Key Points
Disease-causing mutations in GATA1 impair binding to the cofactors FOG1 or TAL1 but not DNA.
Different substitutions at the same residue selectively disrupt FOG1 or TAL1 binding leading to distinct disease phenotypes.
Abstract
Missense mutations in transcription factor GATA1 underlie a spectrum of congenital red blood cell and platelet disorders. We investigated how these alterations cause distinct clinical phenotypes by combining structural, biochemical, and genomic approaches with gene complementation systems that examine GATA1 function in biologically relevant cellular contexts. Substitutions that disrupt FOG1 cofactor binding impair both gene activation and repression and are associated with pronounced clinical phenotypes. Moreover, clinical severity correlates with the degree of FOG1 disruption. Surprisingly, 2 mutations shown to impair DNA binding of GATA1 in vitro did not measurably affect in vivo target gene occupancy. Rather, one of these disrupted binding to the TAL1 complex, implicating it in diseases caused by GATA1 mutations. Diminished TAL1 complex recruitment mainly impairs transcriptional activation and is linked to relatively mild disease. Notably, different substitutions at the same amino acid can selectively inhibit TAL1 complex or FOG1 binding, producing distinct cellular and clinical phenotypes. The structure-function relationships elucidated here were not predicted by prior in vitro or computational studies. Thus, our findings uncover novel disease mechanisms underlying GATA1 mutations and highlight the power of gene complementation assays for elucidating the molecular basis of genetic diseases.
Introduction
Erythrocyte and megakaryocyte development are under the control of transcription factor GATA1.1,2 GATA1 promotes differentiation by activating all known erythroid- and megakaryocyte-specific genes and silencing genes associated with the immature, proliferative state and alternative lineages (for review, see Ferreira et al3).
GATA1 contains 2 highly conserved zinc finger (ZF) domains. The C-terminal ZF primarily binds to the sequence (A/T)GATA(A/G) while the N-terminal ZF (NF) stabilizes DNA interactions by contacting noncanonical GATC and palindromic ATC(A/T)GATA(A/G) motifs.4-6 The NF also binds coregulators, including the multi-ZF protein FOG1.7 Like GATA1, FOG1 is required for erythroid and megakaryocyte development, and disrupting the GATA1-FOG1 interaction impairs maturation of these lineages.8-10 Activation and repression of most GATA1-regulated genes requires FOG1,11,12 as does silencing of mast cell–specific genes.13-15 FOG1 also modulates GATA1 chromatin occupancy at a subset of genomic sites.15-17 Additionally, the TAL1 complex, composed of TAL1, E2A, LMO2, and Ldb1, interacts via LMO2 with the GATA1 NF.18,19 TAL1, LMO2, and Ldb1 are essential for erythrocyte and megakaryocyte differentiation.20-22 TAL1 complex recruitment occurs predominantly at GATA1-activated genes and tends to be depleted at sites where GATA1 functions as a repressor.23,24 The distinct interaction surfaces of GATA1 that contact DNA, FOG1, and LMO2 have been defined previously.19,25,26
Missense mutations in the GATA1 NF cause distinct forms of congenital anemia and thrombocytopenia. Although similarly located, the 7 reported mutations produce a wide spectrum of phenotypes27-38 (for review, see Ciovacco et al39) (supplemental Table 1, available on the Blood website). Clinical severity depends on the site and type of substitution, and different substitutions at the same amino acid position produce disparate phenotypes. Broadly, the diseases fall into 2 categories: severe thrombocytopenia with pronounced anemia (V205M, G208R, D218Y) and moderate thrombocytopenia with minimal or no anemia (G208S, R216Q, R216W, D218G). There is also 1 case of congenital erythropoietic porphyria (CEP) associated with a R216W substitution. Five mutations lie on defined surfaces: R216Q and R216W sit on the DNA-binding face, while V205M, G208S, and G208R cluster on the FOG1-binding face (Figure 1A). D218G and D218Y fall outside these surfaces but diminish FOG1 binding in glutathione S-transferase–pulldown experiments.37,38,40 Structural and in vitro studies categorized GATA1 mutations into 2 groups, affecting either DNA or FOG1 binding. However, this classification fails to fully explain the degree of phenotypic variation caused by mutations on the same interaction face. For example, both R216Q and R216W are thought to disrupt DNA binding but the latter causes erythroid porphyria while the former does not.33,36 Similarly, it is unknown whether the disparate clinical phenotypes caused by different substitutions at residues G20828,29 and D21837,38 simply disrupt interaction with FOG1 to different extents or affect GATA1 function in qualitatively different ways. Moreover, D218 falls outside the known FOG1-binding surface, raising the possibility that this residue might connect to other GATA1 cofactors. Understanding how GATA1 mutations produce human diseases might enhance our understanding of molecular hematopoiesis and refine clinical care by linking prognosis and potential therapies to patient genotypes.
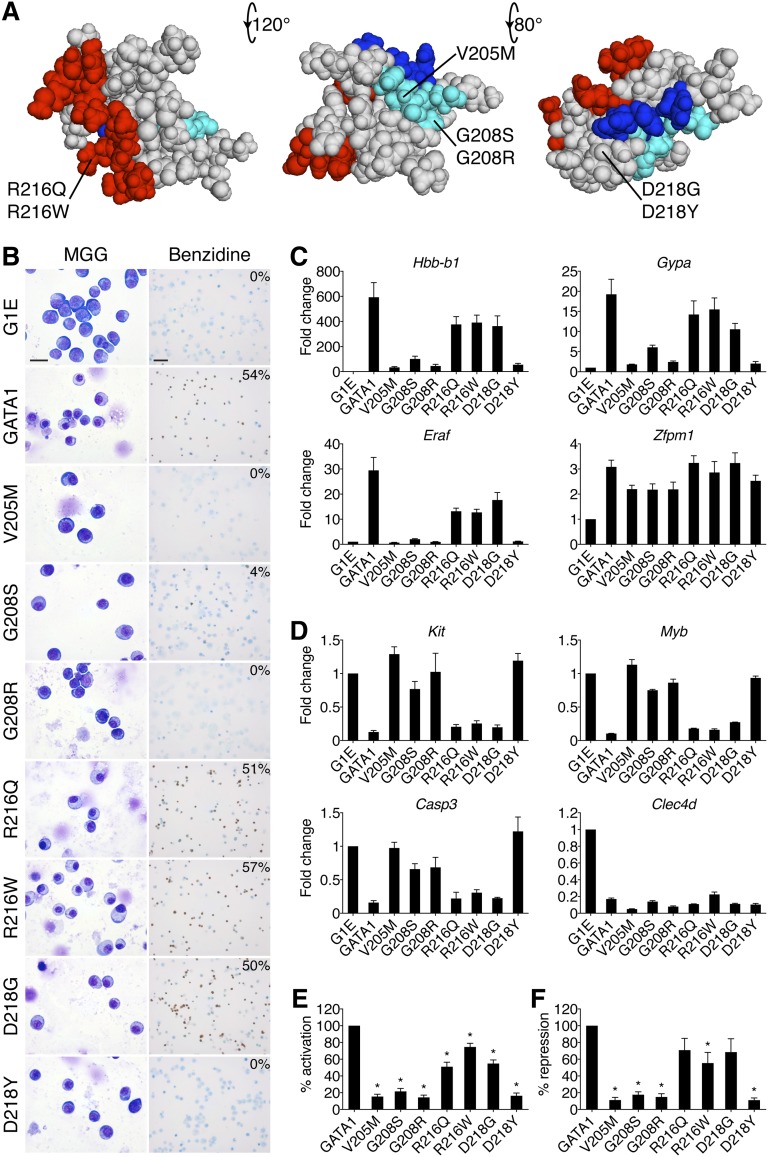
Figure 1.
Impairment of erythroid differentiation by GATA1 mutations. (A) Space-filling model of the GATA1 NF from PDB code 1Y0J with DNA-binding residues in red (based on PDB code 1GAT), FOG1-binding residues in cyan, and LMO2-interacting residues in blue. The locations of disease-associated mutations are noted. The middle structure has been rotated 120 degrees around a horizontal axis from the leftmost model, and the rightmost structure is rotated a further 80 degrees. (B) MGG and benzidine staining of G1E cells expressing wild-type or mutant GATA1 after 72 hours of E2 treatment. The percentage of hemoglobin-positive cells is indicated in the upper right corner of each benzidine panel. Scale bars, 20 μm (left panels) and 50 μm (right panels). (C-D) Expression of (C) GATA1-activated and (D) GATA1-repressed genes after 24 hours of E2 treatment as determined by RT-qPCR, normalized to β-actin and plotted as fold change from uninfected samples. (E-F) Average transcriptional profiles after 24 hours of E2 treatment of all activated (E, n = 24) and repressed (F, n = 6) genes examined. Note that the reduction in transcriptional repression by R216Q and D218G is not statistically significant. *P < .05. All error bars denote SEM (n = 3) unless otherwise noted. MGG, May-Grünwald-Giemsa; PDB, Protein Data Bank.
We examined mechanisms by which missense mutations alter GATA1 function using the G1E and G1ME systems, which enable the study of GATA1 mutants in their natural context at physiological expression levels. G1E cells are erythroid precursors that fail to mature owing to a lack of GATA1.41 Introducing a conditional form of GATA1 (GATA1 fused to the ligand-binding domain of the estrogen receptor [GATA1-ER]) imparts estradiol (E2)-dependent erythroid maturation in a manner largely reproducing that of wild-type cells.42,43 G1ME cells are GATA1-null bipotential megakaryocyte-erythroid progenitors that undergo terminal differentiation toward the erythroid and megakaryocytic lineages when reconstituted with GATA1 and grown in the appropriate cytokine environment.44
Using these cell-based systems, complemented by structural, biochemical, and transcriptome analyses, we comprehensively characterize the effect of missense mutations on GATA1 function, and uncover novel pathways underlying GATA1-mediated hematologic disorders.
Methods
Cell culture
G1E cells were cultured as described41 and treated with 100 nM E2 for 24 to 72 hours where indicated. G1ME cells were maintained as described44 in 1% thrombopoietin-conditioned medium. HEK-293 cells were cultured in Dulbecco modified Eagle medium with 10% fetal bovine serum, 2% penicillin-streptomycin, 1% glutamine, and 1% sodium pyruvate.
Retroviral infections
Retroviral infections were carried out as described.23,44 Briefly, viral particles were generated via transient transfection of Plat-E retrovirus packaging cells. Four milliliters of viral supernatant was mixed with 4 × 106 cells in the presence of 8 μg/mL polybrene and 10 mM HEPES (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid) and spun at 3200 rpm for 90 minutes at room temperature. Erythropoietin (2 U/mL) was added to G1ME transductions to support erythromegakaryocytic differentiation.
Morphologic analysis
Cells were stained with May-Grünwald-Giemsa (Sigma-Aldrich), benzidine for hemoglobin, or acetylcholinesterase, a megakaryocyte marker. Images were acquired with a Zeiss Axioskop 2 microscope, Zeiss Axiocam camera, and Zeiss AxioVision 4.8 software (Carl Zeiss MicroImaging).
Real-time quantitative polymerase chain reaction
RNA was extracted with TRIzol (Invitrogen) and reverse transcription performed using Superscript II (Invitrogen). Results were quantified using SYBR Green dye (Applied Biosystems) on an ABI Prism 7900HT. For primers, see supplemental Methods.
ChIP
Chromatin immunoprecipitation (ChIP) was performed as described.23 For anti-FOG1 ChIP, cells were crosslinked in 1.5 mM EGS for 20 minutes at room temperature before formaldehyde treatment. Antibodies used were GATA1 (sc-265; Santa Cruz Biotechnology), FOG1 (sc-9361; Santa Cruz Biotechnology), LMO2 (AF2726; R&D Systems), TAL1 (sc-12984; Santa Cruz Biotechnology). For primers, see supplemental Methods.
Gene expression profiling
TRIzol (Invitrogen) extracted RNA was purified with RNeasy mini columns (Qiagen). Hybridization (GeneChip Mouse Gene 1.0 ST Array; Affymetrix) and data analysis were performed at the University of Pennsylvania Microarray Core Facility. Data were imported into the Partek Genomics Suite (version 6.6; Partek Inc.) and normalized using RMA. After excluding control probe sets, principal components analysis and hierarchical clustering were performed. One-way analysis of variance across the 5 conditions (each with 3 replicates) and all 10 possible pairwise comparisons was performed, yielding a P value and fold change for all genes. P values were corrected for false discovery rate by the method of Benjamini and Hochberg.45 Genes with fold change ≥1.5 and P < .05 were considered differentially expressed. Microarray data are deposited at Gene Expression Omnibus under accession number GSE43356.
Results
Erythroid defects caused by GATA1 mutations are recapitulated in an erythroid gene complementation system
GATA1-ER fusions (from here on simply referred to as GATA1) were stably introduced into G1E cells. All proteins were expressed equivalently and at levels comparable to endogenous GATA1 (supplemental Figure 1A-B). Upon E2 treatment, wild-type GATA1 induced erythroid maturation as evidenced by morphology and staining with the hemoglobin dye benzidine (Figure 1B). The V205M, G208R, and D218Y versions of GATA1 were inactive while G208S displayed residual activity. The R216Q, R216W, and D218G mutations produced only subtle deficiencies (supplemental Figure 1C).
GATA1-induced morphologic transitions were reflected in gene expression changes (Figure 1C-F). Real-time quantitative polymerase chain reaction (RT-qPCR) analysis of erythroid GATA1 target genes after 24 hours of E2 treatment showed that generally, the V205M, G208S, G208R, and D218Y mutations reduced transcriptional activation (Figure 1C,E) and repression (Figure 1D,F) to <20% of wild type, while R216Q, R216W, and D218G caused more subtle defects. The G208R mutation impaired transcription more than its paired mutant G208S. Fundamentally similar gene expression patterns were observed at >30 genes (supplemental Table 2). Notably, the R216Q and D218G mutations impaired activation more than repression (Figure 1E-F). Gene expression changes were not simply a reflection of delayed maturation because they were also observed at 48 hours of E2 treatment (supplemental Figure 1E-F). Importantly, all mutant proteins properly regulated a subset of genes, including Zfpm1 and Clec4d, suggesting that missense mutations do not trigger a global misfolding of the NF. To examine this assumption more rigorously, we obtained 1-dimensional 1H NMR spectra of all mutants for which such analyses had not been performed. They revealed substantial tertiary structure in all mutant NFs (supplemental Figure 1D and Liew et al26), indicating that the functional consequences of missense mutations are not due to global misfolding.
Among GATA1 missense mutations, R216W is uniquely associated with CEP. Expression analysis of genes encoding heme biosynthetic enzymes revealed a ≥50% decrease in Alas2 and Uros levels in cells harboring the R216W mutation (supplemental Figure 2). Although reduced UROS activity causes CEP, to what extent the observed defect accounts for the patient phenotype remains unclear.46
In summary, the effects of GATA1 mutations on cellular morphology, hemoglobin concentrations, and gene expression profiles in G1E cells essentially mimic clinical erythroid phenotypes, validating G1E cells as an informative model system.
Megakaryocyte defects caused by GATA1 mutations are recapitulated in a megakaryocyte-erythroid gene complementation system
To test GATA1 mutants in a megakaryocytic context, we expressed them in G1ME cells (GATA1-ER fusions are not functional in these cells) and exposed cells to thrombopoietin and erythropoietin to support erythromegakaryocytic differentiation. All proteins were expressed equivalently and at levels comparable to endogenous GATA1 (supplemental Figure 3A). Wild-type GATA1 promoted megakaryocyte maturation, evidenced by large CD42-positive cells with multilobular nuclei and acetylcholinesterase staining (Figure 2A, supplemental Figure 3B-C). The V205M, G208S, G208R, and D218Y mutations caused a marked deficiency in megakaryocyte maturation, while R216Q, R216W, and D218G displayed only mild defects.
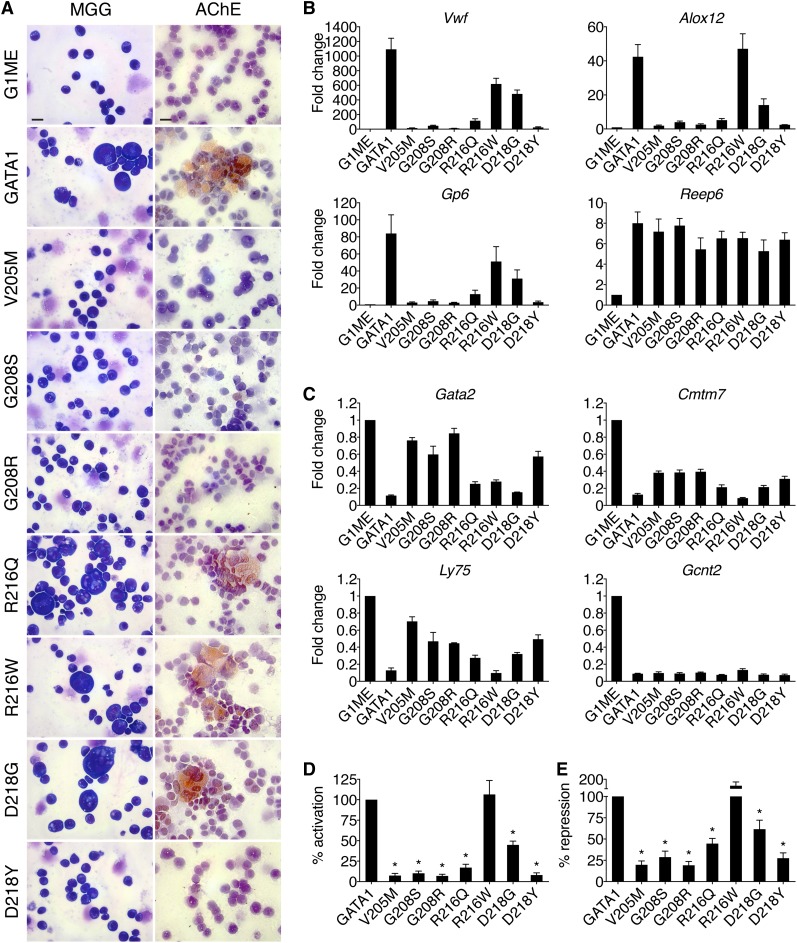
Figure 2.
Effects of GATA1 mutations on megakaryocytic maturation. (A) MGG and AChE staining of G1ME cells 72 hours after infection with wild-type or mutant GATA1-expressing vector. Scale bars, 20 μm. (B-C) Expression of (C) GATA1-activated and (D) GATA1-repressed genes in FACS-purified CD42-positive megakaryocytes as determined by RT-qPCR, normalized to β-actin and plotted as fold change from uninfected samples. (D-E) Average transcriptional activities (72 hours following transduction) of all examined activated (D, n = 10) and repressed (E, n = 6) genes. *P < .05. All error bars denote SEM (n = 3) unless otherwise noted. AChE, acetylcholinesterase.
Megakaryocytic GATA1 target genes were measured by RT-qPCR of fluorescence-activated cell sorter (FACS)-purified CD42-positive cells (Figure 2B-E). The V205M, G208S, G208R, and D218Y mutations reduced both activation (Figure 2B,D) and repression (Figure 2C,E) to ∼20% of that achieved by wild-type GATA1, while R216W had only a mild, gene-specific effect on activation. Although cells harboring the R216Q and D218G mutations displayed no gross morphological deficiencies, there were notable changes in transcriptional activities. Specifically, both mutants displayed an ∼60% loss of activation but only a 40% reduction in repression (Figure 2D-E). These gene expression changes may explain the thrombocytopenia reported in these patients and suggest that defects due to R216Q and D218G mutations manifest themselves at a later stage in megakaryocytic maturation or platelet production, which are not detected in this assay. The R216Q and D218G mutations affect transcription in megakaryocytes more strongly than in erythroid cells, suggesting differential sensitivities to diminished GATA1 function between the 2 cell lineages. This is consistent with the presentation of thrombocytopenia but minimal anemia in these patients. Essentially, the same patterns of misregulation were seen at >20 genes (supplemental Table 3). Importantly, all mutant proteins properly regulated a subset of target genes, including Reep6 and Gcnt2, indicating that no mutation completely abrogated GATA1 function in megakaryocytes.
The degree of thrombocytopenia in patients generally parallels the ability of mutant GATA1 proteins to promote terminal megakaryocyte maturation in G1ME cells as assessed by morphology, acetylcholinesterase staining, CD42 expression, and transcriptional profiles. In conclusion, in lieu of primary cells from patients, G1E and G1ME cells provide convenient, faithful, and robust systems in which to study GATA1 mutants.
A subset of GATA1 mutations diminish FOG1 binding
Except for V205M, studying the impact of GATA1 missense mutations on cofactor binding has been limited to in vitro protein association assays using select FOG1 ZFs but not the entire molecule.27,28,33,37,38,40 We compared the binding of all GATA1 mutants to full-length FOG1 by coimmunoprecipitation (co-IP) following expression in HEK-293 cells. The V205M, G208R, and D218Y mutations diminished FOG1 binding by ∼70%, 80%, and 50%, respectively, while G208S caused only a ∼20% reduction (Figure 3A-B). Proteins containing R216Q, R216W, or D218G mutations bound indistinguishably from wild type. Similar results were obtained with purified NF proteins examined by isothermal titration calorimetry (ITC)26 (supplemental Figure 4A, supplemental Table 4).
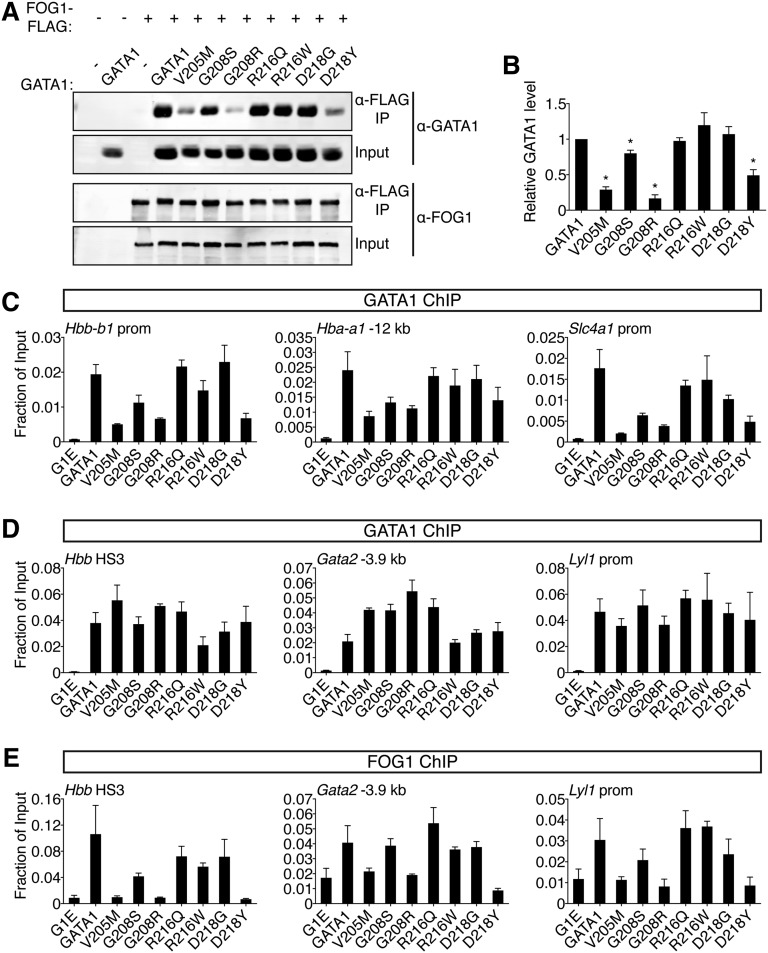
Figure 3.
Comparative analysis of GATA1 mutations on FOG1 binding. (A) Wild-type or mutant GATA1 was coexpressed with FLAG-tagged FOG1 in HEK-293 cells and analyzed by anti-FLAG IP followed by anti-GATA1 or anti-FOG1 western blotting. Input represents 5% of lysate. (B) Quantification of western blot signals. (C-D) Anti-GATA1 ChIP in G1E cells expressing indicated GATA1 versions after 24 hours of E2 treatment. (C) FOG1-dependent binding sites and (D) FOG1-independent binding sites that contain single (Hbb HS3, Gata2 -3.9 kb) or palindromic (Lyl1 prom) motifs and regulate activated (Hbb) or repressed (Gata2, Lyl1) genes. (E) Anti-FOG1 ChIP at FOG1-independent GATA1 binding sites. *P < .05. All error bars denote SEM (n = 3).
Because cofactor association is influenced by cellular context, we examined the ability of GATA1 mutants to recruit FOG1 to target genes in erythroid cells by ChIP. Importantly, the V205M mutation has been reported to diminish GATA1 chromatin occupancy at select sites.16 We found that V205M, G208S, G208R, and D218Y reduced GATA1 binding at these sites while the remaining mutations were innocuous in this regard (Figure 3C). To assay FOG1 association independently of GATA1 loading, we examined sites where GATA1 chromatin occupancy was FOG1-independent, including single and palindromic motifs near active and repressed genes (Figure 3D). Consistent with co-IP data, V205M, G208R, and D218Y severely diminished FOG1 recruitment, G208S had a mild impact, while R216Q, R216W, and D218G had little to no effect (Figure 3E, supplemental Figure 4B). Similar observations were made at >20 target sites (supplemental Table 5). Furthermore, as previously described,13-15 mutations that diminish the GATA1-FOG1 interaction caused aberrant activation of mast cell genes (supplemental Figure 4C).
Two results are especially notable. First, G208S and G208R differ in the degree to which they disrupt binding to FOG1, which mirrors the associated disease severity in patients and supports the notion that inhibition of FOG1 binding, and not another undefined GATA1 cofactor, accounts for the phenotype. Second, D218Y, which falls outside the known FOG1 interaction domain, inhibits FOG1 binding, while a glycine substitution at the same residue does not (see also below), implicating critical features in the mode of NF interactions that were not predicted by structural studies. In summary, disruption of FOG1 binding in vitro is matched by failure to recruit FOG1 in vivo. This diminishes GATA1 chromatin occupancy at select sites, affects both gene activation and repression, and generally produces the most pronounced defects in erythroid and megakaryocytic differentiation in cell-based assays and in patients.
GATA1 harboring DNA-binding surface mutations occupies target genes normally
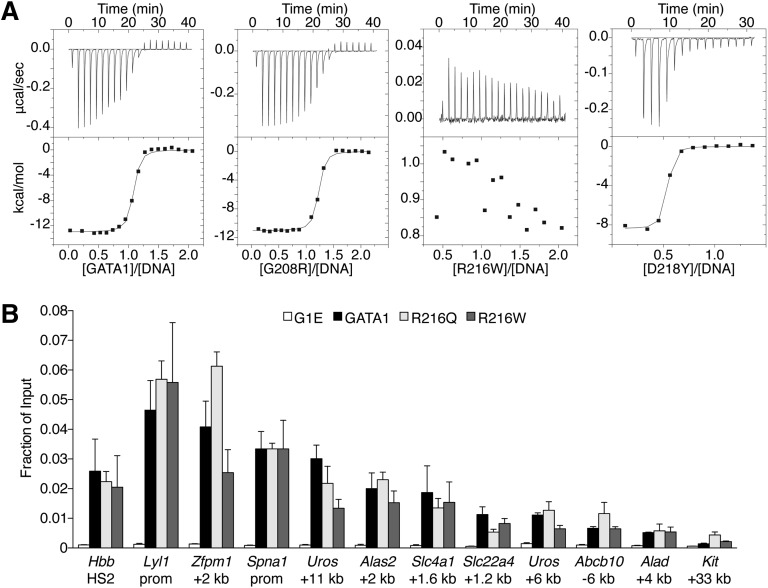
Because R216 contacts DNA in structural studies,25 it has been proposed that R216Q and R216W cause disease by disrupting GATA1 DNA binding.33,36 We measured by ITC the affinities of mutant NFs for a 16-bp oligonucleotide containing a GATC site26 (Figure 4A, supplemental Table 4). NF proteins with V205M, G208S, G208R, D218G, or D218Y mutations bound DNA with similar affinities as wild type. In contrast, R216Q or R216W mutants showed no measureable binding to DNA. Based on these and previous data,6,26,33 R216Q and R216W would be expected to disrupt GATA1 binding to palindromic and GATC motifs in vivo. Surprisingly, ChIP analysis revealed that GATA1 harboring R216Q or R216W mutations bound all examined target sites normally, including those containing palindromic motifs and regardless of whether strong or weaker GATA1 occupied sites were considered (Figures 3C-D, 4B). In accordance with in vitro results, V205M, G208S, G208R, D218G, and D218Y mutants showed normal GATA1 chromatin occupancy at all sites except those at which association with FOG1 is required (see above). Thus, in contrast to in vitro observations, mutations on the DNA-binding surface of the NF do not significantly impair GATA1 target site occupancy in vivo. Therefore, these mutations likely cause human disease through alternate mechanisms.
Figure 4.
Analysis of GATA1 mutations for DNA binding. (A) ITC data showing the titration of wild-type or indicated mutant versions of the GATA1 NF into a 16-bp oligonucleotide containing a GATC motif. (B) Anti-GATA1 ChIP in G1E cells expressing wild-type or mutant GATA1 after 24 hours of E2 treatment using primers spanning single (Hbb HS2) or palindromic (all others) motifs. All error bars denote SEM (n = 3).
R216Q and D218G mutations specifically diminish TAL1 complex binding resulting in overlapping gene expression signatures
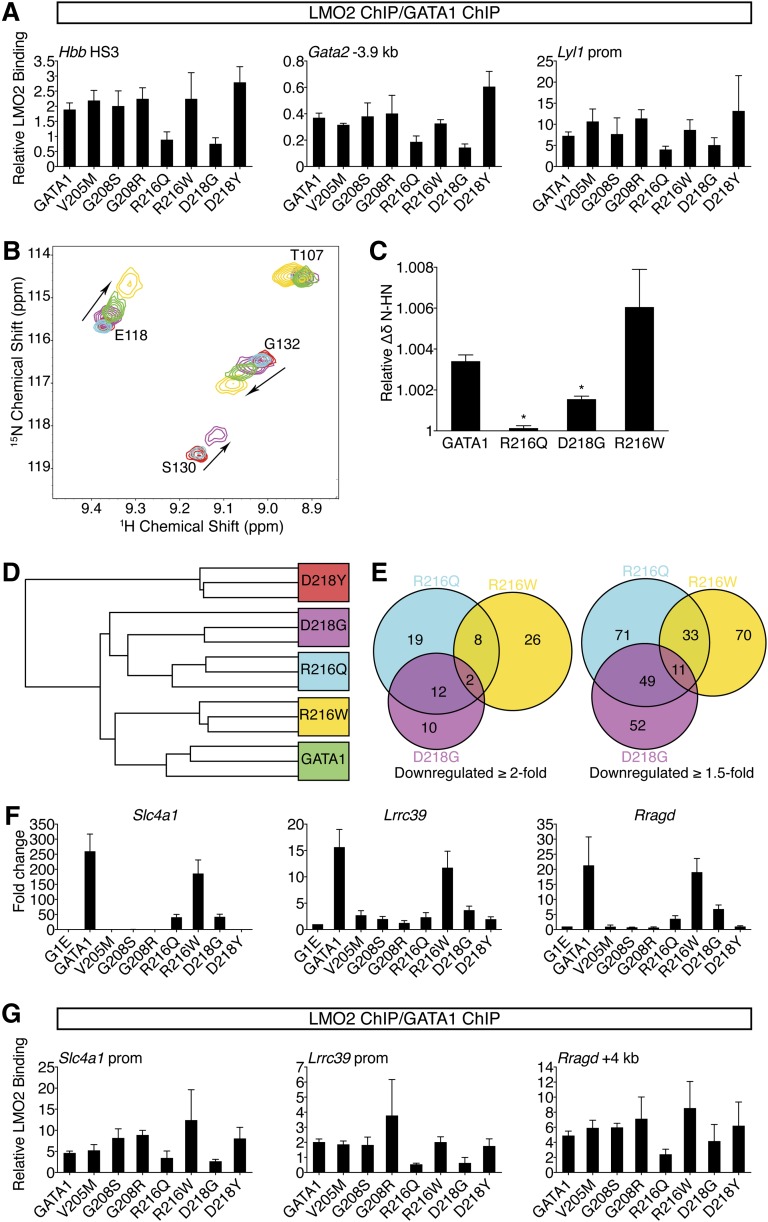
Because the R216Q, R216W, and D218G mutations do not measurably disrupt FOG1 recruitment or DNA binding in vivo, we examined whether they affect association with LMO2, a member of the TAL1 complex, because it interacts directly with the NF in a manner permitting simultaneous FOG1 binding.19 Although no NF mutations affect residues known to bind LMO2, R217, which is flanked by R216 and D218, contributes to the GATA1-LMO2 interaction.19 ChIP analysis revealed that R216Q and D218G mutations diminished LMO2 occupancy at GATA1 target genes (supplemental Figure 5A-B). This trend was more apparent when the LMO2 ChIP signals were normalized to those of GATA1 (Figure 5A). None of the remaining mutations measurably impaired LMO2 recruitment. The same binding profiles were observed for TAL1 (supplemental Figure 5C), suggesting the entire TAL1 complex is recruited less efficiently by the R216Q or D218G mutants. Similar occupancy patterns were observed at >20 target sites (supplemental Table 5). These results were surprising given that R216Q falls on the DNA-binding surface and D218Y disrupts FOG1 binding. However, a shared molecular defect is consistent with the similar clinical phenotypes caused by R216Q and D218G mutations. Furthermore, a qualitative difference in LMO2 binding between R216Q and R216W, and between D218G and D218Y, might explain each pair’s divergent patient presentations. Notably, disruption of LMO2 binding by R216Q and D218G did not reduce GATA1 chromatin occupancy at any site (Figures 3C-D, 4B), suggesting that unlike FOG1, association with LMO2 is not required for stable binding of GATA1 to its target genes. Disruption of LMO2 recruitment impinges on gene activation more so than on repression (Figures 1E-F, 2D-E, supplemental Figure 4C), consistent with the TAL1 complex predominantly occupying GATA1-activated genes.23,24
Figure 5.
R216Q and D218G mutations disrupt LMO2 binding and produce unique transcriptional signatures. (A) Anti-GATA1 and anti-LMO2 ChIP in G1E cells expressing wild-type or mutant GATA1 after 24 hours of E2 treatment using primers as in Figure 3D and E. LMO2 ChIP signals were normalized to GATA1 ChIP signals at each site. Error bars denote SEM (n = 3). (B) A portion of the 15N-HSQC spectra of 15N-LMO2LIM2-Ldb1LID (red peaks) following addition of 1 equivalent of either GATA1 NF (green), R216Q (cyan), R216W (gold), or D218G (purple). R216W caused peak shifts similar to those induced by wild type, while D218G induced qualitatively similar shifts that were smaller in magnitude, and R216Q did not result in significant shifts to any peaks. (C) Relative weighted average change in chemical shift position of resonances from LMO2LIM2-Ldb1LID (B) following addition of wild-type or mutant GATA1 NF. Shown are the average shifts (±SD) of 3 separate resonances for each series of titrations. *P < .05. (D) Unsupervised hierarchical clustering of G1E cells expressing wild-type or mutant GATA1 based on expression profiling with microarrays. One D218G replicate is indistinguishable from the R216Q replicates. (E) Venn diagrams of direct GATA1-activated target genes significantly downregulated when compared with wild-type GATA1. (F) Expression of GATA1-regulated genes highly sensitive to TAL1 complex disruption was validated by RT-qPCR, normalized to β-actin, and plotted as fold change from uninfected samples. Error bars denote SEM (n = 3). (G) Anti-GATA1 and anti-LMO2 ChIP in G1E cells expressing wild-type or mutant GATA1 after 24 hours of E2 treatment using primers against genes significantly impaired in response to R216Q and D218G mutations. LMO2 ChIP signals were normalized to GATA1 ChIP signals at each site. Error bars denote SEM (n = 3).
Due to the low affinity of the GATA1-LMO2 interaction,19 co-IP was unreliable for comparing the GATA1 mutants (not shown), prompting us to turn to 15N-HSQC spectroscopy. While R216W and D218Y did not affect the GATA1 NF-LMO2 interaction, D218G reduced, and R216Q markedly reduced, binding affinity (Figure 5B-C and data not shown). This supports our in vivo results showing that R216Q and D218G inhibit recruitment of LMO2 to GATA1-bound genes.
To strengthen our finding that R216Q and D218G affect LMO2 binding, we compared them to 2 GATA1 substitutions (R202Q and R217M) previously shown to diminish LMO2 binding in vitro.19 In G1E cells, GATA1 harboring R202Q or R217M induced erythroid maturation and transcriptional activities almost perfectly matching the R216Q and D218G mutants (supplemental Figure 6A-D). ChIP confirmed that R202Q and R217M impaired recruitment of LMO2 but not FOG1 (supplemental Figure 6E-F). This strongly supports the idea that LMO2 disruption by R216Q and D218G mutations accounts for their disease-causing effects, implicating for the first time the TAL1 complex in the pathogenesis of disorders caused by GATA1 mutations.
To obtain a broad and unbiased comparison of wild-type GATA1 and paired mutants that affect LMO2 binding (R216Q and D218G) and those that do not (R216W and D218Y), we examined transcriptomes by microarray. Hierarchical clustering analysis revealed that R216Q and D218G were highly similar (Figure 5D). Notably, there was substantial overlap among genes affected at least twofold by the R216Q or D218G mutations that exceeded the R216Q and R216W overlap (Figure 5E). ChIP confirmed diminished LMO2 occupancy at genes affected by the R216Q and D218G mutations (Figure 5F-G). Thus, gene profiling cemented a shared mechanism of action for the R216Q and D218G mutations, namely disruption of the TAL1 complex interaction.
Global expression changes induced by GATA1 harboring R216W are most similar to those of wild type (Figure 5D), suggesting that alterations caused by this mutation are subtle. We note that genes severely misregulated by R216W mutation show no defect in GATA1 occupancy (supplemental Figure 6G), further supporting the conclusion that the R216W mutation impairs functions other than DNA binding.
Discussion
We used gene complementation systems accompanied by structural, biochemical, and transcriptional studies to examine mechanisms by which missense mutations in the NF alter GATA1 function and lead to hematologic disease (for a summary of all results, see supplemental Table 6). Earlier work on GATA1 mutations relied mostly on in vitro studies,27,28,33,37,38,40 with only 2 mutants (V205M and R216Q) examined in cell-based systems.16,27,33 The G1E and G1ME gene complementation assays mimic patient phenotypes quite faithfully. They enable ChIP and biochemical experiments by affording a homogenous cellular and genetic background allowing detection of subtle defects that may be masked in studies of heterogeneous patient material. Despite their usefulness, these systems have some limitations as reflected in subtle phenotypic discrepancies between cells and patients. The G208S mutation causes mild dyserythropoiesis without anemia in patients but in G1E cells produces more pronounced defects (this study and Mehaffey et al28). Furthermore, mild thalassemia seen in some R216Q patients is not recapitulated in G1E cells (this study and Yu et al33). These variations might be rooted in species differences or the fact that these systems do not recapitulate all stages of erythromegakaryocytic maturation. Nonetheless, through these systems we were able to uncover previously unappreciated mechanisms for GATA1 missense mutations that were not predicted from in vitro studies and structural considerations. We attempted generating induced pluripotent stem cells from select patients to be studied by in vitro erythroid differentiation. However, this approach is hampered by the failure to generate sufficient quantities of definitive erythroid cells and substantial variation among induced pluripotent stem cell clones, rendering the study of subtle phenotypic variations a challenge.
Given strong in vitro evidence that mutations in the NF DNA-binding surface (R216Q and R216W) impair binding to palindromic or GATC motifs (this study and Yu et al33), it was surprising that they did not affect in vivo GATA1 target site occupancy. Similar discrepancies in the context of a distinct GATA1 mutant have been noted.47 It remains possible that subtle deficiencies in DNA binding, should they exist, are missed by ChIP-qPCR or are revealed only at sites not included in our analysis. Genome-wide localization of these mutants should address these points in future studies. It is also possible that cofactor complexes surrounding GATA1 help stabilize it within chromatin, thereby compensating for the loss of direct DNA binding by the NF, or that the NF simply does not contribute to DNA binding in vivo as much as originally assumed. Nevertheless, this apparent discordance highlights the importance of validating in vitro studies with in vivo experiments. Importantly, our studies provide an alternative explanation for R216Q, which does not impair DNA binding as previously predicted, but rather inhibits TAL1 complex recruitment to GATA1-occupied genes.
How R216W affects the function of GATA1 remains unclear, as we failed to observe any measurable impact of R216W on DNA, FOG1, or TAL1 complex binding. Also, there was no measurable impact on the occupancy of GATA1 cofactors CBP/p300 and Cdk9 (data not shown). Gene expression analyses provided no immediate clue as to the causes of CEP other than an ∼50% decrease in Uros expression. However, because CEP in humans is associated with much less than 50% UROS activity, this is unlikely to account for the patient phenotype. Although it is possible that a lack of any major identifiable deficiencies in R216W reflects a limitation of our assays, it is important to note that the R216W mutation was identified in a single patient with additional mutations and comorbidities, leaving open the possibility that the effects of the R216W mutation are influenced by genetic modifiers.
An unexpected finding was that R216Q and D218G disrupt the interaction between GATA1 and the TAL1 cofactor complex. Relatively few genes are misregulated by these mutations, in agreement with prior work demonstrating limited alterations upon disruption of this complex23 and in contrast to FOG1 disruption which alters the expression of substantial numbers of genes (data not shown and Johnson et al12). An explanation for the few changes in gene expression and hence mild clinical presentation is that they do not impair GATA1 chromatin occupancy or trigger lineage-inappropriate gene expression.
A remarkable finding was that distinct substitutions at a single residue can lead to not only quantitative changes (G208S vs G208R) but also qualitative differences (D218G vs D218Y and R216Q vs R216W). We speculate that each substitution in R216 or D218 elicits unique phenotypes as a result of different physiochemical properties. Although both R216 mutations remove the positively charged side chain, glutamine substitution trims down the side chain whereas tryptophan substitution introduces a bulky residue that has the potential to form stabilizing cation-π interactions. Thus, the smaller R216Q likely results in minor rearrangements that shift the GATA1-binding surface away from LMO2, whereas the bulkier R216W is able to maintain the surface. Similarly, although both D218 mutations eliminate key hydrogen bonds that likely cause focal changes to the NF structure, conversion to glycine introduces backbone flexibility that could disrupt the LMO2-binding site, whereas mutation to tyrosine introduces a bulky side chain that could preserve the LMO2-binding surface but would necessitate some repacking of the surrounding side chains, disrupting the nearby FOG1-binding surface (supplemental Figure 7).
Notably, GATA1 mutations that affect TAL1 complex binding neither measurably diminish nor increase FOG1 recruitment and vice versa, arguing that these proteins do not mutually stabilize or interfere with each other as has been suggested by co-IP experiments48 and consistent with in vitro data showing simultaneous binding of FOG1 and LMO2 to the GATA1 NF.19
Our work sheds new light on the pathophysiology and mechanisms underlying congenital hematologic diseases caused by GATA1 mutations. This deeper understanding might improve classification, refine clinical management of affected patients, and allow prediction of disease progression based on molecular defect. More broadly, we provide a paradigm for better understanding disease-causing mutations through a combined modality approach. Additional congenital alterations in GATA1 outside the NF have been described.49-51 These affect the N-terminal and C-terminal portions of GATA1 but potential cofactor interactions of these domains as well as contributions of these alterations to protein stability remain unclear. Because mechanisms predicted by in vitro studies may not always reflect events in biologically relevant contexts, in vivo studies such as those reported here serve to not only validate predictions but also provide entirely new and unexpected insights. Our approach could be applied in other contexts where different substitutions at the same amino acid position lead to variable clinical presentations and underlying effects on protein function are incompletely understood.
Supplementary Material
Acknowledgments
The authors thank Dr Mitch Weiss and members of the Blobel laboratory for helpful comments on the manuscript. The authors are grateful to Dr Robert J. Desnick for his insights regarding porphyria.
This work was supported by grants from the National Institutes of Health (5R37DK058044 [G.A.B.] and T32 DK07780 [A.E.C.]) and Senior Research Fellowships from the Australian National Health and Medical Research Council (J.P.M. and J.M.M).
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: A.E.C. and G.A.B. conceived the study; A.E.C., L.W.-W., J.P.M., J.M.M., and G.A.B. designed experiments and analyzed data; A.E.C. and L.W.-W. performed experiments; and A.E.C., J.M.M., and G.A.B. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Gerd A. Blobel, The Children’s Hospital of Philadelphia, Abramson Building, Suite 316H, 3615 Civic Center Blvd, Philadelphia, PA 19104-4318; e-mail: blobel@email.chop.edu.
References
- 1.Fujiwara Y, Browne CP, Cunniff K, Goff SC, Orkin SH. Arrested development of embryonic red cell precursors in mouse embryos lacking transcription factor GATA-1. Proc Natl Acad Sci U S A. 1996;93(22):12355–12358. doi: 10.1073/pnas.93.22.12355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shivdasani RA, Fujiwara Y, McDevitt MA, Orkin SH. A lineage-selective knockout establishes the critical role of transcription factor GATA-1 in megakaryocyte growth and platelet development. EMBO J. 1997;16(13):3965–3973. doi: 10.1093/emboj/16.13.3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ferreira R, Ohneda K, Yamamoto M, Philipsen S. GATA1 function, a paradigm for transcription factors in hematopoiesis. Mol Cell Biol. 2005;25(4):1215–1227. doi: 10.1128/MCB.25.4.1215-1227.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martin DI, Orkin SH. Transcriptional activation and DNA binding by the erythroid factor GF-1/NF-E1/Eryf 1. Genes Dev. 1990;4(11):1886–1898. doi: 10.1101/gad.4.11.1886. [DOI] [PubMed] [Google Scholar]
- 5.Trainor CD, Omichinski JG, Vandergon TL, Gronenborn AM, Clore GM, Felsenfeld G. A palindromic regulatory site within vertebrate GATA-1 promoters requires both zinc fingers of the GATA-1 DNA-binding domain for high-affinity interaction. Mol Cell Biol. 1996;16(5):2238–2247. doi: 10.1128/mcb.16.5.2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Newton A, Mackay J, Crossley M. The N-terminal zinc finger of the erythroid transcription factor GATA-1 binds GATC motifs in DNA. J Biol Chem. 2001;276(38):35794–35801. doi: 10.1074/jbc.M106256200. [DOI] [PubMed] [Google Scholar]
- 7.Tsang AP, Visvader JE, Turner CA, et al. FOG, a multitype zinc finger protein, acts as a cofactor for transcription factor GATA-1 in erythroid and megakaryocytic differentiation. Cell. 1997;90(1):109–119. doi: 10.1016/s0092-8674(00)80318-9. [DOI] [PubMed] [Google Scholar]
- 8.Tsang AP, Fujiwara Y, Hom DB, Orkin SH. Failure of megakaryopoiesis and arrested erythropoiesis in mice lacking the GATA-1 transcriptional cofactor FOG. Genes Dev. 1998;12(8):1176–1188. doi: 10.1101/gad.12.8.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crispino JD, Lodish MB, MacKay JP, Orkin SH. Use of altered specificity mutants to probe a specific protein-protein interaction in differentiation: the GATA-1:FOG complex. Mol Cell. 1999;3(2):219–228. doi: 10.1016/s1097-2765(00)80312-3. [DOI] [PubMed] [Google Scholar]
- 10.Chang AN, Cantor AB, Fujiwara Y, et al. GATA-factor dependence of the multitype zinc-finger protein FOG-1 for its essential role in megakaryopoiesis. Proc Natl Acad Sci U S A. 2002;99(14):9237–9242. doi: 10.1073/pnas.142302099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Muntean AG, Crispino JD. Differential requirements for the activation domain and FOG-interaction surface of GATA-1 in megakaryocyte gene expression and development. Blood. 2005;106(4):1223–1231. doi: 10.1182/blood-2005-02-0551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson KD, Boyer ME, Kang J-A, Wickrema A, Cantor AB, Bresnick EH. Friend of GATA-1-independent transcriptional repression: a novel mode of GATA-1 function. Blood. 2007;109(12):5230–5233. doi: 10.1182/blood-2007-02-072983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cantor AB, Iwasaki H, Arinobu Y, et al. Antagonism of FOG-1 and GATA factors in fate choice for the mast cell lineage. J Exp Med. 2008;205(3):611–624. doi: 10.1084/jem.20070544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gregory GD, Miccio A, Bersenev A, et al. FOG1 requires NuRD to promote hematopoiesis and maintain lineage fidelity within the megakaryocytic-erythroid compartment. Blood. 2010;115(11):2156–2166. doi: 10.1182/blood-2009-10-251280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chlon TM, Doré LC, Crispino JD. Cofactor-mediated restriction of GATA-1 chromatin occupancy coordinates lineage-specific gene expression. Mol Cell. 2012;47(4):608–621. doi: 10.1016/j.molcel.2012.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Letting DL, Chen Y-Y, Rakowski C, Reedy S, Blobel GA. Context-dependent regulation of GATA-1 by friend of GATA-1. Proc Natl Acad Sci U S A. 2004;101(2):476–481. doi: 10.1073/pnas.0306315101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pal S, Cantor AB, Johnson KD, et al. Coregulator-dependent facilitation of chromatin occupancy by GATA-1. Proc Natl Acad Sci U S A. 2004;101(4):980–985. doi: 10.1073/pnas.0307612100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wadman IA, Osada H, Grütz GG, et al. The LIM-only protein Lmo2 is a bridging molecule assembling an erythroid, DNA-binding complex which includes the TAL1, E47, GATA-1 and Ldb1/NLI proteins. EMBO J. 1997;16(11):3145–3157. doi: 10.1093/emboj/16.11.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilkinson-White L, Gamsjaeger R, Dastmalchi S, et al. Structural basis of simultaneous recruitment of the transcriptional regulators LMO2 and FOG1/ZFPM1 by the transcription factor GATA1. Proc Natl Acad Sci U S A. 2011;108(35):14443–14448. doi: 10.1073/pnas.1105898108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Porcher C, Swat W, Rockwell K, Fujiwara Y, Alt FW, Orkin SH. The T cell leukemia oncoprotein SCL/tal-1 is essential for development of all hematopoietic lineages. Cell. 1996;86(1):47–57. doi: 10.1016/s0092-8674(00)80076-8. [DOI] [PubMed] [Google Scholar]
- 21.Yamada Y, Warren AJ, Dobson C, Forster A, Pannell R, Rabbitts TH. The T cell leukemia LIM protein Lmo2 is necessary for adult mouse hematopoiesis. Proc Natl Acad Sci U S A. 1998;95(7):3890–3895. doi: 10.1073/pnas.95.7.3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li L, Lee JY, Gross J, Song SH, Dean A, Love PE. A requirement for Lim domain binding protein 1 in erythropoiesis. J Exp Med. 2010;207(12):2543–2550. doi: 10.1084/jem.20100504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tripic T, Deng W, Cheng Y, et al. SCL and associated proteins distinguish active from repressive GATA transcription factor complexes. Blood. 2009;113(10):2191–2201. doi: 10.1182/blood-2008-07-169417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheng Y, Wu W, Kumar SA, et al. Erythroid GATA1 function revealed by genome-wide analysis of transcription factor occupancy, histone modifications, and mRNA expression. Genome Res. 2009;19(12):2172–2184. doi: 10.1101/gr.098921.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Omichinski JG, Clore GM, Schaad O, et al. NMR structure of a specific DNA complex of Zn-containing DNA binding domain of GATA-1. Science. 1993;261(5120):438–446. doi: 10.1126/science.8332909. [DOI] [PubMed] [Google Scholar]
- 26.Liew CK, Simpson RJY, Kwan AHY, et al. Zinc fingers as protein recognition motifs: structural basis for the GATA-1/friend of GATA interaction. Proc Natl Acad Sci U S A. 2005;102(3):583–588. doi: 10.1073/pnas.0407511102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nichols KE, Crispino JD, Poncz M, et al. Familial dyserythropoietic anaemia and thrombocytopenia due to an inherited mutation in GATA1. Nat Genet. 2000;24(3):266–270. doi: 10.1038/73480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mehaffey MG, Newton AL, Gandhi MJ, Crossley M, Drachman JG. X-linked thrombocytopenia caused by a novel mutation of GATA-1. Blood. 2001;98(9):2681–2688. doi: 10.1182/blood.v98.9.2681. [DOI] [PubMed] [Google Scholar]
- 29.Del Vecchio GC, Giordani L, De Santis A, De Mattia D. Dyserythropoietic anemia and thrombocytopenia due to a novel mutation in GATA-1. Acta Haematol. 2005;114(2):113–116. doi: 10.1159/000086586. [DOI] [PubMed] [Google Scholar]
- 30.Kratz CP, Niemeyer CM, Karow A, Volz-Fleckenstein M, Schmitt-Gräff A, Strahm B. Congenital transfusion-dependent anemia and thrombocytopenia with myelodysplasia due to a recurrent GATA1(G208R) germline mutation. Leukemia. 2008;22(2):432–434. doi: 10.1038/sj.leu.2404904. [DOI] [PubMed] [Google Scholar]
- 31.Dührsen U, Kratz CP, Flotho C, et al. Long-term outcome of hemizygous and heterozygous carriers of a germline GATA1 (G208R) mutation. Ann Hematol. 2011;90(3):301–306. doi: 10.1007/s00277-010-1088-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raskind WH, Niakan KK, Wolff J, et al. Mapping of a syndrome of X-linked thrombocytopenia with thalassemia to band Xp11-12: further evidence of genetic heterogeneity of X-linked thrombocytopenia. Blood. 2000;95(7):2262–2268. [PubMed] [Google Scholar]
- 33.Yu C, Niakan KK, Matsushita M, Stamatoyannopoulos G, Orkin SH, Raskind WH. X-linked thrombocytopenia with thalassemia from a mutation in the amino finger of GATA-1 affecting DNA binding rather than FOG-1 interaction. Blood. 2002;100(6):2040–2045. doi: 10.1182/blood-2002-02-0387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Balduini CL, Pecci A, Loffredo G, et al. Effects of the R216Q mutation of GATA-1 on erythropoiesis and megakaryocytopoiesis. Thromb Haemost. 2004;91(1):129–140. doi: 10.1160/TH03-05-0290. [DOI] [PubMed] [Google Scholar]
- 35.Tubman VN, Levine JE, Campagna DR, et al. X-linked gray platelet syndrome due to a GATA1 Arg216Gln mutation. Blood. 2007;109(8):3297–3299. doi: 10.1182/blood-2006-02-004101. [DOI] [PubMed] [Google Scholar]
- 36.Phillips JD, Steensma DP, Pulsipher MA, Spangrude GJ, Kushner JP. Congenital erythropoietic porphyria due to a mutation in GATA1: the first trans-acting mutation causative for a human porphyria. Blood. 2007;109(6):2618–2621. doi: 10.1182/blood-2006-06-022848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Freson K, Devriendt K, Matthijs G, et al. Platelet characteristics in patients with X-linked macrothrombocytopenia because of a novel GATA1 mutation. Blood. 2001;98(1):85–92. doi: 10.1182/blood.v98.1.85. [DOI] [PubMed] [Google Scholar]
- 38.Freson K, Matthijs G, Thys C, et al. Different substitutions at residue D218 of the X-linked transcription factor GATA1 lead to altered clinical severity of macrothrombocytopenia and anemia and are associated with variable skewed X inactivation. Hum Mol Genet. 2002;11(2):147–152. doi: 10.1093/hmg/11.2.147. [DOI] [PubMed] [Google Scholar]
- 39.Ciovacco WA, Raskind WH, Kacena MA. Human phenotypes associated with GATA-1 mutations. Gene. 2008;427(1-2):1–6. doi: 10.1016/j.gene.2008.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Freson K, Thys C, Wittewrongel C, Vermylen J, Hoylaerts MF, Van Geet C. Molecular cloning and characterization of the GATA1 cofactor human FOG1 and assessment of its binding to GATA1 proteins carrying D218 substitutions. Hum Genet. 2003;112(1):42–49. doi: 10.1007/s00439-002-0832-1. [DOI] [PubMed] [Google Scholar]
- 41.Weiss MJ, Yu C, Orkin SH. Erythroid-cell-specific properties of transcription factor GATA-1 revealed by phenotypic rescue of a gene-targeted cell line. Mol Cell Biol. 1997;17(3):1642–1651. doi: 10.1128/mcb.17.3.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Welch JJ, Watts JA, Vakoc CR, et al. Global regulation of erythroid gene expression by transcription factor GATA-1. Blood. 2004;104(10):3136–3147. doi: 10.1182/blood-2004-04-1603. [DOI] [PubMed] [Google Scholar]
- 43.Pilon AM, Ajay SS, Kumar SA, et al. NISC Comparative Sequencing Center. Genome-wide ChIP-Seq reveals a dramatic shift in the binding of the transcription factor erythroid Kruppel-like factor during erythrocyte differentiation. Blood. 2011;118(17):e139–e148. doi: 10.1182/blood-2011-05-355107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stachura DL, Chou ST, Weiss MJ. Early block to erythromegakaryocytic development conferred by loss of transcription factor GATA-1. Blood. 2006;107(1):87–97. doi: 10.1182/blood-2005-07-2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57(1):289–300. [Google Scholar]
- 46.Balwani M, Desnick RJ. The porphyrias: advances in diagnosis and treatment. Blood. 2012;120(23):4496–4504. doi: 10.1182/blood-2012-05-423186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lamonica JM, Vakoc CR, Blobel GA. Acetylation of GATA-1 is required for chromatin occupancy. Blood. 2006;108(12):3736–3738. doi: 10.1182/blood-2006-07-032847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rodriguez P, Bonte E, Krijgsveld J, et al. GATA-1 forms distinct activating and repressive complexes in erythroid cells. EMBO J. 2005;24(13):2354–2366. doi: 10.1038/sj.emboj.7600702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hollanda LM, Lima CSP, Cunha AF, et al. An inherited mutation leading to production of only the short isoform of GATA-1 is associated with impaired erythropoiesis. Nat Genet. 2006;38(7):807–812. doi: 10.1038/ng1825. [DOI] [PubMed] [Google Scholar]
- 50.Sankaran VG, Ghazvinian R, Do R, et al. Exome sequencing identifies GATA1 mutations resulting in Diamond-Blackfan anemia. J Clin Invest. 2012;122(7):2439–2443. doi: 10.1172/JCI63597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Singleton BK, Roxby DJ, Stirling JW, et al. A novel GATA1 mutation (Stop414Arg) in a family with the rare X-linked blood group Lu(a-b-) phenotype and mild macrothrombocytic thrombocytopenia. Br J Haematol. 2013;161(1):139–142. doi: 10.1111/bjh.12184. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.