Abstract
Human TopBP1 plays a critical role in the control of DNA replication checkpoint. In this study, we report a specific interaction between TopBP1 and BACH1/FANCJ, a DNA helicase involved in the repair of DNA cross-links. The TopBP1/BACH1 interaction is mediated by the very C-terminal tandem BRCT domains of TopBP1 and S phase-specific phosphorylation of BACH1 at Thr 1133 site. Interestingly, we demonstrate that depletion of TopBP1 or BACH1 attenuates the loading of RPA on chromatin. Moreover, both TopBP1 and BACH1 are required for ATR-dependent phosphorylation events in response to replication stress. Taken together, our data suggest that BACH1 has an unexpected early role in replication checkpoint control. A specific interaction between TopBP1 and BACH1 is likely to be required for the extension of single-stranded DNA regions and RPA loading following replication stress, which is pre-requisite for the subsequent activation of replication checkpoint.
Keywords: TopBP1, BACH1/FNACJ, RPA, replication, checkpoint, ATR, Chk1
INTRODUCTION
DNA damage checkpoints are evolved in eukaryotic cells to coordinate cellular responses to DNA lesions in an attempt to maintain genomic integrity. One major DNA damage checkpoint pathway is the replication checkpoint, which is activated primarily in response to stalled replication forks (Hook et al., 2007). This checkpoint pathway is controlled by the activation of two protein kinases ATR and Chk1. It is generally believed that the assembly of multi-protein complexes including ATR-ATRIP, TopBP1, Rad9/Hus1/Rad1 (dubbed as 9-1-1) at stalled replication forks are important for ATR and subsequently Chk1 activation. Recent studies also highlight the involvement of RPA-coated single-stranded DNA (ssDNA) regions and TopBP1 in connecting ATR-ATRIP and 9-1-1 in response to replication stress (Burrows and Elledge, 2008; Delacroix et al., 2007; Kumagai et al., 2006; Lee et al., 2007; Mordes et al., 2008).
Human TopBP1 possess eight BRCT phospho-peptide recognition motifs and an ATR-activating domain (Bartek and Mailand, 2006; Cimprich and Cortez, 2008; Kumagai et al., 2006; Manke et al., 2003; Yu et al., 2003). The orthologs of TopBP1 in Schizosaccharomyces pombe Rad4/Cut5, Xenopus Cut5, Saccharomyces cerevisiae Dpb11, and Drosophila melanogaster Mus101 all play essential roles in both DNA replication and DNA damage checkpoints (Saka et al., 1994; Van Hatten et al., 2002; Wang and Elledge, 1999; Yamamoto et al., 2000). Like its counterparts in other species, human TopBP1 has also been implicated in both DNA replication and checkpoint signaling (Kim et al., 2005; Makiniemi et al., 2001; Yamane et al., 2002).
It is speculated that multiple BRCT repeats within TopBP1 could explain its diverse functions. Indeed, the fifth BRCT domain (BRCT5) of TopBP1 is required for its focus localization following DNA damage (Yamane et al., 2002). Studies in yeast suggest that the first two BRCT domains of Dpb11 associate with phosphorylated Sld3 and are required for replication initiation (Tanaka et al., 2007; Zegerman and Diffley, 2007). Recently, the N-terminal BRCT domains of TopBP1 are shown to interact with phosphorylated Rad9 tail in the 9-1-1 complex and be required for ATR-mediated Chk1 activation in mammalian cell lines (Delacroix et al., 2007) and Xenopus egg extracts (Lee et al., 2007). Excitingly, a region between the sixth and seventh BRCT repeats of TopBP1 termed the ATR-activating domain (AD) has been identified (Kumagai et al., 2006). This domain is sufficient to activate ATR in vitro and in vivo (Delacroix et al., 2007; Kumagai et al., 2006). One can envision a scenario that, in response to replication stress, ssDNA-coated RPA and other components at stalled replication forks may independently signal the recruitment of ATR-ATRIP complex and trigger Rad17/RFC dependent clamp loading of the 9-1-1 complex. In this model, TopBP1 plays a critical mediator role by binding to 9-1-1 complex via its N terminal tandem BRCT domains and activating ATR through its ATR activation domain (AD), which leads to subsequent ATR-dependent Chk1 phosphorylation and activation. However, it is not yet known whether TopBP1 has several functions during this process, especially given that TopBP1 has multiple protein-protein interaction domains.
In this study, we established a functional connection between TopBP1 and BACH1. BACH1 (BRCA1-associated C-terminal helicase), also known as FANCJ and BRIP1, was first identified as physiological partner of BRCA1 (Cantor et al., 2001). Recent evidence suggest that the BRCA1 BRCT domain directly interacts with phosphorylated BACH1, and this phosphorylation-specific interaction plays a critical role in checkpoint control in response to DNA double-stranded breaks (Yu et al., 2003). More recently, FANCJ was shown to be defective in patients from the FA complementation group J and functions in interstrand cross-link repair (Bridge et al., 2005; Levran et al., 2005; Litman et al., 2005). A role of BACH1 in DNA replication has also been proposed. The helicase activity of BACH1 peaks in S phase and may play a role in S phase progression (Kumaraswamy and Shiekhattar, 2007). This function of BACH1 may be linked with its ability to unwind G4 DNA structures, which could impede DNA replication (London et al., 2008; Wu et al., 2008). However, whether or not BACH1 has a direct role in replication checkpoint control and how it may act in this process have not been elucidated.
Here we report the identification of BACH1 as a TopBP1-binding protein. We provide evidence suggesting that BACH1, together with TopBP1, has an unexpected role at early stage of replication checkpoint control.
MATERIALS AND METHODS
Cell culture and Plasmids
HeLa, 293T and U2OS cells were maintained in RPMI 1640 supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin at 37°C in a humidified incubator with 5% CO2 (v/v). TopBP1 or BACH1 cDNA was cloned using gateway technology (Invitrogen). All mutants were generated by site-directed mutagenesis (Stratagene) and verified by sequencing.
Antibodies
Rabbit polyclonal anti-TopBP1 antibody was described previously (Kim et al., 2005; Yamane et al., 2002). Anti-BACH1pT1133 polyclonal antibody was raised against phospho-peptide CESIYF-(phospho-T)-PELYDPEDT and affinity purified.
Co-immunoprecipitation and Western Blotting
Cells were lysed with NTEN buffer (20 mM Tris-HCl, pH 8.0, 100 mM NaCl, 1 mM EDTA, 0.5% Nonidet P-40) containing protease inhibitors on ice for 20 minutes. The soluble fractions were collected after centrifuge and incubated with either protein A agarose beads coupled with anti-TopBP1, BACH1 antibodies or streptavidin sepharose beads (Amersham Biosciences) for 3 hours at 4 °C. The precipitates were then washed and boiled in 2× SDS loading buffer. Samples were resolved on SDS-PAGE, transferred to PVDF membrane and immunoblottings were carried out with antibodies as indicated.
Chromatin extraction preparation
The preparation of chromatin fraction was performed as described previously (Ward and Chen, 2001) with modifications. Briefly, cells were harvested at indicated time after treatment with 10 mM hydroxyurea (HU) and subsequently lysed with 10 volumes of NTEN buffer with low salt (20 mM Tris-HCl, pH 8.0, 10 mM NaCl, 1.5mM MgCl2, 1 mM EDTA, 0.5% Nonidet P-40, 20mM NaF, 1mM Na3VO4, 1μg/ml aprotinin, and 1μg/ml pepstatin). The chromatin-enriched pellet was washed with PBS three times. The insoluble chromatin fractions were resuspended in 0.2 M HCl for 30 minutes on ice. The resultant soluble extraction was neutralized with 1M Tris-HCl pH 8.5 for further analysis.
RNA interference
Briefly, HeLa cells were transfected twice at 24-hour time intervals with indicated siRNAs using oligofectamine (Invitrogen) according to the manufacturer's instructions. Small interfering RNAs (siRNAs) against human TopBP1 or BACH1 were previously described (Kim et al., 2005; Yu and Chen, 2004; Yu et al., 2003). The sequence of control siRNA is UUCAAUAAAUUCUUGAGGUUU. The smart pool small interfering RNAs (siRNAs) against ATR or Rad9 were purchased from Dharmacon Inc.
RESULTS
TopBP1 interacts with BACH1
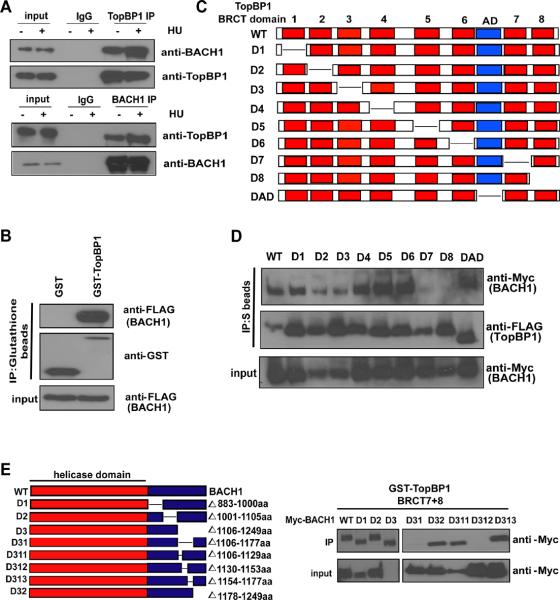
Since we speculate that TopBP1 may exert its functions by interacting with many different binding partners, we established a 293T derivative cell line stably expressing a triple-tagged (S-protein, FLAG and streptavidin binding peptide) TopBP1 and performed tandem affinity purification of TopBP1 complexes. These experiments were performed 24 hours after incubation of cells with 2 mM HU to maximize the chance to identify TopBP1 binding partners involved in replication checkpoint control. Mass spectrometry analysis revealed that one of the major TopBP1 binding proteins is BACH1, a DNA helicase that involves in the repair of DNA cross-links (Taniguchi and D'Andrea, 2006; Wang, 2007). In fact, an interaction between TopBP1 and BACH1 has been previously reported (Greenberg et al., 2006). We confirmed the binding of TopBP1 to BACH1 using co-immunoprecipitation experiments (Fig. 1A). The binding of BACH1 to TopBP1 increased modestly after cells were treated with HU (Fig. 1A). Because TopBP1 interacted with BACH1 in baculovirus-insect expression system (Fig. 1B) and that the TopBP1/BACH1 interaction occurred in cells depleted of BRCA1 or CtIP (Fig. S1A), these two proteins likely interact directly with each other.
FIGURE 1. TopBP1 interacts with BACH1.
(A) Endogenous interaction between TopBP1 and BACH1. HeLa cells were treated with or without HU (1 mM) for 24 hours before collection. Control, anti-TopBP1 immunoprecipitates or anti-BACH1 immunoprecipitates were immunoblotted with indicated antibodies. (B) TopBP1 specifically binds to BACH1. Sf9 cells were co-infected with baculoviruses expressing GST or GST-tagged TopBP1 together with viruses expressing SFB-BACH1. GST pulldowns were immunoblotted with antibodies as indicated. (C) Schematic presentation of wild-type and deletion mutants of TopBP1 used in this study. (D, E) Mapping of the corresponding regions required for the TopBP1/ BACH1 interaction. Immunoprecipitation reactions were performed using S-protein beads and then subjected to Western blot analyses using antibodies as indicated (D). Also see Figure.S1B. (E) Schematic diagram of wild-type and deletion mutants of BACH1 used in this study (left). Beads coated with bacterially expressed GST or GST fusion of 7th and 8th tandem BRCT domains of TopBP1 were incubated with cell lysates containing exogenously expressed Myc-tagged wild-type or deletions of BACH1. Immunoblotting experiments were carried out using indicated antibodies (right).
Using SFB-tagged wild-type TopBP1 and a series of TopBP1 deletion mutants (Fig. 1C), we showed that deletions of either the 7th or the 8th BRCT domains of TopBP1 led to a dramatic decrease in TopBP1/BACH1 interaction (Fig. 1D), indicating that the very C-terminal tandem BRCT domains of TopBP1 is important for its binding to BACH1. Conversely, we generated and purified recombinant GST-fused TopBP1-BRCT7/8 protein from bacteria. Using a series of BACH1 deletion mutants, we were able to map the minimal TopBP1-binding region to residues 1130 to 1153 of BACH1 (Fig. 1E).
Phosphorylation of BACH1 at Thr1133 is required for its interaction with TopBP1
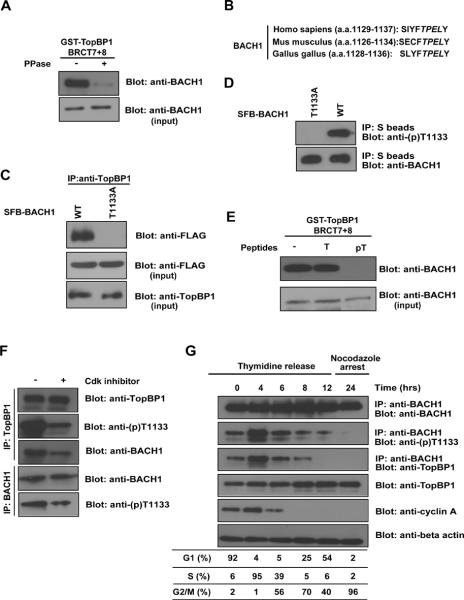
BRCT domain is a phospho-protein binding domain (Manke et al., 2003; Yu et al., 2003). Based on above results, we reasoned that the very C-terminal tandem BRCT domains of TopBP1 might bind specifically to phosphorylated BACH1. Indeed, while TopBP1 bound specifically to endogenous BACH1 in cell lysate, this interaction was abolished by pre-treatment of cell lysate with λ protein phosphatase (PPase) (Fig. 2A).
FIGURE 2.
Phosphorylation of BACH1 at Thr1133 is required for its interaction with TopBP1. (A) TopBP1 interacts with phosphorylated BACH1. Whole cell extract prepared from HeLa cells were mock treated or treated with Lambda PPase, and then incubated with beads coated with bacterially expressed GST-TopBP1-BRCT7&8. Immunoblotting was conducted using anti-BACH1 antibody. (B) Alignment of potential phospho-motif on BACH1 among species. (C, D) In vivo recognition of Thr1133 phosphorylated BACH1 by TopBP1. Cell lysates containing SFB-tagged wild-type or T1133A mutant were subjected to immunoprecipitation using anti-TopBP1 antibodies or S protein beads and immunoblotted with indicated antibodies. Also see Figure S2. (E) Phosphorylated BACH1 peptide competes with endogenous BACH1 for TopBP1 binding. Extracts prepared from 293T cells were incubated with beads containing GST-TopBP1-BRCT7&8 fusion proteins in the presence of phosphorylated BACH1 peptides (pT) or unphosphorylated control peptides (T). Beads were washed and associated BACH1 were detected by immunoblotting. (F) Phosphorylation of BACH1 at Thr1133 might be regulated by cyclin-dependent kinases. HeLa cells were treated with CDK inhibitor for 10 hours before collection. Anti-TopBP1 immunoprecipitates or anti-BACH1 immunoprecipitates were immunoblotted with indicated antibodies. (G) Phosphorylation of BACH1 at Thr1133 site is cell cycle regulated. HeLa Cells were synchronized by double thymidine block as described previously (Yu and Chen, 2004), and then released in fresh medium without thymidine and collected at the indicated timepoints. Alternatively, HeLa cells were synchronized at mitosis with nocodazole (0.5 μg/ml) treatment for 24 hours. Cell lysates were prepared, and immunoprecipitaion and immunoblotting experiments were performed using antibodies as indicated.
BACH1 is DNA helicase known to be phosphorylated and function in S phase (Kumaraswamy and Shiekhattar, 2007; Yu et al., 2003). Many cell cycle regulated phosphorylation events are regulated by cyclin-dependent kinases, which normally phosphorylate serine or threonine residues followed by proline. Within the TopBP1-binding region of BACH1, the residue following Thr1133 of BACH1 is a proline (Fig. 2B). In addition, the residues surrounding Thr1133 of human BACH1 is conserved in other species, suggesting this motif may be important for BACH1 function (Fig. 2B). Indeed, in contrast to wild-type BACH1, the T1133A mutant of BACH1 failed to bind to endogenous TopBP1 in vivo (Fig. 2C), suggesting that residue Thr1133 of BACH1 is critical for the BACH1-TopBP1 interaction.
We generated a phospho-specific antibody raised against a peptide containing phospho-Thr1133 of BACH1. This antibody specifically recognized wild-type BACH1, but failed to recognize the T1133A mutant (Fig. 2D), indicating that Thr1133 of BACH1 is indeed phosphorylated in vivo. Moreover, only the phosphorylated BACH1 peptide, but not the unphosphorylated control peptide, competed with endogenous BACH1 for binding to the C terminal tandem BRCT domains of TopBP1 (Fig. 2E), further confirming that TopBP1 binds selectively to the phosphorylated Thr1133 site of BACH1. In addition, we observed that the BRCT7&8 of TopBP1 can specifically bind pT1133 of BACH1 by peptides pulldown and fluorescence polarization assay (Fig. S2). These results support that TopBP1 interacts with BACH1 in a phosphorylation-dependent manner.
Since the Thr1133 site appears to be a CDK site, we examined and observed that phosphorylation of BACH1 and its interaction with TopBP1 decreased in cells treated with CDK inhibitor (Fig. 2F). As shown in Fig. 2G, while BACH1 protein level remained constant throughout cell cycle, phosphorylation of BACH1 at Thr1133 site was significantly enriched during S phase, which appears to correlate with its interaction with TopBP1. Take together, these data indicate that BACH1 is specifically phosphorylated at Thr1133 site in S phase and only BACH1 that is phosphorylated at Thr1133 interact with TopBP1.
Both TopBP1 and BACH1 contribute to chromatin loading of RPA, which is pre-requisite for ATR activation in response to replication stress
RPA-bound ssDNA is known to be necessary for ATR activation following DNA damage (Zou and Elledge, 2003). Since ATR activation also requires ATRIP, TopBP1, and probably 9-1-1 complex, the interesting question is which factors arrive at sites of stalled replication fork firstly and how they facilitate the activation of ATR on RPA-coated ssDNA. In order to answer this question, we decided to first examine whether TopBP1 or BACH1 would be involved in the chromatin loading of RPA in response to replication stress.
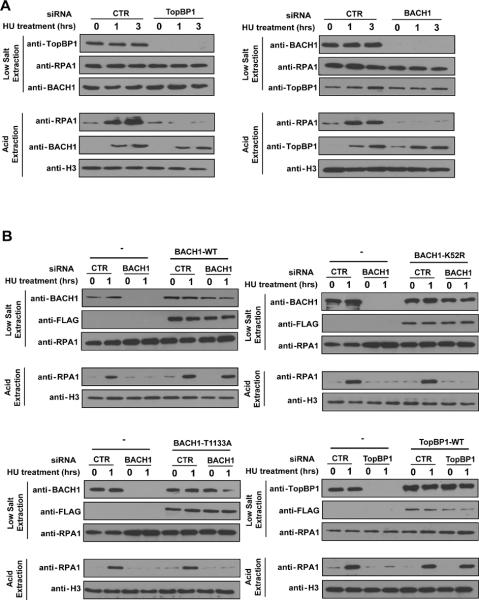
Compared with cells transfected with control siRNA, cells with TopBP1 or BACH1 depletion displayed a substantial decrease of chromatin loading of RPA following HU treatment, indicating that efficient RPA accumulation at arrested replication forks requires TopBP1 and BACH1 (Fig. 3A). Moreover, the expression of siRNA-resistant wild-type BACH1 or TopBP1 fully restored RPA1 chromatin loading in BACH1 or TopBP1 depleted cells, respectively (Fig. 3B), whereas the expression of siRNA-resistant BACH1 K52R mutant (a helicase inactive mutant of BACH1) failed to accumulate RPA1 on chromatin when endogenous BACH1 was depleted (Fig. 3B), indicating that BACH1 helicase activity is involved in this function of BACH1. This defect of RPA1 chromatin loading was also observed in cells only expressing the T1133A mutant of BACH1 (Fig. 3B), suggesting that the interaction between TopBP1 and BACH1 plays a critical role on RPA chromatin loading following replication stress. In addition, we noticed that the number of HU-induced RPA1 foci reduced dramatically in cells depleted of BACH1 or TopBP1, or in cells only expressing T1133A mutant of BACH1 (Fig. S3A). These data are consistent with the idea that BACH1 and its interaction with TopBP1 are involved in the generation of extended ssDNA regions, which allow the efficient accumulation of RPA molecules.
FIGURE 3.
TopBP1 and BACH1 are involved in the chromatin accumulation of RPA following replication stress. (A) Both TopBP1 and BACH1 are required for RPA loading following HU treatment. U2OS cells transfected with control, TopBP1 or BACH1 siRNAs were mock treated or treated with HU (10 mM). Cells were harvested at the indicated timepoints. The soluble and chromatin fractions were prepared and immunoblotted with antibodies as indicated. Also see Figure S3. (B) The interaction between TopBP1 and BACH1 is required for RPA1 loading on chromatin at stalled replication stress. U2OS cells were infected with retroviruses expressing siRNA-resistant wild-type TopBP1 and then transfected with control or TopBP1 siRNAs. In the other case, U2OS cells stably expressing siRNA resistant wild-type, K52R mutant or T1133A mutant of BACH1 were transfected with control siRNA or BACH1 specific siRNA. 72 hours after initial siRNA transfection, cells were treated with 10mM HU and collected one hour later. Cell lysates were immunoblotted with indicated antibodies.
We further verified that TopBP1 and BACH1 are positioned at early stage of replication checkpoint, since both of them are also involved in the loading of Rad9 and ATR to chromatin at stalled replication forks (Fig. S4A). This is consistent with the recent data shown in Xenopus laevis, which TopBP1 is required for the loading of the 9-1-1 complex onto stalled replication forks (Yan and Michael, 2009). In addition, while cells with ATR or Rad9 depletion exhibited obvious decline of Chk1 and RPA2 phosphorylation following HU treatment, these cells did not display any apparent defect in RPA chromatin loading (Fig. S4B).
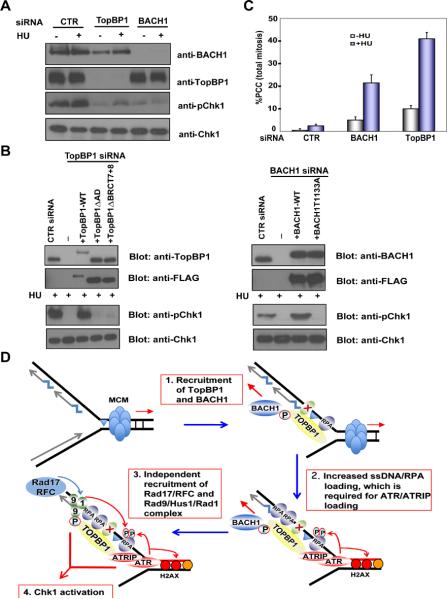
TopBP1 is required for Chk1 activation following replication stress and/or DNA damage (Burrows and Elledge, 2008). Downregulation of BACH1 inhibited HU-induced Chk1 phosphorylation (Fig. 4A). Similarly, cells with TopBP1 downregulation also failed to activate Chk1 (Fig. 4A) as previously reported (Kim et al., 2005). While expression of siRNA-resistant wild-type TopBP1 completely rescued Chk1 activation in cells depleted of endogenous TopBP1, reconstitution with TopBP1 with a deletion of its ATR activation domain (AD) failed to do so (Fig. 4B). This is consistent with previous reports (Delacroix et al., 2007; Kumagai et al., 2006) and suggests that the AD of TopBP1 plays a critical function in replication checkpoint control. Interestingly, TopBP1 mutant deleted of very C terminal tandem BRCT domains also failed to rescue HU-induced Chk1 phosphorylation (Fig. 4B). Conversely, while the expression of siRNA-resistant wild-type BACH1 fully restored Chk1 activation, the T1133A mutant, which does not interact with TopBP1, failed to rescue Chk1 activation defect following HU treatment (Fig. 4B). Taken together, these results suggest that the TopBP1/BACH1 interaction is important for the activation of ATR kinase activity in response to replication stress.
FIGURE 4.
The TopBP1/BACH1 interaction is required for replication checkpoint. (A) BACH1 is required for Chk1 activation upon replication stress. U2OS cells transfected with control siRNA, TopBP1 siRNA or BACH1 siRNA were mock treated or treated with HU (10 mM) and harvested one hour later. Cell lysates were prepared and immunoblotted with antibodies as indicated. (B) The interaction between TopBP1 and BACH1 is required for Chk1 activation. Left panel: U2OS cells were infected with retroviruses expressing siRNA-resistant wild-type or deletion mutants of TopBP1 and then transfected with TopBP1 siRNA. Right panel: U2OS cells or U2OS cells stably expressing siRNA resistant wild-type or T1133A mutant of BACH1 were transfected with BACH1 siRNA. 72 hours after initial siRNA transfection, cells were treated with 10 mM HU and collected one hour later. Cell lysates were immunoblotted with indicated antibodies. (C) Both TopBP1 and BACH1 prevent PCC following replication stress. HeLa cells were transfected with control siRNA, TopBP1 siRNA or BACH1 siRNA and then treated with HU (2 mM) and nocodazole (0.2 μg/μl) for 20 hours. Mitotic spreads were prepared and percentages of cells containing PCC were evaluated under the microscope. The results were the average of three independent experiments and were presented as mean ± SD. (D) A revised model of replication checkpoint control that implicates BACH1 at early stage of this checkpoint response.
It has been suggested that the replication checkpoint, which is mediated by ATR and Chk1 kinases, delays mitotic entry and thus prevents premature chromosome condensation (PCC) (Nghiem et al., 2001). We observed a significant increase of PCC in cells transfected with either TopBP1 or BACH1 siRNA following HU treatment when compared to cells transfected with control siRNA (Fig. 4C), again supporting the idea that like TopBP1, BACH1 is also involved in replication checkpoint control.
DISCUSSION
In this study, we demonstrated that BACH1 interacts directly with TopBP1. This interaction is mediated by the very C-terminal tandem BRCT domains of TopBP1 and the phosphorylation of BACH1 at Thr1133 site. Given that both TopBP1 and BACH1 have been proposed to function in S phase, it is not surprising that Thr1133 site of BACH1 is mainly phosphorylated during S phase, which correlates with its interaction with TopBP1. The exciting finding presented here is that BACH1 has a previously unrecognized role in replication checkpoint control. We showed that BACH1 and its interaction with TopBP1 are required for Chk1 and RPA phosphorylation following replication stress, suggesting that the interaction between BACH1 and TopBP1 plays an important role in replication checkpoint control. Of course, we cannot rule out the possibility that both BACH1 and TopBP1 may use the same binding motifs to interact with other partners that could also contribute to this checkpoint function.
The checkpoint function of BACH1 is likely linked to its involvement in RPA chromatin loading following replication stress. We showed that both BACH1 and TopBP1 are required for the efficient chromatin loading of RPA following HU treatment. TopBP1 and its binding partner BACH1 seem to be at the top of this replication stress pathway, since both of them are also involved in ATR and Rad9 chromatin loading at stalled replication forks. We would like to propose a revised model of replication checkpoint control (Fig. 4D). We speculate that one of the initial events occurred at stalled replication forks is the retention of TopBP1, which requires the fifth BRCT domain of TopBP1 and may also involve RPA-coated ssDNA and other unknown proteins or structures. Similarly, BACH1 is also recruited to stalled replication forks. While the events required for BACH1 recruitment are still not clear, we know that this can occur independent of TopBP1 (Fig. 3A and Fig. S3B). Although TopBP1 and BACH1 are independently recruited to stalled replication forks, they function together in the activation of replication checkpoint. Since BACH1 is a DNA helicase, we suspect that at least one of the functions of BACH1 is to facilitate the unwinding of double-stranded DNA or other DNA structures, which allows the exposure of large tracts of ssDNAs that are then coated by additional RPA molecules. This efficient accumulation of ssDNA/RPA is important for the amplification of replication stress signals since it leads to the loading of the ATRIP-ATR and the 9-1-1 complex at stalled replication forks, and initiates replication checkpoint. Further refinement of this working hypothesis will provide insights into the mechanisms of replication stress-induced checkpoint control, which is critically important for the maintenance of genomic integrity and tumor suppression.
Supplementary Material
ACKNOWLEDGMENTS
We would like to thank all colleagues in Chen's laboratory for insightful discussion and technical assistance. This work was supported in part by grants from the National Institutes of Health (CA089239, CA092312 and CA100109 to J.C.). J.C is a recipient of an Era of Hope Scholar award from the Department of Defense and a member of the Mayo Clinic Breast SPORE program.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCE
- Bartek J, Mailand N. TOPping up ATR activity. Cell. 2006;124:888–890. doi: 10.1016/j.cell.2006.02.029. [DOI] [PubMed] [Google Scholar]
- Bridge WL, Vandenberg CJ, Franklin RJ, Hiom K. The BRIP1 helicase functions independently of BRCA1 in the Fanconi anemia pathway for DNA crosslink repair. Nat Genet. 2005;37:953–957. doi: 10.1038/ng1627. [DOI] [PubMed] [Google Scholar]
- Burrows AE, Elledge SJ. How ATR turns on: TopBP1 goes on ATRIP with ATR. Genes Dev. 2008;22:1416–1421. doi: 10.1101/gad.1685108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantor SB, Bell DW, Ganesan S, Kass EM, Drapkin R, Grossman S, Wahrer DC, Sgroi DC, Lane WS, Haber DA, Livingston DM. BACH1, a novel helicase-like protein, interacts directly with BRCA1 and contributes to its DNA repair function. Cell. 2001;105:149–160. doi: 10.1016/s0092-8674(01)00304-x. [DOI] [PubMed] [Google Scholar]
- Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol. 2008;9:616–627. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delacroix S, Wagner JM, Kobayashi M, Yamamoto K, Karnitz LM. The Rad9-Hus1-Rad1 (9-1-1) clamp activates checkpoint signaling via TopBP1. Genes Dev. 2007;21:1472–1477. doi: 10.1101/gad.1547007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg RA, Sobhian B, Pathania S, Cantor SB, Nakatani Y, Livingston DM. Multifactorial contributions to an acute DNA damage response by BRCA1/BARD1-containing complexes. Genes Dev. 2006;20:34–46. doi: 10.1101/gad.1381306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hook SS, Lin JJ, Dutta A. Mechanisms to control rereplication and implications for cancer. Curr Opin Cell Biol. 2007;19:663–671. doi: 10.1016/j.ceb.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JE, McAvoy SA, Smith DI, Chen J. Human TopBP1 ensures genome integrity during normal S phase. Mol Cell Biol. 2005;25:10907–10915. doi: 10.1128/MCB.25.24.10907-10915.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai A, Lee J, Yoo HY, Dunphy WG. TopBP1 activates the ATR-ATRIP complex. Cell. 2006;124:943–955. doi: 10.1016/j.cell.2005.12.041. [DOI] [PubMed] [Google Scholar]
- Kumaraswamy E, Shiekhattar R. Activation of BRCA1/BRCA2-associated helicase BACH1 is required for timely progression through S phase. Mol Cell Biol. 2007;27:6733–6741. doi: 10.1128/MCB.00961-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Kumagai A, Dunphy WG. The Rad9-Hus1-Rad1 checkpoint clamp regulates interaction of TopBP1 with ATR. J Biol Chem. 2007;282:28036–28044. doi: 10.1074/jbc.M704635200. [DOI] [PubMed] [Google Scholar]
- Levran O, Attwooll C, Henry RT, Milton KL, Neveling K, Rio P, Batish SD, Kalb R, Velleuer E, Barral S, et al. The BRCA1-interacting helicase BRIP1 is deficient in Fanconi anemia. Nat Genet. 2005;37:931–933. doi: 10.1038/ng1624. [DOI] [PubMed] [Google Scholar]
- Litman R, Peng M, Jin Z, Zhang F, Zhang J, Powell S, Andreassen PR, Cantor SB. BACH1 is critical for homologous recombination and appears to be the Fanconi anemia gene product FANCJ. Cancer Cell. 2005;8:255–265. doi: 10.1016/j.ccr.2005.08.004. [DOI] [PubMed] [Google Scholar]
- London TB, Barber LJ, Mosedale G, Kelly GP, Balasubramanian S, Hickson ID, Boulton SJ, Hiom K. FANCJ is a structure-specific DNA helicase associated with the maintenance of genomic G/C tracts. J Biol Chem. 2008;283:36132–36139. doi: 10.1074/jbc.M808152200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makiniemi M, Hillukkala T, Tuusa J, Reini K, Vaara M, Huang D, Pospiech H, Majuri I, Westerling T, Makela TP, Syvaoja JE. BRCT domain-containing protein TopBP1 functions in DNA replication and damage response. J Biol Chem. 2001;276:30399–30406. doi: 10.1074/jbc.M102245200. [DOI] [PubMed] [Google Scholar]
- Manke IA, Lowery DM, Nguyen A, Yaffe MB. BRCT repeats as phosphopeptide-binding modules involved in protein targeting. Science. 2003;302:636–639. doi: 10.1126/science.1088877. [DOI] [PubMed] [Google Scholar]
- Mordes DA, Glick GG, Zhao R, Cortez D. TopBP1 activates ATR through ATRIP and a PIKK regulatory domain. Genes Dev. 2008;22:1478–1489. doi: 10.1101/gad.1666208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nghiem P, Park PK, Kim Y, Vaziri C, Schreiber SL. ATR inhibition selectively sensitizes G1 checkpoint-deficient cells to lethal premature chromatin condensation. Proc Natl Acad Sci U S A. 2001;98:9092–9097. doi: 10.1073/pnas.161281798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saka Y, Fantes P, Sutani T, McInerny C, Creanor J, Yanagida M. Fission yeast cut5 links nuclear chromatin and M phase regulator in the replication checkpoint control. EMBO J. 1994;13:5319–5329. doi: 10.1002/j.1460-2075.1994.tb06866.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S, Umemori T, Hirai K, Muramatsu S, Kamimura Y, Araki H. CDK-dependent phosphorylation of Sld2 and Sld3 initiates DNA replication in budding yeast. Nature. 2007;445:328–332. doi: 10.1038/nature05465. [DOI] [PubMed] [Google Scholar]
- Taniguchi T, D'Andrea AD. Molecular pathogenesis of Fanconi anemia: recent progress. Blood. 2006;107:4223–4233. doi: 10.1182/blood-2005-10-4240. [DOI] [PubMed] [Google Scholar]
- Van Hatten RA, Tutter AV, Holway AH, Khederian AM, Walter JC, Michael WM. The Xenopus Xmus101 protein is required for the recruitment of Cdc45 to origins of DNA replication. J Cell Biol. 2002;159:541–547. doi: 10.1083/jcb.200207090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Elledge SJ. DRC1, DNA replication and checkpoint protein 1, functions with DPB11 to control DNA replication and the S-phase checkpoint in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1999;96:3824–3829. doi: 10.1073/pnas.96.7.3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W. Emergence of a DNA-damage response network consisting of Fanconi anaemia and BRCA proteins. Nat Rev Genet. 2007;8:735–748. doi: 10.1038/nrg2159. [DOI] [PubMed] [Google Scholar]
- Ward IM, Chen J. Histone H2AX is phosphorylated in an ATR-dependent manner in response to replicational stress. J Biol Chem. 2001;276:47759–47762. doi: 10.1074/jbc.C100569200. [DOI] [PubMed] [Google Scholar]
- Wu Y, Shin-ya K, Brosh RM., Jr. FANCJ helicase defective in Fanconia anemia and breast cancer unwinds G-quadruplex DNA to defend genomic stability. Mol Cell Biol. 2008;28:4116–4128. doi: 10.1128/MCB.02210-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto RR, Axton JM, Yamamoto Y, Saunders RD, Glover DM, Henderson DS. The Drosophila mus101 gene, which links DNA repair, replication and condensation of heterochromatin in mitosis, encodes a protein with seven BRCA1 C-terminus domains. Genetics. 2000;156:711–721. doi: 10.1093/genetics/156.2.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane K, Wu X, Chen J. A DNA damage-regulated BRCT-containing protein, TopBP1, is required for cell survival. Mol Cell Biol. 2002;22:555–566. doi: 10.1128/MCB.22.2.555-566.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan S, Michael WM. TopBP1 and DNA polymerase-alpha directly recruit the 9-1-1 complex to stalled DNA replication forks. J Cell Biol. 2009;184:793–804. doi: 10.1083/jcb.200810185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Chen J. DNA damage-induced cell cycle checkpoint control requires CtIP, a phosphorylation-dependent binding partner of BRCA1 C-terminal domains. Mol Cell Biol. 2004;24:9478–9486. doi: 10.1128/MCB.24.21.9478-9486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Chini CC, He M, Mer G, Chen J. The BRCT domain is a phospho-protein binding domain. Science. 2003;302:639–642. doi: 10.1126/science.1088753. [DOI] [PubMed] [Google Scholar]
- Zegerman P, Diffley JF. Phosphorylation of Sld2 and Sld3 by cyclin-dependent kinases promotes DNA replication in budding yeast. Nature. 2007;445:281–285. doi: 10.1038/nature05432. [DOI] [PubMed] [Google Scholar]
- Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.