Summary
Dynamic changes of chromatin structure facilitate diverse biological events, including DNA replication, repair, recombination, and gene transcription. Recent evidence revealed that DNA damage elicits alterations to the chromatin to facilitate proper checkpoint activation and DNA repair. Here we report the identification of the PWWP domain-containing protein EXPAND1 as an architectural component of the chromatin, which in response to DNA damage, serves as an accessory factor to promote cell survival. Depletion of EXPAND1 or inactivation of its PWWP domain resulted in chromatin compaction. Upon DNA damage, EXPAND1 rapidly concentrates at the vicinity of DNA damage sites via its direct interaction with 53BP1. Ablation of this interaction impaired damage-induced chromatin decondensation, which is accompanied by sustained DNA damage and hypersensitivity to genotoxic stress. Collectively, our study uncovers a chromatin bound factor that serves an accessory role in coupling damage signaling with chromatin changes in response to DNA damage.
Keywords: 53BP1, DNA damage, DNA repair, chromatin, PWWP, EXPAND1, BRCT
Introduction
The eukaryotic chromatin is made up of strings of nucleosome particles, which consist of 146 bp of DNA wrapped around a histone octamer. Further folding and condensation of these linear arrays of nucleosomes result in higher-order chromatin structures that impede many of the activities that require access to the genetic material. To overcome this physical barrier, cells have evolved various mechanisms to mark and modify the chromatin landscape, including histone modifications and local recruitment of chromatin remodeling factors. These mechanisms work together to allow regulated spatial and temporal assess to the genetic material, and thus facilitate and control processes such as DNA replication and gene expression.
Likewise, DNA damage triggers various chromatin modifications and alters chromatin structure to promote DNA repair and cell survival. Recent studies implicated structural components of the chromatin in modulating the DNA damage response. Specifically, cells depleted of the linker histone H1, which normally compacts the chromatin into higher-ordered structure, displayed hypersensitive checkpoint control and were resistant to DNA damage (Murga et al., 2007). Similarly, while the nucleosome binding protein HMGN1 is required for proper ATM chromatin association and activation (Kim et al., 2009), mobilization of the chromatin-binding protein HP1 beta from sites of DNA damage is essential for a full DNA damage response (Ayoub et al., 2008). These and many other examples suggest that genotoxic stress induces numerous dynamic changes of the chromatin architecture, which involve a host of chromatin components that act together and facilitate checkpoint activation and DNA repair.
Several lines of evidence indicated that the major transducer kinase ATM couples DNA damage signaling with chromatin structure changes. A number of ATP-dependent remodeling complexes have been shown, via their binding to the phosphorylated histone H2AX, to localize at the vicinity of DNA breaks and facilitate double-strand break (DSB) repair (Cairns, 2004; Downs et al., 2004; Liang et al., 2007; Morrison et al., 2007). Likewise, the transcription repressor KAP1 has also been reported to facilitate chromatin relaxation in an ATM-dependent manner (Ziv et al., 2006). Together, these studies clearly indicate that damage-induced chromatin change is highly coordinated and involves numerous factors that work in concert to prepare the chromatin for efficient DNA repair.
The tumor suppressor 53BP1 has an established role in mediating non-homologous end joining (Adams and Carpenter, 2006). Specifically, mice and cells deficient in 53BP1 display repair defects, and manifest significant deficits in class-switch recombination and long-range V(D)J recombination (Difilippantonio et al., 2008a; Manis et al., 2004; Ward et al., 2004). Interestingly, 53BP1 was recently implicated in enhancing local dynamics of DNA ends to promote non-homologous end joining (Dimitrova et al., 2008). However, mechanistically how this is achieved remains largely speculative. To better understand how chromatin change is coupled to 53BP1 and to search for novel DNA damage responsive elements, we purified 53BP1-associated factors from the chromatin and identified a PWWP-domain containing protein as a component involved in the DNA damage response. Our data indicate that this newly identified protein is a nucleosome-binding protein that contributes to the maintenance of chromatin state. Therefore, we named this protein as expandere or EXPAND1 to reflect its in vivo function. We propose that EXPAND1, via its interaction with 53BP1, serves as an accessory factor in the DNA damage response pathway to promote chromatin change, which is essential for cell survival.
Results
Identification of EXPAND1 as a 53BP1-associated protein
To identify components involved in the DNA damage response pathway, we generated a 293T derivative stable cell line that expresses streptavidin binding peptide-S peptide-flag (SFB)-tagged 53BP1. Since 53BP1 tightly associates with chromatin (Figure 1a), chromatin-associated 53BP1 complexes were isolated and subjected to mass spectrometry analysis (Figure 1b). Interestingly, a number of DNA damage response factors were identified, including DNA-PK, TOPBP1, NBS1 and RIF1. The functional or physical interactions of 53BP1 with RIF1, NBS1 and TopBP1 were previously reported (Celeste et al., 2003; Silverman et al., 2004; Yamane et al., 2002), whereas the association between 53BP1 and DNA-PK likely reflects the established role of 53BP1 in the NHEJ pathway. While the presence of these known DNA damage repair proteins in 53BP1-containing complexes reconfirmed the role of 53BP1 in the DNA damage response pathway, we were intrigued by the identification of histone H4 and a previously uncharacterized PWWP domain-containing protein (Coulie et al., 1995) in our purification. Since PWWP domain is present amongst many chromatin associated factors, we decided to further our analysis on this PWWP domain-containing protein we named as EXPAND1, and explore its potential functions in the DNA damage response.
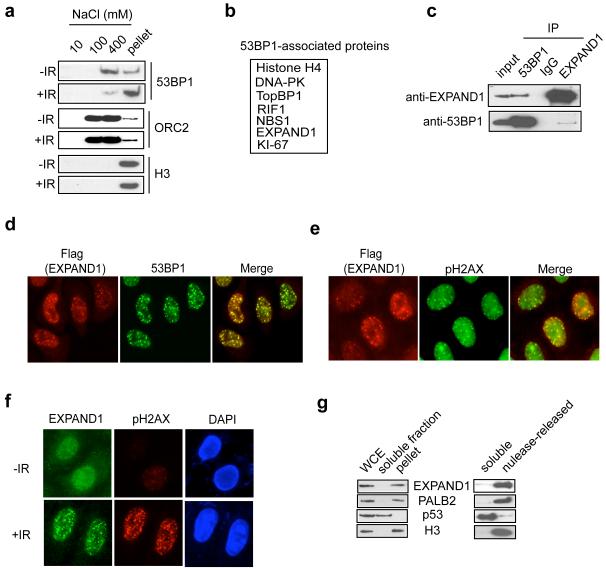
Figure 1.
Identification of EXPAND1 as a damage-response element
a) Untreated or irradiated (10 Gy) HeLa cells were lysed sequentially in NETN buffer containing increasing amount of salt. Cell lysates were then separated by SDS-PAGE and immunoblotted using indicated antibodies, b) Tandem affinity purification identifies EXPAND1 as a 53BP1-associated protein. Table summarizes proteins identified by mass spectrometry analysis of 53BP1 protein complex. Sequence of the identified peptide corresponding to EXPAND1 was LDGSQRPPAVQLEPMAAGAAPSPGPGPGPR, c) Reciprocal immunoprecipitation of 53BP1 and EXPAND1. Nuclease-treated 293T cell lysates were incubated with protein A agarose beads conjugated with indicated antibodies and Western blotting was performed according to standard procedures, d & e) EXPAND1 co-localizes with 53BP1 (d) and pH2AX (e). HeLa cells grown on coverslips were transfected with plasmids encoding flag-tagged EXPAND1, irradiated (10 Gy) and processed as described in Materials and Methods. Indirect immunostaining was performed using indicated antibodies, f) EXPAND1 relocalizes in response to DNA damage. HeLa cells were either irradiated (10 Gy) or left untreated. Immunostaining was conducted using indicated antibodies, g) EXPAND1 is a chromatin associated protein. HeLa cells were biochemically fractionated as described in Materials and Methods. Immunoblotting experiments were performed using indicated antibodies. WCE, whole cell extract.
Using nuclease-treated cell lysate, the interaction between 53BP1 and EXPAND1 was confirmed by reciprocal immunoprecipitations using antibodies against endogenous EXPAND1 and 53BP1, respectively (Figure 1c). We also examined the sub-cellular localization of flag-tagged EXPAND1 after irradiation and found that EXPAND1 colocalized extensively with 53BP1 (Figure 1d). Moreover, exogenous EXPAND1 foci overlapped extensively with those of the DNA damage marker pH2AX (Figure 1e), suggesting that like 53BP1, EXPAND1 may also play a role in the DNA damage response. Furthermore, we affinity-purified rabbit polyclonal antibodies raised against GST-fused EXPAND1 and showed that endogenous EXPAND1 protein displayed a diffuse nuclear staining in unperturbed cells (Figure 1f). Resembling those of tagged version of EXPAND1, IR-induced focal accumulation of endogenous EXPAND1 can be readily detected (Figure 1f), which again overlapped with those of pH2AX. Given that EXPAND1 harbors a PWWP domain and was purified as a 53BP1-associated protein only upon digestion of the chromatin (data not shown), we performed biochemical fractionation and determined that the majority of the EXPAND1 protein remained in the chromatin-enriched fraction (pellet) following NETN extraction, where the chromatin associated protein PALB2 and histone H3 resided (Figure 1g). Moreover, this fraction of EXPAND1, like those of PALB2 and histone H3, can be released upon nuclease treatment, suggesting that EXPAND1 stably associates with chromatin. Together, our results indicate that EXPAND1 is a chromatin component that is involved in the DNA damage response.
Genetic requirement of EXPAND1 focal accumulation at sites of DNA breaks
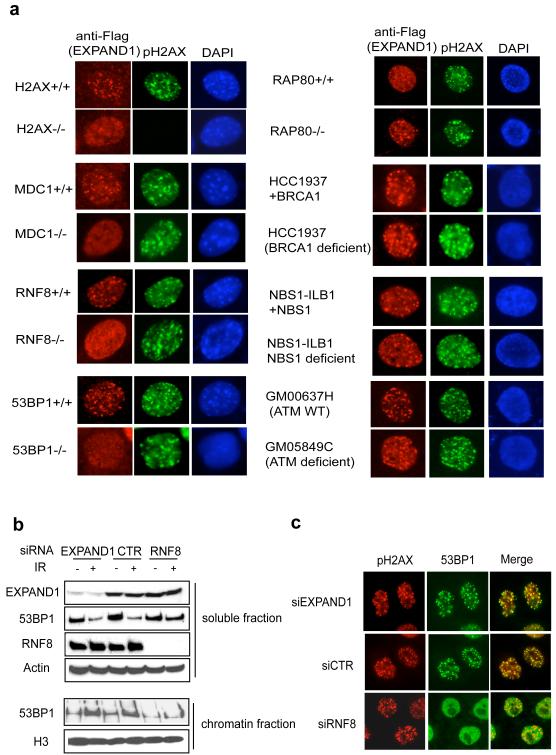
Phosphorylation of the histone variant H2AX is one of the initial DNA damage signals, and is essential for the concentration of various checkpoint and repair proteins at DNA damage sites. Recent studies highlight a DNA damage signaling cascade that entails a step-wise propagation and amplification of the DNA damage signal to elicit proper cellular responses. Since EXPAND1 relocalizes to DNA damage sites, we asked whether ionizing radiation-induced foci formation (IRIF) of EXPAND1 requires these DNA damage response factors. As shown in Figure 2a, EXPAND1 IRIF required H2AX, MDC1, RNF8 and 53BP1 but not RAP80, BRCA1, NBS1 or ATM, suggesting that EXPAND1 plays a role downstream of 53BP1 in the DNA damage response pathway. Since 53BP1 displays enhanced affinity for the damaged chromatin (Figure 1a), we examined whether EXPAND1 might help the chromatin loading of 53BP1 following DNA damage. Fractionating cell lysates derived from siRNA-treated cells revealed that the depletion of RNF8, but not that of EXPAND1, inhibited the damage-induced accumulation of 53BP1 to the chromatin fraction (Figure 2b). In addition, RNF8 but not EXPAND1 depletion abolished 53BP1 IRIF (Figure 2c). Together, these data suggest that RNF8 regulates 53BP1, which in turn governs EXPAND1 localization following DNA damage.
Figure 2.
Genetic requirements of focal accumulation of EXPAND1 to DNA damage sites.
a) EXPAND1 is a component of the DNA damage signaling pathway. Mouse embryonic fibroblasts and patient cells with deficiencies in various components of the DNA damage signaling pathway were retrovirally infected with constructs encoding flag-tagged EXPAND1. Cells were irradiated (10 Gy) and processed 6 hr later. Immunostaining experiments were performed using indicated antibodies, b) EXPAND1 does not affect 53BP1 loading to the chromatin. siRNA-treated HeLa cells were lysed on ice for 10 minutes in NETN (soluble fraction). Soluble fraction was clarified by centrifugation and the pellet (chromatin fraction) was resuspended in sampling buffer and boiled. Western blotting was performed using indicated antibodies, c) EXPAND1 does not affect 53BP1 IRIF. siRNA-treated HeLa cells were irradiated and processed. Immunofluorescent studies were done using indicated antibodies.
EXPAND1 IRIF and its interaction with 53BP1 requires its very N-terminus
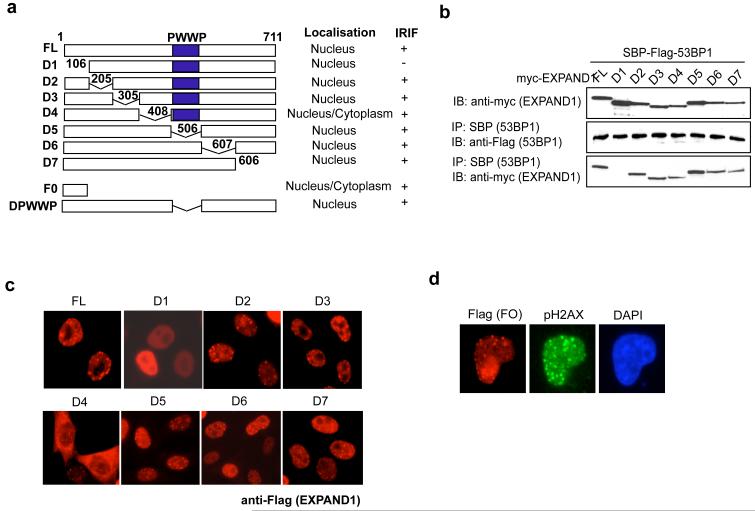
Given that EXPAND1 interacts with 53BP1, we next mapped the 53BP1 binding domain on EXPAND1. A series of deletion mutants that span the entire coding region of EXPAND1 were generated (Figure 3a). Co-immunoprecipitation experiments revealed that EXPAND1 associated with 53BP1 via its very N terminus, since deletion mutant lacking the N-terminal 105 amino acid (D1) failed to co-precipitate with 53BP1 (Figure 3b). Similarly, the D1 mutant, which does not bind to 53BP1, failed to relocalize to DNA breaks (Figure 3c), whereas full-length EXPAND1 and the other mutants retained the ability to concentrate at pH2AX-marked DNA damage sites. Moreover, we found that the EXPAND1 N-terminus alone (F0) was sufficient in localizing to sites of DNA breaks (Figure 3d). Taken together, these data suggest that the binding of EXPAND1 to 53BP1 is important for its proper localization in response to DNA damage.
Figure 3.
The binding of EXPAND1 to 53BP1 requires its N-terminus.
a) Schematic illustration of EXPAND1 full-length (FL) and deletion mutants, b) Co-immunoprecipitation experiments using streptavidin binding peptide-flag-tagged 53BP1 and myc-tagged EXPAND1 or its mutants. 293T cells were transfected with constructs encoding tagged 53BP1 and EXPAND1. Precipitation was conducted using streptavidin beads and immunoblotting was performed using indicated antibodies. c & d) EXPAND1 IRIF requires its 53BP1-binding domains. 293T cells were transfected with constructs encoding flag-tagged EXPAND1 or its mutants. Cells were irradiated 24 hr post transfection and immunostaining was performed to visualize EXPAND1 localization.
53BP1 recruits EXPAND1 to DNA damage sites via its BRCT domain
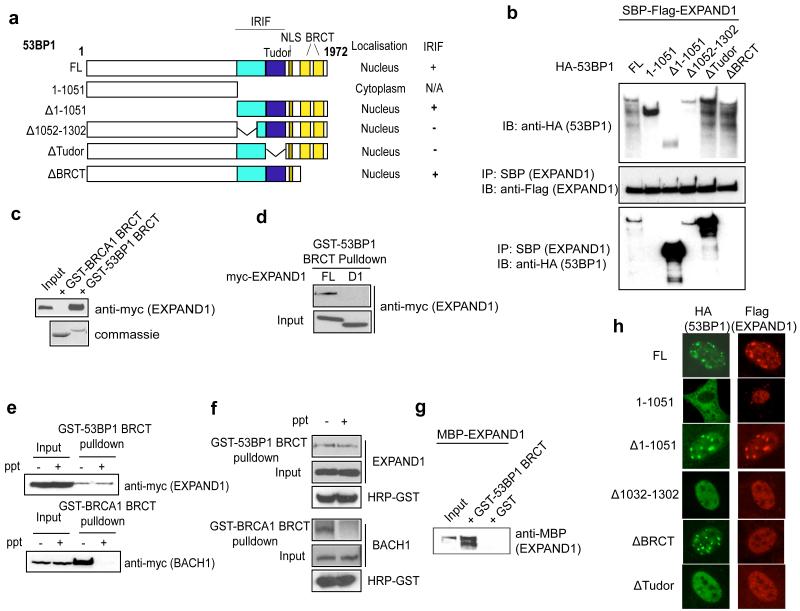
To understand how 53BP1 interacts with EXPAND1 and facilitates its concentration at the vicinity of DNA damage sites, we used a panel of 53BP1 constructs (Figure 4a; Ward et al., 2006). Results indicated that 53BP1 associated with EXPAND1 via its BRCT domain, since the mutants that lack the BRCT domain (mutant 1-1051 and delBRCT) failed to interact with EXPAND1 (Figure 4b). To further corroborate this finding with an independent assay, we performed pulldown experiments using GST-53BP1 BRCT and included GST-BRCA1 BRCT as control. We found that overexpressed EXPAND1 specifically interacted with the 53BP1 BRCT domain but not the BRCA1 BRCT domain (Figure 4c). Furthermore, pulldown experiments consolidated the above notion that EXPAND1 interacts with 53BP1 via its very N-terminus (Figure 4d).
Figure 4.
53BP1 recruits EXPAND1 to sites of DNA damage via its BRCT domain.
a) Schematic illustration of 53BP1 full-length (FL) and deletion mutants, b) Co-immunoprecipitation experiments were performed using SBP-flag-tagged EXPAND1 and HA-tagged 53BP1 full length (FL) or its mutants (see (a)), c) 53BP1 interacts with EXPAND1 via its BRCT domain. Pulldown experiments were performed using bacterially expressed GST-53BP1 BRCT or GST-BRCA1 BRCT together with lysates prepared from 293T cells expressing myc-tagged EXPAND1, d) Pulldown experiment using 53BP1 BRCT domain together with lysates containing myc-tagged full-length (FL) or D1 mutant of EXPAND1, e & f) 53BP1 BRCT-EXPAND1 interaction is phosphorylation-independent. Lysates pre-treated with lambda phosphatase was incubated with GST-53BP1 BRCT or control GST-BRCA1 BRCT domain. Immunoblotting was performed using indicated antibodies, g) Direct interaction between 53BP1 BRCT and EXPAND1. Bacterially purified MBP-EXPAND1 and GST-53BP1 BRCT proteins were co-incubated and pulldown experiment was performed, h) EXPAND1 IRIF requires 53BP1 BRCT domain. 53BP1 deficient cells expressing flag-tagged EXPAND1 were reconstituted with 53BP1 full-length (FL) or mutants. Cells were irradiated (10 Gy) and processed to visualize EXPAND1 localization.
Since BRCT domains have been shown to be phospho-protein binding motifs (Manke et al., 2003; Yu et al., 2003), we tested whether the EXPAND1-53BP1 interaction might be phosphorylation-dependent. The BRCA1 BRCT domain is known to interact with BACH1 in a phosphorylation-dependent manner, and that this interaction can be impaired by prior treatment of cell lysate using protein phosphatase. Surprisingly, although phosphatase treatment abolished the BRCA1 BRCT-BACH1 interaction, it did not noticeably affect the 53BP1 BRCT-EXPAND1 binding (Figure 4e and 4f), indicating that the 53BP1-EXPAND1 interaction might occur irrespective of EXPAND1 phosphorylation status. To confirm this finding, we obtained bacterially expressed MBP-EXPAND1 and showed that EXPAND1 specifically and directly interacted with GST-53BP1 BRCT but not GST alone (Figure 4g). We also monitored EXPAND1 IRIF in 53BP1 deficient cells that has been reconstituted with either wildtype 53BP1 or its various mutants. Consistent with the requirement of the BRCT domain in the 53BP1-EXPAND1 interaction, cells reconstituted with the 53BP1 delBRCT mutant failed to support EXPAND1 IRIF (Figure 4h). Together, the above data suggest that 53BP1 interacts with EXPAND1 via its BRCT domain, which facilitates the relocalization of EXPAND1 to DNA damage sites.
ATM/ATR signaling occurs in EXPAND1-depleted cells
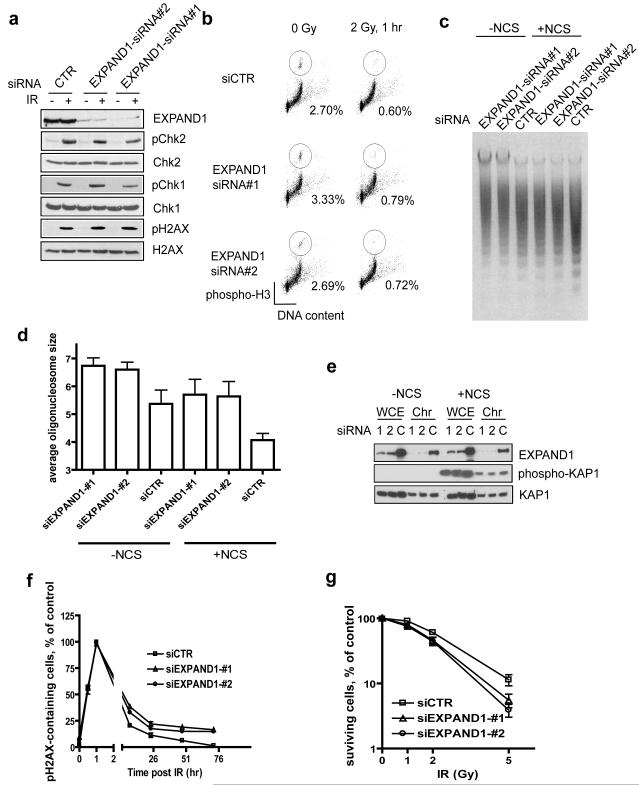
Given that 53BP1 has been reported to play a role in checkpoint control (Abraham, 2002; Fernandez-Capetillo et al., 2002), we next examined whether EXPAND1 might also be involved in damage-induced checkpoint activation. Employing two siRNAs that specifically target EXPAND1, we found that cells depleted of EXPAND1 supported IR-induced Chk1 and Chk2 phosphorylation to similar levels as those observed in control siRNA-treated cells (Figure 5a). In addition, consistent with the notion that EXPAND1 acts downstream of H2AX phosphorylation, EXPAND1 depletion did not affect H2AX phosphorylation (Figure 5a). Together with the fact that EXPAND1-depleted cells arrested at the G2/M checkpoint upon irradiation (Figure 5b), our data imply that EXPAND1 does not play a primary role in IR-induced checkpoint control. Nevertheless, this does not exclude the possibility that EXPAND1 may have an accessory role in checkpoint regulation, especially since siRNA-mediated depletion is never complete. The role of EXPAND1 in checkpoint control should be revisited in the future when genetic knockout mice or cells of EXPAND1 are established.
Figure 5.
Roles of EXPAND1 in chromatin dynamics.
a & b) EXPAND1 is dispensable for checkpoint activation. siRNA-treated HeLa cells were irradiated (10 Gy) or left untreated. Cells were lysed 1 hr post IR and immunoblotting was performed using indicated antibodies (a). EXPAND1-depleted cells properly arrest at the G2/M checkpoint (b). Experiments were repeated three times and similar results were obtained. Representative figure showing mitotic cell population in siRNA-treated cells following DNA damage, c & d) Microccocal nuclease (MNase) sensitivity assay. siRNA-treated cells were incubated with 200 ng/ml NCS or DMSO for 1 hr. Nuclei were extracted as described in Methods and Materials and digested using MNase (c). Bar chart showing average size of nucleosomes (d), e) EXPAND1 depletion does not affect KAP1 expression or its phosphorylation. WCE, whole cell extract; Chr, chromatin-enriched fraction, f & g) EXPAND1 is required for efficient DNA repair and cell survival in response to genotoxic stress. siRNA-treated cells were irradiated (3 Gy) and pH2AX foci were monitored (f). Clonogenic assay showing reduced survival rates in EXPAND1-depleted cells (g). siRNA-treated cells were irradiated with indicated dose of IR. The number of colonies was determined 14 days post irradiation. Results represent mean ±S.D. Experiments were performed twice each in triplicate.
EXPAND1 is required for chromatin relaxation, DNA repair and cell survival
Recent evidence implicated a role of 53BP1 in promoting chromatin dynamics (Dimitrova et al., 2008). However, mechanistically how this is achieved remains largely unexplored. Since EXPAND1 harbors a PWWP domain, which is present amongst many chromatin remodeling factors, we speculated whether EXPAND1 might serve as a component of the chromatin, and in response to DNA damage, facilitate the 53BP1-dependent chromatin changes. DNA damage is known to trigger chromatin relaxation or decondensation and facilitate efficient DNA repair. Therefore, we adopted the microccocal nuclease sensitivity assay to evaluate whether EXPAND1 might participate in the opening of the chromatin structures before and after DNA damage. EXPAND1 depletion partially inhibited nuclease digestion of chromatin as compared to that observed in control cells (Figure 5c). Moreover, while DNA damage resulted in further increase in the digestion of chromatin in control cells, which reflects chromatin relaxation following DNA damage, EXPAND1 siRNA-treated cells were again resistant to this damage-induced relaxation (Supplemental Figure 1a-b). These data suggest that EXPAND1 might play a role in the maintenance of chromatin structure, and that EXPAND1 is required for proper and early chromatin changes in response to genotoxic stress (Supplemental Figure 1c). Although the transcriptional repressor KAP1 was previously shown to mediate chromatin relaxation in response to DNA damage (Ziv et al., 2006), we did not observe detectable changes in the localization and phosphorylation of KAP1 in EXPAND1-depleted cells as compared to control cells (Figure 5e), implying that the KAP1 and EXPAND1-mediated chromatin changes might represent distinct events.
Early studies suggested that chromatin relaxation in response to DNA damage is likely to facilitate efficient DNA repair. We used residual pH2AX foci formation as readout to assess DNA repair efficiency. As depicted in Figure 5f, EXPAND1 depletion, which compromised chromatin relaxation, resulted in significant portion of cells with unrepaired DNA damage. Furthermore, these cells also displayed increased sensitivity to ionizing radiation (Figure 5g). Together, our data suggest that EXPAND1 is involved in modulation of the chromatin structure, which is required for efficient DNA repair and cell survival following DNA damage.
EXPAND1 targets to chromatin via its PWWP-mediated binding to nucleosomes
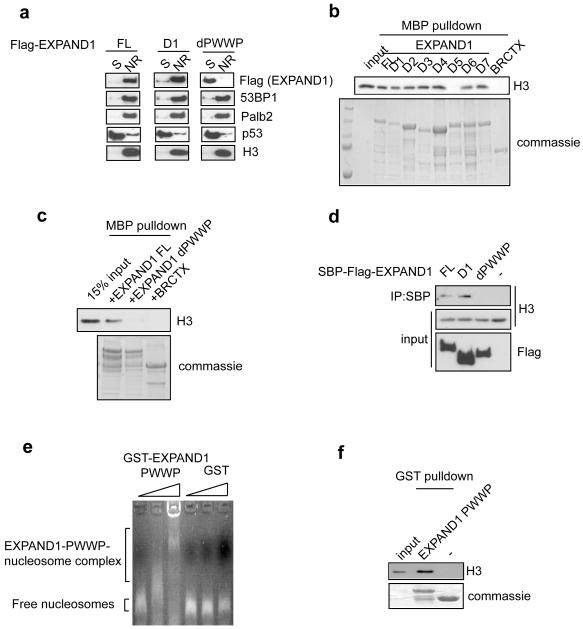
Since PWWP domain has previously been reported to be important for chromatin targeting, we decided to determine whether the association of EXPAND1 with chromatin requires its PWWP domain. Cells stably expressing tagged full-length (FL) EXPAND1, its D1 or its PWWP deletion (dPWWP) mutants were generated. Using the methodology as described in Figure 1g, we found that deletion of the PWWP domain resulted in a shift of EXPAND1 distribution, from that residing in the nuclease-released chromatin-enriched fraction to the soluble fraction (Figure 6a), indicating that the PWWP domain might target EXPAND1 to chromatin structure in vivo. Our data also indicated that the ablation of 53BP1 binding, while inhibits the damage-induced focus localization of EXPAND1, did not result in noticeable defect in EXPAND1 chromatin association (Figure 6a), suggesting that these two events are independently regulated.
Figure 6.
EXPAND1 associates with the chromatin via its PWWP-mediated nucleosome interaction.
a) Biochemical fractionation of EXPAND1 full-length (FL), D1 and dPWWP mutants. Immunoblotting was done using indicated antibodies, b & c) Nucleosome binding experiments using MBP-fused EXPAND1 and its mutants. Interaction was assessed by immunoblotting for histone H3, d) EXPAND1 interacts with components of the chromatin in vivo. Nuclease-treated lysates derived from cells expressing SBP-Flag-tagged EXPAND1 were immunoprecipitated using streptavidin beads. Immunoblotting was performed using indicated antibodies, e & f) EXPAND1 PWWP domain is sufficient for nucleosome interactions. Purified GST proteins were incubated with nucleosomes, e) Protein-nucleosome complex was separated by agarose gel electrophoresis and nucleosomes were stained with ethidium bromide, f) GST pulldown experiments using GST-EXPAND1 PWWP or GST alone. Interaction with nucleosomes was assessed by immunoblotting with histone H3 antibodies.
To determine whether EXPAND1 binds directly to chromatin, we used bacterially expressed and purified MBP-tagged full-length EXPAND1 (and its various mutants) and incubated them with mononucleosomes prepared from 293T cells. Apart from the D5 mutant, which failed to precipitate nucleosomes as judged by histone H3 immunoblotting, all other mutations and full-length EXPAND1 associated with nucleosomes (Figure 6b). Interestingly, this region missing in the D5 mutant partially overlaps with the PWWP domain (Figure 3a). We generated a precise deletion of the PWWP domain and showed that this mutation impaired the EXPAND1-nucleosome binding (Figure 6c), suggesting that the chromatin targeting of EXPAND1 is likely to occur via its PWWP-mediated interaction with nucleosomal structures. To further confirm that EXPAND1 interacts with chromatin components in vivo, nuclease-treated lysates derived from cells expressing tagged full-length (FL) EXPAND1, its D1 and PWWP deletion mutants were precipitated and immunoblotted for histone H3. In line with the requirement of the PWWP domain for EXPAND1 chromatin association, only the dPWWP mutant failed to precipitate histone H3 (Figure 6d).
We also examined whether the PWWP domain of EXPAND1 is sufficient for nucleosome binding. We incubated nucleosomes with bacterially expressed and purified GST-PWWP domain of EXPAND1 or GST alone. As shown in Figure 6e and 6f, EXPAND1 PWWP domain alone was sufficient in nucleosome binding. Thus, we conclude that the in vivo chromatin-association of EXPAND1 relies on its PWWP domain-mediated binding to nucleosomes.
The PWWP domain is indispensable for EXPAND1 function in the maintenance of chromatin state
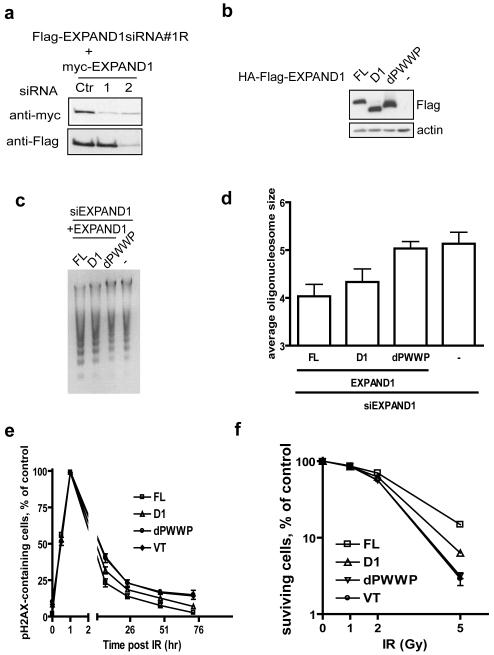
Having established that EXPAND1 targets the chromatin via its PWWP domain, and that it is recruited to DNA damage sites via its N-terminus-mediated binding to 53BP1, we next examined whether EXPAND1 function in chromatin decondensation requires both of these properties. We generated siRNA#1-resistant constructs that encode the full-length, D1 and dPWWP EXPAND1 (Figure 7a). Cells stably expressing each of these siRNA-resistant tagged EXPAND1 constructs were transfected with EXPAND1 specific siRNA#1 to deplete endogenous EXPAND1. Consistent with the requirement of the PWWP domain in EXPAND1 chromatin association, reconstitution with the dPWWP mutant failed to restore the EXPAND1-dependent chromatin relaxation (Figure 7b-d). Interestingly, the D1 mutant also exhibited a partial defect in damage-induced chromatin decondensation. In addition, the D1 mutant only partially restored the EXPAND1-depletion-associated deficits in DNA repair and hypersensitivity towards DNA damaging agent, whereas cells reconstituted with the dPWWP mutant displayed phenotypes comparable to EXPAND1-depleted cells (Figure 7e and 7f). These data suggest that EXPAND1 is involved in the maintenance of proper chromatin structure in the cell, and that EXPAND1, through its interaction with 53BP1, facilitates chromatin relaxation following DNA damage and promotes cell survival.
Figure 7.
The PWWP domain is indispensable for EXPAND1 functions.
a) Generation of siRNA-resistant EXPAND1 constructs. 293T cells were co-transfected with indicated constructs and siRNAs. 24 post transfection, cells were lysed and immunoblotting experiment was done using indicated antibodies, b-d) PWWP is required for EXPAND1-dependent chromatin condensation. Stable cells expressing siRNA#1-resistant EXPAND1 full-length (FL), its D1 or dPWWP mutants were treated with siRNA#1 twice at 24 hr interval. Cells were incubated with 200 ng/ml NCS for 1 hr before nuclei were extracted. Microccocal nuclease sensitivity was assessed as described in Methods and Materials. Expression of siRNA-resistant EXPAND1-expressing cells are shown (b), Microccocal nuclease sensitivity assay was displayed in (c) and average size of nucleosomes is shown in (d), e) Stable cells as described in (b) were treated with EXPAND siRNA#1 twice at 24 hr intervals. 24 hr after the second transfection, cells were seeded onto coverslips and irradiated (3 Gy) 24 hr later. At indicated time points cells were fixed and processed for immunostaining using pH2AX antibodies, f) clonogenic assay of EXPAND1-expressing cells following radiation. EXPAND1 siRNA-transfected cells were seeded onto 60 mm dish and were irradiated with indicated dose. Cells were incubated for 14 days before being fixed and counted. Results represent mean±S.D. from two independent experiments each done in triplicate.
Discussion
In this study, we have identified a damage-responsive element that contributes to the maintenance of chromatin architecture in the cell. Through its direct interaction with the DNA damage mediator protein 53BP1, EXPAND1 concentrates at the vicinity of DNA breaks, modulates DNA repair, and promotes cell survival in response to genotoxic stress.
EXPAND1 is an uncharacterized protein that was first described in a human melanoma as an antigenic peptide recognized by cytolytic T lymphocytes (Coulie et al., 1995). This protein contains a PWWP domain, which is found amongst many chromatin-associated proteins and is important in targeting various proteins to the chromatin. Notably, apart from an ascribed role as a DNA binding fold (Laguri et al., 2008; Lukasik et al., 2006; Qiu et al., 2002), the PWWP domain has also been reported to interact with histones (Laue et al., 2008). Our findings that EXPAND1 chromatin association requires its PWWP domain and that its PWWP domain was sufficient in binding to nucleosomes is supportive of a general function of this module in chromatin tethering. Interestingly, the yeast pdp1 PWWP was recently found to preferentially bind methylated histone H4K20 (Wang et al., 2009). Whether the EXPAND1 PWWP domain targets a subset of nucleosomes or histones requires further investigation. Nevertheless, the observations that the PWWP domain is critical for EXPAND1 function clearly illustrate the strict requirement for regulated sub-cellular localization in ensuing protein function in vivo.
A scenario is emerging in which the dynamic equilibrium of the eukaryotic chromatin is dictated by a balance between histone modifications, recruitment of chromatin remodeling factors and chromatin-associated proteins. Our identification of EXPAND1 as a chromatin component involved in the DNA damage response clearly indicates the complexity of the chromatin landscape. Intriguingly, EXPAND1 chromatin association and its focal accumulation at DNA damage sites require distinct domains, suggesting that the general function of EXPAND1 in the maintenance of chromatin structure is exploited by the DNA damage response pathway. We speculate that the EXPAND1 PWWP domain allows constitutive chromatin-tethering, which would facilitate swift damage-induced accumulation at the damage modified chromatin via its direct interaction with 53BP1, and further enhances EXPAND1 chromatin association. Given that 53BP1 directly concentrates EXPAND1 to the damage-modified chromatin, our results revealed yet an additional layer of integration between DNA damage signaling and chromatin remodeling. Mechanistically how EXPAND1 contributes to chromatin architecture regulation requires further work. Nevertheless, since 53BP1 was recently implicated in enhancing DNA repair by promoting chromatin dynamics (Dimitrova et al., 2008), it is tempting to speculate that EXPAND1, as a stable component of the chromatin, may serve as one of the means via which 53BP1 promotes DNA repair and cell survival. Consistent with the observed overlapping but not redundant functions of MUM1 and 53BP1 in the DNA damage response, co-depletion of these DNA damage response factors further sensitized cells to IR treatment (Supplemental Figure 2a and 2b). Similarly, abrogation of EXPAND1-binding contributed partially to the overall capacity of 53BP1 in promoting cell survival following DNA damage (Supplemental Figure 3a and 3b).
The chromatin fiber is decorated with a host of chromatin-binding factors, which allow site-specific control of chromatin structure to facilitate access of enzymatic activities, in particular transcriptional machineries. In this sense, the linker histone H1 and PARP1, which bind to nucleosomes with similar properties in vitro, were previously shown to exhibit reciprocal pattern of chromatin binding, where PARP1 occupancy results in displacement of H1 and was associated with promoter activation (Krishnakumar et al., 2008). Similarly, HMGN proteins have also been reported to compete with histone H1 to destabilize nucleosomal structures (Catez et al., 2002). Our findings that EXPAND1 interacts with nucleosomal structures highlight the possibility that EXPAND1 might contribute to the maintenance of chromatin state by coordinating with other nucleosome-binding proteins. In line with this possibility, co-depletion of EXPAND1 and KAP1 resulted in a marginal increase in cellular sensitivity to IR (Supplemental Figure 2a and 2c), suggesting that the concerted actions of these proteins may provide an optimal chromatin environment important for cell survival in response to DNA damage.
As cell cycle progression is tightly coupled with chromatin changes, it is tempting to speculate that the EXPAND1-dependent chromatin modulation not only is required for efficient DNA repair per se, but may be important for the reassembly of the chromatin and checkpoint recovery. In addition, the sustained DNA damage observed in EXPAND1-depleted cells may also reflect delayed DNA repair due to defective checkpoint (Goodarzi et al., 2008). Interestingly, our preliminary studies indicated that EXPAND1 may play an anti-recombinogenic role (Supplemental Figure 4a and 4b), similar to that reported for 53BP1 (Tripathi et al., 2008; Tripathi et al., 2007). Since deficiencies in EXPAND1 and 53BP1 did not significantly affect rates of genome integration of linearised plasmid DNA (Supplemental Figure 4c), we speculate that EXPAND1, like 53BP1, may only be essential for certain specialized repair processes, including class switch and long-range V(D)J recombination (Difilippantonio et al., 2008b; Manis et al., 2004; Reina-San-Martin et al., 2007; Ward et al., 2004). Further work will be needed to address these possibilities.
In summary, our study reveals a new link between DNA damage signaling and chromatin remodeling. Our results support that the PWWP-domain containing protein EXPAND1 contributes to the chromatin architecture, and via its direct interaction with the DNA damage mediator 53BP1, plays an accessory role to facilitate damage-induced chromatin changes, and is important for efficient DNA repair and cell survival following DNA damage.
Experimental Procedures
Antibodies
The EXPAND1 polyclonal antibody was raised against GST-EXPAND1 fusion protein and affinity purified using column coated with MBP-EXPAND1 fusion protein. Antibodies specifically recognizing the myc epitope (9E10), pH2AX, BRCA1, MDC1, Palb2, 53BP1 and BACH1 were previously described (Huen et al., 2007; Sy et al., 2009b; Yu et al., 2003). The anti-H3 and anti-ORC2 antibodies were obtained from Upstate Cell Signaling. Anti-HA and anti-p53 antibodies were purchased from Covance and Santa Cruz, respectively. Anti-actin and anti-Flag (M2) were obtained from Sigma. Anti-KAP1 and anti-phospho-KAP1 were obtained from Bethyl Laboratories.
Tandem affinity purification
293T cells expressing streptavidin binding peptide-S peptide-Flag-tagged 53BP1 were irradiated and lysed in NETN for 10 minutes on ice. The pellet obtained was resuspended in nuclease digestion buffer (15 mM Tris pH 7.4, 60 mM KCl, 15 mM NaCl, 0.25 M sucrose, 1 mM CaCl2, 0.5 mM DTT) and microccocal nuclease (Sigma; 200U/ml) was added. After incubation at 37°C water bath for an additional 10 minutes, equal volume of NETN was added and the supernatant was clarified by centrifugation. The supernatant containing the released 53BP1 protein complexes were subjected to tandem affinity purification as described previously (Huen et al., 2008).
Cell Culture and Transfection
Cell lines were cultured in RPMI 1640 or DMEM media supplemented with 10% fetal bovine serum (FBS) and maintained in 5% CO2 at 37°C. Cell transfection was performed using Lipofectamine 2000 (Invitrogen) following manufacturer’s protocol.
SiRNA
SiRNAs targeting RNF8, 53BP1 (Smartpool), KAP1 (Smartpool), EXPAND1 and a non-targeting control siRNA were purchased from Dharmacon. SiRNA transfection was performed according to the manufacturer’s protocol. Sequences of siRNA#1 and siRNA#2 against EXPAND1 are 5′ CUA AUU CCA UGC GUU CUA UUU 3′ and 5′ GAU UAU GGC CUG CGA AGG UUU 3′, respectively. siRNA against RNF8 was previously described (Huen et al., 2007). Sequence of the 53BP1-specific siRNA used for reconstitution experiments is 5′ GAG CUG GGA AGU AUA AAU UUU 3′.
Generation of wild-type and mutant EXPAND1 and 53BP1 expression constructs
EXPAND1 cDNA was obtained from OpenBiosystems (IMAGE: 4933735; Homo sapiens melanoma associated antigen (mutated) 1; GenBank: BC110874.1). PCR amplified DNA fragment containing wild-type EXPAND1 was subcloned into the pDONR201 vector using Gateway Technology (Invitrogen). Site-directed mutagenesis was performed according to standard procedures to obtain the EXPAND1 mutants. For transient expression of EXPAND1 and its mutants, the corresponding fragments in the entry vectors were transferred into Gateway compatible destination vectors which harbor either an N-terminal triple-epitope tag (S protein tag, Flag epitope tag and Streptavidin binding peptide tag), an N-terminal myc tag, or an N-terminal HA-Flag epitope tag for retrovirus packaging. To generate the siRNA#1-resistant EXPAND1 expression constructs, site-directed mutagenesis was performed within the EXPAND1 coding region that is targeted by EXPAND1 siRNA#1. Sequences of the primers used are 5′ CCG AAG AGT CCA TGG GGT CTA ACT CGA TGC GTT CTA TCC TGG AGG AAG ACG 3′ and 5′ CCT CCA GGA TAG AAC GCA TCG AGT TAG ACC CCA TGG ACT CTT CGG AAG ACT G 3′. All clones were sequenced to verify desired mutations. HA-tagged 53BP1 and its mutants were previously described (Ward et al., 2006).
Culture of MEFs and retroviral infection
Mouse embryonic fibroblasts (MEFs) derived from various knockout strains were cultured in DMEM supplemented with 15% FBS and maintained in 5% CO2 at 37°C. For viral particle packaging, BOSC23 cells were transiently transfected with the pclampho and expression constructs. Viral supernatant was collected 48 hours post-transfection and was used for infection. Stable pools of infected MEFs were selected in the presence of 2 μg/ml puromycin.
Immunostaining procedure
To visualize ionizing radiation induced foci (IRIF), cells were cultured on coverslips and treated with 10 Gy IR followed by recovery for 6 hrs. Cells were then washed in PBS, incubated in 3% paraformaldehyde for 20 minutes and permeabilized in 0.5% triton solution for 3 minutes at room temperature. Samples were blocked with 5% goat serum and then incubated with primary antibodies for 30 minutes. Samples were washed three times and incubated with secondary antibodies for 30 minutes. Cells were then stained with DAPI to visualize nuclear DNA. The coverslips were mounted onto glass slides with anti-fade solution and visualized using a Nikon ECLIPSE E800 fluorescence microscope.
Micrococcal nuclease sensitivity assay
Micrococcal nuclease (MNase) sensitivity was assessed as described with modifications (Ziv et al., 2006). Briefly, HeLa cells were incubated with 200 ng/ml NCS or left untreated for 1 hr. Cells were then lysed with nuclei extraction (NE) buffer (10 mM Tris, 1.5 mM MgCl2, 1 mM CaCl2, 0.25M Sucrose) on ice for 8 minutes, followed by three additional washes. The nuclei pellet was resuspended each time by gentle tapping. Nuclei were then washed with nuclease digestion buffer once before incubating in digestion buffer with MNase (2.5 U/250 ml). Digestion was terminated using stop buffer (20 mM Tris, 5 mM EDTA, 2 % SDS, 150 mM NaCl) and DNA was extracted by phenol-chloroform extraction method, resuspended in TE buffer and separated by 1.2% agarose gel electrophoresis. Each lane of the ethidium bromide-stained gels was scanned and profiles representing band intensity were obtained using the NIH IMAGE J software. Average size of oligonucleosomes was calculated as described (Ziv et al., 2006).
Nucleosome preparation and binding assay
To prepare nucleosomes, 293T cells were lysed in NE Buffer for 8 minutes on ice. After centrifugation, the pellet was washed once with nuclease digestion buffer and was subsequently resuspended in nuclease digestion buffer containing MNase. The mixture was incubated at 37C for 1 hr before a final concentration of 1 mM EDTA was added to terminate the reaction. The resultant nuclease-digested nuclei pellet was resuspended in NETN (with 400 mM NaCl) and incubated on ice for an additional 30 minutes. The mixture was then centrifuged to remove cell debris. The salt concentration of the supernatant was then adjusted to 150 mM NaCl by addition of NETN drop-wise. The resulting H1-chromatin-enriched precipitate was cleared by centrifugation and the supernatant containing mononucleosomes were used for binding studies. MBP and GST-fusion proteins were incubated with mononucleosomes at 4C for 2 hr with shaking. Thereafter, beads containing the protein-nucleosome complex was washed 4 times with NETN buffer and boiled in sampling buffer. Binding was detected using western blotting for the presence of histone H3. Alternatively, the protein-nucleosome complex was loaded onto 1% agarose gel to detect mobility of mononucleosomes by ethidium bromide staining.
IR sensitivity, G2/M checkpoint assay and Chromatin fractionation
IR sensitivity and G2/M checkpoint assays were performed as described previously (Huen et al., 2007). Preparation of chromatin fractions were described previously with modifications. Briefly, cells were harvested at indicated times after treatment and washed once with PBS. Cell pellets were subsequently resuspended in NETN (and protease inhibitors; soluble fraction) and incubated on ice for 10 min. The resulting pellet, enriched in chromatin-bound proteins, was sonicated in sampling buffer and boiled. For nuclease-release experiments, the NETN extracted pellet was resuspsended in NETN in the presence of 1 mM MgCl2 and incubated with benzonase (EMD chemicals) for 30 minutes on ice. After centrifugation, the supernatant containing the nuclease-released chromatin components were obtained and boiled in sampling buffer.
Gene conversion and genome integration assays
The mutant GFP reporter assay was used to assess gene conversion rates and was performed essentially as described previously (Sy et al., 2009a). To assay for NHEJ, siRNA-treated cells were electroporated with 1 μg of linearised plasmid DNA that encodes the puromycin resistant gene at 275V, 500 μF using a Bio-Rad Gene Pulser Xcell system. 48 hours after electroporation the cells were serially diluted and incubated in media supplemented with or without puromycin. The number of colonies that formed after a10-day incubation was then counted to determine the relative efficiency of genome integration of foreign DNA.
Supplementary Material
Acknowledgements
This work was supported in part by grants from the National Institutes of Health (CA089239, CA092312 and CA100109 to J.C.), from Startup Fund (Department of Anatomy, HKU to M.S.Y.H.) and Seed Funding for Basic Research (HKU to M.S.Y.H.). M.S.Y.H. would like to thank Drs Zhiwei Chen and Henggui Liu (AIDS Institute, HKU) for help with flow cytometry analysis, and is grateful to J.C. for his continuous support and mentoring. J.C is a recipient of an Era of Hope Scholar award from the Department of Defence and a member of the Mayo Clinic Breast SPORE program (P50 CA116201).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference
- Abraham RT. Checkpoint signalling: focusing on 53BP1. Nature cell biology. 2002;4:E277–279. doi: 10.1038/ncb1202-e277. [DOI] [PubMed] [Google Scholar]
- Adams MM, Carpenter PB. Tying the loose ends together in DNA double strand break repair with 53BP1. Cell Div. 2006;1:19. doi: 10.1186/1747-1028-1-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayoub N, Jeyasekharan AD, Bernal JA, Venkitaraman AR. HP1-beta mobilization promotes chromatin changes that initiate the DNA damage response. Nature. 2008;453:682–686. doi: 10.1038/nature06875. [DOI] [PubMed] [Google Scholar]
- Cairns BR. Around the world of DNA damage INO80 days. Cell. 2004;119:733–735. doi: 10.1016/j.cell.2004.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catez F, Brown DT, Misteli T, Bustin M. Competition between histone H1 and HMGN proteins for chromatin binding sites. EMBO reports. 2002;3:760–766. doi: 10.1093/embo-reports/kvf156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celeste A, Fernandez-Capetillo O, Kruhlak MJ, Pilch DR, Staudt DW, Lee A, Bonner RF, Bonner WM, Nussenzweig A. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nature cell biology. 2003;5:675–679. doi: 10.1038/ncb1004. [DOI] [PubMed] [Google Scholar]
- Coulie PG, Lehmann F, Lethe B, Herman J, Lurquin C, Andrawiss M, Boon T. A mutated intron sequence codes for an antigenic peptide recognized by cytolytic T lymphocytes on a human melanoma. Proc Natl Acad Sci U S A. 1995;92:7976–7980. doi: 10.1073/pnas.92.17.7976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Difilippantonio S, Gapud E, Wong N, Huang CY, Mahowald G, Chen HT, Kruhlak MJ, Callen E, Livak F, Nussenzweig MC, et al. 53BP1 facilitates long-range DNA end-joining during V(D)J recombination. Nature. 2008a doi: 10.1038/nature07476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Difilippantonio S, Gapud E, Wong N, Huang CY, Mahowald G, Chen HT, Kruhlak MJ, Callen E, Livak F, Nussenzweig MC, et al. 53BP1 facilitates long-range DNA end-joining during V(D)J recombination. Nature. 2008b;456:529–533. doi: 10.1038/nature07476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrova N, Chen YC, Spector DL, de Lange T. 53BP1 promotes non-homologous end joining of telomeres by increasing chromatin mobility. Nature. 2008 doi: 10.1038/nature07433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downs JA, Allard S, Jobin-Robitaille O, Javaheri A, Auger A, Bouchard N, Kron SJ, Jackson SP, Cote J. Binding of chromatin-modifying activities to phosphorylated histone H2A at DNA damage sites. Molecular cell. 2004;16:979–990. doi: 10.1016/j.molcel.2004.12.003. [DOI] [PubMed] [Google Scholar]
- Fernandez-Capetillo O, Chen HT, Celeste A, Ward I, Romanienko PJ, Morales JC, Naka K, Xia Z, Camerini-Otero RD, Motoyama N, et al. DNA damage-induced G2-M checkpoint activation by histone H2AX and 53BP1. Nat Cell Biol. 2002;4:993–997. doi: 10.1038/ncb884. [DOI] [PubMed] [Google Scholar]
- Goodarzi AA, Noon AT, Deckbar D, Ziv Y, Shiloh Y, Lobrich M, Jeggo PA. ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Mol Cell. 2008;31:167–177. doi: 10.1016/j.molcel.2008.05.017. [DOI] [PubMed] [Google Scholar]
- Huen MS, Grant R, Manke I, Minn K, Yu X, Yaffe MB, Chen J. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell. 2007;131:901–914. doi: 10.1016/j.cell.2007.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huen MS, Sy SM, van Deursen JM, Chen J. Direct interaction between SET8 and proliferating cell nuclear antigen couples H4-K20 methylation with DNA replication. The Journal of biological chemistry. 2008;283:11073–11077. doi: 10.1074/jbc.C700242200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YC, Gerlitz G, Furusawa T, Catez F, Nussenzweig A, Oh KS, Kraemer KH, Shiloh Y, Bustin M. Activation of ATM depends on chromatin interactions occurring before induction of DNA damage. Nature cell biology. 2009;11:92–96. doi: 10.1038/ncb1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnakumar R, Gamble MJ, Frizzell KM, Berrocal JG, Kininis M, Kraus WL. Reciprocal binding of PARP-1 and histone H1 at promoters specifies transcriptional outcomes. Science (New York, NY. 2008;319:819–821. doi: 10.1126/science.1149250. [DOI] [PubMed] [Google Scholar]
- Laguri C, Duband-Goulet I, Friedrich N, Axt M, Belin P, Callebaut I, Gilquin B, Zinn-Justin S, Couprie J. Human mismatch repair protein MSH6 contains a PWWP domain that targets double stranded DNA. Biochemistry. 2008;47:6199–6207. doi: 10.1021/bi7024639. [DOI] [PubMed] [Google Scholar]
- Laue K, Daujat S, Crump JG, Plaster N, Roehl HH, Kimmel CB, Schneider R, Hammerschmidt M. The multidomain protein Brpf1 binds histones and is required for Hox gene expression and segmental identity. Development (Cambridge, England) 2008;135:1935–1946. doi: 10.1242/dev.017160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang B, Qiu J, Ratnakumar K, Laurent BC. RSC functions as an early double-strand-break sensor in the cell’s response to DNA damage. Curr Biol. 2007;17:1432–1437. doi: 10.1016/j.cub.2007.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukasik SM, Cierpicki T, Borloz M, Grembecka J, Everett A, Bushweller JH. High resolution structure of the HDGF PWWP domain: a potential DNA binding domain. Protein Sci. 2006;15:314–323. doi: 10.1110/ps.051751706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manis JP, Morales JC, Xia Z, Kutok JL, Alt FW, Carpenter PB. 53BP1 links DNA damage-response pathways to immunoglobulin heavy chain class-switch recombination. Nature immunology. 2004;5:481–487. doi: 10.1038/ni1067. [DOI] [PubMed] [Google Scholar]
- Manke IA, Lowery DM, Nguyen A, Yaffe MB. BRCT repeats as phosphopeptide-binding modules involved in protein targeting. Science. 2003;302:636–639. doi: 10.1126/science.1088877. [DOI] [PubMed] [Google Scholar]
- Morrison AJ, Kim JA, Person MD, Highland J, Xiao J, Wehr TS, Hensley S, Bao Y, Shen J, Collins SR, et al. Mec1/Tel1 phosphorylation of the INO80 chromatin remodeling complex influences DNA damage checkpoint responses. Cell. 2007;130:499–511. doi: 10.1016/j.cell.2007.06.010. [DOI] [PubMed] [Google Scholar]
- Murga M, Jaco I, Fan Y, Soria R, Martinez-Pastor B, Cuadrado M, Yang SM, Blasco MA, Skoultchi AI, Fernandez-Capetillo O. Global chromatin compaction limits the strength of the DNA damage response. The Journal of cell biology. 2007;178:1101–1108. doi: 10.1083/jcb.200704140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu C, Sawada K, Zhang X, Cheng X. The PWWP domain of mammalian DNA methyltransferase Dnmt3b defines a new family of DNA-binding folds. Nature structural biology. 2002;9:217–224. doi: 10.1038/nsb759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reina-San-Martin B, Chen J, Nussenzweig A, Nussenzweig MC. Enhanced intra-switch region recombination during immunoglobulin class switch recombination in 53BP1−/− B cells. Eur J Immunol. 2007;37:235–239. doi: 10.1002/eji.200636789. [DOI] [PubMed] [Google Scholar]
- Silverman J, Takai H, Buonomo SB, Eisenhaber F, de Lange T. Human Rif1, ortholog of a yeast telomeric protein, is regulated by ATM and 53BP1 and functions in the S-phase checkpoint. Genes & development. 2004;18:2108–2119. doi: 10.1101/gad.1216004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sy SM, Huen MS, Chen J. PALB2 is an integral component of the BRCA complex required for homologous recombination repair. Proc Natl Acad Sci U S A. 2009a;106:7155–7160. doi: 10.1073/pnas.0811159106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sy SM, Huen MS, Zhu Y, Chen J. PALB2 regulates recombinational repair through chromatin association and oligomerization. The Journal of biological chemistry. 2009b doi: 10.1074/jbc.M109.016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi V, Kaur S, Sengupta S. Phosphorylation-dependent interactions of BLM and 53BP1 are required for their anti-recombinogenic roles during homologous recombination. Carcinogenesis. 2008;29:52–61. doi: 10.1093/carcin/bgm238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi V, Nagarjuna T, Sengupta S. BLM helicase-dependent and -independent roles of 53BP1 during replication stress-mediated homologous recombination. J Cell Biol. 2007;178:9–14. doi: 10.1083/jcb.200610051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Reddy B, Thompson J, Wang H, Noma K, Yates JR, 3rd, Jia S. Regulation of Set9-mediated H4K20 methylation by a PWWP domain protein. Molecular cell. 2009;33:428–437. doi: 10.1016/j.molcel.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward I, Kim JE, Minn K, Chini CC, Mer G, Chen J. The tandem BRCT domain of 53BP1 is not required for its repair function. The Journal of biological chemistry. 2006;281:38472–38477. doi: 10.1074/jbc.M607577200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward IM, Reina-San-Martin B, Olaru A, Minn K, Tamada K, Lau JS, Cascalho M, Chen L, Nussenzweig A, Livak F, et al. 53BP1 is required for class switch recombination. J Cell Biol. 2004;165:459–464. doi: 10.1083/jcb.200403021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane K, Wu X, Chen J. A DNA damage-regulated BRCT-containing protein, TopBP1, is required for cell survival. Molecular and cellular biology. 2002;22:555–566. doi: 10.1128/MCB.22.2.555-566.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Chini CC, He M, Mer G, Chen J. The BRCT domain is a phospho-protein binding domain. Science. 2003;302:639–642. doi: 10.1126/science.1088753. [DOI] [PubMed] [Google Scholar]
- Ziv Y, Bielopolski D, Galanty Y, Lukas C, Taya Y, Schultz DC, Lukas J, Bekker-Jensen S, Bartek J, Shiloh Y. Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM- and KAP-1 dependent pathway. Nature cell biology. 2006;8:870–876. doi: 10.1038/ncb1446. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.