Abstract
In response to sodium butyrate and trichostatin A treatment in erythroid cells, p38 mitogen activated protein kinase (MAPK) mediates fetal hemoglobin induction by activating cAMP response element binding protein 1 (CREB1). To expand on this observation, we completed studies to determine the role of p38 MAPK in steady-state γ-globin regulation. We propose that p38 signaling regulates Gγ-globin transcription during erythroid maturation through its downstream effector CREB1 which binds the Gγglobin cAMP response element (G-CRE). We demonstrated that a loss of p38 or CREB1 function by siRNA knockdown resulted in target gene silencing. Moreover, gain of p38 or CREB1 function augments γ-globin transcription. These regulatory effects were conserved under physiological conditions tested in primary erythroid cells. When the G-CRE was mutated in a stable chromatin environment Gγ-globin promoter activity was nearly abolished. Furthermore, introduction of mutations in the G-CRE abolished Gγ-globin activation via p38 MAPK/CREB1 signaling. Chromatin immunoprecipitation assays (ChIP) demonstrated that CREB1 and its binding partner CREB binding protein (CBP) co-localize at the G-CRE region. These data support the role of p38 MAPK/CREB1 signaling in Gγ-globin gene transcription under steady-state conditions.
Keywords: γ-globin, p38 MAPK, CREB1, globin gene regulation
INTRODUCTION
A family of Ser/Thr kinases called the MAPKs has members including p38, ERK and cJun amino terminal kinase (JNK) [1; 2]. Although initially identified as a protein kinase activated by stress, p38 MAPK signaling coordinates cellular responses during erythropoiesis [3]. For instance, p38 MAPK is essential for the synthesis of hemoglobin [4] and p38α-/- mice exhibit severe anemia and die in utero owing to defects in angiogenesis, and placental insufficiency [5]. Furthermore, the proliferation and differentiation of erythroid progenitors is controlled by erythropoietin (Epo) through the p38 MAPK and JNK signaling cascades [6]; finally, p38 is required for Epo mRNA stability [7].
In general, p38 MAPK augments gene transcription by the phosphorylation of CREB1 and ATF-2 [8]. CREB1 is a 43kDa protein which belongs to the bZIP superfamily of transcription factors. It is highly conserved through evolution and is ubiquitously expressed. When phosphorylated at Ser 133, CREB1 binds as a homo-dimer to the CRE to mediate transcriptional activation [9]. CREB1 is essential for the transcription of hematopoietic genes and critical for the development of the myeloid lineage [10]. It has been previously demonstrated that histone deacetylase inhibitors induce γ-globin expression by p38 MAPK activation [11; 12]. Treatment with butyrate and trichostatin A induces p38, CREB1 and ATF-2 phosphorylation [13]. We expanded upon these findings to determine the role of p38-mediated CREB1 activation in steady-state Gγ-globin regulation in normal erythroid progenitors.
In order to mediate trans-activation, CREB1 recruits CBP which forms complexes with other co-activators like PCAF and PCIP [14]. CBP/p300 acts as a bridge by recruiting the basal transcriptional machinery to the promoter resulting in transcriptional activation by CREB1 [15]. The CREB1 interaction domain of CBP is essential for hematopoiesis since mutations in this region severely affect erythropoiesis [16]. CBP is also crucial for erythropoiesis since it stimulates transcription factors such as cMyb, GATA1, NF-E2 and EKLF by direct physical interactions [17; 18; 19; 20]. It has been proposed that an NF-E2, GATA1 and EKLF complex linked by CBP is formed to activate hemoglobin synthesis [21].
These studies formed the basis of our study to investigate the role of p38/CREB signaling in Gγglobin regulation. Silencing p38 MAPK or CREB1 using siRNA knockdown had a deleterious effect on γ-globin transcription at steady-state. On the contrary, direct activation of p38 MAPK and CREB1 enforced expression augmented γ-globin synthesis. These effects were confirmed in primary erythroid progenitors. Site-directed mutagenesis of the G-CRE resulted in reduced Gγ-globin promoter activity and the abolishment of p38MAPK/CREB1 mediated Gγ-globin transcription. ChIP assays demonstrated CREB1 and CBP co-localize to the G-CRE in vivo. These data confirm a crucial role for p38 MAPK/CREB1 signaling in γ-globin gene regulation.
EXPERIMENTAL PROCEDURES
Tissue culture
K562 erythroleukemia cells were maintained in Iscove's Modified Dulbecco's Medium (IMDM; Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS; Atlanta Biologicals, Atlanta GA); cell viability was measured using 0.4% trypan blue stain.
siRNA mediated gene silencing
Cells were transfected with SMARTpool p38 MAPK siRNA (M-003555-04, Dharmacon) using Oligofectamine™ (Invitrogen) per the manufacturer's instructions. 50,000 cells were suspended in Opti-MEM I in 24-well plates in an siRNA-oligofectamine mix. Subsequently, IMDM with 30% FBS was added and cells were incubated for 48 hrs. CREB1 siRNA (siCREB1; M-003619-01, Dharmacon) treatment was completed using the same system. Control reactions included siGenome Non-Targeting siRNA (scrambled; D-001210-01-05, 100nM) or oligofectamine without siRNA molecules (mock).
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was isolated using RNA Stat-60™ (TEL-TEST “B” Inc., Friendswood, TX) and used for RT-qPCR analysis as previously published [13]. Briefly, cDNA was generated from total RNA (1μg) using the Improm-II reverse transcriptase system and oligo (dT) primers (Promega, Madison, WI). qPCR was performed on an iCycler (Bio-Rad, Hercules, CA) using the Sybergreen iQ Supermix (Bio-Rad) and 10μM of gene-specific primers (Table 1). Standard curves for different genes were generated using 500 to 0.5 ng of genomic DNA or Topo7 plasmids carrying the cDNA sequences for γ-globin, GAPD and β-globin. For p38 MAPK and CREB1 gene profiling, standard curves were generated with genomic DNA and p38 MAPK (PPH00715B, SABiosciences, Frederick, MD) or CREB1 primers (Table 1). All mRNA levels were normalized to GAPD or actin.
TABLE 1.
SUMMARY OF PRIMERS USED FOR QUANTITATIVE PCR ANALYSIS.
| GAPD | Forward: 5′ GAAGGTGAAGGTCGGAGT 3′ |
| Reverse: 5′ GAAGATGGTGATGGGATTTC 3′ | |
| Actin | Forward: 5’ GGCATGGGTCAGAAGGATT 3’ |
| Reverse: 5’ CACACGCAGCTCATTGTAGA 3’ | |
| Gγ-globin | Forward: 5’ AAGCCTTACACAGGATTATGAAGTCTG 3’ |
| Reverse: 5’ ACATGGCAGGAAGTATTCATGCTG 3’ | |
| Aγ-globin | Forward: 5’ AAGCTTTACACAGGATCATGAAGG 3’ |
| Reverse: 5’ CAGGGTAGGAAGTATTTATGGTGG 3’ | |
| γ-globin | Forward: 5′ GGCAACCTGTCCTCTGCCTC 3′ |
| Reverse: 5′ GAAATGGATTGCCAAAACGG 3′ | |
| β-globin | Forward: 5′ CTCATGGCAAGAAAGTGCTCG 3′ |
| Reverse: 5′ AATTCTTTGCCAAAGTGATGGG 3′ | |
| CREB1 | Forward: 5′ ATTACCCAGGGAGGAGCAAT 3′ |
| Reverse: 5′ TGGTTGCTGGGCACTAAGAT 3′ | |
| G-CRE | Forward: 5’ AAGCCTTACACAGGATTATGAAGTCTG 3’ |
| Reverse: 5’ AAGCCTTACACAGGATTATGAAGTCTG 3’ | |
| pGγLuc2 G-CRE | Forward: 5’TACGCGTGCTAGCCCGGGCTCGAG 3’ |
| Reverse: 5’ ACATGGCAGGAAGTATTCATGCTG 3’ | |
| γ-globin promoter | Forward:5′AAGCCTTACACAGGATTATGAAGTCTG 3′ |
| Reverse: 5′ ACATGGCAGGAAGTATTCATGCTG 3′ |
Western blot (WB) analysis
Cells were mixed with lysis buffer (Promega) to isolate total protein (150μg) which was separated by a 10% sodium dodecyl sulfate polyacrylamide gel. Antibodies targeting phosporylated-p38 (p-p38) and total p38 MAPK (t-p38) (12F8 and 9212, Cell Signaling) were used for WB. Anti-hemagglutinin (HA) antibody (2362, Cell Signaling) was used to quantify MKK3 and MKK6 protein. Rabbit anti-CREB1 antibody (48H2, Cell Signaling) was used to quantify CREB1 levels and the loading control actin (MAB1501, Chemicon Millipore, Billerica, MA). Secondary antibodies horseradish peroxidase-conjugated anti-rabbit (sc2004, Santa Cruz Biotechnology, Santa Cruz, CA) and anti-mouse (31430, Pierce Biotechnology, Rockford, IL) were used to visualize proteins in the ECL system (Amersham, Piscataway NJ); band intensities were measured on the ChemiDoc System (BioRad).
MKK3/MKK6 and CREB1 K562 stable cell lines
The pcDNA-MKK6b(E) constitutively active plasmid was established in a pcDNA3 vector from wild-type MKK6 by mutating Ser208/Thr212 to Glu208/Glu212 and the HA epitope was added [22]. Likewise, pcDNA-MKK3b(E) was produced from wild-type MKK3 by mutating Ser189/Thr193 to Glu189/Glu193 [23]. Previously, these vectors along with pcDNA3, pCMV and pCMV-CREB1 vectors were used to establish stable cell lines [12; 24]. Subsequently, single cell clones were isolated by serial dilutions and selected using the G418 selectable marker.
Fluorescent immunocytochemistry
Cytospin cell preparations were fixed with 4% paraformaldehyde and then permeabilized with 0.3% Triton-X-100. Immunostaining was performed overnight with anti-HbF fluorescein isothiocyanate (FITC) conjugated antibody (Bethyl Laboratories Inc., TX). Cell nuclei were stained with 4, 6-diamidino-2-phenylindole (DAPI; Santa Cruz Biotechnology) and visualized with an Olympus BX 51 epifluorescent microscope (Olympus America Inc, Center Valley, PA). Images were acquired with a Nikon DXM 1200F CCD camera (Nikon Instruments, Inc. Melville, NY). Images were acquired at 400x magnification and imported into NISelements software (Nikon Instruments).
Enzyme-linked immunoassay (ELISA)
Protein extracts (200ng) were used to quantify total (t-Hb) and fetal hemoglobin (HbF) levels using the human Hemoglobin and Hemoglobin F ELISA Quantitation kits respectively (Bethyl Laboratory). Briefly, 96-well plates were coated with sheep anti-human HbF antibody, or goat total anti-human Hb antibody, blocked with 1% bovine serum albumin and treated with horse radish peroxidase-conjugated secondary antibody; hemoglobin level were detected at 450nm. Raw data were analyzed using PRISM GraphPad (GraphPad Software, Inc., La Jolla, CA) and HbF levels were calculated as a ratio of t-hemoglobin corrected for total protein concentrations.
siRNA rescue experiment
For combined siRNA gene knockdown and rescue experiments, K562 cells were simultaneously transfected with siCREB1 and pCMV-CREB1 or pMT3-HDAC10 expression vectors; the respective empty vector controls were also tested. Transfections were performed using a K562-Nucleofector kit intended for use with a Nucleofector device (Amaxa Inc., Gaithersburg, MD) per the manufacturer's instructions. Cells were electroporated using the T-016 Amaxa program, incubated for 48hrs and then RNA was isolated for RT-qPCR analysis.
Primary erythroid cells
Human buffy coats were purchased from Carter Blood Care (Fort Worth, TX) and mononuclear cells were isolated using Histopaque-1077 (Sigma) and liquid cultures established using the Fibach method [25]. During phase 1 (day 0), erythroid progenitors were cultured in Minimum Essential Medium-Alpha (MEM-α) supplemented with 10% FBS and 50ng/mL each of granulocyte monocyte colony stimulating factor, interleukin-3, interleukin-6 and stem cell factor. At phase two (day 7), cells were switched to Epo (4U/mL) and stem cell factor.
Progenitors were transfected using the CD34+ Nucleofector kit (Amaxa); briefly, 3 mil cells were isolated on day 11 and resuspended in 100μl of Nucleofector solution with siRNA targeting p38 or siCREB1; 2μg of the control pMaxGFP reporter was added to all reactions to monitor transfection efficiency. The cells were electroporated on the U-008 setting and then grown for 72hrs. Similar experimental settings were used for pCMV or pCMV-CREB1 expression vector studies.
Luciferase K562 stable cell lines
The Gγ-globin promoter (-1500 to +36) was cloned into the pGL4.17 Luc2/neo vector to produce the wild-type pGγLuc2 construct. Three mutant plasmids including pGγLuc2(m1) with a -1225 G→A mutation, pGγLuc2(m2) with a -1227 AC→TG mutation and pGγLuc2(m3s) containing a scrambled G-CRE, were produced by site-directed mutagenesis (Bio S&T, Montreal, Canada). K562 cells (10×106) were transfected with 10μg of plasmid by electroporation at 270V, 975μF (Gene pulser, Bio-Rad). The cells were cultured in IMDM and 10% FBS for 48hrs, followed by the addition of G418 (400μg/mL). For drug inductions, 2 mil cells were treated for 48hrs with 2mM sodium butyrate or 0.5μM trichostatin A. Drug inducers and antibiotics were purchased from Sigma. For enforced expression studies pCMV or pCMV-CREB1 (20-50μg) were transfected into K562 stable cells and luciferase assay was performed after 24hrs using the luciferase assay kit (Promega) and a TD-20/20 luminometer (Turner Biosystems, San Diego, CA).
The plasmid copy number was measured in the stable lines using a qPCR based method as previously published [24; 26]. The primers used to determine Gγ-globin gene copy number are shown in Table 1. The relative copy number (Q) of target gene (Gγ-globin) was calculated as previously published [26].
Transient Transfection of Aγ-globin reporter constructs
The Aγ-globin promoter (-1500 to +36) was cloned into the pGL4.17Luc2/neo vector to produce the pAγLuc2 construct. The G-CRE “TGACGTCA” sequence was introduced into the -1222 position of pAγLuc2 by site-directed mutagenesis (Bio S&T) to produce the hybrid vector pA/GγLuc2. All constructs were verified by direct sequencing. The pGγLuc2, pAγLuc2, and pA/GγLuc2 constructs (10μg) were transfected into K562 cells. Total protein was isolated at 24hrs and luciferase activity was measured; all results were normalized by total protein.
ChIP assay and sequential ChIP
ChIP assays were performed in the KGγLuc2 stable cells as previously described [26] using the Upstate (Lake Placid, NY) protocol. DNA was sonicated using a Sonicator S-4000 (Misonix, Farmingdale, NY) to generate 500-600bp DNA fragments; input DNA was used to generate standard curves for qPCR. ChIP reactions were performed using phosphorylated CREB1 (pCREB1) or CBP antibodies (sc7879, sc369, Santa Cruz Biotechnology). Acetylated histone H3 (ac-H3, 07-081, Upstate), IgG (Sigma) and no antibody reactions were set up as controls. DNA-protein complexes were eluted and cross links were reversed by heating samples at 65°C. The precipitated chromatin was analyzed by qPCR.
For enforced expression ChIP assays, K562 cells were transfected with 120μg of pCMV-CREB1 or pRC/RSV-CBP. After 48hrs, cells were harvested and ChIP assay performed as described above. For sequential ChIP, the first immunoprecipitation reaction was carried out as described above then the DNA-protein complexes were eluted and treated with 0.1M DTT. Then the second IP was performed for 24hrs with a second antibody. For each IP IgG and ac-H3 were used as controls.
Statistical analysis
The data are reported as the mean ± standard error of the mean (SEM) from at least three data points generated from independent experiments. Data were analyzed by a two-tailed student's t test and values of p<0.05 were considered statistically significant. Statistical analyses were performed using Microsoft Excel (Redmond, WA).
RESULTS
Steady-state γ-globin expression involves p38 MAPK signaling
To elucidate cellular mechanisms mediating γ-globin gene regulation, dose-dependent p38 silencing [50 to 200nM] was performed for 48hrs. We observed significant 95% p38 silencing at the 200nM concentration (Fig. 1A). WB analysis confirmed a 43% decrease in p38 MAPK protein. These conditions produced a dose-dependent 81% decrease in Gγ-globin gene transcription compared to 47% silencing of the Aγ-globin gene (Fig. 1B).
Fig. 1.
p38 MAPK signaling regulates γ-globin transcription. (A) p38 MAPK mRNA levels were quantified by RT-qPCR analysis using gene specific primers (Table 1); non-targeting scrambled (Scr) siRNA was used as a control. Data were calculated as the mean ± standard error of the mean (SEM), p<0.05 was considered significant. WB analysis was performed using 150μg of total protein and an anti-total p38 (t-p38) antibody, actin was used as a loading control. (B) The effect of p38 MAPK silencing on Gγ-globin and Aγ-globin transcription were quantified using gene specific primers for RT-qPCR analysis. (C) Three stable lines including PC, kMKK3, and kMKK6 were established in K562 cells using the pcDNA, pcDNA3-MKK3 and pcDNA3-MKK6 expression vectors respectively and then clones were isolated (see “Experimental procedures”). WB was performed with 150μg of protein using an anti-HA antibody to confirm gene expression. Levels of phosphophyrolated-p38 (p-p38) and t-p38 were determined by WB. (D) The effect of stable enforced activation of p38 MAPK on γ-globin transcription was determined by RT-qPCR analysis. (E) ELISA was performed to quantify the effect of activating p38 MAPK on HbF expression (see “Experimental procedures”). HbF levels were normalized by total hemoglobin (t-Hb) and total protein (t-Protein). (F) HbF positive cells were visualized by immunostaining with FITC conjugated anti-γ-globin antibody (HbF-FITC); DAPI staining was performed to visualize cell nuclei (magnification ×400). At least 500 DAPI positive cells were counted along with the number of FITC positive cells in the same field and then used to calculate the percentage of HbF positive cells.
To determine the direct effect of constitutively active p38 MAPK on γ-globin regulation, two K562 cell lines, kMKK3 and kMKK6, were established with the expression vectors pcDNA-MKK3b(E) and pcDNA-MKK6b(E). MKK3 is capable of activating p38α, p38δ and p38γ while MKK6 preferentially activates p38β [27; 28]. K562 cell lines expressing the empty vector pcDNA3 (PC) were used as a control. Three stable lines were established from individual clones and enforced expression of MKK3 and MKK6 was confirmed by WB using anti-HA antibody (Fig 1C). Interestingly, WB analysis of p-p38 revealed 70% activation in the kMKK6 clones, while no effect was observed in the PC or kMKK3 clones (Fig 1C). This lack of p-p38 accumulation may be attributed to the differential activation of p38 isoforms. While MKK3 preferentially activates p38α, p38δ and p38γ, MKK6 activates all p38 isoforms. RT-qPCR analysis demonstrated a 3-fold (p<0.01) increase in γ-globin mRNA in the kMKK6 clones (Fig 1D). This effect was further confirmed by direct quantification of HbF by ELISA where a 2-fold induction of HbF was observed in kMKK6 clones (Fig 1E). Cytospin preparations demonstrated a 2.3-fold increase in HbF positive cells in kMKK6 clones compared to the PC clones (Fig 1F). These data demonstrate a role for p38 MAPK signaling in γ-globin expression at steady-state in K562 cells.
CREB1 trans-activates γ-globin gene expression
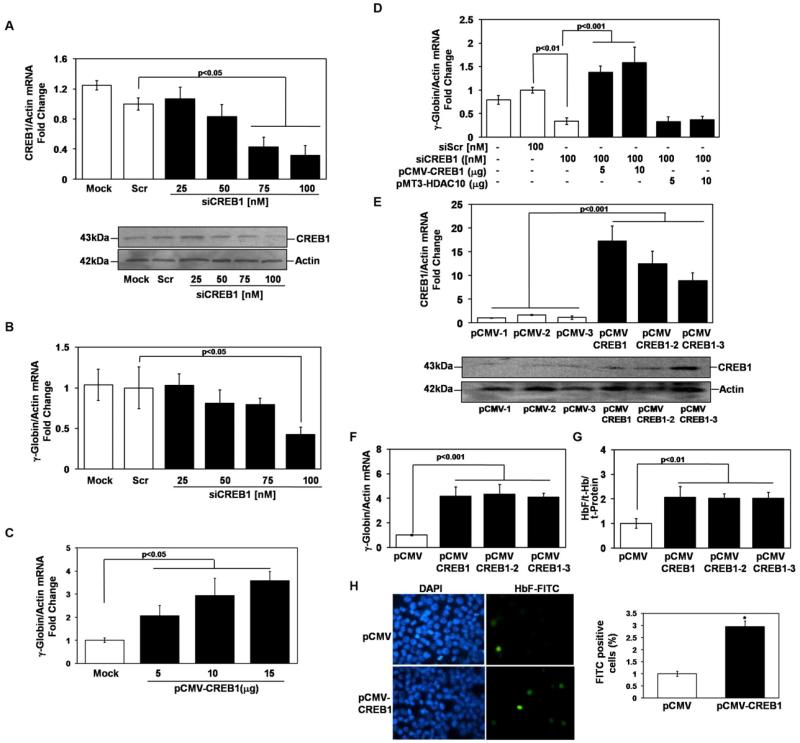
After induction with butyrate and trichostatin A, the p38 MAPK signaling cascade activates CREB1 and ATF-2 to induce HbF [13]. To gain further insight we completed experiments to examine whether CREB1 plays a direct role in γ-globin regulation independent of drug induction. Dose-dependent silencing experiments were performed using siCREB1 molecules [25 to 100nM] for 48hrs which produced a 53% decrease in CREB1 protein (Fig 2A). When the effect on γglobin transcription was analyzed, we observed a 68% loss of transcription (Fig 2B). To support these observations transient enforced expression of CREB1 (5μg and 15μg) was performed which produced a dose-dependent activation of γ-globin (2 to 3.5- fold) (Fig 2C).
Fig. 2.
CREB1 trans-activates γ-globin expression. (A) The effect of siCREB1 treatment on target gene transcription was quantified by RT-qPCR analysis using gene specific primers (Table 1); non-targeting scrambled (Scr) siRNA was used as a control. Data were calculated as the mean ± SEM; p<0.05 was considered significant. WB analysis was performed using 150μg of total protein to confirm CREB1 silencing. (B) The effect of CREB1 silencing on γ-globin transcription was quantified using RT-qPCR. (C) Transient enforced expression of CREB1 was performed in K562 cells for 48hrs. The effect on γglobin transcription was quantified using RT-qPCR (D) Rescue experiments were performed by the addition of siCREB1 alone or combined with pCMV-CREB1 or pMT3-HDAC10 expression vectors (see “Experimental procedures”). For each condition the amount of plasmid added is shown. Represented is the fold change for the expression vectors after subtracting the values obtained from empty vector control transfections. (E) K562 stable lines were established using the pCMV or pCMV-CREB1 expression vectors and then clones were isolated (see “Experimental procedures”). Enforced CREB1 expression was confirmed by RT-qPCR (bar graph) and WB analysis. (F) The effect of stable enforced CREB1 expression on γ-globin transcription was determined by RT-qPCR analysis. (G) ELISA was performed to quantify the effect on HbF levels. (H) HbF positive cells were visualized by immunostaining with FITC conjugated anti-γ-globin antibody and DAPI staining to visualize nuclei.
To confirm that gene silencing was not due to non-specific effects, rescue experiments were performed. K562 cells were transfected with siCREB alone or combined with pCMV-CREB1. The 65% silencing of γ-globin transcription observed with 100nM of siCREB1 was reversed by pCMV-CREB1 producing a 1.5-fold trans-activation of γ-globin (Fig 2D); on the contrary, pMT3-HDAC10 was not able to rescue γ-globin expression. Enforced expression of CREB1 and HDAC10 was confirmed by RT-qPCR analysis of the target genes (data not shown).
To test whether γ-globin gene expression could be stimulated directly by CREB1, cell lines that stably express this protein were established with the pCMV and pCMV-CREB1 expression vectors; subsequently three individual clones were isolated. We observed 8- to 17-fold increase in CREB1 gene expression (Fig 2E) and a corresponding increase in CREB1 protein was observed by WB. In the clones, 4-fold γ-globin activation was produced (Fig. 2F) and this effect was corroborated by a 2-fold induction of HbF measured by ELISA (Fig. 2G) and a 3-fold increase in HbF positive cells measured by immunostaining (Fig 2H).
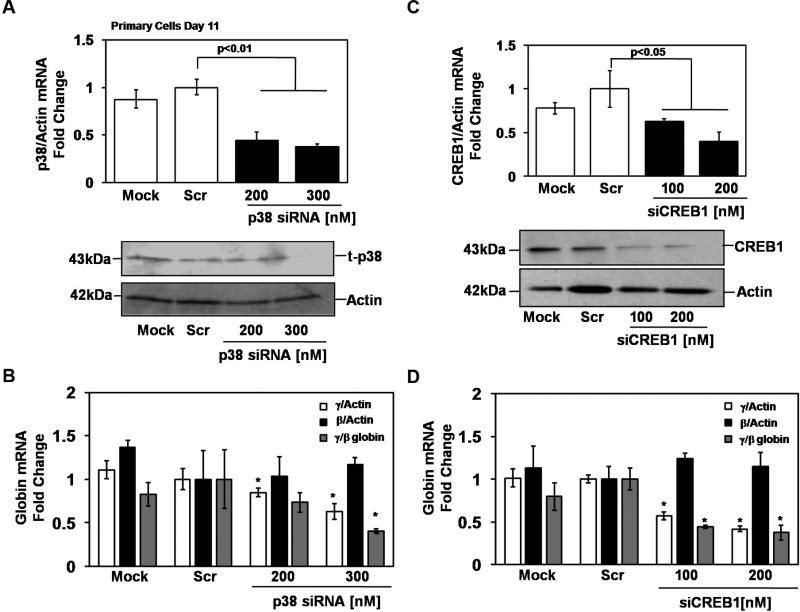
γ-Globin expression during normal erythroid maturation requires activated p38 MAPK
To confirm the physiological relevance of p38 signaling in γ-globin regulation, human erythroid progenitors derived from peripheral blood mononuclear cells were used. In the first set of experiments, siRNA silencing of p38 MAPK [200 and 300nM] was performed for 72hrs at day 11 of culture. Transfection efficiency monitored by pMaxGFP remained greater than 35% for all replicates (data not shown). After siRNA treatment, p38 MAPK protein levels decreased 40% (Fig 3A). When compared with the scrambled siRNA control, p38 MAPK silencing repressed γ-globin transcription 38% without a significant effect on β-globin expression producing an overall 60% decrease in the γ/β ratio (Fig 3B). Similarly, siCREB1 [100nM and 200nM] treatment caused 40-60% target gene silencing and a 54% decrease in CREB1 protein (Fig 3C). Specifically, γ-globin transcription was repressed 59% which produced a 64% decrease in the γ/β ratio after siCREB1 treatment (Fig 3D).
Fig. 3.
p38 MAPK/CREB1 signaling regulates γ-globin expression in human erythroid progenitors. Peripheral blood mononuclear cells were grown in the two-phase liquid culture system. On day 11, progenitors were transfected with siRNA targeting p38 MAPK or CREB1 using a Nucleofector device transfection system (see “Experimental procedures”). Cells were harvested after 72hrs and RNA and protein were isolated for further analysis. (A) p38 MAPK silencing was confirmed by RT-qPCR and WB analysis. (B) The levels of γ-globin, β-globin and actin gene expression were measured by RT-qPCR. The effect of p38 MAPK silencing on γ-globin and β-globin transcription as well as the ratio of γ/β globin was calculated after the expression of each gene was normalized by actin. (C) In cells treated with siCREB1, target gene silencing was confirmed by RT-qPCR and WB analysis. (D) The effects of CREB1 silencing on γ-globin and β-globin expression and the γ/β ratio were analyzed as described in panel B.
Complementary enforced expression studies performed at day 11 with pCMV-CREB1 showed highly significant CREB1 activation and protein synthesis (Fig 4A). Transfection efficiency monitored by pMaxGFP remained greater than 40% for all replicates (data not shown). RT-qPCR demonstrated 4-fold γ-globin trans-activation (Fig 4B), establishing the ability of CREB1 to enhance γ-globin gene expression in a physiological environment. These data were validated by immunostaining cells for HbF using anti-γ-FITC antibody (Fig 4C). A 20% increase in HbF positive cells was observed in CREB1 transfected progenitors (Fig 4D) which translated into a 2-fold increase in HbF (Fig 4E).
Fig. 4.
CREB1 enforced expression activates γ-globin in erythroid progenitors. Erythroid progenitors grown in the two phase liquid culture system were transfected with pCMV or pCMV-CREB1 on day 11. For all panels, the data shown was obtained from expression vector transfections after subtracting the values obtained from empty vector control transfections. Data were calculated as the mean ± SEM; p<0.05 was considered significant. (A) CREB1 enforced expression was confirmed by RT-qPCR and WB analysis. (B) The effect of enforced CREB1 expression of increasing concentrations of CREB1on γglobin was calculated as a ratio of γ/β mRNA as described in Fig. 3B. (C) HbF positive cells were visualized by immunostaining with FITC conjugated anti-γ-globin antibody (HbF-FITC) after enforced CREB1expression. DAPI staining was performed to visualize nuclei and to determine cell counts (×400 magnification). (D) At least 500 DAPI positive cells were counted along with the number of FITC positive cells in the same field and then used to calculate the percentage of HbF positive cells. (E) ELISA was performed to quantify the effects of enforced CREB1 expression on HbF levels.
The G-CRE is required for Gγ-globin promoter activation
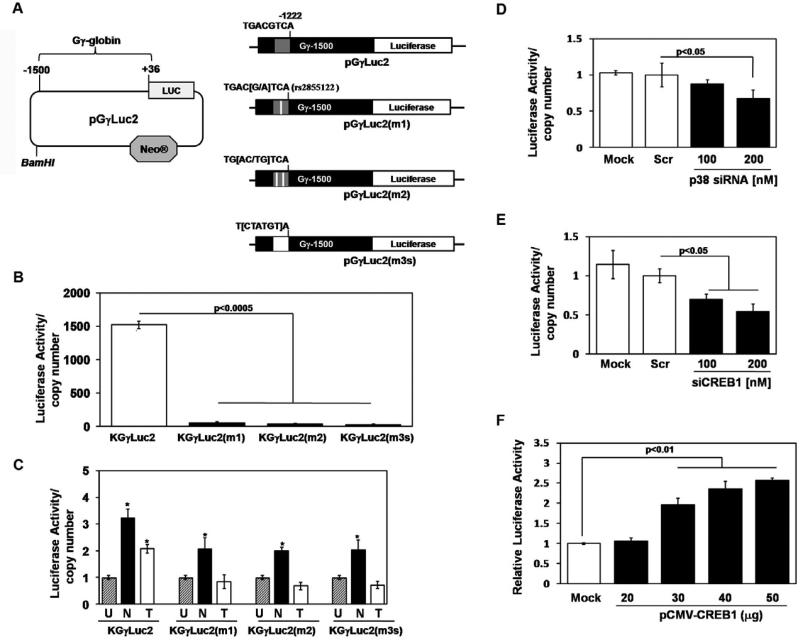
The combined K562 and primary erythroid cells data demonstrate that p38 MAPK and CREB1 are required for γ-globin gene transcription at steady-state. Our next experiments were designed to explore mechanisms by which CREB1 activates γglobin expression. A target binding site for CREB1 was identified in the Gγ-globin promoter called the G-CRE which is a palindromic sequence (5’-TGACGTCA-3’) located between -1222 and -1229 relative to the cap site. We wanted to study the role of the G-CRE in a native chromatin context. For this purpose, the Gγ-globin promoter (-1500 to +36) was cloned upstream of a luciferase reporter in the pGL4.17luc2/neo vector to create the wild-type pGγLuc2 construct. Three mutant constructs including pGγLuc2(m1), pGγLuc2(m2), and pGγLuc2(m3s) were created using site-directed mutagenesis (Fig. 5A); five K562 stable cell lines were established.
Fig. 5.
The G-CRE is required for steady-state promoter activity. (A) Shown are schematics of the reporter plasmids established with the pGL4.17Luc2/neo vector and wild-type or mutant Gγ-globin promoters. (B) Stable lines were established in K562 with reporter plasmids, along with a corresponding empty vector control (see “Experimental procedures”). The luciferase activity of the stable lines was determined by normalizing luciferase activity to total protein and correcting for the copy number of the stable line (luciferase/copy number). Data was calculated as the mean ± SEM; p<0.05 was considered significant. (C) The ability of sodium butyrate (N) or trichostatin A (T) to activate Gγ-promoter activity in the stable lines was tested; cells were treated with each drug for 48hrs and luciferase activity was measured. (D) The effect of p38 MAPK silencing on γ-globin promoter activity was tested in the KGγLuc2 stable line. Cells were treated with p38 MAPK siRNA for 48hrs after which luciferase activity was analyzed. (E) Similar experiments to determine the effects of siCREB1 treatment on γ-globin activity were performed. (F) CREB1 enforced expression studies were performed in the KGγLuc2 line. Protein was harvested after 48hrs and luciferase assay was performed. Shown in the graph is the luciferase activity obtained at the different CREB1 concentrations after subtracting the values obtained for empty vector transfections.
The first set of studies was performed to evaluate the effect of the different G-CRE mutations on steady-state promoter transcription. The luciferase activity of the KGγLuc2 line was 1500-fold higher than the empty vector control (Fig 5B), demonstrating that the promoter was intact and functional. Interestingly, all three mutant lines showed greater than 90% loss of luciferase activity indicating that the G-CRE is required, and any loss of sequence integrity is deleterious to promoter activity. To establish that the Gγ-globin promoter inserted in the stable lines responds in the same manner as the endogenous γglobin promoter, drug induction studies were completed. Treatment with sodium butyrate for 48hrs resulted in a significant 3-fold induction of luciferase activity in the KGγLuc2 line (Fig 5C); a lower fold induction was observed in the mutant lines. The ability of sodium butyrate to induce γ-globin even with mutations in the G-CRE might be attributed to the previously demonstrated butyrate response elements at other positions on the γ-globin promoter [29; 30], which mediate induction independent of the G-CRE. Similar studies were performed with trichostatin A; while the wild type KGγLuc2 line was induced 2-fold, this effect was lost in the three mutants (Fig 5C) suggesting trichostatin A mainly targets the GCRE.
To determine if the G-CRE is required for Gγ-globin activation via p38 MAPK signaling, we performed siRNA silencing of p38 MAPK and CREB1 in the luciferase stable lines. p38 MAPK siRNA treatments in the KGγLuc2 line, produced a 35% (p<0.05) loss of luciferase activity (Fig 5D) whereas in the mutant lines no change was produced (data not shown). Similarly, the KGγLuc2 line showed a 30-45% loss of luciferase activity with siCREB1 treatment (Fig 5E), an effect lost in the mutant lines (data not shown). Enforced expression of CREB1 in KGγLuc2 mediated a dose-dependent 3-fold increase in luciferase activity (Fig 5F). These data demonstrate that an intact G-CRE is essential for the regulation of Gγ-globin activity though the p38 MAPK/CREB1 signaling cascade.
The G-CRE mediates Aγ-globin activation by CREB1
We next examined the ability the CRE to activate the Aγ-globin promoter which does not carry a CRE element, through CREB1 trans-activation. We cloned the Aγ-globin promoter (-1500 to +36) in the pGL4.17luc2/neo vector to create the pAγLuc2 construct. Site-directed mutagenesis was performed to introduce a CRE between -1222 and -1229 (Fig 6A). Transient transfections of K562 cells were performed using 10μg of the pGγLuc2, pAγLuc2 and pA/GγLuc2 constructs. The pAγLuc2 and pA/GγLuc2 constructs displayed 25% lower luciferase activity than the pGγLuc2 construct (Fig 6B). This suggested the G-CRE was not detrimental to Aγ-globin promoter activity; however, we did not observe greater promoter transcription for the hybrid promoter at steady-state.
Fig. 6.
The G-CRE mediates activation of the Aγ-globin promoter by CREB1. (A) Shown are schematics of the reporter plasmids established with wild-type Aγ-globin and hybrid promoter with the G-CRE inserted at base -1222 (see “Experimental procedures”). (B) Transient transfection assays were performed in K562 cells. Shown is the luciferase activity of the three reporters after normalization by total protein. Data were calculated as the mean ± SEM; p<0.05 was considered significant. (C) Transient cotransfections of the three constructs with pCMV or pCMV-CREB1 were performed. For each condition the presence (+) or absence (-) of the various reagents is shown.
We next tested the ability of CREB1 to trans-activate the pA/GγLuc2 construct. As anticipated, CREB1 induced luciferase activity 2.3-fold for the control pGγLuc2 construct (Fig 6C). By contrast, pAγLuc2 was not activated by CREB1 while the hybrid promoter, pA/GγLuc2 construct was trans-activated 2-fold (p<0.01) comparable to the control reporter. These data support the ability of the G-CRE to confer CREB1 inducibility to the Aγ-globin promoter.
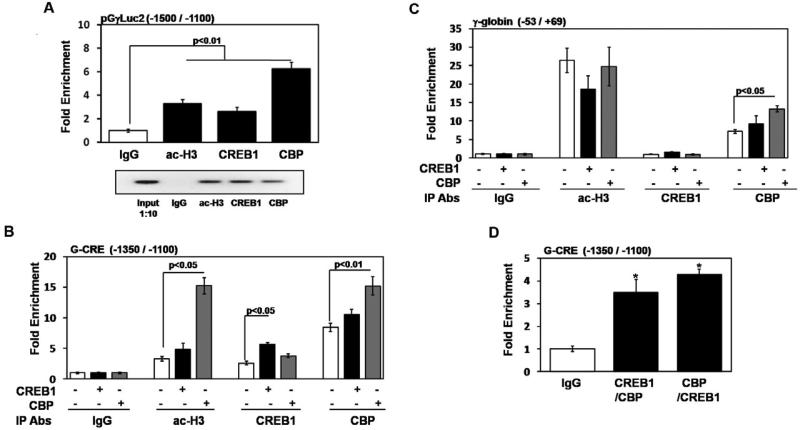
CREB1 and CBP co-localize to the G-CRE region
Our data suggests the importance of the G-CRE to enable CREB1 trans-activation of γ-globin. We know that CREB1 binds to its target CRE as a dimer and when phosphorylated, co-activators such as CBP and p300 are recruited to the promoter [31] to regulate transcription. To elucidate mechanisms by which CREB1 interacts with the G-CRE in vivo, we performed ChIP assays in the KGγLuc2 stable line to determine whether pCREB1 and CBP are capable of binding to the G-CRE region. A unique primer was designed for qPCR to distinguish chromatin enrichment in the G-CRE region of the pGγLuc2 stably integrated construct from the endogenous G-CRE region (Table 1). The proximal region of the γ-globin promoter was also analyzed to evaluate a second unrelated region by ChIP assay and ac-H3 and IgG were used as controls.
Initially studies confirmed that CREB1 and CBP bind under steady-state conditions. Analysis of the pGγLuc2 G-CRE regions showed 2.6-fold and 6.0-fold chromatin enrichment with CREB1 and CBP respectively compared to IgG (Fig. 7A); PCR products generated using input DNA and chromatin from the different immunoprecipitation reactions was analyzed by gel electrophoresis as well. Similar studies for the endogenous G-CRE are shown in Fig. 7B. Comparable 2.6-fold pCREB1 and 8.5-fold CBP mediated chromatin enrichment was observed. Control studies at the proximal γ-globin promoter demonstrated no pCREB1 binding while 3-fold CBP and 26-fold ac-H3 chromatin enrichments were observed (Fig. 7C). All subsequent ChIP assays were analyzed using only the endogenous G-CRE since both regions were bound by CREB1 and CBP in vivo.
Fig. 7.
CREB1 and CBP co-localize to the G-CRE region in vivo. (A) ChIP assays were performed in the KGγLuc2 stable line with CREB1, CBP, acetylated histone H3 (ac-H3) and IgG antibodies; chromatin enrichment was normalized to IgG. qPCR analysis was performed using primers unique to the pGγLuc2 construct or endogenous G-CRE regions (Table 1). Data was calculated as the mean ± SEM; p<0.05 was considered significant. Shown in the agarose gel is PCR products of representative samples from each ChIP assay condition; 1:10 diluted input DNA was used as a control. (B) ChIP assays were performed using the KGγLuc2 stable line after CREB1 or CBP enforced expression; untreated cells were used as a control. The antibodies (Abs) used for the different immunoprecipitation (IP) reactions are shown in the graph. For each condition the presence (+) or absence (-) of the various constructs is shown. qPCR analysis was performed using primers specific to the endogenous G-CRE region (Table 1); chromatin enrichment is represented as fold change relative to IgG. (C) For the same chromatin described in Fig.7B, qPCR analysis was also performed using proximal γ-globin promoter primers as a control (Table 1). (D) Sequential ChIP assay was performed in the KGγLuc2 stable line (see “Experimental procedures”). Experiments were performed with CREB1 antibody followed by CBP and vice versa. Enrichment obtained after sequential ChIP was normalized relative to values obtained for IgG.
To gain additional evidence for CREB1 binding in vivo ChIP assays were performed following enforced gene expression. pCREB1 binding was enriched 5.6-fold after enforced expression compared to 2.6-fold chromatin enrichment at baseline (Fig. 7B). Interestingly, CBP enforced expression increased CREB1 binding to 3.8-fold but this was not statistically significant. Similarly, CBP enforced expression produced 15.2-fold chromatin enrichment compared to 10.6-fold baseline enrichment (Fig. 7B); note that CBP expression also increased the level of ac-H3 from 3.2-fold to 15.2-fold. This latter observation supports the ability of CBP to act as a histone acetyl transferase. At the proximal γ-promoter region, no CREB1 binding was observed at baseline or following CREB1 or CBP enforced expression (Fig. 7C). However, increased enforced CBP expression produced up to 13.3-fold chromatin enrichment.
While we our data demonstrate that pCREB1 and CBP can independently bind to the G-CRE region, it can be argued that co-localization does not occur therefore sequential ChIP assays were completed. When immunoprecipitation was performed with CREB1 antibody followed by CBP, 3.5-fold chromatin enrichment was observed (Fig. 7D); for the reverse reactions 4.3-fold chromatin enrichment was produced, demonstrating co-occupancy of CREB1 and CBP at the G-CRE region in the Gγ-globin promoter.
DISCUSSION
Sickle cell anemia is a β-hemoglobinopathy which affects millions of people worldwide. The severity of the disease is determined by clinical symptoms such as pain, infection and strokes. Individuals with increased HbF levels (>8.6%) show ameliorated symptoms and improved survival [32], demonstrating the therapeutic potential of HbF induction. Of the currently available treatment options, the most feasible is the induction of HbF through pharmacological agents. These agents have been shown to act by hyperacetylation or hypomethylation of the local chromatin environment of the γ-globin promoters [11; 33; 34; 35; 36; 37]. An alternative mechanism of HbF induction through cell signaling pathways was elucidated by our laboratory and others. The signaling pathways implicated in γ-globin regulation include cGMP, cAMP, NO, and p38 MAPK [12; 38; 39; 40; 41].
Interestingly, p38 signaling is implicated in γ-globin reactivation by a wide variety of compounds [11; 41; 42]. However, Hydroxyurea is the only FDA approved drug for the treatment of sickle cell anemia. HbF induction by this drug involves nitric oxide signaling through the cGMP [43] and p38 MAPK pathways [42; 44]. Treatment of K562 cells with butyrate resulted in p38 MAPK activation and increased γ-globin transcription [13; 41]. More recently, Aerbajinai et al. [45] demonstrated that thalidomide induces γ-globin expression through a similar mechanism in human erythroid progenitor cells.
Previous work from our laboratory has demonstrated role for reactive oxygen species as an upstream activator of p38 MAPK signaling to achieve γ-globin activation by trichostatin A and butyrate [11; 12; 44]. In the current study, we explored the role of p38 MAPK signaling in γ-globin regulation at steady-state in the absence of pharmacological stimulation. We demonstrated that p38 and CREB1 silencing mediated Gγ-globin gene repression, supporting a positive regulatory role for both proteins. Stable enforced expression of MKK3 and MKK6 resulted in the constitutive activation of p38 MAPK accompanied by Gγ-globin trans-activation and HbF induction; moreover, stable CREB1 enforced expression produced the same effect. To confirm these findings, studies were conducted in primary erythroid progenitors. We demonstrated that loss of p38 MAPK and CREB1 function by siRNA silencing resulted in significant γ-globin repression in the absence of changes in β-globin. Furthermore, direct trans-activation by CREB1 augmented HbF levels to confirm a positive regulatory role for p38 MAPK/CREB1 signaling in a physiological system.
The downstream effectors activated by p38 include CREB1, CREB2, CREM and ATF1-4 [8; 46]. Members of the CREB family recognize and bind to the CRE as homo- or heterodimers to regulate gene expression. The G-CRE is a palindromic sequence located between -1222 and -1229 of the Gγ-globin promoter. Pissard et al. [47] described four Pre-γ frameworks characterized by polymorphisms in the GCRE (-1225 G/A), -1280 GATA and -1450 TTATT (T/G) sites. The -1225 G/A mutation is associated with lower HbF levels in sickle cell patients with the Benin β-locus haplotype [47]. In vitro binding studies have demonstrated that CREB1 binding to the G-CRE is abolished if two or more base mutations are introduced [13]. A possible mechanism for the abolishment of promoter activity is the introduction of a 5’-TGTGGT-3’ sequence by the two base mutation which produces a consensus motif for the transcriptional repressor acute myeloid leukemia [48].
To gain greater insights into the role of the G-CRE in regulating Gγ-promoter activity in a chromatin context, stable luciferase reporter lines were established. One or more base mutations in the G-CRE abolished promoter functionality and promoter silencing produced by p38 and CREB1 siRNA treatment. These findings indicate that mutating the G-CRE abrogates the ability of p38 MAPK/CREB1 signaling to activate Gγ-globin expression.
The CRE is an enhancer element capable of regulating transcription in a position and orientation independent manner [49]. To further explore the ability of the -1222 G-CRE to regulate a heterologous promoter, we introduced a CRE into the Aγ-globin promoter to create a hybrid promoter. Although no increase in steady state promoter activity was observed there was increased hybrid promoter activity after CREB1 enforced expression demonstrating the ability of the G-CRE to activate a heterologous promoter.
CREB1 is capable of binding the CRE in the unphosphorylated state, but requires activation to mediate gene trans-activation. This molecule is activated by phosphorylation at Ser133 which facilitates interaction with CBP/p300 which acts as a molecular bridge permitting recruitment of DNA binding proteins to the basal transcription machinery [15]. To study the chromatin binding of CREB1 and CBP, we performed ChIP assays. Both CREB1 and CBP were bound in the upstream G-CRE region and enforced CREB1 expression increased pCREB1 and CBP binding. However, enforced CBP expression only increased CBP binding in the G-CRE region. Subsequently, sequential ChIP assays demonstrated co-occupancy of pCREB1 and CBP the in the G-CRE region in vivo. Our data support a novel mechanism of Gγ-globin regulation by p38 MAPK/CREB1 signaling and the functional relevance of the G-CRE in the formation of a CREB1/CBP complex to regulate Gγ-globin transcription under steady-state conditions.
ACKNOWLEDGEMENTS
We give special thanks to Dr. Li Liu in the Pace laboratory for cloning the pGγLuc2 reporter plasmid and TzuFang Lou for technical assistance with the two-phase liquid culture system. We greatly appreciate the assistance of Dr. Santosh D'Mello and Dr. Carole Mikoryak at the University of Texas at Dallas, with the fluorescence microscope studies. This work was supported by grant HL69234 from the National Heart Lung and Blood Institute to Dr. Betty Pace.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Neiman AM. Conservation and reiteration of a kinase cascade. Trends Genet. 1993;9:390–4. doi: 10.1016/0168-9525(93)90139-9. [DOI] [PubMed] [Google Scholar]
- 2.Nishida E, Gotoh Y. The MAP kinase cascade is essential for diverse signal transduction pathways. Trends Biochem Sci. 1993;18:128–31. doi: 10.1016/0968-0004(93)90019-j. [DOI] [PubMed] [Google Scholar]
- 3.Martin-Blanco E. p38 MAPK signalling cascades: ancient roles and new functions. Bioessays. 2000;22:637–45. doi: 10.1002/1521-1878(200007)22:7<637::AID-BIES6>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 4.Steinbrech DS, Mehrara BJ, Saadeh PB, et al. VEGF expression in an osteoblast-like cell line is regulated by a hypoxia response mechanism. Am J Physiol Cell Physiol. 2000;278:C853–60. doi: 10.1152/ajpcell.2000.278.4.C853. [DOI] [PubMed] [Google Scholar]
- 5.Tamura K, Sudo T, Senftleben U, et al. Requirement for p38alpha in erythropoietin expression: a role for stress kinases in erythropoiesis. Cell. 2000;102:221–31. doi: 10.1016/s0092-8674(00)00027-1. [DOI] [PubMed] [Google Scholar]
- 6.Nagata Y, Takahashi N, Davis RJ, Todokoro K. Activation of p38 MAP kinase and JNK but not ERK is required for erythropoietin-induced erythroid differentiation. Blood. 1998;92:1859–69. [PubMed] [Google Scholar]
- 7.Dean JL, Brook M, Clark AR, Saklatvala J. p38 mitogen-activated protein kinase regulates cyclooxygenase-2 mRNA stability and transcription in lipopolysaccharide-treated human monocytes. J Biol Chem. 1999;274:264–9. doi: 10.1074/jbc.274.1.264. [DOI] [PubMed] [Google Scholar]
- 8.Krebs FC, Goodenow MM, Wigdahl B. Neuroglial ATF/CREB factors interact with the human immunodeficiency virus type 1 long terminal repeat. J Neurovirol. 1997;3(Suppl 1):S28–32. [PubMed] [Google Scholar]
- 9.Montminy MR, Bilezikjian LM. Binding of a nuclear protein to the cyclic-AMP response element of the somatostatin gene. Nature. 1987;328:175–8. doi: 10.1038/328175a0. [DOI] [PubMed] [Google Scholar]
- 10.Sakamoto KM, Fraser JK, Lee HJ, Lehman E, Gasson JC. Granulocyte-macrophage colony-stimulating factor and interleukin-3 signaling pathways converge on the CREB-binding site in the human egr-1 promoter. Mol Cell Biol. 1994;14:5975–85. doi: 10.1128/mcb.14.9.5975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson J, Hunter R, McElveen R, et al. Fetal hemoglobin induction by the histone deacetylase inhibitor, scriptaid. Cell Mol Biol (Noisy-le-grand) 2005;51:229–38. [PubMed] [Google Scholar]
- 12.Pace BS, Qian XH, Sangerman J, et al. p38 MAP kinase activation mediates gamma-globin gene induction in erythroid progenitors. Exp Hematol. 2003;31:1089–96. doi: 10.1016/s0301-472x(03)00235-2. [DOI] [PubMed] [Google Scholar]
- 13.Sangerman J, Lee MS, Yao X, et al. Mechanism for fetal hemoglobin induction by histone deacetylase inhibitors involves gamma-globin activation by CREB1 and ATF-2. Blood. 2006;108:3590–9. doi: 10.1182/blood-2006-01-023713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Korzus E, Torchia J, Rose DW, et al. Transcription factor-specific requirements for coactivators and their acetyltransferase functions. Science. 1998;279:703–7. doi: 10.1126/science.279.5351.703. [DOI] [PubMed] [Google Scholar]
- 15.Nakajima T, Uchida C, Anderson SF, Parvin JD, Montminy M. Analysis of a cAMP-responsive activator reveals a two-component mechanism for transcriptional induction via signal-dependent factors. Genes Dev. 1997;11:738–47. doi: 10.1101/gad.11.6.738. [DOI] [PubMed] [Google Scholar]
- 16.Kasper LH, Boussouar F, Ney PA, et al. A transcription-factor-binding surface of coactivator p300 is required for haematopoiesis. Nature. 2002;419:738–43. doi: 10.1038/nature01062. [DOI] [PubMed] [Google Scholar]
- 17.Dai P, Akimaru H, Tanaka Y, et al. CBP as a transcriptional coactivator of c-Myb. Genes Dev. 1996;10:528–40. doi: 10.1101/gad.10.5.528. [DOI] [PubMed] [Google Scholar]
- 18.Blobel GA, Nakajima T, Eckner R, Montminy M, Orkin SH. CREB-binding protein cooperates with transcription factor GATA-1 and is required for erythroid differentiation. Proc Natl Acad Sci U S A. 1998;95:2061–6. doi: 10.1073/pnas.95.5.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheng X, Reginato MJ, Andrews NC, Lazar MA. The transcriptional integrator CREB-binding protein mediates positive cross talk between nuclear hormone receptors and the hematopoietic bZip protein p45/NF-E2. Mol Cell Biol. 1997;17:1407–16. doi: 10.1128/mcb.17.3.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang W, Bieker JJ. Acetylation and modulation of erythroid Kruppel-like factor (EKLF) activity by interaction with histone acetyltransferases. Proc Natl Acad Sci U S A. 1998;95:9855–60. doi: 10.1073/pnas.95.17.9855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Merika M, Orkin SH. Functional synergy and physical interactions of the erythroid transcription factor GATA-1 with the Kruppel family proteins Sp1 and EKLF. Mol Cell Biol. 1995;15:2437–47. doi: 10.1128/mcb.15.5.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Han J, Lee JD, Jiang Y, et al. Characterization of the structure and function of a novel MAP kinase kinase (MKK6). J Biol Chem. 1996;271:2886–91. doi: 10.1074/jbc.271.6.2886. [DOI] [PubMed] [Google Scholar]
- 23.Raingeaud J, Whitmarsh AJ, Barrett T, Derijard B, Davis RJ. MKK3- and MKK6-regulated gene expression is mediated by the p38 mitogen-activated protein kinase signal transduction pathway. Mol Cell Biol. 1996;16:1247–55. doi: 10.1128/mcb.16.3.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kodeboyina S, Balamurugan P, Liu L, Pace BS. cJun modulates Ggamma-globin gene expression via an upstream cAMP response element. Blood Cells Mol Dis. 2009 doi: 10.1016/j.bcmd.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fibach E, Rachmilewitz EA. The two-step liquid culture: a novel procedure for studying maturation of human normal and pathological erythroid precursors. Stem Cells. 1993;11(Suppl 1):36–41. doi: 10.1002/stem.5530110608. [DOI] [PubMed] [Google Scholar]
- 26.Yao X, Kodeboyina S, Liu L, et al. Role of STAT3 and GATA-1 interactions in gamma-globin gene expression. Exp Hematol. 2009;37:889–900. doi: 10.1016/j.exphem.2009.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reuter CW, Morgan MA, Bergmann L. Targeting the Ras signaling pathway: a rational, mechanism-based treatment for hematologic malignancies? Blood. 2000;96:1655–69. [PubMed] [Google Scholar]
- 28.Boosalis MS, Bandyopadhyay R, Bresnick EH, et al. Short-chain fatty acid derivatives stimulate cell proliferation and induce STAT-5 activation. Blood. 2001;97:3259–67. doi: 10.1182/blood.v97.10.3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ikuta T, Kan YW, Swerdlow PS, Faller DV, Perrine SP. Alterations in protein-DNA interactions in the gamma-globin gene promoter in response to butyrate therapy. Blood. 1998;92:2924–33. [PubMed] [Google Scholar]
- 30.Pace BS, Chen YR, Thompson A, Goodman SR. Butyrate-inducible elements in the human gamma-globin promoter. Exp Hematol. 2000;28:283–93. doi: 10.1016/s0301-472x(99)00153-8. [DOI] [PubMed] [Google Scholar]
- 31.Kwok RP, Lundblad JR, Chrivia JC, et al. Nuclear protein CBP is a coactivator for the transcription factor CREB. Nature. 1994;370:223–6. doi: 10.1038/370223a0. [DOI] [PubMed] [Google Scholar]
- 32.Platt OS, Brambilla DJ, Rosse WF, et al. Mortality in sickle cell disease. Life expectancy and risk factors for early death. N Engl J Med. 1994;330:1639–44. doi: 10.1056/NEJM199406093302303. [DOI] [PubMed] [Google Scholar]
- 33.Perrine SP, Miller BA, Faller DV, et al. Sodium butyrate enhances fetal globin gene expression in erythroid progenitors of patients with Hb SS and beta thalassemia. Blood. 1989;74:454–9. [PubMed] [Google Scholar]
- 34.Cao H, Stamatoyannopoulos G, Jung M. Induction of human gamma globin gene expression by histone deacetylase inhibitors. Blood. 2004;103:701–9. doi: 10.1182/blood-2003-02-0478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Atweh GF, Sutton M, Nassif I, et al. Sustained induction of fetal hemoglobin by pulse butyrate therapy in sickle cell disease. Blood. 1999;93:1790–7. [PMC free article] [PubMed] [Google Scholar]
- 36.Ley TJ, DeSimone J, Noguchi CT, et al. 5-Azacytidine increases gamma-globin synthesis and reduces the proportion of dense cells in patients with sickle cell anemia. Blood. 1983;62:370–80. [PubMed] [Google Scholar]
- 37.Lavelle DE. The molecular mechanism of fetal hemoglobin reactivation. Semin Hematol. 2004;41:3–10. doi: 10.1053/j.seminhematol.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 38.Haby C, Lisovoski F, Aunis D, Zwiller J. Stimulation of the cyclic GMP pathway by NO induces expression of the immediate early genes c-fos and junB in PC12 cells. J Neurochem. 1994;62:496–501. doi: 10.1046/j.1471-4159.1994.62020496.x. [DOI] [PubMed] [Google Scholar]
- 39.Kuroyanagi Y, Kaneko Y, Muta K, et al. cAMP differentially regulates gamma-globin gene expression in erythroleukemic cells and primary erythroblasts through c-Myb expression. Biochem Biophys Res Commun. 2006;344:1038–47. doi: 10.1016/j.bbrc.2006.03.203. [DOI] [PubMed] [Google Scholar]
- 40.King SB. A role for nitric oxide in hydroxyurea-mediated fetal hemoglobin induction. J Clin Invest. 2003;111:171–2. doi: 10.1172/JCI17597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Witt O, Sand K, Pekrun A. Butyrate-induced erythroid differentiation of human K562 leukemia cells involves inhibition of ERK and activation of p38 MAP kinase pathways. Blood. 2000;95:2391–6. [PubMed] [Google Scholar]
- 42.Park JI, Choi HS, Jeong JS, Han JY, Kim IH. Involvement of p38 kinase in hydroxyurea-induced differentiation of K562 cells. Cell Growth Differ. 2001;12:481–6. [PubMed] [Google Scholar]
- 43.Cokic VP, Andric SA, Stojilkovic SS, Noguchi CT, Schechter AN. Hydroxyurea nitrosylates and activates soluble guanylyl cyclase in human erythroid cells. Blood. 2008;111:1117–23. doi: 10.1182/blood-2007-05-088732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hsiao CH, Li W, Lou TF, Baliga BS, Pace BS. Fetal hemoglobin induction by histone deacetylase inhibitors involves generation of reactive oxygen species. Exp Hematol. 2006;34:264–73. doi: 10.1016/j.exphem.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 45.Aerbajinai W, Zhu J, Gao Z, Chin K, Rodgers GP. Thalidomide induces gamma-globin gene expression through increased reactive oxygen species-mediated p38 MAPK signaling and histone H4 acetylation in adult erythropoiesis. Blood. 2007;110:2864–71. doi: 10.1182/blood-2007-01-065201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Foulkes NS, Laoide BM, Schlotter F, Sassone-Corsi P. Transcriptional antagonist cAMP-responsive element modulator (CREM) down-regulates c-fos cAMP-induced expression. Proc Natl Acad Sci U S A. 1991;88:5448–52. doi: 10.1073/pnas.88.12.5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pissard S, Beuzard Y. A potential regulatory region for the expression of fetal hemoglobin in sickle cell disease. Blood. 1994;84:331–8. [PubMed] [Google Scholar]
- 48.Okuda T, van Deursen J, Hiebert SW, Grosveld G, Downing JR. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell. 1996;84:321–30. doi: 10.1016/s0092-8674(00)80986-1. [DOI] [PubMed] [Google Scholar]
- 49.Hoeffler JP, Meyer TE, Yun Y, Jameson JL, Habener JF. Cyclic AMP-responsive DNA-binding protein: structure based on a cloned placental cDNA. Science. 1988;242:1430–3. doi: 10.1126/science.2974179. [DOI] [PubMed] [Google Scholar]