proportion of sulfate-reducing bacteria in the bacterial population is only 2.7% in Needles as compared with 28% in Dead Dog and 53% at Chowder Hill.
Microbial diversity
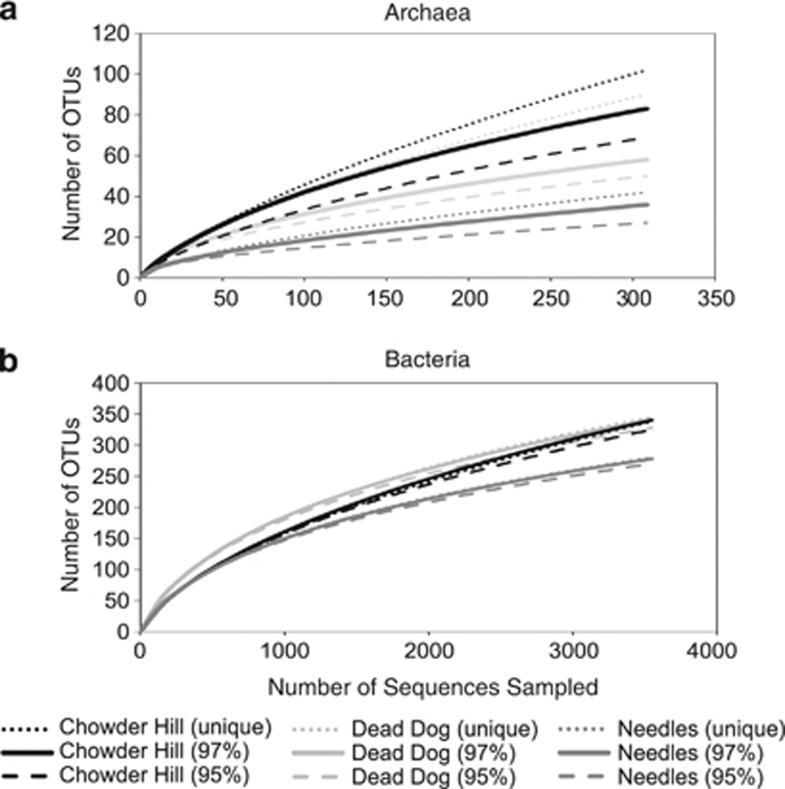
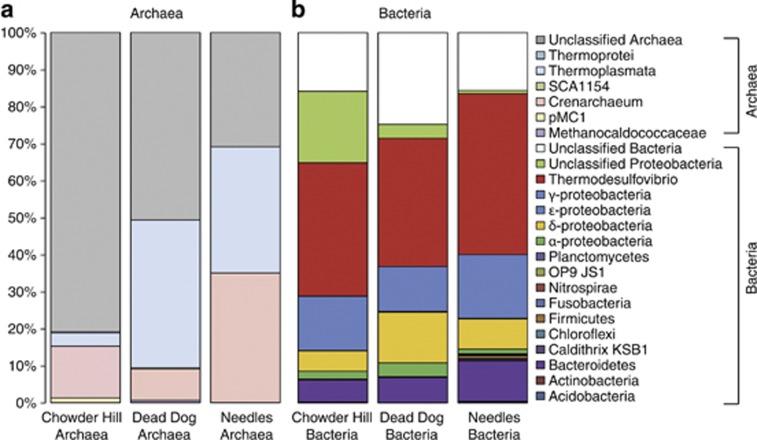
454 pyrotag sequencing, rarefaction analyses, and diversity metrics all revealed measureable differences in microbial community composition among the three hydrothermal deposits (Table 3). Via these assessments, Needles hosts the least diverse assemblage of bacteria and archaea, while Chowder Hill and Dead Dog host communities of comparable diversity. Examination of OTUs at 97%, 95%, and 92% sequence similarity further reveal differences in microbial community composition among the three sites. Among archaea at the 97% level, only two archaeal OTUs (1% of all archaeal OTUs classified) are shared among the hydrothermal deposits. The sequences classified to these OTUs represent 69%, 48%, and 18% of all library sequences from Needles, Dead Dog, and Chowder Hill, respectively. One of these OTUs is allied to the ammonium-oxidizing archaeal Candidatus Cenarchaeum in the phylum Thaumarchaeota, and accounts for 35% of Needles and <5.0% of Dead Dog and Chowder Hill library sequences. The other OTU is allied to a thermophilic sulfur-respiring archaeon within the class Thermoplasmata. Nearly 40% of the archaeal sequences from Dead Dog were allied to this archaeon. Methanogens allied to Methanocaldococus comprised about 1.0% of the total archaeal sequences from Dead Dog and were not represented in the libraries from Chowder Hill or Needles. Most of the archaeal diversity at Chowder Hill (80% of sequences) and Dead Dog (50% of sequences) was unclassified. No sequences allied to true sulfate-reducing archaeal lineages such as Archaeoglobus fugilis or Aciduliprofundus boonei were recovered. However, the potential diversity of thermophilic sulfate-reducing archaea in these samples is likely much greater than suggested here. This may be explained in part by biases in primer binding, sequencing, or even DNA extractions. For example, the archaeal sequencing primers used in this study only target about 34% of the Archaeoglobus-like sequences contained in the RDP database (as assayed by Probe Match; Cole et al., 2005). Furthermore, the primers may miss members of the dominant Thermoplasmatales, as in silico analysis only returns 48% (1715/3558 sequences) of the RDP reported sequences. Together, these archaeal sequencing primers (349F–806R) miss 42% of the total archaeal sequences (67 713/117 373 sequences) in the RDP database. Similar bias has been reported in other studies in the deep-sea and deep-subsurface biotopes (for example, Dhillon et al., 2003, 2005, Teske et al., 2008).
Table 3. 16S rRNA sequence tag and alpha diversity characteristics among sites.
| Sample | Trimmed reads | Sampled reads | OTU (97%) | Coverage | Npshannon | Simpson | Chao |
|---|---|---|---|---|---|---|---|
| Chowder Hill-bacteria | 3997 | 3544 | 340 | 0.948 | 3.30 | 0.168 | 647.23 |
| Dead Dog-bacteria | 3544 | 3544 | 341 | 0.957 | 3.69 | 0.141 | 572.12 |
| Needles-bacteria | 3983 | 3544 | 278 | 0.966 | 3.19 | 0.203 | 458.02 |
| Chowder Hill-archaea | 783 | 308 | 76 | 0.877 | 3.59 | 0.095 | 134.58 |
| Dead Dog-archaea | 308 | 308 | 58 | 0.903 | 3.34 | 0.071 | 97.55 |
| Needles-archaea | 2587 | 308 | 35 | 0.938 | 2.24 | 0.233 | 63.5 |
Abbreviation: OTU, operational taxonomic unit.
Among bacteria at the 97% similarity level, 54 of the classified bacterial OTUs (7.0%) were shared among all hydrothermal deposits and account for 84%, 80%, 71% of the sequences from Chowder Hill, Needles, and Dead Dog, respectively (Figure 4). One of these OTUs accounted for 44%, 36%, and 25% of the sequences from Chowder Hill, Dead Dog, and Needles, respectively. Aligning representative sequences from this OTU via Blastn (Altschul et al., 1997) revealed a best match to Thermodesulfovibrio, an anaerobic, thermophilic, sulfate-reducing bacterium from the phylum Nitrospira (81% identity). Given its abundance, we postulate that it likely contributes substantially to the high thermophilic SRRs. Furthermore, the dsrB gene library was dominated by sequences phylogenetically allied to Thermodesulfovibrio (Supplementary Table S1). Other dominant groups of bacteria include members of the Gammaproteobacteria, Bacteroidetes, Deltaproteobacteria, and Alpha proteobacteria. Via Blastn, most of the unclassified sequences matched to partial 16S rRNA gene sequences from hydrothermal vent fluid communities (Nunoura et al., 2010; Sylvan et al., 2012). Sequences classified as Deltaproteobacteria comprised 5.5%, 8.2%, and 14% of the total population of Chowder Hill, Needles, and Dead Dog, respectively. Although the Dead Dog library had the highest proportion of Deltaproteobacteria, sequences related to known sulfate reducers within the Deltaproteobacteria (Desulfobacteraceae, Desulfobulbus rhabdoformis, and Desulfovibrio) were only found at Needles and comprised 1.1% of the 16S rRNA gene library. The majority of the sequences classified as Deltaproteobacteria in each of the three sites were from one unclassified Deltaproteobacterial OTU and comprised of 4.4%, 5.3%, and 13% of the sequences from Chowder Hill, Needles, and Dead Dog, respectively.
Discussion
SRRs measured in deposits recovered from the Middle Valley vent field reveal the potential for active sulfate reduction within hydrothermal deposits. The magnitude of all measured rates (from 15.7 nmol g−1 day−1 at Dead Dog at 60 °C to 2670 nmol g−1 day−1 at Needles at 90 °C) is comparable in magnitude to those previously observed in hydrothermally influenced sediments (for example, Guaymas basin or Lake Tanganyika; Elsgaard et al., 1994a, 1994b; Weber and Jorgensen, 2002; Kallmeyer and Boetius, 2004), although the availability of organic carbon is markedly higher in those hydrothermal vent sediments, with Guaymas having up to 200-times greater concentrations of organic carbon (Chen et al., 1993; Cruse and Seewald, 2006; Lang et al., 2006). However, these rates are measurably higher than those typically observed in non hydrothermal deep-sea sediments (0.1–10 nmol g−1 day−1, converted here for comparison by assuming an average sediment density of 2 g cm−3; Elsgaard et al., 1994a; Weber and Jorgensen, 2002; Joye et al., 2004). To date, the only other measurement of sulfate reduction from sulfide deposits along the East Pacific Rise exhibited rates comparable to those reported here, (Bonch-Osmolovskaya et al., 2011), but it should be noted that their samples were incubated under a pure H2 atmosphere and likely represented autotrophic sulfate reduction.
Notably, the maximum rates of sulfate reduction in Middle Valley sulfides occurred at 90 °C in all three deposits. This is in contrast to measurements of sulfate reduction in hydrothermal sediments, where the greatest rates are often observed between 40 °C and 70 °C, and more modest rates of sulfate reduction have been reported between 80 °C and 91 °C (Elsgaard et al., 1994a, 1994b; Weber and Jorgensen, 2002). The relatively low or insignificant SRRs between 4 °C and 80 °C suggest that Middle Valley deposits harbor a high proportion of hyperthermophilic sulfate-reducing microbes.
The significant differences in the rates we observed among deposits (Kruskal–Wallis, P<0.0001) are likely due to differences in biomass and the composition of microbial communities. These differences in density and composition are, in turn, a reflection of the availability of relevant substrates and the physicochemical conditions at each site. Indeed, microbial biomass (as estimated by 16S rRNA genes) directly correlates to rates of activity and is likely one of the strongest factors affecting the observed rates of sulfate reduction (Pearson correlation coefficient r=0.879, P<0.0005). Needles had both the highest observed rates as well as the highest cell density (Figures 2 and 3). Of all the deposits sampled, Needles had the lowest venting fluid temperature (123 °C) resulting in the largest zone of microbial habitability. Consistent with this observation, Needles also had the greatest abundance of dsrA and aprA genes per gram, suggesting a larger potential sulfate-reducing community. Here, Deltaproteobacteria allied to Desulfovibrio, Desulfobulbus, Desulfobacteria, and Desulfuromonas accounted for 25.7% of the bacterial community. These clades of Deltaproteobacteria were not observed at Chowder Hill or Dead Dog by either qPCR or pyrosequencing. Cultured representatives from some of these Deltaproteobacterial clades (Desulfovibrio vulgaris and Desulfovibrio desulfuricans) have been shown to reduce sulfate at high rates (ranging from 10 to 1340 nmol min−1 mg−1 protein) with varying electron donors (Cypionka and Konstanz, 1989; Fitz and Cypionka, 1991).
Figure 3.
Rarefaction analysis of archaeal (a) and bacterial (b) sequences from each site at OTU clustering at the 95%, 97% and unique level. All 16S rRNA libraries were randomly sampled down to the smallest sample size, (a) n=308 (Dead Dog), (b) n=3544 (Dead Dog). For both bacteria and archaea, Needles had the least diverse populations.
Thermodesulfovibrio-like organisms dominated the bacterial communities within each hydrothermal deposit (35–44% Figure 4). Thermodesulfovibrio sp. are considered obligately anaerobic, thermophilic bacteria that can reduce sulfate and other sulfur compounds (Garrity and Holt, 2001). In pure cultures, members of this genus are able to link growth with hydrogen and a limited range of organic carbon molecules (formate, pyruvate, and lactate), maintaining optimal growth between 55 °C and 70 °C (Sekiguchi et al., 2008). Needles had a greater proportion of sequences (from pyrosequencing) related to Thermodesulfovibrio-like species than the other two deposits. The combination of sequences related to a thermophilic sulfate-reducing bacteria and higher rates of sulfate reduction at elevated temperatures (90 °C) suggests that Thermodesulfovibrio-like organisms may have a substantial role in sulfate reduction in warmer environments. However, constraining the relative proportion of sulfate reduction by Thermodesulfovibrio-like organisms in these mixed communities was beyond the scope of this study.
Figure 4.
Archaeal (a) and Bacterial (b) taxonomic distribution among the different hydrothermal deposits revealed differences in microbial community composition. Bacterial V1–V3 region and archaeal V3–V4 regions of the 16S were sequenced by 454, analyzed with Mothur, and all libraries were randomly sampled down to the smallest sample size, (a) n=308 (Dead Dog), (b) n=3544 (Dead Dog).
It is unclear why Chowder Hill and Dead Dog exhibited large differences in rates of sulfate reduction despite other similarities in geochemistry and biomass. One plausible explanation might be that different types of biological interactions (e.g., syntrophy or competition) occur because of differences in the composition and distribution of microbial communities within the mineral matrix of each deposit. Slight differences in community composition, such as Dead Dog having a higher representation of sequences related to sulfur-respiring (Thermoplasmata) and methanogenic (Methanocaldococus-like) archaea than Chowder Hill, may reflect differences in biological interactions, which have implications for rates of sulfate reduction in each deposit. Also, substrate competition for H2 or consumption of locally produced DOC (Oremland and Polcin, 1982; Lovley and Phillips, 1987) may be more prevalent in one deposit over another. Future experiments should aim to better resolve how specific interactions between populations, for example, syntrophy or competition for a common substrate, may influence sulfate reduction.
The potential role of heterotrophic sulfate reduction in productivity and biogeochemistry
Heterotrophic sulfate reduction is likely a prominent metabolic mode within Middle Valley sulfides and sediments, and the SRR data herein (which solely measure hetrotrophic sulfate reduction) support that supposition. Sedimented vent fields typically contain allochthonous organic carbon that could readily support heterotrophy. Indeed, at Middle Valley, bottom waters contain 3.5 mg DOC•L–1 (about seven-fold higher than the overlying surface seawater), whereas porewater concentrations range from 0.1 to 84.0 mg DOC•L–1 at sediment depths to 200 mbsf (Ran and Simoneit, 1994). Based on data from culture studies of Desulfovibrio strains, including the H+/H2 ratio of 1.0 for Desulfovibrio vulgarius Marburg (Fitz and Cypionka, 1991), a P/2e− ratio of 1/3 for Desulfovibrio gigas (this is the number of ATPs produced for every two electrons transferred to an electron acceptor (Barton et al., 2003), and the assumption that 10% of ATP production supports growth (20 mmol ATP per gram biomass), our estimates suggest that—at our maximum empirically measured rates—heterotrophic sulfate reduction could support 140 g biomass yr−1 (∼1.5 × 1014 cells) at Chowder Hill (volume=109 900 cm3), 16 g biomass yr−1 (∼1.7 × 1013 cells) at Needles (volume=5495 cm3), and 2.1 g biomass yr−1 (∼2.2 × 1012 cells) at Dead Dog (volume=12 560 cm3). These values are modest in comparison to global estimates of chemoautotrophic biomass production on the global ridge system (1010–1013 g of biomass yr−1; McCollom and Shock, 1997; Bach and Edwards, 2003), yet may be significant in the context of local secondary productivity. Moreover, with respect to the sulfur cycle, the sulfide produced by these heterotrophic sulfate reducers could represent up to 3% of the H2S flux from Middle Valley deposits (given previously published vent fluid flow rates from the Main Endeavor field; Wankel et al., 2011), which can influence, for example, sulfur isotope biogeochemistry. Additional rate measurements that represent the diversity of physicochemical conditions found within deposits or ridge systems are necessary to better constrain the contribution of heterotrophic sulfate reducers to global vent biomass and geochemistry.
Hydrothermal vents are dynamic environments where carbon and sulfur cycling are intimately linked. Both autotrophic and heterotrophic sulfate-reducing microbes have been isolated from vents, and the data shown here are among the first to constrain the potential for heterotorophic sulfate reduction at vents (in particular those with higher organic carbon loads), as well as the relationship between SRRs, temperature, microbial biomass, and community density and composition. These data, as well as the vent field estimates of sulfate reduction, underscore the relevance of sulfate reduction in hydrothermal ecosystems and further indicate the need for continued studies of sulfur cycling along ridge systems.
Acknowledgments
We are grateful for the expert assistance of the R/V Atlantis crews and the pilots and team of the DSV Alvin for enabling the collections of hydrothermal deposits used in our experiments. We also thank Steve Sansone, Dr Joseph Ring, Ms Julie Hanlon, Dr Kathleen Scott, Dr Vladimir Samarkin, Dr David Johnston and Dr Jan Amend for providing assistance with various technical aspects of the experiments. We are also very thankful for the constructive feedback from the reviewers. Financial support for this research was provided by the National Science Foundation (NSF OCE-0838107 and NSF OCE-1061934 to PR Girguis), and the National Aeronautic and Space Administration (NASA-ASTEP NNX09AB78G to C Scholin and PR Girguis and NASA-ASTEP NNX07AV51G to A Knoll and PR Girguis).
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Alazard D, Dukan S, Urios A, Verhe F, Bouabida N, Morel F, et al. Desulfovibrio hydrothermalis sp nov., a novel sulfate-reducing bacterium isolated from hydrothermal vents. Int J Syst Evol Microbiol. 2003;53:173–178. doi: 10.1099/ijs.0.02323-0. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amend JP, Shock EL. Energetics of overall metabolic reactions of thermophilic and hyperthermophilic archaea and bacteria. Fems Microbiology Reviews. 2001;25:175–243. doi: 10.1111/j.1574-6976.2001.tb00576.x. [DOI] [PubMed] [Google Scholar]
- Audiffrin C, Cayol JL, Joulian C, Casalot L, Thomas P, Garcia JL, et al. Desulfonauticus submarinus gen. nov., sp nov., a novel sulfate-reducing bacterium isolated from a deep-sea hydrothermal vent. Int J Syst Evol Microbiol. 2003;53:1585–1590. doi: 10.1099/ijs.0.02551-0. [DOI] [PubMed] [Google Scholar]
- Bach W, Edwards KJ. Iron and sulfide oxidation within the basaltic ocean crust: implications for chemolithoautotrophic microbial biomass production. Geochimica et Cosmochimica. 2003;67:3871–3887. [Google Scholar]
- Barton LL, Hamilton WA, Thauer RK, Stackebrandt E, Legall J, Odom JM, et al. 2003Energy metabolism and phylogenetic diversity of sulphate-reducing bacteriaIn Hamilton WA, Barton LL (eds)Sulphate-Reducting Bacteria: Environmental and Engineered Systems Cambridge University Press; 1–10. [Google Scholar]
- Blöchl E, Rachel R, Burggraf S, Hafenbradl D, Jannasch HW, Stetter KO. Pyrolobus fumarii, gen. and sp. nov., represents a novel group of archaea, extending the upper temperature limit for life to 113 degrees C. Extremophiles. 1997;1:14–21. doi: 10.1007/s007920050010. [DOI] [PubMed] [Google Scholar]
- Bonch-Osmolovskaya E, Perevalova A, Kolganova TV, Rusanov II, Jeanthon C, Pimenov NV. Activity and distribution of thermophilic prokaryotes in hydrothermal fluid, sulfidic structures, and sheaths of alvinellids (East Pacific Rise, 13°N) Appl Environ Microbiol. 2011;77:2803–2806. doi: 10.1128/AEM.02266-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostick BC, Fendorf S, Helz GR. Differential adsorption of molybdate and tetrathiomolybdate on pyrite (FeS2) Environ Sci Technol. 2003;37:285–291. doi: 10.1021/es0257467. [DOI] [PubMed] [Google Scholar]
- Butterfield DA, Mcduff RE, Franklin J, Wheat CG. Geochemistry of hydrothermal vent fluids from Middle Valley, Juan de Fuca ridge. Proc ODP Sci Res. 1994a;139:395–410. [Google Scholar]
- Butterfield DA, McDuff RE, Mottl MJ, Lilley MD, Lupton JE, Massoth GJ. Gradients in the composition of hydrothermal fluids from the endeavour segment vent field: phase separation and brine loss. J Geophys Res. 1994b;99:9561–9583. [Google Scholar]
- Campbell BJ, Engel AS, Porter ML, Takai K. The versatile epsilon-proteobacteria: key players in sulphidic habitats. Nat Rev Microbiol. 2006;4:458–468. doi: 10.1038/nrmicro1414. [DOI] [PubMed] [Google Scholar]
- Canfield DE. Sulfate Reduction and oxic respiration in marine- sediments- Implications for organic-carbon preservation in euxinic envrionments. Deep Sea Res Pt A Oceanographic Res Papers. 1989;36:121–138. doi: 10.1016/0198-0149(89)90022-8. [DOI] [PubMed] [Google Scholar]
- Chen RF, Bada JL, Suzuk Y. The relationship between dissolved organic carbon ( DOC ) and fluorescence in anoxic marine porewaters: implications for estimating benthic DOC fluxes. Geochimica Et Cosmochimica Acta. 1993;57:2149–2153. [Google Scholar]
- Christophersen CT, Morrison M, Conlon MA. Overestimation of the abundance of sulfate-reducing bacteria in human feces by quantitative PCR targeting the Desulfovibrio 16S rRNA gene. Appl Environ Microbiol. 2011;77:3544–3546. doi: 10.1128/AEM.02851-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole JR, Chai B, Farris RJ, Wang Q, Kulam S, McGarrell DM, et al. The Ribosomal Database Project (RDP-II): sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res. 2005;33:D294–D296. doi: 10.1093/nar/gki038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruse AM, Seewald JS. Geochemistry of low-molecular weight hydrocarbons in hydrothermal fluids from Middle Valley, northern Juan de Fuca Ridge. Geochimica Et Cosmochimica Acta. 2006;70:2073–2092. [Google Scholar]
- Cypionka H, Konstanz U. Characterization of sulfate transport in Desulfovibrio desulfuricans. Arch Microbiol. 1989;152:237–243. doi: 10.1007/BF00409657. [DOI] [PubMed] [Google Scholar]
- Dittmar W. Report on researches into the composition of ocean water, collected by the HMS Challenger, during the years 1873-1876. Rept Sci Results Voyage HMS Challenger (Phys Chem) 1884;1:1–251. [Google Scholar]
- Dhillon A, Lever M, Lloyd KG, Albert DB, Sogin ML, Teske A. Methanogen diversity evidenced by molecular characterization of methyl coenzyme M reductase A (mcrA) genes in hydrothermal sediments of the Guaymas Basin. Appl Environ Microbiol. 2005;71:4592–4601. doi: 10.1128/AEM.71.8.4592-4601.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhillon A, Teske A, Dillon J, Stahl DA, Sogin ML. Molecular characterization of sulfate-reducing bacteria in the Guaymas Basin. Appl Environ Microbiol. 2003;69:2765–2772. doi: 10.1128/AEM.69.5.2765-2772.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowd SE, Sun Y, Secor PR, Rhoads DD, Wolcott BM, James GA, et al. Survey of bacterial diversity in chronic wounds using Pyrosequencing, DGGE, and full ribosome shotgun sequencing. BMC Microbiol. 2008;8:43. doi: 10.1186/1471-2180-8-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsgaard L, Guezennec J, Benbouzidrollet N, Prieur D. Mesophilic sulfate-reducing bacteria from 3 deep-sea hydrothermal vent sites. Oceanologica Acta. 1995;18:95–104. [Google Scholar]
- Elsgaard L, Isaksen MF, Jorgensen BB, Alayse AM, Jannasch HW. Microbial sulfate reduction in deep-sea sediments at the Guaymas Basin hydrothermal vent area: influence of temperature and substrates. Geochimica Et Cosmochimica Acta. 1994a;58:3335–3343. [Google Scholar]
- Elsgaard L, Prieur D, Mukwaya GM, Jorgensen BB. Thermophilic sulfate reduction in hydrothermal sediment of lake tanganyika, East Africa. Appl Environ Microbiol. 1994b;60:1473–1480. doi: 10.1128/aem.60.5.1473-1480.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitz R, Cypionka H. Generation of a proton gradient in Desulfovibrio vulgaris. Arch Microbiol. 1991;155:444–448. [Google Scholar]
- Fossing H, Jorgensen BB. Chromium reduction method of bacterial sulfate reduction in sediments: measurement reduction of a single-step chromium method evaluation. Biogeochemistry. 1989;8:205–222. [Google Scholar]
- Frias-lopez J, Zerkle AL, Bonheyo GT, Fouke BW. Partitioning of bacterial communities between seawater and healthy, black band diseased, and dead coral surfaces. Appl Environ Microbiol. 2002;68:2214–2228. doi: 10.1128/AEM.68.5.2214-2228.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrity GM, Holt JG.2001The road map to the manualIn Castenholz RW,, Garrity GM (eds)Bergys Manual of Systematic Bacteriology Springer: New York, NY, USA; 119–166. [Google Scholar]
- Hoek J, Banta A, Hubler F, Reysenbach AL. Microbial diversity of a sulphide spire located in the Edmond deep-sea hydrothermal vent field on the Central Indian Ridge. Geobiology. 2003;1:119–127. [Google Scholar]
- Houghton JL, Seyfried WE, Banta AB, Reysenbach AL. Continuous enrichment culturing of thermophiles under sulfate and nitrate-reducing conditions and at deep-sea hydrostatic pressures. Extremophiles. 2007;11:371–382. doi: 10.1007/s00792-006-0049-7. [DOI] [PubMed] [Google Scholar]
- Huber JA, Welch DM, Morrison HG, Huse SM, Neal PR, Butterfield DA, et al. Microbial population structures in the deep marine biosphere. Science. 2007;318:97–100. doi: 10.1126/science.1146689. [DOI] [PubMed] [Google Scholar]
- Jannasch HW, Mottl MJ. Geomicrobiology of deep-sea hydrothermal vents. Science. 1985;229:717–725. doi: 10.1126/science.229.4715.717. [DOI] [PubMed] [Google Scholar]
- Jannasch HW, Wirsen CO, Molyneaux SJ, Langworthy TA. Extremely thermophilic fermentative archaebacteria of the genus Desulfurococcus from deep-sea hydrothermal vents. Appl Environ Microbiol. 1988;54:1203–1209. doi: 10.1128/aem.54.5.1203-1209.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KS, Beehler CL, Sakamoto-Arnold C, Childress J. In situ measurements of chemical distributions in a deep-sea hydrothermal vent field. Science. 1986;231:1139–1141. doi: 10.1126/science.231.4742.1139. [DOI] [PubMed] [Google Scholar]
- Jorgensen BB, Isaksen MF, Jannasch HW. Bacterial sulfate reduction above 100-degrees-C in deep-sea hydrothermal vent sediments. Science. 1992;258:1756–1757. doi: 10.1126/science.258.5089.1756. [DOI] [PubMed] [Google Scholar]
- Joye SB, Boetius A, Orcutt BN, Montoya JP, Schulz HN, Erickson MJ, et al. The anaerobic oxidation of methane and sulfate reduction in sediments from Gulf of Mexico cold seeps. Chem Geol. 2004;205:219–238. [Google Scholar]
- Kallmeyer J, Boetius A. Effects of temperature and pressure on sulfate reduction and anaerobic oxidation of methane in hydrothermal sediments of Guaymas basin. Appl Environ Microbiol. 2004;70:1231–1233. doi: 10.1128/AEM.70.2.1231-1233.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klappenbach J, Saxman PR, Cole JR, Schmidt TM. Rrndb: the ribosomal RNA operon copy number database. Nucleic Acids Res. 2001;29:181–184. doi: 10.1093/nar/29.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein M, Friedrich M, Roger AJ, Hugenholtz P, Fishbain S, Abicht H, et al. Multiple lateral transfers of dissimilatory sulfite reductase genes between major lineages of sulfate-reducing prokaryotes. Appl Environ Microbiol. 2001;183:6028–6035. doi: 10.1128/JB.183.20.6028-6035.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo R, Nedwell DB, Purdy KJ, Silva SQ. Detection and enumeration of sulphate-reducing bacteria in estuarine sediments by competitive PCR. Geomicrobiol J. 2004;21:145–157. [Google Scholar]
- Lang SQ, Butterfield DA, Lilley MD, Johnson HP, Hedges JI. Dissolved organic carbon in ridge-axis and ridge-flank hydrothermal systems. Geochimica et Cosmochimica Acta. 2006;70:3830–3842. [Google Scholar]
- Lee ZM-P, Bussema C, Schmidt TM. rrnDB: documenting the number of rRNA and tRNA genes in bacteria and archaea. Nucleic Acids Res. 2009;37:489–493. doi: 10.1093/nar/gkn689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovley DR, Phillips EJ. Competitive mechanisms for inhibition of sulfate reduction and methane production in the zone of ferric iron reduction in sediments. Appl Environ Microbiol. 1987;53:2636–2641. doi: 10.1128/aem.53.11.2636-2641.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manefield M, Whiteley AS, Griffiths RI, Bailey MJ. RNA stable isotope probing, a novel means of linking microbial community function to phylogeny. Appl Environ Microbiol. 2002;68:5367–5373. doi: 10.1128/AEM.68.11.5367-5373.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCollom TM, Shock EL. Geochemical constraints on chemolithoautotrophic metabolism by microorganisms in seafloor hydrothermal systems. Geochimica et Cosmochimica Acta. 1997;61:4375–4391. doi: 10.1016/s0016-7037(97)00241-x. [DOI] [PubMed] [Google Scholar]
- Muyzer G, Stams AJM. The ecology and biotechnology of sulphate-reducing bacteria. Nat Rev Microbiol. 2008;6:441–544. doi: 10.1038/nrmicro1892. [DOI] [PubMed] [Google Scholar]
- Nakagawa S, Takai K, Inagaki F, Hirayama H, Nunoura T, Horikoshi K, et al. Distribution, phylogenetic diversity and physiological characteristics of epsilon-Proteobacteria in a deep-sea hydrothermal field. Environ Microbiol. 2005;7:1619–1632. doi: 10.1111/j.1462-2920.2005.00856.x. [DOI] [PubMed] [Google Scholar]
- Nakagawa S, Takai K. Deep-sea vent chemoautotrophs: diversity, biochemistry and ecological significance. FEMS Microbiol Ecol. 2008;65:1–14. doi: 10.1111/j.1574-6941.2008.00502.x. [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Nakagawa S, Inagaki F, Takai K, Horikoshi K. Phylogenetic diversity of sulfate-reducing prokaryotes in active deep-sea hydrothermal vent chimney structures. FEMS Microbiol Let. 2004;232:145–152. doi: 10.1016/S0378-1097(04)00044-8. [DOI] [PubMed] [Google Scholar]
- Nercessian O, Bienvenu N, Moreira D, Prieur D, Jeanthon C. Diversity of functional genes of methanogens, methanotrophs and sulfate reducers in deep-sea hydrothermal environments. Environ Microbiol. 2005;7:118–132. doi: 10.1111/j.1462-2920.2004.00672.x. [DOI] [PubMed] [Google Scholar]
- Newport PJ, Nedwell DB. The mechanisms of inhibition of Desulfovibrio and Desulfotomaculum species by selenate and molybdate. J Appl Microbiol. 1988;65:419–423. [Google Scholar]
- Nunoura T, Oida H, Nakaseama M, Kosaka A, Ohkubo SB, Kikuchi T, et al. Archaeal diversity and distribution along thermal and geochemical gradients in hydrothermal sediments at the Yonaguni Knoll IV hydrothermal field in the Southern Okinawa trough. Appl Environ Microbiol. 2010;76:1198–1211. doi: 10.1128/AEM.00924-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley BB, Carbonero F, Dowd Scot E, Hawkins RJ, Purdy KJ. Contrasting patterns of niche partitioning between two anaerobic terminal oxidizers of organic matter. ISME J. 2011;6:905–914. doi: 10.1038/ismej.2011.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oremland RS, Polcin S. Methanogenesis and sulfate reduction: competitive and noncompetitive substrates in estuarine sediments. Appl Environ Microbiol. 1982;44:1270–1276. doi: 10.1128/aem.44.6.1270-1276.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran B, Simoneit BRT. Dissolved organic carbon in interstitial waters from sediments of Middle Valley, Leg 139 1. Proc ODP Sci Res. 1994;139:441–446. [Google Scholar]
- Saleh AM, Macpherson R, Miller JDA. The effect of inhibitors on sulphate reducing bacteria: a compilation. J Appl Microbiol. 1964;27:281–293. [Google Scholar]
- Santelli CM, Orcutt BN, Banning E, Bach W, Moyer CL, Sogin ML, et al. Abundance and diversity of microbial life in ocean crust. Nature. 2008;453:653–657. doi: 10.1038/nature06899. [DOI] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JRA, Hartmann M, Hollister EB, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrenk MO, Kelley DS, Delaney JR, Baross JA. Incidence and diversity of microorganisms within the walls of an active deep-sea sulfide chimney. Appl Environ Microbiol. 2003;69:3580–3592. doi: 10.1128/AEM.69.6.3580-3592.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiguchi Y, Muramatsu M, Imachi H, Narihiro T, Ohashi A, Harada H, et al. Thermodesulfovibrio aggregans sp. nov. and Thermodesulfovibrio thiophilus sp. nov., anaerobic, thermophilic, sulfate-reducing bacteria isolated from thermophilic methanogenic sludge, and emended description of the genus Thermodesulfovibrio. Int J Syst Evol Microbiol. 2008;58:2541–2548. doi: 10.1099/ijs.0.2008/000893-0. [DOI] [PubMed] [Google Scholar]
- Stults JR, Snoeyenbos-West O, Methe B, Lovley DR, Chandler DP. Application of the 5′ fluorogenic exonuclease assay (TaqMan) for quantitative ribosomal DNA and rRNA analysis in sediments. Appl Environ Microbiol. 2001;67:2781–2789. doi: 10.1128/AEM.67.6.2781-2789.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki MT, Beja O, Taylor LT, DeLong EF. Phylogenetic analysis of ribosomal RNA operons from uncultivated coastal marine bacterioplankton. Environ Microbiol. 2001;3:323–331. doi: 10.1046/j.1462-2920.2001.00198.x. [DOI] [PubMed] [Google Scholar]
- Sylvan JB, Toner BM, Edwards KJ. Life and death of deep-sea vents: bacterial diversity and ecosystem succession on inactive hydrothermal sulfides. mBio. 2012;3:e00279–11. doi: 10.1128/mBio.00279-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai K, Horikoshi K. Rapid detection and quantification of members of the archaeal community by quantitative PCR using fluorogenic probes. Appl Environ Microbiol. 2000;66:5066–5072. doi: 10.1128/aem.66.11.5066-5072.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teske A, Sorensen KB. Uncultured archaea in deep marine subsurface sediments: have we caught them all. ISME J. 2008;2:3–18. doi: 10.1038/ismej.2007.90. [DOI] [PubMed] [Google Scholar]
- Wagner M, Roger AJ, Flax JL, Brusseau GA, Stahl DA. Phylogeny of dissimilatory sulfite reductases supports an early origin of sulfate respiration. J Bact. 1998;180:2975–2982. doi: 10.1128/jb.180.11.2975-2982.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wankel SD, Germanovich LN, Lilley Marvin D, Genc G, DiPerna CJ, Bradley AS, et al. Influence of subsurface biosphere on geochemical fluxes from diffuse hydrothermal fluids. Nat Geo. 2011;4:461–468. [Google Scholar]
- Weber A, Jorgensen BB. Bacterial sulfate reduction in hydrothermal sediments of the Guaymas Basin, Gulf of California, Mexico. Deep-Sea Res Pt I-Oceanographic Res Papers. 2002;49:827–841. [Google Scholar]
- Xu N, Christodoulatos C, Braida W. Adsorption of molybdate and tetrathiomolybdate onto pyrite and goethite: effect of pH and competitive anions. Chemosphere. 2006;62:1726–1735. doi: 10.1016/j.chemosphere.2005.06.025. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.