Abstract
Recombinase-mediated cassette exchange, or RMCE, is a clean approach of gene delivery into a desired chromosomal location, as it is able to insert only the required sequences, leaving behind the unwanted ones. RMCE can be mediated by a single site-specific DNA recombinase or by two recombinases with different target specificities (dual RMCE). Recently, using the Flp–Cre recombinase pair, dual RMCE proved to be efficient, provided the relative ratio of the enzymes during the reaction is optimal. In the present report, we analyzed how the efficiency of dual RMCE mediated by the Flp–Int (HK022) pair depends on the variable input of the recombinases—the amount of the recombinase expression vectors added at transfection—and on the order of the addition of these vectors: sequential or simultaneous. We found that both in the sequential and the simultaneous modes, the efficiency of dual RMCE was critically dependent on the absolute and the relative concentrations of the Flp and Int expression vectors. Under optimal conditions, the efficiency of ‘simultaneous’ dual RMCE reached ∼12% of the transfected cells. Our results underline the importance of fine-tuning the reaction conditions for achieving the highest levels of dual RMCE.
INTRODUCTION
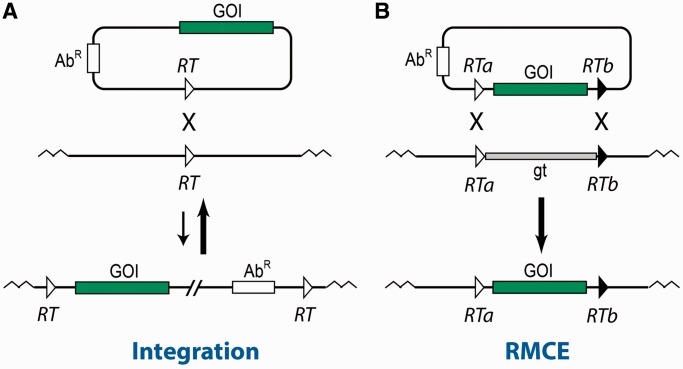
Random DNA insertions in the eukaryotic genome may be undesirable owing to their mutagenic and position effects. Moreover, if the activities of different alleles of a certain gene are to be compared, random integration of each allele may distort the results. Site-specific recombinases that have become efficient tools for site-specific gene manipulations can overcome these problems, as they can promote gene insertions, deletions or inversion at predefined loci of the eukaryotic genome. Among the most popular site-specific recombination systems currently used are the bacteriophage P1 Cre-lox and yeast’s Flp-FRT that belong to the tyrosine family recombinases, as well as the Int-att system of phage ϕC31 that belong to the serine family recombinases. Each family is named after the conserved nucleophylic residue in the active site of the recombinase (1–4). In these systems, the site-specific integrations of desired genes are catalyzed by a recombination reaction between two specific short DNA target sites (∼30–40 bp long), one located in a chromosome and the other one, identical or similar, located on a plasmid that carries the gene(s) to be inserted (Figure 1A). Such integration reaction, however, leads to the insertion of the entire vector plasmid that may include undesired resistance markers and/or replication origins. Moreover, in the case of tyrosine-family recombinases mentioned above, the insertion results in two identical tandem target sites that flank the inserted DNA and are substrates for the more efficient reverse excision reaction (stronger arrow in Figure 1A).
Figure 1.
Comparison between integration and cassette exchange reactions to deliver a gene of interest into a desired genome location. (A) A single site-specific recombination reaction integrates a gene of interest along with entire vector sequences. (B) RMCE integrates just a gene of interest. GOI, gene of interest; gt, genomic target; AbR, antibiotic resistance gene; RT, recombination target site; RTa and RTb, incompatible recombination target sites that can be recognized either by a single recombinase or by two different recombinases.
A more elegant and ‘clean’ approach of gene insertions is the recombinase-mediated cassette exchange (RMCE) reaction (5) that is able to replace a plasmid borne gene of interest with a genomic target leaving out undesired plasmid sequences provided both DNA fragments are flanked by a set of incompatible recombination targets (Figure 1B). Moreover, the reversibility of this cassette exchange reaction is low because the compatible recombination sites are located on different DNA molecules.
The RMCE reaction can be catalyzed by one recombinase. In this, original type of RMCE (6,7), the two incompatible target sites that flank the DNA fragments to be exchanged are usually the respective cognate wild-type recombination target and its incompatible mutated version, both targets being substrates for the same recombinase: homotypic targets. Alternatively, RMCE can be performed by two recombinases. In this type of the replacement reaction, dual RMCE, each target site in the pair of the incompatible recombination targets that flank the replacement cassettes is recognized by the respective recombinase: heterotypic targets (8).
The introduction of new site-specific recombination systems for eukaryotic gene manipulations is important, in particular when these manipulations become more complex, i.e. in cases in which a particular recombinase becomes useless if its target site(s) has remained in the genome owing to a previous manipulation. We have recruited the site-specific recombination system of coliphage HK022 to be active in human cells (9). This system, which is similar to that of the better known coliphage λ, has two different but compatible target sites: the 21 bp attB site that originates from the host (Escherichia coli) chromosome and the longer 230 bp attP site that comes from the phage chromosome. The phage-encoded integrase (Int) catalyzes the integrative attP × attB site-specific recombination reaction that leads to the formation of the recombinant attL and attR sites, which serve as substrates for the Int-catalyzed excisive reaction. In the natural E. coli host, as well as in in vitro reactions, both integration and excision reaction require accessory proteins (10,11), which are dispensable when the system is introduced into human and plant cells (12), though their presence alleviates the reactions (13).
In preliminary dual RMCE reactions that were performed in E. coli cells using HK022’s Int and yeast’s Flp systems, we have shown that the most efficient RMCE reaction occurs when the recombinases are supplied sequentially rather than simultaneously. In this mode, one recombinase was supplied first to catalyze recombination between one pair of target sites that led to the integration of the entire incoming plasmid, followed by the supply of the second recombinase that subsequently completes the RMCE reaction (14).
In this article, we report results of experiments that test the ability of Flp and HK022 Int to mediate a dual RMCE reaction in mammalian cells in the sequential and simultaneous modes. To this end, we created a model Chinese Hamster ovary (CHO) cell line that bears an integrated platform reporter plasmid. The platform plasmid, together with the incoming reporter plasmid, makes up a dual RMCE reporter system designed to detect separate fluorescent signals from the Flp and Int recombination events that allows monitoring of the activity of each recombinase in both the sequential and in the simultaneous dual RMCE modes.
Using the CHO model system, we show that Flp and Int can mediate dual RMCE in both sequential and simultaneous modes. We also show that the replacement reaction performed in the simpler faster simultaneous mode can be more efficient than the one performed in the sequential mode. We found that the sequential ‘Flp-then-Int’ mode, when Flp catalyzed the integration reaction while Int catalyzed the deletion reaction, was less efficient than the ‘Int-then-Flp’ mode, when Int catalyzed the integration reaction while Flp catalyzed the deletion reaction. We also found that the efficiency of the dual RMCE reaction in both sequential and simultaneous modes was critically dependent on the absolute and the relative concentrations of the Int and Flp expression vectors underlining the importance of optimizing the parameters of the reaction for reaching the highest levels of dual RMCE.
MATERIALS AND METHODS
Cell line and transfection
Derivatives of CHO cells, CHO TD-In cells (15), were used as model mammalian cells. The cells were propagated in F12-K medium. The medium for the intact CHO TD-In cells was supplemented with zeocin (100 mg/l). On integration of the platform reporter plasmid pNA1372plat, the resultant CHO TD-1372 cells were grown in the medium supplemented with hygromycin (550 mg/l). Cell transfections were performed using Polyfect (Qiagen).
Vectors
Expression vectors
The expression vector for Flp recombinase, pOG-Flp, is a derivative of pOG44 (Flp-In system, Invitrogen), in which Flp(F70L) is replaced with Flpe. The construction of pOG-Flp is described in Anderson et al. (15).
The expression vectors for the human-optimized HK022 oInt, IHF and Xis were pNA979oInt, pIHF2cP and pNA998, respectively (13).
Reporter vectors
Both the platform and the incoming reporter vectors are based on the vector pcTD (15), which is a derivative of the pcDNA5/FRT vector of the Flp-In system (Invitrogen).
To construct the platform plasmid pNA1372plat, the CMV promoter of pcTD was replaced by an MluI-KpnI fragment carrying the EF-1α promoter, following by cloning of a polymerase chain reaction (PCR)-generated KpnI-EcoRV FRT-neoR fragment and an EcoRV-NotI STOP-attL-DsRed fragment.
To construct the incoming pNA1345inc plasmid, the NheI-MluI CMV fragment was deleted from the pcTD vector, followed by cloning of a PCR-generated FRT-EGFP fragment between the NheI and XhoI sites and a CMV-attR fragment between the RsrII and SacII sites.
Plasmid CMV-LEF (15) that expresses EGFP under the CMV promoter was used to estimate the rate of transfection in the RMCE experiments. CMV-LEF has the same backbone and similar size as the incoming reporter vector pNA1345inc.
Construction of CHO TD-1372 cell line
To construct CHO TD-1372 cell line, CHO TD-In cells were co-transfected with the platform reporter pNA1372plat and pOG-TD1-40 (15), which expresses the TD1-40 variant of the TD recombinase (16). The transfection was done in six-well plates. The amount of the reporter and the expression plasmids added at transfection was 0.2 and 2 µg, respectively. Forty-eight hours after transfection, one-sixth of the cells were transferred into a 100-mm plate into the medium supplemented with hygromycin. After ∼10 days, several hygromycin resistant colonies were transferred into 96-well plate and their sensitivity to zeocin and neomycin was tested. The colonies that were sensitive to zeocin and resistant to neomycin were used in the RMCE experiments.
RMCE experiments
Sequential dual RMCE
Sequential dual RMCE experiments were performed either in the ‘Flp-then-Int’ mode or in the ‘Int-then-Flp’ mode. In both cases, the first integrative step of the sequential RMCE was initiated by co-transfecting cells that carried the integrated platform reporter (CHO TD-1372) with the respective expression vector—either pOG-Flp or pNA979oInt—and with the incoming reporter vector pNA1345inc.
The experiments were performed in six-well plates. The amount of the incoming reporter pNA1345inc at transfection was kept at 0.5 µg. The amount of the expression vectors pOG-Flp and pNA979oInt at transfection is indicated where the respective experiments are described. The number of the transfected cells in the RMCE experiments was estimated by calculating the transfection efficiency in the control experiments, which were always performed in parallel with each experimental transfection. The control transfections were done by transfecting the platform reporter cells with the EGFP-expressing plasmid CMV-LEF. The amount of CMV-LEF added at transfection was 0.5 µg; the ratio of green cells to the total number of the cells was calculated 24 hr after transfection. The range of the efficiency of the control transfections was between 20 and 25%.
Forty-eight hours after transfection, one-sixth of the cells from each experimental well were transferred into 100-mm plates, the cells were allowed to become confluent and the number of the green (in the ‘Flp-then-Int’ mode) or red (in the ‘Int-then-Flp’ mode) colonies was counted. Several green or red colonies from these plates were expanded and analyzed. For the second excisive step of RMCE, the green cells were transfected with various amounts of pNA979oInt, and the red cells were transfected with various amounts of pOG-Flp. The transfections were done in six-well plates. The number of the transfected cells was estimated as above. Forty-eight hours after transfection, one-sixth of the cells were transferred into 100-mm plates, the cells were allowed to become confluent, and the number of the red (in the ‘Flp-then-Int’ mode) or green (in the ‘Int-then-Flp’ mode) colonies was counted. Several green or red colonies from these plates were expanded and analyzed.
Simultaneous dual RMCE
Simultaneous dual RMCE experiments were performed essentially as described above except that the CHO TD-1372 cells that harbor the platform reporter were co-transfected with the incoming reporter pNA1345inc and with both expression vectors: pOG-Flp and pNA979oInt. The replacement reactions were done in six-well plates. The number of the transfected cells was estimated as in the sequential RMCE experiments. The amount of the incoming reporter pNA1345inc at transfection was kept at 0.5 µg. The various concentrations of the expression vectors pOG-Flp and pNAo979Int added at transfection are indicated in the respective parts of the ‘Results’ section. Forty-eight hours after transfection, one-sixth of the cells were transferred into 100-mm plates, the cells were allowed to become confluent and the number of the green, red and green-red colonies was counted. Several colonies that were both green and red were expanded and analyzed.
In all RMCE experiments with Int, 0.6 µg of the IHF expression plasmid and 0.6 µg of the Xis expression plasmid were also included.
Other methods
Plasmid DNA was isolated using GeneJET Plasmid Miniprep Kit (Thermo). Amplification of the DNA fragments used for cloning was performed using Pfu-Ultra polymerase (Agilent Technologies). PCR analysis of the mammalian genomic DNA was performed using Taq polymerase (New England Biolabs). Genomic DNA from cultured mammalian cells was isolated using GeneJET Genomic DNA Purification Kit (Thermo). General genetic engineering experiments were performed as described in Sambrook and Russell (17).
RESULTS
Reporter vectors for Flp-Int dual RMCE
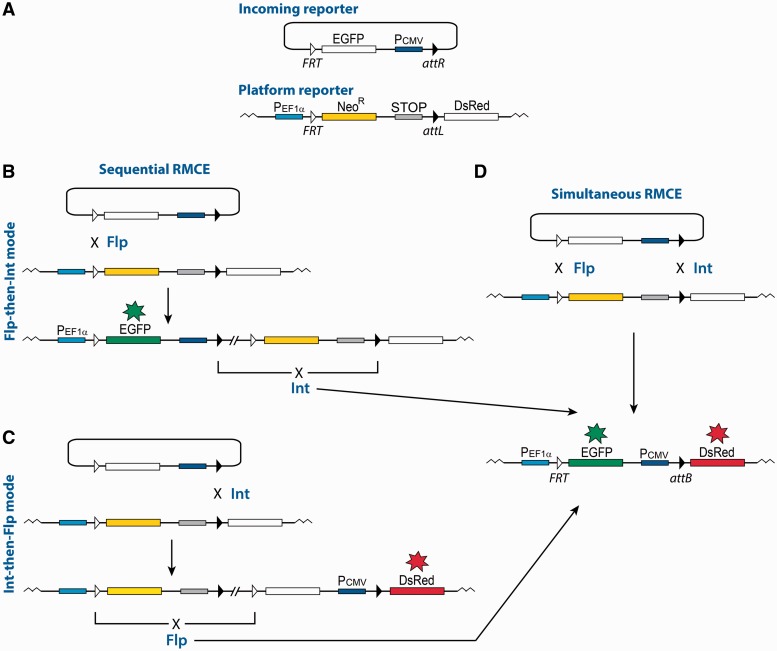
To monitor the activity of each recombinase during dual RMCE, we have constructed a set of two reporter plasmids that, via activating the expression of two different fluorescent markers, can assess the efficiency of a replacement reaction catalyzed by Flp and Int in the absence of a selection force (Figure 2). Moreover, with this set of reporters one can compare the efficiency of the dual RMCE reaction when the recombinases are supplied sequentially or simultaneously. One of the two reporter plasmids serves as a platform that is integrated into an actively transcribed locus of the CHO genome, and the other is an incoming reporter (Figure 2A).
Figure 2.
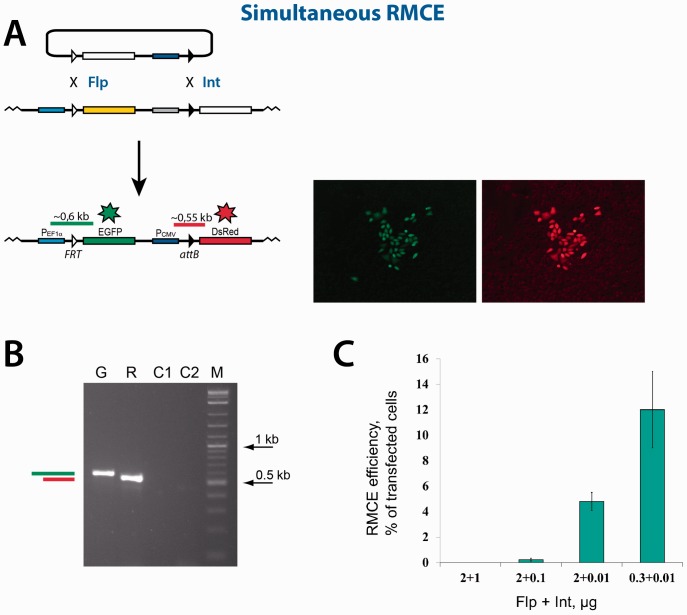
Schematics of the sequential and simultaneous RMCE reactions. (A) The incoming and the chromosomally integrated platform reporters used in this study. Only essential elements of the reporters are shown. The reporters are designed to activate the expression of the EGFP gene from the EF1α promoter on recombination between the FRT sites, and to activate the expression of the DsRed gene from the CMV promoter on recombination between the attL and attR sites. NeoR, neomycin resistance gene; STOP, transcription terminator. (B and C) Sequential RMCE. (B) In the ‘Flp-then-Int’ mode of the sequential RMCE reaction, the first integration reaction is mediated by Flp. The successful integration can be detected by the appearance of the green cells. The second deletion reaction is mediated by Int. The successful deletion can be detected by the appearance of the red cells that also maintain the expression of the EGFP gene. (C) ‘Int-then-Flp’ mode of the sequential RMCE reaction. In this mode of the replacement reaction, the integration reaction is mediated by Int. The successful integration can be detected by the appearance of the red cells. The deletion reaction is mediated by Flp. The successful deletion can be detected by the appearance of the green cells that also maintain the expression of the DsRed gene. (D) Simultaneous RMCE. The replacement of the NeoR-STOP cassette with the EGFP-Pcmv cassette proceeds via a seemingly one-step reaction. The successful replacement can be detected by the appearance of the cells that are both green and red.
The reporter cassette in the platform plasmid pNA1372plat contains the NeoR gene under the control of the EF1α promoter. The NeoR gene is followed by the transcription terminator STOP (18) and the promoterless DsRed gene (Figure 2A). The Flp cognate sequence FRT is located between the EF1α promoter and the NeoR gene; the Int cognate sequence attL is located between STOP and the DsRed gene. The platform reporter pNA1372plat, which is a derivative of the pcTD plasmid of the TD-In system (15), was integrated into the TDRT site located in the genome of the CHO TD-In cells using the TD-40 variant of TD recombinase to obtain the CHO TD-1372 cell line. The incoming plasmid pNA1345inc carries a reporter cassette composed of the promoterless EGFP gene followed by the CMV promoter. FRT and attR that can recombine with their counterparts in the plasmid pNA1372plat, flank the EGFP-CMV reporter cassette (Figure 2A).
The attL and attR sites in the platform and incoming reporters were chosen as substrates for Int because this pair of recombination targets supports the most efficient recombination in human cells (13,14).
Flp-catalyzed recombination between the FRT sites located on the platform and the incoming reporters leads to the swap between the NeoR and the EGFP genes and therefore activates the expression of the EGFP gene (Figure 2B–D). Int-catalyzed recombination between the attL and attR sites leads to the swap between the transcription terminator STOP and the CMV promoter thus activating the expression of the DsRed gene (Figure 2B–D).
In a sequential mode of dual RMCE, the first integrative step is catalyzed by supplying either Flp or Int (Figure 2B and C). A successful integration reaction is signaled by the appearance of the green or red cell, respectively. The second step of the sequential dual RMCE—deletion—is catalyzed by supplying the other recombinase: either Int or Flp, respectively (Figure 2B and C). A successful deletion is indicated by the activation of the expression of the second fluorescent gene, thus allowing the expression of both the EGFP and DsRed genes.
A ‘simultaneous’ dual RMCE reaction between the reporter cassettes located in the incoming and the platform plasmids is catalyzed by a simultaneous supply of both Flp and Int recombinases (Figure 2D). Successful ‘simultaneous’ dual RMCE is expected to replace the NeoR-STOP cassette in the integrated platform reporter with the EGFP-CMV cassette in the incoming plasmid. As a result, the expression of both EGFP and DsRed genes is activated, which can be detected by the appearance of cells that are both green and red (Figure 2D). The ‘simultaneous’ dual RMCE proceeds as a seemingly one-step reaction although it is proposed to progress in a sequential integration-then-excision mode (6). In this work, we refer to the ‘simultaneous’ dual RMCE only in a context of the simultaneous supply of Flp and Int.
Sequential dual RMCE reactions catalyzed by Flp and Int
Sequential dual RMCE is performed as follows: one of the two recombinases is used to integrate the entire incoming plasmid into the respective recombination site of the chromosomally located platform plasmid and the cells, in which the successful integration occurs, are identified and expanded (Figure 2B and C). After that, the second recombinase is used to perform the excision recombination between its cognate sites, and the cells with correctly replaced reporters are identified and expanded. To compare the efficiency of the sequential dual RMCE initiated by Flp or Int, we performed sequential dual RMCE in two modes: ‘Flp-then-Int’ and ‘Int-then-Flp’ (Figures 3 and 4).
Figure 3.
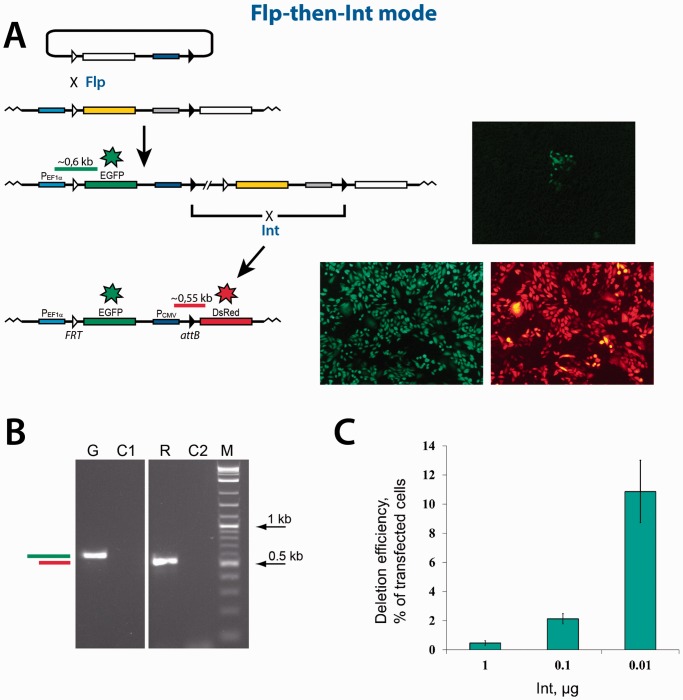
Sequential dual RMCE reaction in the ‘Flp-then-Int’ mode. (A) Schematics of the reaction. The color scheme of the functional elements shown is the same as in Figure 2. Typical group of green cells formed as a result of the first integration reaction mediated by Flp, are shown to the right of the respective integration product. The expanded green/red cells formed as a result of the second deletion reaction mediated by Int, are shown to the right of the respective deletion product. (B) The PCR analyses of typical expanded green and green/red colonies. The horizontal green and red bars in panels (A) and (B) schematically represent the control PCR products that can be generated if the respective recombination reactions are successful. The sequencing of the PCR products obtained confirmed their identity. G, PCR analysis of the expanded green cells obtained as a result of the Flp recombination. C1, control PCR analysis of the cells with the integrated platform reporter using the same primers as in lane ‘G’. R, PCR analysis of the expanded green/red cells obtained as a result of the Int recombination. C2, control PCR analysis of the green cells obtained as a result of the Flp recombination using the same primers as in lane ‘R’. M, 2-log DNA ladder (New England Biolabs). (C) The efficiency of the Int-mediated deletion reaction inversely depends on the amount of the Int expression vector added at transfection. The amount of the vector added is indicated. The efficiency of the deletion reaction is represented by green bars. The green bars show the mean value of three experiments; the error bars indicate standard deviation.
Figure 4.
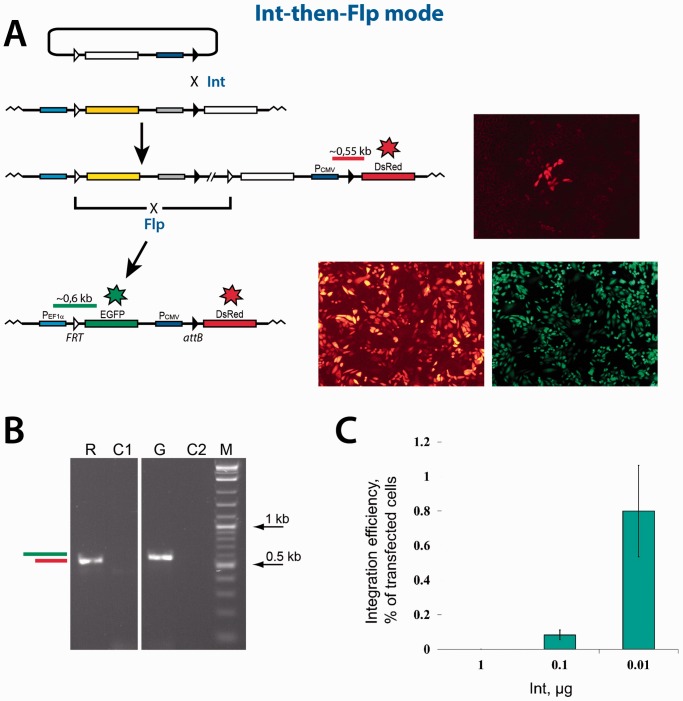
Sequential dual RMCE reaction in the ‘Int-then-Flp’ mode. (A) Schematics of the reaction. Typical group of red cells formed as a result of the integration reaction mediated by Int, are shown to the right of the respective integration product. The expanded red/green cells formed as a result of the deletion reaction mediated by Flp, are shown to the right of the respective deletion construct. (B) The PCR analyses of typical expanded red and red/green colonies. The horizontal green and red bars in panels (A) and (B) schematically represent the PCR products that are expected if the respective recombination reactions are successful. The sequencing of the PCR products obtained confirmed their identity. R, PCR analysis of the expanded red cells obtained as a result of the Int recombination. C1, control PCR analysis of the cells with the integrated platform reporter using the same primers as in lane ‘R’. G, PCR analysis of the expanded red/green cells obtained as a result of the Flp recombination. C2, control PCR analysis of the red cells obtained as a result of the Int recombination using the same primers as in lane ‘G’. M, 2-log DNA ladder (New England Biolabs). (C) The efficiency of the Int-mediated integration reaction inversely depends on the amount of the Int expression vector added at transfection. The amount of the vector added is indicated. The efficiency of the integration reaction is represented by green bars. The green bars show the mean value of three experiments; the error bars indicate standard deviation.
To perform the reaction in the Flp-then-Int mode (Figure 3), the Flp-expressing plasmid and the incoming reporter pNA1345inc were co-transfected into the CHO TD-1372 cells. As in our recent Flp-Cre dual RMCE experiments (15), we considered only those green cells as positive integrants that were able to form groups after the post-transfection expansion (Figure 3A). When the amount of the Flp-expressing plasmid added at transfection was 2 µg, a fraction of the groups of the green cells constituted ∼0.1% of the transfected cells. When the amount of the Flp-expressing plasmid added at transfection was lowered, the fraction of the groups of the green cells was lower (data not shown). Several groups of the green cells were expanded and analyzed by PCR and sequencing confirming that the pNA1345inc reporter plasmid was integrated into the CHO TD-1372 cells correctly (Figure 3B). At the next step, the expanded green cells were transfected with the Int-expressing plasmid to complete the RMCE reaction by an excision of the DNA fragment between the attL and attR sites. The transfected cells were expanded and groups of the red cells that formed were counted. In these experiments, we noticed that the fraction of the transfected cells that were able to form groups of the red cells was inversely dependent on the amount of the Int-expressing plasmid added at transfection: 1 µg of the Int-expressing plasmid was able to generate groups of the red cells in ∼0.5% of the transfected cells, while 0.01 µg of the Int-expressing plasmid generated groups of red cells in ∼10% of the transfected cells (Figure 3C).
The sequential Int-then-Flp dual RMCE experiments (Figure 4) were performed methodologically similar to the Flp-then-Int one described above. The CHO TD-1372 cells were co-transfected with the Int-expressing plasmid and the pNA1345inc reporter, and the groups of the red cells that formed after the expansion of the transfected cells were counted (Figure 4A). When the amount of the Int-expressing plasmid at transfection was 2 or 1 µg, no groups of the red cells were observed. Rare groups of the red cells started to form only when the amount of the Int-expressing plasmid at transfection was lowered. The fraction of the red cell groups reached ∼0.8% of the transfected cells when the amount of the plasmid was lowered to 0.01 µg (Figure 4C). Several groups of the red cells were expanded and analyzed by PCR and sequencing confirming that Int integrated the pNA1345inc reporter plasmid into the CHO TD-1372 cells correctly (Figure 4B). The expanded red cells were then transfected with the Flp-expressing plasmid to delete the DNA fragment between the FRT sites in the pNA1372plat-pNA1345inc co-integrant thus activating the expression of the EGFP gene (Figure 4A). When the ‘standard’ amount of the Flp-expressing plasmid (2 µg) was added at transfection, the fraction of the green cells that were able to form groups after expansion was ∼50%. Lower amounts of the Flp-expressing plasmid added at transfection generated fewer groups of the green cells.
Successful simultaneous dual RMCE requires a balanced supply of Int and Flp
Simultaneous dual RMCE is performed by co-transfecting the platform reporter cells CHO TD-1372 with both recombinase-expressing plasmids and the incoming reporter. The expected positive outcome of this type of RMCE is the appearance of cells that are both green and red (Figures 2D and 5). The experiments on sequential dual RMCE described above suggested that high amounts of the Int-expressing plasmid added at transfection may not be optimal neither for the integration reaction nor for the deletion reaction. If the same holds true for the simultaneous dual RMCE, then there will be few, if any, positive cells when the amounts of the Flp- and Int-expressing plasmids added at transfection are ∼2 µg. Indeed, when the initial experiments on the simultaneous dual RMCE were performed by adding 2 µg and 1 µg of the Flp- and Int-expressing plasmids, respectively, no red/green or just red cells were observed. Only when the amount of the Int-expressing plasmid was significantly lowered (to ∼0.1 µg), we started to see red/green cells (Figure 5C). When the amount of the Int-expressing plasmid was lowered to 0.01 µg, a fraction of the colony-forming red/green cells grew to ∼5% of the transfected cells. This number became even larger—∼12% of the transfected cells—when the amount of the Flp-expressing plasmid added at transfection was decreased to 0.3 µg (Figure 5C). Under these optimized conditions, just green and just red groups of cells, which reflect the integration of the incoming reporter plasmid into FRT and attL, respectively, constituted ∼1.5% of the transfected cells each. Several groups of the red/green cells were expanded and analyzed by PCR and sequencing confirming their nature as those that underwent dual RMCE (Figure 5B).
Figure 5.
Simultaneous dual RMCE. (A) Schematics of the reaction. Typical group of green/red cells formed when the expression vectors that code for Flp and Int are simultaneously added to the transfection mixture, are shown to the right of the replacement product. (B) The PCR analyses of a typical expanded green/red colony. The horizontal green and red bars in panels (A) and (B) represent the expected PCR products characteristic of a successful simultaneous dual RMCE reaction. The sequencing of the PCR products obtained confirmed their identity. G and R, PCR analysis of the expanded green/red cells with the primers that anneal on the EF1α promoter and the EGFP gene and on the CMV promoter and the DsRed gene, respectively. C1 and C2, control PCR analysis of the cells with the integrated platform reporter using the same set of primers as in lanes ‘G’ and ‘R’. M, 2-log DNA ladder (New England Biolabs). (C) The efficiency of the simultaneous dual RMCE reaction depends on the amount of the Flp and Int expression vectors added at transfection. The amounts of the vectors added are indicated. The efficiency of the replacement reaction is represented by green bars. The green bars show the mean value of three experiments; the error bars indicate standard deviation.
DISCUSSION
In this report, we presented an analysis of the dual RMCE reaction catalyzed by Flp and HK022 Int in a model setting of CHO cells. The analysis was aimed to identify the conditions that permit and maximize the efficiency of the replacement reaction performed in a sequential (Flp-then-Int and Int-then-Flp) and in a simultaneous (Flp + Int) mode. We demonstrate that HK022 Int paired with Flp can mediate efficient replacement reaction that does not require a selection force to identify the desired DNA rearrangements. We found that low amounts of the Int-expressing plasmid added at transfection is a key for the efficient RMCE in both the sequential and the simultaneous dual RMCE modes, while lowering the amount of the Flp-expressing plasmid helps improve the efficiency of dual RMCE in the simultaneous mode.
To simplify the analysis of the dual RMCE reaction, we developed a reporter system that detects the recombination events mediated by each recombinase (Figure 2). The detection of the recombination events is accomplished by the independent activation of the expression of the EGFP and DsRed genes on recombination at FRT and att sites, respectively. Our dual RMCE reporter system is functionally complete: in a population of cells transfected with the incoming reporter and the Flp and Int expression plasmids, the system can differentiate between the cells that underwent a complete dual RMCE reaction and those in which an incomplete RMCE reaction, that is, just integration of the incoming reporter into either FRT or att sites, has occurred. Our reporter system can be modified to analyze the dual RMCE reaction mediated by other recombinases by replacing FRT and att sites with the respective recombination targets.
It was rather unexpected that the efficient simultaneous dual RMCE reaction required a low amount of the Int-expressing plasmid. This amount was significantly lower than the amount of the Flp-expressing plasmid: 0.01 µg versus 0.33 µg, respectively, for the reaction performed in six-well plates (Figure 5). This low amount of the Int-expressing plasmid mirrors the conditions that are optimal for the sequential dual RMCE reaction (Figures 3 and 4) where Int acts alone and needs to perform either deletion (Flp-then-Int mode) or integration (Int-then-Flp mode). Overall, these results suggest that the inability of Int to support simultaneous dual RMCE when the amount of the Int-expressing plasmid added at transfection is high may be mainly due to its inability to perform any type of recombination reaction under these conditions. This contrasts sharply with the properties of Flp that mediates the highest level of recombination when the amount of the Flp expression plasmid is high.
One explanation for the difference in the functional properties of the recombinases is based on the difference in the complexity of the synaptic complexes they form. Flp is a simple recombinase that assembles a simple synaptic complex (19), while Int is a complex recombinase that requires additional factors to assemble its synaptic complex. It is possible that the results on the Int activity described here reflect the fact that Int can assemble a productive synaptic complex only when the concentration of Int in a cell is below certain threshold. If the concentration of Int is above that threshold, a synaptic complex formed becomes unproductive, which might result from Int aggregation. Indeed, in a bacterial assay, using a regulatable promoter, we observed that insolubility of Int increases as the level of its expression increases (unpublished data). This property of Int suggests an additional or alternative explanation for the inverse correlation between the amount of the Int expression plasmid added at transfection and the activity of Int: Int aggregates can sequester Int molecules, thus limiting its ability to form functional synaptic complexes.
In the model setting tested here, Flp, when it acts alone, exhibits the highest activity when the amount of the Flp-expressing plasmid is the highest tested (2 µg per six-well plate). It is interesting to note that this is not the case in the simultaneous dual RMCE reaction, when both recombinases are present (Figure 5). We found that the efficiency of the replacement reaction is the highest at a lower amount of the Flp-expressing plasmid added at transfection (0.33 µg per six-well plate). One possible explanation of this phenomenon is that at higher concentrations Flp and Int mediated reactions or their synaptic complexes may interfere with each other.
Our earlier work on dual RMCE, aimed to identify the conditions that maximize the efficiency of the reaction performed by simple recombinases Flp and Cre (15), determined that the low input of Cre helps improve the replacement reaction. It was suggested that such input increases the integration activity of Cre (without significantly decreasing its deletion activity), which, in turn, improves the overall efficiency of RMCE. In the present study, we observed that lowering the input of Int improves the efficiency of the Flp-Int dual RMCE. Despite the apparent similarity between the conditions that increase the efficiency of the replacement reaction in the Flp-Cre and Flp-Int systems, it is unlikely that the mechanisms behind the phenomena are the same. In contrast to Cre, both the integration and the deletion activities of Int are equally dependent on the input of its expression plasmid (Figures 3–5), which, we believe, reflects the assembly of the productive synaptic complex discussed above.
In our previous work on dual Flp-Int RMCE reaction performed in E. coli, we have shown that the replacement reaction performed in the sequential modes was by far more efficient than the one performed in the simultaneous mode (14). As the results of the present work show, the reason might be because in the bacterial experiments we were unable to control the levels of Flp and Int expression.
In the sequential dual RMCE modes, the integrative trans reactions mediated by Flp and Int was somewhat inefficient (∼0.1% and ∼1%, respectively) owing to the known high reversibility rate of the integration reaction [(3) and Figure 1A]. In contrast, the subsequent cis excision reactions (mediated by Int and Flp, respectively) led to the final RMCE product and were efficient (∼10% and ∼50%, respectively). Optimized conditions of the simultaneous dual RMCE mode led to the complete RMCE products in respectful 12% of the transfected cells. The progenitors of RMCE (6) envisioned the reaction as a fast succession of integration and excision. If that is correct, the relatively high yield of the dual RMCE products seen in the simultaneous mode, despite the considerably inefficient integrative reaction, is probably due to the presence of both recombinases that thermodynamically enable the high potential of the efficient excision reaction to overcome the instability of the integrative intermediate. The successive integration-excision model is also supported by the detection of small fraction of each intermediate incoming plasmid integrant in the simultaneous RMCE mode.
In summary, in mammalian cells, the simpler simultaneous supply of both Flp and Int in a dual RMCE reaction is preferable over the sequential protocol. Our work also shows that Int-HK022, as an efficient recombinase, can be added to the repertoire of recombinases used in mammalian genome engineering.
FUNDING
National Institutes of Health (NIH) [R01GM085848 to Y.V.]; German Israeli Foundation for Scientific Research and Development (G.I.F) [1062-62.3/2008]; Israel Science Foundation [702/11 to E.Y. and M.K.]. Funding for open access charge: NIH.
Conflict of interest statement. None declared.
REFERENCES
- 1.Branda CS, Dymecki SM. Talking about a revolution: the impact of site-specific recombinases on genetic analyses in mice. Dev. Cell. 2004;6:7–28. doi: 10.1016/s1534-5807(03)00399-x. [DOI] [PubMed] [Google Scholar]
- 2.Coates CJ, Kaminski JM, Summers JB, Segal DJ, Miller AD, Kolb AF. Site-directed genome modification: derivatives of DNA-modifying enzymes as targeting tools. Trends Biotechnol. 2005;23:407–419. doi: 10.1016/j.tibtech.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 3.Sorrell DA, Kolb AF. Targeted modification of mammalian genomes. Biotechnol. Adv. 2005;23:431–469. doi: 10.1016/j.biotechadv.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 4.Wirth D, Gama-Norton L, Riemer P, Sandhu U, Schucht R, Hauser H. Road to precision: recombinase-based targeting technologies for genome engineering. Curr. Opin. Biotechnol. 2007;18:411–419. doi: 10.1016/j.copbio.2007.07.013. [DOI] [PubMed] [Google Scholar]
- 5.Turan S, Galla M, Ernst E, Qiao J, Voelkel C, Schiedlmeier B, Zehe C, Bode J. Recombinase-mediated cassette exchange (RMCE): traditional concepts and current challenges. J. Mol. Biol. 2011;407:193–221. doi: 10.1016/j.jmb.2011.01.004. [DOI] [PubMed] [Google Scholar]
- 6.Schlake T, Bode J. Use of mutated FLP recognition target (FRT) sites for the exchange of expression cassettes at defined chromosomal loci. Biochemistry. 1994;33:12746–12751. doi: 10.1021/bi00209a003. [DOI] [PubMed] [Google Scholar]
- 7.Bethke B, Sauer B. Segmental genomic replacement by Cre-mediated recombination: genotoxic stress activation of the p53 promoter in single-copy transformants. Nucleic Acids Res. 1997;25:2828–2834. doi: 10.1093/nar/25.14.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lauth M, Spreafico F, Dethleffsen K, Meyer M. Stable and efficient cassette exchange under non-selectable conditions by combined use of two site-specific recombinases. Nucleic Acids Res. 2002;30:e115. doi: 10.1093/nar/gnf114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harel-Levi G, Goltsman J, Tuby CN, Yagil E, Kolot M. Human genomic site-specific recombination catalyzed by coliphage HK022 integrase. J. Biotechnol. 2008;134:46–54. doi: 10.1016/j.jbiotec.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 10.Weisberg RA, Gottesmann ME, Hendrix RW, Little JW. Family values in the age of genomics: comparative analyses of temperate bacteriophage HK022. Annu. Rev. Genet. 1999;33:565–602. doi: 10.1146/annurev.genet.33.1.565. [DOI] [PubMed] [Google Scholar]
- 11.Azaro MA, Landy A. Integrase and the λ Int Family. Washington, DC: ASM Press; 2002. [Google Scholar]
- 12.Malchin N, Tuby CN, Yagil E, Kolot M. Arm site independence of coliphage HK022 integrase in human cells. Mol. Genet. Genomics. 2011;285:403–413. doi: 10.1007/s00438-011-0614-3. [DOI] [PubMed] [Google Scholar]
- 13.Malchin N, Goltsman J, Dabool L, Gorovits R, Bao Q, Droge P, Yagil E, Kolot M. Optimization of coliphage HK022 Integrase activity in human cells. Gene. 2009;437:9–13. doi: 10.1016/j.gene.2009.02.013. [DOI] [PubMed] [Google Scholar]
- 14.Malchin N, Molotsky T, Borovok I, Voziyanov Y, Kotlyar AB, Yagil E, Kolot M. High efficiency of a sequential recombinase-mediated cassette exchange reaction in Escherichia coli. J. Mol. Microbiol. Biotechnol. 2010;19:117–122. doi: 10.1159/000321497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anderson RP, Voziyanova E, Voziyanov Y. Flp and Cre expressed from Flp-2A-Cre and Flp-IRES-Cre transcription units mediate the highest level of dual recombinase-mediated cassette exchange. Nucleic Acids Res. 2012;40:e62. doi: 10.1093/nar/gks027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blaisonneau J, Sor F, Cheret G, Yarrow D, Fukuhara H. A circular plasmid from the yeast Torulaspora delbrueckii. Plasmid. 1997;38:202–209. doi: 10.1006/plas.1997.1315. [DOI] [PubMed] [Google Scholar]
- 17.Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. 3rd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 18.Sauer B. Manipulation of transgenes by site-specific recombination: use of Cre recombinase. Methods Enzymol. 1993;225:890–900. doi: 10.1016/0076-6879(93)25056-8. [DOI] [PubMed] [Google Scholar]
- 19.Chen Y, Narendra U, Iype LE, Cox MM, Rice PA. Crystal structure of a Flp recombinase-Holliday junction complex: assembly of an active oligomer by helix swapping. Mol. Cell. 2000;6:885–897. [PubMed] [Google Scholar]