Abstract
Invasions by non-native plants can alter ecosystem functions and reduce native plant diversity, but relatively little is known about their effect on belowground microbial communities. We show that invasions by knapweed (Centaurea stoebe) and leafy spurge (Euphorbia esula, hereafter spurge)—but not cheatgrass (Bromus tectorum)—support a higher abundance and diversity of symbiotic arbuscular mycorrhizal fungi (AMF) than multi-species native plant communities. The higher AMF richness associated with knapweed and spurge is unlikely due to a co-invasion by AMF, because a separate sampling showed that individual native forbs hosted a similar AMF abundance and richness as exotic forbs. Native grasses associated with fewer AMF taxa, which could explain the reduced AMF richness in native, grass-dominated communities. The three invasive plant species harbored distinct AMF communities, and analyses of co-occurring native and invasive plants indicate that differences were partly driven by the invasive plants and were not the result of pre-invasion conditions. Our results suggest that invasions by mycotrophic plants that replace poorer hosts can increase AMF abundance and richness. The high AMF richness in monodominant plant invasions also indicates that the proposed positive relationship between above and belowground diversity is not always strong. Finally, the disparate responses among exotic plants and consistent results between grasses and forbs suggest that AMF respond more to plant functional group than plant provenance.
Keywords: arbuscular mycorrhizal fungi, community composition, diversity, native community, plant invasion, 454-sequencing
Introduction
Plants associate with diverse groups of microorganisms and the nature of their interactions depend upon the evolutionary histories of each plant–microbe pair and the context of the surrounding biotic and abiotic environment (Johnson et al., 1997; Lambers et al., 2009; Porras-Alfaro and Bayman, 2011). Non-native plant invasions are often associated with shifts in plant community composition and ecosystem processes (Levine et al., 2003), but the interactions between exotic plants and soil microbes encountered in their non-native range are not well understood. Here we focus on one group, the arbuscular mycorrhizal fungi (AMF) in the phylum Glomeromycota, which colonize the great majority of terrestrial plant roots. These obligate biotrophic fungi receive plant carbon in exchange for phosphorus and other putative services (Smith and Read, 2008). Due to their ubiquity and location in the root–soil interface, AMF have been referred to as keystone mutualists (O'Neill et al., 1991), although parasitic interactions have been observed under controlled conditions (Johnson et al., 1997). Several invaders, such as garlic mustard (Allaria petiolata), are non-mycorrhizal, which has led to suggestions that invasive plants are either non-mycorrhizal or have a low inherent or evolved mycorrhizal dependency (van der Putten et al., 2007; Seifert et al., 2009; Vogelsang and Bever, 2009). On the other hand, AMF increase growth and competitiveness of spurge and knapweed—two of the most invasive plants in the intermountain west of the USA—under controlled conditions (Marler et al., 1999; Klironomos, 2002). Three main hypotheses have been proposed to explain the role of mycorrhizal fungi in plant invasions; (1) the degraded mutualism hypothesis, which states that non-mycorrhizal invaders reduce the fungal abundance, which, in turn, negatively affect mycotrophic native plants, (2) the enhanced mutualist hypothesis, whereby invasiveness is enhanced due to an encounter of better mutualists in the exotic than native range, and finally, (3) the mutualist hypothesis, in which invasions are prohibited due to the absence of specific mutualists (see review by Shah et al., 2009). Overall, the role of AMF for plant invasive success may be determined by the mycorrhizal dependency of the invader relative to the native plants, as well as the ability of the invasive plant to change the abundance and composition of AMF in the community being invaded (Pringle et al., 2009).
Quite a few studies have investigated the role of AMF on plant invasive success, but less is known about how invasive plants affect AMF communities. Shifts in both AMF abundance and community composition have been observed with several invaders (Al-Qawari, 2002; Mummey et al., 2005; Hawkes et al., 2006; Busby et al., 2012). However, the focus on single plant species and locations has limited comparisons among invasive plants, and the reproducibility across independent invasions has rarely been assessed (but see Busby et al., 2012).
We used 454-sequencing and visual assessments of AMF root colonization (hereafter referred to as AM colonization) to determine if and how AMF communities differ among native remnant plants and three exotic invaders knapweed (Centaurea stoebe), spurge (Euphorbia esula) and cheatgrass (Bromus tectorum) in six locations in western Montana, USA. This approach allowed us to replicate among independent invasions and to directly compare the effects of multiple invasive plant species. We hypothesized that fungal abundance would differ among vegetation types (native, cheatgrass, knapweed and spurge-dominated communities) due to variations in host quality among plant species (Baylis, 1975; indirectly assessed here by AM colonization). We also predicted that all plant invasions would decrease AMF richness due to the dominance of single plant species and reduction of AMF niches. Finally, due to the shift in life history strategy, root architecture and rooting depth with the three invaders combined with fungal host preference (for example, Vandenkoornhuyse et al., 2003, but see Stukenbrock and Rosendahl, 2005), we hypothesized that plant invasions would cause reproducible shifts in AMF communities.
To elucidate underlying mechanisms that may have caused differences observed in the main survey, we returned to one location (MPG Ranch) and conducted three additional surveys (survey 2–4). We measured concentrations of AMF biomarker lipids in soils harboring native and invasive plants to determine whether differences in AM colonization among vegetation types correspond with overall shifts in AMF soil abundance (survey 2). To address whether the higher AMF richness in knapweed and spurge invasions is due to a co-invasion by plants and AMF or a loss of AMF in native plant communities, we characterized AMF richness in individual grasses and forbs (survey 3). Finally, we sampled co-occurring native and invasive plants to evaluate whether the invasive plants drive observed shifts in AMF community composition, or if invasive plants and AMF respond instead to similar, pre-existing environmental conditions (survey 4).
Materials and methods
Sampling 1—comparison of AMF communities among vegetation types
Locations, vegetation types and sampling procedures
This study took place in rangelands around the Missoula and Bitterroot Valleys of western Montana (USA). The area encompassed approximately 255 km2 and sampling locations were located up to 50 km from each other. We sampled at six locations that each harbored plant communities (>100 m2) dominated by either native plants, cheatgrass, knapweed or spurge (hereafter referred to as vegetation types) within 500 m of each other (see Supplementary Table S1 for GPS (Global Positioning System) coordinates, invasion and herbicide history). Most invasions were older than 20 years, and all had a focal invasive coverage >60% based on visual observations, whereas the native plant communities harbored both grasses and forbs and had invasive cover of <5% (see Supplementary Table S2 for plant species info). A solid spurge invasion could not be found at one location (Indreland), and another location (Sapphire) lacked a cheatgrass-dominated community, leaving a total of 22 sites sampled across six locations and four vegetation types. We sampled roots and rhizosphere soil (0–15 cm depth) on 13–18 May 2010, when all plants were actively growing. Four plants were destructively harvested and pooled from within six areas (1 m2) for a total of 24 plants per site. We collected only the focal exotic plant from the invaded plant communities, but we included representative samples of grasses and forbs from native plant communities. We washed roots and took sub-samples for DNA extraction (2 ml micro-centrifuge tube). AM colonization measures (Brundrett et al., 1996) and soil analyses (AgSource Harris, Lincoln, NE, USA) were conducted on pooled samples from the six areas per site (Supplementary Table S3). Soils were analyzed for organic matter, available nitrogen, phosphorus (Bray-1 P), potassium, magnesium, calcium, cation exchange capacity and pH.
DNA extraction and amplification
We extracted DNA from freeze-dried, ground roots (15 mg) using CTAB lysis buffer (Gardes and Bruns, 1993), and we amplified optimized template concentrations (1:40–1:160) with the LSU (large sub-unit) rRNA primer set FLR3 and FLR4 (Gollotte et al., 2004), which targets the nLSU-D2 region that is suitable for high-throughput sequencing (Stockinger et al., 2010). We added linkers A and B, and MID (molecular identifier)-tag sequences (one for each vegetation type and location for a total of 22 individual MID-tags) to primers according to Roche's recommendations. A proofreading hot-start Phusion polymerase was used (New England Biolabs, Ipswich, MA, USA) with the following thermocycling parameters: 1 min 98 °C, 34 cycles of 1 min at 98 °C, 40 s of 58 °C and 11 s at 72 °C, with a final extension phase of 2 min at 72 °C. We pooled PCR products (4–6 amplicons, one from each of the six, 1 m2 areas sampled) within each vegetation type and location based on band intensity run on a 2% agarose gel. The 425–475 bp bands were purified using the QIAEX II kit (Qiagen, Valencia, CA, USA) and eluted with 20 μl 10 mℳ Tris buffer. This pooling was justified because our replicate was the vegetation type within each location not individual plants. Amplicons from all the vegetation types and locations were combined before sequencing based on DNA concentrations (NanoDrop 1000, Thermo Scientific, Asheville, NC, USA) to ensure similar representations from all the 22 sites sampled. To reduce problems with PCR bias, we conducted a second PCR amplification, cleaning and pooling of all the samples. The two PCR runs were pooled based on DNA concentrations and sent for 454-sequencing to the UC Denver School of Medicine on a 1/2 PicoTiterPlate using a GS-FLX pyrosequencing system according to the manufacturer's instructions (Roche, Branford, CT, USA).
Sequence analyses and fungal operational taxonomic unit (OTU) designation
Raw sequence files for all the 22 sites are accessible at the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA037764.2). We removed sequences <200 bp and with a quality score <30. To standardize the number of reads, we resampled 3023 sequences from each of the 22 sites (the lowest number of sequences from one site). We divided the remaining sequences into preliminary OTUs using the cd-hit-est clustering algorithm (Li and Godzik, 2006) at 97% similarity (default settings, except aS=0.8), and blasted representatives from each cluster against NCBI GenBank (Altschul et al., 1990). All non-AMF clusters were removed (identified based on closest BLAST hit not annotated as ‘glomeromycota') as well as clusters with <5 reads to reduce the risk of artificially inflating richness due to sequencing error. Sampling effort curves were calculated with Past (http://folk.uio.no/ohammer/past/) and indicated that the sampling intensity was sufficient to identify the taxa present in all the sites, within the restrictions of our PCR protocol (Supplementary Figure S1). We constructed a neighbor joining tree in MEGA v5 (Tamura et al., 2011) to identify OTUs as monophyletic clades, blasted representative sequences against NCBI GenBank to obtain the most similar sequences from other studies (Supplementary Table S4) and counted their relative read numbers (Supplementary Table S5). A Neighbor Net was constructed on all remaining sequences using SplitsTree4 v. 4.10 (Huson and Bryant, 2006) (Supplementary Figure S2). Community sampling effort curves were calculated in EstimateS (Supplementary Figure S3) using default settings and the Mao Tau richness function (Version 8.2, R. K. Colwell, http://purl.oclc.org/estimates). Representative sequences of our 46 fungal OTUs are archived at NCBI (JN180868—JN180913). The selected sequences were the most frequent sequence types.
Statistical analyses
We calculated Shannon diversity and evenness for each location and vegetation type using fungal OTU read numbers as a measure of abundance (Supplementary Table S6) and conducted an analysis of phylogenetic diversity by estimating average pair-wise nucleotide differences between the fungal OTUs in the program DnaSP v. 5.1 (Librado and Rozas, 2009; Supplementary Table S6). To increase the information, sequences were extended into the more conserved region at the 5′ end using sequence information from the closest matches in Genbank (alignment available upon request).
We analyzed differences in AM colonization, fungal OTU richness, fungal evenness and diversity among vegetation types and locations using a two-factor analysis of variance in Minitab (Minitab Inc., State College, PA, USA) with location as a random factor. The fungal community composition was analyzed using multivariate analyses in CANOCO 4.54 (ter Braak and Šmilauer, 2002), with fungal OTU read numbers as measures of relative abundance. All analyses had samples as scaling focus and all species data were Hellinger-transformed using the program PrCoord 1.0 (ter Braak and Šmilauer, 2002). Detrended Correspondence Analysis of fungal OTU abundance data showed axes lengths <3.0, suggesting a linear treatment of the data. Spatial relationships among the 22 sites were based on geographical x–y coordinates according to suggestions in Legendre and Legendre (2012). We used Distance-based Redundancy Analysis (CANOCO 4.54) with manual forward selection (999 permutations) to evaluate correlations among vegetation types, spatial distribution, soil chemical parameters (Supplementary Table S6), focal exotic plant dominance (Supplementary Table S1) and fungal OTU composition (Supplementary Table S5). All samples were plotted on the first two principal coordinate axes, with significant explanatory variables (as indicated by forward selection) overlaid as supplementary data. Finally, we conducted a network analysis using QIIME v 1.5.0 (Caporaso et al., 2010) and cytoscape software packages (Lopes et al., 2010) to assess how individual fungal OTUs were distributed among the four vegetation types.
Survey 2—measurements of AMF biomass in soil
We collected rhizosphere soil (five 10 cm deep soil cores, pooled) from 5–7 independent and interspersed knapweed, spurge and cheatgrass invasions and remnant native plant communities on MPG Ranch on 27 October 2011. We restricted our sampling to MPG Ranch due to limited access to other locations. Sampling was conducted at the end—rather than the beginning—of the growing season to ensure that values reflected living mycelia that had accumulated during the growing season. Invasions were chosen using the same criteria as in our main survey. The soil samples were immediately sieved and frozen (−20 °C), and lipids were extracted from 3 g of freeze-dried soil following the instructions in van Aarle and Olsson (2003). We used the neutral lipid fatty acid 16:1ω5 as a biomarker for AMF as it is superior to the more commonly used phospholipid fatty acid when extracting from soil (Frostegård et al., 2010). The fatty acid residues in neutral lipids were converted into free fatty acid methyl esters and analyzed as in van Aarle and Olsson (2003). Also, 16:1ω5 concentrations were calculated relative to the internal standard 19:0, and differences among vegetation types were assessed using a one-way analysis of variance.
Survey 3—comparison of AMF communities among plant individuals
We sampled 10 individual plants each of knapweed and spurge, representatives of five native forb species and four native grass species from MPG Ranch on 6 October 2011 to measure AM colonization and fungal OTU richness per plant (Supplementary Table S7). The same procedures were used as outlined in Survey 1, except we resampled to 265 sequences per plant individual that saturated all samples (sampling effort curves not shown). All unique, unclustered reads and all non-AMF were removed. Differences in AM colonization and fungal OTU richness among plants (knapweed, spurge, native grasses, native forbs) were analyzed using a one-way analysis of variance, and dominance was calculated based on the ratio of the most abundant fungal OTU to the total number of sequences per sample. We used correlation analyses in Minitab to quantify relationships among measured variables.
Survey 4—assessment of AMF host specificity
We sampled co-occurring native (10 individuals of representative grasses and forbs) and spurge plants (10 individuals) for a total of 20 plants on 3 June 2011, within a 50 × 50 m recent spurge invasion (5–15% spurge cover) on MPG Ranch. DNA was extracted from eight, 1-cm root fragments per plant individual using chelex and TE-buffer (Rosendahl and Stukenbrock, 2004) and amplified with the primer pair FLR3-FLR4 (Gollotte et al., 2004) using the same PCR conditions as outlined above. All positive PCR products were sent for Sanger sequencing at the High-throughput Genomics Center, Seattle, WA, USA. Seventy-eight successful sequences (that is, not mixed or failed) were aligned with OTUs from Survey 1 to assess their identity (sequences >200 bp are archived at NCBI under accession numbers KC533027–KC533092). These originated from six out of the ten native plants and six out of the ten spurge plants. The fungal OTU distribution between the co-occurring native and spurge plants was compared with the 454 data (resampled to the same sampling intensity) using the same ordination methods as outlined above.
Results
Our main survey documented AM colonization and AMF community composition in remnant native communities and knapweed, spurge and cheatgrass invasions. We observed large differences in AM colonization among vegetation types; cheatgrass roots were poorly colonized, whereas knapweed and spurge had a higher AM colonization than native plants (Table 1). We identified 46 fungal OTUs across all the 22 sites sampled (Supplementary Figure S2 and Supplementary Table S4). Invasion did not reduce fungal OTU richness, which was similar in cheatgrass invasions and native plant communities, but almost twice as high in knapweed and spurge invasions (Table 1, Supplementary Tables S5 and S6). AMF α-diversity was also highest in knapweed and spurge invasions, and there was a positive correlation between average AM colonization and fungal OTU richness (R=0.77, P<0.001), and between AM colonization and fungal diversity (R=0.74, P<0.001), but not between plant richness and fungal OTU richness (R=0.03, P=0.89). The fungal β-diversity (γ/α), on the other hand, was highest in the native communities and cheatgrass invasions (P=0.042; Supplementary Table S6; Supplementary Figure S3).
Table 1. Mean (±s.e.) of AM colonization (%), fungal operational taxonomic units (OTU) richness, evenness, diversity and average genetic diversity in four vegetation types (native, cheatgrass, knapweed and spurge).
| Plant community | n | AM colonization (%) | OTU richness | Evenness (EH) | Shannon (H) | Genetic diversity (pi) |
|---|---|---|---|---|---|---|
| Native | 6 | 39.9 (8.05)b | 13.0 (1.03)c | 0.730 (0.020)a | 1.85 (0.03)b | 0.152 (0.007)b |
| Cheatgrass | 5 | 12.6 (5.29)c | 14.6 (0.87)c | 0.556 (0.066)b | 1.49 (0.19)c | 0.189 (0.005)a |
| Knapweed | 6 | 78.9 (2.48)a | 24.2 (1.08)a | 0.713 (0.009)a | 2.27 (0.06)a | 0.152 (0.006)b |
| Spurge | 5 | 87.0 (3.22)a | 20.4 (1.47)b | 0.698 (0.021)a | 2.10 (0.10)ab | 0.136 (0.003)b |
Abbreviation: AM, arbuscular mycorrhizal.
OTU evenness and diversity were calculated using sequence numbers as a measure of abundance (Supplementary Tables S4 and S5). Super-scripted letters indicate significant differences (P⩽0.05) among means using Fisher's lsd. There was no significant difference among locations for any variable.
AMF communities clustered according to vegetation type, with cheatgrass and spurge invasions harboring the most dissimilar AMF communities (Figure 1). Cheatgrass invasions also harbored more phylogenetically dispersed AMF than the other three vegetation types (Table 1). Although the four most abundant fungal OTUs were found in all the vegetation types (see network analysis in Supplementary Figure S4), their prevalence differed. OTU 10 (a member of the genus Glomus) was found in higher abundance in spurge invasions, OTU 40 (most likely a Claroideoglomus) was predominately found in cheatgrass invasions and OTUs 26 and 28 (members of the Rhizophagus) were in higher abundances in native and knapweed communities, respectively (Supplementary Figure S5). There were no significant differences in AM colonization, fungal OTU richness, evenness and Shannon diversity among the locations. Also, the spatial distribution of sites did not influence the fungal OTU composition. Soil-available P differed among vegetation types (highest under cheatgrass and lowest under native plants) as did available K (highest under spurge and lowest under native plants). Soil-available P also differed among locations (highest at Indreland Ranch and lowest at Granite Creek) as did soil pH (highest at University and lowest at Granite Creek, Supplementary Table S3).
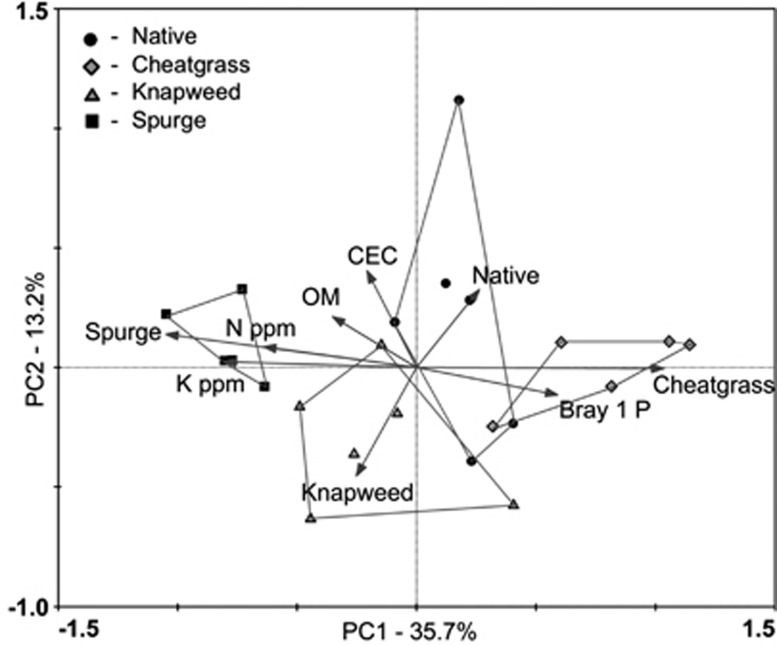
Figure 1.
Principal coordinate (PC) plot of Hellinger-transformed OTU composition in roots of native plants, cheatgrass, knapweed and spurge. For clarity, samples are partitioned within convex hulls according to vegetation type. The vectors show variables that correlate significantly with AMF community structure (as determined using a Monte Carlo permutation test, N=999, via forward selection in a constrained Distance-based Redundancy Analysis; P<0.05). CEC, cation exchange capacity; K, potassium; N, nitrogen; OM, organic matter; P, phosphorus; p.p.m., parts per million.
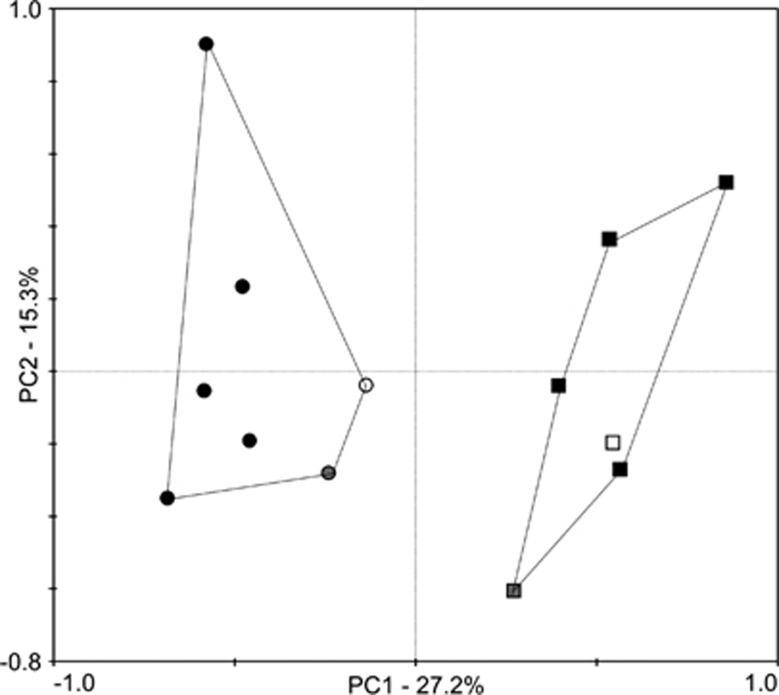
We observed significant (P<0.001) differences in soil 16:1ω5 concentration among vegetation types in our second survey, with knapweed invasions having the highest—and cheatgrass the lowest—16:1ω5 concentrations (Table 2). Our third survey, in which we documented the AM colonization and AMF richness in individual plants, revealed a similar fungal OTU richness in native forbs (five plant species sampled and listed in Supplementary Table S6) and knapweed and spurge (Table 3). Individual native grasses (four species sampled and listed in Supplementary Table S6), on the other hand, had a lower AM colonization and on average 2–3 fewer fungal OTUs (Table 3). We found a negative relationship between fungal OTU dominance (most abundant OTU/total sequence number) and fungal OTU richness (R=−0.37, P=0.026) and a similar trend between fungal OTU dominance and AM colonization (R=−0.30, P=0.07). Finally, co-occurring native and spurge plants harbored different AMF communities in our fourth survey, and the two communities clustered within their respective vegetation type from our main sampling (Figure 2).
Table 2. Mean (±s.e.) of 16:1ω5 NLFA concentrations in soil collected from replicated knapweed, spurge and cheatgrass invasions, as well as remnant native plant communities on MPG Ranch.
| Plant community | n | 16:1ω5 NLFA (nmol g per soil) |
|---|---|---|
| Native | 7 | 34.5 (3.19)b |
| Cheatgrass | 6 | 18.8 (3.71)c |
| Knapweed | 5 | 59.0 (8.91)a |
| Spurge | 7 | 39.1 (4.58)b |
Abbreviation: NLFA, neutral lipid fatty acid.
Super-scripted letters indicate significant differences (P⩽0.05) among means using Fisher's lsd.
Table 3. Mean (±s.e.) of AM root colonization (%), fungal operational taxonomic units (OTU) richness and cumulative richness (mean Mao Tau±s.d., standardized to eight plants) within individual native grasses (four species), native forbs (five species), knapweed and spurge plants (Supplementary Table S6).
| Plant community | n | AM colonization (%) | OTU richness | Cumulative richness |
|---|---|---|---|---|
| Native grasses | 10 | 46.2 (4.33)c | 7.80 (0.44)b | 13.7 (0.38) |
| Native forbs | 9 | 81.3 (1.99)ab | 10.3 (0.67)a | 21.3 (2.02) |
| Knapweed | 8 | 76.1 (4.61)b | 9.25 (1.08)ab | 21.0 (1.66) |
| Spurge | 10 | 90.7 (2.15)a | 10.0 (0.70)a | 21.2 (3.51) |
Abbreviation: AM, arbuscular mycorrhizal.
Super-scripted letters indicate significant differences (P⩽0.05) among means using Fisher's lsd. Two knapweed plants and one native forb contained too few sequences to analyze.
Figure 2.
Principal coordinate (PC) analysis of AMF communities (first two axes) associated with spurge (squares) and native (circles) plants that co-occur in a young spurge invasion (white color-filled square and circle), in separate, adjacent patches in the same location (gray color-filled square and circle) and in other locations (black color-filled squares and circles). The shaded symbols are sites from our original 454 sequencing but resampled to the same sampling intensity as the co-occurring spurge and native plants.
Discussion
We conducted four surveys to assess if and how plant invasions affect AMF communities. Our results support the first hypothesis and show that AM colonization differs predictably among vegetation types (Table 1). Differences in AM colonization could indicate significant disparities in C-allocations to AMF. However, a substantial portion of the fungal biomass is in the soil (Hart and Reader, 2002), and we therefore supplemented our fungal root colonization data with measurements of fungal biomarker concentrations in soil. Cheatgrass roots were poorly colonized by AMF (Table 1), and rhizosphere soil had the lowest concentration of AMF biomarker (Table 2), which supports previous findings that cheatgrass invasions can reduce overall AMF abundances (Al-Qawari, 2002; Busby et al., 2012). Knapweed, on the other hand, was highly colonized by AMF, and soil in knapweed invasions had the highest biomarker concentrations, which shows that invasive mycorrhizal forbs can be good hosts and enhance AMF abundances relative to native mixed plant communities (Greipsson and DiTommaso, 2006; Shah et al., 2010). We propose that invasion-mediated shifts in AMF abundance can be estimated based on changes in forb/grass ratios combined with some knowledge about the mycorrhizal status of the plants. Comparisons from about 100 prairie plant species corroborate our findings and show that forbs have a higher AM colonization and benefit more from AMF than cool season grasses (Wilson and Hartnett, 1998).
Contrary to our second hypothesis, the replacement of multi-species native plant communities with mono-dominant invasive plants did not reduce fungal OTU richness. We found almost twice as many fungal OTUs in knapweed and spurge invasions relative to cheatgrass invasions and remnant native plant communities (Table 1, Supplementary Tables S5 and S6), even though we sampled single plant species in the invasions and multiple plant species in the native communities. Fungal community evenness was as high in knapweed and spurge invasions as native plant communities (Table 1) which suggest that these invaders did not change the degree of fungal OTU dominance. Findings by Stephan et al. (2001) indicate that our results are not unprecedented and may extend to other microbial groups, because bacterial diversity was comparable in the rhizosphere of a single plant species and in a community of 32 plant species. Previous work has suggested that resource supply—not plant diversity—drives soil microbial diversity (Waldrop et al., 2006). We found no relationship between plant species richness and fungal OTU numbers but a good correlation between AM colonization and fungal OTU numbers. This suggests that the positive relationship between plant and AMF richness (van der Heijden et al., 1998; Landis et al., 2004) is not always strong and that carbon allocation could be one factor that determines AMF richness. Additional experimental work with altered C-allocation to AMF is required to test this hypothesis and exclude alternative explanations. For example, plants often associate with different and diverse assemblages of endophytes that could interact directly as well as indirectly with AMF taxa by producing secondary metabolites (see review by Porras-Alfaro and Bayman, 2011). It is interesting to note that secondary metabolites can be more abundant and diverse in forbs than grasses (as discussed in Mole and Joern, 1993), and research into more complex interactions involving AMF, plants and other microbial groups could be fruitful.
Irrespective of how knapweed and spurge maintain a higher AMF richness, there are three possible explanations for how it originated. First, AMF richness may just be historically higher in areas where knapweed and spurge invade. We find this to be unlikely because AMF responses to environmental shifts are rapid (Drigo et al., 2010) and historical differences are probably not maintained over time. More likely, the higher AMF richness is due to either a co-invasion by AMF, knapweed and spurge or a loss of AMF taxa in remnant native communities. Co-invasion with plants and their mycorrhizal fungi have been shown previously (Dickie et al., 2010) but is an unlikely explanation here for two main reasons. First, the greater richness was driven by rare taxa that differed among locations (Supplementary Table S5), whereas a co-invasion would be more supported by a few, abundant and widespread fungal OTUs. Second, individual native forb plants harbored a similar fungal OTU richness as the exotic forbs in our supplemental sampling, whereas grasses were hosts to on average 2–3 fewer fungal OTUs per individual. We therefore propose that the difference in AMF community richness among knapweed, spurge and native communities is due to a loss (or at least a very spotty and infrequent distribution) of rare taxa in the native communities. We argue that this is driven by the abundance of grasses in the native communities that can be high in some communities due to non-target effects from the repeated use of dicot-specific herbicides (Ortega and Pearson, 2011). Ironically, the exotic forb invasions that provoke these herbicide applications appear to boost AMF communities. Shah et al. (2010) reported a lower AMF richness associated with two invasive forbs (Anthemis cotula and Conyza canadensis) relative to the native communities that appear contradictory to our findings. However, spore surveys were used to assess differences that may have a reduced ability to detect rare taxa. Also, the AM colonization and structure of the native plant community are unknown in Shah et al. (2010) that complicate direct comparisons with our study.
AMF communities clustered according to vegetation type irrespective of location that supports our third prediction (Figure 1) and suggests that community shifts are reproducible and predictable and that host communities impose a strong environmental filter on AMF. Because AMF communities differed between co-occurring native and spurge plants (survey 4) and clustered with their respective vegetation type in our main survey (Figure 2), differences in AMF communities appear to be, at least partly, host mediated. Host preference is found in some (for example, Vandenkoornhuyse et al., 2003) but not all (for example, Stukenbrock and Rosendahl, 2005) studies and may depend on the degree of phenological differences among plants as well as their abundance and distribution pattern. Indeed, cheatgrass and spurge harbored the most dissimilar AMF communities, and they are also the most phenotypically disparate plants of the ones surveyed here; cheatgrass is a shallow-rooted, facultative mycorrhizal annual grass, whereas spurge is a deep-rooted, mycotrophic perennial forb. In addition to direct host effects, shifts in AMF communities could be exacerbated by changes in abiotic factors (Johnson, 1993), because plant invasions have been shown to alter soil nutrient availabilities (Liao et al., 2008). Available N, P and K correlated with the fungal OTU distribution here (Figure 1).
Moora et al. (2011) found that an exotic plant associated with generalist AMF to a greater extent than native plants and argued that this may facilitate invasions. Cheatgrass, knapweed and spurge all associated with different AMF communities, and knapweed and spurge harbored a greater number of rare taxa than the adjacent native plants, which is not what one might expect if these invaders behaved as the focal plant Trachycarpus fortunei in Moora et al, (2011). Also, OTU 26—closely related to Rhizophagus intraradices (former Glomus intraradices)—was slightly more abundant in native plant communities here (Supplementary Figure S5), and this fungus is often considered to be a cosmopolitan generalist (Öpik et al., 2006). Overall, comparisons of knapweed, spurge and cheatgrass in their native and exotic ranges would be informative.
We calculated the average phylogenetic distance among co-occurring fungal OTUs within communities, because phylogenetic trait conservatism has been shown within AMF (Powell et al., 2009), and relatedness can determine ecological function and suggest underlying forces that structure communities (Maherali and Klironomos, 2007). For example, a community that has high phylogenetic dispersal (that is, harbors more distantly related taxa than expected by chance) is thought to be structured by competition, because only dissimilar taxa that occupy different niches can co-exist. A community of closely related taxa, on the other hand, suggests a strong habitat filter. Cheatgrass invasions harbored the most divergent fungal communities (Table 1), and we propose that this may be a result of competitive exclusion of closely related fungal taxa driven by the small root system and poor host quality by cheatgrass. Interestingly, Busby et al. (2012) also found a greater phylogenetic dispersal of fungi colonizing cheatgrass than those colonizing the more mycotrophic sagebrush (Artemisia tridentata). The negative relationships between fungal OTU dominance and richness, and between fungal OTU dominance and AM colonization, suggest that competitive fungal taxa may reduce overall AMF richness and that these fungal taxa are more abundant in poorly colonized plants, such as grasses (Supplementary Table S6). Similar to Busby et al. (2012), we found a higher AMF β-diversity in cheatgrass invasions and the grass-dominated native communities than in knapweed and spurge invasions (Supplementary Table S6, Supplementary Figure S3), which suggest a greater turnover among locations and an influence of stochastic processes in AMF community assembly within these plant communities.
Our findings could have functional implications for plant invasive success. For example, fungal richness influences overall productivity and interactions among plants (van der Heijden et al., 1998; Maherali and Klironomos, 2007), and the ability of knapweed and spurge to associate with many fungal taxa could aid invasions into a wide variety of environments. The observed shifts in AMF communities could further increase invasiveness because positive feedbacks between belowground communities and exotic plants have been observed and invasive plants can alter the AMF community to increase their competitiveness toward native plants (Klironomos, 2002; Zhang et al., 2010). These altered fungal communities can extend to native neighboring plants (Mummey et al., 2005; Hawkes et al., 2006) and may help generate alternative stable states that allow long-term persistence of invasive plants (Suding et al., 2004). Attempts have been made to overcome invasion legacies by reintroducing soil biota from native areas during restoration (Kardol et al., 2009). However, if our results extend to other locations and constituents of the microbial community, we should not assume that grass-dominated native communities are islands of diversity. On the contrary, knapweed and spurge invasions may harbor the ecological memory of fungal taxa once found in the pristine, native plant communities they replaced, and restoration aspects of this should be explored. Because AMF can be the single most important carbon source for other soil microbes (Olsson et al., 1999), differences observed here could cascade throughout the belowground ecosystem and alter functionality.
In summary, we show that AMF richness can be high in severe mono-dominant invasions, which suggests that plant diversity is not always a strong predictor of AMF richness. By sampling from multiple exotic species and independent, replicated invasions, we were able to detect unprecedented patterns and show that while invasive plants share many traits that promote invasiveness (van Kleunen et al., 2010), they differ substantially in their interactions with other biota. In fact, we found more similarities within functional groups (forbs vs grasses) than place of origin, which support recent pleas to move away from the native/alien dichotomy in favor of more trait-based approaches in invasion biology (Davis et al., 2011).
Acknowledgments
We thank land managers and ranchers for providing sample sites, Lauren Stoffel and Alexii Cornell for help during sampling, PCR and plant identification and Jill Castoe, Scott Hunicke-Smith, Weizhong Li and Sitao Wu for guidance with 454-sequencing and associated analyses. We also thank Roger Koide, Dean Pearson, John Maron, Lauren Waller and three anonymous reviewers for their comments on the manuscript, Dan Mummey for advice regarding PCR and statistics and MPG Operations for funding.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Al-Qawari AA. PhD dissertation. Colorado State University: Fort Collins, Colorado, USA; 2002. Relationships among nitrogen availability, vesicular-arbuscular mycorrhizae, and Bromus tectorum in disturbed rangeland sites in Colorado. [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Baylis GTS.1975The magnolioid mycorrhiza and mycotrophy in root systems derived from itIn: Sanders F-E, Mosse B, Tinker P-B eds.Endomycorrhizas Academic Press: London, UK; 373–389. [Google Scholar]
- Brundrett M, Bougher N, Dell B, Grove T, Malajczuk N.1996Working with Mycorrhizas in Forestry and Agriculture ACIAR Monograph; Issue 32Canberra, Australia [Google Scholar]
- Busby RR, Stromberger ME, Rodriguez G, Gebhart DL, Paschke MW. Arbuscular mycorrhizal fungal community differs between coexisting native shrub and introduced annual grass. Mycorrhiza. 2012;23:129–141. doi: 10.1007/s00572-012-0455-x. [DOI] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis MA, Chew MK, Hobbs RJ, Lugo AE, Ewel JJ, Vermeij GJ, et al. Don't judge species on their origins. Nature. 2011;474:153–154. doi: 10.1038/474153a. [DOI] [PubMed] [Google Scholar]
- Dickie IA, Bolstridge N, Cooper JA, Peltzer DA. Co-invasion by Pinus and its mycorrhizal fungi. New Phytol. 2010;187:475–484. doi: 10.1111/j.1469-8137.2010.03277.x. [DOI] [PubMed] [Google Scholar]
- Drigo B, Pijl AS, Duyts H, Kielak AM, Gamper HA, Houtekamer MJ, et al. Shifting carbon flow from roots into associated microbial communities in response to elevated atmospheric CO2. Proc Natl Acad Sci. 2010;107:10938–10942. doi: 10.1073/pnas.0912421107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frostegård Å, Tunlid A, Bååth E. The use and misuse of PLFA measurements in soils. Soil Biol Biochem. 2010;43:1621–1625. [Google Scholar]
- Gardes M, Bruns TD. ITS primers with enhanced specificity for basidiomycetes—application to the identification of mycorrhizae and rusts. Mol Ecol. 1993;2:113–118. doi: 10.1111/j.1365-294x.1993.tb00005.x. [DOI] [PubMed] [Google Scholar]
- Gollotte A, van Tuinen D, Atkinson D. Diversity of arbuscular mycorrhizal fungi colonising roots of the grass species Agrostis capillaris and Lolium perenne in a field experiment. Mycorrhiza. 2004;14:111–117. doi: 10.1007/s00572-003-0244-7. [DOI] [PubMed] [Google Scholar]
- Greipsson S, DiTommaso A. Invasive non-native plants alter the occurrence of arbuscular mycorrhizal fungi and benefit from this association. Ecological Restoration. 2006;24:236–241. [Google Scholar]
- Hart MM, Reader RJ. Taxonomic basis for the variation in the colonization strategy of arbuscular mycorrhizal fungi. New Phytol. 2002;153:335–344. [Google Scholar]
- Hawkes CV, Belnap J, D'Antonio C, Firestone MK. Arbuscular mycorrhizal assemblages in native plant roots change in the presence of invasive exotic grasses. Plant Soil. 2006;281:369–380. [Google Scholar]
- Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 2006;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- Johnson NC. Can fertilization of soil select less mutualistic mycorrhizae. Ecol Appl. 1993;3:749–757. doi: 10.2307/1942106. [DOI] [PubMed] [Google Scholar]
- Johnson NC, Graham JH, Smith FA. Functioning of mycorrhizal associations along the mutualism-parasitism continuum. New Phytol. 1997;135:575–585. [Google Scholar]
- Kardol P, Bezemer MT, van der Putten WH. Soil organism and plant introductions in restoration of species-rich grassland communities. Restor Ecol. 2009;17:258–269. [Google Scholar]
- Klironomos JN. Feedback with soil biota contributes to plant rarity and invasiveness in communities. Nature. 2002;417:67–70. doi: 10.1038/417067a. [DOI] [PubMed] [Google Scholar]
- Lambers H, Mougel C, Jaillard B, Hinsinger P. Plant-microbe-soil interactions in the rhizosphere: an evolutionary perspective. Plant Soil. 2009;321:83–115. [Google Scholar]
- Landis FC, Gargas A, Givnish TJ. Relationships among arbuscular mycorrhizal fungi, vascular plants and environmental conditions in oak savannas. New Phytol. 2004;164:493–504. [Google Scholar]
- Legendre P, Legendre L. Numerical Ecology (Developments in Environmental Modelling) Elsevier; 2012. [Google Scholar]
- Levine JM, Vilà M, D'Antonio CM, Dukes JS, Grigulis K, Lavorel S. Mechanisms underlying the impacts of exotic plant invasions. Proc R Soc London B Biol Sci. 2003;270:775–781. doi: 10.1098/rspb.2003.2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- Liao C, Peng R, Luo Y, Zhou X, Fang C, Cheng J, et al. Altered ecosystem carbon and nitrogen cycles by plant invasion: a meta-analysis. New Phytol. 2008;177:706–714. doi: 10.1111/j.1469-8137.2007.02290.x. [DOI] [PubMed] [Google Scholar]
- Librado PJ, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- Lopes CT, Franz M, Kazi F, Donaldson SL, Morris Q, Bader GD. Cytoscape web: an interactive web-based network browser. Bioinformatics. 2010;15:2347–2348. doi: 10.1093/bioinformatics/btq430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maherali H, Klironomos JN. Influence of phylogeny on fungal community assembly and ecosystem functioning. Science. 2007;316:1746–1748. doi: 10.1126/science.1143082. [DOI] [PubMed] [Google Scholar]
- Marler MJ, Zabinski CA, Callaway RM. Mycorrhizae indirectly enhance competitive effects of an invasive forb on a native bunchgrass. Ecology. 1999;80:1180–1186. [Google Scholar]
- Mole S, Joern A. The foliar phenolics if Nebraska sandhills prairie graminoids: between years, seasonal and interspecific variation. J Chem Ecol. 1993;19:1861–1874. doi: 10.1007/BF00983792. [DOI] [PubMed] [Google Scholar]
- Moora M, Berger S, Davison J, Öpik M, Bommarco R, Bruelheide H, et al. Alien plants associate with widespread generalist arbuscular mycorrhizal fungal taxa: evidence from a continental-scale study using massively parallel 454 sequencing. J Biogeogr. 2011;38:1305–1317. [Google Scholar]
- Mummey DL, Rillig MC, Holben WE. Neighboring plant influences on arbuscular mycorrhizal fungal community composition as assessed by T-RFLP analysis. Plant Soil. 2005;271:83–90. [Google Scholar]
- Olsson PA, Thingstrup I, Jakobsen I, Bååth E. Estimation of the biomass of arbuscular mycorrhizal fungi in a linseed field. Soil Biol Biochem. 1999;31:1879–1887. [Google Scholar]
- O'Neill EG, O'Neill RV, Norby RJ. Hierarchy theory as a guide to mycorrhizal research on large-scale problems. Environ Pollut. 1991;73:271–284. doi: 10.1016/0269-7491(91)90054-z. [DOI] [PubMed] [Google Scholar]
- Öpik M, Moora M, Liira J, Zobel M. Composition of root-colonizing arbuscular mycorrhizal fungal communities in different ecosystems around the globe. J Ecol. 2006;94:778–790. [Google Scholar]
- Ortega YK, Pearson DE. Long-term effects of weed control with picloram along a gradient of spotted knapweed invasion. Rangeland Ecol Manag. 2011;64:67–77. [Google Scholar]
- Powell JR, Parrent JL, Hart MM, Klironomos JN, Rillig MC, Maherali H. Phylogenetic trait conservatism and the evolution of functional trade-offs in arbuscular mycorrhizal fungi. Proc R Soc London B Biol Sci. 2009;276:4237–4245. doi: 10.1098/rspb.2009.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porras-Alfaro A, Bayman P. Hidden fungi, emergent properties: endophytoes and microbiomes. Annu Rev Phytopathol. 2011;49:291–315. doi: 10.1146/annurev-phyto-080508-081831. [DOI] [PubMed] [Google Scholar]
- Pringle A, Bever JD, Gardes M, Parrent JL, Rillig MC, Klironomos JN. Mycorrhizal symbioses and plant invasions. Annu Rev Ecol Evol Syst. 2009;40:699–715. [Google Scholar]
- Rosendahl S, Stukenbrock EH. Community structure of arbuscular mycorrhizal fungi in undisturbed vegetation revealed by analyses of LSU rDNA sequences. Mol Ecol. 2004;13:3179–3186. doi: 10.1111/j.1365-294X.2004.02295.x. [DOI] [PubMed] [Google Scholar]
- Seifert EK, Bever JD, Maron JL. Evidence for the evolution of reduced mycorrhizal dependence during plant invasion. Ecology. 2009;90:1055–1062. doi: 10.1890/08-0419.1. [DOI] [PubMed] [Google Scholar]
- Shah MA, Reshi ZA, Khasa DP. Arbuscular mycorrhizas: drivers or passengers of alien plant invasions. Bot Rev. 2009;75:397–417. [Google Scholar]
- Shah MA, Reshi ZA, Rasool N. Plant invasion induce a shift in Glomalean spore diversity. Trop Ecol. 2010;51:317–323. [Google Scholar]
- Smith SE, Read DJ. Mycorrhizal Symbiosis. Academic Press and Elsevier: London, UK; 2008. [Google Scholar]
- Stephan A, Meyer AH, Schmid B. Plant diversity affects culturable soil bacteria in experimental grassland communities. J Ecol. 2001;88:988–998. [Google Scholar]
- Stockinger H, Kruger M, Schüßler A. DNA barcoding of arbuscular mycorrhizal fungi. New Phytol. 2010;187:461–474. doi: 10.1111/j.1469-8137.2010.03262.x. [DOI] [PubMed] [Google Scholar]
- Stukenbrock EH, Rosendahl S. Distribution of dominant arbuscular mycorrhizal fungi among five plant species in undisturbed vegetation of a coastal grassland. Mycorrhiza. 2005;15:497–503. doi: 10.1007/s00572-005-0357-2. [DOI] [PubMed] [Google Scholar]
- Suding KN, Gross KL, Houseman GR. Alternative states and positive feedbacks in restoration ecology. Trends Ecol Evol. 2004;19:46–53. doi: 10.1016/j.tree.2003.10.005. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ter Braak CJF, Šmilauer P.2002CANOCO Reference Manual and CanoDraw for Windows User's Guide: Software for Canonical Community Ordination5th ednvol. 4Microcomputer Power: Ithaca, NY, USA [Google Scholar]
- van Aarle IM, Olsson PA. Fungal lipid accumulation and development of mycelial structures by two arbuscular mycorrhizal fungi. Appl Environ Microbiol. 2003;69:6762–6767. doi: 10.1128/AEM.69.11.6762-6767.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Heijden MGA, Klironomos JN, Ursik M, Moutoglis P, Streitwolf-Engel R, Boller T, et al. Mycorrhizal fungal diversity determines plant biodiversity, ecosystem variability and productivity. Nature. 1998;396:69–72. [Google Scholar]
- Vandenkoornhuyse P, Ridgway KP, Watson IJ, Fitter AH, Young JPW. Co-existing grass species have distinctive arbuscular mycorrhizal communities. Mol Ecol. 2003;12:3085–3095. doi: 10.1046/j.1365-294x.2003.01967.x. [DOI] [PubMed] [Google Scholar]
- van der Putten WH, Klironomos JN, Wardle DA. Microbial ecology of biological invasions. ISME J. 2007;1:28–37. doi: 10.1038/ismej.2007.9. [DOI] [PubMed] [Google Scholar]
- van Kleunen M, Weber E, Fischer M. A meta-analysis of trait differences between invasive and non-invasive plant species. Ecol Lett. 2010;13:235–245. doi: 10.1111/j.1461-0248.2009.01418.x. [DOI] [PubMed] [Google Scholar]
- Vogelsang KM, Bever JD. Mycorrhizal densities decline in association with nonnative plants and contribute to plant invasion. Ecology. 2009;90:399–407. doi: 10.1890/07-2144.1. [DOI] [PubMed] [Google Scholar]
- Waldrop MP, Zak DR, Blackwood CB, Curtis CD, Tillman D. Resource availability controls fungal diversity across a plant diversity gradient. Ecol Lett. 2006;9:1127–1135. doi: 10.1111/j.1461-0248.2006.00965.x. [DOI] [PubMed] [Google Scholar]
- Wilson GW, Hartnett DC. Interspecific variation in plant responses to mycorrhizal colonization in tallgrass prairie. Am J Bot. 1998;85:1732–1738. [PubMed] [Google Scholar]
- Zhang Q, Yang R, Tang J, Yang H, Hu S, Chen X. Positive feedback between mycorrhizal fungi and plants influences plant invasion success and resistance to invasion. PLoS ONE. 2010;5:e12380. doi: 10.1371/journal.pone.0012380. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.