Abstract
The healthy intestine is characterized by a low level of oxygen and by the presence of large bacterial communities of obligate anaerobes. Dysbiosis of the gut microbiota has been reported in patients suffering from inflammatory bowel diseases (IBDs), but the mechanisms causing this imbalance remain unknown. Observations have included a decrease in obligate anaerobes of the phylum Firmicutes and an increase in facultative anaerobes, including members of the family Enterobacteriaceae. The shift of bacterial communities from obligate to facultative anaerobes strongly suggests a disruption in anaerobiosis and points to a role for oxygen in intestinal dysbiosis. Proposals to evaluate this hypothesis of a role for oxygen in IBD dysbiosis are provided. If this hypothesis is confirmed, decreasing oxygen in the intestine could open novel means to rebalance the microbiota and could provide novel preventative or therapeutic strategies for IBD patients in whom current treatments are ineffective.
Keywords: anaerobiosis, dysbiosis, IBD, gut microbiota
Introduction
Inflammatory bowel diseases (IBDs) include Crohn's disease (CD) and ulcerative colitis (UC) that occur in the distal ileum or colon. The gut microbiota representing the commensal bacteria inhabiting the gastrointestinal tract have a central role in the pathogenesis of IBD, in combination with genetic loci of hosts that present risk factors for inflammation in the gastrointestinal tract. The commensal bacteria are associated with the induction, perpetuation and reactivation of IBD through an altered interaction with the host, triggering inappropriate immune reactions (Xavier and Podolsky, 2007). Dysbiosis of the gut microbiota has been reported in patients suffering from IBDs, but the mechanisms causing this imbalance remain unknown. The hypothesis proposed in this article is that an increase in oxygen tension in the gut is the cause of the dysbiosis in IBD patients. This hypothesis is based on the combination of the physiological anaerobic conditions of the distal intestine in healthy individuals and the microbiological features of the dysbiosis of gut microbiota observed in IBD patients.
Gut microbiota in health
The gastrointestinal tract is germ free at birth and is then colonized sequentially by a myriad of microorganisms. The sequence of events that lead to the succession of species colonizing the human large intestine has been reviewed by Mackie et al. (1999) and Adlerberth and Wold (2009). In the ‘classical' pattern of colonization in infants, the initial step involves facultative anaerobes such as Escherichia coli and Enterococcus. The colonization of the gastrointestinal tract by facultative anaerobes occurs immediately after birth in an intestinal environment where the redox potential (Eh) is high or electropositive (Grutte et al., 1965). The increase in facultatively anaerobic bacteria caused the consumption of the available oxygen, which produces a reduced environment that favors the arrival of obligate anaerobes a few days after birth. The Eh associated with the concentration of oxygen then decreases to a low level, as observed in the large intestine of healthy adults (Lind Due et al., 2003). The oxidation-reduction state has been proposed to participate in the resistance to colonization of the gut by creating a physiological environment that is inhibitory to competitors and potential intestinal pathogens. Meynell (1963) elucidated the role of Eh and fatty acids in the antibacterial mechanisms in the gut of mice. He reported that Eh in the large intestine of untreated mice was about −200 mV. After a treatment with streptomycin, the level of fatty acids decreased and the redox state shifted to +200 mV, concomitant with an increase in the Salmonella population. In healthy adults, the distal part of the gastrointestinal tract is characterized by a low level of oxygen tension and usually harbors large communities of obligate anaerobes: 107−108 g−1 in the distal ileum and 109−1011 g−1 in the colon. Most of the bacteria of the gastrointestinal tract have not yet been cultured ex vivo, due in part to their anaerobic physiology or sensitivity to oxygen; those that have been cultured require long and laborious techniques (Duncan et al., 2007).
Over the last 15 years, the composition of the gut microbiota in healthy humans has been explored by culture-independent techniques (Sekirov et al., 2010; Lozupone et al., 2012) and further described by metagenomic analysis (Qin et al., 2010). Analyses of the gene encoding 16S ribosomal RNA by pyrosequencing have identified no more than 10 phyla in the microbiota. The two dominant phyla, Firmicutes and Bacteroidetes, representing 90% of the gut bacteria, are mainly obligate anaerobes. At a lower taxonomic level, up to a thousand species are generally present in one individual. Metagenomic analysis has revealed, within the interindividual variation, the presence of fewer than 60 species shared among individuals. These species represent the core gut microbiota. In a study of colonic samples from different countries (Arumugam et al., 2011), three clusters of hosts were described and identified as enterotypes. The microbiotas of these enterotypes vary in species and genetic composition but are not specific to countries or continents. The enterotypes may represent groups of humans that will respond differently to drug treatment or nutrition. Wu et al. (2011) reported an association between long-term dietary patterns and two of the enterotypes. The definition of the enterotypes, however, has recently been questioned (Yong, 2012), the bacterial communities of the gut microbiota forming more a spectrum rather than belonging to distinct groups.
Gut microbiotic dysbiosis in IBD
The gut microbiota in patients with IBD and in healthy individuals have been compared to determine whether microbiotic signatures were associated with IBD, in particular if groups, species, strains, genes or even bacterial epitopes could be associated with the diseases. Imbalance of the gut microbiota, or dysbiosis, was observed in samples from CD and UC patients (Seksik et al., 2003; Sokol et al., 2006, Swidsinski et al., 2007; Sartor, 2008, Sokol et al., 2008, 2009). A reduction in the diversity of the phylum Firmicutes was repeatedly observed, that is, fewer species of this phylum of obligate anaerobes were present in IBD patients (Ott et al., 2004; Manichanh et al., 2006; Frank et al., 2007). In agreement with this reduction in diversity, a metagenomic analysis revealed that the microbiota of IBD patients had 25% fewer genes than the microbiota of healthy individuals (Qin et al., 2010). The metaproteomic approach further demonstrated a depletion of proteins and functional pathways (Erickson et al., 2012). Within the phylum Firmicutes, a decrease or even the disappearance was reported for the extremely oxygen-sensitive Faecalibacterium prausnitzii, a dominant species in the gut microbiota of healthy subjects (Sokol et al., 2008, 2009). In parallel, the pullulation, or overgrowth, of subdominant facultative anaerobes from the family Enterobacteriaceae, including E. coli, was observed (Darfeuille-Michaud et al., 2004; Baumgart et al., 2007; Kotlowski et al., 2007; Lupp et al., 2007). Aerobes and unusual bacteria were also detected in the gut, the best example being Mycobacterium avium subspecies paratuberculosis (Naser et al., 2004), an obligate aerobe. The main observations from unrelated individuals were similar to those from studies on pairs of twins, performed to avoid interindividual variation in the microbiotic composition and the influence of host genotype (Willing et al., 2009, Lepage et al., 2011).
In a study of a cohort of twins concordant or discordant for CD or UC, pyrosequencing was used to determine the compositions of bacterial communities in feces (Willing et al., 2010). The gut microbiota of patients differed from those of healthy individuals, especially in CD involving the ileum compared with those involving the colon. Changes in patients with ileal CD included the disappearance of bacteria from the core microbiota, such as Faecalibacterium, and increased levels of Enterobacteriaceae and Ruminococcus gnavus. The compositions reported for individuals with UC were similar to those for healthy individuals. In another pyrosequencing study on a cohort of twins with UC, however, marked differences were observed and were characterized by less bacterial diversity and more Actinobacteria and Proteobacteria (Lepage et al., 2011). The differences between the studies, although, may be due to sampling differences, as the patients were either in remission or had active disease.
Nagalingam and Lynch (2012) have documented an extensive list of microbial modifications in IBD patients, and Mukhopadhya et al. (2012) have reviewed proteobacteria associated with IBD. Whether the observed modifications in the microbiota are a cause or a consequence of the development of IBD, although, remains a debate (Manichanh et al., 2012).
Is dysbiosis a ‘dysanaerobiosis'?
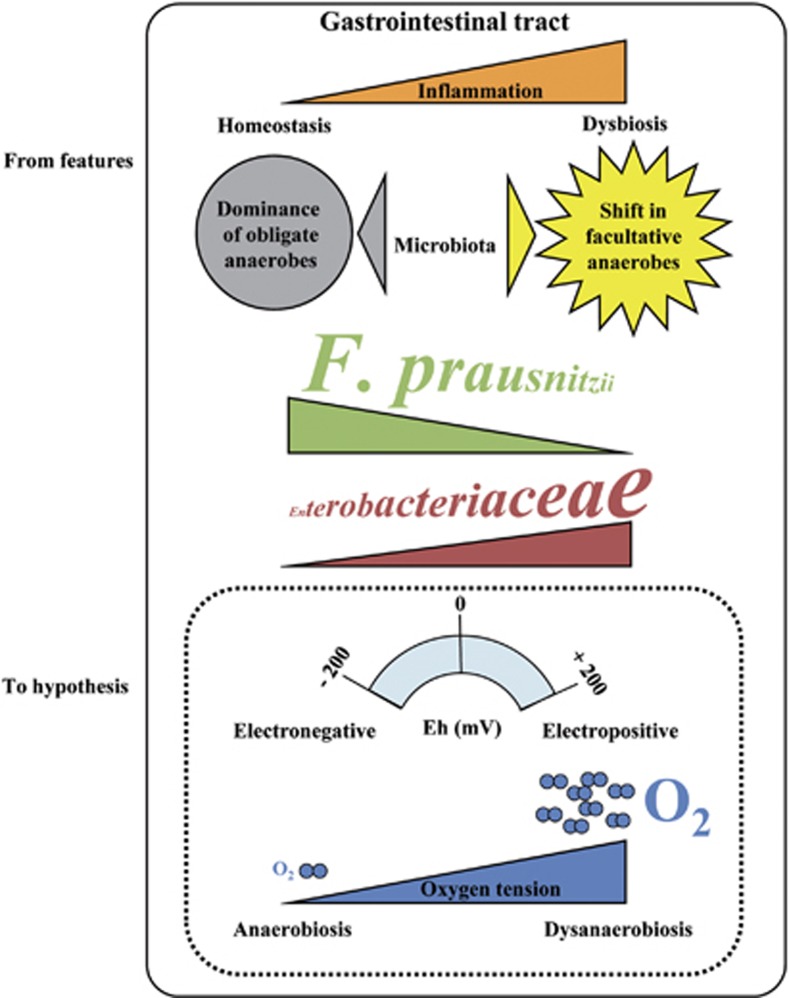
Intestinal dysbiosis generally shows a reduction in diversity and decrease of dominant obligate anaerobes in parallel with an increase of subdominant facultative anaerobes or the appearance of unusual aerobes. These features have surprisingly not been discussed in the context of specific gut physiology, particularly in relation to the normal conditions of anaerobiosis encountered by the bacteria in the distal intestine. The microbiological observations combined with the normal physiological conditions in the gut led to the hypothesis of a role for oxygen in the intestinal dysbiosis of IBD patients (Figure 1). The increase of oxygen causing a disruption in anaerobiosis would confer an ecological selective advantage to facultative anaerobes or even to aerobes, allowing them to become more competitive and able to overgrow. Conversely, the obligate anaerobes sensitive to oxygen and harmed by its presence would be at a disadvantage. Interestingly, Mondot et al. (2011) noted that the microbiotas present during active CD are adapted to high Ehs, but these authors did not associate the shift to an increase of oxygen. Anaerobic Bacteroidetes are less often reported to be affected in the dysbiosis of IBD patients than is F. prausnitzii. Interestingly, some Bacteroides can grow in the presence of nanomolar concentrations of oxygen (Baughn and Malamy, 2004). Survival in the presence of oxygen has even been used as a means of counterselection to isolate Bacteroides in fecal samples (Corthier et al., 1996). The lower sensitivity of these bacteria to oxygen compared with extremely oxygen-sensitive bacteria (Faecalibacterium) would also support the hypothesis of a role for oxygen.
Figure 1.
The imbalance of the gut microbiota, or dysbiosis, in IBDs: a role for oxygen? Dysbiosis is observed in patients suffering from IBDs. Repeated observations indicate a decrease in obligate anaerobes (F. prausnitzii), an increase of facultative anaerobes (Enterobacteriaceae) and even the appearance of aerobes. Superimposed with the normal anaerobic gut physiology of the distal intestine, these observations support the hypothesis of a shift in bacterial communities caused by an increase of oxygen that changes the Eh in the gastrointestinal tract.
The biostructure of fecal microbiota in healthy subjects reported by Swidsinski et al. (2008) provides information on the in situ location of Enterobacteriaceae and F. prausnitzii in relation to oxygen availability. Enterobacteriaceae are observed at the interface between feces and mucus, close to epithelial cells, and are absent or in lower proportions in the feces. Bacteroides and Firmicutes, including F. prausnitzii, are observed at high concentrations in the feces inside the lumen but also occasionally at low concentrations in the mucus. Despite being extremely sensitive to oxygen, F. prausnitzii can be found adhering to the gut mucosa where oxygen diffuses from epithelial cells. To explain this paradox, Khan et al. (2012) recently reported that F. prausnitzii could grow in the presence of low levels of oxygen. They showed that F. prausnitzii employed an extracellular electron shuttle of flavins and thiols to transfer electrons to oxygen. Both flavins and thiols present in the healthy human gut, however, are needed to allow F. prausnitzii to survive in the presence of oxygen, which may not be the case in the injured tissue of IBD patients.
Hartman et al. (2009) reported an oxygen-dependent shift in the gut, where the microbiotic composition was monitored after transplantation of the small bowel combined with an ileostomy. The gut bacterial communities shifted from obligate anaerobes to communities dominated by facultative anaerobes, and Enterobacteriaceae increased, as in the microbiota of IBD patients. After closure of the ileostomy, which was hypothesized as providing an entry for oxygen, the shift was reversed, and obligate anaerobes again dominated. The authors proposed that oxygen was the cause of the shift in bacterial communities from obligate to facultative anaerobes or aerobes. Although Enterobacteriaceae increased, inflammation was not observed in the majority of the patients. Of note, Lactobacillus also thrived, in contrast to IBD dysbiosis, where increases in lactobacilli have rarely been reported. Studies have shown that lactobacilli and their cell wall components have anti-inflammatory properties (for a review see Macho Fernandez et al. (2011)), suggesting that a modulation or a supplementation of lactobacilli may limit inflammatory reactions in IBD patients.
In IBD, an increase in the concentration of oxygen could be caused by the entry of blood into the gastrointestinal tract often observed during chronic inflammation, provoking the release of hemoglobin carrying oxygen in the intestinal mucosa and lumen where the gut bacteria reside. A mouse model indicated that inflammation may cause the dysbiosis of the microbiota and promote the overgrowth of Enterobacteriaceae (Lupp et al., 2007). A pilot study evaluated the effect of anti-tumor necrosis factor-α antibody therapy in CD on the composition and activity of fecal microbiota, and reported a modest increase of Faecalibacterium (Van Nuenen, 2005), suggesting that blocking the inflammatory response may promote this group. The changing oxygen conditions may also be due to the inflammatory reaction itself, causing an oxidative burst in the intestinal tissue, for example, by the release of reactive oxygen species by neutrophils. Interestingly, Zhu and Li (2012) recently reviewed the experimental and clinical evidence supporting oxidative stress as a component of IBD and as a proposed mechanism underlying the pathophysiology. Winter et al. (2010) elegantly showed that reactive oxygen species could react with endogenous luminal sulfuric compounds to form a respiratory electron acceptor that is used by Salmonella enterica serotype Typhimurium (an aerobic species and intestinal pathogen belonging to the Enterobacteriaceae), conferring a growth advantage to Salmonella over the competing gut microbiota of obligate anaerobes.
Perspectives and proposals to evaluate the oxygen hypothesis
Human clinical studies and animal models of intestinal inflammation have already provided a better understanding of the pathogenesis of IBD and could be used to evaluate the oxygen hypothesis. Animal models would enable an easier application of methods for tracing oxygen in the gut to monitor in vivo the variation of oxygen levels during inflammation. The noninvasive measurement of oxygen developed by He et al. (1999) would be preferred. Treatment with an oxygen sensor, such as Pd-porphyrin developed by Handa et al. (2010), could also be used to quantify local oxygen tension with the intravital phosphorescence assay. These measurements of oxygen should be combined with the so-called ‘-omics' strategies targeting both host and gut bacteria to determine how the potential increase of oxygen impacts the interaction between host and bacteria. The critical step will be to select animal models with an anaerobic intestinal physiology most similar to the conditions in humans. In human clinical studies, similar approaches could be applied combining in vivo measurements of the effects of oxygen on host and bacteria. Longitudinal studies with individuals acting as their own controls at different stages of the disease (in remission or with active disease) are suggested. To overcome variation in host genotypes and composition of microbiota, studies with pairs of twins should also be considered. The characterization of dysbiosis and the measurement of oxygen could be combined to determine the dysbiotic differences between ileal and colonic CD and UC. The data from such studies would provide a better understanding of why alterations in the microbiota are different between UC and CD. They would also allow us to determine whether oxygen is present only in the small intestine, where inflammation occurs, or along the gastrointestinal tract affected in ileal CD, where dysbiosis occurs from the small intestine to the rectum.
If a role for oxygen is established in dysbiosis, the modulation of oxygen levels in the colons of IBD patients could open new areas of research and means of proposing new preventative or therapeutic strategies for the well-being of IBD patients, providing alternative treatments when current treatments are ineffective. An attractive strategy will be to counterbalance the dysbiosis by limiting the oxygen in the distal part of the gastrointestinal tract. Oral supplements of pharmacological agents, such as antioxidants, or of probiotic microorganisms selected for their ability to capture or consume oxygen could prove to be effective.
Acknowledgments
I thank Dr William Blackhall for the English editing and revision of the text.
The author declares no conflict of interest.
References
- Adlerberth I, Wold AE. Establishment of the gut microbiota in Western infants. Acta Paediatr. 2009;98:229–238. doi: 10.1111/j.1651-2227.2008.01060.x. [DOI] [PubMed] [Google Scholar]
- Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baughn AD, Malamy MH. The strict anaerobe Bacteroides fragilis grows in and benefits from nanomolar concentrations of oxygen. Nature. 2004;427:441–444. doi: 10.1038/nature02285. [DOI] [PubMed] [Google Scholar]
- Baumgart M, Dogan B, Rishniw M, Weitzman G, Bosworth B, Yantiss R, et al. Culture independent analysis of ileal mucosa reveals a selective increase in invasive Escherichia coli of novel phylogeny relative to depletion of Clostridiales in Crohn's disease involving the ileum. ISME J. 2007;1:403–418. doi: 10.1038/ismej.2007.52. [DOI] [PubMed] [Google Scholar]
- Corthier G, Muller MC, L'Haridon R. Selective enumeration of Bacteroides vulgatus and B. distasonis organisms in the predominant human fecal flora by using monoclonal antibodies. Appl Environ Microbiol. 1996;62:735–738. doi: 10.1128/aem.62.2.735-738.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darfeuille-Michaud A, Boudeau J, Bulois P, Neut C, Glasser AL, Barnich N, et al. High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn's disease. Gastroenterology. 2004;127:412–421. doi: 10.1053/j.gastro.2004.04.061. [DOI] [PubMed] [Google Scholar]
- Duncan SH, Louis P, Flint HJ. Cultivable bacterial diversity from the human colon. Lett Appl Microbiol. 2007;44:343–350. doi: 10.1111/j.1472-765X.2007.02129.x. [DOI] [PubMed] [Google Scholar]
- Erickson AR, Cantarel BL, Lamendella R, Darzi Y, Mongodin EF, Pan C, et al. Integrated metagenomics/metaproteomics reveals human host-microbiota signatures of Crohn's disease. PLoS One. 2012;7:e49138. doi: 10.1371/journal.pone.0049138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank DN, St, Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci USA. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grutte FK, Horn R, Haenel H. Ernahrung und biochemischmikrookologische voorgange in enddarm von sauglingen (Nutrition and biochemical microecology processes occuring in the colon of infants) Z Kinderheikld. 1965;93:28–39. [PubMed] [Google Scholar]
- Handa K, Ohmura M, Nishime C, Hishiki T, Nagahata Y, Kawai K, et al. Phosphorescence-assisted microvascular O(2) measurements reveal alterations of oxygen demand in human metastatic colon cancer in the liver of superimmunodeficient NOG mice. Adv Exp Med Biol. 2010;662:423–429. doi: 10.1007/978-1-4419-1241-1_61. [DOI] [PubMed] [Google Scholar]
- Hartman AL, Lough DM, Barupal DK, Fiehn O, Fishbein T, Zasloff M, et al. Human gut microbiome adopts an alternative state following small bowel transplantation. Proc Natl Acad Sci USA. 2009;106:17187–17192. doi: 10.1073/pnas.0904847106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He G, Shankar RA, Chzhan M, Samouilov A, Kuppusamy P, Zweier JL. Noninvasive measurement of anatomic structure and intraluminal oxygenation in the gastrointestinal tract of living mice with spatial and spectral EPR imaging. Proc Natl Acad Sci USA. 1999;96:4586–4591. doi: 10.1073/pnas.96.8.4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan MT, Duncan SH, Stams AJ, van Dijl JM, Flint HJ, Harmsen HJ. The gut anaerobe Faecalibacterium prausnitzii uses an extracellular electron shuttle to grow at oxic-anoxic interphases. ISME J. 2012;6:1578–1585. doi: 10.1038/ismej.2012.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotlowski R, Bernstein CN, Sepehri S, Krause DO. High prevalence of Escherichia coli belonging to the B2+D phylogenetic group in inflammatory bowel disease. Gut. 2007;56:669–675. doi: 10.1136/gut.2006.099796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepage P, Hasler R, Spehlmann ME, Rehman A, Zvirbliene A, Begun A, et al. Twin study indicates loss of interaction between microbiota and mucosa of patients with ulcerative colitis. Gastroenterology. 2011;141:227–236. doi: 10.1053/j.gastro.2011.04.011. [DOI] [PubMed] [Google Scholar]
- Lind Due V, Bonde J, Kann T, Perner A. Extremely low oxygen tension in the rectal lumen of human subjects. Acta Anaesth Scand. 2003;47:372. doi: 10.1034/j.1399-6576.2003.00542.x. [DOI] [PubMed] [Google Scholar]
- Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489:220–230. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupp C, Robertson ML, Wickham ME, Sekirov I, Champion OL, Gaynor EC, et al. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe. 2007;2:204. doi: 10.1016/j.chom.2007.08.002. [DOI] [PubMed] [Google Scholar]
- Macho Fernandez E, Pot B, Grangette C. Beneficial effect of probiotics in IBD: are peptidogycan and NOD2 the molecular key effectors. Gut Microbes. 2011;2:280–286. doi: 10.4161/gmic.2.5.18255. [DOI] [PubMed] [Google Scholar]
- Mackie RI, Sghir A, Gaskins HR. Developmental microbial ecology of the neonatal gastrointestinal tract. Am J Clin Nutr. 1999;69:1035S–1045S. doi: 10.1093/ajcn/69.5.1035s. [DOI] [PubMed] [Google Scholar]
- Manichanh C, Rigottier-Gois L, Bonnaud E, Gloux K, Pelletier E, Frangeul L, et al. Reduced diversity of faecal microbiota in Crohn's disease revealed by a metagenomic approach. Gut. 2006;55:205–211. doi: 10.1136/gut.2005.073817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manichanh C, Borruel N, Casellas F, Guarner F. The gut microbiota in IBD. Nature reviews Gastroenterology & hepatology. 2012;9:599–608. doi: 10.1038/nrgastro.2012.152. [DOI] [PubMed] [Google Scholar]
- Meynell GG. Antibacterial mechanisms of the mouse gut. II. The role of Eh and volatile fatty acids in the normal gut. Br J Exper Pathol. 1963;44:209–219. [PMC free article] [PubMed] [Google Scholar]
- Mondot S, Kang S, Furet JP, Aguirre de Carcer D, McSweeney C, Morrison M, et al. Highlighting new phylogenetic specificities of Crohn's disease microbiota. Inflamm Bowel Dis. 2011;17:185–192. doi: 10.1002/ibd.21436. [DOI] [PubMed] [Google Scholar]
- Mukhopadhya I, Hansen R, El-Omar EM, Hold GL. IBD-what role do Proteobacteria play. Nat Rev Gastroenterol Hepatol. 2012;9:219–230. doi: 10.1038/nrgastro.2012.14. [DOI] [PubMed] [Google Scholar]
- Nagalingam NA, Lynch SV. Role of the microbiota in inflammatory bowel diseases. Inflamm Bowel Dis. 2012;18:968–984. doi: 10.1002/ibd.21866. [DOI] [PubMed] [Google Scholar]
- Naser SA, Ghobrial G, Romero C, Valentine JF. Culture of Mycobacterium avium subspecies paratuberculosis from the blood of patients with Crohn's disease. Lancet. 2004;364:1039–1044. doi: 10.1016/S0140-6736(04)17058-X. [DOI] [PubMed] [Google Scholar]
- Ott SJ, Musfeldt M, Wenderoth DF, Hampe J, Brant O, Folsch UR, et al. Reduction in diversity of the colonic mucosa associated bacterial microflora in patients with active inflammatory bowel disease. Gut. 2004;53:685–693. doi: 10.1136/gut.2003.025403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartor RB. Microbial influences in inflammatory bowel diseases. Gastroenterology. 2008;134:577–594. doi: 10.1053/j.gastro.2007.11.059. [DOI] [PubMed] [Google Scholar]
- Sekirov I, Russell SL, Antunes LC, Finlay BB. Gut microbiota in health and disease. Physiol Rev. 2010;90:859–904. doi: 10.1152/physrev.00045.2009. [DOI] [PubMed] [Google Scholar]
- Seksik P, Rigottier-Gois L, Gramet G, Sutren M, Pochart P, Marteau P, et al. Alterations of the dominant faecal bacterial groups in patients with Crohn's disease of the colon. Gut. 2003;52:237–242. doi: 10.1136/gut.52.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol H, Seksik P, Rigottier-Gois L, Lay C, Lepage P, Podglajen I, et al. Specificities of the fecal microbiota in inflammatory bowel disease. Inflamm Bowel Dis. 2006;12:106–111. doi: 10.1097/01.MIB.0000200323.38139.c6. [DOI] [PubMed] [Google Scholar]
- Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermudez-Humaran LG, Gratadoux JJ, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci USA. 2008;105:16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol H, Seksik P, Furet JP, Firmesse O, Nion-Larmurier I, Beaugerie L, et al. Low counts of Faecalibacterium prausnitzii in colitis microbiota. Inflamm Bowel Dis. 2009;15:1183–1189. doi: 10.1002/ibd.20903. [DOI] [PubMed] [Google Scholar]
- Swidsinski A, Loening-Baucke V, Theissig F, Engelhardt H, Bengmark S, Koch S, et al. Comparative study of the intestinal mucus barrier in normal and inflamed colon. Gut. 2007;56:343–350. doi: 10.1136/gut.2006.098160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swidsinski A, Loening-Baucke V, Verstraelen H, Osowska S, Doerffel Y. Biostructure of fecal microbiota in healthy subjects and patients with chronic idiopathic diarrhea. Gastroenterology. 2008;135:568–579. doi: 10.1053/j.gastro.2008.04.017. [DOI] [PubMed] [Google Scholar]
- Van Nuenen HMC.2005. Composition and Activity of Gut Microbiota in Inflammatory Bowel Disease. PhD thesis, Erasmus University Rotterdam, The Netherlands.
- Willing B, Halfvarson J, Dicksved J, Rosenquist M, Jarnerot G, Engstrand L, et al. Twin studies reveal specific imbalances in the mucosa-associated microbiota of patients with ileal Crohn's disease. Inflamm Bowel Dis. 2009;15:653–660. doi: 10.1002/ibd.20783. [DOI] [PubMed] [Google Scholar]
- Willing BP, Dicksved J, Halfvarson J, Andersson AF, Lucio M, Zheng Z, et al. A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology. 2010;139:1844–1854e1. doi: 10.1053/j.gastro.2010.08.049. [DOI] [PubMed] [Google Scholar]
- Winter SE, Thiennimitr P, Winter MG, Butler BP, Huseby DL, Crawford RW, et al. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature. 2010;467:426–429. doi: 10.1038/nature09415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xavier RJ, Podolsky DK. Unravelling the pathogenesis of inflammatory bowel disease. Nature. 2007;448:427–434. doi: 10.1038/nature06005. [DOI] [PubMed] [Google Scholar]
- Yong E.2012Gut microbial ‘enterotypes' become less clear-cut Nature News(e-pub ahead of print 21 March 2012doi: 10.1038/nature.2012.10276) [DOI]
- Zhu H, Li YR. Oxidative stress and redox signaling mechanisms of inflammatory bowel disease: updated experimental and clinical evidence. Exp Biol Med. 2012;237:474–480. doi: 10.1258/ebm.2011.011358. [DOI] [PubMed] [Google Scholar]