Abstract
The indoor microbiome is a complex system that is thought to depend on dispersal from the outdoor biome and the occupants' microbiome combined with selective pressures imposed by the occupants' behaviors and the building itself. We set out to determine the pattern of fungal diversity and composition in indoor air on a local scale and to identify processes behind that pattern. We surveyed airborne fungal assemblages within 1-month time periods at two seasons, with high replication, indoors and outdoors, within and across standardized residences at a university housing facility. Fungal assemblages indoors were diverse and strongly determined by dispersal from outdoors, and no fungal taxa were found as indicators of indoor air. There was a seasonal effect on the fungi found in both indoor and outdoor air, and quantitatively more fungal biomass was detected outdoors than indoors. A strong signal of isolation by distance existed in both outdoor and indoor airborne fungal assemblages, despite the small geographic scale in which this study was undertaken (<500 m). Moreover, room and occupant behavior had no detectable effect on the fungi found in indoor air. These results show that at the local level, outdoor air fungi dominate the patterning of indoor air. More broadly, they provide additional support for the growing evidence that dispersal limitation, even on small geographic scales, is a key process in structuring the often-observed distance–decay biogeographic pattern in microbial communities.
Keywords: bioaerosol, built environment microbiome, community assembly, isolation by distance, ITS, 454 pyrosequencing
Introduction
Microbial components to indoor air are receiving increased attention, but the sources of the microbial communities and the processes that affect them are not well understood (Corsi et al., 2012). Exposure to bioaerosols, including fungal ones, has been linked to a range of detrimental health effects (Douwes et al., 2003); for example, molds are associated with the onset of asthma in both infants (Jaakkola et al., 2010) and adults (Karvala et al., 2010). Conversely, the hygiene hypothesis, which posits that exposure to microbial material early in life can actually be preventative in developing disease later in life (Strachan, 1989), continues to find empirical support (for example, Iossifova et al., 2007). Because of this link to human health, the complex microbial habitat of the built environment continues to be explored, and new technologies allow for a richer understanding of the ecological context of microorganisms indoors.
In particular, early work directed at indoor microbial ecology was based primarily on culture-based methods, but in the last several years it has been facilitated by sequence-based methods. Together, these approaches have identified fungi that grow indoors when water is available (for example, Ara et al., 2004; Anderson et al., 2011; Zalar et al., 2011), that are shed from humans or pets (Paulino et al., 2006), and that have obvious outdoor origins (Pitkaranta et al., 2008). From recent studies that relied on PCR-based techniques to assess total fungal assemblages in two pairs of buildings with and without water damage, we know that fungi in settled dust are diverse, that variation between buildings can be large and fluctuate with the outdoor air, and that, as a result, there may not be a typical profile of indoor fungi that distinguishes the two types of buildings (Huttunen et al., 2008; Pitkaranta et al., 2011). We also know that on a global scale, the geographical patterning of communities dominates over the effect of individual buildings, which suggests that the outdoor environment is a primary factor at the broadest scale (Amend et al., 2010).
Elucidating which microorganisms are present in indoor air is fundamentally a question of community assembly. Vellend (2010) recently posited that issues in community ecology would benefit from an approach emphasizing four fundamental processes that have analogies in population genetics: selection, dispersal, community drift and speciation. Adopting this approach to explain the biogeographic patterns now commonly observed in microorganisms, Hanson et al. (2012) recently argued that selection and community drift generate the distance–decay relationship, and dispersal counteracts it, espousing the ‘everything is everywhere, but the environment selects' paradigm (Baas Becking 1934; Finlay 2002), while also allowing for random demographic fluctuations. However, this view that microbes are not dispersal limited is not substantiated by direct measures of dispersal limitation in microorganisms. Most often, statistical methods are used to separate the effects of geographic distance and habitat heterogeneity (Martiny et al., 2006). In an attempt to experimentally disentangle the effect of habitat filtering with geographic distance, Peay et al. (2012) measured dispersal directly for ectomycorrhizal fungi from a forest edge and found decreases of spore numbers from individual species and decreasing species richness of spores, with distances of <1 km. Although obviously more studies are needed, this study suggests that dispersal limitation, independent of selection, can be an important process in creating biogeographic patterns of fungi.
In the current study, we use a high-throughput sequence approach and focus on a local scale, by sampling the airborne indoor fungi at one locality and one building type with high replication, thus holding constant many of the variables that might otherwise effect fungal content of these buildings. We sample air indirectly as settled dust, but do so within a defined, 4-week time window. The settled dust approach has been used extensively in indoor air studies (Rintala et al., 2012), although typically it is taken from surfaces (vacuum samples from floor, swab from shelf and so on) intentionally as a time-integrated sample of airborne material (Institute of Medicine, 2004). However, samples of particular time windows are necessary to understand the processes structuring that long-term accumulation, and Würtz et al. (2005) developed a dustfall collector to sample dust for a given duration of time. This kind of empty, sterile sampler has another advantage in addition to collecting for a distinct time period—and in this way, it is similar to active air samplers that pump air through a filter—in that, it only measures the airborne dispersed propagules of that environment.
Our goals in the current study are to catalog the composition of the aeromycoflora in ‘healthy' buildings (that is, no reported mold problems) and to identify the processes that determine the fungal assemblages, and specifically the processes that influence the dispersal and selection regimes of microbes in the indoor space. We hypothesized that the majority of indoor air fungi are derived from outdoor air. Second, we hypothesized that variation in microbial communities between residences would be large and partly attributable to selection pressures imposed by resident behavior. Third, we hypothesized that airborne fungal communities would vary among rooms within a building based on room use and water availability. We expected that cleaning and cooking patterns and exposure to the outdoors would have a predictable effect on the composition of the indoor fungal communities and that the air in rooms with water sources (bathrooms and kitchens) would be marked by a resident fungal population.
Materials and methods
Study location, sample collection and metadata
To explore variation between buildings while minimizing variation in building construction and design, we sampled the uniform housing of a university-sponsored family housing complex (Aerial photo, Supplementary Figure 1). The residences have a similar floor plan, were constructed between 1999 and 2008, and have a mean time of occupancy per family of about 2 years. The exterior siding is fiber-cement (HardiPlank) with vinyl window sash and frames, and the interior is painted sheetrock. The kitchen and bathrooms are floored with vinyl tiles, whereas the remaining areas are carpeted. Each of the buildings has its own forced-air ventilation system with heating but no air conditioning, and the entire housing complex occupies less than a square kilometer. Volunteer involvement was regulated under the University of California's Committee for the Protection of Human Subject Protocol ID 2011-03-2947.
As mentioned, airborne dust was collected using a simple, passive, sampling scheme that involved suspending a sterile, empty, 9 cm petri dish, 0.3 m from the ceiling (Supplementary Figure 2). This petri-dish sampler is a variation on the ‘pizza box' sampling scheme (the dustfall collector; Würtz et al., 2005), favors high replication and has the advantage of being smaller and less obtrusive. At each apartment (hereafter referred to as a unit), a total of four petri-dish samplers were situated indoors (kitchen, living room, bathroom and bedroom), and one was placed outdoors (on the patio or deck). Sampling was carried on for 4 weeks at two seasons, summer (July 2011, 11 units) and winter (January 2012, 8 of those same units). We interviewed residents to complete a questionnaire on house characteristics (Table 1); three measured factors—the use of a humidifier, the presence of houseplants and the frequency of cooking—were invariant and therefore not included in analyses. We measured air temperature and relative humidity for the duration of the sampling period (HOBO U10; Onset Computer Corporation, Bourne, MA, USA).
Table 1. Significance and variance in community composition in indoor airborne fungal communities explained by different environmental and behavioral factors.
| Factor |
Single factor |
Multiple factors |
||||||
|---|---|---|---|---|---|---|---|---|
|
Summer |
Winter |
Summer |
Winter |
|||||
| P-value | R2 | P-value | R2 | P-value | R2 | P-value | R2 | |
| Categorical | ||||||||
| Unit | 0.001 | 0.48 | 0.001 | 0.35 | — | — | — | — |
| Room type | — | — | — | — | — | — | — | — |
| Unit age | — | — | 0.027 | 0.14 | — | — | — | — |
| Floor number | 0.004 | 0.15 | 0.008 | 0.15 | — | — | — | — |
| Number of bedrooms | 0.016 | 0.11 | — | — | — | — | — | — |
| Number of bathrooms | — | — | — | — | — | — | — | — |
| Number of residents | — | — | 0.030 | 0.18 | — | — | — | — |
| Frequency of cleaning | 0.001 | 0.26 | 0.018 | 0.15 | — | — | — | — |
| Continuous | ||||||||
| Geographic distance | 0.001 | 0.20 | 0.003 | 0.19 | 0.001 | 0.33 | 0.011 | 0.24 |
| Temperature | 0.035 | 0.21 | — | — | — | — | — | — |
| Relative humidity | — | — | — | — | — | — | — | — |
| Temperature variance | — | — | — | — | — | — | — | — |
| Relative humidity variance | — | — | — | — | — | — | — | — |
Community composition was calculated using the β-sim distance metric. Single-factor analyses were completed with ADONIS (permutational multivariate analysis of variance) for categorical variables and Mantel correlations for continuous variables. Multi-factor analyses considered all variables in ADONIS. For simplicity, insignificant factors (that is, P>0.05) are excluded.
DNA extraction and amplification
For nucleotide extraction from the petri-dish sampler, a cotton swab was moistened with sterile water and used to wipe the surface. The tip of the swab was then cut directly into a tube containing lysing matrix E beads (MP Biomedicals, Burlingame, CA, USA). A volume of 400 μl each of Miller Phosphate Buffer and Miller SDS lysis buffer (Miller et al., 1999) and 450 μl of 25:24:1 phenol: chloroform: isoamyl alcohol were added. Tubes were homogenized using a Biospec Mini 8 bead mill (Biospec, Bartlesville, OK, USA) at full speed for 1 min. The supernatant, approximately 560 μl, was isolated after centrifuging at 10 000 g for 5 min at 4 °C and mixed with an equal volume of chloroform. The supernatant was again isolated, approximately 480 μl, after centrifuging at 10 000 g for 5 min at 4 °C, and then processed further with the MoBio Power Soil DNA Extraction Kit (Carlsbad, CA, USA), starting with two volumes of solution C4 to isolate DNA onto the spin filters. Genomic DNA was further cleaned with the C5 ethanol-based wash solution and finally eluted in 100 μl warm C6 elution buffer run twice through the filter. Negative controls of sterile cotton swabs were also processed, and any taxa detected were filtered out of the other samples.
Pyrosequencing
Amplification targeted the internal transcribed spacer (ITS) region 1 of the nuclear ribosomal coding cistron, part of the region formally proposed as a universal DNA barcode for fungi (Schoch et al., 2012). Forward primers comprised the 454 Fusion Primer A-adaptor, a specific 8-bp multiplex identifier (MID) barcode, and the ITS1F primer (5′-CTTGGTCATTTAGAGGAAGTAA-3′ Gardes and Bruns, 1993), while the reverse primer was composed of the B-adapter and ITS2 primer (5′-GCTGCGTTCTTCATCGATGC-3′ White et al., 1990).
Pyrosequencing PCR mixtures contained 0.25 μl of HotStarTaq polymerase (Qiagen, Valencia, CA, USA), 2.5 μl of 10 × PCR buffer supplied by manufacturer, 2.5 μl 10 × each dNTPs (200 μℳ), 0.2 μl of 50 μℳ reverse primer, 1 μl of 10 μℳ forward primer, 0.25 μl of 100 mg ml−1 BSA, 5 μl DNA template (some samples diluted 1:10 to overcome inhibitors) and water up to 25 μl. Following an initial denaturation at 95 °C for 15 min to activate the polymerase, samples were amplified by 35 cycles of 94 °C for 1 min, 51 °C for 1 min and 72 °C for 1 min, and subjected to a final extension at 72 °C for 10 min. Samples were PCR-amplified in triplicate and pooled before cleaning using AMPure magnetic beads (Beckman Coulter Genomics, Danvers, MA, USA). Amplicon samples were then individually quantified using the Qubit flourometer (Invitrogen, Carlsbad, CA, USA) and pooled to an equimolar concentration. Sequences were run on 1/8th of a 454 FLX Titanium pico-titer plate at the Duke Institute for Genomic Sciences and Policy (Durham, NC, USA) and submitted to the National Center for Biotechnology Information Sequence Read Archive under accession number SRA059097.
Quantitative PCR
To compare differences in fungal community composition with fungal biomass, we employed quantitative PCR (qPCR). The assay used universal fungal primers FF2 and FR1 that amplify a 425-bp region of the small subunit rRNA (Zhou et al., 2000). PCR mixtures were run on a Real-Time PCR System (Applied Biosystems 7300, Carlsbad, CA, USA) containing 5 μl 1:10 dilution genomic DNA, 10 μl iTAQ SYBR Green Supermix with ROX (Bio-Rad, Hercules, CA, USA), 0.2 μl of 100 mg ml−1 BSA, 0.15 μl of each 50 μℳ primer and water to 20 μl. To explore variation in fungal biomass estimation based on specific DNA regions, a subset of samples was amplified in parallel with the primers used for pyrosequencing (that is, ITS1F and ITS2), which target a locus that varies in length across taxa. The correlation of estimates of fungal biomass between the two primer pairs was high (R2=0.978), and results are reported only for the small subunit primers, as the target is of invariable length across taxa, and therefore less likely to bias the qPCR technique. Thermal cycling conditions consisted of an initial denaturation at 95 °C for 3 min, followed by 40 cycles of 15 s of denaturation at 95 °C and 1 min of annealing and extension at 60 °C, finishing with a dissociation stage of 95 °C for 15 s, 60 °C for 30 s and 95 °C for 15 s. Standard curves were developed using a six-order of magnitude dilution series (ranging from 5 × 10−1–105 spores) from a known concentration of Penicillium spinulosum spores counted using a 1400-mm hemocytometer (Hausser Scientific, Horsham, PA, USA). The dissociation phase ensured that the melt curve values of the samples fell within the values of the standard curve. To confirm that inhibitory effects in airborne samples were not impeding amplification (Alvarez et al., 1995), a quarter of the samples were also run with a PCR mix containing a known amount of spores; no inhibition was observed for this subset at the 1:10 dilution in which all samples were processed. All qPCR reactions were replicated three times per sample, and the analysis relied on the mean value.
Analysis of pyrosequencing data
Pyrosequence data were processed using the Quantitative Insights Into Microbial Ecology (QIIME) pipeline. Sequences were trimmed to 100–400 bp for this primer combination and had to meet a mean minimum quality score of 25. Of the 129 768 raw input sequences, 62 478 remained for downstream analysis (most dropping out due to lengths <100 bp), resulting in a mean of 664 sequences for each of the 94 samples. Sequences were denoised in one batch using the flowgram clustering algorithm (Reeder and Knight, 2010), and the ITS1 spacer was extracted from the sequence (Nilsson et al., 2010) and used as the basis for comparison. We then used USEARCH (Edgar, 2010) as implemented in the QIIME pipeline to remove chimeric sequences relying on the UCHIME algorithm (Edgar et al., 2011) and referenced against the UNITE database, and to cluster sequences into operational taxonomic units (OTUs) to 97% similarity (via UCLUST). We removed singleton OTUs, and the resulting OTU table was rarefied to 100 sequences per sample, thus reducing the sample size to 84. An even sampling depth of 100 sequences struck a balance between retaining the greatest number of samples while eliminating samples with low coverage, and increasing the sampling depth to greater values did not change results (data not shown). Community analyses relied on the vegan (Oksanen et al., 2012), labdsv (Roberts, 2012) and sp (Pebesma and Bivand, 2012) packages in R (R Development Core Team, 2012). The resulting OTU table was converted to presence/absence rather than relying on read abundance for subsequent analyses. OTU richness was estimated using the Chao 1 function (Chao, 1987) and calculated in EstimateS (Colwell, 2006) based on 100 randomizations.
Community composition was visualized using nonmetric multidimensional scaling and relied on the βsim distance metric (Lennon et al., 2001), which controls for large differences in richness between samples, as seen between our indoor and outdoor samples. Environmental and behavioral factors predicting community composition were analyzed with ADONIS, a permutational multivariate analysis of variance (Oksanen et al., 2012). Mantel correlations were used to directly compare various distances with community-composition differences. Initially factors were considered individually, but because unit explained such a large portion of the variance in fungal community composition, we also measured the variance of environmental and behavioral factors after accounting for the variance explained by unit in a multifactorial ADONIS model. To consider geographic distances in the multifactorial models, geographic locations were converted into a principal coordinates of neighborhood matrix (Oksanen et al., 2012).
Indicator taxa were identified within QIIME, and taxonomy was assigned by running the BLAST algorithm against the UNITE fungal database (Abarenkov et al., 2010), version updated on 13 April 2012. The BLAST output was analyzed in MEGAN (Huson et al., 2011) and FHiTINGS (Dannemiller et al., in review). To explore whether the assemblages of fungi inside represented a biased sample on those outside or were more likely a stochastic subset, we compared the observed community compositional distance to those estimated from series of simulated random communities. Specifically, we used the ‘oecosimu' function in the vegan package (Oksanen et al., 2012) of R to evaluate the βsim distance metric (Lennon et al., 2001) between the true outdoor and indoor samples, and compared that with simulations (n=999) in which communities were permuted using a predefined algorithm (‘swsh_samp') that works by, first, swapping smaller matrices within the larger one, and, second, shuffling samples among the new non-zero cells (Oksanen et al., 2012). We rejected the null hypothesis of no signal for selective filtering of the outdoor taxa by the indoor environment if the observed distance was outside the 95% quantile of simulated distances.
Results
Taxa richness and community composition
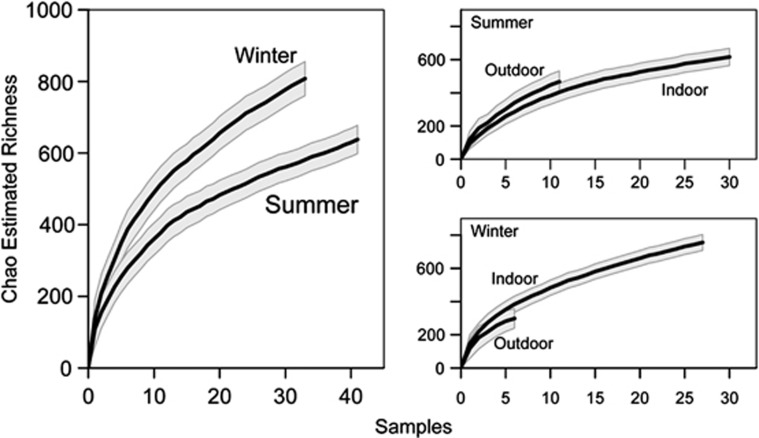
We found a total of 986 fungal OTUs across all sampling locations and seasons. Richness of individual indoor samples ranged from 17 to 271 OTUs, and individual outside samples ranged from 78 to 263 OTUs. Across seasons, 643 OTUs appeared in both winter and summer, and across localities, 619 OTUs appeared in both indoor and outdoor samples. However, although the majority of OTUs were cosmopolitan, they were not frequent, and our data showed the long tail of low-abundance taxa common in microbial communities (Reeder and Knight, 2009; Huse et al., 2010), even after implementing strict standards of denoising and removing singleton OTUs. In the 84 samples, 10 OTUs appeared in 65 or more samples (Table 2; Supplementary Figure 3), whereas 100 OTUs appeared in only one (Supplementary Figure 3). The Chao richness estimators did not approach asymptotes (Figure 1), and Good's coverage (Good 1953) indicates that results detected 20.7–58.1% (median 36.8%) of the total OTUs (Supplementary Table 1). Estimated richness was higher for the winter samples than the summer samples. Within seasons, however, there was not a consistent pattern of higher richness outdoors or indoors, and the outdoor and indoor estimations are statistically indistinguishable (Figure 1).
Table 2. The 25-most abundant fungal taxa in all airborne samples identified by the top BLAST match.
| OTU rank | BLAST-assigned taxonomy | Length (bp) | % Sequence similarity |
Frequency |
||
|---|---|---|---|---|---|---|
| Indoor | Outdoor | Total | ||||
| 1 | Cryptococcus victoriae | 131 | 99.2 | 94.0 (63/67) | 100 (17/17) | 80 |
| 2 | Cladosporium sp. | 84 | 98.8 | 91.0 (61/67) | 100 (17/17) | 78 |
| 3 | Epicoccum sp. | 156 | 93.6 | 88.1 (59/67) | 100 (17/17) | 76 |
| 4 | Penicillium sp. | 36 | 97.2 | 89.6 (60/67) | 94.1 (16/17) | 76 |
| 5 | Cryptoccocus sp. | 155 | 96.8 | 88.1 (59/67) | 100 (17/17) | 76 |
| 6 | Cladosporium sp. | 70 | 98.6 | 80.6 (54/67) | 100 (17/17) | 71 |
| 7 | Epicoccum sp. | 30 | 100 | 79.1 (53/67) | 100 (17/17) | 70 |
| 8 | none | 103 | — | 76.1 (51/67) | 100 (17/17) | 68 |
| 9 | Aureobasidum pullulans | 51 | 100 | 74.6 (50/67) | 100 (17/17) | 67 |
| 10 | Alternaria sp. | 171 | 99.4 | 71.6 (48/67) | 100 (17/17) | 65 |
| 11 | Cladosporium sp. | 87 | 95.4 | 70.1 (47/67) | 100 (17/17) | 64 |
| 12 | Phoma sp. | 143 | 91.6 | 58.2 (39/67) | 100 (17/17) | 56 |
| 13 | Epicoccum sp. | 156 | 93.6 | 56.7 (38/67) | 100 (17/17) | 55 |
| 14 | Cladosporium sp. | 90 | 98.9 | 56.7 (38/67) | 100 (17/17) | 55 |
| 15 | Stemphylium sp. | 96 | 96.9 | 55.2 (37/67) | 100 (17/17) | 54 |
| 16 | Cladosporium sp. | 85 | 100 | 53.7 (36/67) | 100 (17/17) | 53 |
| 17 | Cryptoccocus carnescens | 132 | 100 | 50.7 (34/67) | 100 (17/17) | 51 |
| 18 | Penicillium brevicompactum | 182 | 98.9 | 58.2 (39/67) | 70.6 (12/17) | 51 |
| 19 | none | 80 | — | 49.3 (33/67) | 94.1 (16/17) | 49 |
| 20 | Sporobolomyces sp. | 149 | 95 | 47.8 (32/67) | 94.1 (16/17) | 48 |
| 21 | Epicoccum sp. | 34 | 100.0 | 50.7 (34/67) | 82.4 (14/17) | 48 |
| 22 | Stemphylium sp. | 120 | 90.0 | 46.3 (31/67) | 94.1 (16/17) | 47 |
| 23 | Aspergillus sp. | 91 | 97.8 | 47.8 (32/67) | 64.7 (11/17) | 43 |
| 24 | Aureobasidum pullulans | 167 | 98.2 | 41.8 (28/67) | 88.2 (15/17) | 43 |
| 25 | Rhodotorula mucilaginosa | 148 | 96.0 | 50.7 (34/67) | 52.9 (9/17) | 43 |
Frequency of the taxon in indoor and outdoor samples is given as percentage followed by proportion of samples, followed by the total number in all samples. Seasons are pooled, and the maximum number of samples is 84.
Figure 1.
Rarefaction curves of estimated OTU richness across seasons, and within each season across outdoor and indoor samples. The estimated richness with a solid black line and the s.d. in gray shading are shown. Richness was estimated to be greater in winter than in summer, while within seasons, the indoor and outdoor samples accumulate OTUs at a similar rate.
Fungal community composition, as visualized by nonmetric multidimensional scaling (stress=0.29), shows a strong temporal pattern in that summer and winter samples are distinct (Supplementary Figure 4). Moreover, outdoor fungal communities are clustered relative to indoor ones and largely distinct from the indoor communities.
Fungal biomass
Fungal biomass, as determined by qPCR, largely mirrored results of richness and compositional differences (Supplementary Figure 5). Fungal biomass outdoors tended to be greater in winter than in summer, though the difference was not significant (analysis of variance, P=0.30), and was significantly greater than indoor fungal biomass within each of the two seasons (summer, P=0.03; winter, P<0.01). There was no significant difference in biomass across the indoor rooms (summer, P=0.33; winter, P=0.71).
Predictors of community composition
Several potential predictors of fungal community composition were collected for each unit (Table 1). These included both physical parameters of the space, for example, the location and the number of bedrooms and bathrooms, and behavioral characteristics of the residents of the unit. Considered individually, several of these factors were significant predictors of fungal community composition (P<0.01; R2 0.11–0.48; Table 1). These significant predictors included: unit, floor level and frequency of cleaning, all of which were significant across seasons. Also included was the continuous variable of geographic distance.
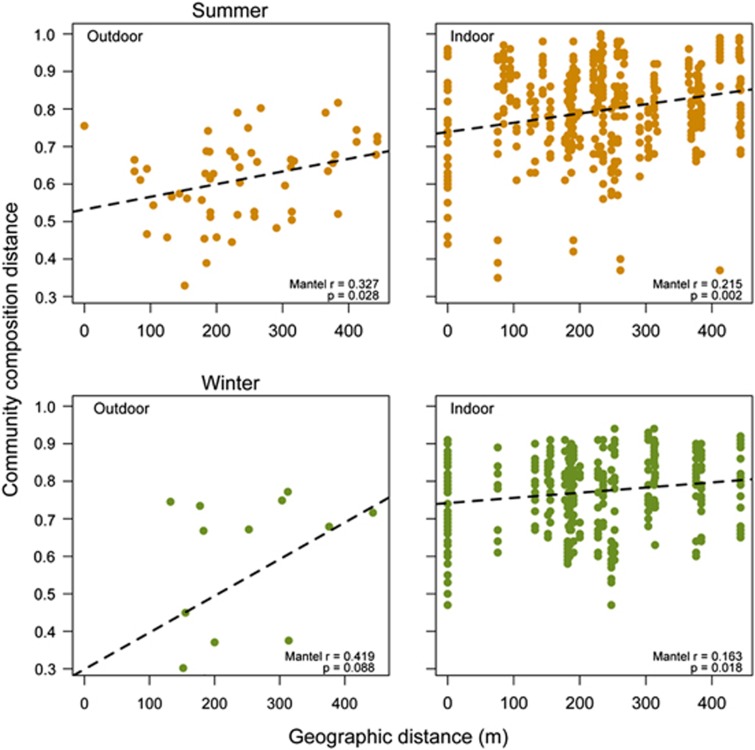
However, when considered as factors in a multifactor model, the linked factors of geographic distance or unit remained the only significant predictor. The analysis of variance method partitions variations in the order in which the factors are entered into the model (Oksanen et al., 2012). The first factor in the model, whether it was unit or geographic distance, was the only one that explained a significant portion of the variation in fungal community composition. (For this reason, in Table 1, we report the result from geographic distance solely.) Thus, the single factor in our model that structures indoor airborne fungal communities is location in space, whether described by geographic distance or unit (Table 1). In the multi-factored model, the R2 for geographic distance was at least 0.24 in winter and 0.33 in summer. This strong signal of isolation by distance is apparent across seasons in both outdoor and indoor samples (Figure 2) and occurs on the spatial scale of hundreds of meters. The strength of the correlation was stronger outdoors than indoors.
Figure 2.
Community-composition difference as a function of geographic distance. Distances in (Bray–Curtis) community-composition differences increase with geographic distance for indoor and outdoor samples across both seasons.
Taxonomy and indicator taxa
The most abundant fungal taxa were molds and yeasts and were dominant in both indoor and outdoor samples (Table 2). Sixteen of the twenty-five most abundant taxa were present in every outdoor sample and appeared in over half of the indoor ones. Many of the molds and yeasts like Rhotodorula are recognized from culture-based surveys (Ara et al., 2004), whereas the diversity within and abundance of Cryptococcal yeasts have largely been missed by culture-based surveys (Table 2; order Tremellomycetes in Table 3). Overall, pyrosequencing revealed at least 18 orders of fungi encompassing distinct evolutionary lineages and a diversity of morphologies (Table 3). In addition to fungi expected indoors—for example, saprotrophic Dothideomycete and Wallemiomycete molds—we also detected many taxa with a clear outdoor origin: plant pathogens, lichenized fungi, mushrooms and puffballs. In fact, the order with the most taxon richness was Agaricomycetes—outdoor fungi that are predominately mushrooms and polypores that decay plants. Approximately half of the most abundant taxa matched a known specimen with >98% similarity (Table 2). Two of the most abundant taxa did not have a BLAST match (Table 2), and overall 130 OTUs (16%) were unclassified (Table 3).
Table 3. Orders of fungi and their representation.
| Order | Number of OTUs | Description | Primary source | |
|---|---|---|---|---|
| 1 | Agaricomycetes | 238 | Mushrooms and polypores | Outdoor |
| 2 | Dothideomycetes | 208 | Molds | Mixed |
| 3 | Tremellomycetes | 111 | Yeasts | Mixed |
| 4 | Leotiomycetes | 54 | Plant pathogens and saprobes | Outdoor |
| 5 | Eurotiomycetes | 48 | Molds | Mixed |
| 6 | Sordariomycetes | 39 | Plant pathogens and saprobes | Mixed |
| 7 | Microbotryomycetes | 35 | Yeasts | Mixed |
| 8 | Saccharomycetes | 19 | Yeasts | Mixed |
| 9 | Chytridiomycetes | 9 | Aquatic saprobes and pareasites | Outdoor |
| 10 | Wallemiomycetes | 8 | Molds | Mixed |
| 11 | Lecanoromycetes | 7 | Lichenized fungi | Outdoor |
| 12 | Pezizomycetes | 6 | Mushrooms and molds | Outdoor |
| 13 | Agaricostilbomycetes | 5 | Yeasts | Outdoor |
| 14 | Glomeromycetes | 3 | Plant root biotrophs | Outdoor |
| 15 | Taphrinomycetes | 3 | Plant pathogens | Outdoor |
| 16 | Orbiliomycetes | 2 | Saprobes | Outdoor |
| 18 | Cystobasidiomycetes | 1 | Yeasts | Outdoor |
| 19 | Ambiguous | 44 | ||
| 20 | Incertae sedis | 16 | ||
| 21 | Unclassified | 130 |
The abundance of OTUs, which were identified to order by BLAST match, were summed. The representative ecology for those OTUs are given, and their proposed source, either outdoor or mixed indoor and outdoor, are also given.
Indicator taxa analysis identified few taxa that clustered with a particular category, and the ones that did cluster were indicative of the winter or the outdoors. For example, an OTU with a BLAST-assigned identity of Stereum hirsutum clustered with the winter samples, which is when this wood-decay fungus primarily produces fruiting bodies and sporulates. Likewise, clustering with the outdoor samples was a likely member of Stemphylium, a genus associated with plants. Some species of this genus cause allergies in humans (Simon-Nobbe et al., 2008), and their growth indoors is known to be limited. However, no taxa were indicative of the indoor environment or a particular unit or room.
Simulating indoor assemblages from outdoor sources
Simulations showed that observed distances in the communities between indoor and outdoor samples for each unit were within the range of randomized distances (Supplementary Figures 6 and 7).
Discussion
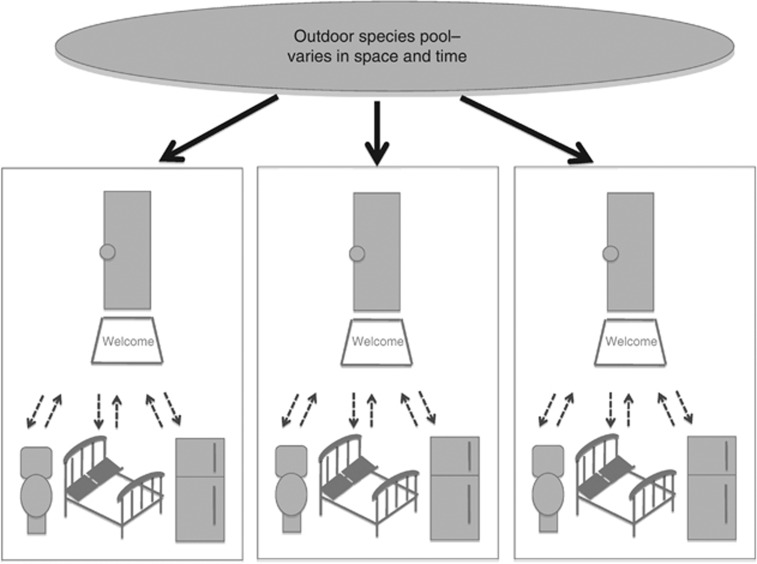
Indoor air microbial communities are thought to be a function of dispersal from the outdoors, and growth and resuspension from the indoor environment. Our results indicate that the movement of fungal material from the outdoors is sufficient to explain the observed indoor fungal air assemblages, and the fungal growth and resuspension that occurs indoors is not quantitatively large enough or qualitatively different enough (or does not become airborne in significant amounts) to give individual rooms or buildings a distinct signature that cannot be explained simply by isolation by distance. We conclude this for the following reasons: (1) indoor communities differ by season, just as the outdoor samples; (2) geographic distance (and the proxy for that, unit) is the single significant determinant of community structure when considered in a multifactor model, and not, for example, room function or resident behavior; (3) no fungi were identified as indicators of indoor air or a particular room type. Thus, as the outdoor species pool varies, so will the indoor airborne fungal communities. Moreover, the fungal taxa we detected indoors of healthy buildings appear to be stochastic subsets of the available immigrant from outdoors, as demonstrated using simulations. We summarize the main results from this study in Figure 3.
Figure 3.
A working model for the structure of airborne fungal communities. Indoor air microbial communities are thought to be a function of dispersal from the outdoors (heavy black arrows), and growth and resuspension from the indoor environment (gray dashed arrows). Our results indicate that dispersal from the outdoor species pool, which changes with both geographic distance and seasonal variation, is a stronger determinant of the diversity of fungal exposure indoors than growth and resuspension associated with the function of individual rooms.
As with all air-sampling approaches, what is being sampled is not an established community or assemblage in the usual ecological sense, but rather the parts of such communities that are shed into the air. The ultimate source for microbes that grow indoors is from outside the built environment (that is, the outdoors and residents), and only subsets of these organisms are capable of finding suitable growth conditions indoors. However, these endogenous populations are known to associate with distinct household surfaces, and there is little evidence that microorganisms utilize the air itself, particularly indoor air, as a matrix for growth (but see Womack et al., (2010) for a case on the atmosphere). In other words, no habitat filtration or selection has occurred here because there is no requirement to grow within the placement of samplers. Thus, in taking a biogeographic approach to study this assemblages, we would argue that the indoor air microbiome is a community in which the four processes have been stripped to one, dispersal, and the correlation of community dissimilarity and geographic distance can only be ascribed to dispersal limitation. Thus, sampling the air not only measures airborne exposure for humans but also provides a unique opportunity to further understand the dispersal characteristics of microbes.
The small size and abundant production of fungal propagules might lead one to believe that dispersal limitation is rare in fungi, or at least limited to large geographic scales. Here we show that despite the size and abundance of fungal propagules, dispersal limitation occurs within a scale of less than a square kilometer. This is approximately the same scale that Peay et al. (2012) found in the study of ectomycorrhizal fungal dispersal at the margins of a forest border. Thus, our current results add to evidence that many fungi may be dispersal limited at relatively fine scales, at least within the time windows of a climatic season or year. Indeed, in addition to the spatial scales at which dispersal limitation occurs, more work is needed on temporal scale. Interestingly, Bell (2010) found evidence of dispersal limitation in soil bacteria at a similar spatial scale but a much shorter time scale than here (a few days). This indicates that the temporal scale of community assembly, and specifically the time interval between so-called rare long-distance dispersal events, may be very different for different microorganisms.
Our sampling scheme intentionally removed many sources of variation in buildings, including but not limited to, building design and broad age differences, construction material, the particular type of heating, ventilation and air-conditioning system, and resident characteristics, such as the inclusion of pets. Many of these factors are known to affect microbial growth and abundance in built environments (Fujimura et al., 2010; Andersen et al., 2011). Moreover, the mild climate of our sampling locality increases the exposure of building interiors to the outdoors in both winter and summer in this study system. However, a global survey of fungi in vacuum samples also detected an effect of distance and not building type (Amend et al., 2010), thus indicating that the changes in outdoor air drive corresponding changes in indoor air at both broad and local spatial scales. Such a pattern of the strong influence of the outdoor had been suggested in other indoor air studies specifically looking at contaminated buildings, although the limited replication of buildings (two paired buildings) and the geographic distance between paired buildings (100 km in one pairing) constrained generalizations (Huttunen et al., 2008; Pitkaranta et al., 2011). A strong influence of the outdoors has also been observed for bacterial assemblages indoors (Kembel et al., 2012), whereas the contribution of residents and their behaviors appear to play a stronger role indoors in bacteria than in fungi (Hospodsky et al., 2012). Kembel et al., (2012) (showed that the effect of different environments (indoor mechanical ventilation, indoor window ventilation and outdoor) on bacteria had an R2 of 0.57, whereas here geographic distance showed an R2 of at most 0.33. Future studies that reintroduce sources of variation in buildings and residents and that sample different climatic regions will be key to determine the relative effect size of different processes, including isolation by distance, on indoor air fungal communities.
Indoor air mycobiome sampling and analysis
Total fungal diversity in this system was not completely sampled (Figure 1), and most assuredly increased sampling depth both within and across units would detect further (likely rare) sequence diversity. The elevated accumulation of taxa in the winter relative to the summer is expected in this sampling locality, where the Mediterranean climate leads to more fungal sporulation during the wet winter months than the dry summer ones. Indeed, rarefied samples visualized in nonmetric multidimensional scaling show that fungal composition is largely structured by season, as well as location outdoors or indoors. Outdoor samples appear to be more similar in composition to each other than indoor samples (Supplementary Figure 4), and we speculate that this clustering of the outdoor sampling is because of a bias in richness between the indoor and outdoor samples such that there are more rare taxa detected in the indoor samples. Specifically, the mean observed richness in the indoor samples (μ=78.5) is less than outdoor samples (μ=179.7), whereas the number of indoor singletons (n=235) is greater than outdoor singletons (n=124). From qPCR, we estimate that outdoor samples have approximately 20–100-fold greater fungal biomass than indoor samples. Thus, because all the samples were sequenced at equimolar amplicon concentrations, the likelihood of detecting rare taxa in indoor samples is higher than in outdoor samples because of the consequence that the outdoor samples are effectively sequenced to a lesser depth. If indoor samples are essentially random subsamples of the outdoor air, as the simulation suggest (Supplementary Figures 6 and 7), then the rare species randomly assembled and detected in individual indoor samples could drive the dispersion seen in nonmetric multidimensional scaling space, even though it is likely that these rare indoor taxa are also present outdoors and go undetected.
The passive settling of dust on sterile, inert, petri dishes was an effective sampling method for capturing a distinct window of time (as demonstrated from the clear separation of winter and summer samples), compared with, for example, collecting settled dust, which is accumulative. Although the placement of the sampler at a particular height has the potential to introduce bias because the dynamics of particles are known to vary depending on their size (Nazaroff, 2004), and fungal taxonomic composition can be linked with certain aerodynamic diameters (Yamamoto et al., 2012), we detected both small- (for example, the mold Penicillium sp, 3 μm) and large-spored taxa (for example, the puffball Pisolithus, 9 μm). Further work is required to determine the individual bias introduced with this particular sampling device. In any case, it is an inexpensive and simple technique that makes large-scale collection of air samples feasible.
Advances in technology allow a broader and, in many cases, more basic set of questions to be asked about the indoor microbiome. In this way, nonculturing techniques and now next-generation pyrosequencing have greatly expanded our ability to detect the microbial component to ambient environment samples. Interestingly, many of the fungal taxa frequently detected using pyrosequencing by and large match taxa frequently detected in culture-based surveys (Ara et al., 2004; Pitkaranta et al., 2008; Oliveira et al., 2009; Amend et al., 2010). These are taxa known to grow on moist surfaces indoors, such as Aureobasidium on bathroom tiles and Penicillium on foodstuffs. However, the diversity of fungal taxa and ecologies, particularly the many that are plant-associated and do not grow indoors, is essentially unknown from culture-based sampling of indoor air (Ara et al., 2004) but is well represented in the sequence data here (Table 3) and in prior sequence-based studies (Pitkaranta et al., 2008, 2011; Amend et al., 2010). On the other hand, an inability to separate living from dead tissue and cell fragments, or the nature of the starting material in general (spore versus fragment), in all PCR-based studies potentially biases our understanding of what is the true microbial community. In the indoor air, fungal fragments can outnumber intact spores 1000:1 (Reponen et al., 2007), and both entities have the potential to be detected with molecular techniques.
Analysis of pyrosequenced data relies on many parameter settings and assumptions. For our analyses, we required a minimum sequence length of 100 bp. We repeated analyses at varying minimum lengths, therefore including different number of sequences into the analysis, and qualitative conclusions do not change. Although the number of OTUs changes with different settings, most of the variation comes within the ‘rare biosphere,' although the pattern of isolation by distance remains even when only considering abundant taxa. Therefore, we would argue that community analysis is largely robust to slight variations in the abundance table. Perhaps, most important is the OTU-picking process of the pyrosequencing pipeline (Lee et al., 2012), which groups sequences into OTUs according to a user-defined threshold. In our study, we found that some of the ‘distinct' OTUs were given identical taxon assignments from BLAST matches. This could indicate true variation in taxa—for example, there are probably different strains of Cladosporium mold present in this one locality, and some individual stains could have multiple ITS sequence variants—but it could also result from a limitation in the analysis software to accurately pool taxa (Reeder and Knight, 2009). Interestingly, ‘denoising' proved to be highly important, as analysis done without this error-reducing step detected nearly 4 × more OTUs.
In conclusion, taking an ecological approach to investigate the processes of fungal community assembly in the indoor environment has shown that, when building type is held constant, it is dispersal from the outdoors to the built environment, and not resident behavior, that structures fungal composition. Moreover, fungi show signs of dispersal limitation at the geographic scale of hundreds of meters, indicating that dispersal is a process like selection and drift that can strengthen the distance–decay relationship in microbial communities.
Acknowledgments
We thank the Sloan Foundation for funding and for facilitating collaborations and discussions. We acknowledge the input of fellow members of the Berkeley Indoor Microbial Ecology Research Consortium, particularly, Steven Lindow, Seema Bhangar and Bill Nazaroff, and the input of Kabir Peay, Andy Rominger and the Bruns and Taylor labs for providing valuable discussion of methods. We also thank the volunteers and staff of our study site.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Abarenkov K, Nilsson RH, Larsson KH, Alexander IJ, Eberhardt U, Erland S, et al. The UNITE database for molecular identification of fungi—recent updates and future perspectives. New Phytol. 2010;186:281–285. doi: 10.1111/j.1469-8137.2009.03160.x. [DOI] [PubMed] [Google Scholar]
- Alvarez AJ, Buttner MP, Stetzenbach LD. PCR for bioaerosol monitoring: sensitivity and environmental interference. Appl Environ Microbiol. 1995;61:3639–3644. doi: 10.1128/aem.61.10.3639-3644.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amend AS, Seifert KA, Samson R, Bruns TD. Indoor fungal composition is geographically patterned and more diverse in temperate zones than in the tropics. Proc Natl Acad Sci USA. 2010;107:13748–13753. doi: 10.1073/pnas.1000454107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen B, Frisvad JC, Sondergaard I, Rasmussen IS, Larsen LS. Associations between fungal species and water-damaged building materials. Appl Environ Microbiol. 2011;77:4180–4188. doi: 10.1128/AEM.02513-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson TH, Heinemeyer O, Weigel HJ. Changes in the fungal-to-bacterial respiratory ratio and microbial biomass in agriculturally managed soils under free-air CO2 enrichment (FACE)—a six-year survey of a field study. Soil Biol Biochem. 2011;43:895–904. [Google Scholar]
- Ara K, Aihara M, Ojima M, Toshima Y, Yabune C, Tokuda H, et al. Survey of fungal contamination in ordinary houses in Japan. Allergol Int. 2004;53:369–377. [Google Scholar]
- Bell T. Experimental tests of the bacterial distance-decay relationship. ISMEJ. 2010;4:1357–1365. doi: 10.1038/ismej.2010.77. [DOI] [PubMed] [Google Scholar]
- Baas Becking LGM. W.P. Van Stockum & Zoon: The Hague, the Netherlands; 1934. Geobiologie of inleiding tot de milieukunde. [Google Scholar]
- Chao A. Estimating the population size for capture-recapture data with unequal catchability. Biometrics. 1987;43:783–791. [PubMed] [Google Scholar]
- Colwell RK.2006EstimateS: statistical estimation of species richness and shared species from samplesVersion 8.0 edn http://www.purl.oclc.org/estimates .
- Corsi RL, Kinney KA, Levin H. Microbiomes of built environments: 2011 symposium highlights and workgroup recommendations. Indoor Air. 2012;22:171–172. doi: 10.1111/j.1600-0668.2012.00782.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dannemiller KC, Reeves D, Bibby K, Yamamoto N, Peccia J.in reviewFungal High- throughput Taxonomic Identification tool for use with Next-Generation Sequencing (FHiTINGS) http://sourceforge.net/projects/fhitings/ . [DOI] [PubMed]
- Douwes J, Thorne P, Pearce N, Heederik D. Bioaerosol health effects and exposure assessment: progress and prospects. Ann Occup Hyg. 2003;47:187–200. doi: 10.1093/annhyg/meg032. [DOI] [PubMed] [Google Scholar]
- Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlay BJ. Global dispersal of free-living microbial eukaryote species. Science. 2002;296:1061–1063. doi: 10.1126/science.1070710. [DOI] [PubMed] [Google Scholar]
- Fujimura KE, Johnson CC, Ownby DR, Cox MJ, Brodie EL, Havstad SL, et al. Man's best friend? The effect of pet ownership on house dust microbial communities. J Allergy Clin Immunol. 2010;126:410–412.e413. doi: 10.1016/j.jaci.2010.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardes M, Bruns T. ITS primers with enhanced specificity for basidiomycetes—application to the identification of mycorrhizae and rusts. Mol Ecol. 1993;2:113–118. doi: 10.1111/j.1365-294x.1993.tb00005.x. [DOI] [PubMed] [Google Scholar]
- Good IJ. The population frequencies of species and the estimation of population parameters. Biometrika. 1953;40:237–264. [Google Scholar]
- Hanson CA, Fuhrman JA, Horner-Devine MC, Martiny JBH. Beyond biogeographic patterns: processes shaping the microbial landscape. Nat Rev Microbiol. 2012;10:497–506. doi: 10.1038/nrmicro2795. [DOI] [PubMed] [Google Scholar]
- Hospodsky D, Qian J, Nazaroff WW, Yamamoto N, Bibby K, Rismani-Yazdi H, et al. Human cccupancy as a source of indoor airborne bacteria. PLoS One. 2012;7:e34867. doi: 10.1371/journal.pone.0034867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huse SM, Welch DM, Morrison HG, Sogin ML. Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Environ Microbiol. 2010;12:1889–1898. doi: 10.1111/j.1462-2920.2010.02193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson DH, Mitra S, Ruscheweyh H-J, Weber N, Schuster SC. Integrative analysis of environmental sequences using MEGAN4. Genome Res. 2011;21:1552–1560. doi: 10.1101/gr.120618.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttunen K, Rintala H, Hirvonen MR, Vepsalainen A, Hyvarinen A, Meklin T, et al. Indoor air particles and bioaerosols before and after renovation of moisture-damaged buildings: the effect on biological activity and microbial flora. Environ Res. 2008;107:291–298. doi: 10.1016/j.envres.2008.02.008. [DOI] [PubMed] [Google Scholar]
- Institute of Medicine . Damp Indoor Space and Health. National Academies of Science: Washington, DC; 2004. [Google Scholar]
- Iossifova YY, Reponen T, Bernstein DI, Levin L, Kalra H, Campo P, et al. House dust (1-3)-beta-D-glucan and wheezing in infants. Allergy. 2007;62:504–513. doi: 10.1111/j.1398-9995.2007.01340.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaakkola JJK, Hwang B-F, Jaakkola MS. Home dampness and molds as determinants of allergic rhinitis in childhood: A 6-Year, population-based cohort study. Am J Epidemiol. 2010;172:451–459. doi: 10.1093/aje/kwq110. [DOI] [PubMed] [Google Scholar]
- Karvala K, Toskala E, Luukkonen R, Lappalainen S, Uitti J, Nordman H. New-onset adult asthma in relation to damp and moldy workplaces. Int Arch Occup Environ Health. 2010;83:855–865. doi: 10.1007/s00420-010-0507-5. [DOI] [PubMed] [Google Scholar]
- Kembel SW, Jones E, Kline J, Northcutt D, Stenson J, Womack AM, et al. Architectural design influences the diversity and structure of the built environment microbiome. ISME J. 2012;6:1469–1479. doi: 10.1038/ismej.2011.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CK, Herbold CW, Polson SW, Wommack KE, Williamson SJ, McDonald IR, et al. Groundtruthing next-gen sequencing for microbial ecology—biases and errors in community structure estimates from PCR amplicon pyrosequencing. PLoS One. 2012;7:e44224. doi: 10.1371/journal.pone.0044224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennon JJ, Koleff P, Greenwood JJD, Gaston KJ. The geographical structure of British bird distributions: diversity, spatial turnover and scale. J Anim Ecol. 2001;70:966–979. [Google Scholar]
- Martiny JBH, Bohannan BJM, Brown JH, Colwell RK, Fuhrman JA, Green JL, et al. Microbial biogeography: putting microorganisms on the map. Nature Reviews Microbiology. 2006;4:102–112. doi: 10.1038/nrmicro1341. [DOI] [PubMed] [Google Scholar]
- Miller DN, Bryant JE, Madsen EL, Ghiorse WC. Evaluation and optimization of DNA extraction and purification procedures for soil and sediment samples. Appl Environ Microbiol. 1999;65:4715–4724. doi: 10.1128/aem.65.11.4715-4724.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazaroff WW. Indoor particle dynamics. Indoor Air. 2004;14:175–183. doi: 10.1111/j.1600-0668.2004.00286.x. [DOI] [PubMed] [Google Scholar]
- Nilsson RH, Veldre V, Hartmann M, Unterseher M, Amend A, Bergsten J, et al. An open source software package for automated extraction of ITS1 and ITS2 from fungal ITS sequences for use in high-throughput community assays and molecular ecology. Fungal Ecol. 2010;3:284–287. [Google Scholar]
- Oksanen J, Blanchet FG, Kindt R, Legendre R, Minchin PR, O'Hara RB, et al. 2012. Vegan: community ecology package, R Package Version 2.1-17 edn.
- Oliveira M, Ribeiro H, Delgado JL, Abreu I. Aeromycological profile of indoor and outdoor environments. J Environ Monitor. 2009;11:1360–1367. doi: 10.1039/b820736d. [DOI] [PubMed] [Google Scholar]
- Paulino LC, Tseng CH, Strober BE, Blaser MJ. Molecular analysis of fungal microbiota in samples from healthy human skin and psoriatic lesions. J Clin Microbiol. 2006;44:2933–2941. doi: 10.1128/JCM.00785-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peay KG, Schubert MG, Nguyen NH, Bruns TD. Measuring ectomycorrhizal fungal dispersal: macroecological patterns driven by microscopic propagules. Mol Ecol. 2012;21:4122–4136. doi: 10.1111/j.1365-294X.2012.05666.x. [DOI] [PubMed] [Google Scholar]
- Pebesma EJ, Bivand RS.2012. sp: classes and methods for spatial data, R Package Version 0.9-100 edn.
- Pitkaranta M, Meklin T, Hyvarinen A, Nevalainen A, Paulin L, Auvinen P, et al. Molecular profiling of fungal communities in moisture damaged buildings before and after remediation—a comparison of culture-dependent and culture-independent methods. BMC Microbiol. 2011;11:235. doi: 10.1186/1471-2180-11-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitkaranta M, Meklin T, Hyvarinen A, Paulin L, Auvinen P, Nevalainen A, et al. Analysis of fungal flora in indoor dust by ribosomal DNA sequence analysis, quantitative PCR, and culture. Appl Environ Microbiol. 2008;74:233–244. doi: 10.1128/AEM.00692-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team 2012R: A Language and Environment for Statistical Computing R Foundation for Statistical Computing; R version 2.15.1 edn http://www.R-project.org Vienna, Austria [Google Scholar]
- Reeder J, Knight R. The ‘rare biosphere': a reality check. Nat Methods. 2009;6:636–637. doi: 10.1038/nmeth0909-636. [DOI] [PubMed] [Google Scholar]
- Reeder J, Knight R. Rapidly denoising pyrosequencing amplicon reads by exploiting rank-abundance distributions. Nat Methods. 2010;7:668–669. doi: 10.1038/nmeth0910-668b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reponen T, Seo SC, Grimsley F, Lee T, Crawford C, Grinshpun SA. Fungal fragments in moldy houses: a field study in homes in New Orleans and Southern Ohio. Atmos Environ. 2007;41:8140–8149. doi: 10.1016/j.atmosenv.2007.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts DW.2012. labdsv: ordination and multivariate analysis for ecology, R Package Version 1.5-0 edn.
- Rintala H, Pitkaranta M, Taubel M. Microbial communities associated with house dust. Adv Appl Microbiol. 2012;78:75–120. doi: 10.1016/B978-0-12-394805-2.00004-X. [DOI] [PubMed] [Google Scholar]
- Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, Levesque CA, et al. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc Natl Acad Sci USA. 2012;109:6241–6246. doi: 10.1073/pnas.1117018109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon-Nobbe B, Denk U, Poll V, Rid R, Breitenbach M. The spectrum of fungal allergy. Int Arch Allergy Immunol. 2008;145:58–86. doi: 10.1159/000107578. [DOI] [PubMed] [Google Scholar]
- Strachan DP. Hay-fever, hygiene, and household size. Br Med J. 1989;299:1259–1260. doi: 10.1136/bmj.299.6710.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vellend M. Conceptual synthesis in community ecology. Quaterly Review of Biology. 2010;85:183–206. doi: 10.1086/652373. [DOI] [PubMed] [Google Scholar]
- White T, Bruns T, Lee S, Taylor J.1990Amplification and direct sequencing of fungal ribosomal RNA genes for phylogeneticsIn: Innis N, Gelfand D, Sninsky J, White T (eds)PCR: Protocols and Applications—A Laboratory Manual Academic Press: New York; 315–322. [Google Scholar]
- Womack AM, Bohannan BJM, Green JL. Biodiversity and biogeography of the atmosphere. Philos T R Soc B. 2010;365:3645–3653. doi: 10.1098/rstb.2010.0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Würtz H, Sigsgaard T, Valbjørn O, Doekes G, Meyer HW. The dustfall collector—a simple passive tool for long-term collection of airborne dust: a project under the Danish Mould in Buildings program (DAMIB) Indoor Air. 2005;15:33–40. doi: 10.1111/j.1600-0668.2005.00342.x. [DOI] [PubMed] [Google Scholar]
- Yamamoto N, Bibby K, Qian J, Hospodsky D, Rismani-Yazdi H, Nazaroff WW, et al. Particle-size distributions and seasonal diversity of allergenic and pathogenic fungi in outdoor air. ISME J. 2012;6:1801–1811. doi: 10.1038/ismej.2012.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalar P, Novak M, De Hoog GS, Gunde-Cimerman N. Dishwashers—a man-made ecological niche accommodating human opportunistic fungal pathogens. Fungal Biol. 2011;115:997–1007. doi: 10.1016/j.funbio.2011.04.007. [DOI] [PubMed] [Google Scholar]
- Zhou G, Whong WZ, Ong T, Chen B. Development of a fungus-specific PCR assay for detecting low-level fungi in an indoor environment. Mol Cell Probes. 2000;14:339–348. doi: 10.1006/mcpr.2000.0324. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.