Abstract
Here, we show a novel molecular mechanism promoted by the DEAD-box RNA helicase DDX3 for translation of the HIV-1 genomic RNA. This occurs through the adenosine triphosphate-dependent formation of a translation initiation complex that is assembled at the 5′ m7GTP cap of the HIV-1 mRNA. This is due to the property of DDX3 to substitute for the initiation factor eIF4E in the binding of the HIV-1 m7GTP 5′ cap structure where it nucleates the formation of a core DDX3/PABP/eIF4G trimeric complex on the HIV-1 genomic RNA. By using RNA fluorescence in situ hybridization coupled to indirect immunofluorescence, we further show that this viral ribonucleoprotein complex is addressed to compartmentalized cytoplasmic foci where the translation initiation complex is assembled.
INTRODUCTION
Most of the RNA polymerase II (RNAPII)-transcribed eukaryotic mRNAs assemble into a messenger ribonucleoprotein (mRNP) that is processed before nuclear export by the NFX1/p15-dependent pathway (1,2). Capping and splicing of the mRNA occur co-transcriptionally on the nascent mRNA and are critical for nuclear export and efficient translation in the cytoplasm (2–4). In the nucleus, the m7GpppN (where N is the first transcribed nucleotide) cap structure is bound by the nuclear cap-binding complex (CBC), which is composed of the CBP20/80 heterodimer (5). Early recruitment of CBP20/80 to the cap is important for transcription elongation and nuclear export (6,7). Once in the cytoplasm, exported mRNAs undergo a CBP20/80-dependent pioneer round of translation that is essential for the quality control of the transcript (8,9). However, CBP20/80-bound mRNAs are only associated with few ribosomes, and replacement of the CBC by the translation initiation factor eIF4E is necessary to engage the association of the mRNA with polysomes and the bulk of protein synthesis (9,10). Interestingly, certain mRNAs are routed and accumulate in specific subcellular locations where temporal and spatial control of translation is achieved (11,12).
More than 40 transcripts have been identified during HIV-1 replication (13). These viral mRNA species are transcribed by the host RNAPII and include the 2-kb fully spliced, the 4-kb partially spliced and the 9-kb full-length genomic RNA (gRNA), which is not spliced and associates with the virally encoded protein Rev to promote nuclear export and the cytoplasmic accumulation of the gRNA and other intron-containing viral mRNAs (14). Rev-mediated nuclear export does not rely on the canonical NFX1/p15 pathway but rather uses the alternative CRM1-dependent cargo pathway (15). As a consequence, the composition of the exported viral mRNP differs from that of a classical cellular mRNA (16,17); therefore, the mechanisms used by the HIV-1 gRNA to reach the host translational machinery after nuclear export may be different from the one used by viral and cellular spliced transcripts.
There is in vitro evidence suggesting that capping of the HIV-1 mRNAs is facilitated by the interaction of the viral protein Tat with cellular capping enzymes (18,19). Along this line, recent data have shown that CBP20/80 is required for HIV-1 Tat-mediated trans-activation of transcription, and likewise in cellular mRNAs, the HIV-1 gRNA is associated to the nuclear CBC (7,20). Interestingly, HIV-1 expression induces the de-phosphorylation and the consequent inactivation of eIF4E raising several questions about the mechanism by which the HIV-1 gRNA could be efficiently translated by a cap-dependent mechanism in the cytoplasm (20).
We have recently shown that the DEAD-box helicase DDX3 is essential for translation of a subset of specific mRNAs; among those is the HIV-1 gRNA (21). As such, DDX3 directly binds to the HIV-1 5′-UTR and interacts with translation initiation factors eIF4G and PABP (21). In this study, we extend these data on the role of DDX3 in driving HIV-1 gRNA translation in the context of viral replication. Our results show that DDX3 drives the adenosine triphosphate (ATP)-dependent assembly of a pre-translation initiation intermediate essential for viral protein synthesis. As a consequence, the HIV-1 gRNA is compartmentalized in large cytoplasmic RNA granules composed of the gRNA associated with DDX3 together with eIF4G and PABP but lacking the major cap-binding proteins CBP20/80 and eIF4E. Interestingly, we observed that cytoplasmic DDX3 could be retained on an m7GTP affinity matrix independently of eIF4E, suggesting that DDX3 can assemble a pre-initiation complex on the HIV-1 gRNA in the cytoplasm in the absence of the major eIF4E cap-binding protein. This provides evidence for a novel cap-binding pre-initiation complex, which is assembled by DDX3 in an ATP-dependent manner to prepare the HIV-1 gRNA to enter translation initiation.
MATERIALS AND METHODS
DNA constructs
The pNL4-3 vector was described previously (22). The pNL4-3R vector was previously described (21,23). The pNL4-3 stop vector was kindly provided by Dr Denis Gerlier (INSERM U758, France). The pNL4-3R ΔRev vector was generated by inserting the SpeI fragment of the pNL4-3R vector into the pNL4-3fB vector kindly provided by Dr Barbara Felber (Frederick National Laboratory for Cancer Research, USA), which was previously described (24). The pCIneo-HA-eIF4E, eIF4G, eIF4A and eIF4B were previously described previously (21). cDNAs for CRM1 and DDX1 were obtained by reverse transcriptase–polymerase chain reaction (RT–PCR) using HeLa cells total RNAs as template and inserted into the EcoRI/NotI or MluI/NotI sites of the pCIneo-HA vector as previously described (21). The pCMV-CBP80 vector was described previously (25) and was kindly provided by Dr Yoon Ki Kim (Korea University, Republic of Korea). A PCR fragment corresponding to the Gag-Pol fragment obtained from pNL4-3 proviral DNA was inserted into the EcoRV-digested pBluescript vector to generate the pBSKGag-Pol vector used to generate the probes targeting the gRNA. The sequences 5′-GCAGCACGACTTCTTCAAGTTCAAGAGACTTGAAGAAGTCGTGCTGC-3′ and 5′-GATGCTGGCTCGTGATTTCTTTCAAGAGAAGAAATCACGAGCCAGCATC-3′ (target sequence in italic) were inserted between the BglII/HindIII sites of the pRetroSuper vector (OligoEngine) following supplier’s instructions to generate pRS-shCtrl and pRS-shDDX3 vectors, respectively. The target sequence corresponds to the same sequence used in the siRNAs previously described (21).
Cell culture and nucleic acid transfections
T-lymphocytes (Jurkat cells) were transfected with the pRS-shCtrl or pRS-shDDX3 vectors using a MicroPorator (Digital Bio) and the Neon™ system (Invitrogen) following supplier’s indications and then maintained in RPMI growth media (Gibco, BRL) supplemented with 10 μg/ml puromycin, 10% FCS and 1% l-glutamine. shCtrl and shDDX3 cells were infected with HIV-1 NL4-3 virus at an MOI of 10, and cells extracts were recovered at 48 h post-infection for western blot and cytoplasmic RNA extraction and RT–qPCR. Cell culture supernatants were used for reverse transcriptase assay as described previously (23).
Renilla activity
Renilla activity was measured using the Renilla Luciferase Assay System (Promega Co, Madison, WI, USA) in a Veritas Luminometer (TurnerBiosystems) as previously described (21,23,26).
RNA extraction and RT–qPCR
Cytoplasmic RNA extraction and RT–qPCR from cytoplasmic RNA were performed as previously described (23). Briefly, cells were washed intensively with phosphate-buffered saline (PBS), recovered with PBS–ethylenediaminetetraacetic acid 10 mM and lysed for 1–2 min at room temperature with 200 µl of buffer [10 mM Tris–HCl, pH 8.0, 10 mM NaCl, 3 mM MgCl2, 1 mM DTT, 0.5% NP-40 and 15 U/ml of RNaseOUT (Life Technologies)]. Cell lysates were centrifuged at 13 000 rpm for 4 min; supernatant containing the cytoplasmic fraction was recovered; and RNA extraction was carried out by adding 1 ml of TRIzol® Reagent (Life Technologies) as indicated by the manufacturer. Cytoplasmic RNAs (300 ng) were reverse-transcribed using the High Capacity RNA-to-cDNA Master Mix (Life Technologies). For quantitative PCR, a 20 µl reaction mix was prepared with 5 µl of template cDNAs (previously diluted to 1/10), 10 µl of FastStart Universal SYBR Green Master (Rox) (Roche), 0.2 µM of sense and antisense primers and subjected to amplification using a fluorescence thermocycler (Applied Biosystems 7000 Real-time PCR, Foster City, CA, USA). The GAPDH or RPS24 housekeeping genes were amplified in parallel to serve as a control reference. Relative copy numbers of Renilla luciferase cDNAs were compared with GAPDH or RPS24 using x−DCt (where x corresponds to the experimentally calculated amplification efficiency of each primer couple).
Western blot
Samples were subjected to 7.5 or 10% sodium dodecyl sulphate–polyacrylamide gel electrophoresis, transferred to PVDF membrane and blotted using anti-HIV-1 p24 and Nef (NIH Reagents program), Vif (kindly provided by Dr Denis Gerlier, INSERM U758, France), DDX3 (Abcam), eIF2α (Cell Signalling), eIF2α-P Ser51 (US Biologicals), eIF4G, PABP, eIF4A, eIF4E, eIF4E-P, eIF4B and eIF3 (kindly provided by Dr Simon Morley, University of Sussex, UK) and actin (Santa Cruz Biotechnologies) as previously described (27).
Synthesis of 11-digoxigenin-UTP probes
Before in vitro transcription, the pBSKGag-Pol and the pBSKNef vectors were linearized with EcoRI and XhoI, respectively. In vitro transcription was carried out as described previously (26) with the following modifications: rNTPs mix was composed of 10 mM GTP, ATP, CTP and 6.5 mM UTP/3.5 mM 11-digoxigenin-UTP (Roche). In vitro transcription using pBSKGag-Pol as template was performed with T3 RNA polymerase (Promega), and the resulting 5-kb RNA was partially digested with 0.2 M NaOH during 90 min on ice to generate ∼100 nt-long RNA fragments. pBSKNef transcription was carried out with T7 RNA polymerase (Promega). Digoxigenin-labelled RNAs were treated with RQ1 DNAse and precipitated ethanol.
Fluorescent in situ hybridization, immunofluorescence and confocal microscopy
HeLa cells were cultured in Lab-Tek™ Chamber Slides (Nunc™) and maintained and transfected with 0.5 μg of pNL4-3 or 0.2 μg of the corresponding HA vectors as indicated. At 24 hpt, cells were washed twice with 1× PBS and fixed for 10 min at room temperature with 4% paraformaldehyde at room temperature. Cells were subsequently permeabilized for 5 min at room temperature with 0.2% Triton X-100 and hybridized overnight at 37°C in 200 µl of hybridization mix (10% dextran sulphate, 2 mM vanadyl–ribonucleoside complex, 0.02% RNase-free bovine serum albumin, 50% formamide, 300 µg of tRNA and 120 ng of 11-digoxigenin-UTP probes) in a humid chamber. Cells were washed with 0.2× SSC/50% formamide during 30 min at 50°C and then incubated three times with antibody dilution buffer (2× SSC, 8% formamide, 2 mM vanadyl–ribonucleoside complex and 0.02% RNase-free bovine serum albumin). Mouse anti-digoxin and rabbit anti-HA (Sigma Aldrich) primary antibodies diluted to 1/100 in antibody dilution buffer were added for 2 h at room temperature. After three washes with antibody dilution buffer, cells were incubated for 90 min at room temperature with anti-mouse Alexa 488 and anti-rabbit Alexa 565 antibodies (Molecular Probes) diluted at 1/1000. Cells were washed three times in wash buffer (2× SSC, 8% formamide and 2 mM vanadyl–ribonucleoside complex), twice with 1× PBS, incubated with Hoescht (1/10000, Invitrogen Life Technologies) for 5 min at room temperature, washed three times with 1× PBS, three times with water and mounted with Fluoromount (Invitrogen Life Technologies).
Images representative from several cells (usually >100) obtained in independent experiments were obtained with a TCS SP5 AOBS Spectral Confocal Microscope (Leica Microsystems) and recovered and merged using the LAS AF Lite software (Leica Microsystems). Colour profiles and co-localized points were obtained with ImageJ software (NIH) using the colour function and the co-localization analysis plugins, respectively.
m7GTP cap affinity matrix
Analyses were carried out as previously described (21). Briefly, cytoplasmic extracts from HeLa cells (100 μg of total protein) were previously treated with S7 nuclease (Roche) and then incubated for 10 min at 37°C in cap-binding buffer (50 mM Tris–HCl, pH 7.5, 30 mM NaCl, 1 mM DTT, 2.5 mM MgCl2 and 0.5% glycerol) in the absence or presence of 5 mM m7GpppG cap analogue (New England Biolabs) as control. Extracts were incubated over night at 4°C with 15–20 μl of m7GTP–Sepharose 4B (GE Healthcare) under gentle shaking. Retained proteins were washed three times with cap washing buffer (50 mM HEPES, pH 7.5, 40 mM NaCl, 2 mM ethylenediaminetetraacetic acid and 0.1% Triton), eluted by boiling in sodium dodecyl sulphate-loading buffer and analysed by western blot as indicated earlier in the text.
RESULTS
DDX3 is required for HIV-1 gRNA translation in the context of viral replication
A meta-analysis of genome-wide screenings identified DDX3 as a host factor required for efficient HIV-1 replication (28). We have recently shown that DDX3 is critical for HIV-1 gRNA translation during replication of a full-length provirus and that this process involves its ATPase activity (21). By using an mRNA transfection strategy, we further identified the TAR RNA motif present at the very 5′-end of the 5′-UTR to be the molecular target for DDX3 activity during translation initiation (21).
To extend these observations to viral replication during infection, we have created a Jurkat cell line stably expressing a control shRNA (shCtrl) or an shRNA directed against the DDX3 transcript (shDDX3). These cells lines were infected with the HIV-1 NL4-3 strain, and western blot analysis revealed a reduction of the intracellular levels of DDX3 and the viral proteins Gag, Vif and Nef but not β-actin in shDDX3 (Figure 1A), confirming the role of DDX3 on HIV-1 gene expression during infection in CD4+ T-cells. Quantification of the level of the corresponding viral transcripts (gRNA, Vif and Nef) present in the cytoplasm by RT–qPCR revealed that they were unchanged on DDX3 depletion (Figure 1B), confirming that DDX3 promotes viral mRNA translation. Consistent with reduced intracellular Gag levels, virion production was also diminished (up to 3-fold) in shDDX3 cells, confirming that reduced levels of DDX3 negatively impacted viral replication (Figure 1C). This confirms and extends our previous results showing that DDX3 is needed for HIV-1 gRNA translation in the context of viral infection.
Figure 1.
DDX3 is required for HIV-1 gRNA translation. (A) Jurkat shCtrl (lane 1) and shDDX3 (lane 2) cells were infected with the HIV-1 NL4-3 virus at an MOI of 10, and cell extracts were prepared for western blot analysis using DDX3, actin, Gag, Nef and Vif antibodies or (B) cytoplasmic RNA extraction and viral mRNA quantification by RT–qPCR analysis. (C) At 48 h post-infection, reverse transcriptase activity from the cell supernatant was quantified. Western blot is representative of three independent experiments, and RT–qPCR data are presented as mean ± SD of three independent experiments. ***P < 0.001; (non directional t-test).
DDX3 and the HIV-1 gRNA form large cytoplasmic granules
Our goal was then to determine the molecular mechanism by which DDX3 can affect translation of the HIV-1 gRNA. In particular, we wanted to investigate in which compartment of the cell the gRNA and the DDX3 protein can be found. For this, we carried out RNA in situ hybridization coupled to indirect immunofluorescence [RNA fluorescence in situ hybridization (FISH)–IF] and laser scanning confocal microscopy (LSCM) analyses. It should be mentioned that given the limited cytoplasmic space present in T-cells, we have used the HeLa cells model system. In preliminary experiments, we have checked that siRNA knockdown of DDX3 in HeLa cells resulted in the same effects on HIV-1 translation than those obtained in Jurkat (data not shown). Thus, we first transfected HeLa cells with the NL4-3 molecular clone together with a vector coding HA-DDX3. Interestingly, we observed two major patterns of cytoplasmic localization for both components (Figure 2A). In the first pattern, the HIV-1 gRNA was found to disperse through the cytoplasm forming very small dots (Figure 2A, left). Although DDX3 was also observed to disperse throughout the cytoplasm, a closer inspection suggests that both signals rather do not co-localize (Figure 2A, see co-localized points and intensity plot). The patterns of localization for the HIV-1 gRNA and DDX3 observed earlier in the text are consistent with previous reports and may reflect their respective localizations at steady-state (29–33). Interestingly, the second pattern of localization revealed the assembly of large cytoplasmic granules in which the gRNA and DDX3 co-localized perfectly (Figure 2A, right), suggesting that their physical interaction results in the assembly of these particular cytoplasmic viral mRNPs. It should be noted that these granules can be formed in HIV-1-expressing cells even in the absence of ectopically added DDX3 (Supplementary Figure S1).
Figure 2.
HIV-1 gRNA and DDX3 are localized in large cytoplasmic RNA granules (A) HeLa cells were transfected with 0.5 μg of pNL4-3 and 0.2 μg of pCIneo-HA-DDX3 for 24 h and prepared for LSCM as indicated in ‘Materials and Methods’ section. The HIV-1 gRNA (green staining) and DDX3 (red staining) were observed either disperse throughout the cytoplasm (left, scale bar 25 μm) or co-localizing in large cytoplasmic granules (right, scale bar 10 μm). White arrowheads indicate cytoplasmic granules. Co-localized points are indicated in white. Merge images include nucleus staining with Hoechst (blue). Colour profiles from a region of interest (ROI) are also shown below. (B) HeLa cells were transfected with 0.5 μg of pNL4-3 and 0.2 μg of pCIneo-HA-CRM1 for 24 h and prepared for LSCM. Green staining corresponds to the gRNA, and red staining identifies the CRM1 protein. White arrowheads indicate HIV-1 gRNA granules. Merge images include nucleus staining with Hoechst (blue). Scale bar 25 μm. (C) Control (siCtrl) or DDX3-depleted (siDDX3) HeLa cells were transfected with 0.3 μg of pNL4-3R or pNL4-3R ΔRev proviral DNA, and gRNA translation (Gag–Renilla luciferase activity normalized to the amount of cytoplasmic gRNA as measured by RT–qPCR) was determined as described in ‘Materials and Methods’ section. Results were normalized to siCtrl (set to 100%) and are presented as mean ± SD of three independent experiments. **P < 0.01; ***P < 0.001 (non-directional t-test). (D) HeLa cells were transfected with 0.5 μg of pNL4-3 Gag-Stop and 0.2 μg of pCIneo-HA-DDX3 for 24 h and prepared for LSCM as indicated in ‘Materials and Methods’ section. Green staining corresponds to the gRNA, and red staining identifies DDX3. White arrowheads indicate cytoplasmic granules. Co-localized points are indicated in white. Merge image includes nucleus staining with Hoechst (blue). Scale bar 25 μm.
As DDX3 was found to be associated with the HIV-1 gRNA nuclear export components (34,35), we next examined whether the gRNA granules corresponded to a cytoplasmic nuclear export intermediate. For this, we looked for the presence of the gRNA export factors CRM1 (15) and DDX1 (36). However, neither CRM1 (Figure 2B) nor DDX1 (data not shown) was found in these gRNA granules, suggesting that these cytoplasmic structures are assembled once the exported viral mRNP has been remodelled.
Thus, we have also investigated a possible interplay between the specific viral nuclear export factor Rev and DDX3. For this, we took advantage of our recently described pNL4-3R vector, which possesses the Renilla luciferase gene inserted in frame within the Gag-coding region and allows for the quantification of gRNA translation (23). We also have created the pNL4-3R ΔRev proviral DNA, a derivate of the pNL4-3fB vector that does not express the Rev protein (24). As expected, cytoplasmic quantification of the gRNA present in the cytoplasm of transfected cells revealed a 4.5-fold reduction, which is consistent with the role of Rev in nuclear export (data not shown). Then, the pNL4-3R and the pNL4-3R ΔRev reporter proviruses were transfected in siCtrl and siDDX3 cells, and gRNA translation was quantified as indicated (Figure 2C). Interestingly, we observed that DDX3 knockdown inhibited gRNA translation with the same magnitude whether Rev was present. This confirms that DDX3 can play a role on gRNA translation independently from its function on Rev-mediated nuclear export.
Cytoplasmic mRNPs containing the HIV-1 gRNA and the Gag polyprotein have been previously described and were shown to correspond to cytoplasmic intermediate of viral particle assembly (37). To determine whether the DDX3-containing mRNPs could correspond to any of these structures, we used a modified pNL4-3 proviral DNA in which the Gag-coding region is interrupted by several premature stop codons. As a consequence, the gRNA enters initiation and elongation of translation, but ribosomes are prematurely disassembled from the coding region, and the Gag polyprotein is not produced. RNA FISH–IF and LSCM analysis revealed that the gRNA from this provirus (named gRNA Stop) was fully able to form cytoplasmic granules together with DDX3 (Figure 2D) despite the absence of Gag production. This result indicates that the mRNPs formed between the gRNA and DDX3 are not scaffolded by the Gag polyprotein; thus, they unlikely correspond to a viral particle assembly intermediate.
DDX3 assembles the gRNA granules in an ATP-dependent manner
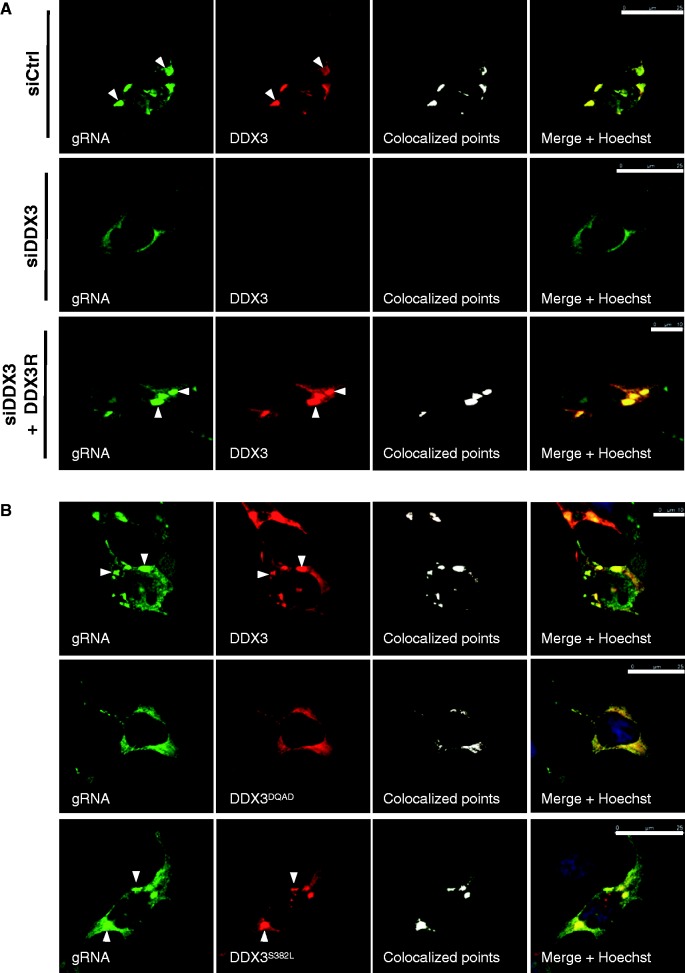
Given the fact that DEAD-box proteins promote the assembly and remodelling of large mRNPs (38), it was of interest to examine whether DDX3 could be at the onset of the assembly of gRNA granules. For this, we first looked at gRNA granules formation in control and DDX3 knockdown cells (Figure 3A). Interestingly, we observed that although DDX3-containing gRNA granules were assembled in control cells (Figure 3A, see siCtrl), they completely disappeared in cells that lacked DDX3 (Figure 3A, see siDDX3), suggesting an active role of the latter in gRNA granules formation. Consistent with this notion, expression of a siRNA-resistant DDX3 (DDX3R) in siDDX3 cells was sufficient to restore the assembly of gRNA granules, confirming the critical role of DDX3 in driving the assembly of these viral mRNPs.
Figure 3.
DDX3 drives the ATP-dependent assembly of gRNA translation granules. (A) Control (siCtrl) or DDX3-depleted (siDDX3) HeLa cells were transfected with 0.5 μg of pNL4-3 and 0.2 μg of pCIneo-HA-DDX3 (top and middle) or 0.2 μg of pCIneo-HA-DDX3R, which codes a siRNA-resistant DDX3 (bottom). At 24 hpt, cells were prepared for LSCM as indicated in ‘Materials and Methods’ section. Green staining corresponds to the gRNA, and red staining identifies DDX3. White arrowheads indicate cytoplasmic granules. Co-localized points are indicated in white. Merge images include nucleus staining with Hoechst (blue). Scale bars 25 μm (siCtrl/siDDX3) and 10 μm (siDDX3 + DDX3R). (B) HeLa cells were transfected with 0.5 μg of pNL4-3, 0.2 μg of pCIneo-HA-DDX3 (top) or pCIneo-HA-DDX3DQAD (middle) or pCIneo-HA-DDX3S382L (bottom) for 24 h and prepared for LSCM as indicated in ‘Materials and Methods’ section. Green staining corresponds to the gRNA, and red staining identifies DDX3. White arrowheads indicate cytoplasmic granules. Co-localized points are indicated in white. Merge images include nucleus staining with Hoechst (blue). Scale bars 10 μm (upper image) and 25 μm (middle and bottom images).
We then examined which of the catalytic activities of DDX3 were required for granule formation. As such, we performed RNA FISH–IF and LSCM analyses using wild-type DDX3 or the DDX3DQAD or the DDX3S382L mutants, which are deficient in the ATPase and helicase activities, respectively (39). Interestingly, we observed that although the wild-type and the mutant in the helicase activity (DDX3S382L) were able to co-localize with the gRNA in cytoplasmic granules, the DDX3DQAD mutant failed to drive the assembly of HIV-1 gRNA granules, indicating that this process was ATP dependent (Figure 3B).
HIV-1 gRNA granules correspond to a novel pre-translation initiation intermediate
We next reasoned that the cytoplasmic foci could correspond to structures reminiscent to stress granules that are sites of mRNA storage during translational arrest (40). Such a hypothesis is reinforced by the fact that DDX3 is a key structural component of cytoplasmic stress granules (21,33,41). Interestingly, the assembly of stress granules can be triggered by some viral infections (42). Although granules formation occurred in a fraction of HIV-1 expressing cells (18%, Supplementary Figure S1), we believe that any modifications or cleavage would be detected by western blotting. Thus, we have analysed the phosphorylation status of the α subunit of the translation initiation factor eIF2, as it is one of the first proteins modified on cellular stress or viral infections (40,43). Consistent with two recent reports (20,29), we were not able to detect the phosphorylated form of eIF2α or variations in the overall levels of eIF2α in HIV-1 expressing cells (Supplementary Figure S2). However, stress granules assembly does not necessarily require the phosphorylation of eIF2α and could rather involve the inactivation or reduced levels of other eIFs, such as eIF4G, eIF4A, eIF4B and PABP (44,45). Therefore, we also checked the status of several eIFs involved in translation initiation, and we did not observe any modification on HIV-1 expression (Supplementary Figure S2), suggesting that the virus does not induce major changes in the integrity of the host translation initiation machinery (at least at 24 h after DNA transfection). Thus, this suggests that these gRNA/DDX3 granules might not correspond to stress granules, at least those that were described to be assembled on modifications of components of the translation initiation machinery.
Then, we investigated this further and went on to search for translation initiation components that could localize together with the gRNA in granules. Obvious candidates were eIF4G and PABP, as they were recently shown to physically interact with DDX3 and to co-localize with the RNA helicase in stress granules (21,41). Thus, we first examined whether these translation initiation factors could localize together with the gRNA in the absence of induced stress. Interestingly, we observed that this was indeed the case as both initiation factors accumulated and co-localized with the gRNA in cytoplasmic granules (Figure 4A and B, respectively). We also observed the HIV-1 gRNA, DDX3 and eIF4G co-localized together in cytoplasmic granules, indicating that they accumulate together in the same mRNP granule (Figure 4C).
Figure 4.
gRNA granules are a novel pre-initiation intermediate. HeLa cells were transfected with 0.5 μg of pNL4-3 and 0.2 μg of (A) pCIneo-HA-eIF4G, (B) pCIneo-HA-DDX3, (C) pEGFP-DDX3 and pCIneo-HA-eIF4G (D) pCMV-myc-CBP80, (E) pCIneo-HA-eIF4E or (F) pCIneo-HA-eIF4A for 24 h and prepared for LSCM as indicated in ‘Materials and Methods’ section. White arrowheads indicate cytoplasmic granules. Co-localized points are indicated in white. Merge images includes nucleus staining with Hoechst (blue). Scale bars 25 and 10 μm in (E).
As eIF4G and PABP are present on mRNPs bound to the cap-binding proteins CBP80/20 and eIF4E (9,10), we examined whether gRNA granules could correspond to one of these two different translation initiation mRNPs. As observed, CBP80 presented its typical nuclear localization and was not present in cytoplasmic gRNA granules (Figure 4D). This is consistent with the nuclear CBC-bound mRNP being rapidly remodelled and re-imported to the nucleus by importins (46,47). To most of our surprise, eIF4E was found to disperse through the cytoplasm, and we did not observe an enrichment in gRNA granules similar to that observed with DDX3, eIF4G or PABP (Figure 4E). This is consistent with recent data showing that HIV-1 replication, through the viral protein Vpr, could inactivate eIF4E (20), thus providing a rationale for the absence of this cap-binding protein in gRNA granules (see ‘Discussion’ section). Interestingly, we observed a partial co-localization between the gRNA and eIF4A (Figure 4F), eIF4B and eIF3g (Supplementary Figure S3). Thus, it seems to indicate that PABP and eIF4G are core components of these gRNA granules, but they can also, to a lesser extent, contain eIF4A, eIF4B and eIF3. However, none of the major cap-binding proteins (CBP80 and eIF4E) were found to co-localize with the gRNA in these granules.
DDX3 can substitute for eIF4E during cap binding
Assembly of a pre-initiation complex in the absence of eIF4E suggests that another protein must mediate m7GTP attachment on the HIV-1 gRNA. Therefore, we next went on to examine whether DDX3 could fulfil this function. Thus, we prepared a cytoplasmic S7 nuclease-treated HeLa cells extract that was incubated together with the m7GTP affinity matrix in the presence of buffer as control or m7GpppG cap analogue. Consistent with our previous observations, a small fraction of cytoplasmic DDX3 was retained together with eIF4E on the m7GTP affinity matrix (Figure 5A, lane 2). Interestingly, we repeatedly observed that addition of the cap analogue completely abolished retention of eIF4E; yet, it was not completely able to prevent DDX3 binding (Figure 5A, compare lane 2 and 3). These results indicate that retention of DDX3 can still occur in the physical absence of the major cytoplasmic cap-binding protein on the column, and that retention of DDX3 on m7GTP (mononucleotide cap) is more resistant to competition with m7GpppG (dinucleotide cap) than eIF4E. This is not without precedents, as cap-binding proteins showed different affinities towards different cap analogues (48,49). Remarkably, addition of recombinant His-DDX3 to the cell extract chased away eIF4E but increased the retention of both PABP and eIF4G, further indicating that DDX3 together with PABP and eIF4G can be bound on the cap affinity matrix independently of eIF4E (Figure 5B, compare lanes 2 and 3).
Figure 5.
DDX3 substitutes for eIF4E. (A) Hundred micrograms of S7 nuclease-treated HeLa cell cytoplasmic extracts was passed through m7GTP affinity matrix in the presence of buffer (lane 2) or an excess of m7GpppG cap analogue (lane 3). After elution of the column, the presence of eIF4E and DDX3 in the retained material was analysed by western blot. Input (lane 1) corresponds to 50 μg of cell extract. (B) Hundred micrograms of S7 nuclease-treated HeLa cell cytoplasmic extracts was passed through m7GTP affinity matrix in the presence of an excess of m7GpppG cap analogue (lane 1), buffer (lane 2) or recombinant His-tagged DDX3 (lane 3). After elution of the column, the presence of eIF4E, DDX3, PABP and eIF4G in the retained material was analysed by western blot. (C) Hundred micrograms of S7 nuclease-treated HeLa cells cytoplasmic extracts from mock transfected cells (lane 1) or cells transfected with pCineo-HA-DDX3 (lane 2), -HA-DDX3W60A (included as a control, lane 3) or -HA-DDX3Y38A/L43A (lane 4) was incubated with an m7GTP affinity matrix, and the presence of the HA-tagged proteins in the input (corresponding to 50 μg of extract, left) or in the retained material (right) was analysed by western blot. (D) HeLa cells were transfected with 0.5 μg of pNL4-3 and 0.2 μg of pCIneo-HA-DDX3Y38A/L43A for 24 h and prepared for LSCM as indicated in ‘Materials and Methods’ section. Green staining corresponds to the gRNA, and red staining identifies DDX3Y38A/L43A. White arrowheads indicate cytoplasmic granules. Co-localized points are indicated in white. Merge image includes nucleus staining with Hoechst (blue). Scale bar 25 μm. (E) HeLa cells were transfected with 0.3 μg of pNL4-3R and 1 μg of pCIneo-HA-vector (EGFP, DDX3, DDX3DQAD or DDX3Y38A/L43A), and gRNA translation (Gag–Renilla luciferase activity normalized to the amount of cytoplasmic gRNA as measured by RT–qPCR) was determined as indicated. Results are normalized to the control (set to 100%) and are presented as mean ± SD of three independent experiments. (F) DDX3-depleted HeLa cells were transfected with 0.3 μg of pNL4-3R and 1 μg of pCIneo-HA-vector (EGFP, DDX3R, DDX3RDQAD or DDX3RY38A/L43A), and gRNA translation was determined as indicated. Results are normalized to the rescue by DDX3R (set to 100%) and are presented as mean ± SD of three independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001 (non directional t-test).
However, as it could still be argued that retention of DDX3 on the cap-column may be mediated by an interaction with eIF4E, we have used a previously described DDX3 mutant in which the putative YxxxxLϕ (38YIPPHLR44 in human DDX3) eIF4E-binding motif has been mutated and was shown to be unable to interact with eIF4E anymore (50). We observed that the DDX3Y38A/L43A mutant was retained as efficiently as the wild-type protein (or the W60A control mutant) on the m7GTP affinity matrix (Figure 5C), confirming that DDX3 is retained on the cap structure independently of its ability to interact with eIF4E.
The next step was then to investigate whether this DDX3 mutant could promote cytoplasmic mRNPs assembly and support translation initiation from the HIV-1 genomic RNA in cells. To answer the first question, we analysed mRNP formation in the presence of a DDX3Y38A/L43A mutant (Figure 5D) and observed that the latter was incorporated in the gRNA granules just as efficiently as the wild-type DDX3.
These data were confirmed by showing that ectopic expression of the DDX3Y38A/L43A mutant in control cells did not interfere with HIV-1 genomic RNA translation (Figure 5E). Moreover, expression of the DDX3Y38A/L43A mutant was able to fully rescue HIV-1 translation in DDX3-depleted cells as observed with the wild-type protein (Figure 5F). It is noteworthy that although this mutant is unable to interact with eIF4E, it is fully competent to bind and hydrolyse ATP (41,50). In contrast, the DDX3DQAD mutant, which failed to drive granule assembly (Figure 3B), was also unable to rescue HIV-1 translation from knocked down cells (Figure 5F) and significantly interfered when added ectopically (Figure 5E).
Taken together these data confirm that DDX3 can promote HIV-1 gRNA translation initiation and mRNPs assembly in an ATP-dependent, but in an eIF4E-independent manner.
DISCUSSION
All HIV-1 mRNAs posses long and structured 5′-UTRs that start with the TAR motif, which is a 57 nt-long stem–loop in which the m7GTP cap moiety is embedded at the basis of the duplex. Because of its position within the 5′-UTR, TAR interferes with the assembly of a pre-initiation complex at the 5′ cap structure (21,23,51). However, despite this physical constraint, the HIV-1 gRNA is efficiently translated in infected cells, as it produces massive amounts of the structural protein Gag, which is the structural basis for viral particle assembly.
It has recently been shown that efficient translation on the HIV-1 gRNA occurred by a 5′ cap-dependent mechanism (23,52). By using a synthetic mRNA containing the HIV-1 5′-UTR, we could show that this was due to the specific recruitment of the DEAD-box RNA helicase DDX3 to the 5′ TAR structure (21).
In this study, we further confirm the critical role that DDX3 plays in translation of HIV-1 mRNAs in the context of viral replication in CD4+ human cells (Figure 1). Interestingly, our cell imaging analysis reveals that DDX3 and the viral gRNA were co-localized in the form of a novel pre-translation initiation complex also containing translation initiation factors eIF4G and PABP (Figure 4). However, when we looked for the presence of other eIFs, such as CBP80, eIF4E, eIF4A, eIF4B or eIF3, we were surprised to find that we could virtually not detect any of the cap-binding proteins (CBP80 and eIF4E) and only a weak, if anything, co-localization of eIF4A, eIF4B and eIF3 (Figure 4). This suggests that these foci are not active sites of translation but rather cytoplasmic spots where a minimal pre-translation initiation complex (composed of eIF4G, DDX3 and PABP) is assembled on the gRNA.
Such a viral mRNP was visualized in HeLa cells as large cytoplasmic RNA granules that were functionally different from mammalian stress granules, as (i) they were assembled in the absence of any evident modification of the host translation initiation machinery (Supplementary Figure S2); (ii) they did not require the stress granules assembly domain 38YIPPHLR43 of DDX3 (Figure 5); and (iii) they were deprived from eIF4E (Figure 4), which is usually found in mammalian stress granules (21,53). Consistent with this is the ability of HIV-1 to interfere with stress granules assembly induced by arsenite treatment (29).
Interestingly, translation initiation factors eIF4G and PABP interact with DDX3 (21,41) and have been previously shown to play a role in HIV-1 viral replication. Indeed, eIF4G expression is increased during viral replication and is required for HIV-1 cap-dependent translation (54–57), and PABP was shown to bind the gRNA in a nuclear export-dependent manner to enhance translation of intron-containing viral transcripts (58,59).
Although none of the major cap-binding proteins (eIF4E and CBP80) was observed to be efficiently recruited to gRNA granules (Figure 4), we showed that cytoplasmic DDX3 has the ability to be retained on an m7GTP cap structure independently of eIF4E. Moreover, addition of recombinant DDX3 to a nuclease-treated cytoplasmic cell extract prevented the retention of eIF4E while stimulated the binding of its interacting partners eIF4G and PABP (Figure 5), suggesting that DDX3 could be (or be associated to) a novel cytoplasmic cap-binding protein that is bound to the HIV-1 gRNA and recruited to cytoplasmic granules. Consistent with this idea, we have also observed that recombinant DDX3 protein could bind the m7GTP sepharose matrix in the absence of any other partner (data not shown). Although these data clearly show that DDX3 can substitute for eIF4E and CBP80, further work is required to determine which proportion of the HIV-1 RNAs is associated with DDX3 or any of the aforementioned cap-binding proteins.
Interestingly, overexpression of the yeast homologue of DDX3, Ded1, was shown to suppress the deleterious effects of allelic mutations in eIF4E (60). Moreover, it was recently shown that a small fraction of recombinant Ded1 could be retained on an m7GTP affinity matrix in the absence of the eIF4E/eIF4G complex (61). Indeed, other cellular proteins, including 4E-HP, Gemin5 and Pumilio2, were previously shown to bind immobilized cap structures (62–64).
Interestingly, retention of DDX3 on the m7GTP cap affinity matrix was more resistant than eIF4E to competition with the m7GpppG dinucleotide cap analogue (Figure 5), suggesting different affinities between eIF4E and DDX3 for the cap. This is particularly interesting given the fact that the HIV-1 gRNA cap structures could be trimethylated by the PIMT protein (65). Although expression from the gRNA was enhanced by PIMT (65), it has been shown that translation of an mRNA carrying a trimethylated cap is inhibited at least in vitro (66,67). Indeed, the affinities of the CBC and eIF4E for a trimethylated cap are, 100- and 760-fold lower when compared with a classical monomethylated cap (48,49). These data suggest that trimethylation of the gRNA may be responsible of the absence of both CBP80 and eIF4E from gRNA/DDX3 granules, as trimethylation of the HIV-1 cap may strongly decrease the affinity of the latter for the gRNA. As PIMT is associated to the viral protein Rev (65), it is tempting to speculate that trimethylation of the HIV-1 gRNA cap structure occurs once CBP20/80 has been dissociated from the viral mRNP during or early after nuclear export. Then, DDX3 may be recruited at the 5′-end of the gRNA to recruit eIF4G and PABP while driving the ATP-dependent assembly of cytoplasmic granules. Therefore, DDX3 may provide a molecular anchor that prepares the HIV-1 gRNA to enter translation initiation early after nuclear export and before the 43S pre-initiation complex is recruited onto the 5′-UTR. Thus, gRNA/DDX3 granules may represent a highly dynamic viral mRNP intermediate that assembles and dissembles between nuclear export and translation in initiation.
Interestingly, Ded1 was shown to act in a similar way by driving the assembly and resolution of large mRNPs before the mRNA enters in translation (61). As such, these granules would not be active sites of translation but rather foci where pre-initiation complex formation is taking place.
Although localized translation is a feature of highly polarized cells, such as oocytes and differentiated neurons, where spatial and temporal control of gene expression is required, compartmentalization of the HIV-1 gRNA translation initiation may present several advantages. In fact, assembly of these granules may be a way to sequester and protect the gRNA that has not yet acquired all components needed for efficient translation initiation. In addition, accumulation in these cytoplasmic structures may also serve to locally concentrate the gRNA and translation initiation factors that could, in turn, enhance the efficiency of ribosome recruitment and polysome association. This may also ensure that the gRNA reaches the host translational machinery and is not prematurely sequestered for viral particle assembly by the Gag protein. Such a hypothesis is consistent with the fact that these granules can form in the absence of the Gag polyprotein (Figure 2).
Further work will be needed to address the importance of these issues for viral translation and its impact on replication and assembly.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–3.
FUNDING
CONICYT-Chile/French Embassy doctoral fellowship and Agence Nationale des Recherches sur le SIDA et les Hépatites Virales (ANRS) post-doctoral fellowship (to R.S.R.); ‘Becas Chile/CONICYT’ doctoral fellow (to P.S.R.), ANRS and ‘Contrat d’Interface’ with the Hospices Civils de Lyon (to T.O.). Funding for open access charge: INSERM.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Dr Denis Gerlier (ENS-Lyon, France), Dr Simon Morley (University of Sussex, UK), Dr Barbara Felber (Frederick National Laboratory for Cancer Research, USA), Dr Yoon Ki Kim (Korea University, Republic of Korea) and the NIH AIDS Reagents program for reagents.
REFERENCES
- 1.Moore MJ. From birth to death: the complex lives of eukaryotic mRNAs. Science. 2005;309:1514–1518. doi: 10.1126/science.1111443. [DOI] [PubMed] [Google Scholar]
- 2.Moore MJ, Proudfoot NJ. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell. 2009;136:688–700. doi: 10.1016/j.cell.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 3.Le Hir H, Nott A, Moore MJ. How introns influence and enhance eukaryotic gene expression. Trends Biochem. Sci. 2003;28:215–220. doi: 10.1016/S0968-0004(03)00052-5. [DOI] [PubMed] [Google Scholar]
- 4.Topisirovic I, Svitkin YV, Sonenberg N, Shatkin AJ. Cap and cap-binding proteins in the control of gene expression. Wiley Interdiscip Rev. RNA. 2011;2:277–298. doi: 10.1002/wrna.52. [DOI] [PubMed] [Google Scholar]
- 5.Izaurralde E, Lewis J, McGuigan C, Jankowska M, Darzynkiewicz E, Mattaj IW. A nuclear cap binding protein complex involved in pre-mRNA splicing. Cell. 1994;78:657–668. doi: 10.1016/0092-8674(94)90530-4. [DOI] [PubMed] [Google Scholar]
- 6.Cheng H, Dufu K, Lee CS, Hsu JL, Dias A, Reed R. Human mRNA export machinery recruited to the 5' end of mRNA. Cell. 2006;127:1389–1400. doi: 10.1016/j.cell.2006.10.044. [DOI] [PubMed] [Google Scholar]
- 7.Lenasi T, Peterlin BM, Barboric M. Cap-binding protein complex links pre-mRNA capping to transcription elongation and alternative splicing through positive transcription elongation factor b (P-TEFb) J. Biol. Chem. 2011;286:22758–22768. doi: 10.1074/jbc.M111.235077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ishigaki Y, Li X, Serin G, Maquat LE. Evidence for a pioneer round of mRNA translation: mRNAs subject to nonsense-mediated decay in mammalian cells are bound by CBP80 and CBP20. Cell. 2001;106:607–617. doi: 10.1016/s0092-8674(01)00475-5. [DOI] [PubMed] [Google Scholar]
- 9.Maquat LE, Tarn WY, Isken O. The pioneer round of translation: features and functions. Cell. 2010;142:368–374. doi: 10.1016/j.cell.2010.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Czaplinski K, Singer RH. Pathways for mRNA localization in the cytoplasm. Trends Biochem. Sci. 2006;31:687–693. doi: 10.1016/j.tibs.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 12.Martin KC, Ephrussi A. mRNA localization: gene expression in the spatial dimension. Cell. 2009;136:719–730. doi: 10.1016/j.cell.2009.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stoltzfus CM. Chapter 1. Regulation of HIV-1 alternative RNA splicing and its role in virus replication. Adv. Virus Res. 2009;74:1–40. doi: 10.1016/S0065-3527(09)74001-1. [DOI] [PubMed] [Google Scholar]
- 14.Pollard VW, Malim MH. The HIV-1 Rev protein. Annu. Rev. Microbiol. 1998;52:491–532. doi: 10.1146/annurev.micro.52.1.491. [DOI] [PubMed] [Google Scholar]
- 15.Fornerod M, Ohno M, Yoshida M, Mattaj IW. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 16.Cochrane AW, McNally MT, Mouland AJ. The retrovirus RNA trafficking granule: from birth to maturity. Retrovirology. 2006;3:18. doi: 10.1186/1742-4690-3-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Swanson CM, Malim MH. Retrovirus RNA trafficking: from chromatin to invasive genomes. Traffic. 2006;7:1440–1450. doi: 10.1111/j.1600-0854.2006.00488.x. [DOI] [PubMed] [Google Scholar]
- 18.Chiu YL, Coronel E, Ho CK, Shuman S, Rana TM. HIV-1 Tat protein interacts with mammalian capping enzyme and stimulates capping of TAR RNA. J. Biol. Chem. 2001;276:12959–12966. doi: 10.1074/jbc.M007901200. [DOI] [PubMed] [Google Scholar]
- 19.Chiu YL, Ho CK, Saha N, Schwer B, Shuman S, Rana TM. Tat stimulates cotranscriptional capping of HIV mRNA. Mol. Cell. 2002;10:585–597. doi: 10.1016/s1097-2765(02)00630-5. [DOI] [PubMed] [Google Scholar]
- 20.Sharma A, Yilmaz A, Marsh K, Cochrane A, Boris-Lawrie K. Thriving under Stress: Selective Translation of HIV-1 structural protein mRNA during Vpr-mediated impairment of eIF4E translation activity. PLoS Pathog. 2012;8:e1002612. doi: 10.1371/journal.ppat.1002612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Soto-Rifo R, Rubilar PS, Limousin T, de Breyne S, Decimo D, Ohlmann T. DEAD-box protein DDX3 associates with eIF4F to promote translation of selected mRNAs. EMBO J. 2012;31:3745–3756. doi: 10.1038/emboj.2012.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Adachi A, Gendelman HE, Koenig S, Folks T, Willey R, Rabson A, Martin MA. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Soto-Rifo R, Limousin T, Rubilar PS, Ricci EP, Decimo D, Moncorge O, Trabaud MA, Andre P, Cimarelli A, Ohlmann T. Different effects of the TAR structure on HIV-1 and HIV-2 genomic RNA translation. Nucleic Acids Res. 2012;1:2653–2667. doi: 10.1093/nar/gkr1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sodroski J, Goh WC, Rosen C, Dayton A, Terwilliger E, Haseltine W. A second post-transcriptional trans-activator gene required for HTLV-III replication. Nature. 1986;321:412–417. doi: 10.1038/321412a0. [DOI] [PubMed] [Google Scholar]
- 25.Choe J, Oh N, Park S, Lee YK, Song OK, Locker N, Chi SG, Kim YK. Translation initiation on mRNAs bound by nuclear cap-binding protein complex CBP80/20 requires interaction between CBP80/20-dependent translation initiation factor and eukaryotic translation initiation factor 3g. J. Biol. Chem. 2012;287:18500–18509. doi: 10.1074/jbc.M111.327528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Soto-Rifo R, Ricci EP, Decimo D, Moncorge O, Ohlmann T. Back to basics: the untreated rabbit reticulocyte lysate as a competitive system to recapitulate cap/poly(A) synergy and the selective advantage of IRES-driven translation. Nucleic Acids Res. 2007;35:e121. doi: 10.1093/nar/gkm682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Soto-Rifo R, Limousin T, Rubilar PS, Ricci EP, Decimo D, Moncorge O, Trabaud MA, Andre P, Cimarelli A, Ohlmann T. Different effects of the TAR structure on HIV-1 and HIV-2 genomic RNA translation. Nucleic Acids Res. 2011;40:2653–2667. doi: 10.1093/nar/gkr1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bushman FD, Malani N, Fernandes J, D'Orso I, Cagney G, Diamond TL, Zhou H, Hazuda DJ, Espeseth AS, Konig R, et al. Host cell factors in HIV replication: meta-analysis of genome-wide studies. PLoS Pathog. 2009;5:e1000437. doi: 10.1371/journal.ppat.1000437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abrahamyan LG, Chatel-Chaix L, Ajamian L, Milev MP, Monette A, Clement JF, Song R, Lehmann M, DesGroseillers L, Laughrea M, et al. Novel Staufen1 ribonucleoproteins prevent formation of stress granules but favour encapsidation of HIV-1 genomic RNA. J. Cell Sci. 2010;123:369–383. doi: 10.1242/jcs.055897. [DOI] [PubMed] [Google Scholar]
- 30.Monette A, Ajamian L, Lopez-Lastra M, Mouland AJ. Human immunodeficiency virus type 1 (HIV-1) induces the cytoplasmic retention of heterogeneous nuclear ribonucleoprotein A1 by disrupting nuclear import: implications for HIV-1 gene expression. J. Biol. Chem. 2009;284:31350–31362. doi: 10.1074/jbc.M109.048736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Milev MP, Brown CM, Mouland AJ. Live cell visualization of the interactions between HIV-1 Gag and the cellular RNA-binding protein Staufen1. Retrovirology. 2010;7:41. doi: 10.1186/1742-4690-7-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee CS, Dias AP, Jedrychowski M, Patel AH, Hsu JL, Reed R. Human DDX3 functions in translation and interacts with the translation initiation factor eIF3. Nucleic Acids Res. 2008;36:4708–4718. doi: 10.1093/nar/gkn454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lai MC, Lee YH, Tarn WY. The DEAD-box RNA helicase DDX3 associates with export messenger ribonucleoproteins as well as tip-associated protein and participates in translational control. Mol. Biol. Cell. 2008;19:3847–3858. doi: 10.1091/mbc.E07-12-1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yedavalli VS, Neuveut C, Chi YH, Kleiman L, Jeang KT. Requirement of DDX3 DEAD box RNA helicase for HIV-1 Rev-RRE export function. Cell. 2004;119:381–392. doi: 10.1016/j.cell.2004.09.029. [DOI] [PubMed] [Google Scholar]
- 35.Naji S, Ambrus G, Cimermancic P, Reyes JR, Johnson JR, Filbrandt R, Huber MD, Vesely P, Krogan NJ, Yates JR, III, et al. Host cell interactome of HIV-1 rev includes RNA helicases involved in multiple facets of virus production. Mol. Cell Proteomics. 2012;11 doi: 10.1074/mcp.M111.015313. M111 015313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Edgcomb SP, Carmel AB, Naji S, Ambrus-Aikelin G, Reyes JR, Saphire AC, Gerace L, Williamson JR. DDX1 is an RNA-dependent ATPase involved in HIV-1 Rev function and virus replication. J. Mol. Biol. 2012;415:61–74. doi: 10.1016/j.jmb.2011.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kutluay SB, Bieniasz PD. Analysis of the initiating events in HIV-1 particle assembly and genome packaging. PLoS Pathog. 2010;6:e1001200. doi: 10.1371/journal.ppat.1001200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Linder P, Jankowsky E. From unwinding to clamping—the DEAD box RNA helicase family. Nat. Rev. Mol. Cell. Biol. 2011;12:505–516. doi: 10.1038/nrm3154. [DOI] [PubMed] [Google Scholar]
- 39.Cordin O, Banroques J, Tanner NK, Linder P. The DEAD-box protein family of RNA helicases. Gene. 2006;367:17–37. doi: 10.1016/j.gene.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 40.Anderson P, Kedersha N. Stress granules: the Tao of RNA triage. Trends Biochem. Sci. 2008;33:141–150. doi: 10.1016/j.tibs.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 41.Shih JW, Wang WT, Tsai TY, Kuo CY, Li HK, Wu Lee YH. Critical roles of RNA helicase DDX3 and its interactions with eIF4E/PABP1 in stress granule assembly and stress response. Biochem. J. 2011;1:119–129. doi: 10.1042/BJ20110739. [DOI] [PubMed] [Google Scholar]
- 42.Valiente-Echeverria F, Melnychuk L, Mouland AJ. Viral modulation of stress granules. Virus Res. 2012;169:430–437. doi: 10.1016/j.virusres.2012.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gale M, Jr, Tan SL, Katze MG. Translational control of viral gene expression in eukaryotes. Microbiol. Mol. Biol. Rev. 2000;64:239–280. doi: 10.1128/mmbr.64.2.239-280.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mokas S, Mills JR, Garreau C, Fournier MJ, Robert F, Arya P, Kaufman RJ, Pelletier J, Mazroui R. Uncoupling stress granule assembly and translation initiation inhibition. Mol. Biol. Cell. 2009;20:2673–2683. doi: 10.1091/mbc.E08-10-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dang Y, Kedersha N, Low WK, Romo D, Gorospe M, Kaufman R, Anderson P, Liu JO. Eukaryotic initiation factor 2alpha-independent pathway of stress granule induction by the natural product pateamine A. J. Biol. Chem. 2006;281:32870–32878. doi: 10.1074/jbc.M606149200. [DOI] [PubMed] [Google Scholar]
- 46.Sato H, Maquat LE. Remodeling of the pioneer translation initiation complex involves translation and the karyopherin importin beta. Genes Dev. 2009;23:2537–2550. doi: 10.1101/gad.1817109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dias SM, Wilson KF, Rojas KS, Ambrosio AL, Cerione RA. The molecular basis for the regulation of the cap-binding complex by the importins. Nat. Struct. Mol. Biol. 2009;16:930–937. doi: 10.1038/nsmb.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Worch R, Niedzwiecka A, Stepinski J, Mazza C, Jankowska-Anyszka M, Darzynkiewicz E, Cusack S, Stolarski R. Specificity of recognition of mRNA 5' cap by human nuclear cap-binding complex. RNA. 2005;11:1355–1363. doi: 10.1261/rna.2850705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Niedzwiecka A, Marcotrigiano J, Stepinski J, Jankowska-Anyszka M, Wyslouch-Cieszynska A, Dadlez M, Gingras AC, Mak P, Darzynkiewicz E, Sonenberg N, et al. Biophysical studies of eIF4E cap-binding protein: recognition of mRNA 5' cap structure and synthetic fragments of eIF4G and 4E-BP1 proteins. J. Mol. Biol. 2002;319:615–635. doi: 10.1016/S0022-2836(02)00328-5. [DOI] [PubMed] [Google Scholar]
- 50.Shih JW, Tsai TY, Chao CH, Wu Lee YH. Candidate tumor suppressor DDX3 RNA helicase specifically represses cap-dependent translation by acting as an eIF4E inhibitory protein. Oncogene. 2008;27:700–714. doi: 10.1038/sj.onc.1210687. [DOI] [PubMed] [Google Scholar]
- 51.Parkin NT, Cohen EA, Darveau A, Rosen C, Haseltine W, Sonenberg N. Mutational analysis of the 5' non-coding region of human immunodeficiency virus type 1: effects of secondary structure on translation. EMBO J. 1988;7:2831–2837. doi: 10.1002/j.1460-2075.1988.tb03139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Berkhout B, Arts K, Abbink TE. Ribosomal scanning on the 5'-untranslated region of the human immunodeficiency virus RNA genome. Nucleic Acids Res. 2011;39:5232–5244. doi: 10.1093/nar/gkr113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kedersha N, Anderson P. Mammalian stress granules and processing bodies. Methods Enzymol. 2007;431:61–81. doi: 10.1016/S0076-6879(07)31005-7. [DOI] [PubMed] [Google Scholar]
- 54.Krishnan V, Zeichner SL. Alterations in the expression of DEAD-box and other RNA binding proteins during HIV-1 replication. Retrovirology. 2004;1:42. doi: 10.1186/1742-4690-1-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ricci EP, Soto-Rifo R, Herbreteau CH, Decimo D, Ohlmann T. Lentiviral RNAs can use different mechanisms for translation initiation. Biochem. Soc. Trans. 2008;36:690–693. doi: 10.1042/BST0360690. [DOI] [PubMed] [Google Scholar]
- 56.Castello A, Franco D, Moral-Lopez P, Berlanga JJ, Alvarez E, Wimmer E, Carrasco L. HIV- 1 protease inhibits Cap- and poly(A)-dependent translation upon eIF4GI and PABP cleavage. PLoS One. 2009;4:e7997. doi: 10.1371/journal.pone.0007997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.de Breyne S, Chamond N, Decimo D, Trabaud MA, Andre P, Sargueil B, Ohlmann T. In vitro studies reveal that different modes of initiation on HIV-1 mRNA have different levels of requirement for eukaryotic initiation factor 4F. FEBS J. 2012;279:3098–3111. doi: 10.1111/j.1742-4658.2012.08689.x. [DOI] [PubMed] [Google Scholar]
- 58.Campbell LH, Borg KT, Haines JK, Moon RT, Schoenberg DR, Arrigo SJ. Human immunodeficiency virus type 1 Rev is required in vivo for binding of poly(A)-binding protein to Rev-dependent RNAs. J. Virol. 1994;68:5433–5438. doi: 10.1128/jvi.68.9.5433-5438.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Marsh K, Soros V, Cochrane A. Selective translational repression of HIV-1 RNA by Sam68DeltaC occurs by altering PABP1 binding to unspliced viral RNA. Retrovirology. 2008;5:97. doi: 10.1186/1742-4690-5-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.de la Cruz J, Iost I, Kressler D, Linder P. The p20 and Ded1 proteins have antagonistic roles in eIF4E-dependent translation in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 1997;94:5201–5206. doi: 10.1073/pnas.94.10.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hilliker A, Gao Z, Jankowsky E, Parker R. The DEAD-Box protein Ded1 modulates translation by the formation and resolution of an eIF4F-mRNA complex. Mol. Cell. 2011;43:962–972. doi: 10.1016/j.molcel.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rosettani P, Knapp S, Vismara MG, Rusconi L, Cameron AD. Structures of the human eIF4E homologous protein, h4EHP, in its m7GTP-bound and unliganded forms. J. Mol. Biol. 2007;368:691–705. doi: 10.1016/j.jmb.2007.02.019. [DOI] [PubMed] [Google Scholar]
- 63.Bradrick SS, Gromeier M. Identification of gemin5 as a novel 7-methylguanosine cap-binding protein. PLoS One. 2009;4:e7030. doi: 10.1371/journal.pone.0007030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cao Q, Padmanabhan K, Richter JD. Pumilio 2 controls translation by competing with eIF4E for 7-methyl guanosine cap recognition. RNA. 2010;16:221–227. doi: 10.1261/rna.1884610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yedavalli VS, Jeang KT. Trimethylguanosine capping selectively promotes expression of Rev-dependent HIV-1 RNAs. Proc. Natl Acad. Sci. USA. 2010;107:14787–14792. doi: 10.1073/pnas.1009490107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cai A, Jankowska-Anyszka M, Centers A, Chlebicka L, Stepinski J, Stolarski R, Darzynkiewicz E, Rhoads RE. Quantitative assessment of mRNA cap analogues as inhibitors of in vitro translation. Biochemistry. 1999;38:8538–8547. doi: 10.1021/bi9830213. [DOI] [PubMed] [Google Scholar]
- 67.Darzynkiewicz E, Stepinski J, Ekiel I, Jin Y, Haber D, Sijuwade T, Tahara SM. Beta-globin mRNAs capped with m7G, m2.7(2)G or m2.2.7(3)G differ in intrinsic translation efficiency. Nucleic Acids Res. 1988;16:8953–8962. doi: 10.1093/nar/16.18.8953. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.