Abstract
Although the right-handed double helical B-form DNA is most common under physiological conditions, DNA is dynamic and can adopt a number of alternative structures, such as the four-stranded G-quadruplex, left-handed Z-DNA, cruciform and others. Active transcription necessitates strand separation and can induce such non-canonical forms at susceptible genomic sequences. Therefore, it has been speculated that these non-B DNA motifs can play regulatory roles in gene transcription. Such conjecture has been supported in higher eukaryotes by direct studies of several individual genes, as well as a number of large-scale analyses. However, the role of non-B DNA structures in many lower organisms, in particular proteobacteria, remains poorly understood and incompletely documented. In this study, we performed the first comprehensive study of the occurrence of B DNA–non-B DNA transition-susceptible sites (non-B DNA motifs) within the context of the operon structure of the Escherichia coli genome. We compared the distributions of non-B DNA motifs in the regulatory regions of operons with those from internal regions. We found an enrichment of some non-B DNA motifs in regulatory regions, and we show that this enrichment cannot be simply explained by base composition bias in these regions. We also showed that the distribution of several non-B DNA motifs within intergenic regions separating divergently oriented operons differs from the distribution found between convergent ones. In particular, we found a strong enrichment of cruciforms in the termination region of operons; this enrichment was observed for operons with Rho-dependent, as well as Rho-independent terminators. Finally, a preference for some non-B DNA motifs was observed near transcription factor-binding sites. Overall, the conspicuous enrichment of transition-susceptible sites in these specific regulatory regions suggests that non-B DNA structures may have roles in the transcriptional regulation of specific operons within the E. coli genome.
INTRODUCTION
Inactive, non-transcribed DNA exists predominantly in the stable right-handed B-DNA form (1,2). However, DNA can also adopt a number of alternative non-B DNA structures under specific conditions (3,4). These structures include not only the well-described four-stranded G-quadruplex, but also cruciform, triple-stranded H-DNA, left-handed Z-DNA and a variety of looped-out or slipped single-stranded DNA conformations. Such non-B DNA conformations occur within specific sequences, referred to here as non-B DNA motifs, but require energy to form. Thus, they may occur under circumstances where DNA experiences a high degree of torsion or stress (5–8). Some non-B DNA structures are favoured by repeat elements, and they may play significant evolutionary roles via events such as genomic inversion, recombination, mutation, deletion or expansion (9,10). All such processes create genetic instability and as a result may be implicated in many human diseases (11,12). It has also been suggested that transitions from B-form to non-B form DNA structures might be involved in important regulatory processes, such as open-complex formation, transcription factor recruitment, initiation, repression, activation, stalling or termination (13). However, transition-susceptible sequences may occur for reasons other than transitions. Motifs compatible with structures that form in a single strand, such as quadruplex and cruciform, may be present to enable the structure to occur in a transcribed RNA rather than the encoding DNA. A bidirectional process that requires sequence-specific protein binding will have these binding sites arranged in an inverted repeat arrangement suggestive of cruciform susceptibility. Specific sequence biases, such as CpG islands, might lead to a prevalence of Z-susceptible regions upstream of transcription that may or may not be involved in transitions (14).
In eukaryotes, a variety of studies have corroborated roles of non-B DNA in transcriptional regulation. An important example is the mammalian oncogene c-MYC, which has several non-B DNA motifs in its promoter (7,15,16). Transcription-driven superhelicity melts the far upstream element (FUSE), which is an SIDD (superhelically induced duplex destabilization) sites (7,17). The SIDD formation (melting) enables binding of the FUSE-binding protein, which acts bimodally to either activate or inhibit the next initiation event (7,18). Studies have also shown that a G-quadruplex structure can form at the CT element (direct repeats of sequence CCCTCCCCA) in the promoter region, and it may function as a transcriptional repressor (19,20). The same CT element can also form an H-DNA motif, which has been proposed to act as a positive transcriptional regulator through interactions with ribonucleoproteins and other factors (21,22). Further, three discrete Z-susceptible elements in the c-MYC promoter region have been associated with its transcription (23,24).
A substantial amount of computational work has been deployed to complement the biochemical characterization of non-B DNA structures in eukaryotes. For example, genome-wide bioinformatics surveys have shown that G-quadruplex motifs are conserved across different eukaryotic organisms and enriched in gene promoter regions (25–33). In certain eukaryotes, cruciform sequences are enriched in intergenic regions and are closely clustered at the 3′-ends of genes (34), whereas Z-DNA motifs have been found near their 5′-end (35,36). Importantly, the occurrence of a motif does not imply that the structure has to form in vivo, and in addition, some motifs (cruciform, G-quadruplex) may correspond to formation of structures in mRNA rather than in DNA.
Although a primary strategy in such genome-wide analyses is to extrapolate from experimentally determined motif sequences (e.g. four proximal guanine runs used to predict a G-quadruplex). A deeper analysis examines the statistical mechanics of these transitions, including the competitions among them that arise. In this approach, the free energy of each alternate conformation available to the DNA sequence is determined, and then the equilibrium distribution among available states is calculated at a specified level of imposed superhelicity. This approach explicitly treats the competitive nature of these transitions (37). Comparisons of its results with experiments have shown that this approach provides quantitatively accurate results (38). However, such calculations can only be applied to treat transitions whose energetics have been well characterized. At present this limits its use to strand separation (superhelically induced duplex destabilization or SIDD) and B–Z transitions (superhelically induced B–Z transition or SIBZ). This approach has uncovered the presence of SIDD motifs in gene regulatory regions of several eukaryotes (37,39,40).
Because prokaryotes lack a well-defined chromatin-like nucleosome structure, the occurrence and importance of non-B DNA structures may be rather different in these organisms than in eukaryotes. In this context, the biophysics of the polynucleotide chain itself have been shown to play important roles in genetic regulation. However, bacterial genomes are gene dense, and short intragenic regions might induce specific restrictions on the composition of these regions. In addition, sequence repeats that often underline non-B DNA structures are far less frequent than in eukaryotic genomes. Still, recent studies showed that simple sequence repeats, even if less abundant, are present in Escherichia coli genome (41). In addition, the well-documented operon architecture of E. coli, in which multiple genes are segmentally co-transcribed, might be susceptible to these secondary structure transitions. For example, AT-rich regions in the promoter regions (42) are compatible with formation of SIDDs. Early studies of E. coli showed that lower expression level of a gene can be achieved through cruciform extrusion (43). Specific roles for non-B DNA have been illustrated in a few cases, such as the ilvGMEDA, leuV and ilvYC operons (44,45). More recently, in vitro studies have provided evidence for G-quadruplex formation in a transcription-dependent manner (46). A genome-wide view would be helpful in expanding on these anecdotal instances of non-B DNA in the bacterial genome.
To date, bioinformatics analysis of non-B DNA motifs in the E. coli genome have been limited. Pioneering work has examined SIDD sites and G-quadruplex motifs globally and, in both cases, shown them to be associated with regions upstream of start codons (47,48). Z-DNA formation in this organism has been proposed to be strongly suppressed at both ends of genes (37,49), in contrast to the case in the human genome (49). However, to date only some alternative structures and motifs have been analysed in E. coli, and none of the analyses has accounted for the operon organization of this prokaryotic genome. The operon-based transcriptional organization of the E. coli genome provides a unique opportunity to investigate whether distributions of non-B DNA motifs are consistent with possible regulatory/functional roles of their alternate structures. Specifically, if non-B DNA structures play broad transcriptional regulatory roles, then susceptible sites should be enriched in the regulatory regions of operons (the promoter region of the first gene or termination region of the last gene of operons) relative to the corresponding regions of internal genes. In addition, previous studies typically did not rigorously compare observed enrichment or depletion relative to the expectation arising from the base composition properties of the corresponding region.
In this study, we have performed a comprehensive genome-wide analysis of the distribution of non-B DNA and susceptible sites in the E. coli genome that focuses explicitly on operon structure. We have documented enrichment of SIDD sites, cruciform and H-DNA motifs in the regulatory regions of operons, and we showed that this is not by chance because of the base composition bias in these regions. In contrast, the previously observed depletion of Z-DNA motifs here is shown to be actually consistent with the expectation implied by the base composition of the genomic regions involved. We also found higher densities of SIDD sites and H-DNA motifs in intergenic regions separating divergent operon pairs, whereas cruciform motifs have higher densities in intergenic regions separating convergent operon pairs. Cruciform motifs also underline formation of hairpins in mRNA structures and are known to play important roles in Rho-independent transcription termination in prokaryotes. This type of termination region consists of a G + C-rich hairpin structure followed by a sequence enriched in thymine residues (50) and occurs at approximately half of the E. coli genes (51). We also found an enrichment of cruciform motifs at Rho-dependent termination stop sites. Finally, we observed a preference of cruciform, SIDD and H-DNA to occur near transcription factor-binding sites (TFBS). Taken together, our analysis provides novel insights into possible regulatory/functional roles of non-B DNA structures in E. coli.
MATERIALS AND METHODS
Genome data and regulatory elements
Genome sequence for E. coli (strain K12, substrain MG1655) was downloaded from the National Center for Biotechnology Information (NCBI) RefSeq database (accession number NC_000913.2) (52,53). Gene product, operon structures, transcriptional start sites (TSS) and TFBS for E. coli were obtained from the RegulonDB database (54) (in case of multiple TSS, the closest TSS to the start codon was taken). In the RegulonDB database, an operon is defined as a sequence of contiguous co-transcribed genes, whereas a transcription unit (TU) is defined as a sequence of one or more genes transcribed from a single promoter. Thus, a complex operon with several promoters contains several TUs, but at least one TU must include all the genes of the operon, and a gene can belong to more than one TU. According to RegulonDB 8.1 (released 17 December 2012), there are 2650 operons and 3202 TUs in the E. coli genome. There are 3118 non-redundant genes that can be considered as first genes of TUs, and 2759 non-redundant genes that can be considered as last genes of TUs. In consequence, 1403 non-redundant genes are not first genes of any TUs (non-first gene of TU), and 1762 non-redundant genes are not last genes of any TUs (non-last gene of TU). A pair of operons is divergent if they have overlapping promoter region; operon pair with overlapping termination region is considered as convergent operon pair. Adjacent operon pair in the same direction is described as tandem operon pair. The information in the Regulon DB annotates 671 convergent operon pairs, in 563 of which the operon pairs do not overlap. Also, there are 671 divergent operon pairs, 655 without overlap and 1307 tandem operon pairs, 1269 of which do not overlap. TFBS are partitioned into activator sites and repressor sites based on the effect of transcription factor binding at that site.
Identification of non-B DNA motifs
Susceptibilities to non-B DNA structures, such as G-quadruplex, Z-DNA, SIDD, cruciform, H-DNA and slipped DNA structure, are sequence dependent. A DNA region must have a specific sequence pattern to allow formation of a non-B DNA structure. We call these patterns non-B DNA motifs. We stress that, unlike typical sequence searches for motifs, the SIDD and SIBZ calculations take into account both the larger sequence context and the competitive nature of superhelical transitions, which cannot be summarized by a short sequence pattern.
The G-quadruplex motif can fold into a four-stranded DNA structure that comprises a square co-planar array of four guanine bases stabilized by hydrogen-bonding between them (55). Potential G-quadruplex motifs were predicted using QuadParser (56). In general, G-quadruplex motif was defined as Gn-NL1-Gn-NL2-Gn-NL3-Gn, where G is guanine and N is any nucleotide, including G, L stands for the loop length and the number of guanines constituting the stem is given by n. In this study, two definitions of a G-quadruplex motif were used. The standard definition assumes n = 3 and L varying between 1 and 7 (stringent G-quadruplex motif) (57). A relaxed definition of G-quadruplex motif where n is between 2 and 5 and L is between 1 and 5 (relaxed G-quadruplex motif) has also been used and shown to be biologically relevant (47). Here, we used both definitions, as the number of the stringent G-quadruplex motifs was often too small for statistical analysis. B–Z transition sites were identified using SIBZ algorithm as maximal sequences of consecutive base pairs that have transition probability >0.5 at temperature 310 K and superhelical density σ = −0.06. Cruciforms are formed by perfect or imperfect inverted repeats adopting symmetric hairpin loops in the DNA molecule (58). Cruciform motifs were predicted using Inverted Repeat Finder (59) with threshold scores exceeding 16 and loop length between 1 and 10 bp. SIDD motifs are sites where strand separation in a DNA sequence is favoured at equilibrium under negative superhelical stress (60). They often correspond to AT-rich regions but are generally context dependent. SIDD profiles were calculated using SIDD algorithm at temperature 310 K and superhelical density σ = −0.055 (48). SIDD sites (48) with minimum destabilization energy <4.0 kcal/mol were used in this study. H-DNA is a triple helical DNA where a third oligonucleotide strand binds to the already existing double helix through non-canonical hydrogen bonds (61). They require long homopurine (or homopyrimidine) runs with mirror symmetry. H-DNA motifs were predicted using Triplex program with P-value <0.01 (62). Slipped DNA structures are formed by direct repeats where the strands pair in a misaligned slipped fashion (63). Slipped DNA motif was predicted using Tandem Repeat Finder program (64) with threshold scores >40 and repeat length between 8 and 50 bp. According to these criteria we found the E. coli genome to contain 52 stringent G-quadruplex motifs, 6673 relaxed G-quadruplex motifs, 1091 Z-DNA sites, 2139 cruciform-susceptible inverted repeats, 3311 SIDD sites, 2265 H-DNA motifs and 2181 slipped DNA motifs. All overlapping motifs were counted as one.
Rho-dependent terminator and Rho-independent terminator
E. coli genes with a Rho-dependent terminator were obtained from the work of Peters et al. (65). Here, we only used genes that are the last genes of the TUs and have Rho-dependent terminator in their 3′-end (intergenic) region. In total, 68 genes were assigned to be Rho-dependent. Rho-independent terminators were predicted using the TransTermHP program (66).
Statistical analysis
The genomic location of each non-B DNA motif or transition site was defined as the position of its central base. The probability density distribution function for each non-B DNA motif or site was determined for 1-kb regions centred at either the start codon or the stop codon and oriented so transcription occurs to the right. This was done for each group of genes using the Gaussian-kernel smoothing method (67). The significance of the difference between pairs of distribution functions found in this way was evaluated using Kolmogorov–Smirnov test.
To assess the contribution of the local base composition profile to the distribution of non-B DNA motifs, we generated randomized sequences for each group of genes preserving position dependent composition bias in the E. coli genome. To do so, we first aligned DNA sequences from each group of genes at either start or stop codons. Then, for each position of the alignment, we randomly shuffled nucleotides at that position among the aligned genes. This randomization procedure was carried out 100 times. Distribution functions were determined as above for the randomized sequences. As the randomized sequences preserve the average base composition, this allowed us to assess the statistical significances of any deviations observed in the real data from those arising from the nucleotide-bias preserving random sequences. This procedure was done for each group of genes.
Although there are 1762 genes that are non-last genes, there are only 68 genes with Rho-dependent terminator that are also last genes of TUs. To increase the statistical power, we calculated the distribution based on an artificial group of 1762 genes generated from the 68 genes using bootstrapping by re-sampling these genes 1762 times with repetition. The densities of non-B DNA motifs were defined as number of bases involved in the motif normalized by the length of the respective region. This was done for all intergenic regions separating divergent, convergent or tandem operon pairs. To test any preference, we might observe for non-B DNA motifs to occur near regulatory elements, their densities near TSS or TFBS (also separated into activator sites and repressor sites) were calculated within a 50-bp window centred on the site involved. As a reference, densities in promoter regions were calculated using 50-bp runs that were randomly selected from intergenic regions separating divergent operon pairs without overlap (the whole region was used if the intergenic region was <50-bp long). The Wilcoxon signed-rank test was used to test whether there are significant differences between two groups of density values.
RESULTS
Distribution of non-B DNA motifs in the regulatory region of TUs
We first investigated the distributions of non-B DNA motifs within TUs. Because all genes within a TU are transcribed together, we hypothesized that there should be a difference between the distribution of non-B DNA motifs in regulatory regions and their distribution in internal gene regions. We first compared the distribution of motifs in the promoter region of first genes of TUs with the regions near the start codons of non-first genes (non-first gene control region). We also compared the distribution of non-B DNA motifs in the termination region of last genes of TUs with the regions near stop codons of non-last genes (non-last gene control region). To avoid false enrichment because of overlapping promoters and terminators, we first focused on promoter regions of divergent operon pairs and termination regions of convergent operon pairs.
In the promoter regions of TUs, we found a significant enrichment of SIDD sites, and of cruciform and H-DNA motifs, and a significant depletion of G-quadruplexes (relaxed definition) (Figure 1A and Supplementary Figure S1). We also observed a depletion of Z-DNA motifs, although the depletion is not statistically significant (Figure 1A). Enrichment was not significant for slipped DNA structure or for stringently defined G-quadruplex motifs (Figure 1A and Supplementary Figure S1). However, the latter might be due to poor statistical power arising from the small number of such sites, only 52 of which were found. The cruciform motif was the only one that was significantly enriched in termination regions, whereas SIDDs were strongly depleted in these regions (Figure 1B).
Figure 1.
Distribution of non-B DNA motifs in the regulatory region of divergent or convergent operon pairs. Probability densities (A) in the promoter region of first genes of operons from divergent operon pairs and (B) in the termination region of last genes of operons from convergent operon pairs. Probability densities (distribution functions) were generated through Gaussian-kernel smoothing method based on positions of central base of stringent G-quadruplex, relaxed G-quadruplex, Z-DNA, cruciform, SIDD, H-DNA and slipped DNA structure motifs within 1-kb region centred at either start codon (A) or stop codon (B) (dashed lines). Significance level of difference between distributions was given based on the Kolmogorov–Smirnov test, as follows: *P < 0.05; **P < 0.01; ***P < 0.001.
We next broadened our analysis to include all operons, whether divergently, convergently or tandemly oriented relative to their neighbours. To address any predisposition towards non-B DNA motifs that might arise from local base composition biases in the E. coli genomic DNA, real sequences were compared with base randomized sets. To obtain randomized sequences, the corresponding real sequences were aligned according to the start codon (main text), transcription start site (Supplementary Materials) or stop codon (main text). Then we permuted the nucleotides in each column of the alignment (see ‘Materials and Methods’ section for specifics on protocol for base randomization). The results obtained using start codon and transcription start site were consistent (Figure 2 and Supplementary Figure S2).
Figure 2.
Distribution of non-B DNA motifs in the regulatory region of TUs. Probability densities for (A) stringent G-quadruplex and (B) relaxed G-quadruplex motifs were presented in the promoter region of first genes of TUs and compared with non-first gene control region on both strands: template strand or non-template strand. (C) Probability densities in the promoter region of first genes of TUs compared with non-first gene control regions for Z-DNA, cruciform, SIDD and H-DNA motifs. (D) Probability densities of cruciform in the termination region of TUs compared with non-last gene control regions. Solid and dashed curves represent real and randomized data, respectively. See Figure 1 for detailed legend description.
G-quadruplexes were examined rigorously under these revised conditions, as our initial results seemingly contradicted previous evidence for enrichment of G-quadruplexes in regulatory regions (47). Given the small number of stringent G-quadruplex motifs, their enrichment near promoter regions remained slight in this all-operon analysis. However, their enrichment was statistically significant when compared with occurrences in the randomized sequences, where stringent G-quadruplex motifs were in fact depleted (Figure 2A and Supplementary Figure S2A). Moreover, the abundance of stringent G-quadruplexes in non-first gene regulatory regions was consistent with expectations based only on local base composition (solid versus dotted green curves in Figure 2A and Supplementary Figure S2A). We also compared their occurrence on template and non-template strands, as G-quadruplex motifs in non-template strands (coding strands) can be transcribed in mRNA. Here, a similar pattern was observed, although the enrichment of stringent G-quadruplexes in non-template strands was not statistically significant when compared with the randomized sequences (Figure 2A and Supplementary Figure S2A). Relaxed G-quadruplex motifs were significantly depleted in promoter regions when compared with non-first gene regions, but enriched compared with randomized sequences in which the base composition was preserved at each position (Figure 2B and Supplementary Figure S2B). In contrast, the distribution of relaxed G-quadruplex motifs in the non-first gene control regions is consistent with random when one accounts for base composition effects (Figure 2B and Supplementary Figure S2B). Here also a similar pattern of depletion was observed when comparing relaxed G-quadruplexes motifs on template strands with those on non-template strands (Figure 2B and Supplementary Figure S2B).
We observed a significant depletion of Z-DNA motifs in the promoter region compared with non-first gene control regions (Figure 2C and Supplementary Figure S2C). However, this depletion can be entirely accounted for by the base composition effects (Figure 2C and Supplementary Figure S2C). We also observed significant enrichment of SIDD sites, and cruciform and H-DNA motifs in the promoter region of first genes, as compared with non-first gene control regions (Figure 2C and Supplementary Figure S2C). These enrichments exceeded expectations based on the base composition of the E. coli genome (Figure 2C and Supplementary Figure S2C). With the exception of cruciform, the distribution of motifs and sites in non-first gene control regions all correspond to expectations from the base composition of the E. coli genome (Figure 2C and Supplementary Figure S2C). There was also enrichment of cruciform motifs in the termination regions of last genes relative to non-last gene control regions (Figure 2D). This termination region enrichment also exceeds that expected from the base composition profile of the E. coli genome (Figure 2D).
As most previous Z-DNA analyses were performed using Z-hunt program rather than the newer SIBZ approach, we also preformed analysis with Z-hunt (49,68). Consistently with the SIBZ analysis, we observed significant depletion of Z-hunt detected Z-DNA motifs in promoter region, and, as in the case of regions identified the SIBZ program, the permutation test showed that this depletion pattern could be explained by the base composition of the E. coli genome (Supplementary Figure S3).
Preference for non-B DNA motifs in intergenic regions
The intergenic regions of divergent and convergent operon pairs are regions with overlapping regulatory elements, promoter regions in the case of divergent operon pairs and termination regions for convergent operon pairs. This suggests that the distribution of non-B DNA in these regions may shed additional light on their possible regulatory roles.
We observed a higher density of cruciform motifs in intergenic regions of convergent operon pairs than in intergenic regions separating tandem operon pairs, which in turn was higher than in intergenic regions separating divergent operon pairs (Figure 3A and Supplementary Figure S4A). Consistent with previous observations (48,69), the density of SIDD motifs was higher in intergenic regions separating divergent operon pairs than in intergenic regions separating tandem operon pairs, which again was higher than in intergenic regions separating convergent operon pairs (Figure 3B and Supplementary Figure S4B). There was a similar pattern for H-DNA motifs, except that the density difference was not significant for H-DNA motifs in intergenic regions separating divergent operon pairs when compared with the intergenic region separating tandem operon pairs (Figure 3C and Supplementary Figure S4C). We observed also enrichment of relaxed G-quadruplex in convergent operon pairs relative to other arrangements of operons (Supplementary Figure S5); however, as there is no difference between the density of the G-quadruplex motif at the stop codons of internal genes comparing with the stop codons of last genes of operons, the observed enrichment is likely simply a reflection of the depletion of such structures in promoter regions (Figure 1A). In summary, cruciform motifs were enriched in intergenic regions separating convergent operon pairs, whereas SIDD and H-DNA motifs were enriched in intergenic regions separating divergent operon pairs.
Figure 3.

Preference of non-B DNA motifs in the intergenic region separating convergent, divergent or tandem operon pairs. Densities for (A) cruciform, (B) SIDD and (C) H-DNA motifs were calculated as proportion of base pairs involved in the non-B DNA motifs to the whole base pairs in intergenic regions of convergent operon pairs (operon pairs with overlapping termination region), divergent operon pairs (operon pairs with overlapping promoter region) or tandem operon pairs (operon pairs in the same direction). Mean and standard deviation of the density are given. Statistical significance levels were calculated based on the Wilcoxon signed-rank test between pair of different intergenic regions: *P < 0.05; **P < 0.01; ***P < 0.001.
Cruciform motifs in transcription termination regions
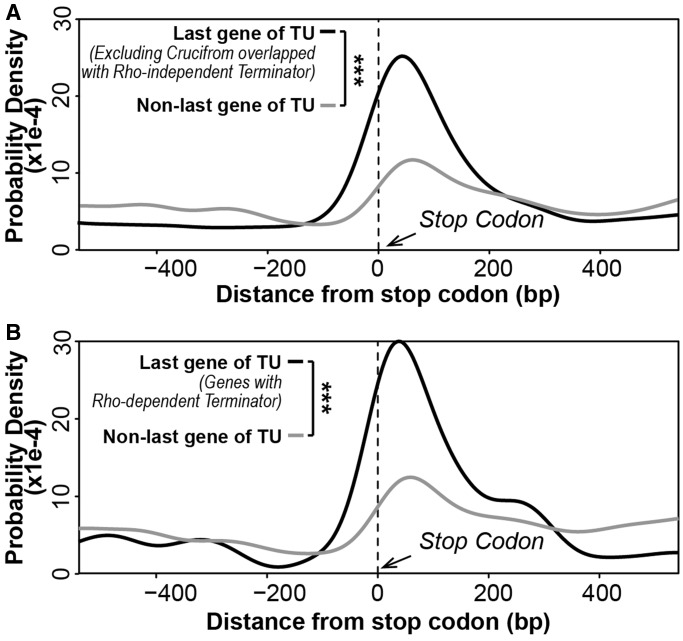
From the aforementioned analyses, cruciform motifs showed significant enrichment in the termination regions of last genes and also had higher density in intergenic regions separating convergent operon pairs. It is known that Rho-independent terminators contain hairpins, and thus should display patterns consistent with cruciform motifs (50). Given lack of an obvious source of negative supercoiling at the termination region of convergent operons, a functional role of a cruciform motif in this region is more likely associated with formation of such hairpin structures than with formation of non-B DNA cruciform structures. Hairpin structure plays important role in Rho-independent termination. Interestingly, after excluding predicted cruciform motifs that overlapped with Rho-independent terminators, there was still an observed enrichment of cruciform motifs in the termination region of last genes (Figure 4A). This enrichment might, at least in part, reflect false negatives of the algorithm used to predict Rho-independent termination sites.
Figure 4.
Distribution of cruciform motifs in the termination region of TUs. (A) Distribution of cruciform motifs in the termination region (1-kb region centred at the stop codon) of last genes of TUs after excluding cruciform motifs overlapped with predicted Rho-independent terminators (black curve) was compared with distribution of cruciform motifs in non-last gene control regions (grey curve). Distributions were based on positions of the central base of cruciform motifs. (B) Distribution of cruciform motifs in the termination region (1-kb region centred at the stop codon) of last genes of TUs with Rho-dependent terminator (black curve) was compared with distribution of cruciform motifs in non-last gene control regions (grey curve). Only the last genes of TUs with intergenic terminator were used. Distribution difference was calculated based on the Kolmogorov–Smirnov test: *P < 0.05; **P < 0.01; ***P < 0.001.
We also examined whether cruciform/hairpin motifs were enriched in the termination region of genes that have been experimentally determined to undergo Rho-dependent termination. To test this, the distribution of cruciform motifs in termination region of last genes undergoing Rho-dependent termination was compared with the distribution of cruciform motifs in non-last gene control regions. Results of this comparison showed that there was indeed enrichment of cruciform motifs in termination region of last genes even after correction for Rho-independent terminators, as well as in the termination region of last genes that undergo Rho-dependent termination (Figure 4B).
Preference for non-B DNA motifs near transcription factor-binding sites
Finally, we examined whether the non-B DNA motifs that were enriched near promoter regions were specifically concentrated near predicted TFBSs. Such an analysis could suggest possible roles for non-B DNA structures in recruiting or blocking transcription factor binding in E. coli. To this end, the densities of non-B DNA motifs near TFBSs were compared with their densities near either TSSs or promoter regions (see ‘Materials and Methods’ section).
We found a preference for cruciform motifs near TFBS when compared with the promoter region reference (Figure 5A and Supplementary Figure S6A). This preference was evident regardless of the activating or repressive role of the predicted TFBS. When comparing TFBSs with TSSs, however, the preference for cruciform motifs was only observed in repressor sites. More cruciform motifs were predicted near TSSs than in the promoter reference. For SIDD and H-DNA, there was a significant preference for TFBSs (regardless of activator or repressor sites) when compared with either promoter regions or TSSs (Figure 5B and C and Supplementary Figure S6B and C). For H-DNA, in addition to the aforementioned pattern, promoter regions exhibited more structures than TSS (Figure 5C and Supplementary Figure S6C).
Figure 5.

Preference of non-B DNA motifs near TFBS. Densities for (A) cruciform, (B) SIDD and (C) H-DNA were calculated as proportion of base pairs involved in non-B DNA motifs within 50-bp window near TFBS (further divided into activator sites and repressor sites) and compared with densities near TSS and promoter region. For promoter region, 50-bp randomly selected base pairs between divergent operon pairs were used to calculate the density (all the base pairs were used if the intergenic region separating divergent operon pair is <50-bp long). Mean and the standard deviation of the density are given. Statistical significance levels were calculated based on the Wilcoxon signed-rank test: *P < 0.05; **P < 0.01; ***P < 0.001.
DISCUSSION
The operon structure of the E. coli genome was used as a framework to analyse the potential impact of non-B DNA motifs on regulation of transcription. Specifically, we reasoned that if non-B DNA motifs indeed play regulatory roles, we should observe differences in the distribution of sequence motifs required for non-B DNA formation in the regulatory regions of TUs and/or operons compared with control regions. Upstream regions of genes that are not first genes of TUs were used as control regions in the analysis of promoter regions (non-first gene control regions), and the downstream regions of genes that are not last genes of transcriptional units were used as control regions for transcription termination (non-last gene control region).
Using this analysis in E. coli, we observed significant depletion of relaxed G-quadruplexes, and significant enrichment of cruciform, SIDD and H-DNA motifs in promoter regions based on divergent operon pairs. We also observed significant enrichment of cruciform motifs near termination regions of convergent operon pairs. Based on all the TUs in E. coli, with the exception of Z-DNA motifs, those patterns cannot be simply explained by the base composition profile of the E. coli genome. The regulatory roles of non-B DNA motifs were further evinced by the fact that the overlapped regulatory regions of operon pairs indeed had higher densities of non-B DNA motifs: a higher density of cruciform motifs in the overlapped termination region of convergent operon pairs and a higher density of SIDD and H-DNA motifs in the overlapped promoter region of divergent operon pairs. For cruciform, there was still enrichment in the termination region of the last gene of TUs even after excluding for cruciform motifs overlapping with predicted Rho-independent terminators. There was also enrichment in the termination region of last gene of TUs with experimentally determined Rho-dependent terminators. Finally, we observed significant preferences for cruciform, SIDD and H-DNA motifs near TFBS.
This unique analysis framework combined with systematic genome-wide analysis of all well-known non-B DNA motifs provides many new insights. For example, G-quadruplex motifs have been widely studied in both eukaryotes (e.g. Homo sapiens) and prokaryotes (e.g. E. coli) and have been suggested to play important roles in transcriptional regulation (27,33,70–73). Although there were fewer stringent G-quadruplex motifs in E. coli than in typical eukaryotic organisms and a depletion of relaxed G-quadruplex motifs in promoter regions, there were in fact more G-quadruplexes in regulatory regions when compared with the randomized sequences used as controls. This may suggest an evolutionary advantage and, as a result, regulatory role for relaxed G-quadruplex motifs in E. coli (27,31,47). Similarly, although Z-DNA motifs are enriched in the promoter region of genes of many eukaryotic organisms (35,36,39,49,74), we observed depletion of Z-DNA motifs in regulatory regions in E. coli. This depletion corresponds to the base composition profile of the E. coli genome, reflecting a possible incompatibility of Z-DNA motifs with regulatory regions in E. coli. For cruciform motifs and H-DNA motifs, although previous studies found that they are not abundant in the E. coli genome compared with eukaryotic genomes (75), we indeed observed enrichment of cruciform motifs and H-DNA motifs in the regulatory regions of transcriptional units. Complementing previously observed preference of SIDD in promoter region and their enrichment in intergenic regions separating divergent operon pairs (48,69), we observed significant preference of SIDD sites near TFBS. Although initial analyses based on divergent or convergent operon pairs showed no enrichment of slipped DNA structures near the regulatory region of operons, there is a significant enrichment in the promoter regions of first genes of TUs when all operons are considered and this enrichment cannot be simply explained by the base composition (Supplementary Figure S7A). Of particular interest, with regard to slipped motifs, there seems to be a higher density near TFBS (Supplementary Figure S7B). This suggests that DNA slippage is not just a contributor to genetic instability, but it may also be pre-encoded to play a role in regulation of transcription initiation structurally or sequentially through tandemly repeated TFBSs.
Our analysis also suggests that transcription termination in E. coli may involve secondary structure in previously unanticipated ways. Prokaryotes use two different types of transcription termination, termed Rho-dependent and Rho-independent or intrinsic termination. Intrinsic termination is known to involve the formation of hairpins in the transcribed mRNA, which must be encoded by inverted repeats (i.e. cruciform motifs) in the DNA. But we observed that cruciform motifs were enriched in the termination regions of last genes, even after excluding for predicted Rho-independent terminators, as well as in the termination regions of last genes predicted to undergo Rho-dependent termination. This result may indicate that the current definition of Rho-independent termination used in this analysis may need to be refined. At the same time, the enrichment of cruciform motifs in genes with Rho-dependent terminators points to a more complex termination system in E. coli. Different systems (Rho-dependent terminators or cruciform motifs) may be recruited at different times, in different locations or under different conditions, or there may be a common, as yet unknown, underlying process.
Transcriptional regulation is a complex and dynamic process, possibly involving competitions between different DNA structures, as well as between proteins targeting those structures in DNA (37,76,77). In addition, dynamic changes in gene expression may also be controlled by nutritional and environmental conditions (78). This complex interplay requires a tightly responsive regulatory system that can make use of the intrinsic biophysics of DNA itself (8,15,17,18). Take for example, in eukaryotes, regulation of c-MYC, which contains multiple interlaced secondary structural motifs, subordinate to different regulatory controls (7,15,16). Under sufficient negative supercoiling, FUSE presents sequential SIDDs, which in this form can be bound by the FUSE-binding protein to control expression of c-MYC (7,17). In the case of nuclease hypersensitive element (NHE III1) of c-MYC, competition between protein binding and G-quadruplex formation is capable of controlling the c-MYC expression (15). Moreover, the existence of Z-DNA in portions of the c-MYC promoter can upregulate gene expression during transcription (23). Similarly, in E. coli, which lacks chromatin structure, transcription may be more reliant on the inherent attributes of its genetic material. The existence of alternative non-B DNA motifs within the short regulatory regions in E. coli could contribute in several ways to precise transcriptional regulation. SIDD motifs could facilitate the opening of duplex DNA, resulting in recruitment of necessary factors to initiate transcription (78). Non-B DNA structures that have either enrichment or depletion in promoter regions of transcriptional units can function as either binding sites for transcription factors or other proteins involved in transcription (21,22,27,46,47,58,61,79) or as energy sinks necessary for responding to high levels of transcription-induced supercoiling (7,8,18,80). Additionally, the non-B DNA structures can function as blockers to stall replication forks (33,81,82), which could in turn be counteracted by other proteins to subsequently facilitate transcription. Indeed, the observed preference for cruciform, SIDD and H-DNA motifs near TFBS is indicative of this response. In the case of cruciform, the intricate coordination with Rho-dependent and intrinsic terminators may contribute an additional dimension of control in transcriptional termination.
CONCLUSION
In this study, we have performed a comprehensive analysis of non-B DNA motifs in regulatory regions in E. coli based on its unique operon structure. Our findings suggest that non-B DNA motifs are indeed preferentially located in the regulatory regions of operons. Based on our genome-wide analyses of non-B DNA motifs in E. coli, compared with current knowledge of non-B DNA motifs in other organisms, we observed differences in non-B DNA motifs distributions between prokaryotes and eukaryotes. This may indicate differences in the transcription regulation systems of prokaryotes compared with eukaryotes. In particular, the formation of non-B DNA motifs may be differently constrained in eukaryotes. Even in the smaller genome of E. coli, with fewer influencing partners than the mammalian genome, it is still unclear how exactly non-B DNA motifs work cooperatively and dynamically during transcriptional regulation. Further studies are needed to understand the function of the non-B DNA motifs and structures, and their cooperation with other factors in regulation of DNA transaction. The results of this analysis provide important preliminary information for the systematic elucidation of regulatory roles of non-B DNA motifs in E. coli, and they can serve as a prelude for future experimental work that directly assesses and roles of non-B DNA structures.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–7.
ACKNOWLEDGEMENTS
The authors thank Sally Madden, UC Davis, for running the SIDD and B–Z analysis programs.
FUNDING
Intramural Research Program of the US National Institute of Health; National Library of Medicine; National Cancer Institute; Center for Cancer Research. Funding for access charge: Intramural Research Program of the US National Institute of Health; National Library of Medicine.
Conflict of interest statement. None declared.
REFERENCES
- 1.Watson JD, Crick FH. Genetical implications of the structure of deoxyribonucleic acid. Nature. 1953;171:964–967. doi: 10.1038/171964b0. [DOI] [PubMed] [Google Scholar]
- 2.Watson JD, Crick FH. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature. 1953;171:737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- 3.Mirkin SM. Discovery of alternative DNA structures: a heroic decade (1979-1989) Front. Biosci. 2008;13:1064–1071. doi: 10.2741/2744. [DOI] [PubMed] [Google Scholar]
- 4.Wells RD. Unusual DNA structures. J. Biol. Chem. 1988;263:1095–1098. [PubMed] [Google Scholar]
- 5.Paleček E. Local Supercoil-Stabilized DNA Structure. Crit. Rev. Biochem. Mol. Biol. 1991;26:151–226. doi: 10.3109/10409239109081126. [DOI] [PubMed] [Google Scholar]
- 6.Travers A, Muskhelishvili G. DNA supercoiling - a global transcriptional regulator for enterobacterial growth? Nat. Rev. Microbiol. 2005;3:157–169. doi: 10.1038/nrmicro1088. [DOI] [PubMed] [Google Scholar]
- 7.Kouzine F, Levens D. Supercoil-driven DNA structures regulate genetic transactions. Front. Biosci. 2007;12:4409–4423. doi: 10.2741/2398. [DOI] [PubMed] [Google Scholar]
- 8.Levens D, Benham CJ. DNA stress and strain, in silico, in vitro and in vivo. Phys. Biol. 2011;8:035011. doi: 10.1088/1478-3975/8/3/035011. [DOI] [PubMed] [Google Scholar]
- 9.Zhao J, Bacolla A, Wang G, Vasquez KM. Non-B DNA structure-induced genetic instability and evolution. Cell Mol. Life Sci. 2010;67:43–62. doi: 10.1007/s00018-009-0131-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Damas J, Carneiro J, Gonçalves J, Stewart JB, Samuels DC, Amorim A, Pereira F. Mitochondrial DNA deletions are associated with non-B DNA conformations. Nucleic Acids Res. 2012;40:7606–7621. doi: 10.1093/nar/gks500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wells RD. Non-B DNA conformations, mutagenesis and disease. Trends Biochem. Sci. 2007;32:271–278. doi: 10.1016/j.tibs.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 12.Wells RD. Discovery of the role of Non-B DNA structures in mutagenesis and human genomic disorders. J. Biol. Chem. 2009;284:8997–9009. doi: 10.1074/jbc.X800010200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dai X, Rothman-Denes LB. DNA structure and transcription. Curr. Opin. Microbiol. 1999;2:126–130. doi: 10.1016/S1369-5274(99)80022-8. [DOI] [PubMed] [Google Scholar]
- 14.Rich A, Nordheim A, Wang AHJ. The chemistry and biology of left-handed Z-DNA. Annu. Rev. Biochem. 1984;53:791–846. doi: 10.1146/annurev.bi.53.070184.004043. [DOI] [PubMed] [Google Scholar]
- 15.Brooks TA, Hurley LH. The role of supercoiling in transcriptional control of MYC and its importance in molecular therapeutics. Nat. Rev. Cancer. 2009;9:849–861. doi: 10.1038/nrc2733. [DOI] [PubMed] [Google Scholar]
- 16.Michelotti GA, Michelotti EF, Pullner A, Duncan RC, Eick D, Levens D. Multiple single-stranded cis elements are associated with activated chromatin of the human c-myc gene in vivo. Mol. Cell. Biol. 1996;16:2656–2669. doi: 10.1128/mcb.16.6.2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kouzine F, Liu J, Sanford S, Chung HJ, Levens D. The dynamic response of upstream DNA to transcription-generated torsional stress. Nat. Struct. Mol. Biol. 2004;11:1092–1100. doi: 10.1038/nsmb848. [DOI] [PubMed] [Google Scholar]
- 18.Kouzine F, Sanford S, Elisha-Feil Z, Levens D. The functional response of upstream DNA to dynamic supercoiling in vivo. Nat. Struct. Mol. Biol. 2008;15:146–154. doi: 10.1038/nsmb.1372. [DOI] [PubMed] [Google Scholar]
- 19.Sun D, Hurley LH. The Importance of Negative Superhelicity in Inducing the Formation of G-Quadruplex and i-Motif Structures in the c-Myc Promoter: Implications for Drug Targeting and Control of Gene Expression. J. Med. Chem. 2009;52:2863–2874. doi: 10.1021/jm900055s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qin Y, Hurley LH. Structures, folding patterns, and functions of intramolecular DNA G-quadruplexes found in eukaryotic promoter regions. Biochimie. 2008;90:1149–1171. doi: 10.1016/j.biochi.2008.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davis TL, Firulli AB, Kinniburgh AJ. Ribonucleoprotein and protein factors bind to an H-DNA-forming c-myc DNA element: possible regulators of the c-myc gene. Proc. Natl Acad. Sci. USA. 1989;86:9682–9686. doi: 10.1073/pnas.86.24.9682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kinniburgh AJ. A cis-acting transcription element of the c-myc gene can assume an H-DNA conformation. Nucleic Acids Res. 1989;17:7771–7778. doi: 10.1093/nar/17.19.7771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wittig B, Wolfl S, Dorbic T, Vahrson W, Rich A. Transcription of human c-myc in permeabilized nuclei is associated with formation of Z-DNA in three discrete regions of the gene. EMBO J. 1992;11:4653–4663. doi: 10.1002/j.1460-2075.1992.tb05567.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rich A, Zhang S. Timeline: Z-DNA: the long road to biological function. Nat. Rev. Genet. 2003;4:566–572. doi: 10.1038/nrg1115. [DOI] [PubMed] [Google Scholar]
- 25.Mullen MA, Olson KJ, Dallaire P, Major F, Assmann SM, Bevilacqua PC. RNA G-Quadruplexes in the model plant species Arabidopsis thaliana: prevalence and possible functional roles. Nucleic Acids Res. 2010;38:8149–8163. doi: 10.1093/nar/gkq804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Capra JA, Paeschke K, Singh M, Zakian VA. G-quadruplex DNA sequences are evolutionarily conserved and associated with distinct genomic features in Saccharomyces cerevisiae. PLoS Comput. Biol. 2010;6:e1000861. doi: 10.1371/journal.pcbi.1000861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Du Z, Zhao Y, Li N. Genome-wide colonization of gene regulatory elements by G4 DNA motifs. Nucleic Acids Res. 2009;37:6784–6798. doi: 10.1093/nar/gkp710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Verma A, Halder K, Halder R, Yadav VK, Rawal P, Thakur RK, Mohd F, Sharma A, Chowdhury S. Genome-wide computational and expression analyses reveal G-quadruplex DNA motifs as conserved cis-regulatory elements in human and related species. J. Med. Chem. 2008;51:5641–5649. doi: 10.1021/jm800448a. [DOI] [PubMed] [Google Scholar]
- 29.Hershman SG, Chen Q, Lee JY, Kozak ML, Yue P, Wang LS, Johnson FB. Genomic distribution and functional analyses of potential G-quadruplex-forming sequences in Saccharomyces cerevisiae. Nucleic Acids Res. 2008;36:144–156. doi: 10.1093/nar/gkm986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eddy J, Maizels N. Conserved elements with potential to form polymorphic G-quadruplex structures in the first intron of human genes. Nucleic Acids Res. 2008;36:1321–1333. doi: 10.1093/nar/gkm1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huppert JL, Balasubramanian S. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 2007;35:406–413. doi: 10.1093/nar/gkl1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Du Z, Kong P, Gao Y, Li N. Enrichment of G4 DNA motif in transcriptional regulatory region of chicken genome. Biochem. Biophys. Res. Commun. 2007;354:1067–1070. doi: 10.1016/j.bbrc.2007.01.093. [DOI] [PubMed] [Google Scholar]
- 33.Eddy J, Vallur AC, Varma S, Liu H, Reinhold WC, Pommier Y, Maizels N. G4 motifs correlate with promoter-proximal transcriptional pausing in human genes. Nucleic Acids Res. 2011;39:4975–4983. doi: 10.1093/nar/gkr079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Strawbridge EM, Benson G, Gelfand Y, Benham CJ. The distribution of inverted repeat sequences in the Saccharomyces cerevisiae genome. Curr. Genet. 2010;56:321–340. doi: 10.1007/s00294-010-0302-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schroth GP, Chou PJ, Ho PS. Mapping Z-DNA in the human genome. Computer-aided mapping reveals a nonrandom distribution of potential Z-DNA-forming sequences in human genes. J. Biol. Chem. 1992;267:11846–11855. [PubMed] [Google Scholar]
- 36.Hamada H, Petrino MG, Kakunaga T. A novel repeated element with Z-DNA-forming potential is widely found in evolutionarily diverse eukaryotic genomes. Proc. Natl Acad. Sci. USA. 1982;79:6465–6469. doi: 10.1073/pnas.79.21.6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhabinskaya D, Benham CJ. Theoretical Analysis of Competing Conformational Transitions in Superhelical DNA. PLoS Comput. Biol. 2012;8:e1002484. doi: 10.1371/journal.pcbi.1002484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.He L, Liu J, Collins I, Sanford S, O'Connell B, Benham CJ, Levens D. Loss of FBP function arrests cellular proliferation and extinguishes c-myc expression. EMBO J. 2000;19:1034–1044. doi: 10.1093/emboj/19.5.1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhabinskaya D, Benham CJ. Theoretical analysis of the stress induced B-Z transition in superhelical DNA. PLoS Comput. Biol. 2011;7:e1001051. doi: 10.1371/journal.pcbi.1001051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang H, Benham CJ. Superhelical destabilization in regulatory regions of stress response genes. PLoS Comput. Biol. 2008;4:e17. doi: 10.1371/journal.pcbi.0040017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gur-Arie R, Cohen CJ, Eitan Y, Shelef L, Hallerman EM, Kashi Y. Simple Sequence Repeats in Escherichia coli: Abundance, Distribution, Composition, and Polymorphism. Genome Res. 2000;10:62–71. [PMC free article] [PubMed] [Google Scholar]
- 42.Hawley DK, McClure WR. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983;11:2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Horwitz M, Loeb L. An E. coli promoter that regulates transcription by DNA superhelix-induced cruciform extrusion. Science. 1988;241:703–705. doi: 10.1126/science.2456617. [DOI] [PubMed] [Google Scholar]
- 44.Opel ML, Hatfield GW. DNA supercoiling-dependent transcriptional coupling between the divergently transcribed promoters of the ilvYC operon of Escherichia coli is proportional to promoter strengths and transcript lengths. Mol. Microbiol. 2001;39:191–198. doi: 10.1046/j.1365-2958.2001.02249.x. [DOI] [PubMed] [Google Scholar]
- 45.Sheridan SD, Benham CJ, Hatfield GW. Inhibition of DNA supercoiling-dependent transcriptional activation by a distant B-DNA to Z-DNA transition. J. Biol. Chem. 1999;274:8169–8174. doi: 10.1074/jbc.274.12.8169. [DOI] [PubMed] [Google Scholar]
- 46.Mela I, Kranaster R, Henderson RM, Balasubramanian S, Edwardson JM. Demonstration of ligand decoration, and ligand-induced perturbation, of G-quadruplexes in a plasmid using atomic force microscopy. Biochemistry. 2012;51:578–585. doi: 10.1021/bi201600g. [DOI] [PubMed] [Google Scholar]
- 47.Rawal P, Kummarasetti VBR, Ravindran J, Kumar N, Halder K, Sharma R, Mukerji M, Das SK, Chowdhury S. Genome-wide prediction of G4 DNA as regulatory motifs: role in Escherichia coli global regulation. Genome Res. 2006;16:644–655. doi: 10.1101/gr.4508806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang H, Noordewier M, Benham CJ. Stress-induced DNA duplex destabilization (SIDD) in the E. coli genome: SIDD sites are closely associated with promoters. Genome Res. 2004;14:1575–1584. doi: 10.1101/gr.2080004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Champ PC, Maurice S, Vargason JM, Camp T, Ho PS. Distributions of Z-DNA and nuclear factor I in human chromosome 22: a model for coupled transcriptional regulation. Nucleic Acids Res. 2004;32:6501–6510. doi: 10.1093/nar/gkh988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wilson KS, von Hippel PH. Transcription termination at intrinsic terminators: the role of the RNA hairpin. Proc. Natl Acad. Sci. USA. 1995;92:8793–8797. doi: 10.1073/pnas.92.19.8793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cardinale CJ, Washburn RS, Tadigotla VR, Brown LM, Gottesman ME, Nudler E. Termination factor Rho and its cofactors NusA and NusG silence foreign DNA in E. coli. Science. 2008;320:935–938. doi: 10.1126/science.1152763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sayers EW, Barrett T, Benson DA, Bolton E, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2011;39:D38–D51. doi: 10.1093/nar/gkq1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gama-Castro S, Salgado H, Peralta-Gil M, Santos-Zavaleta A, Muñiz-Rascado L, Solano-Lira H, Jimenez-Jacinto V, Weiss V, García-Sotelo JS, López-Fuentes A, et al. RegulonDB version 7.0: transcriptional regulation of Escherichia coli K-12 integrated within genetic sensory response units (Gensor Units) Nucleic Acids Res. 2011;39:D98–D105. doi: 10.1093/nar/gkq1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lipps HJ, Rhodes D. G-quadruplex structures: in vivo evidence and function. Trends Cell Biol. 2009;19:414–422. doi: 10.1016/j.tcb.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 56.Huppert JL, Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005;33:2908–2916. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Todd AK, Johnston M, Neidle S. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res. 2005;33:2901–2907. doi: 10.1093/nar/gki553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brazda V, Laister RC, Jagelska EB, Arrowsmith C. Cruciform structures are a common DNA feature important for regulating biological processes. BMC Mol. Biol. 2011;12:33. doi: 10.1186/1471-2199-12-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Warburton PE, Giordano J, Cheung F, Gelfand Y, Benson G. Inverted repeat structure of the human genome: the X-chromosome contains a preponderance of large, highly homologous inverted repeats that contain testes genes. Genome Res. 2004;14:1861–1869. doi: 10.1101/gr.2542904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Benham CJ. Sites of predicted stress-induced DNA duplex destabilization occur preferentially at regulatory loci. Proc. Natl Acad. Sci. USA. 1993;90:2999–3003. doi: 10.1073/pnas.90.7.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jain A, Wang G, Vasquez KM. DNA triple helices: biological consequences and therapeutic potential. Biochimie. 2008;90:1117–1130. doi: 10.1016/j.biochi.2008.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lexa M, Martinek T, Burgetova I, Kopecek D, Brazdova M. A dynamic programming algorithm for identification of triplex-forming sequences. Bioinformatics. 2011;27:2510–2517. doi: 10.1093/bioinformatics/btr439. [DOI] [PubMed] [Google Scholar]
- 63.Sinden RR, Pytlos-Sinden MJ, Potaman VN. Slipped strand DNA structures. Front. Biosci. 2007;12:4788–4799. doi: 10.2741/2427. [DOI] [PubMed] [Google Scholar]
- 64.Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Peters JM, Mooney RA, Kuan PF, Rowland JL, Keles S, Landick R. Rho directs widespread termination of intragenic and stable RNA transcription. Proc. Natl Acad. Sci. USA. 2009;106:15406–15411. doi: 10.1073/pnas.0903846106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kingsford CL, Ayanbule K, Salzberg SL. Rapid, accurate, computational discovery of Rho-independent transcription terminators illuminates their relationship to DNA uptake. Genome Biol. 2007;8:R22. doi: 10.1186/gb-2007-8-2-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Parzen E. On estimation of a probability density function and mode. Ann. Math. Stat. 1962;33:1065–1076. [Google Scholar]
- 68.Ho PS, Ellison MJ, Quigley GJ, Rich A. A computer aided thermodynamic approach for predicting the formation of Z-DNA in naturally occurring sequences. EMBO J. 1986;5:2737–2744. doi: 10.1002/j.1460-2075.1986.tb04558.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang H, Kaloper M, Benham CJ. SIDDBASE: a database containing the stress-induced DNA duplex destabilization (SIDD) profiles of complete microbial genomes. Nucleic Acids Res. 2006;34:D373–D378. doi: 10.1093/nar/gkj007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Johnson JE, Cao K, Ryvkin P, Wang LS, Johnson FB. Altered gene expression in the Werner and Bloom syndromes is associated with sequences having G-quadruplex forming potential. Nucleic Acids Res. 2010;38:1114–1122. doi: 10.1093/nar/gkp1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Verma A, Yadav VK, Basundra R, Kumar A, Chowdhury S. Evidence of genome-wide G4 DNA-mediated gene expression in human cancer cells. Nucleic Acids Res. 2009;37:4194–4204. doi: 10.1093/nar/gkn1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fernando H, Sewitz S, Darot J, Tavaré S, Huppert JL, Balasubramanian S. Genome-wide analysis of a G-quadruplex-specific single-chain antibody that regulates gene expression. Nucleic Acids Res. 2009;37:6716–6722. doi: 10.1093/nar/gkp740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Du Z, Zhao Y, Li N. Genome-wide analysis reveals regulatory role of G4 DNA in gene transcription. Genome Res. 2008;18:233–241. doi: 10.1101/gr.6905408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Khuu P, Sandor M, DeYoung J, Ho PS. Phylogenomic analysis of the emergence of GC-rich transcription elements. Proc. Natl Acad. Sci. USA. 2007;104:16528–16533. doi: 10.1073/pnas.0707203104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schroth GP, Ho PS. Occurrence of potential cruciform and H-DNA forming sequences in genomic DNA. Nucleic Acids Res. 1995;23:1977–1983. doi: 10.1093/nar/23.11.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Edwards SF, Sirito M, Krahe R, Sinden RR. A Z-DNA sequence reduces slipped-strand structure formation in the myotonic dystrophy type 2 (CCTG) x (CAGG) repeat. Proc. Natl Acad. Sci. USA. 2009;106:3270–3275. doi: 10.1073/pnas.0807699106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Leng F, McMacken R. Potent stimulation of transcription-coupled DNA supercoiling by sequence-specific DNA-binding proteins. Proc. Natl Acad. Sci. USA. 2002;99:9139–9144. doi: 10.1073/pnas.142002099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hatfield GW, Benham CJ. DNA topology-mediated control of global gene expression in Escherichia coli. Annu. Rev. Genet. 2002;36:175–203. doi: 10.1146/annurev.genet.36.032902.111815. [DOI] [PubMed] [Google Scholar]
- 79.Oh DB, Kim YG, Rich A. Z-DNA-binding proteins can act as potent effectors of gene expression in vivo. Proc. Natl Acad. Sci. 2002;99:16666–16671. doi: 10.1073/pnas.262672699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schon E, Evans T, Welsh J, Efstratiadis A. Conformation of promoter DNA: Fine mapping of S1-hypersensitive sites. Cell. 1983;35:837–848. doi: 10.1016/0092-8674(83)90116-2. [DOI] [PubMed] [Google Scholar]
- 81.Wang G, Vasquez KM. Z-DNA, an active element in the genome. Front. Biosci. 2007;12:4424–4438. doi: 10.2741/2399. [DOI] [PubMed] [Google Scholar]
- 82.Grabczyk E, Fishman MC. A long purine-pyrimidine homopolymer acts as a transcriptional diode. J. Biol. Chem. 1995;270:1791–1797. doi: 10.1074/jbc.270.4.1791. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.