Abstract
The importance of bacteria in the anaerobic bioremediation of groundwater polluted with organic and/or metal contaminants is well recognized and in some instances so well understood that modeling of the in situ metabolic activity of the relevant subsurface microorganisms in response to changes in subsurface geochemistry is feasible. However, a potentially significant factor influencing bacterial growth and activity in the subsurface that has not been adequately addressed is protozoan predation of the microorganisms responsible for bioremediation. In field experiments at a uranium-contaminated aquifer located in Rifle, CO, USA, acetate amendments initially promoted the growth of metal-reducing Geobacter species, followed by the growth of sulfate reducers, as observed previously. Analysis of 18S rRNA gene sequences revealed a broad diversity of sequences closely related to known bacteriovorous protozoa in the groundwater before the addition of acetate. The bloom of Geobacter species was accompanied by a specific enrichment of sequences most closely related to the ameboid flagellate, Breviata anathema, which at their peak accounted for over 80% of the sequences recovered. The abundance of Geobacter species declined following the rapid emergence of B. anathema. The subsequent growth of sulfate-reducing Peptococcaceae was accompanied by another specific enrichment of protozoa, but with sequences most similar to diplomonadid flagellates from the family Hexamitidae, which accounted for up to 100% of the sequences recovered during this phase of the bioremediation. These results suggest a prey–predator response with specific protozoa responding to increased availability of preferred prey bacteria. Thus, quantifying the influence of protozoan predation on the growth, activity and composition of the subsurface bacterial community is essential for predictive modeling of in situ uranium bioremediation strategies.
Keywords: Geobacter, Breviata, Peptococcaceae, Hexamitidae, uranium bioremediation, protozoan predation
Introduction
Anaerobic protozoa are important members of microbial communities inhabiting the anoxic zone of most aquatic environments. They have been found in a variety of pristine and contaminated freshwater subsurface habitats (Sinclair et al., 1993; Novarino et al., 1997; Ellis et al., 1998; Zarda et al., 1998; Kinner et al., 2002; Luo et al., 2005; Brad et al., 2008; Yagi et al., 2010; Lin et al., 2012) as deep as 200 m below the surface (Sinclair et al., 1993; Nagaosa et al., 2008). Many of these protozoa are bacteriovorous, actively grazing on bacteria in the subsurface (Fenchel, 1982; Linley et al., 1983; Fenchel, 1986; Fenchel and Ramsing, 1992; Kinner et al., 1998; Decamp et al., 1999; Kinner et al., 2002) and having a major role in top-down biological control of bacteria and the regeneration of nutrients, such as nitrogen, phosphorus, trace metals and organics, in these ecosystems (Caron et al., 1988; Caron, 1994; Eccleston-Parry and Leadbeater, 1995).
Although protozoan grazing removes bacterial cells, the release of growth-limiting nutrients secreted by grazing protozoa can stimulate bacterial metabolism. For example, nitrification, methane production and sulfate reduction all increased in the subsurface in the presence of protozoan grazing, and the specific growth rate of bacteria was twofold higher in the presence of protozoa (Bloem et al., 1988; Verhagen et al., 1995; Strauss and Dodds, 1997; Biagini et al., 1998). This enhanced metabolism can improve the efficiency of biodegradation of organic compounds, such as anaerobic sludge from dairy wastewater treatment plants (Priya et al., 2008); hydrocarbons (Rogerson and Berger, 1983; Mattison and Harayama, 2001; Mattison et al., 2005); and plant material (Biagini et al., 1998; Ribblett et al., 2005). Grazing by protozoa can also help prevent reduction in hydrological conductivity that might otherwise result from microbial biomass plugging aquifer pore spaces (Sinclair et al., 1993; DeLeo and Baveye, 1997; Kinner et al., 2002; Mattison et al., 2002).
Alternatively, in some instances protozoan grazing can have a negative impact on groundwater bioremediation by critically reducing the number of contaminant-degrading microorganisms (Kota et al., 1999). For example, protozoa inhibited trichloroethylene and BTEX (benzene, toluene, ethylbenzene, and o-, m- and p-xylenes) degradation in laboratory microcosms constructed with contaminated aquifer sediments (Kota et al., 1999; Cunningham et al., 2009) and when protozoan predation was not considered in a computer model that simulated bioremediation of trichloroethylene by a methanotrophic community, rates of trichloroethylene degradation were overestimated by 25% (Travis and Rosenberg, 1997).
Efforts to model the in situ bioremediation of uranium-contaminated water have become increasingly sophisticated with the introduction of genome-scale metabolic models to predict the growth and metabolic activity of the microorganisms thought to influence the bioremediation process (Scheibe et al., 2009; Fang et al., 2011; Lovley et al., 2011; Mahadevan et al., 2011; Zhuang et al., 2011; Barlett et al., 2012). However, these modeling efforts have not considered the potential role of protozoa in influencing microbial community dynamics. Here we report that stimulating the growth of the bacterial community with acetate to promote U(VI) reduction results in specific enrichment of protozoa with different protozoan genera responding to the growth of Geobacter or sulfate-reducing bacteria.
Materials and methods
Site and description of field site
In 2010, a small-scale in situ bioremediation experiment was conducted on the grounds of a former uranium ore-processing facility in Rifle, CO, USA, during the months of August–October as described previously (Miletto et al., 2011; Giloteaux et al., 2012). This same plot was biostimulated by acetate additions during the months of August–October in 2011. This research was part of the Uranium Mill Tailings Remedial Action (UMTRA) program of the US Department of Energy. The plot used in both field experiments was adjacent to a previously studied larger experimental plot at the site (Anderson et al., 2003; Vrionis et al., 2005). The monitoring array consisted of an injection gallery with six injection wells, nine downgradient wells and one background monitoring well-located upstream from the injection gallery (see Supplementary Material and Supplementary Figure S1). Groundwater for the experiment was collected from well CD-04 in 2010 and CD-01 in 2011.
The Old Rifle site is located on a flood plain of the Colorado River. Groundwater moves primarily in the topmost hydrostratigraphic unit of the unconfined aquifer, a sandy gravel, gravelly sand alluvium. The upper permeable layer (hydraulic conductivity ca. 37 m d−1) is underlain by a relatively impermeable silty shale layer (conductivity ca. 0.005 m d−1) from the weathered Wasatch formation (DOE, 1999; Anderson et al., 2003; Yabusaki et al., 2007).
Porosity of the Rifle sediments is ca. 25%, and only 38% of the aquifer material is contained in the <2 mm size fraction and the majority is gravel size (Fox et al., 2012).
During the field experiment, a concentrated acetate/bromide solution (50/20 mℳ) mixed with native groundwater was injected into the subsurface to provide approximately 5 mℳ acetate to the groundwater over the course of 30 days in 2010 and 68 days in 2011as described previously (Anderson et al., 2003; Williams et al., 2011). Bromide was utilized as a non-reactive tracer.
Analytical techniques.
Samples for geochemical analyses were collected after purging 12 l of groundwater from the wells with a peristaltic pump. Ferrous iron was measured spectrophotometrically immediately after sampling using the phenanthroline method (AccuVac ampules; Hach Company, Loveland, CO, USA) for ferrous iron. After filtration through a 0.2 μm pore size polytetrafluoroethylene (Teflon) filter (Alltech Associates Inc., Deerfield, IL, USA), acetate concentrations were measured with a Dionex ICS-1000 ion chromatograph equipped with an IonPac AS22 column, an ASRS 300 suppressor and 4.5 mℳ carbonate/1.4 mℳ bicarbonate eluent (Dionex Corporation, Sunnyvale, CA, USA).
Extraction of nucleic acids from samples
DNA and RNA were extracted from groundwater collected from the U(VI)-contaminated aquifer during the bioremediation field experiments. To obtain sufficient biomass from the groundwater, it was necessary to concentrate 50 l of groundwater by impact filtration on 293 mm diameter Supor membrane disc filters (Pall Life Sciences, Port Washington, NY, USA), which took about 3 min. All filters were placed into whirl-pack bags, flash frozen in a dry ice/ethanol bath and shipped back to the laboratory where they were stored at −80 °C. RNA was extracted from filters as described previously (Holmes et al., 2005) and DNA was extracted with the FastDNA SPIN Kit for Soil (MP Biomedicals, Santa Ana, CA, USA).
Absorbance readings with the NanoDrop spectrophotometer (Thermo Scientific, Wilmington, DE, USA) and agarose gel electrophoresis showed that high-quality DNA and RNA were extracted from the groundwater samples. To ensure that RNA samples were not contaminated with DNA, polymerase chain reaction (PCR) amplification with primers targeting the 16S rRNA gene was conducted on RNA samples that had not undergone reverse transcription.
A DuraScript enhanced avian RT single-strand synthesis kit (Sigma, Sigma-Aldrich, St Louis, MO, USA) was used to generate cDNA as described previously (Giloteaux et al., 2012).
PCR amplification parameters and clone library construction
Several previously described primer pairs were used for the amplification of 16S rRNA, 18S rRNA and β-tubulin gene fragments from genomic DNA and cDNA constructed from mRNA extracted from groundwater. Gene fragments from the 16S rRNA and 18S rRNA genes were amplified with 8F (Eden et al., 1991) and 519R (Lane et al., 1985), and 515F (Giovannoni et al., 1988) and 1209R (Reysenbach et al., 1992); respectively; BT107F and BT261R (Baker et al., 2004) were used to amplify the β-tubulin gene. The 18S rRNA and β-tubulin primer sets were both nonspecific and amplified both protozoan and non-protozoan gene sequences. Some of the non-protozoan gene sequences detected at this site came from plant, fungal and animal species, which accounted for ca. 5% and 25% of the 18S rRNA and β-tubulin clone libraries (see Supplementary Material, Supplementary Figure S2 and Supplementary Table S1). These studies focused exclusively on the protozoan sequences detected in these eukaryotic libraries.
Degenerate primers targeting the gene coding for the α-subunit of the dissimilatory sulfite reductase protein (dsrA) from Peptococcaceae species (dsrPept_380F and dsrPept_740R) (Supplementary Table S2) were designed from various Desulfitobacteria, Desulfosporosinus and Desulfotomaculum dsrA nucleotide sequences obtained from the NCBI GenBank website (http://www.ncbi.nlm.nih.gov).
A 50 μl PCR reaction consisted of the following solutions: 10 μl Q buffer (Qiagen, Valencia, CA, USA), 0.4 mℳ of each dNTP, 1.5 mℳ MgCl2, 0.2 μℳ of each primer, 5 μg bovine serum albumin, 2.5 U Taq DNA polymerase (Qiagen) and 10 ng of PCR template. Amplification was performed with a minicycler PTC 200 (MJ Research, Waltham, MA, USA) starting with 5 min at 94 °C, followed by 35 cycles consisting of denaturation (45 s at 94 °C), annealing (see Supplementary Table S1), extension (90 s at 72 °C) and a final extension at 72 °C for 10 min.
After PCR amplification of these gene fragments, PCR products were purified with the Gel Extraction Kit (Qiagen), and cloned into the TOPO TA cloning vector, version M (Invitrogen, Carlsbad, CA, USA). In all, 100 plasmid inserts from each of these clone libraries were sequenced with the M13F primer at the University of Massachusetts Sequencing Facility.
Calculation of diversity indices
The Shannon–Wiener and Simpson indices of diversity were used to determine the diversity of taxa present in groundwater collected from the site. The Shannon–Wiener diversity index (H′) was calculated as follows (Margalef, 1958; Dunbar et al., 1999):
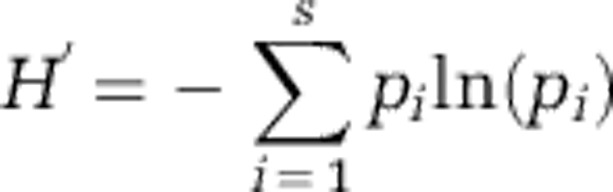 |
Simpson's diversity index (D) was calculated with the following equation (Simpson, 1949):
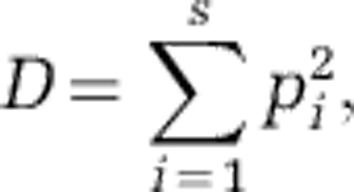 |
where pi in both of these equations represented the proportion of the ith phylotype.
Species evenness was represented by Pielou's evenness index (J′), which was calculated from H′/H′max, where H′max is equal to ln(s) and s is the total number of phylotypes (Pielou, 1966).
Testing and design of qPCR primers
The following primer sets were used to quantify 16S rRNA and citrate synthase (gltA) gene and mRNA transcript copies found in groundwater collected during both field experiments by quantitative (q)PCR: 16S rRNA was amplified with 338F (Weisburg et al., 1991) and 518R (Muyzer et al., 1993), and gltA was amplified with CS375F and CS598R (Holmes et al., 2005). New qPCR primer sets targeting dsrA and β-tubulin genes were designed according to the manufacturer's specifications (Applied Biosystems, Carlsbad, CA, USA) and had amplicon sizes ranging from100 to 200 bp. qPCR primers targeting all β-tubulin protozoan genes found in the groundwater (qbetGen_260F/qbetGen_340R) were designed from sequences found in our clone libraries and from representative protozoan β-tubulin sequences obtained from the GenBank database.
Primers for qPCR were also designed to target specifically Breviata and Hexamita β-tubulin genes, and Desulfosporosinus dsrA genes in the groundwater (Supplementary Table S2). The Breviata β-tubulin primer pair (qbetBrev_521F/qbetBrev_610R) was designed from a β-tubulin clone (clone RB) that had 86% nucleotide identity to β-tubulin from Breviata anathema and accounted for 65% of the β-tubulin clone library assembled from groundwater collected on day 14 during the 2010 field experiment. The Geobacter gltA primer pair (CS375F/CS598R) was previously designed from a Geobacter sequence most similar to Geobacter sp. M18 (Holmes et al., 2005) and accounted for 79% of the Geobacter gltA clone library assembled with groundwater collected at the peak of Fe(III) reduction during the 2010 field experiment. The Hexamita primer pair (qbetHex_309F/qbetHex_542R) was designed from a β-tubulin clone (clone RH) that was 84% identical to the nucleotide sequence of β-tubulin from Hexamita inflata and accounted for 86% of the β-tubulin sequences detected on day 46 in 2011. The Desulfosporosinus dsrA primer pair (qdsrA_56F/qdsrA_217R) was designed from a dsrA clone (clone RD) that was 98% identical to Desulfosporosinus youngiae and accounted for 95% of the clone library in groundwater collected on day 39 in 2011.
Quantification of gene and transcript abundance by qPCR
qPCR amplification and detection were performed with the 7500 Real-Time PCR System (Applied Biosystems) using genomic DNA and cDNA made by reverse transcription from mRNA extracted from groundwater collected during the bioremediation experiment.
All qPCR assays were run in triplicate. Each reaction mixture consisted of a total volume of 25 μl and contained 1.5 μl of the appropriate primers (stock concentrations, 1.5 μℳ), 5 ng cDNA and 12.5 μl Power SYBR Green PCR Master Mix (Applied Biosystems). Standard curves covering eight orders of magnitude were constructed with serial dilutions of known amounts of purified cDNA quantified with a NanoDrop ND-1000 spectrophotometer (Thermo Scientific) at an absorbance of 260 nm. Transcript abundances and qPCR efficiencies (95–99%) were calculated from appropriate standard curves.
Optimal thermal cycling parameters consisted of an activation step at 50 °C for 2 min, an initial 10 min denaturation step at 95 °C, followed by 50 cycles of 95 °C for 15 s and 60 °C for 1 min. After 50 cycles of PCR amplification, dissociation curves were made for all qPCR products by increasing the temperature from 60–95 °C at a ramp rate of 2%. The curves all yielded a single predominant peak, further supporting the specificity of the PCR primer pairs.
Cell number estimates from qPCR results
Bacterial and protozoan cell numbers were estimated from 16S rRNA and β-tubulin gene copies determined by qPCR with 338F/518R and qbetGen_2260f/340r. Before estimates could be made, the average number of 16S rRNA and β-tubulin gene copies found within bacterial and protozoan genomes were determined. Analysis of all available bacterial genomes indicated that bacteria have an average of 4.3 copies of the 16S rRNA gene per cell (Lee et al., 2009) (http://rrndb.mmg.msu.edu/search.php), while the number of 18S rRNA gene copies can vary considerably among protists and individuals can have anywhere from 1to 1000 copies of this gene in their genome (Saito et al., 2002; Galluzzi et al., 2004; Zhu et al., 2005; Terrado et al., 2011). This enormous variability among 18S rRNA gene copy number would make it difficult to extrapolate cell numbers based on qPCR results. Therefore, the β-tubulin gene was selected for qPCR analysis. Analysis of 74 different protozoan genomes showed that protozoa have an average of 1.94 β-tubulin gene copies in their genome (Supplementary Table S3).
Cell numbers were estimated from the following equations:
(1) Weight of the gene for X copies:
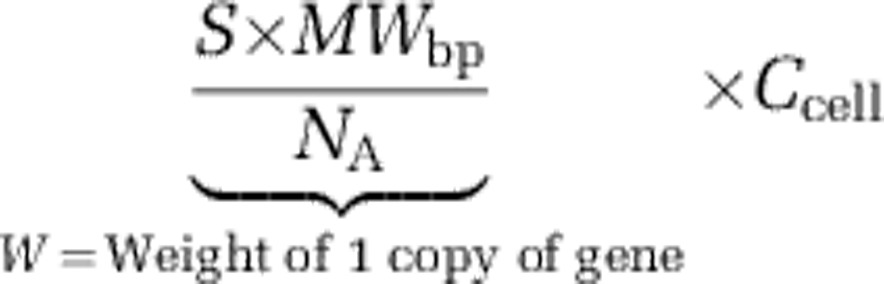 |
where S is the average size of the gene (bp); MWbp is the average weight of a basepair: 660 g mol−1; NA is the Avagadro constant: 6.023 × 1023 mol−1; and Ccell is the average number of gene copies per cell. (In our study Ccell=4.3 for 16S rRNA and 1.94 for β-tubulin).
(2) Quantity of the gene from sample collected:
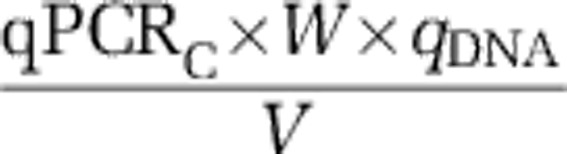 |
where qPCRC is the number of qPCR gene copies; W is the weight of one copy of the gene; qDNA is the quantity of DNA extracted from sample; and V the volume of groundwater collected on filter.
(3) Number of cells per volume of sample:
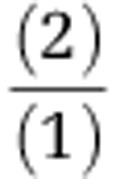 |
Phylogenetic analysis
16S and 18S rRNA and functional gene sequences were assembled with Geneious 5.6 and compared with GenBank nucleotide and protein databases with the blastn and blastx algorithms (Altschul et al., 1998). Alignments were made in ClustalX (Thompson et al., 1997) and corrected with PROSEQ v.2.9 (Filatov, 2002) before phylogenetic trees were constructed with MEGA v.4 (Tamura et al., 2007). The neighbor-joining algorithm was used to construct all phylogenetic trees (Saitou and Nei, 1987). All evolutionary distances were computed with the Poisson correction method with 1000 bootstrap replicates. Homologous coverage calculations and rarefaction analyses of each library were performed as described previously (Giloteaux et al., 2010).
The nucleotide sequences of 18S rRNA, dsrA, β-tubulin and 16S rRNA genes amplified from the uranium-contaminated aquifer have been deposited in the GenBank database under accession numbers HF568845–HF568867 and HF569144–HF569146.
Results and discussion
Specific enrichment of Breviata species in response to Geobacter growth
Addition of acetate to the groundwater during the 2010 field experiment resulted in a significant increase in bacteria, followed by an increase in protozoa (Figure 1a). Bacterial cell numbers estimated from qPCR of the 16S rRNA gene with the equation described in the Materials and methods section were similar to those estimated by fluorescence in situ hybridization analysis (Holmes et al., 2013). In 2010, bacterial cell numbers estimated by qPCR ranged from 2.31 × 104 to 2.02 × 106 cells per ml and the number of cells estimated by fluorescence in situ hybridization ranged from 3.89 × 104 to 2.19 × 106 (Holmes et al., 2013). Protozoan cell numbers estimated from qPCR of the protozoan β-tubulin gene indicated that the number of protozoan cells ranged from ca. 1.0 × 102 to 3.75 × 103 cells per ml.
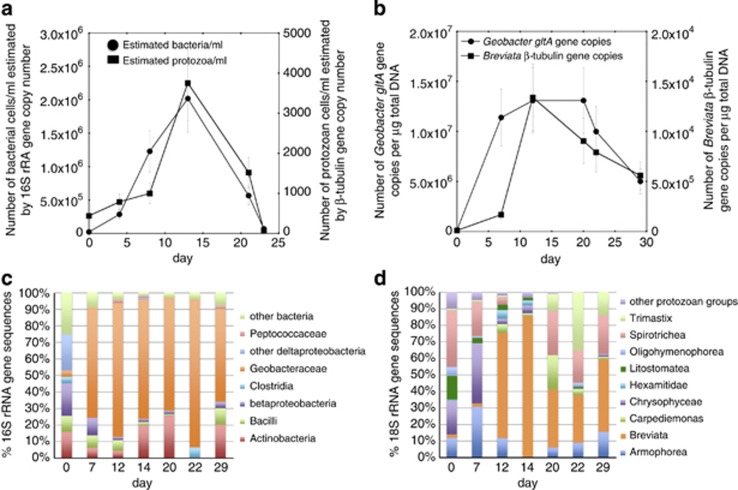
Figure 1.
Microbial community analyses of groundwater samples collected during the Fe(III)-reducing phase of the 2010 field experiment. (a) Estimates of bacterial and protozoan cell numbers calculated from 16S rRNA and protozoan β-tubulin (b2t) gene copy numbers estimated by qPCR (Pearson's correlation, r=0.90, P=0.015). (b) The number of Breviata β-tubulin and Geobacter citrate synthase (gltA) gene copies per μg total DNA (r=0.51, P=0.02). (c) Percentage breakdown of bacterial 16S rRNA gene sequences detected in clone libraries constructed from genomic DNA extracted from groundwater. (d) Percentage breakdown of protozoan 18S rRNA gene sequences detected in clone libraries constructed from genomic DNA extracted from groundwater.
These protozoan abundances were similar to those observed in other pristine and contaminated subsurface aquifers where 102–104 individuals per ml have been detected (Sinclair and Alexander, 1989; Sinclair et al., 1993; Ellis et al., 1998; Zarda et al., 1998; Ekelund et al., 2001). The ratio of protozoan to bacterial cell numbers observed in the groundwater was also consistent with previously reported ratios from other freshwater aquifers (Sinclair and Alexander 1989; Sinclair et al., 1993; Zarda et al., 1998); ca. 1:102 in 2010 and ca. 1:103 in 2011.
As noted in previous studies (Anderson et al., 2003; Holmes et al., 2005; Vrionis et al., 2005; Williams et al., 2011), the increase in bacteria could be attributed to enhanced growth of Geobacter species, as evidenced by an increase in Geobacter gltA gene copies (Figure 1b). Analysis of 16S rRNA gene sequences demonstrated that Geobacter species became the predominant bacterial species in the groundwater, accounting for as much as 89% of the bacterial community (Figure 1c) before a decline in numbers that was coincident with the increased abundance of the protozoa (Figure 1a).
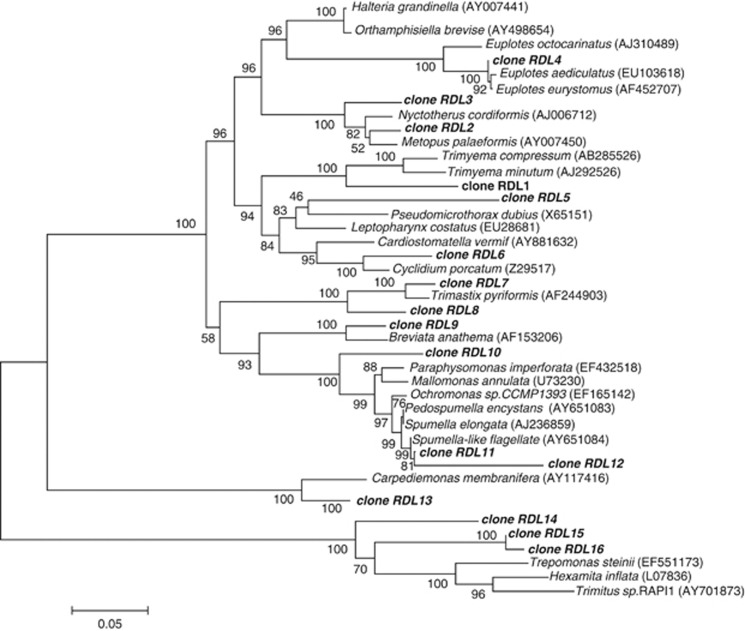
Before acetate injections, the protozoan community was highly diverse with as many as 42 different protozoan 18S rRNA gene sequences detected in the groundwater. Shannon–Wiener (H′) and Simpson diversity (D) indices calculated with formulas described in the Materials and methods section were 2.81 and 0.9 (Supplementary Table S4B). The protozoan community was dominated by species from the classes Spirotrichea (34.2% of the 18S rRNA gene sequences) and Chrysophyceae (20.4% of the 18S rRNA gene sequences). The majority (63.6%) of Spirotrichea 18S rRNA gene sequences were most similar to the ciliated protozoan species Euplotes aediculatus (96% similar) represented by clone RDL11 in Figure 2. The majority of Chrysophyceae sequences (84.8%) were most similar to the Spumella-like flagellate JBM/S11 (98% similar) represented by clone RDL4 as in Figure 2.
Figure 2.
Phylogenetic tree comparing 18S rRNA gene sequences detected in the groundwater to sequences from previously characterized protozoa.
Both of these dominant protozoan species have been detected in other contaminated environments. For example, Spumella species are frequently found in freshwater aquifers contaminated with sewage (Novarino et al., 1994; Kinner et al., 1998); coal-tar waste (Yagi et al., 2010) and polyaromatic hydrocarbons (Lara et al., 2007), and freshwater Euplotes species have been isolated from activated sewage sludge (Salvado et al., 1997) and industrial effluents (Rehman et al., 2008). Studies have also shown that Euplotes species are resistant to high concentrations of heavy metals and can use bioaccumulation to remove up to 90% of these metals from the environment (Schlenk and Moore, 1994; Madoni et al., 1996; Shakoori et al., 2004; Martin-Gonzalez et al., 2006; Chaudhry and Shakoori, 2011). It is possible that the continuous U(VI) removal observed at the Rifle site after acetate additions have stopped (N'Guessan et al., 2008) might be due to bioaccumulation of U(VI) by metal-resistant Euplotes species in the subsurface.
With the addition of acetate, protozoan species richness dropped down to 7 and H′ and D were as low as 0.78 and 0.31 (Figure 1d and Supplementary Table S4B). This decrease in protozoan diversity could be attributed to a specific enrichment of 18S rRNA gene sequences most similar to the bacteriovorous ameboid flagellate, B. anathema (95% similar; Figure 2), which accounted for as much as 86% of the protozoan population (Figure 1d). As acetate and Fe(II) concentrations started to decline after day 23 (Figures 1b and c and Supplementary Figure 3A), the number of Breviata 18S rRNA sequences decreased and overall protozoan diversity increased (H′=1.56, D=0.71) (Figures 1b and d and Supplementary Table S4B). This increase and subsequent decline in numbers of Breviata species tracked with changes in the abundance of Geobacter species (Figure 1b), and the number of Geobacter gltA and Breviata β-tubulin gene copies were correlated (Pearson's correlation, r=0.51, P=0.02).
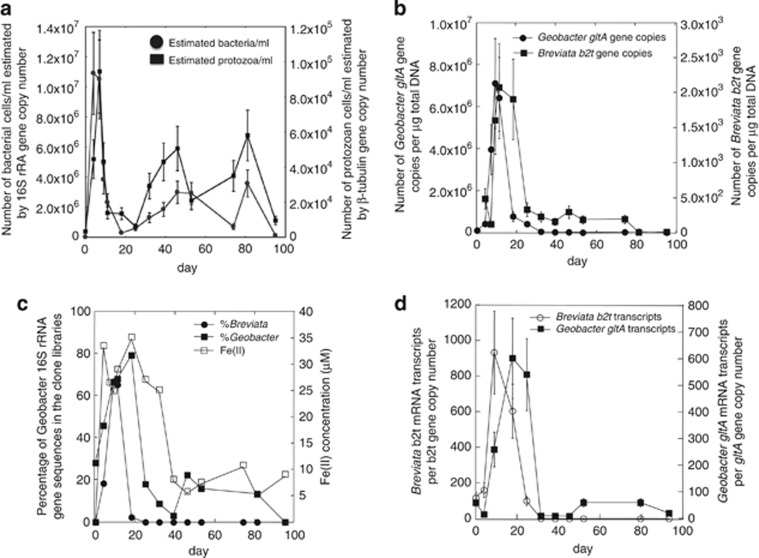
In the 2010 field experiment, sampling was ended as soon as sulfide began accumulating in the water, indicative of the start of sulfate reduction (Supplementary Figure 3A). In 2011, acetate amendments were made in the same well and additions and monitoring were extended through sulfate reduction (Supplementary Figure 3B). Similar to the 2010 field experiment, when acetate was added to the groundwater, a correlation was observed between bacterial and protozoan abundance (r=0.59, P=0.02) (Figure 3a), and bacterial and protozoan diversity decreased significantly during the Fe(III)-reducing phase of the experiment; H′ for the bacterial community decreased from 2.59 on day 0 to 1.82 on day 18 and H′ for the protozoan community decreased from 1.97 to 0.70 on these same days (Supplementary Tables 4C and D). Bacterial cell numbers estimated by qPCR ranged from 2.30 × 104 to 1.09 × 107 cells per ml and protozoan cell numbers ranged from 3.32 × 103 to 5.06 × 104.
Figure 3.
Analysis of bacterial and protozoan communities in the groundwater during the Fe(III)-reducing phase of the 2011 field experiment. (a) Estimates of bacterial and protozoan cell numbers calculated from 16S rRNA and protozoan β-tubulin (b2t) gene copy numbers estimated by qPCR (Pearson's correlation, r=0.59, P=0.02 (b) The number of Breviata β-tubulin (b2t) and Geobacter citrate synthase (gltA) gene copies per μg total DNA (r=0.63, P=0.01). (c) Percentage of Breviata 18S and Geobacter 16S rRNA gene sequences in clone libraries compared to Fe(II) concentrations (r=0.68, P=0.01). (d) The number of Geobacter gltA and Breviata β-tubulin mRNA transcripts normalized against the number of Geobacter gltA and Breviata β-tubulin gene copies (r=0.75, P=0.1).
The initial increase in bacterial and protozoan cell numbers could again be attributed to an enrichment of Geobacter and Breviata species (Figure 3b), with Geobacter and Breviata species accounting for as much as 79% and 67% of the bacterial and protozoan communities detected during this time (Figure 3c).
Quantitative reverse transcription PCR of Breviata β-tubulin gene transcripts demonstrated that relative expression of this gene, and thus presumably Breviata metabolic activity, was high during the increase in Breviata numbers (Figure 3d). Breviata β-tubulin gene transcript abundance correlated well (r=0.75, P=0.007), with relative expression of the Geobacter citrate synthase gene, gtlA, which is known to be an indicator of Geobacter metabolic activity (Holmes et al., 2005).
Enrichment of Hexamitidae in the sulfate reduction phase
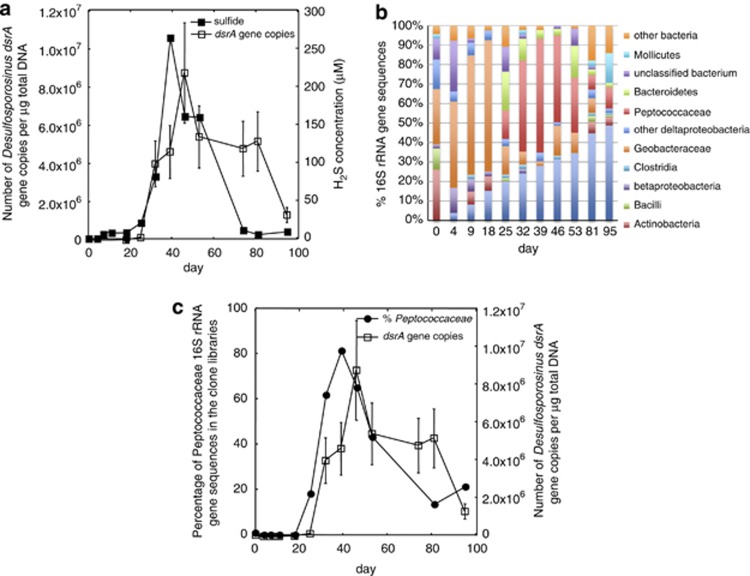
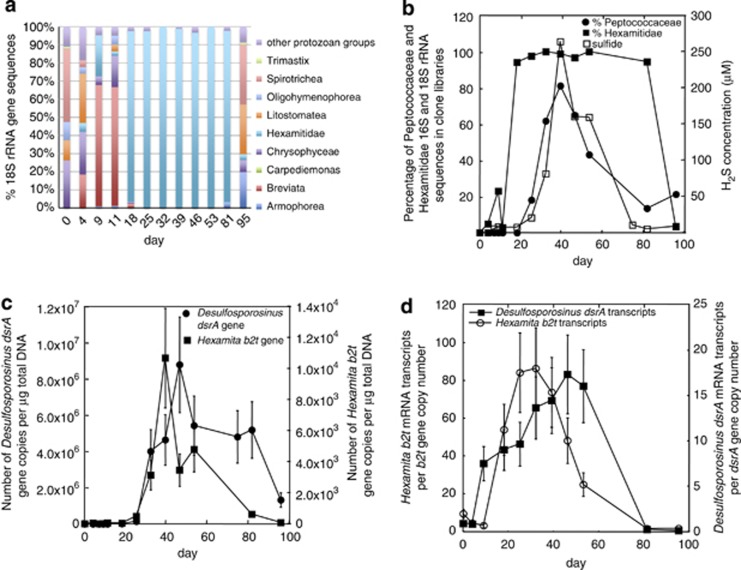
Similar to previous field experiments (Dar et al., submitted for publication; Vrionis et al., 2005; Miletto et al., 2011), continued addition of acetate after the Fe(III)-reducing phase of the experiment promoted sulfate reduction. Sulfate reduction during the 2011 experiment started as early as day 7 when sulfide began to accumulate in the groundwater (Supplementary Figure 3B). A dramatic increase in the number of dsrA gene copies associated with sulfate-reducing bacteria was observed after day 25 (Figure 4a).
Figure 4.
Analysis of the bacterial and protozoan communities found in the groundwater during the sulfate-reducing phase of the 2011 field experiment. (a) H2S concentration (μℳ) and the number of dsrA gene copies per μg total DNA (r=0.67, P=0.002). (b) Percentage breakdown of bacterial 16S rRNA gene sequences detected in clone libraries. (c) Proportion of Peptococcaceae 16S rRNA gene sequences compared with the number of dsrA gene copies per μg total DNA (r=0.79, P=0.002).
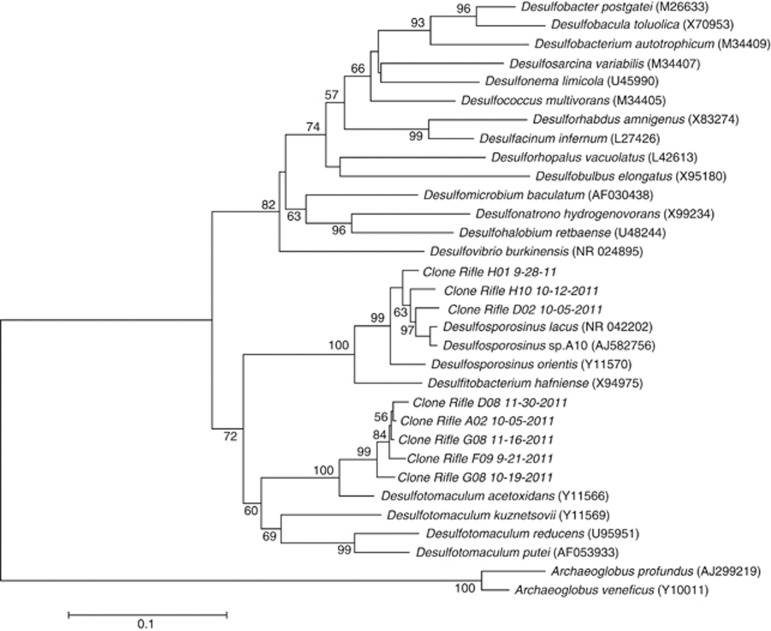
In previous field experiments, it was found that sulfate reducers phylogenetically affiliated with either the family Desulfobacteraceae or Peptococcaceae were significant members of the bacterial community (Anderson et al., 2003; Vrionis et al., 2005; Miletto et al., 2011; Dar et al., submitted for publication). In the 2011 field experiment, Peptococcaceae were predominant, accounting for 89% of the bacterial 16S rRNA gene sequences on day 39 when H2S concentrations were at their peak (Figure 4b). The majority of Peptococcaceae clones were most similar to Desulfotomaculum acetoxidans (95% similar) and Desulfosporosinus sp. A10 (96% similar) (Figure 5). There was a strong correlation between sulfide concentrations in the groundwater and both the proportion of Peptococcaceae sequences in the clone libraries (r=0.72, P=0.007) and the number of Desulfosporosinus dsrA gene copies estimated by qPCR (r=0.67, P=0.002). In addition, there was a strong correlation between the proportion of Peptococcaceae sequences and dsrA gene copies (r=0.79, P=0.002) (Figure 4c).
Figure 5.
Phylogenetic tree comparing 16S rRNA gene sequences from sulfate-reducing Peptococcaceae species detected in the groundwater to sequences from previously described sulfate-reducing prokaryotes.
The rise in sulfide concentrations and Peptococcaceae sequences was associated with a shift in the protozoan community. Species from the family Hexamitidae became predominant and accounted for up to 100% of the 18S rRNA gene sequences detected in the groundwater during this period (Figures 6a and b). All of the Hexamitidae 18S rRNA gene sequences were most similar to H. inflata (84% similar) or Trepomonas sp. steinii (83% similar), represented by clones RDL14-15-16 as in Figure 5. Clone library and qPCR studies showed a direct correlation between the percentage and abundance of sulfate-reducing Peptococcaceae and Hexamitidae species and between Hexamitidae and sulfide concentrations (r=0.64 and 0.67, P=0.02) (Figure 6b). qPCR analysis of Hexamita β-tubulin and Desulfosporosinus dsrA gene copies also demonstrated a strong correlation between the abundance of Hexamitidae and Desulfosporosinus cells in the groundwater (r=0.57, P=0.03) (Figure 6c). Furthermore, quantitative reverse transcription-PCR of Hexamita β-tubulin and Desulfosporosinus dsrA mRNA transcript abundance indicated that Hexamitidae species were metabolically active during the period when Hexamitidae were increasing in abundance and when Desulfosporosinus had high relative expression of dsrA (r=0.53, P=0.05) (Figure 6d).
Figure 6.
Analysis of the protozoan community associated with the sulfate-reducing phase of the 2011 field experiment. (a) Percentage breakdown of protozoan 18S rRNA gene sequences detected in clone libraries. (b) Percentage of Hexamitidae 18S and Peptococcaceae 16S rRNA gene sequences in clone libraries compared with sulfide concentrations. Peptococcaceae vs Hexamitidae rRNA sequences (r=0.64, P=0.02); Hexamitidae vs H2S concentrations (r=0.67, P=0.02). (c) Comparison of Hexamita β-tubulin (b2t) and Desulfosporosinus dsrA gene copies per μg total DNA (P=0.57, P=0.03). (d) The number of Desulfosporosinus dsrA and Hexamita β-tubulin mRNA transcripts normalized against the number of dsrA and β-tubulin gene copies (r=0.53, P=0.05).
Implications
These studies demonstrate that the specific enrichment of bacteria associated with the addition of acetate to the subsurface to promote U(VI) reduction in turn promotes the growth of specific bacteriovorous protozoa. Although it seems likely that the enrichment of different genera of protozoa in response to changes in the most abundant genera of bacteria is related to prey preferences, it is possible that other factors, such as changes in groundwater chemistry, might also be important.
Breviata species that become predominant during the Geobacter bloom are likely to have an impact on the growth and activity of the Geobacter species considered to be important in U(VI) reduction at this site (Lovley et al., 2011; Williams et al., 2011). Breviata have not been noted as dominant members of any previously reported protozoan communities. Breviata sequences were recovered from Dutch agricultural soils (Moon-van der Staay et al., 2006) and closely related ameboid flagellates from the genus Mastigamoeba have been detected in anoxic sediments collected from a sewage-contaminated aquifer (Novarino et al., 1997), a marine tidal flat (Dawson and Pace, 2002) and several freshwater ponds (Bernard et al., 2000). Although previous descriptions of B. anathema have classified this organism as a microaerophile (Klebs, 1892; Walker et al., 2006; Minge et al., 2009), we were able to grow Breviata under strictly anaerobic conditions with Geobacter uraniireducens provided as a food source (data not shown). Protozoa related to the Hexamitidae species that arose with the growth of sulfate reducers following the addition of acetate have been found in other freshwater aquatic environments (Lee et al., 2005; Luo et al., 2005), some of which contain high sulfide concentrations (Horie, 1969; Luo et al., 2005). In addition, Trepomonas species have been seen consuming bacteria in laboratory cultures, which provides support for in situ predation by Hexamitidae (Dujardin, 1841; Proceedings of Societies: Dublin Microscopical Club, 1879; Eyden and Vickerman, 1975).
At present, there is insufficient data to quantify the impact that Breviata and Hexamitidae species grazing on Geobacter and Peptococcaceae species might have on in situ U(VI) reduction. Previous studies have suggested that protozoan grazing can accelerate the metabolism of bacterial communities (Bloem et al., 1988; Verhagen et al., 1995; Strauss and Dodds, 1997; Biagini et al., 1998) and may prevent the reduction of subsurface permeability by preventing the accumulation of bacterial biomass (Kinner et al., 2002; Mattison et al., 2002). However, when the concentrations of electron donor, that is, acetate, are high, as they were during the field experiments, the number of catalytic units capable of reducing U(VI) is likely to be a major factor controlling the rate of U(VI) reduction. Thus, grazing of Geobacter, and possibly other bacterial species, would be expected to lower the rate of U(VI) reduction. These considerations indicate that attempts to model predictively the dynamics of microbial growth and activity during in situ uranium bioremediation (Scheibe et al., 2009; Lovley et al., 2011; Mahadevan et al., 2011) should include protozoan grazing.
Acknowledgments
Research at the University of Massachusetts was funded by the Office of Science (BER), US Department of Energy, Awards no. DE-SC0004080 and DE-SC0004814 and Cooperative Agreement no. DE-FC02-02ER63446. Additional support for field research was equally supported through the Integrated Field Research Challenge Site (IFRC) at Rifle, CO, USA and the Lawrence Berkeley National Laboratory's Sustainable Systems Scientific Focus Area. The US Department of Energy (DOE), Office of Science, Office of Biological and Environmental Research funded the work under contract DE-AC02-05CH11231 (Lawrence Berkeley National Laboratory; operated by the University of California).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Altschul S, Madden T, Schaffer A, Zhang J, Zhang Z, Miller W, et al. Gapped blast and psi-blast: a new generation of protein database search programs. Nucleic Acids Res. 1998;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson RT, Vrionis HA, Ortiz-Bernad I, Resch CT, Long PE, Dayvault R, et al. Stimulating the in situ activity of Geobacter species to remove uranium from the groundwater of a uranium-contaminated aquifer. Appl Environ Microbiol. 2003;69:5884–5891. doi: 10.1128/AEM.69.10.5884-5891.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker BJ, Lutz MA, Dawson SC, Bond PL, Banfield JF. Metabolically active eukaryotic communities in extremely acidic mine drainage. Appl Environ Microbiol. 2004;70:6264–6271. doi: 10.1128/AEM.70.10.6264-6271.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlett M, Zhuang K, Mahadevan R, Lovley D. Integrative analysis of Geobacter spp. and sulfate-reducing bacteria during uranium bioremediation. Biogeosciences. 2012;9:1033–1040. [Google Scholar]
- Bernard C, Simpson AGB, Patterson DJ. Some free-living flagellates (protista) from anoxic habitats. Ophelia. 2000;52:113–142. [Google Scholar]
- Biagini GA, Finlay BJ, Lloyd D. Protozoan stimulation of anaerobic microbial activity: enhancement of the rate of terminal decomposition of organic matter. FEMS Microbiol Ecol. 1998;27:1–8. [Google Scholar]
- Bloem J, Starink M, Bargilissen MJB, Cappenberg TE. Protozoan grazing, bacterial activity and mineralization in 2-stage continuous cultures. Appl Environ Microbiol. 1988;54:3113–3121. doi: 10.1128/aem.54.12.3113-3121.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brad T, Braster M, van Breukelen BM, van Straalen NM, Roling WFM. Eukaryotic diversity in an anaerobic aquifer polluted with landfill leachate. Appl Environ Microbiol. 2008;74:3959–3968. doi: 10.1128/AEM.02820-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron DA. Inorganic nutrients, baceria, and the microbial loop. Microb Ecol. 1994;28:295–298. doi: 10.1007/BF00166820. [DOI] [PubMed] [Google Scholar]
- Caron DA, Goldman JC, Dennett MR. Experimental demonstration of the roles of bacteria and bacterivorous protozoa in plankton nutrient cycles. Hydrobiologia. 1988;159:27–40. [Google Scholar]
- Chaudhry R, Shakoori AR. Characterization of copper resistant ciliates: potential candidates for consortia of organisms used in bioremediation of wastewater. Afr J Biotechnol. 2011;10:9101–9113. [Google Scholar]
- Cunningham JJ, Kinner NE, Lewis M. Protistan predation affects trichloroethene biodegradation in a bedrock aquifer. Appl Environ Microbiol. 2009;75:7588–7593. doi: 10.1128/AEM.01820-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dar SA, Tan H, Peacock A, Jaffe P, N'Guessan L, Williams KH, et al. Spatial distribution of Geobacteraceae and sulfate reducing bacteria during in situ bioremediation of uranium contaminated groundwater Appl Environ Microbiolsubmitted for publication.
- Dawson SC, Pace NR. Novel kingdom-level eukaryotic diversity in anoxic environments. Proc Natl Acad Sci USA. 2002;99:8324–8329. doi: 10.1073/pnas.062169599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decamp O, Warren A, Sanchez R. The role of ciliated protozoa in subsurface flow wetlands and their potential as bioindicators. Water Sci Technol. 1999;40:91–98. [Google Scholar]
- DeLeo PC, Baveye P. Factors affecting protozoan predation of bacteria clogging laboratory aquifer microcosms. Geomicrobiol J. 1997;14:127–12. [Google Scholar]
- DOE . Final Site Observational Work Plan for the UMTRA Old Rifle site GJO-99-88-TAR. DOE: Grand Junction, CO, USA; 1999. [Google Scholar]
- Dujardin F. Histoire naturelle des Zoophytes, Infusoires, comprenant la physiologie et la clasification de ces animaux et la manière de les étudier à l'aide du microscope. vol. i–xii. Librarie Encyclopedique de Roret: Paris, France; 1841. pp. 1–684. [Google Scholar]
- Dunbar J, Takala S, Barns SM, Davis JA, Kuske CR. Levels of bacterial community diversity in four arid soils compared by cultivation and 16S rRNA gene cloning. Appl Environ Microbiol. 1999;65:1662–1669. doi: 10.1128/aem.65.4.1662-1669.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eccleston-Parry JD, Leadbeater BSC. Regeneration of phosphorus and nitrogen by 4 species of heterotrophic nanoflagellates feeding on 3 nutritional states of a single bacterial strain. Appl Environ Microbiol. 1995;61:1033–1038. doi: 10.1128/aem.61.3.1033-1038.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden PA, Schmidt TM, Blakemore RP, Pace NR. Phylogenetic analysis of Aquaspirillum magnetotacticum using polymerase chain reaction amplified 16S ribosomal RNA specific DNA. Int J System Bacteriol. 1991;41:324–325. doi: 10.1099/00207713-41-2-324. [DOI] [PubMed] [Google Scholar]
- Ekelund F, Ronn R, Christensen S. Distribution with depth of protozoa, bacteria and fungi in soil profiles from three Danish forest sites. Soil Biol Biochem. 2001;33:475–481. [Google Scholar]
- Ellis BK, Stanford JA, Ward JV. Microbial assemblages and production in alluvial aquifers of the Flathead River, Montana, USA. J N Am Benthol Soc. 1998;17:382–402. [Google Scholar]
- Eyden BP, Vickerman K. Ultrastructure and vacuolar movements in the free-living diplomonad Trepomonas agilis klebs. J Euk Microbiol. 1975;22:54–66. [Google Scholar]
- Fang YL, Scheibe TD, Mahadevan R, Garg S, Long PE, Lovley DR. Direct coupling of a genome-scale microbial in silico model and a groundwater reactive transport model. J Contam Hydrol. 2011;122:96–103. doi: 10.1016/j.jconhyd.2010.11.007. [DOI] [PubMed] [Google Scholar]
- Fenchel T. Ecology of heterotrophic microflagellates. 4. Quantitative occurence and importance as bacterial consumers. Mar Ecol-Progr Ser. 1982;9:35–42. [Google Scholar]
- Fenchel T. The ecology of heterotrophic microflagellates. Adv Microb Ecol. 1986;9:57–97. [Google Scholar]
- Fenchel T, Ramsing NB. Identification of sulfate-reducing ectosymbiotic bacteria from anaerobic ciliates using 16S ribosomal RNA binding oligonucleotides probes. Arch Microbiol. 1992;158:394–397. doi: 10.1007/BF00276298. [DOI] [PubMed] [Google Scholar]
- Filatov DA. PROSEQ: A software for preparation and evolutionary analysis of DNA sequence data sets. Mol Ecol Notes. 2002;2:621–624. [Google Scholar]
- Fox PM, Davis JA, Hay MB, Conrad ME, Campbell KM, Williams KH, et al. 2012Rate-limited U(VI) desorption during a small-scale tracer test in a heterogeneous uranium-contaminated aquifer Water Resour Res 48W05512doi: 10.1029/2011WR011472 [DOI] [Google Scholar]
- Galluzzi L, Penna A, Bertozzini E, Vila M, Garces E, Magnani M. Development of a real-time PCR assay for rapid detection and quantification of Alexandrium minutum (a dinoflagellate) Appl Environ Microbiol. 2004;70:1199–1206. doi: 10.1128/AEM.70.2.1199-1206.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giloteaux L, Goni-Urriza M, Duran R. Nested PCR and new primers for analysis of sulfate-reducing bacteria in low-cell-biomass environments. Appl Environ Microbiol. 2010;76:2856–2865. doi: 10.1128/AEM.02023-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giloteaux L, Holmes DE, Williams K, Wrighton KC, Wilkins MJ, Smith J, et al. Characterization and transcription of arsenic respiration and resistance genes during in situ uranium bioremediation. ISME J. 2012;7:370–383. doi: 10.1038/ismej.2012.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni SJ, Delong EF, Olsen GJ, Pace NR. Phylogenetic group-specific oligodeoxynucleotide probes for identification of single microbial cells. J Bacteriol. 1988;170:720–726. doi: 10.1128/jb.170.2.720-726.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes DE, Giloteaux L, Barlett M, Chavan MA, Smith JA, Risso C, et al. 2013Molecular analysis of the growth rate of subsurface Geobacter species during in situ uranium bioremediation Appl Environ Microbiol(In press). [DOI] [PMC free article] [PubMed]
- Holmes DE, Nevin KP, O'Neil RA, Ward JE, Adams LA, Woodard TL, et al. Potential for quantifying expression of the Geobacteraceae citrate synthase gene to assess the activity of Geobacteraceae in the subsurface and on current-harvesting electrodes. Appl Environ Microbiol. 2005;71:6870–6877. doi: 10.1128/AEM.71.11.6870-6877.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie G. In Eutrophication: Causes, Consequences, Correctives. Nature Academy of Science: Washington, DC, USA; 1969. pp. 98–123. [Google Scholar]
- Kinner NE, Harvey RW, Blakeslee K, Novarino G, Meeker LD. Size-selective predation on groundwater bacteria by nanoflagellates in an organic-contaminated aquifer. Appl Environ Microbiol. 1998;64:618–625. doi: 10.1128/aem.64.2.618-625.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinner NE, Harvey RW, Shay DM, Metge DW, Warren A. Field evidence for a protistan role in an organically-contaminated aquifer. Environ Sci Technol. 2002;36:4312–4318. doi: 10.1021/es020611m. [DOI] [PubMed] [Google Scholar]
- Klebs G. Flagellatenstudien, Theil 1. Z Wissen Zool. 1892;55:265–445. [Google Scholar]
- Kota S, Borden RC, Barlaz MA. Influence of protozoan grazing on contaminant biodegradation. FEMS Microbiol Ecol. 1999;29:179–189. [Google Scholar]
- Lane DJ, Pace B, Olsen GJ, Stahl DA, Sogin ML, Pace NR. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc Natl Acad Sci USA. 1985;82:6955–6959. doi: 10.1073/pnas.82.20.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara E, Berney C, Ekelund F, Harms H, Chatzinotas A. Molecular comparison of cultivable protozoa from a pristine and a polycyclic aromatic hydrocarbon polluted site. Soil Biol Biochem. 2007;39:139–148. [Google Scholar]
- Lee WJ, Simpson AGB, Patterson DJ. Free-living heterotrophic flagellates from freshwater sites in Tasmania (Australia), a field survey. Acta Protozool. 2005;44:321–350. [Google Scholar]
- Lee ZM-P, Bussema C, Schmidt TM. rrnDB: documenting the number of rRNA and tRNA genes in bacteria and archaea. Nucleic Acids Res. 2009;37:D489–D493. doi: 10.1093/nar/gkn689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, McKinley J, Resch CT, Kaluzny R, Lauber CL, Fredrickson J, et al. Spatial and temporal dynamics of the microbial community in the Hanford unconfined aquifer. ISME J. 2012;6:1665–1676. doi: 10.1038/ismej.2012.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linley EAS, Newell RC, Lucas MI. Quantitative relationships between phytoplnakton, bacteria and heterotrophic microflagellates in shelf waters. Mar Ecol-Progr Ser. 1983;12:77–89. [Google Scholar]
- Lovley DR, Ueki T, Zhang T, Malvankar NS, Shrestha PM, Flanagan KA, et al. 2011Geobacter: the microbe electric's physiology, ecology, and practical applicationsIn: Poole RK, (ed)Advances in Microbial Physiology vol. 591–100. [DOI] [PubMed] [Google Scholar]
- Luo QW, Krumholz LR, Najar FZ, Peacock AD, Roe BA, White DC, et al. Diversity of the microeukaryotic community in sulfide-rich zodletone spring (Oklahoma) Appl Environ Microbiol. 2005;71:6175–6184. doi: 10.1128/AEM.71.10.6175-6184.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madoni P, Davoli D, Gorbi G, Vescovi L. Toxic effect of heavy metals on the activated sludge protozoan community. Water Res. 1996;30:135–141. [Google Scholar]
- Mahadevan R, Palsson BO, Lovley DR. In situ to in silico and back: elucidating the physiology and ecology of Geobacter spp. using genome-scale modelling. Nat Rev Microbiol. 2011;9:39–50. doi: 10.1038/nrmicro2456. [DOI] [PubMed] [Google Scholar]
- Margalef R. Modern orientations in hydrobiology. Scientia. 1958;93:41–46. [Google Scholar]
- Martin-Gonzalez A, Diaz S, Borniquel S, Gallego A, Gutierrez JC. Cytotoxicity and bioaccumulation of heavy metals by ciliated protozoa isolated from urban wastewater treatment plants. Res Microbiol. 2006;157:108–118. doi: 10.1016/j.resmic.2005.06.005. [DOI] [PubMed] [Google Scholar]
- Mattison RG, Harayama S. The predatory soil flagellate Heteromita globosa stimulates toluene biodegradation by a Pseudomonas sp. FEMS Microbiol Lett. 2001;194:39–45. doi: 10.1111/j.1574-6968.2001.tb09443.x. [DOI] [PubMed] [Google Scholar]
- Mattison RG, Taki H, Harayama S. The bacterivorous soil flagellate Heteromita globosa reduces bacterial clogging under denitrifying conditions in sand-filled aquifer columns. Appl Environ Microbiol. 2002;68:4539–4545. doi: 10.1128/AEM.68.9.4539-4545.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattison RG, Taki H, Harayama S. The soil flagellate Heteromita globosa accelerates bacterial degradation of alkylbenzenes through grazing and acetate excretion in batch culture. Microbial Ecol. 2005;49:142–150. doi: 10.1007/s00248-003-0226-5. [DOI] [PubMed] [Google Scholar]
- Miletto M, Williams KH, N'Guessan AL, Lovley DR. Molecular Analysis of the metabolic rates of discrete subsurface populations of sulfate reducers. Appl Environ Microbiol. 2011;77:6502–6509. doi: 10.1128/AEM.00576-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minge MA, Silberman JD, Orr RJS, Cavalier-Smith T, Shalchian-Tabrizi K, Burki F, et al. Evolutionary position of breviate amoebae and the primary eukaryote divergence. Proc R Soc Ser B. 2009;276:597–604. doi: 10.1098/rspb.2008.1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon-van der Staay SY, Tzeneva VA, van der Staay GWM, de Vos WM, Smidt H, JHP Hackstein. Eukaryotic diversity in historical soil samples. FEMS Microbiol Ecol. 2006;57:420–428. doi: 10.1111/j.1574-6941.2006.00130.x. [DOI] [PubMed] [Google Scholar]
- Muyzer G, Dewaal EC, Uitterlinden AG. Profiling of complex microbial populations by denaturing gel electrophoresis analysis of polymerase chain reaction amplified genes coding for 16S ribosomal RNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- N'Guessan AL, Vrionis HA, Resch CT, Long PE, Lovley DR. Sustained removal of uranium from contaminated groundwater following stimulation of dissimilatory metal reduction. Environ Sci Technol. 2008;42:2999–3004. doi: 10.1021/es071960p. [DOI] [PubMed] [Google Scholar]
- Nagaosa K, Maruyama T, Welikala N, Yamashita Y, Saito Y, Kato K, et al. Active bacterial populations and grazing impact revealed by an in situ experiment in a shallow aquifer. Geomicrobiol J. 2008;25:131–141. [Google Scholar]
- Novarino G, Warren A, Butler H, Lambourne G, Boxshall A, Bateman J, et al. Protistan communities in aquifers: a review. FEMS Microbiol Rev. 1997;20:261–275. doi: 10.1111/j.1574-6976.1997.tb00313.x. [DOI] [PubMed] [Google Scholar]
- Novarino G, Warren A, Kinner NE, Harvey RW. Protists from a sewage contaminated aquifer on Cape Cod, Massachusetts. Geomicrobiol J. 1994;12:23–36. [Google Scholar]
- Pielou EC. Measurement of diversity in different types of biological collections. J Theoret Biol. 1966;13:131–144. [Google Scholar]
- Priya M, Haridas A, Manilal VB. Anaerobic protozoa and their growth in biomethanation systems. Biodegradation. 2008;19:179–185. doi: 10.1007/s10532-007-9124-8. [DOI] [PubMed] [Google Scholar]
- Proceedings of Societies: Dublin Microscopical Club Q J Microsc Sci. 1879. pp. 438–448.
- Rehman A, Shakoori FR, Shakoori AR. Heavy metal resistant freshwater ciliate, Euplotes mutabilis, isolated from industrial effluents has potential to decontaminate wastewater of toxic metals. Bioresource Technol. 2008;99:3890–3895. doi: 10.1016/j.biortech.2007.08.007. [DOI] [PubMed] [Google Scholar]
- Reysenbach AL, Giver LJ, Wickham GS, Pace NR. Differential amplification of ribosomal-RNA genes by polymerase chain reaction. Appl Environ Microbiol. 1992;58:3417–3418. doi: 10.1128/aem.58.10.3417-3418.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribblett SG, Palmer MA, Coats DW. The importance of bacterivorous protists in the decomposition of stream leaf litter. Freshwater Biol. 2005;50:516–526. [Google Scholar]
- Rogerson A, Berger J. Enhancement of the microbial degradation of crude oil by the ciliate Colpidium colpoda. J Gen Appl Microbiol. 1983;29:41–50. [Google Scholar]
- Saito K, Drgon T, Robledo JAF, Krupatkina DN, Vasta GR. Characterization of the rRNA locus of Pfiesteria piscicida and development of standard and quantitative PCR-based detection assays targeted to the nontranscribed spacer. Appl Environ Microbiol. 2002;68:5394–5407. doi: 10.1128/AEM.68.11.5394-5407.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Salvado H, Gracia MP, Amigo JM, Rius M. Effects of cadmium on growth and motility in Euplotes aediculatus isolated from activated sludge. Bull Environ Contam Toxicol. 1997;58:838–844. doi: 10.1007/s001289900410. [DOI] [PubMed] [Google Scholar]
- Scheibe TD, Mahadevan R, Fang YL, Garg S, Long PE, Lovley DR. Coupling a genome-scale metabolic model with a reactive transport model to describe in situ uranium bioremediation. Microb Biotechnol. 2009;2:274–286. doi: 10.1111/j.1751-7915.2009.00087.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlenk D, Moore CT. Effect of pH and time on the acute toxicity of copper sulfate to the ciliate protozoan Tetrahymena thermophila. Bull Environ Contam Toxicol. 1994;53:800–804. doi: 10.1007/BF00196207. [DOI] [PubMed] [Google Scholar]
- Shakoori AR, Rehman A, Riaz ul H. Multiple metal resistance in the ciliate protozoan, Vorticella microstoma, isolated from industrial effluents and its potential in bioremediation of toxic wastes. Bull Environ Contam Toxicol. 2004;72:1046–1051. doi: 10.1007/s00128-004-0349-5. [DOI] [PubMed] [Google Scholar]
- Simpson EH. Measurment of diversity. Nature. 1949;163:688–688. [Google Scholar]
- Sinclair JL, Alexander M. Effect of protozoan predation on relative abundance of fast-growing and slow-growing bacteria. Can J Microbiol. 1989;35:578–582. [Google Scholar]
- Sinclair JL, Kampbell DH, Cook ML, Wilson JT. Protozoa in subsurface sediments from sites contaminated with aviation gasoline or jet fuel. Appl Environ Microbiol. 1993;59:467–472. doi: 10.1128/aem.59.2.467-472.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss EA, Dodds WK. Influence of protozoa and nutrient availability on nitrification rates in subsurface sediments. Microb Ecol. 1997;34:155–165. doi: 10.1007/s002489900045. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Terrado R, Medrinal E, Dasilva C, Thaler M, Vincent WF, Lovejoy C. Protist community composition during spring in an Arctic flaw lead polynya. Polar Biol. 2011;34:1901–1914. [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travis BJ, Rosenberg ND. Modeling in situ bioremediation of TCE at Savannah River: effects of product toxicity and microbial interactions on TCE degradation. Environ Sci Technol. 1997;31:3093–3102. [Google Scholar]
- Verhagen FJM, Laanbroek HJ, Woldendorp JW. Competition for ammonium between plant roots and nitrifying and heterotrophic bacteria and the effects of protozoan grazing. Plant Soil. 1995;170:241–250. [Google Scholar]
- Vrionis HA, Anderson RT, Ortiz-Bernad I, O'Neill KR, Resch CT, Peacock AD, et al. Microbiological and geochemical heterogeneity in an in situ uranium bioremediation field site. Appl Environ Microbiol. 2005;71:6308–6318. doi: 10.1128/AEM.71.10.6308-6318.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker G, Dacks JB, Embley TM. Ultrastructural description of Breviata anathema, N. Gen., N. Sp., the organism previously studied as ‘Mastigamoeba invertens'. J Eukaryot Microbiol. 2006;53:65–78. doi: 10.1111/j.1550-7408.2005.00087.x. [DOI] [PubMed] [Google Scholar]
- Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 16S Ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams KH, Long PE, Davis JA, Wilkins MJ, N'Guessan AL, Steefel CI, et al. Acetate availability and its influence on sustainable bioremediation of uranium-contaminated groundwater. Geomicrobiol J. 2011;28:519–539. [Google Scholar]
- Yabusaki SB, Fang Y, Long PE, Resch CT, Peacock AD, Komlos J, et al. Uranium removal from groundwater via in situ biostimulation: field-scale modeling of transport and biological processes. J Contam Hydrol. 2007;93:216–235. doi: 10.1016/j.jconhyd.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Yagi JM, Neuhauser EF, Ripp JA, Mauro DM, Madsen EL. Subsurface ecosystem resilience: long-term attenuation of subsurface contaminants supports a dynamic microbial community. ISME J. 2010;4:131–143. doi: 10.1038/ismej.2009.101. [DOI] [PubMed] [Google Scholar]
- Zarda B, Mattison G, Hess A, Hahn D, Hohener P, Zeyer J. Analysis of bacterial and protozoan communities in an aquifer contaminated with monoaromatic hydrocarbons. FEMS Microbiol Ecol. 1998;27:141–152. [Google Scholar]
- Zhu F, Massana R, Not F, Marie D, Vaulot D. Mapping of picoeucaryotes in marine ecosystems with quantitative PCR of the 18S rRNA gene. FEMS Microbiol Ecol. 2005;52:79–92. doi: 10.1016/j.femsec.2004.10.006. [DOI] [PubMed] [Google Scholar]
- Zhuang K, Izallalen M, Mouser P, Richter H, Risso C, Mahadevan R, et al. Genome-scale dynamic modeling of the competition between Rhodoferax and Geobacter in anoxic subsurface environments. ISME J. 2011;5:305–316. doi: 10.1038/ismej.2010.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.