Abstract
We surveyed the distribution and diversity of fungi associated with eight macroalgae from Antarctica and their capability to produce bioactive compounds. The collections yielded 148 fungal isolates, which were identified using molecular methods as belonging to 21 genera and 50 taxa. The most frequent taxa were Geomyces species (sp.), Penicillium sp. and Metschnikowia australis. Seven fungal isolates associated with the endemic Antarctic macroalgae Monostroma hariotii (Chlorophyte) displayed high internal transcribed spacer sequences similarities with the psychrophilic pathogenic fungus Geomyces destructans. Thirty-three fungal singletons (66%) were identified, representing rare components of the fungal communities. The fungal communities displayed high diversity, richness and dominance indices; however, rarefaction curves indicated that not all of the fungal diversity present was recovered. Penicillium sp. UFMGCB 6034 and Penicillium sp. UFMGCB 6120, recovered from the endemic species Palmaria decipiens (Rhodophyte) and M. hariotii, respectively, yielded extracts with high and selective antifungal and/or trypanocidal activities, in which a preliminary spectral analysis using proton nuclear magnetic resonance spectroscopy indicated the presence of highly functionalised aromatic compounds. These results suggest that the endemic and cold-adapted macroalgae of Antarctica shelter a rich, diversity and complex fungal communities consisting of a few dominant indigenous or mesophilic cold-adapted species, and a large number of rare and/or endemic taxa, which may provide an interesting model of algal–fungal interactions under extreme conditions as well as a potential source of bioactive compounds.
Keywords: Antarctica, marine fungi, seaweeds, diversity, extremophiles
Introduction
Antarctica represents one of the most pristine ecosystems in the world, characterised by short food chains dominated by microorganisms. The fungal mats in the Antarctic exhibit complex communities that are able to survive under extreme environmental conditions, including dehydration, freeze-thaw cycles, low nutrient concentrations, low temperatures, osmotic stress and ultraviolet radiation irradiation (Fell et al., 2006; Gonçalves et al., 2012). The Antarctic fungal communities include representatives of genera and species from the major fungal phyla Ascomycota, Basidiomycota, Zygomycota, Chytridiomycota and Glomeromycota, as well as Oomycetes (Heterokontophyta, Oomycota) and slime moulds (Mycetozoa) traditionally studied by mycologists (Bridge and Spooner, 2012). In recent decades, mycological studies in Antarctica have mainly focused on the fungi present in soil, ice and lakes, and their associations with plants. However, limited attention has thus far been devoted to the fungal diversity of the Antarctic marine environment. Few existing studies address marine Antarctic fungi, which have been detected in water (Fell and Statzel, 1971; Fell et al., 1973), wood baits (Grasso et al., 1997) and water and marine sediments (Vaz et al., 2011). According to Bridge and Spooner (2012), only 42 fungal species have been described from the Antarctic marine ecosystem.
The known marine macroalgal communities from Antarctica are not very diverse in comparison with that of warmer regions. However, this flora is characterised by a high degree of endemism and the presence of cold-adapted species (Wiencke and Clayton, 2002; Oliveira et al., 2009). In addition, macroalgae are important primary producers, producing ∼74 000 tons of wet biomass and having a key role in organic carbon fluxes in Antarctica (Nedzarek and Rakusa-Suszczewski, 2004). Also, macroalgae may shelter large numbers of associated organisms, including microbial mats surviving under extreme conditions (Loque et al., 2010).
According to Bugni and Ireland (2004), fungi recovered from macroalgae represent the second largest source of marine fungi and include parasites, saprobes or mutualistic species. A number of macroalgal species have been studied in detail worldwide with respect to their associated fungal communities, which include the genera Ascophyllum, Ballia, Caulerpa, Ceramium, Ceratiodictyon, Cladophora, Chondrus, Dictyota, Dilsea, Egregia, Enteromorpha, Fucus, Gelidiella, Gracilaria, Grateloupia, Halimeda, Halymenia, Hypnea, Laminaria, Lobophora, Padina, Porphyra, Portieria, Saccorhiza, Sargassum, Stoechospermum, Turbinaria and Ulva (Kohlmeyer and Volkmann-Kohlmeyer, 1991, Stanley, 1992; Zuccaro and Mitchell, 2005; Zuccaro et al., 2008; Suryanarayanan et al., 2010). To the best of our knowledge, except for an initial contribution by Loque et al. (2010), no other data are available regarding the species composition of fungal communities associated with Antarctic macroalgae. In this study, we acquire to present information on the diversity and distribution of fungal communities associated with endemic and cold-adapted macroalgae across latitudinal gradients along the Antarctic Peninsula and their capability to produce bioactive compounds.
Materials and methods
Macroalagae collection
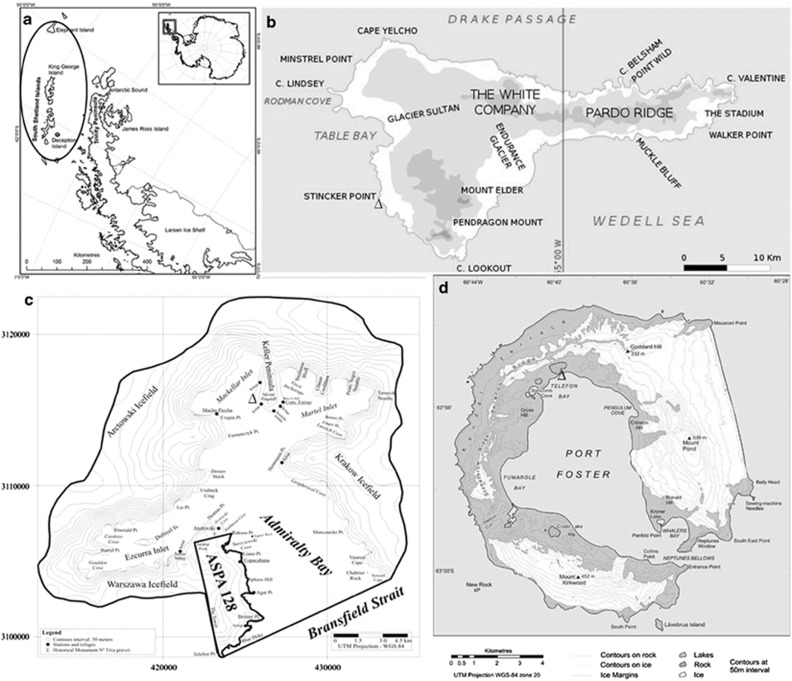
Sixty fresh thalli from each selected macroalgal species (four Phaeophyceae, three Chlorophyta and one Rhodophyta) were collected during December 2010 and January 2011 in intertidal transects along a rocky coastline that becomes ice free during the Antarctic summer. Samples of Adenocystis utricularis (Bory de Saint-Vicent) Skottsberg, Adenocystis species (sp.), Desmarestia menziesii J Agardh, Phaeurus antarcticus Skottsberg, Acrosiphonia arcta (Dillwyn) Gain, Monostroma hariotii Gain, Ulva intestinalis Linnaeus and Palmaria decipiens RW Ricker (Figure 1) were collected onboard the Brazilian Navy Polar Ship Almirante Maximiano (H41) along a 350-km transect through Elephant, King George and Deception Islands, in the Antarctic Peninsula (Figure 2). Physical and chemical water parameters (temperature, salinity, conductivity, dissolved oxygen and pH) were also recorded at each site using a multiparameter probe Hexis TCS (Yellow Springs, OH, USA).
Figure 1.
Macroalgae collected from the Antarctic Peninsula, with information on their distributions given in parenthesis. (a) Acrosiphonia arcta (Dillwyn) Gain (cosmopolitan); (b) Adenocystis sp. (most likely endemic); (c) Adenocystis utricularis (Bory de Saint-Vicent) Skottsberg (Australia, New Zealand and Antarctica); (d) Monostroma hariotii Gain (Antarctic and Subantartic islands); (e) Palmaria decipiens RW Ricker (Antarctic and Subantarctic islands); (f) Ulva intestinalis Linnaeus (cosmopolitan); (g) Phaeurus antarcticus Skottsberg (Antarctic and the Subantarctic islands); (h) Desmarestia menziesii J Agardh (Antarctic and the Subantarctic islands). Bars represent 1 cm.
Figure 2.
Maps showing the positions of the islands sampled on the (a) Antarctic Peninsula across a transect of 350 km. Sampling sites: Δ, Ulva intestinalis, Palmaria decipiens and Phaeurus antarcticus in (b)=Stinker Point (61°07.935'S; 055°25.997'W) at Elephant island; Δ, Acrosiphonia arcta and D. menziesii in (c), Keller Peninsula (62°05.163'S; 058°24.784'W) at Admiralty Bay at King George Island; Δ, Adenocystis utricularis, Monostroma hariotii, Adenocystis sp. in (d), Telefon Bay (62°55.192'S; 060°39.797'W) at Deception Island.
Macroalgae identification
Complete and fertile samples were sorted, washed and preserved in seawater formalin (4%) in the ship's laboratory, with the aim of performing macro- and micromorphological analyses. The identification of the macroalgal specimens was based on the publications of Papenfuss (1964), Ricker (1987), Wiencke and Clayton (2002), Quartino et al. (2005) and Amsler et al. (2009). Nomenclatural updates followed Guiry and Guiry (2012). Exsiccatae vouchers were manufactured for deposition in the SP Herbarium of the Jardim Botânico of São Paulo, Brazil.
Fungal isolation
Five discs 8 mm in diameter were cut from each macroalgal specimen and washed twice using sterile local seawater for 2 min. The discs were inoculated in Petri dishes containing marine agar (Difco, Franklin Lakes, NJ, USA) supplemented with 2% glucose and chloramphenicol (Sigma, St Louis, MO, USA) at a concentration of 200 μg ml−1 for selective isolation of marine fungi. All of the inoculated Petri dishes were incubated for up to 60 days at 10 °C, and individual colonies of fungi were purified on marine agar. Long-term preservation of fungi was carried out at −80 °C using cryotubes with sterile 15% glycerol. All of the fungal isolates examined in this work were deposited in the Culture Collection of Microorganisms and Cells of the Universidade Federal of Minas Gerais, Brazil, under codes UFMGCB.
Fungal identification
The protocol for DNA extraction from filamentous fungi followed Rosa et al. (2009). The internal transcribed spacer (ITS) region was amplified with the universal primers ITS1 and ITS4 (White et al., 1990). Amplification of the ITS region was performed as described by Rosa et al. (2009). Yeasts were characterised via standard methods (Yarrow, 1998), and their identification was carried out using the taxonomic keys of Kurtzman et al. (2011). Yeast identities were confirmed by sequencing the D1-D2 variable domains of the large subunit ribosomal RNA gene using the primers NL1 and NL4, as described by Lachance et al. (1999).
Amplification of the β-tubulin gene was performed with the Bt2a and Bt2b primers (Glass and Donaldson, 1995). PCR assays were conducted in 50 μl reaction mixtures containing 1 μl of genomic DNA (10 ng μl−1), 5 μl of PCR buffer (100 mℳ Tris-HCl, 500 mℳ KCl, pH 8.8), 2 μl of dNTPs (10 mℳ) plus 3 μl of MgCl2 (25 mℳ), 1 μl of each primer (50 pmol μl−1), 1 μl of dimethyl sulfoxide (DMSO; Merck, Billerica, MA, USA), 2 μl betaine (5 ℳ), 0.2 μl of Taq polymerase (5 U μl−1 DNA) and 33.8 μl of ultrapure sterile water. PCR amplifications were performed with the Mastercycler pro (Eppendorf, Hamburg, Germany), programmed for initial denaturation at 94 °C for 5 min, followed by 35 cycles of 1 min of denaturation at 94 °C, primer annealing for 1 min at 59 °C and extension for 1.30 min at 72 °C, with a final 7-min elongation step at 72 °C. After amplification of the β-tubulin template, excess primers and dNTPs were removed from the reaction mixture using a commercial GFX column with the PCR DNA Purification kit (Amersham Bioscience, Roosendaal, Netherlands). Purified PCR fragments were resuspended in 50 μl of Tris-EDTA buffer.
The amplified DNA was concentrated and purified using the Wizard Plus SV Miniprep DNA Purification System (Promega, Madison, WI, USA) and sequenced using an ET Dynamic Terminator Kit in a MegaBACE 1000/Automated 96 Capillary DNA sequencer (GE Healthcare, Piscataway, NJ, USA). The obtained sequences were analysed with SeqMan Π with Lasergene software (DNASTAR Inc., Madison, WI, USA), and a consensus sequence was obtained using Bioedit v. 7.0.5.3 software (Carlsbad, ON, Canada). To achieve species-rank identification based on ITS and β-tubulin data, the consensus sequence was aligned with all sequences from related species retrieved from the NCBI GenBank database using BLAST (Altschul et al., 1997). The consensus sequences of the algicolous fungi were deposited into GenBank (see the accession numbers in the Table 1). However, according to Gazis et al. (2011), sequencing of the ITS region may fail to recognise some fungal genera. For this reason, the β-tubulin sequences, which are considered promising for a one-gene phylogeny (Frisvad and Samson, 2004), were used to elucidate the taxonomic positions of the inconclusive taxa identified using ITS sequences. In addition, the followed criteria were used to interpret the sequences from the GenBank database: for query coverage and sequence identities ⩾99%, the genus and species were accepted; for query coverage and sequence identities showing 98%, the genus and species were accepted, but term ‘cf.' (Latin for confer=compares with), used to indicate that the specimen resembles, but has certain minor features not found in the reference species, was included; for query coverage and sequence identities between 95% and 97%, only the genus was accepted; and for query coverage and sequence identities ⩽95%, the isolates were labelled with the order or family name or as ‘unknown' fungi. However, taxa that displayed query coverage and identities⩽97% or an inconclusive taxonomic position were subjected to phylogenetic ITS and β-tubulin analysis, with estimations conducted using MEGA Version 5.0 (Tamura et al., 2011). The maximum composite likelihood method was employed to estimate evolutionary distances with bootstrap values calculated from 1000 replicate runs. To complete the molecular identification, the sequences of known-type fungal strains or reference sequences obtained from fungal species deposited in international culture collections found in GenBank were added to improve the accuracy of the phylogenetic analysis. The information about fungal taxonomic hierarchical follows the rules established by Kirk et al. (2011) and the MycoBank (http://www.mycobank.org), and Index Fungorum (http://www.indexfungorum.org) databases.
Table 1. Molecular identification of fungi associated with Antarctic macroalgae: identification conducted using BLASTn searches of the of the internal transcribed spacer (ITS) region and β-tubulin gene.
| Macroalgae species | UFMGCBa code | No. of isolates | Top BLAST search results (GenBank accession number) | Query coverage (%) | Identity (%) | No. of bp sequenced and analysed | Proposed species or taxonomic group (GenBank accession number) |
|---|---|---|---|---|---|---|---|
| Adenocystis utricularis | D1N10.34.1 | 1 | Debaryomyces hansenii (HE681104) | 100 | 99 | 524 | Debaryomyces hansenii (KC485456b) |
| D1N10.13.1 | 1 | Meyerozyma caribbica (JQ686909) | 100 | 99 | 547 | Meyerozyma caribbica (KC485457b) | |
| Acrosiphonia arcta | 6063c | 19 | Geomyces pannorum (DQ189229) | 98 | 99 | 729 | Geomyces sp. (KC333881d) |
| K2.13.1 | 6 | Metschnikowia australis (U76526) | 99 | 99 | 484 | Metschnikowia australis (KC485458b) | |
| 6074c,e | 2 | Penicillium chrysogenum (HQ652873) | 99 | 99 | 662 | fPenicillium sp. (KC333882d, KC823167g) | |
| K2.6.1 | 1 | Candida sake (EU326138) | 100 | 99 | 588 | Candida sake (KC485459b) | |
| 6361c,e | 1 | Cladosporium perangustum (JF499836) | 100 | 97 | 486 | fCladosporium sp. (KC341715d, KC823166g) | |
| 6079 | 1 | Cladosporium tenuissimum (JQ246357) | 100 | 99 | 476 | Cladosporium tenuissimum (KC341716d) | |
| K2.9.1 | 1 | Debaryomyces hansenii (HQ860269) | 100 | 99 | 546 | Debaryomyces hansenii (KC485460b) | |
| 6077c | 1 | Mortierella sp. (JX270406) | 90 | 97 | 562 | fMortierella sp. (KC341717d) | |
| 6083c | 1 | Phoma sp. (JQ388278) | 99 | 99 | 468 | fPhoma sp. (KC341718d) | |
| 6318 | 1 | Thelebolus microsporus (GU004196) | 99 | 99 | 495 | Thelebolus globosus (KC341719d) | |
| Desmarestia menziesii | K1.17.1 | 1 | Metschnikowia australis (FJ911872) | 99 | 99 | 446 | Metschnikowia australis (KC485461b) |
| 6084c,e | 1 | Penicillium chrysogenum (JN021549) | 100 | 98 | 486 | fPenicillium sp. (KC341720d, KC823168g) | |
| Ulva intestinalis | 6101c,e | 6 | Penicillium commune (JN676122) | 100 | 100 | 496 | fPenicillium sp. (KC485461d, KC823169g) |
| 6092c,e | 3 | Penicillium solitum (JN642222) | 98 | 99 | 534 | fPenicillium discolor (KC485423d, KC823170g) | |
| 6090 | 3 | Antarctomyces psychrotrophicus (JN104511) | 100 | 99 | 482 | Antarctomyces psychrotrophicus (KC485424d) | |
| CM.01.2 | 1 | Cryptococcus victoriae [AY040653] | 98 | 99 | 605 | Cryptococcus victoriae [KC485462b] | |
| 6096 | 1 | Engyodontium album (HM214540) | 90 | 99 | 566 | Engyodontium sp. (KC485425d) | |
| 6097c | 1 | Geomyces luteus (AJ938164) | 100 | 98 | 451 | Geomyces luteus (KC485426d) | |
| 6328 | 1 | Helotiales sp. (HQ533820) | 90 | 99 | 477 | Helotiales sp. (KC485427d) | |
| 6326 | 1 | Mycoarthris corallinus (AF128440) | 97 | 98 | 458 | Mycoarthris cf. corallines (KC485428d) | |
| 6325c,e | 1 | Penicillium chrysogenum (KC009835) | 98 | 98 | 507 | fPenicillium sp. (KC485429d, KC823171g) | |
| 6095 | 1 | Thelebolus microsporus (GU004196) | 100 | 100 | 485 | Thelebolus globosus (KC485430d) | |
| Monostroma hariotii | 6120c,e | 12 | Penicillium chrysogenum (KC009826) | 100 | 99 | 512 | fPenicillium sp. (KC485431d, KC823175g) |
| 6112c | 7 | Geomyces pannorum (JF311913) | 100 | 99 | 439 | Geomyces cf. destructans (KC485432d) | |
| D1N18.31.1 | 4 | Meyerozyma guilliermondii (JF766631) | 100 | 99 | 537 | Meyerozyma guilliermondii (KC485463b) | |
| 6330 | 1 | Cryptococcus laurentii (FJ743631) | 100 | 98 | 570 | Cryptococcus cf. laurentii (KC485433d) | |
| 6130c | 1 | Lecanicillium araneicola (AB378506) | 99 | 92 | 573 | fCordycipitaceae sp. (KC485434d) | |
| 6138c | 1 | Paecilomyces anatarcticus (AJ879113) | 100 | 96 | 475 | fHelotiales sp. (KC485435d) | |
| 6316c | 1 | Pezizella discreta (JF908571) | 99 | 97 | 487 | fHyaloscyphaceae sp. (KC485436d) | |
| D1N18.42.1 | 1 | Rhodotorula mucilaginosa (JQ277252) | 100 | 99 | 537 | Rhodotorula mucilaginosa (KC485464b) | |
| Palmaria decipiens | 6034c,e | 11 | Penicillium chrysogenum (JN032681) | 100 | 99 | 538 | fPenicillium sp. (JX976546d, KC823172g) |
| 6049c | 8 | Geomyces pannorum (DQ189229) | 91 | 99 | 554 | Geomyces sp. (KC485437d) | |
| 6056 | 2 | Acremonium strictum (AY138844) | 92 | 99 | 565 | Acremonium sp. (KC485438d) | |
| 6050 | 2 | Fusarium oxysporum (HQ248198) | 91 | 99 | 524 | Fusarium sp. (KC485439d) | |
| E2N5.42.1 | 2 | Yamadazyma mexicana (FJ455104) | 100 | 99 | 500 | Yamadazyma mexicana (KC485465b) | |
| 6047 | 1 | Aspergillus candidus (FJ441637) | 93 | 99 | 548 | Aspergillus sp. (KC485440d) | |
| 6053 | 1 | Chaetomium globosum (JN689341) | 66 | 99 | 751 | Chaetomium sp. (KC485441d) | |
| 6140c,e | 1 | Penicillium raistrickii (FR670335) | 78 | 97 | 665 | fPenicillium sp.(KC485442d, KC823173g) | |
| 6048c,e | 1 | Penicillium spinulosum (DQ132828) | 74 | 99 | 697 | fPenicillium spinulosum (KC485443d, KC823174g) | |
| Phaeurus antarcticus | 6141c,e | 13 | Penicillium chrysogenum (JQ665262) | 100 | 100 | 482 | fPenicillium sp. (KC485444d, KC823176g) |
| 6148c | 8 | Geomyces pannorum (DQ189229) | 100 | 99 | 495 | Geomyces sp. (KC485445d) | |
| 6153c,e | 2 | Penicillium raistrickii (FR670335) | 81 | 98 | 631 | fPenicillium sp. (KC485446d, KC823178g) | |
| 6164c,e | 1 | Aspergillus terreus (JQ070071) | 100 | 100 | 536 | fAspergillus terreus (KC485447d, KC823163g) | |
| 6167 | 1 | Eurotium herbariorum (JN942870) | 100 | 99 | 429 | Eurotium herbariorum (KC485448d) | |
| 6327 | 1 | Eurotium repens (AY373890) | 100 | 99 | 482 | Eurotium repens (KC485449d) | |
| 6320c,e | 1 | Penicillium steckii (HM469415) | 100 | 99 | 467 | fPenicillium steckii (KC485450d, KC846137g) | |
| Adenocystis sp. | 6323c,e | 3 | Penicillium chrysogenum (JQ665262) | 95 | 99 | 504 | fPenicillium sp. (KC485451d, KC823164g) |
| 6106 | 1 | Aspergillus conicus (HE578068) | 99 | 99 | 426 | Aspergillus conicus (KC485452d) | |
| 6110c | 1 | Geomyces pannorum (JF311913) | 100 | 99 | 451 | Geomyces sp. (KC485453d) | |
| 6109c,e | 1 | Penicillium citrinum (FJ765031) | 93 | 100 | 507 | fPenicillium citrinum (KC485454d, KC823177g) | |
| 6322c,e | 1 | Penicillium commune (FJ499454) | 100 | 98 | 542 | fPenicillium sp. (KC485455d, KC823165g) |
When the number of isolates was >1, 50% of the isolates were sequenced and the best sequence was deposited in the GenBank database.
UFMGCB=Culture of Microorganisms and Cells from the Federal Universidade of Minas Gerais.
D1/D2 sequences deposited.
Taxa subjected to phylogenetic analysis based on the ITS region for elucidation of taxonomic positions.
ITS sequences deposited.
Taxa subjected to phylogenetic analysis based on the β-tubulin regions for elucidation of taxonomic positions.
Taxonomic position suggested by the phylogenetic analyses.
β-tubulin sequences deposited.
Diversity, richness, dominance and distribution
To quantify species diversity, richness and evenness, we used the following indices: (i) Fisher's α, (ii) Margalef's and (iii) Simpson's, respectively. A rarefaction curve was calculated using the Mao Tao index (Colwell et al., 2004). The similarities among fungal taxa from different areas were estimated using the Sorensen coefficient and Bray-Curtis measures. All of the results were obtained with 95% confidence, and bootstrap values were calculated from 1000 iterations. All diversity and similarity indices, rarefaction curves and the principal components analysis calculations were performed using the computer Program PAST, version 1.90 (Hammer et al., 2001). Further information about these indices can be found in Magurran (2011).
Fungal cultivation and preparation of extracts for biological assays
All fungal isolates were cultivated using solid state fermentation. Five-millimetre-diameter plugs from each filamentous fungus were inoculated into the centres of Petri dishes (60 mm diameter, with 20 ml of marine agar). The plates were incubated at 10±2 °C for 15 days, and the cultured materials from each Petri dish were then cut and transferred to 50-ml vials containing 35 ml of ethanol. After 48 h at room temperature, the organic phase was decanted, and the solvent was removed under a vacuum centrifuge at 35 °C (Santiago et al., 2012). An aliquot of each dried extract was dissolved in DMSO (Merck) to prepare a 100-mg ml−1 stock solution, which was stored at −20 °C.
Assay for antimicrobial activity
Susceptibility testing against Escherichia coli ATCC 11775, Staphylococcus aureus ATCC 12600, Pseudomonas aeruginosa ATCC 10145, Candida albicans ATCC 18804, Candida krusei ATCC 6258 and Cladosporium sphaerospermum CCT 1740 was performed using a protocol established by Carvalho et al. (2012). All extracts (dissolved in DMSO) were diluted to a final concentration of 250 μg ml−1 for use in the antimicrobial assays. The results are expressed as the percent inhibition in relation to controls without drugs. All antimicrobial assays were performed in duplicate.
In vitro assays with intracellular amastigote forms of Trypanosoma cruzi
In vitro assays with amastigote forms of T. cruzi were performed according to protocols established by Buckner et al. (1996) with some modifications. Trypanosoma cruzi (Tulahuen strain) expressing the E. coli β-galactosidase gene were grown on a monolayer of mouse L929 fibroblasts. Cultures to be assayed for β-galactosidase activity were grown in Roswell Park Memorial Institute 1640 medium (pH 7.2-7.4) without phenol red (Gibco BRL, Cergy-Pontoise, France), plus 10% foetal bovine serum and glutamine. L929 fibroblasts were seeded into 96-well tissue culture microplates at a concentration of 4.0 × 103 per well in a volume of 80 μl and incubated overnight. β-galactosidase-expressing trypomastigotes were then added at a concentration of 4.0 × 104 per well in a volume of 20 μl. After 2 h, the medium containing trypomastigotes that did not penetrate into the cells was discarded and replaced with 200 μl of fresh medium. After 48 h, the medium was discarded again and replaced with 180 μl of fresh medium and 20 μl of the test extracts. Each extract was tested in triplicate. After 7 days of incubation, chlorophenol red β-𝒟-galactopyranoside (100 μℳ final concentration) and Nonidet P-40 (Sigma-Aldrich, St. Louis, MO, USA) at final concentration of 0.1% were added to the plates, followed by incubation overnight at 37 °C, and the absorbance was measured at 570 nm in an automated microplate reader. Benznidazole at its 50% inhibitory concentration (IC50; 1 μl ml−1) was used as a positive control. The results were expressed as the percentage of growth inhibition. All assays were performed in triplicate.
Minimal inhibitory concentration and IC50
The crude antimicrobial extracts were subjected to determination of a minimal inhibitory concentration (MIC) and the trypanocidal extracts were subjected to determination of a IC50. MIC and IC50 represent the lowest concentrations of a crude extract (or compound) that inhibits the functions of a target (in this study, microorganisms and T. cruzi) by 100% and 50%, respectively. This quantitative measure indicates how much of a particular crude extract is needed to inhibit a given biological process by 50%. SoftMax Pro 5.3 (Sunnyvale, CA, USA) was used to calculate MIC values via nonlinear curve fitting of two or more independent experimental data sets to a four-parameter logistic dose–response model. No constraints were applied to the curve-fitting calculations. All assays were performed in duplicate.
Nuclear magnetic resonance spectroscopy
Bioactive extracts were analysed via nuclear magnetic resonance (NMR) spectroscopy in a Varian INOVA 600 MHz spectrometer (Varian Inc., Palo Alto, CA, USA). 1H-NMR spectra were recorded in DMSO-d6. The samples were prepared at a concentration of ∼10 mg ml−1, and the results were recorded in 3 mm NMR tubes using a standard 1H pulse programme.
Results
Macroalgae collection and fungal identification
A total of 148 fungal isolates were recovered from 391 tissue fragments from the eight macroalgal species, which were identified by DNA sequences of the ITS region and β-tubulin gene in 21 different genera within of the phyla Ascomycota, Basidiomycota and subphylum Mortierellomycotina (Zygomycota; Table 1). All of the screened macroalgae harboured associated fungi, and 50 taxa were identified. However, the number of fungal taxa and diversity indices differed among the macroalgae. Among the phyla characterised, the most represented orders were Eurotiales (43.24%), Helotiales (32.43%), Saccharomycetales (11.48%), Hypocreales (6%) and Thelebolales (4.05%). In contrast, taxa of the orders Tremellales, Capnodiales, Mortierellales, Pleosporales, Sordariales and Sporidiobolales were found as minority components of the fungal communities with an abundance of ⩽1.35%.
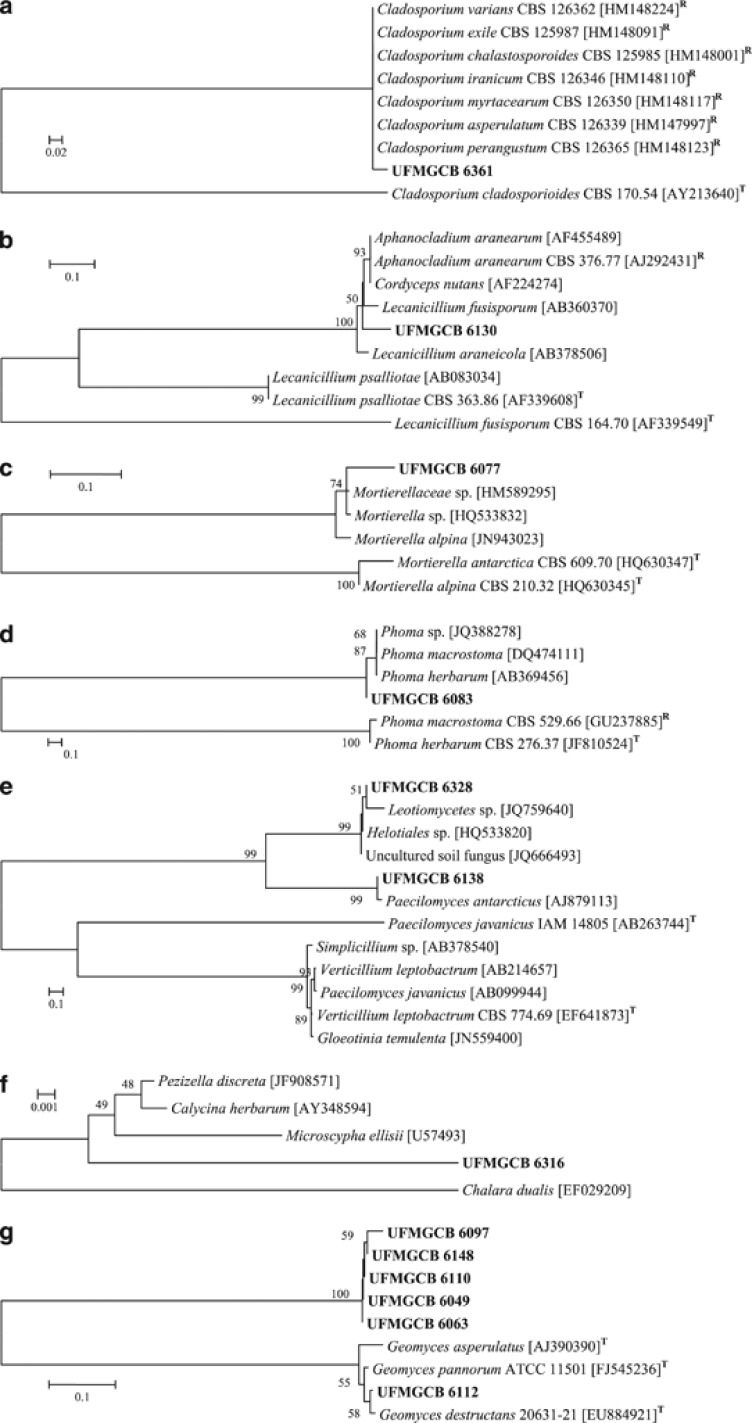
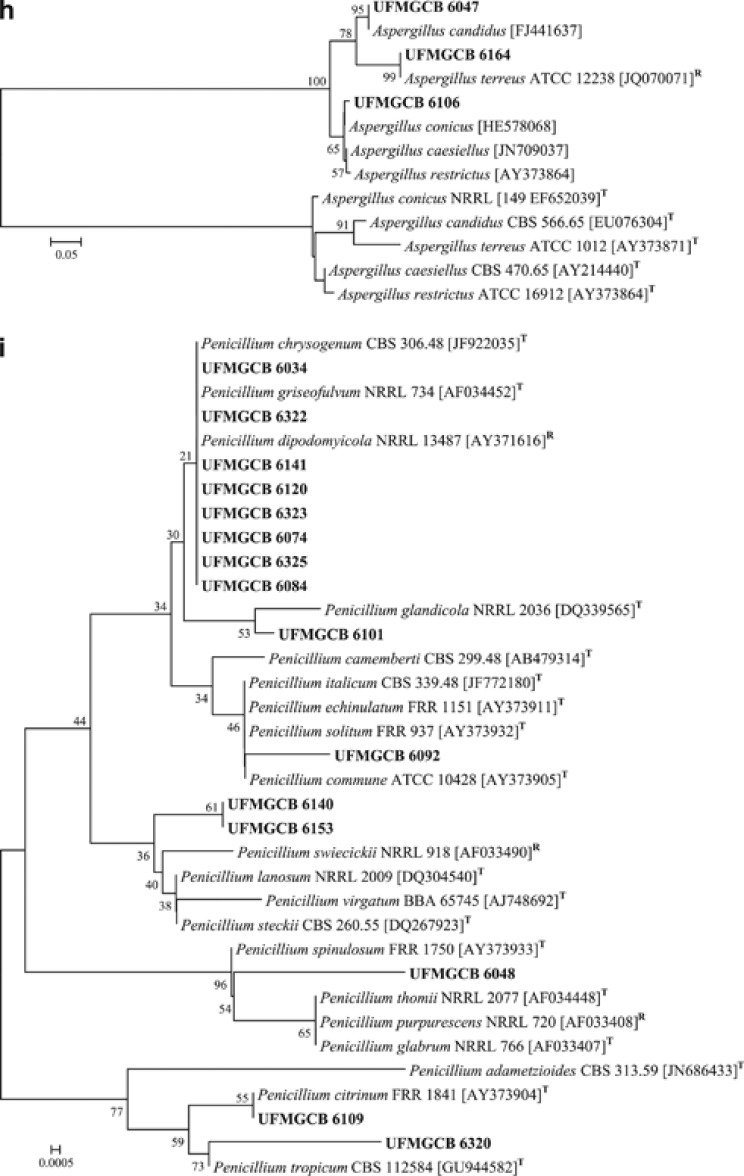
Twenty-eight fungal taxa presented low molecular similarities or inconclusive information in comparison with known fungal ITS sequences deposited in the GenBank database. To identify these fungi, phylogenetic trees of the ITS (Figure 3) and/or β-tubulin (Figure 4) genes were constructed to illustrate their relationship with GenBank sequences. All ITS (Figure 3i) and β-tubulin (Figure 4c) sequences of the Penicillium isolates were analysed and the taxa P. spinulosum, P. steckii, P. citrinum and P. chrysogenum were identified. The other Penicillium taxa displayed inconclusive sequence analyses when compared with the sequences of the type species, and were identified as Penicillium sp.
Figure 3.
Phylogenetic analysis (a–i) of the sequences of fungi (in bold) associated with Antarctic macroalgae in comparison with type (T) or reference (R) sequences of the closest species, following BLAST analysis, deposited in the GenBank database. The trees were constructed based on the ITS region sequences using the maximum composite likelihood method.
Figure 4.
Phylogenetic analysis (a–c) of the β-tubulin sequences of the UFMGCB fungi (in bold) in comparison with type (T) or reference (R) sequences of the closest species, following BLAST analysis, deposited in the GenBank database. The trees were constructed based on the β-tubulin gene sequences using the maximum composite likelihood method.
Thirty-six fungal isolates displayed ITS sequence similarities with Geomyces sp. Among them, the seven isolates were identified as G. destructans, which showed 100% of query coverage and 98% identity; also these fungal isolates presented only 1.1% of sequence difference in comparison with the type species G. destructans (GenBank Access number EU884921; Table 1; Figure 3g). All other Geomyces isolates formed a separated cluster when compared with sequences of the type species of G. pannorum (GenBank Access number FJ545236) and G. asperulatus (GenBank Access number AJ390390; Figure 3g); these taxa were identified as Geomyces sp.
In addition, after ITS (Figure 3) and β-tubulin (Figure 4) sequence analysis, 27 fungal taxa were identified as belonging to the genera Acremonium, Aspergillus, Chaetomium, Cladosporium, Engyodontium, Fusarium, Geomyces, Mortierella, Penicillium, Phoma as well as the families Cordycipitaceae, Hyaloscyphaceae and order Helotiales, which displayed low sequence similarities when compared with fungal sequences deposited in the GenBank.
Diversity, richness, dominance and distribution
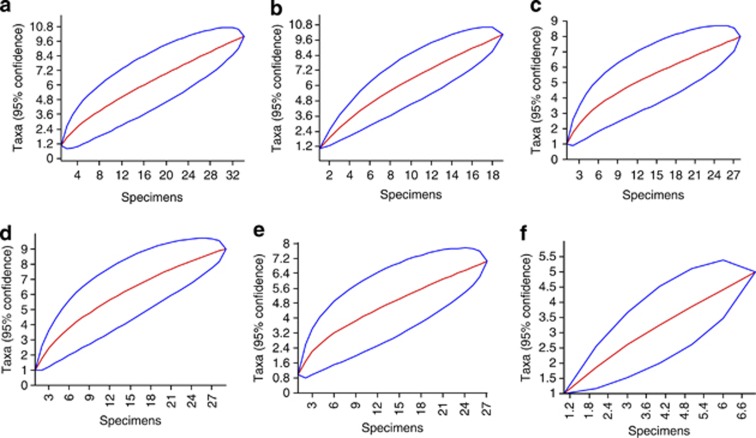
The most frequent fungal taxa identified were Penicillium sp. (35.8%), Geomyces sp. (24.32%) and M. australis (4.72%). However, in general, the fungal communities associated with the Antarctic macroalgae presented high values of the diversity (Fisher's α=22.0), richness (Margalef's=10.41) and dominance (Simpson's=0.94) indices. The phylum Chlorophyta displayed a Fisher's α=5.67 and 9.33 fungal taxa per macroalgal species, which was followed by Rhodophyta (Fisher's α=4.47 and 9 fungal taxa/macroalgal species) and Phaeophyta (Fisher's α=2.7 and 4 fungal taxa/macroalgal species). However, the values of the indices differed among the macroalgae (Table 2). The fungal communities associated with U. intestinalis and Adenocystis sp. showed high diversity and dominance values, whereas the communities corresponding to A. utricularis and D. menziesii displayed the lowest diversity, richness and dominance values. In addition, the A. arcta fungal community showed the highest richness, but moderate diversity and dominance. Only two fungal species were isolated from the macroalgae A. utricularis and D. menziesii, which exhibited the lowest Fisher's α, Margalef's and Simpson's indices. In addition to the high fungal diversity (Fisher's α values) found in the sampled macroalgae, the Mao Tao rarefaction curves continued to rise and did not reach an asymptote (Figure 5), indicating that not all fungal diversity had been recovered. Thirty-three fungal taxa (66%) occurred as singletons corresponding to a single macroalgal species, representing rare components of the fungal communities.
Table 2. Physico-chemical parameters of water and diversity indices of fungal communities associated with the Antarctic macroalgae.
| Parameters/diversity indices |
Phyla of macroalgae host species |
|||||||
|---|---|---|---|---|---|---|---|---|
|
Phaeophyta |
Chlorophyta |
Rhodophyta |
||||||
| Adenocystis utricularis | Adenocystis sp. | Desmarestia menziesii | Phaeurus antarcticus | Acrosiphonia arcta | Monostroma hariotii | Ulva intestinalis | Palmaria decipiens | |
| Temperature (°C) | 3.7 | 3.7 | 0.5 | 2.1 | 0.5 | 3.7 | 2.1 | 2.1 |
| Conductivity (mS cm−1) | 49.89 | 49.89 | 27.23 | 50.6 | 27.23 | 49.89 | 50.6 | 50.6 |
| Salinity (p.p.t.) | 32.1 | 32.1 | 32.8 | 33.0 | 32.8 | 32.1 | 33.0 | 33.0 |
| pH | 7.49 | 7.49 | 7.74 | 7.74 | 7.74 | 7.49 | 7.74 | 7.74 |
| Optical Density (mg l−1) | 79.9 | 79.9 | 100 | 34.6 | 100 | 79.9 | 34.6 | 34.6 |
| Number of fungal taxa | 2 | 5 | 2 | 7 | 10 | 8 | 10 | 9 |
| Fisher's α | 0 (0—0.8)a | 7.8 (0.93—7.82) | 0 (0—0.8) | 3 (0.86—3.1) | 4.77 (1.61—4.0) | 3.7 (1.27—3.74) | 8.54 (2.21—8.54) | 4.47 (1.74—4.47) |
| Margalef's | 1.44 (0—1.44) | 2 (0.5—2.1) | 1.44 (0—1.44) | 1.82 (0.6—1.82) | 2.55 (1.13—2.26) | 2.1 (0.9—2.1) | 0.03 (1.35—3.05) | 2.3 (1.18—2.37) |
| Simpson's | 0.5 (0—0.5) | 0.73 (0.2—0.77) | 0.5 (0—0.5) | 0.66 (0.5—0.75) | 0.64 (0.46—0.75) | 0.72 (0.55—0.8) | 0.83 (0.65—0.86) | 0.7 (0.6—0.82) |
The numbers in parentheses represent the lower and upper diversity values, respectively, with 95% of confidence and bootstrap values calculated from 1000 iterations.
Figure 5.
Species accumulation curves for the fungal communities associated with Antarctic macroalgae. Acrosiphonia arcta (a), Ulva intestinalis (b), Monostroma hariotii (c), Palmaria decipiens (d), Phaeurus antarcticus (e) and Adenocystis sp. (f).
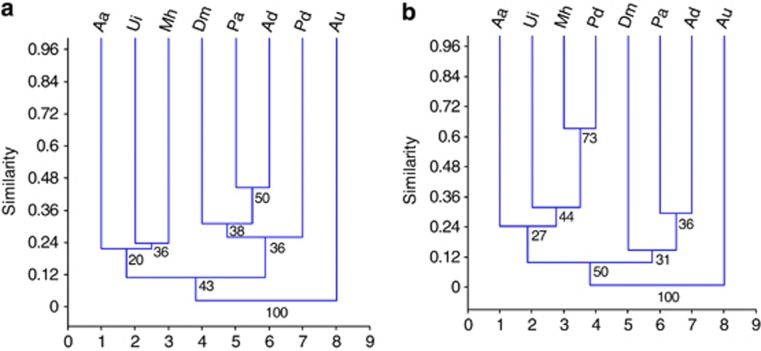
The fungal composition was variable among the macroalgal species, which was confirmed by the values of the Sorensen and Bray-Curtis similarity indices (Figure 6). The Sorensen index showed that the most similar fungal communities were found in P. antarcticus and Adenocystis sp., both of which are brown macroalgal species, but with different morphologies. However, the Bray-Curtis similarity between the fungal communities associated with the endemic macroalgae P. decipiens and M. hariotii indicated the highest similarity with the presence and abundance of the Geomyces and Penicillium taxa associated with both macroalgae. In addition, Penicillium taxa occurred in association with seven other macroalgal species, followed by Geomyces (six macroalgae).
Figure 6.
Dendrograms showing the Sorensen (a) and (b) Bray-Curtis similarity measures for the fungal communities associated with the Antarctic macroalgae. The results were obtained with 95% confidence and bootstrap values calculated from 1000 iterations. The sampled macroalgae were Aa, Acrosiphonia arcta; Ad, Adenocystis sp.; Au, Adenocystis utricularis; Dm, Desmarestia menziesii; Pa, Phaeurus antarcticus; Mh, Monostroma hariotii; Pd, Palmaria decipiens; and Ui, Ulva intestinalis.
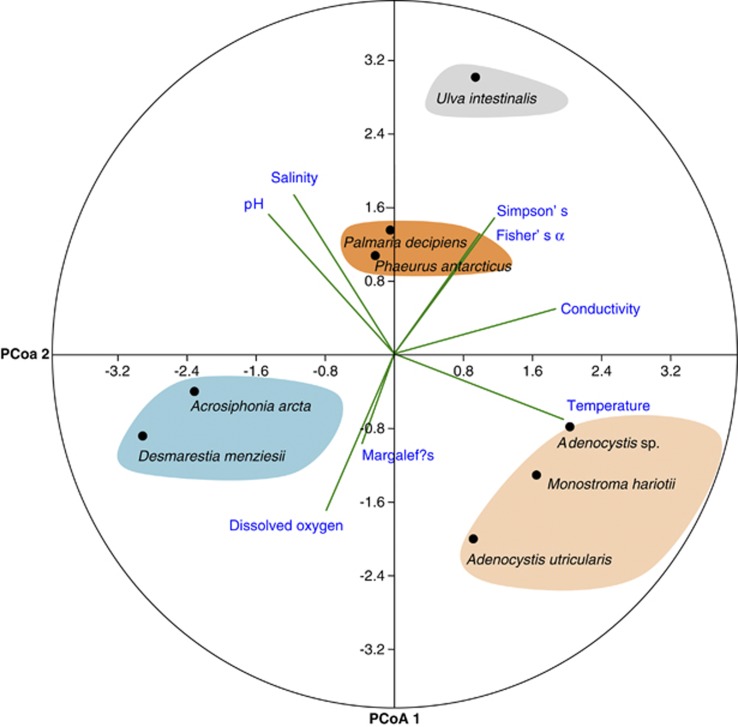
The principal components analysis of the physical and chemical parameters of the water where the macroalgae were sampled related to the fungal diversity indices (Figure 7) revealed that conductivity exhibited a positive correlation with the Fisher's α and Simpson's indices for the fungal assemblages associated with U. intestinalis; dissolved oxygen with Margalef's index for A. arcta and D. menziesii; temperature with Adenocystis sp., M. hariotii and A. utricularis; and salinity and pH with P. decipiens and P. antarcticus.
Figure 7.
Principal component analysis plot calculated among the physicochemical water parameters (temperature, salinity, pH, dissolved oxygen and conductivity) obtained where the macroalgae were collected and Fisher's β (diversity), Margalef's (richness) and Simpson's (dominance) indices of the fungal communities associated with Antarctic macroalgae.
Biological activities
All fungal isolates were grown using solid state fermentation techniques to obtain crude extracts, which were then screened against bacteria, fungi and parasite targets to detect bioactive compounds. The extracts of Penicillium sp. UFMGCB 6034 and Penicillium sp. UFMGCB 6120 recovered from the endemic macroalgae P. decipiens and M. hariotii, respectively, were able to produce bioactive extracts. Penicillium sp. UFMGCB 6120 displayed antifungal activity against the filamentous fungus Cladosporium sphaerospermum, producing 96% inhibition and an MIC value of 250 μg ml−1. In addition, Penicillium sp. UFMGCB 6034 and Penicillium sp. UFMGCB 6120 exhibited 100% inhibition of trypomastigote forms of T. cruzi, with IC50 values of 1.28 and 0.45 μg ml−1 being obtained, respectively, which were close to or lower than the IC50 (1 μg ml−1) of the control drug (Benznidazole) used in the experiments. In addition, these two trypanocidal extracts showed low toxicity against normal fibroblast cells.
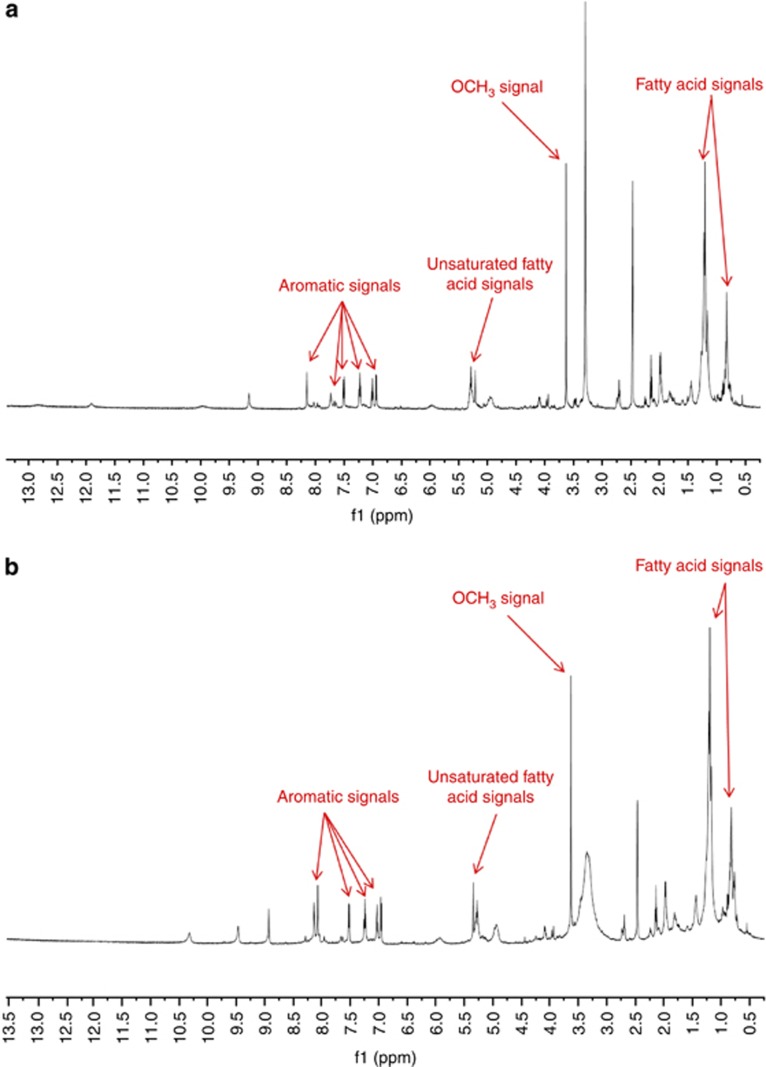
1H NMR (600 MHz, DMSO-d6) analyses of the ethanol extracts from the freeze-dried culture media of Penicillium sp. UFMGCB 6034 and Penicillium sp. UFMGCB 6120 were performed. Preliminary analysis indicated the presence of functional groups associated with aromatic protons, methoxy protons, unsaturated fatty-acid olefinic protons, and fatty-acid methylene and methyl protons, as indicated in Figure 8. Both Penicillium sp. UFMGCB 6034 and Penicillium sp. UFMGCB 6120 clearly contain the same major components, as demonstrated by the similar 1H NMR signals observed.
Figure 8.
1H-NMR spectrum (600 MHz, DMSO-d6) of bioactive ethanol extracts from the freeze-dried culture media of (a) Penicillium sp. UFMGCB 6034 and (b) Penicillium sp. UFMGCB 6120. Regions of interest are labelled above the corresponding signals.
Discussion
Macroalgae collection and fungal identification
Marine algae are known to shelter fungal species, but few taxa have been studied in detail. The initial work of Loque et al. (2010) and the present study, which addresses the fungal communities associated with eight macroalgae sampled along a latitudinal gradient over 350 km on the Antarctica Peninsula, represent the first systematic analyses of this kind. Similar to other mycological studies conducted in the Antarctic that have characterised fungi in soil (Fell et al., 2006), wood debris (Arenz et al., 2006), lakes (Gonçalves et al., 2012) and in associations with plants (Rosa et al., 2009), the fungal taxa associated with the Antarctic macroalgae examined in this study comprised a few dominant taxa. Our results agree with those of Suryanarayanan et al. (2010), who found few dominant fungal species associated with 25 macroalgae occurring along the coast of southern India. However, algicolous fungi remain relatively unexplored as a group. The presence of Geomyces sp., Penicillium sp. and M. australis as dominant species associated with the investigated Antarctic macroalgae suggests that they may exhibit an interesting ecological relationship with their hosts.
Diversity, richness, dominance and distribution
Studies on fungal diversity in Antarctica show inconsistencies in the discrimination of endemic, indigenous and cosmopolitan species. Endemic species are characterised as true psychrophilic fungi that are able to actively grow and reproduce only in Antarctica (Ruisi et al., 2007). According to Onofri et al. (2007), indigenicity is indicated by the observation of a large number of isolates over time as well as being commonly recorded in Antarctica from different sites and substrata. In addition, the majority of Antarctic fungi consist of ecotypes of cosmopolitan species that show mesophilic-psychrotolerant behaviour as an adaptation to the cold Antarctic climate (Zucconi et al., 1996). On the basis of the above criteria, the fungal species M. australis is indigenous to Antarctica, and Penicillium is a cosmopolitan mesophilic-psychrotolerant group that shows adaptation to the cold Antarctic climate. In contrast, Antarctomyces psychrotrophicus (not found outside of Antarctica), which was part of the minority component associated with the macroalga U. intestinalis, has been reported to be endemic to Antarctica (Stchigel et al., 2001).
Geomyces (Helotiales, Ascomycota), found as the most abundant taxa associated with Antarctic macroalgae, includes about 10 named species, which show a wide distribution (http://www.mycobank.org/). Geomyces sp. have been frequently recorded in Antarctica, are a celluloytic, keratinophilic, psychrophilic and halotolerant fungi with a ubiquitous distribution in the soils of cold regions that is able to colonise and to utilise different carbon sources (Mercantini et al., 1989). In addition, Geomyces taxa were found on the thalli of the macroalgae A. utricularis and D. anceps (Loque et al., 2010), leaves of Colobanthus quitensis (Rosa et al., 2010), mosses (Tosi et al., 2002) and in freshwater lakes (Gonçalves et al., 2012). Seven fungal isolates found associated with the endemic macroalgae M. hariotii displayed high sequence similarity with G. destructans. Geomyces destructans is characterised as a psychrophilic pathogenic fungus able to decrease bats' population in temperate regions (Lorch et al., 2011). According to Lorch et al. (2012), the diversity of Geomyces taxa, also classified as Gymnostellatospora and Pseudogymnosascus, is far greater than previously recognised based on traditional taxonomic methods, and can be underestimated. In addition, we found in association with Antarctic macroalgae 36 isolates identified as Geomyces sp., which may include new species and will be subject to further studies to elucidate their taxonomic positions.
The yeast M. australis, which has been reported to be endemic to Antarctica, has been isolated from Antarctic seawater (Fell and Hunter, 1968), the stomach of the Antarctic krill species Euphausia superba (Donachie and Zdanowski, 1998) and, recently, at a high abundance from the algal thalli of A. utricularis (Loque et al., 2010). In addition, M. australis has been found in marine sediment and freshwater in Antarctica (Vaz et al., 2011). The recovery of M. australis in association with the macroalgae A. arcta and D. menziesii reinforces the notion that this yeast may exhibit a specific association with Antarctic macroalgae.
The cosmopolitan genus Penicillium has been recovered from alpine and tundra soils as well as permafrost layers (Gunde-Cimerman et al., 2003). In Antarctica, species of Penicillium have been described from the soil (Azmiá and Seppelt, 1998), lakes (Ellis-Evans, 1996), wood (Arenz et al., 2006) and on the macroalga A. utricularis (Loque et al., 2010). As an extremophile, P. chrysogenum has been isolated as a dominant species from Arctic subglacial ice (Gunde-Cimerman et al., 2003, Sonjak et al., 2005). In addition, according to Bugni and Ireland (2004) Penicillium represents one of the more common genera isolated from macroalgae. In the present study, we found different Penicillium taxa associated with the Antarctic macroalgae. According to Frisvad and Samson (2004), Scott et al. (2004) and Houbraken et al. (2011), the taxonomy of the group Penicillium has been regarded to be especially difficult and needs a polyphasic study including physiological, morphological, multilocus sequence typing and chemical methods. For these reasons, the Penicillium sp. associated with the Antarctic macroalgae were identified using the term ‘cf.' or in genus rank. These Penicillium taxa will be the subject of further taxonomic evaluations.
The non-asymptotic species accumulation curves (Mao Tao analysis) generated for the fungal communities found in associations with the examined Antarctic macroalgae showed that a large number of single individuals (singletons) were recovered, which are considered rare species and often represent more than half of the species within these communities. Similar results were described for the fungal communities of A. utricularis, D. anceps and P. decipiens collected at King George Island, Antarctica (Loque et al., 2010).
Twenty-seven fungal taxa associated with the Antarctic macroalgae displayed a low molecular similarity with the sequences of known fungi deposited in GenBank, suggesting that they are candidates for new fungal species. However, not all described fungal species are deposited in the GenBank database, and further taxonomic analyses including detailed physiological and macro- and micromorphological characterisation and molecular sequencing of other DNA regions, together with new phylogenetic analyses, will be needed to describe these potentially new species. According to the recent checklist of Antarctic fungi published by Bridge and Spooner (2012), the specimens of Meyerozyma caribbica, Mycoarthris coralline and Yamadazyma mexicana, which were found as minor components within the fungal communities associated with Antarctic macroalgae, represent the first records of these species in Antarctica.
According to Ruisi et al. (2007), the distribution of fungi in Antarctica is related to the distribution of their hosts or substrates, such as bird feathers and dung, invertebrates, vegetation, soils, rocks and lichens. However, marine fungal dispersal remains unknown. In the present study, the calculated similarity indices (Sorensen and Bray-Curtis) revealed the presence of some species (D. hansenii, Geomyces sp., Helotiales sp., M. australis, Penicillium sp. and T. globosus) in association with different macroalgae and from distinct Antarctic islands. The high values of the Bray-Curtis similarity index (which has priority regarding the abundance of common species) between the fungal communities associated with endemic macroalgae P. decipiens and M. hariotii indicate that Geomyces and Penicillium sp. might show a specific association with macroalgae adapted to extreme conditions.
Fungi are one of the largest and most diverse kingdoms of eukaryotes and are important biological components of terrestrial ecosystems. According to Bridge and Spooner (2012), there are many records of marine fungi from Antarctica, although there is little available information about their ecological roles. In the present study, some of the fungal taxa found to be associated with Antarctic macroalgae were identified as species known to be mutualists, saprobes and parasites. Bridge and Spooner (2012) suggest that Antarctic fungi occupy many different ecological niches, but their significance in these niches remains poorly understood. Understanding how mutualistic, saprobic and pathogenic fungi achieve their lifestyles is crucial for understanding their ecological functions and their impact on Antarctic macroalgae under the extreme conditions of this continent. In addition, the abiotic seawater parameters in the areas where the macroalgae were collected revealed low temperatures and high salinities, demonstrating that the fungi found in association with the macroalgae may exhibit specific mechanisms that allow them to survive in extreme conditions.
Biological activities
In general, fungi have been reported to represent prolific sources of various compounds, including several bioactive molecules. However, the potential of extremophile fungi to produce bioactive compounds is poorly understood. According to Santiago et al. (2012), the ability of Antarctic fungi to survive in extreme conditions suggests that they may display unusual biochemical pathways that allow them to generate specific or new molecules that could be used to develop new drugs. Our bioprospecting results show that the extracts from Penicillium sp. UFMGCB 6034 and Penicillium sp. UFMGCB 6120, which were recovered as dominant species from endemic macroalgae, P. decipiens and M. hariotii, displayed high and selective antifungal and/or trypanocidal activities with low MIC and IC50 values. The combination of low MIC and IC50 values together with the 1H NMR spectral data suggesting the presence of functionalised aromatic compounds warrants extensive bioassay-directed fractionation to specifically identify the constituents responsible for the observed biological activity.
Marine fungi have been described as a promising source of antimicrobial compounds. Marine Penicillium sp. are known to produce a large variety of compounds with a wide range of biological and pharmacological activities. Penicillium sp. isolated from the alga U. intestinalis were observed to produce cytotoxic metabolites including communesins (Numata et al., 1993), penochalasius A-C (Numata et al., 1995), penostatins A-I (Takahashi et al., 1996, Iwamoto et al., 1998) and penochalasius D-H (Iwamoto et al., 2001). The antibacterial and cytotoxic compounds di(2-ethyl hexayl) phthalate and fungisterol were isolated from P. brevicompactum, which was isolated from the associated marine alga Pterocladia sp. (Atalla et al., 2011). The extracts of Penicillium sp. UFMGCB 6034 and Penicillium sp. UFMGCB 6120 will be subjected to further systematic chemical bioactivity-guided fractionation to isolate their bioactive compounds.
Conclusion
Although marine fungi have been studied for the last 100 years (Jones, 2011), many aspects regarding their taxonomy, ecology and potential biotechnological applications remain poorly documented. Our results are consistent with a suggestion by Bridge and Spooner (2012) that the true diversity of Antarctic fungi may be far greater than is currently estimated. Our results also suggest that macroalgae can act as a substrate to shelter fungi in Antarctica and contribute to the knowledge of the cryptic and unknown algicolous Antarctic mycota. We showed that endemic and cold-adapted macroalgae living under the extreme conditions of Antarctica shelter rich, diverse and complex fungal communities consisting of few dominant indigenous or mesophilic cold-adapted species. In addition, we detected a large number of rare and/or endemic taxa, which may provide an interesting model of algal–fungal interactions in extreme conditions as well as a potential source of bioactive compounds. Our results reinforce the call for further studies of fungal communities present across the Antarctic marine environment as well as their phylogenetic relationships with species that occur in other cold regions. Finally, the present work suggests that examination of the fungi associated with macroalgae in Antarctica may provide new insights into the biological mechanisms underlying tolerance and adaptation to extreme marine polar conditions.
Acknowledgments
This study had financial and logistic support from the Brazilian Antarctic Program (PROANTAR/MCT/CNPq—No. 23/2009), Marine of Brazil and Francisco Petrone from the Clube Alpino Paulista (CAP). We acknowledge the financial support from Fundação de Amparo a Pesquisa do Estado de Minas Gerais (FAPEMIG), Health-PDTIS-FIOCRUZ, PRPq-UFMG, CAPES/PGCI 036/13 and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq). This study represents the Pos-doctoral degree of Dr VM Godinho under the supervision of Dr LH Rosa. Also, the results of this study are part of the Master Science degree of LE Furbino within the Programa of Pós-Graduação in Biotecnologia of UFOP. We thank Isis V Galliza and Fernanda LM Francisco for performing the biological assays. We thank Solomon Green III for technical assistance. We also thank the anonymous reviewers for helpful comments to improve the quality of the manuscript.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z, Miller W, et al. Gapped BLASTand PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amsler CD, Amsler MO, McClintock JB, Baker BJ. Filamentous algal endophytes in macrophytic Antarctic algae: prevalence in hosts and palatability to mesoherbivores. Phycologia. 2009;48:324–334. [Google Scholar]
- Arenz BE, Held BW, Jurgens JA, Farrell RL, Blanchette RA. Fungal diversity in soils and historic wood from the Ross Sea Region of Antarctica. Soil Biol Bioch. 2006;38:3057–3064. [Google Scholar]
- Atalla MM, Zeinab HK, Eman RH, Amani AY, Abeer AAEA.2011Physiological Studies on Some Biologically Active Secondary Metabolites from Marine. Derived Fungus Penicillium brevicompactum 11–15. ; http://www.GAte2Biotech.com .
- Azmiá OR, Seppelt RD. The broad-scale distribution of microfungi in the Windmill Islands region, continental Antarctica. Polar Biol. 1998;19:92–100. [Google Scholar]
- Bridge PD, Spooner BM. Non-lichenized Antarctic fungi: transient visitors or members of a cryptic ecosystem. Fungal Ecol. 2012;5:381–394. [Google Scholar]
- Buckner FS, CLMJ Verlinde, La Flamme AC, Van Voorkhis WC. Efficient Technique for screening drugs for activity against Trypanosoma cruzi using parasites expressing β-galactosidase. Antimicrob Agent Chemother. 1996;40:2592–2597. doi: 10.1128/aac.40.11.2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugni TS, Ireland CM. Marine-derived fungi: a chemically and biologically diverse group of microorganisms. Nat Prod Rep. 2004;21:143–163. doi: 10.1039/b301926h. [DOI] [PubMed] [Google Scholar]
- Carvalho CR, Gonçalves VN, Pereira CB, Johann S, Galliza IV, Alves TMA, et al. The diversity, antimicrobial and anticancer activity of endophytic fungi associated with the medicinal plant Stryphnodendron adstringens (Mart.) Coville (Fabaceae) from the Brazilian savannah. Symbiosis. 2012;57:95–107. [Google Scholar]
- Colwell RK, Mao CX, Chang J. Interpolating, extrapolating, and comparing incidence-based species accumulation curves. Ecology. 2004;85:2717–2727. [Google Scholar]
- Donachie SP, Zdanowski MK. Potential digestive function of bacteria in krill, Euphausia superba stomach. Aquat Microb Ecol. 1998;14:129–136. [Google Scholar]
- Ellis-Evans JC. Microbial diversity and function in Antarctic freshwater ecosystems. Biodivers Conserv. 1996;5:1395–1431. [Google Scholar]
- Fell JW, Hunter IL, Tallman AS. Marine basidiomycetous yeasts (Rhodosporidium spp. n.) with tetrapolar and multiple alletic bipolar mating systems. Can J Microbiol. 1973;19:643–657. doi: 10.1139/m73-106. [DOI] [PubMed] [Google Scholar]
- Fell JW, Hunter IL. Isolation of heterothallic yeast strains of Metschnikowia Kamienski and their mating reactions with Chlamydozyma Wickerham spp. Antonie van Leeuwenhoek. 1968;34:365–376. doi: 10.1007/BF02046459. [DOI] [PubMed] [Google Scholar]
- Fell JW, Scorzetti G, Connell L, Craig S. Biodiversity of micro-eukaryotes in Antarctic Dry Valley soils with <5% soil moisture. Soil Biol Biochem. 2006;38:3107–3119. [Google Scholar]
- Fell JW, Statzel AC. Sympodiomycetes gen. n., a yeast-like organism from southern marine waters. Antonie van Leeuwenhoek. 1971;37:359–367. doi: 10.1007/BF02218506. [DOI] [PubMed] [Google Scholar]
- Frisvad JC, Samson RA. Polyphasic taxonomy of Penicillium subgenus Penicillium: a guide to identification of food and air-borne terverticillate Penicillia and their mycotoxins. Stud Mycol. 2004;49:1–174. [Google Scholar]
- Gazis R, Rehner S, Chaverri P. Species delimitation in fungal endophyte diversity studies and its implications in ecological and biogeographic inferences. Mol Ecol. 2011;20:3001–3013. doi: 10.1111/j.1365-294X.2011.05110.x. [DOI] [PubMed] [Google Scholar]
- Glass NL, Donaldson GC. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl Environ Microbiol. 1995;61:1323–1330. doi: 10.1128/aem.61.4.1323-1330.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonçalves VN, Vaz ABM, Rosa CA, Rosa LH. Diversity and distribution of fungal communities in lakes of Antarctica. FEMS Microbiol Ecol. 2012;82:459–471. doi: 10.1111/j.1574-6941.2012.01424.x. [DOI] [PubMed] [Google Scholar]
- Grasso S, Bruni V, Maio G. Marine fungi in Terra Nova Bay (Ross Sea, Antarctica) Microbiologica. 1997;20:371–376. [PubMed] [Google Scholar]
- Guiry MD, Guiry GM.AlgaeBase World-wide electronic publication National University of Ireland: Galway. , http://www.algaebase.org (searched on 11 October 2012). [Google Scholar]
- Gunde-Cimerman N, Sonjak S, Zalar P, Frisvad JC, Diderichsen B, Plemenitas A. Extremophilic fungi in arctic ice: a relationship between adaptation to low temperature and water activity. Phys Chem Earth. 2003;28:1273–1278. [Google Scholar]
- Hammer Ø, Harper DAT, Ryan PD. PAST: paleontological statistics software package for education and data analysis. Palaeontol Electron. 2001;4:9. [Google Scholar]
- Houbraken J, Frisvad JC, Samson RA. Fleming's penicillin producing strain is not Penicillium chrysogenum but P. rubens. IMA Fungus. 2011;2:87–95. doi: 10.5598/imafungus.2011.02.01.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamoto C, Minoura K, Hagishita S, Nomoto K, Numata AJ. Penostatins F-I, novel cytotoxic metabolites from a Penicillium species separated from na Enteromorpha marine alga. J Chem Soc Perkin. 1998;1:449–456. [Google Scholar]
- Iwamoto C, Yamada T, Ito Y, Minoura K, Numata A. Cytotoxic cytochalasans from a Penicillium species separated from a marine alga. Tetrahedron. 2001;57:2997–3004. [Google Scholar]
- Jones EB. Fifty years of marine mycology. Fungal Div. 2011;50:73–112. [Google Scholar]
- Kirk PM, Cannon PF, Minter DW, Stalpers JA.2011Dictionary of the Fungi10th edn.CAB International: Wallingford, UK [Google Scholar]
- Kohlmeyer J, Volkmann-Kohlmeyer B. Illustrated key to the filamentous higher marine fungi. Botanica Marina. 1991;34:1–61. [Google Scholar]
- Kurtzman CP, Fell JW, Boekhout T.2011The Yeasts: A Taxonomic Study5th edn.Elsevier: Amsterdam, The Netherlands [Google Scholar]
- Lachance MA, Bowles JM, Starmer WT, Barker JS. Kodamaea kakaduensis and Candida tolerans, two new ascomycetous yeast species from Australian hibiscus flowers. Can J Microbiol. 1999;45:172–177. doi: 10.1139/w98-225. [DOI] [PubMed] [Google Scholar]
- Loque CP, Medeiros AO, Pellizzari FM, Oliveira EC, Rosa CA, Rosa LH. Fungal community associated with marine macroalgae from Antarctica. Polar Biol. 2010;33:641–648. [Google Scholar]
- Lorch JM, Lindner DL, Gargas A, Muller LK, Minnis AM, Blehert DS. A culture-based survey of fungi in soil from bat hibernacula in the eastern United States and its implications for detection of Geomyces destructans, the causal agent of bat white-nose syndrome. Mycologia. 2012;105:237–252. doi: 10.3852/12-207. [DOI] [PubMed] [Google Scholar]
- Lorch JM, Meteyer CU, Behr JM, Boyles JG, Cryan PM, Hicks AC, et al. Experimental infection of bats with Geomyces destructans causes white-nose syndrome. Nature. 2011;480:376–378. doi: 10.1038/nature10590. [DOI] [PubMed] [Google Scholar]
- Magurran AE. Measuring Biological Diversity. Blackwell Science: USA; 2011. [Google Scholar]
- Mercantini R, Marsella R, Cervellati MC. Keratinophilic fungi isolated from Antarctic soil. Mycopathologia. 1989;106:47–52. doi: 10.1007/BF00436926. [DOI] [PubMed] [Google Scholar]
- Nedzarek A, Rakusa-Suszczewski S. Decomposition of macroalgae and the release of nutrients in Admiralty Bay, King George Island, Antarctica. Polar Biosci. 2004;17:16–35. [Google Scholar]
- Numata A, Takaheshic, Ito Y, Minoura K, Yamada T, Matsuda C, et al. Penochalasins. A novel class of cytotoxic cytochalasans from a Penicillium species separated from a marine alga: structure determination and solution confirmation. J Chem Soc Perkin. 1995;1:239–245. [Google Scholar]
- Numata A, Takaheshic, Ito Y, Takada T, Kawaik K, Usami Y, et al. Communesins, cytotoxic metabolites of a fungus isolated from a marine alga. Tetrahedron Lett. 1993;34:2355–2358. [Google Scholar]
- Oliveira EC, Absher TM, Pellizzari FM, Oliveira MC. The seaweed flora of Admiralty Bay, King George Island, Antarctic. Polar Biol. 2009;32:1630–1647. [Google Scholar]
- Onofri S, Zucconi L, Tosi S. Continental Antarctic Fungi. IHW-Verlag: Eching bei München; 2007. [Google Scholar]
- Papenfuss GF.1964Catalogue and bibliography of Antarctic and Sub-Antarctic benthic marine algaeIn: Lee MO (eds)Bibliography of the Antarctic Seas American Geophysical Union: Washington DC; vol. 11–76. [Google Scholar]
- Quartino ML, Zaixso HE, Boraso de Zaixso AL. Biological and environmental characterization of marine macroalgal assemblages in Potter Cove, South Shetland Islands, Antarctica. Botanica Marina. 2005;48:187–197. [Google Scholar]
- Ricker RW. Taxonomy and biogeography of Macquarie Island seaweeds. British Museum Natural History: London; 1987. [Google Scholar]
- Rosa LH, Vaz ABM, Caligiorne RB, Campolina S, Rosa CA. Endophytic fungi associated with the Antarctic Grass Deschampsia antarctica Desv. (Poaceae) Polar Biol. 2009;32:161–167. [Google Scholar]
- Rosa LH, Vieira MLA, Santiago IF, Rosa CA. Endophytic fungi community associated with the dicotyledonous plant Colobanthus quitensis (Kunth) Bartl. (Caryophyllaceae) in Antarctica. FEMS Microbiol Ecol. 2010;73:178–189. doi: 10.1111/j.1574-6941.2010.00872.x. [DOI] [PubMed] [Google Scholar]
- Ruisi S, Barreca D, Selbmann L, Zucconi L, Onofri S. Fungi in Antarctica. Rev Environment Science Biotechnol. 2007;6:127–141. [Google Scholar]
- Santiago IF, Alves TMA, Rabello A, Sales-Júnior PA, Romanha AJ, Zani CL, et al. Leishmanicidal and antitumoral activities of endophytic fungi associated with the Antarctic angiosperms Deschampsia antarctica Desv. and Colobanthus quitensis (Kunth) Bartl. Extremophiles. 2012;16:95–103. doi: 10.1007/s00792-011-0409-9. [DOI] [PubMed] [Google Scholar]
- Scott J, Untereiner WA, Wong B, Strauss NA, Malloch D. Genotypic variation in Penicillium chrysogenum from indoor environments. Mycologia. 2004;96:1095–1105. [PubMed] [Google Scholar]
- Sonjak S, Frisvad JC, Gunde-Cimerman N. Comparison of secondary metabolite production by Penicillium crustosum strains, isolated from Arctic and other various ecological niches. FEMS Microbiol Ecol. 2005;53:51–60. doi: 10.1016/j.femsec.2004.10.014. [DOI] [PubMed] [Google Scholar]
- Stanley SJ. Observation on the seasonal occurrence of marine endophytic and parasitic fungi. Can J Bot. 1992;70:2089–2096. [Google Scholar]
- Stchigel AM, Cano J, MacCormack CW. Antarctomyces psychrotrophicus gen. et sp. nov., a new ascomycete from Antarctica. Mycol Res. 2001;105:377–382. [Google Scholar]
- Suryanarayanan TS, Venkatachalam A, Thirunavukkarasu N, Ravishankar JP, Doble M, Geetha V. Internal mycobiota of marine macroalgae from the Tamilnadu coast: distribution, diversity and biotechnological potential. Botanica Marina. 2010;53:457–468. [Google Scholar]
- Takahashi C, Numata A, Yamada T, Minoura K, Enemoto S, Konishi K. Penostatins, a novel cytotoxic metabolites from a Penicillium species separated from a green alga. Tetrahedron Lett. 1996;37:655–658. [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosi S, Casado B, Gerdol R, Caretta G. Fungi isolated from Antarctic mosses. Polar Biol. 2002;25:262–268. [Google Scholar]
- Vaz ABM, Rosa LH, Vieira MLA, Garcia V, Brandão LR, Teixeira LCRS, et al. The diversity, extracellular enzymatic activities and photoprotective compounds of yeasts isolated in Antarctica. Braz J Microbiol. 2011;42:937–947. doi: 10.1590/S1517-838220110003000012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White TJ, Bruns TD, Lee SB.1990Amplification and direct sequencing of fungal ribosomal RNA genes for phylogeneticsIn: Innis NA, Gelfand J, Sninsky J et al. (eds)PCR protocols: A guide to methods and applications Academic Press: San Diego; 315–322. [Google Scholar]
- Wiencke C, Clayton MN. Antarctic Seaweeds. Ruggell: A.R.G. Gantner Verlag KG; 2002. p. 239. [Google Scholar]
- Yarrow D.1998Methods for the isolation, maintenance and identification of yeastIn: Kurtzman CP, Fell JW (eds)The Yeasts: A Taxonomic Study Elsevier: Amsterdam [Google Scholar]
- Zuccaro A, Mitchell JI.2005Fungal Communities of SeaweedsIn: Dighton J, White JF, Oudemans P (eds)The Fungal Community: Its Organization and Role in the Ecosystem CRC Press: Boca Raton FL/Taylor and Francis, New York; 533–579. [Google Scholar]
- Zuccaro A, Schoch CL, Spatafora JW, Kohlmeyer J, Draeger S, Mitchell JI. Detection and identification of fungi intimately associated with the brown seaweed Fucus serratus. Appl Environ Microbiol. 2008;74:931–941. doi: 10.1128/AEM.01158-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zucconi L, Pagano S, Fenice M, Selbmann L, Tosi S, Onofri S. Growth temperature preferences of fungal strains from Victoria Land, Antarctica. Polar Biol. 1996;16:53–61. [Google Scholar]