Abstract
Advances in next-generation sequencing technologies are providing longer nucleotide sequence reads that contain more information about phylogenetic relationships. We sought to use this information to understand the evolution and ecology of bacterioplankton at our long-term study site in the Western Sargasso Sea. A bioinformatics pipeline called PhyloAssigner was developed to align pyrosequencing reads to a reference multiple sequence alignment of 16S ribosomal RNA (rRNA) genes and assign them phylogenetic positions in a reference tree using a maximum likelihood algorithm. Here, we used this pipeline to investigate the ecologically important SAR11 clade of Alphaproteobacteria. A combined set of 2.7 million pyrosequencing reads from the 16S rRNA V1–V2 regions, representing 9 years at the Bermuda Atlantic Time-series Study (BATS) site, was quality checked and parsed into a comprehensive bacterial tree, yielding 929 036 Alphaproteobacteria reads. Phylogenetic structure within the SAR11 clade was linked to seasonally recurring spatiotemporal patterns. This analysis resolved four new SAR11 ecotypes in addition to five others that had been described previously at BATS. The data support a conclusion reached previously that the SAR11 clade diversified by subdivision of niche space in the ocean water column, but the new data reveal a more complex pattern in which deep branches of the clade diversified repeatedly across depth strata and seasonal regimes. The new data also revealed the presence of an unrecognized clade of Alphaproteobacteria, here named SMA-1 (Sargasso Mesopelagic Alphaproteobacteria, group 1), in the upper mesopelagic zone. The high-resolution phylogenetic analyses performed herein highlight significant, previously unknown, patterns of evolutionary diversification, within perhaps the most widely distributed heterotrophic marine bacterial clade, and strongly links to ecosystem regimes.
Keywords: BATS, ecotype, pyrosequencing, SAR11
Introduction
The diversity and abundance of the SAR11 clade has been studied by hybridization of radiolabeled oligonucleotides to ribosomal RNA (rRNA) molecules (Field et al., 1997; Morris et al., 2005), terminal restriction fragment length polymorphism (T-RFLP; (Carlson et al., 2009)), denaturing gradient gel electrophoresis (Tujula et al., 2010) and fluorescence in situ hybridization (Schattenhofer et al., 2009). Since its discovery in a 16S rRNA gene clone library from the Bermuda Atlantic Time-series Study (BATS) site (Giovannoni et al., 1990), SAR11 has been shown to be the dominant clade of bacteria in marine environments (Morris et al., 2005), but the disparity of methods in use has made it difficult to draw general conclusions about their ecology by comparing data from different studies. Varying approaches have resolved SAR11 into multiple ecotypes. To date, a universal system has not emerged for naming SAR11 ecotypes, or for comparing SAR11 diversity between sites (Field et al., 1997; Fuhrman and Steele, 2008; Carlson et al., 2009; Brown et al., 2012).
Ecotypes are closely related groups of bacteria that differ in physiological details that determine niche specificity (Cohan 2006; Cohan and Perry, 2007). Although there is a presumed relationship between ecotype classification and taxonomy, this relationship is informal. The ecotype concept's broadest application has been the interpretation of phylogenetic structure in environmental data. Early examples of environmental nucleic sequencing included reports of unexpected fine-scale phylogenetic structure that were surprising at the time and were postulated to result from population genetic processes such as clonal divergence, microgeographical adaptation, and diversifying selection from phage predation (Giovannoni et al., 1990). Cohan (2006) applied an essential clonal model of bacterial reproduction to interpret similar observations, theorizing that fine-scale phylogenetic structure could be a consequence of periodic selection acting on populations that are diversifying into specialists that differ in minor phenotypic characters. Correlations between fine-scale phylogenetic structure, spatial partitioning of the environment and major phenotypic differences were demonstrated in the marine cyanobacterium Prochlorococcus, making it perhaps the most well-known example of bacterial ecotypes (Rocap et al., 2003).
Approaches that combine next-generation sequencing with the use of short, unique oligonucleotide sequences incorporated in the primer (barcodes) to distinguish between pooled samples (Binladen et al., 2007) provide data with the statistical power needed to link diversity information with spatial temporal patterns. These methods are relatively fast and cheap. They have now been applied to many different environments, including marine ecosystems (Kirchman et al., 2010), sponges (Webster et al., 2010), soil (Roesch et al., 2007) and the human gastrointestinal tract (McKenna et al., 2008). Methods for taxonomically assigning OTUs have been under continual improvement (Hamady et al., 2010; Huse et al., 2010; White et al., 2010), although phylogenetic analysis was initially restricted due to the limited phylogenetic signal from the V6 region of the 16S rRNA gene. Advances in sequencing technologies resulting in longer average read lengths of pyrosequences have opened possibilities for investigation of other 16S rRNA gene regions that are well-suited to phylogenetic analysis (Jeraldo et al., 2011). Methods used to classify next-generation sequencing data vary widely, including those that use k-mer methods (mothur (Schloss et al., 2009), RDP classifier (Wang et al., 2007)), alignment-based pairwise methods (BLAST (Altschul et al., 1990), MEGAN last common ancestor (Huson et al., 2011) and GAST (Huse et al., 2008) implemented in VAMPS (VAMPS, 2012)), as well as phylogenetic methods (AMPHORA (Wu and Eisen, 2008), STAP (Wu et al., 2008), pplacer (Matsen et al., 2010) and EPA, implemented in RAxML (Berger et al., 2011)). The programs mothur, RDP classifier, BLAST and pplacer are all implemented in QIIME (Caporaso et al., 2010). Method selection depends on the data type and the nature of the questions asked. Thus far, most studies have classified microbial diversity into phylum/class (Roesch et al., 2007; McKenna et al., 2008; Kirchman et al., 2010; Webster et al., 2010). Classification to families or finer scales is rare.
The V1–V3 region of the 16S rRNA gene, located at the 5′-end, has long been used in molecular ecology studies of amplified, cloned and sequenced environmental 16S rRNA libraries, and is therefore well represented in large sequence databases, such as SILVA (Pruesse et al., 2007). The 454 FLX sequencing technology yields approximately a 250-bp read length, enough to span the V1 and V2 region, and provide sufficient information for reliable phylogenetic assignments (Chakravorty et al., 2007; Wang et al., 2007; Hamp et al., 2009). This enables comparison of sequence data from new and previous studies using classification systems that are central to microbial systematics.
The need for automated phylogenetic placement of amplicon sequence data and metagenomic studies has been widely recognized, and led to the development of several software tools designed to address this issue (Hamady et al., 2010). Wu and Eisen (2008) developed a pipeline for protein sequence analysis (using profile alignment and placement upon a pre-defined reference phylogeny using maximum parsimony; AMPHORA), as well as one for 16S rDNA analysis (STAP). Similar to STAP, Monier et al. (2008) implemented a workflow using the maximum likelihood criterion. Although the approach is among the best (Felsenstein, 2003), it has not scaled well because it requires evaluation of the likelihood of all possible placement positions on a tree, which is unrealistic with current computational capabilities. Two software tools, pplacer (Matsen et al., 2010) and RAxML (Stamatakis et al., 2005), now provide improved heuristics for likelihood-based phylogenetic placement of sequences in a reference phylogeny.
Here, we use 454 FLX sequencing of the 5′ (V1–V2) region of bacterial 16S rRNA genes and a new bioinformatics pipeline, PhyloAssigner, to assess Alphaproteobacteria diversity in almost 400 samples from the well-characterized BATS site in the highly oligotrophic Western Sargasso Sea (Steinberg et al., 2001). This area is subject to cooling and storms in winter that result in deep mixing (down to 300 m), bringing nutrients such as phosphorus and nitrogen to the surface. The surface layer then heats and stratifies in summer, resulting in stable communities including a surface community, a deep chlorophyll maximum community typically between 80 and 120 m, and a mesopelagic community typically below 160 m (Giovannoni and Rappé, 2000; Treusch et al., 2009). Three previous studies have used T-RFLP to assess microbial diversity in a highly similar sample set (Carlson et al., 2009; Treusch et al., 2009, 2011). One of these studies revealed that three major phylogenetic subclades of SAR11 present at BATS have distinctive spatiotemporal distributions, indicating a relationship between subdivision of niche space and early evolutionary events (Carlson et al., 2009).
Previous reports of correlations between phylogenetic substructure in major bacterioplankton clades and spatiotemporal distributions strongly supported the conclusion that ecotype theory could be a source of insight into adaptive variation in microbial ecology. To explore this idea in further detail, we conceived and designed PhyloAssigner to take advantage of high throughput sequencing technologies while preserving as much phylogenetic detail as possible. We found that phylogenetic classification of all sequences, instead of grouping sequences by OTU and classifying a single representative sequence, improved the resolution of SAR11 diversity. Use of a reference tree fostered comparisons between samples by making bin placement independent of context, that is, independent of other sequences in the sample. The analysis expanded the number of putative SAR11 ecotypes detected at BATS from five to nine. It supports a complex interpretation of SAR11 evolution in which spatiotemporal niches within the water column have been subdivided by re-colonization that crosses deep branches of the clade, rather than by an orderly process of subfunctionalization.
Materials and methods
Sample collection and nucleic acid isolation
Samples used in this analysis are listed in Supplementary Table S1. Briefly, 454 FLX pyrosequence data were generated from 384 monthly samples from the BATS site (representing ∼9 years) collected at the surface and 200 m, as well as 35 depth profile samples (about 32% of the total number of months sampled). Depth profiles included at least two additional depths of 40, 80, 100, 120, 160, 250 and 300 m. Details for sample collection and processing are referenced (Treusch et al., 2009).
PCR amplification and pyrosequencing
Approximately 5 ng DNA from each sample, measured with a Shimadzu spectrophotometer (Columbia, MD, USA), was used as template for PCR. Reactions consisted of (final concentrations) 1 × manufacturer's buffer, 300 μℳ dNTP, 1.6 mℳ Mg2+, 200 nℳ of each primer and 1 U of Platinum Taq DNA polymerase High Fidelity (Invitrogen, Carlsbad, CA, USA), and were cycled 25–30 times (to obtain similar, minimal yields) at the following temperatures and times: 94 °C for 15 s, 55 °C for 30 s and 68 °C for 1 min. Primers used included 338RPL (Daims et al., 1999) appended with the 454 B primer (GCCTTGCCAGCCCGCTCAGTGCWGCCWCCCGTAGGWGT), and 27FB (Morris et al., 2004) appended with the 454 A primer (GCCTCCCTCGCGCCATCAGXXXXXXXXAGRGTTYGATYMT-GGCTCAG; X's represent unique 8 bp barcodes (Hamady et al., 2008)). Positive- and negative control PCR reactions were cleaned with a Qiagen PCR Purification Kit (Valencia, CA, USA) and quantified with a Nanodrop (Thermo Scientific, Wilmington, DE, USA). 50 ng of each PCR reaction, along with the entire negative control (amplified with a unique barcode), was pooled in groups of 60–70 samples and sequenced using Roche 454 (Branford, CT, USA) FLX technology (Margulies et al., 2005). Sequences are available from the CAMERA website (http://camera.calit2.net/).
Data set description and processing
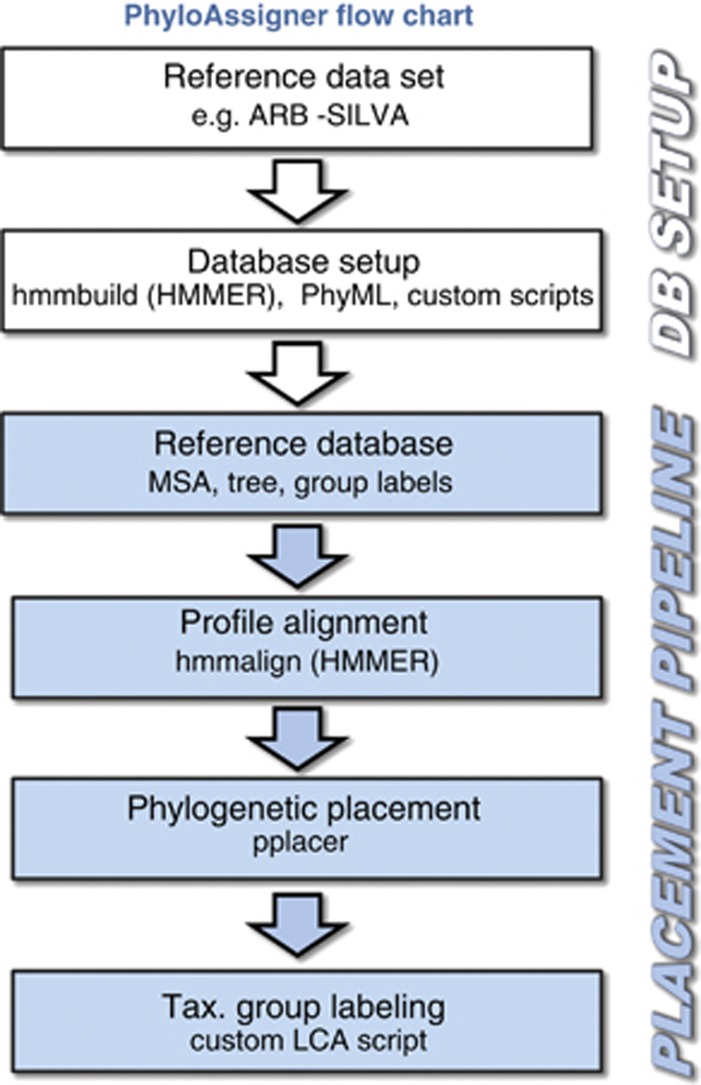
For each of the 384 samples, an average of 6684 reads was generated, with the average read lengths of 257 bases. Sequences spanned the V1 and V2 regions of the 5′-end of the 16S rRNA gene. The 454 sequences were binned and filtered for quality using a published pipeline (Hamady et al., 2008) and then placed in predefined phylogenetic groups using PhyloAssigner (Figure 1; see Supplemental Material for details and reference data sets). In a preliminary analysis, pyrosequencing reads were classified on a reference phylogeny representing a well-curated selection of bacterial taxa previously observed in marine planktonic samples. Based on this analysis, we extracted all reads placed within or at the root of the Alphaproteobacteria. These reads were re-classified, using the same procedure, with a reference tree that represented alphaproteobacterial diversity in detail. Numbers of sequences associated with each node, herein termed nodal taxonomic units (NTUs), were determined for all samples and placed in a matrix used for further analyses. Relative NTU abundances were determined by comparing raw abundance for each NTU to total abundance of Alphaproteobacteria for each sample. Samples were compared using the non-metric multidimensional scaling tool in the ecological software package, Primer 6.0 (Clarke and Gorley, 2006), by transforming relative abundance using log (X+1) and determining similarity using Bray–Curtis. For comparison to the T-RFLP data set used in a different study (Treusch et al., 2009), samples common between the two data sets were selected and identical data transformations (log (X+1)) were performed. The subroutine RELATE with the Spearman correlation option was used to compare the resulting similarity matrices. Draftsman plot analysis was used in Primer 6.0 to determine correlations between NTU abundance and environmental factors (http://bats.bios.edu/).
Figure 1.
Schematic representation of steps in the PhlyoAssigner pipeline. The first two steps (white boxes) correspond to the database setup (DB setup) and the last four (blue boxes) to the placement pipeline. ARB-SILVA, ARB program and SILVA database located online at www.arb-silva.de; MSA, multiple sequence alignment; Tax., taxonomic; LCA, last common ancestor.
Alphaproteobacteria sequence extraction and reference phylogenetic tree
To target the SAR11 clade specifically, pyrosequencing reads were placed on a custom Alphaproteobacteria reference tree composed of sequences representing 328 taxa, including 271 sequences from the overall reference tree, genome-sequenced SAR11 organisms, deep-branching taxa of the Rickettsiales and mitochondrial sequences (Supplementary Figures S1 and S4). The latter were included because a recent study indicated SAR11 and mitochondria share a last common ancestor as a sister group to the Rickettsiales (Thrash et al., 2011). Therefore, mitochondrial and Rickettsiales sequences were added to help prevent erroneous placement of 16S rRNA pyrosequences that might be more closely related to these groups rather than SAR11 strains. In all, the Alphaproteobacteria reference tree included 328 taxa. The reference tree was constructed by first aligning full-length sequences using MUSCLE (Edgar 2004) and performing the phylogenetic reconstruction with RAxML (Stamatakis et al., 2005) using the GTRCAT algorithm and 1000 bootstrapping cycles. The subclade structure of the overall SAR11 clade topology was highly supported (Supplementary Figure S1).
Comparison of SAR11 ecotype structure
SAR11 ecotype structure was illustrated largely as described previously (Carlson et al., 2009). In brief, annually sample months were adjusted to align with the month of deepest mixing (Carlson et al., 2009; Treusch et al., 2011). Relative SAR11 abundances were calculated by adding together the standardized values for nodes comprising each subclade and dividing by the total Alphaproteobacteria abundance for each sample. Adjusted months were averaged (Carlson et al., 2009; Treusch et al., 2011) to yield a composite 1-year time series for each depth. Results were plotted using Ocean Data View (http://odv.awi.de/en/home/) with scales adjusted to show the region and period of maximum abundance for each subclade.
SAR11 subclades were compared using Primer 6.0 by averaging and transforming (log (X+1)) NTUs comprising each subclade after standardization of all NTUs. Subclades were then compared by generating a matrix of Spearman correlations based on abundances in all samples and using that matrix in an MDS plot. Cluster analysis was performed on the similarity matrix using group averages.
Average SAR11 subclade abundance was calculated for all surface and 200 m samples, and samples from the same months were summed and transformed (log(X+1)). Seven complete or nearly complete years were selected for comparison and a similarity matrix was calculated based on monthly subclade patterns. Cyclical models for months and seasons were generated as described (Clarke and Gorley, 2006), and compared to the similarity matrix for subclade patterns using the Relate function and 100 000 random permutations.
Results
Analysis of time-series data using PhyloAssigner
Altogether, 2 660 612 16S rRNA gene pyrosequence reads from a 9-year time series covering the top 300 m water column at BATS were analyzed. Of these, 929 036 pyrosequence reads were placed in the clade, Alphaproteobacteria. In all, 93.7% of the sequences were assigned to known clades of bacterioplankton; 29.8% of these were assigned to terminal nodes on the reference tree and the remaining 70.2% of sequences were assigned to internal nodes, indicating that close relatives had not yet been recovered in previous surveys. Throughout, last common ancestor (Supplementary Figure S2) analysis was used to identify reads with poor support at terminal nodes and assign them to internal nodes.
Samples compared using relative Alphaproteobacteria abundances show depth-dependent variation
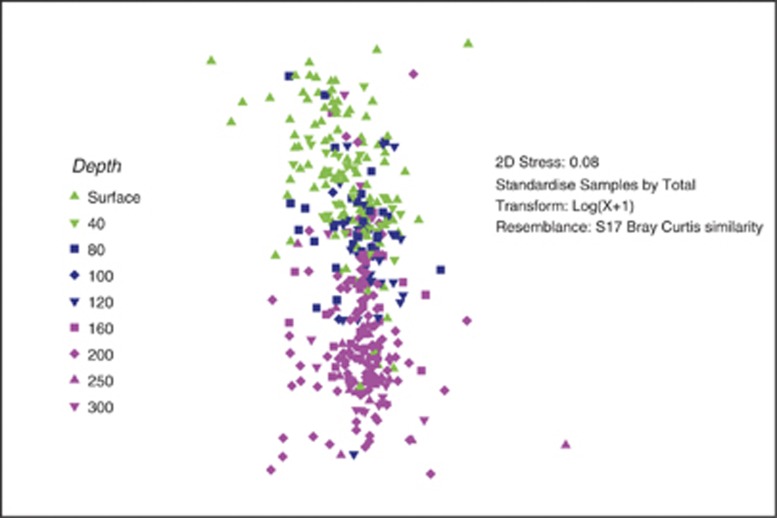
Non-metric multidimensional scaling was used to compare samples grouped together by depth (Figure 2). This ordination was compared to an ordination made from the same samples by T-RFLP (Treusch et al., 2009), using the RELATE function in Primer 6.0 (Clarke and Gorley, 2006). A Spearman rank correlation, which compares the rank order of all the samples and then compares that ranking to one from a permutated data set, indicated that the two ordinations were highly correlated (0.568; P<0.001). To test for the possibility of depth bias in the results, the analysis was repeated using surface samples and 200 m samples only, and the relationship was still highly significant (0.318, P<0.001 for surface samples only and 0.372, P<0.001 for 200 m samples only).
Figure 2.
Non-metric multidimensional scaling plot of 384 samples from the BATS time series as characterized by the Alphaproteobacteria community. Samples from the surface (0 and 40 m), mid-level (80, 100 and 120 m) and deep (160, 200, 250 and 300 m) are shown in green, blue and magenta, respectively.
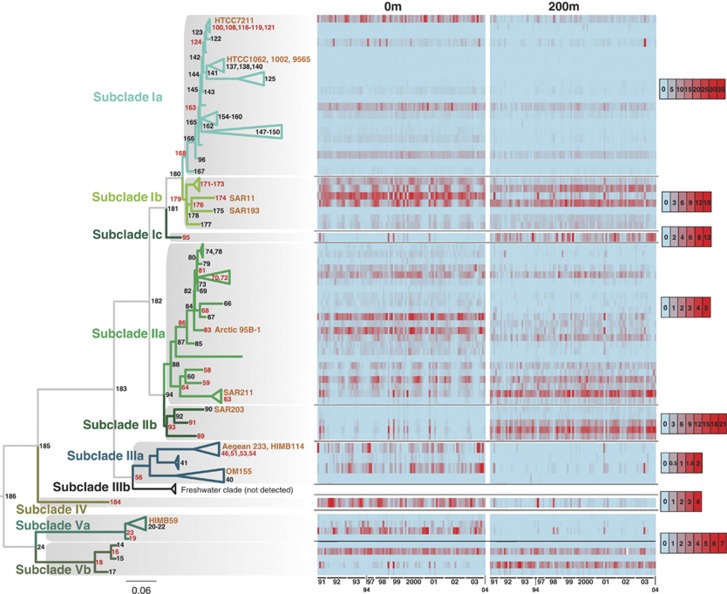
High-resolution phylogenetic analysis reveals ecotype novelty within and between established groups of SAR11
The large number of SAR11 sequences in the reference Alphaproteobacteria phylogeny enabled detection of a wider diversity of putative ecotypes than previously observed (Figure 3). We analyzed the relative abundance of sequences for each node in a comparison of surface (0 m) and upper mesopelagic (200 m) samples. Nine monophyletic subclades, each having distinctive distribution patterns, were identified using this approach. Subclade Ia sequences were placed largely near the reference sequence for HTCC7211 (NTU 112), a strain isolated from BATS, as well as several non-terminal NTUs (124, 163 and 168; Figure 3). NTUs 128, 132 and 139, the nodes containing sequence from strains isolated from the Oregon Coast (HTCC1062, 1002 and 9565, respectively), had little or no representation at BATS. Subclade Ib sequences clustered with the original SAR11 sequence at NTU 174 and nearby NTUs (171-173, 176 and 179). A previously unrecognized subclade, Ic, represented by NTU 95, had relative abundances approaching 3% in deep water, but was much less abundant at the surface. The evolutionary distance of Ic genomes from other SAR11, and their ecological partitioning to the mesopelagic zones, were confirmed in a follow-up study that relied on single amplified genomes and metagenomic data (Thrash, unpublished data). Subclade II sequences showed ecotype partitioning based on depth. Subclade IIa was more abundant at the surface than at 200 m. In contrast, although subclade IIb showed higher relative abundances at the surface than IIa, it was most abundant at 200 m. Subclade IIIa sequences were generally much less abundant and were constrained to about five main NTUs corresponding to the AEGEAN 233/HIMB114 subgroup, and basal NTU 56. OM155 subgroup sequences were poorly represented at BATS. Moreover, subclade IIIb, the freshwater SAR11 clade, was not observed at BATS. Another previously unrecognized subclade, group IV, represented by NTU 184, was highly abundant at the surface. Although the inclusion of subclade V in the SAR11 clade was recently questioned (Rodriguez-Ezpeleta and Embley, 2012), its monophyletic origin with the other SAR11 sequences was supported in our results and is consistent with phylogenomic analyses (Supplementary Figure S1; Thrash et al., 2011). Subclade V formed two subclades segregated by depth, Va and Vb. While sequences assigned to NTU 16 were represented at both the surface and 200 m, subclade Vb NTU sequences were overall more abundant at 200 m, whereas those belonging to subclade Va NTUs were predominantly observed in high abundance at the surface. The strengths of these depth distributions were assessed using the Student's t-test (Supplementary Table S3). In addition, monthly averages with s.d.'s were computed (Supplementary Figure S3).
Figure 3.
SAR11 subclade dynamics at BATS. Each node or collapsed group of nodes on the reference tree has a corresponding row in the heat plots. Heat plot columns represent individual sampling dates, summarized by year (x axis). All information for each node is displayed for 0 and 200 m. Red bars correspond to percent abundance of pyrosequences localized to a node for a given depth and sample date, as given by the scale bars on the right. Note that subclades are on different scales to provide resolution for subclades with lower abundances than Ia. Nodes with particularly high abundance are colored red. Reference sequences from cultures or previously published subclade identification sequences (for example, SAR11 and OM155) are indicated in orange. Scale bar indicates 0.06 changes per position.
SAR11 ecotypes have spatial and temporal specificity
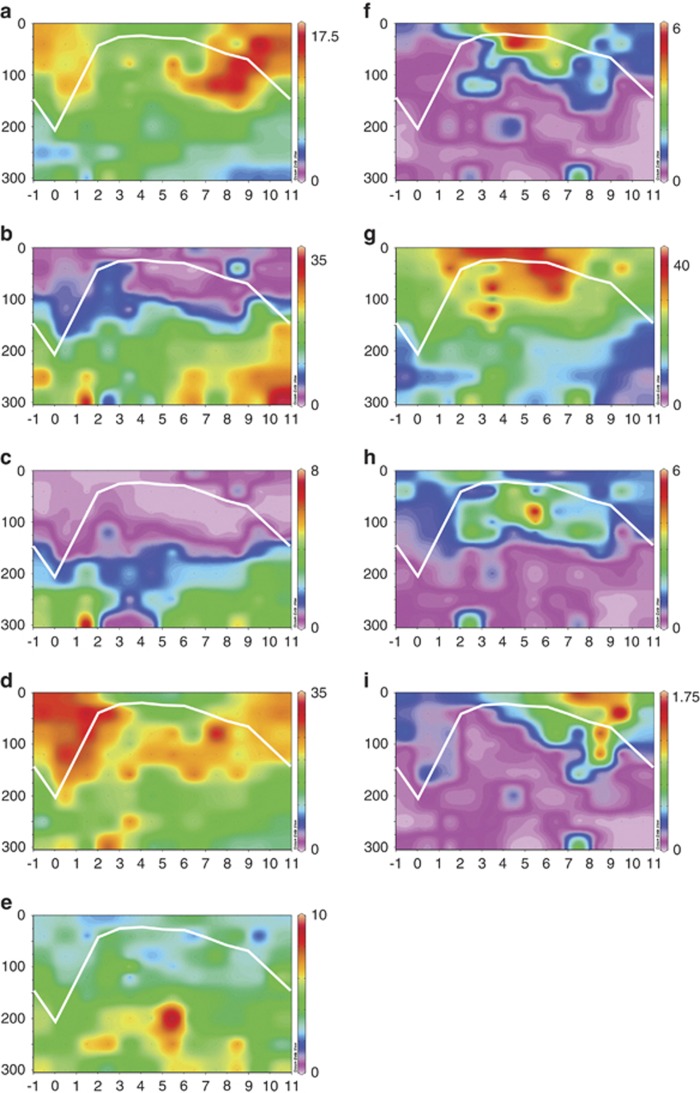
Previous reports (Field et al., 1997; Morris et al., 2002; Carlson et al., 2009) suggested that SAR11 subclades have adapted to specific vertical and temporal ranges at BATS. Our results confirm these earlier findings, but with much higher data resolution. Environmental distributions of the subclades at BATS suggest a seasonal succession of subclades (Figure 4). For example, starting at the annual deep-mixing event, subclade II resolved into two subclades. Sequences from subclade IIa had higher abundance at the surface (Figure 4a), whereas subclade IIb was more abundant at 200 m (Figure 4b). Both subclades became more abundant at the start of the deep-mixing period and remained elevated at the peak of deep mixing. Subclade Ic had higher abundances below 160 m just after the peak of deep mixing (Figure 4c). Subclade Ib is abundant year-round but increased in abundance during the deep-mixing period (Figure 4d). Interestingly, we observed two previously unknown, well-supported distinct groups within subclade V (Supplementary Figures S1 and S4). Subclade Vb is more prevalent at lower depths starting after deep mixing (Figure 4e), whereas subclade Va was predominantly found in the upper 40 m during summer (Figure 4f). Subclade Ia was found throughout the surface waters during the summer months (Figure 4g). Subclade IV was also found primarily in the summer but was lower in the surface layer, predominantly around 80 m (Figure 4h). Subclade IIIa was a low abundance group found in surface waters in the fall when they responded to the beginning of the deep-mixing period (Figure 4i).
Figure 4.
Ocean Data View plots of SAR11 subclades and depth-specific distributions. Data from the upper 300 m during a 9 year time series were averaged over a 1 year time frame after adjusting to the month of deepest mixing (month 0) to determine relative subclade abundances. White lines indicate average mixed layer depth. Heat maps (scale to the right of each plot) are adjusted to show the spatiotemporal maximum for each subclade. (a) Subclade IIa; (b) subclade IIb; (c) subclade Ic; (d) subclade Ib; (e) subclade Vb; (f) subclade Va; (g) subclade Ia; (h) subclade IV; and (i) subclade IIIa.
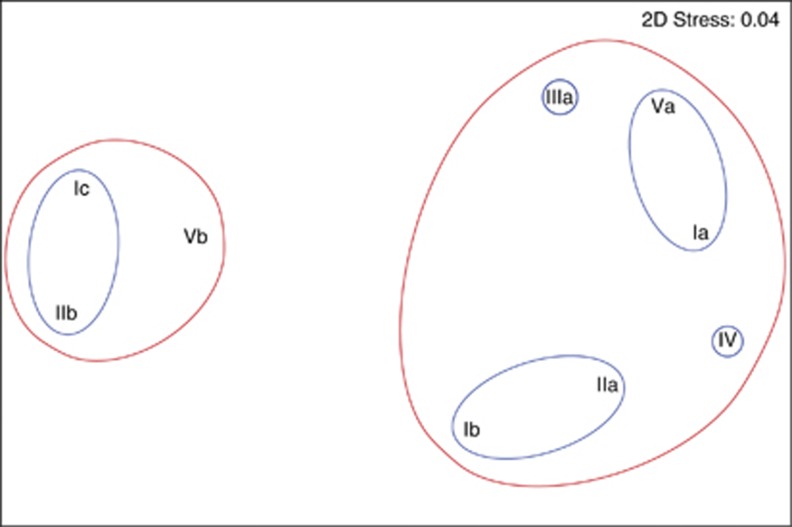
Non-metric multidimensional scaling of the SAR11 data supported the distinction between deep and surface ecotypes, separating deep subclades Ic, IIb and Vb from the surface subclades at a 0.26 similarity threshold (Figure 5, red circles). Subtle differences in depth and season of occurrence emerged at a 0.65 similarity threshold (Figure 5, blue circles); for example, Ic and IIb clustered apart from Vb because, while all three of these subclades bloom during mixing events, Vb is also present in the upper 100 m year-round (Figure 4b, c and e). Similarly, of the summer subclades, IV was distinguished by its slightly deeper distribution (80 m; Figures 4f–h). Non-parametric Mantel tests supported the conclusion that there was a significant component of seasonality driving the ordination of the subclades. Similarity matrices from surface and 200 m data were significantly similar to monthly and seasonal models in comparisons (ρ>0.11, P<0.0001; Figure 6), strongly supporting the conclusion that seasonal factors are a driver of niche partitioning for subclades.
Figure 5.
Non-metric multidimensional scaling plots for Spearman correlations among the nine SAR11 subclades based on average read abundance data in all samples. Correlations of 0.26 and 0.65 are indicated by red and blue lines, respectively.
Figure 6.
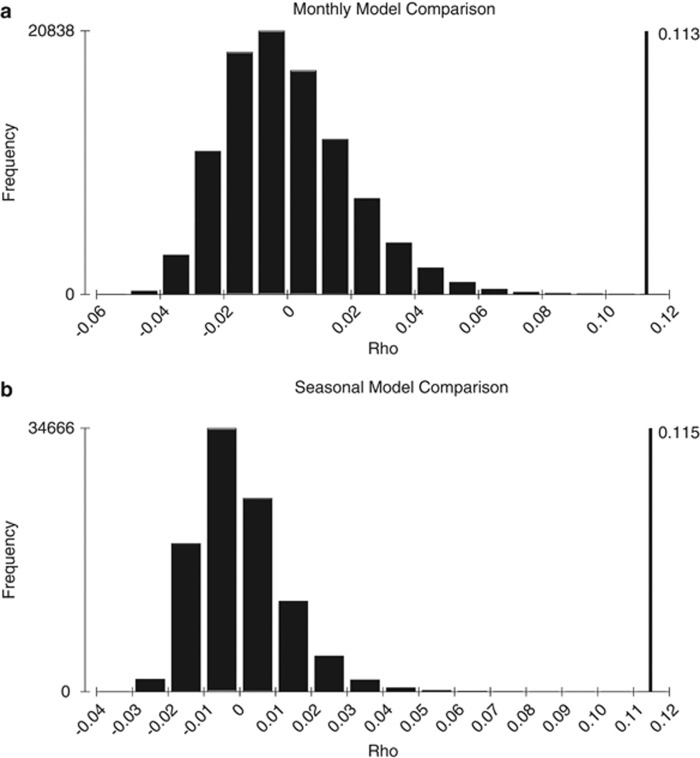
Comparison of a similarity matrix based on SAR11 subclade profiles for each month over 7 years versus modeled matrices for monthly (a), or seasonal (b) cyclical transitions. Histogram plots represent the results of 100 000 random permutations of the data matrix compared with the respective model matrix. Spearman correlations for data comparisons to the model were similar and significantly non-random (ρ>0.11, significance level below 0.0001), indicating a good fit to both models.
Discussion
Pyrosequencing allied with procedures for phylogenetically binning a large volume of sequences by maximum likelihood placement provided strong support for previous studies while nearly doubling the number of SAR11 ecotypes resolved at BATS. These results are an important step towards understanding the evolutionary diversification of the SAR11 clade, and their modern role in marine water columns. By describing in detail the natural history of SAR11 ecotypes, these findings make it possible to recognize and sample SAR11 ecotypes so that comparative genomics can be applied in studying their evolution (Grote et al., 2012).
This study used a new bioinformatics pipeline that was developed when no similar pipelines were available. Pyrosequencing methods were rapidly assimilated into microbial ecology because of significant advantages in the sheer volume of data collected. This advantage generally outweighed the loss of phylogenetic resolution associated with short sequences, and the implementation of BLAST- or kmer-based methods that proved practical for identifying and binning voluminous data. However, classification schemes based on evolutionary patterns of sequence divergence are useful for many applications (Koski and Golding, 2001; Wu et al., 2008). With increasing read lengths, it became possible to apply phylogenetic methods to pyrosequence analysis (Matsen et al., 2010). PhyloAssigner implements this idea practically, replacing OTUs based on sequence divergence with NTUs based on phylogenetic inference, and assigning NTUs to basal nodes when a more recent placement is not justified by likelihood scores. This refinement retained the precise identification of previously classified diversity, while clustering novel sequences and placing these clusters in phylogenetic context with an estimate of reliability.
One important advantage of this approach is that data from different studies can be aligned to a common reference tree, facilitating comparisons. In essence, this concept was widely adopted in microbiology when 16S rRNA phylogeny became the ‘gold standard' (Ludwig and Schleifer, 1999) for classification. By placing sequences at specific nodes (NTUs) in a reference tree, massive data sets from high throughput, deep-sequencing efforts can be projected on established 16S rRNA phylogenies made from the nearly full-length genes of cultured organisms and well-characterized but uncultured taxa. Pyrosequences from SAR11 subclade Ia illustrate the resolution that is attainable with this approach; most Ia pyrosequences from BATS were placed in the NTU assigned to Sargasso Sea isolate, HTCC7211, but not with closely related isolates from the Oregon Coast (HTCC1062, HTCC1002 and HTCC9565), which differ by only 15 bases (1%) over the entire 16S rRNA gene (Figure 3).
A novel clade of bacteria related to the Rhodospirillales was detected in upper mesopelagic samples (NTU 201; Supplementary Figure S4), illustrating how PhyloAssigner classifies examples of diversity that are not represented by near neighbors in the reference tree. NTU 201, which contains over 1200 pyrosequencing reads that have a high average pairwise similarity, was placed deeply in the tree near the outgroup sequences by last common ancestor analysis. BLAST analyses of metagenomic databases (GOS, (Rusch et al., 2007)) revealed examples of sequences related to NTU 201 in other Ocean data (data not shown). The majority of BATS sequences binning to NTU 201 came from 200 m, therefore we give this novel clade the name SMA-1 (Sargasso Mesopelagic Alphaproteobacteria, group 1, Supplementary Figure S5). Interestingly, we found no evidence for the novel marine alphaproteobacterial clade, ‘OMAC', which was described as intermediate to the Rickettsiales and the mitochondria (Brindefalk et al., 2011). Only 269 sequences (<0.03%) from the entire data set were placed in NTUs (638, 574) at the expected branching position of the OMAC clade between the Rickettsiales/mitochondria/SAR11 group clade. Among the sequences placed in proximity to the Rickettsiales/mitochondria/SAR11 group clade, pairwise similarities were low, suggesting that they are artifacts, such as chimeras (Supplementary Figure S4).
Carlson et al. (2009) provided evidence of three major subclades of SAR11 in T-RFLP data. They found that SAR11 Ia was highly abundant at the surface in the summer, group Ib bloomed in the spring throughout the water column and group II favored deeper waters year-round. Two other subclades were identified, but could not be resolved by T-RFLP methods, therefore their distributions were not known. The updated model shown in Figure 4 illustrates that subclades III and V have a very low relative abundance. However, the 454 pyrosequencing data not only distinguished Va from Vb and IIIa from IIIb, but clearly showed that subclade Va is coincident with Ia, with peak abundances in the summer near the surface, whereas IIIa is a very low abundance group that favors autumn surface conditions. The data reveal an annual succession of SAR11 subclades starting with Ib and IIa in the spring, Ia, IV and Va in the summer, IIIa in the autumn, and Ic, IIb, and Vb in deeper waters (Figures 4, 5, 6).
Our results indicate that SAR11 diversification was not an orderly progression of subdivision of the water column niche by depth and season. This contrasts with previous hypotheses that led to the development of the subfunctionalization model (Supplementary Figure S6A) by analogy to a model used to understand enzyme evolution (Lynch and Force, 2000). Specifically, in previous studies, the spatiotemporal distributions of major SAR11 subclades supported the conclusion that early events in SAR11 evolution were correlated with the subdivision of niche space on a vertical axis (Field et al., 1997; Rappé and Giovannoni, 2003; Carlson et al., 2009). This idea is consistent with fundamental knowledge about the ocean surface layer, which spans the euphotic and aphotic zones, and corresponding clines in geochemical processes. However, this new and more detailed analysis revealed at least three different instances in which the major SAR11 subclades were further subdivided into surface and deep ecotypes: major subclades I, II and V each encompass subclades with differing depth specificity.
A large set of oceanographic data from BATS, taken from the same cruises and depths from which DNA samples were collected for pyrosequencing, did not reveal any clear insight into the specific factors that drive ecological specialization among members of the SAR11 clade. Only weak correlations between subclades and broad environmental measures such as DOC, DON, Chl a and other metadata were found using multivariate statistical analyses (Supplementary Table S4). Many studies support the conclusion that major patterns of environmental variation, particularly depth and season, are highly correlated with plankton community structure (Fuhrman et al., 2006; Gilbert et al., 2012). The fact that little compelling evidence has emerged for specific links is testimony to the difficulty of teasing apart complex interactions using multivariate statistical tools in a manner that distinguishes between indirect correlation and causation.
Conclusions
The combination of time-series sampling, deep sequencing and phylogenetic binning resulted in high resolution of SAR11 ecotypes at BATS, and revealed a previously unrecognized clade of Alphaproteobacteria, SMA-1, in the upper mesopelagic zone. SAR11 variation and microbial diversity in general has been studied at BATS extensively. This provided a platform for evaluating a new pipeline designed to extract the maximum amount of information on phylogenetic diversity, whereas preserving the ability to detect and place deeply branching new taxa. The pipeline achieves these goals and preserves the flexibility to use standardized phylogenetic classification systems.
Our findings indicate that the subdivision of niche space at BATS is not consistent with a model in which environmental variation with season and depth alone drives SAR11 evolution. Instead, a complex picture of ecotype dynamics has emerged that suggests unidentified factors have an important role in SAR11 evolution. We propose a model, referred to as the re-colonization model (Supplementary Figure S6B), that is based on the hypothesis that evolutionary innovations lead to competition between ecotypes and niche expansion in the dimensions of time and space. Future comparative genomic and physiology studies should be performed in the context of SAR11 diversity and phylogenetic structure discovered herein.
Acknowledgments
We thank the officers and crew of the R/V Weatherbird II for their assistance and support, and the BATS chief scientists and technicians for assistance in water collection and accommodating wire time requests. We thank Rachel Parsons, Rachel Poage and Dan Smith for technical assistance. We thank Jesse Zaneveld for helpful discussions. AHT was supported by a Feodor Lynen Fellowship of the Alexander von Humboldt Foundation. Major support was provided by independent grants from the Marine Microbiology Initiative of the Gordon and Betty Moore Foundation to SJG and AZW, NSF IOS-0843119 (AZW), and NSF Microbial Observatory grants MCB-0237713 and OCE-0802004 (SJG).
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Berger SA, Krompass D, Stamatakis A. Performance, accuracy, and web server for evolutionary placement of short sequence reads under maximum likelihood. Syst Biol. 2011;60:291–302. doi: 10.1093/sysbio/syr010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binladen J, Gilbert MTP, Bollback JP, Panitz F, Bendixen C, Nielsen R, et al. The use of coded PCR primers enables high-throughput sequencing of multiple homolog amplification products by 454 parallel sequencing. PLoS One. 2007;2:e197. doi: 10.1371/journal.pone.0000197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brindefalk B, Ettema TJ, Viklund J, Thollesson M, Andersson SG. A phylometagenomic exploration of oceanic alphaproteobacteria reveals mitochondrial relatives unrelated to the SAR11 clade. PLoS One. 2011;6:e24457. doi: 10.1371/journal.pone.0024457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MV, Lauro FM, DeMaere MZ, Muir L, Wilkins D, Thomas T, et al. Global biogeography of SAR11 marine bacteria. Mol Syst Biol. 2012;8:595. doi: 10.1038/msb.2012.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson CA, Morris R, Parsons R, Treusch AH, Giovannoni SJ, Vergin K. Seasonal dynamics of SAR11 populations in the euphotic and mesopelagic zones of the northwestern Sargasso Sea. ISME J. 2009;3:283–295. doi: 10.1038/ismej.2008.117. [DOI] [PubMed] [Google Scholar]
- Chakravorty S, Helb D, Burday M, Connell N, Alland D. A detailed analysis of 16S ribosomal RNA gene segments for the diagnosis of pathogenic bacteria. J Microbiol Methods. 2007;69:330–339. doi: 10.1016/j.mimet.2007.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke KR, Gorley RN. PRIMER v6: User manual/tutorial. PRIMER-E Ltd.: Plymouth: United Kingdom; 2006. [Google Scholar]
- Cohan FM. Towards a conceptual and operational union of bacterial systematics, ecology, and evolution. Philos Trans R Soc Lond B Biol Sci. 2006;361:1985–1996. doi: 10.1098/rstb.2006.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohan FM, Perry EB. A systematics for discovering the fundamental units of bacterial diversity. Curr Biol. 2007;17:R373–R386. doi: 10.1016/j.cub.2007.03.032. [DOI] [PubMed] [Google Scholar]
- Daims H, Bruhl A, Amann R, Schleifer KH, Wagner M. The domain-specific probe EUB338 is insufficient for the detection of all Bacteria: development and evaluation of a more comprehensive probe set. Syst Appl Microbiol. 1999;22:434–444. doi: 10.1016/S0723-2020(99)80053-8. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Inferring Phylogenies. Sinauer Associates: Sunderland, MA, USA; 2003. [Google Scholar]
- Field KG, Gordon D, Wright T, Rappé MS, Urbach E, Vergin KL, et al. Diversity and depth-specific distribution of SAR11 cluster rRNA genes from marine planktonic bacteria. Appl Environ Microbiol. 1997;61:63–70. doi: 10.1128/aem.63.1.63-70.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrman JA, Hewson I, Schwalbach MS, Steele JA, Brown MV, Naeem S. Annually reoccurring bacterial communities are predictable from ocean conditions. Proc Natl Acad Sci USA. 2006;103:13104–13109. doi: 10.1073/pnas.0602399103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrman JA, Steele JA. Community structure of marine bacterioplankton: patterns, networks, and relationships to function. Aquat Microb Ecol. 2008;53:69–81. [Google Scholar]
- Gilbert JA, Steele JA, Caporaso JG, Steinbruck L, Reeder J, Temperton B, et al. Defining seasonal marine microbial community dynamics. ISME J. 2012;6:298–308. doi: 10.1038/ismej.2011.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni SJ, Britschgi TB, Moyer CL, Field KG. Genetic diversity in Sargasso sea bacterioplankton. Nature. 1990;345:60–63. doi: 10.1038/345060a0. [DOI] [PubMed] [Google Scholar]
- Giovannoni SJ, Rappé MS.2000Evolution, diversity and molecular ecology of marine prokaryotesIn: Kirchman DL, (ed)Microbial Ecology of the Oceans Wiley-Liss: New York; 47–85. [Google Scholar]
- Grote J, Thrash JC, Huggett MJ, Landry ZC, Carini P, Giovannoni SJ, et al. Streamlining and core genome conservation among highly divergent members of the SAR11 clade. mBio. 2012;3:e00252-12. doi: 10.1128/mBio.00252-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamady M, Walker JJ, Harris JK, Gold NJ, Knight R. Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nat Methods. 2008;5:235–237. doi: 10.1038/nmeth.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamady M, Lozupone C, Knight R. Fast UniFrac: facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME J. 2010;4:17–27. doi: 10.1038/ismej.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamp TJ, Jones WJ, Fodor AA. Effects of experimental choices and analysis noise on surveys of the ‘rare biosphere'. Appl Environ Microb. 2009;75:3263–3270. doi: 10.1128/AEM.01931-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huse SM, Dethlefsen L, Huber JA, Welch DM, Relman DA, Sogin ML. Exploring microbial diversity and taxonomy using SSU rRNA hypervariable tag sequencing. PLoS Genet. 2008;4:e1000255. doi: 10.1371/journal.pgen.1000255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huse SM, Welch DM, Morrison HG, Sogin ML. Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Environ Microbiol. 2010;12:1889–1898. doi: 10.1111/j.1462-2920.2010.02193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson DH, Mitra S, Ruscheweyh HJ, Weber N, Schuster SC. Integrative analysis of environmental sequences using MEGAN4. Genome Res. 2011;21:1552–1560. doi: 10.1101/gr.120618.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeraldo P, Chia N, Goldenfeld N. On the suitability of short reads of 16S rRNA for phylogeny-based analyses in environmental surveys. Environ Microbiol. 2011;13:3000–3009. doi: 10.1111/j.1462-2920.2011.02577.x. [DOI] [PubMed] [Google Scholar]
- Kirchman DL, Cottrell MT, Lovejoy C. The structure of bacterial communities in the western Arctic Ocean as revealed by pyrosequencing of 16S rRNA genes. Environ Microbiol. 2010;12:1132–1143. doi: 10.1111/j.1462-2920.2010.02154.x. [DOI] [PubMed] [Google Scholar]
- Koski LB, Golding GB. The closest BLAST hit is often not the nearest neighbor. J Mol Evol. 2001;52:540–542. doi: 10.1007/s002390010184. [DOI] [PubMed] [Google Scholar]
- Ludwig W, Schleifer K-H. Phylogeny of bacteria beyond the 16S rRNA standard. ASM News. 1999;65:752–757. [Google Scholar]
- Lynch M, Force A. The probability of duplicate gene preservation by subfunctionalization. Genetics. 2000;154:459–473. doi: 10.1093/genetics/154.1.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsen FA, Kodner RB, Armbrust EV. pplacer: linear time maximum-likelihood and Bayesian phylogenetic placement of sequences onto a fixed reference tree. BMC Bioinformatics. 2010;11:538. doi: 10.1186/1471-2105-11-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna P, Hoffmann C, Minkah N, Aye PP, Lackner A, Liu Z, et al. The macaque gut microbiome in health, lentiviral infection, and chronic enterocolitis. PLoS Pathog. 2008;4:e20. doi: 10.1371/journal.ppat.0040020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monier A, Claverie JM, Ogata H. Taxonomic distribution of large DNA viruses in the sea. Genome Biol. 2008;9:R106. doi: 10.1186/gb-2008-9-7-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris RM, Rappé MS, Connon SA, Vergin KL, Siebold WA, Carlson CA, et al. SAR11 clade dominates ocean surface bacterioplankton communities. Nature. 2002;420:806–810. doi: 10.1038/nature01240. [DOI] [PubMed] [Google Scholar]
- Morris RM, Rappé MS, Urbach E, Connon SA, Giovannoni SJ. Prevalence of the Chloroflexi-related SAR202 bacterioplankton cluster throughout the mesopelagic zone and deep ocean. Appl Environ Microbiol. 2004;70:2836–2842. doi: 10.1128/AEM.70.5.2836-2842.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris RM, Vergin KL, Cho JC, Rappé MS, Carlson CA, Giovannoni SJ. Temporal and spatial response of bacterioplankton lineages to annual convective overturn at the Bermuda Atlantic Time-series Study site. Limnol Oceanogr. 2005;50:1687–1696. [Google Scholar]
- Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig WG, Peplies J, et al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappé MS, Giovannoni SJ. The uncultured microbial majority. Ann Rev Microbiol. 2003;57:369–394. doi: 10.1146/annurev.micro.57.030502.090759. [DOI] [PubMed] [Google Scholar]
- Rocap G, Larimer FW, Lamerdin J, Malfatti S, Chain P, Ahlgren NA, et al. Genome divergence in two Prochlorococcus ecotypes reflects oceanic niche differentiation. Nature. 2003;424:1042–1047. doi: 10.1038/nature01947. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Ezpeleta N, Embley TM. The SAR11 group of alpha-proteobacteria is not related to the origin of mitochondria. PLoS One. 2012;7:e30520. doi: 10.1371/journal.pone.0030520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roesch LF, Fulthorpe RR, Riva A, Casella G, Hadwin AKM, Kent AD, et al. Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J. 2007;1:283–290. doi: 10.1038/ismej.2007.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusch DB, Halpern AL, Sutton G, Heidelberg KB, Williamson S, Yooseph S, et al. The Sorcerer II Global Ocean Sampling expedition: northwest Atlantic through eastern tropical Pacific. Plos Biol. 2007;5:398–431. doi: 10.1371/journal.pbio.0050077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schattenhofer M, Fuchs BM, Amann R, Zubkov MV, Tarran GA, Pernthaler J. Latitudinal distribution of prokaryotic picoplankton populations in the Atlantic Ocean. Environ Microbiol. 2009;11:2078–2093. doi: 10.1111/j.1462-2920.2009.01929.x. [DOI] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microb. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A, Ludwig T, Meier H. RAxML-III: a fast program for maximum likelihood-based inference of large phylogenetic trees. Bioinformatics. 2005;21:456–463. doi: 10.1093/bioinformatics/bti191. [DOI] [PubMed] [Google Scholar]
- Steinberg DK, Carlson CA, Bates NR, Johnson RJ, Michaels AF, Knap AH. Overview of the US JGOFS Bermuda Atlantic Time-series Study (BATS): a decade-scale look at ocean biology and biogeochemistry. Deep Sea Res. 2001;48:1405–1447. [Google Scholar]
- Thrash JC, Boyd A, Huggett MJ, Grote J, Carini P, Yoder RJ, et al. Phylogenomic evidence for a common ancestor of mitochondria and the SAR11 clade. Sci Rep. 2011;1:13. doi: 10.1038/srep00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treusch AH, Vergin KL, Finlay LA, Donatz MG, Burton RM, Carlson CA, et al. Seasonality and vertical structure of microbial communities in an ocean gyre. ISME J. 2009;3:1148–1163. doi: 10.1038/ismej.2009.60. [DOI] [PubMed] [Google Scholar]
- Treusch AH, Demir E, Vergin KL, Worden AZ, Carlson CA, Donatz MG, et al. Phytoplankton distribution patterns in the northwestern Sargasso Sea revealed by small subunit rRNA genes from plastids. ISME J. 2011;6:481–492. doi: 10.1038/ismej.2011.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tujula NA, Crocetti GR, Burke C, Thomas T, Holmstrom C, Kjelleberg S. Variability and abundance of the epiphytic bacterial community associated with a green marine Ulvacean alga. ISME J. 2010;4:301–311. doi: 10.1038/ismej.2009.107. [DOI] [PubMed] [Google Scholar]
- VAMPS (2012 ). http://vamps.mbl.edu/index.php .
- Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microb. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster NS, Taylor MW, Behnam F, Lücker S, Rattei T, Whalan S, et al. Deep sequencing reveals exceptional diversity and modes of transmission for bacterial sponge symbionts. Environ Microbiol. 2010;12:2070–2082. doi: 10.1111/j.1462-2920.2009.02065.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JR, Navlakha S, Nagarajan N, Ghodsi M-R, Kingsford C, Pop M. Alignment and clustering of phylogenetic markers - implications for microbial diversity studies. BMC Bioinformatics. 2010;11:152. doi: 10.1186/1471-2105-11-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu DY, Hartman A, Ward N, Eisen JA. An Automated phylogenetic tree-based small subunit rRNA taxonomy and alignment pipeline (STAP) PLoS One. 2008;3:e2566. doi: 10.1371/journal.pone.0002566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, Eisen JA. A simple, fast, and accurate method of phylogenomic inference. Genome Biol. 2008;9:R151. doi: 10.1186/gb-2008-9-10-r151. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.