Abstract
The emergence and future of mammalian synthetic biology depends on technologies for orchestrating and custom tailoring complementary gene expression and signaling processes in a predictable manner. Here, we demonstrate for the first time multi-chromatic expression control in mammalian cells by differentially inducing up to three genes in a single cell culture in response to light of different wavelengths. To this end, we developed an ultraviolet B (UVB)-inducible expression system by designing a UVB-responsive split transcription factor based on the Arabidopsis thaliana UVB receptor UVR8 and the WD40 domain of COP1. The system allowed high (up to 800-fold) UVB-induced gene expression in human, monkey, hamster and mouse cells. Based on a quantitative model, we determined critical system parameters. By combining this UVB-responsive system with blue and red light-inducible gene control technology, we demonstrate multi-chromatic multi-gene control by differentially expressing three genes in a single cell culture in mammalian cells, and we apply this system for the multi-chromatic control of angiogenic signaling processes. This portfolio of optogenetic tools enables the design and implementation of synthetic biological networks showing unmatched spatiotemporal precision for future research and biomedical applications.
INTRODUCTION
The emergence of mammalian synthetic biology is tightly associated with the development of technologies for controlling target gene expression in a time-resolved manner. The functional interconnection of such individual genetic switches enabled the design and implementation of genetic networks showing complex kinetic behavior like tightly regulated expression control (1) with bi-stable (2), hysteretic (3), time-delayed (4) or oscillating (5) expression characteristics. Such control network topologies translated directly into first mammalian synthetic biology applications like molecular ‘bio-computers’ (6), novel drug discovery strategies (7) and closed loop-controlled molecular prostheses for autonomous management of gouty arthritis (8) or artificial insemination (9). These synthetic biological networks relied on genetic control technologies, responsive to small molecular stimuli, such as antibiotics (10–12), metabolites or vitamins (13–15), enabling tight gene control. However, molecule-inherent drawbacks, like difficulties in removing the inducer, prevented rapid reversibility or diffusion-based transport, which in turn prevented spatially controlled transgene expression. These drawbacks can be overcome by optogenetic tools enabling light-adjustable gene expression and control of cell function (16–21). First mammalian light-inducible gene expression control was based on cryptochromes (22), light, oxygen, voltage motif proteins (23,24) or transmembrane receptors (25), all responsive to blue light (450–495 nm), thus preventing the light-induced differential activation of multiple genes as required for the design of synthetic biological networks or for controlling complex mammalian growth and differentiation processes. This limitation was recently addressed by the development of a mammalian red/far-red light-responsive bi-stable switch enabling the spatiotemporal induction of genes in cell culture and an animal model of angiogenesis (26). To fully harness the potential of optical gene control for mammalian synthetic biology and to enable a similar boost of this field as initially triggered by the chemical gene switches, we set out to expand the spectral range of mammalian gene control technology by developing an ultraviolet B (UVB)-inducible gene expression system. Plants sense UVB light by UVB-induced monomerization of the photoreceptor protein UV resistance locus 8 (UVR8) that forms homo-dimers in the absence of UVB (27–29). Downstream signaling events, for instance through the interaction of monomeric UVR8 with the E3-ubiquitin ligase CONSTITUTIVELY PHOTOMORPHOGENIC 1 (COP1) via its WD40 domain (30), are subsequently terminated when UVR8 dimers are re-formed in the absence of UVB (31).
Based on Arabidopsis thaliana UVR8, we designed a UVB-responsive mammalian transcription factor that showed high gene induction in human, monkey, hamster and mice cells. A model-based quantitative analysis of the system enabled the adjustment and fine-tuning of expression characteristics in a predictable manner. By combining UVB, blue and red light-inducible gene switches, we demonstrated for the first time multi-chromatic multi-gene control in mammalian cells and applied it for the control of angiogenic signaling processes.
MATERIALS AND METHODS
DNA cloning
The construction of expression vectors is given in detail in Table 1.
Table 1.
Expression vectors and oligonucleotides designed and used in this study
| Plasmid | Description | Ref. or source |
|---|---|---|
| pFR-LUC | Plasmid encoding FLuc under control of Gal4-UAS5-TATA (Gal4-UAS5-TATA-FLuc-pA) | Agilent |
| pKM001 | Vector encoding SEAP under the control of PTet harboring eight repeats of the etr operator site between the heptameric tetO operator and the minimal promoter (tetO7-etr8-PhCMVmin-SEAP-pA) | (26) |
| pKM006 | Vector encoding SEAP under the control of a modified PTet harboring a 422-bp spacer between the 13mer tetO operator and the minimal promoter (tetO13-422 bp-PhCMVmin-SEAP-pA) | (26) |
| pKM022 | Bicistronic vector encoding PhyB(1–650)-VP16-NLS and TetR-PIF6(1–100)-HA under control of PSV40 [PSV40-PhyB(1–650)-VP16-NLS-IRESPV-TetR-PIF6(1–100)-HA-pA] | (26) |
| pKM081 | Vector encoding SEAP under control of a modified PETR (etr8-PCMVmin-SEAP-pA). pKM001 was digested (NruI/EcoRV) and the backbone was ligated (Supplementary Table S1). | This work |
| pKM085 | Vector encoding Gal4(65)-VVD-p65 under control of PEF1α [PEF1α -Gal4(65)-VVD-p65-pA]. Codon optimized Gal4(65)-VVD-p65 [GAVPO (24)] was chemically synthesized (Supplementary Table S1), digested (KpnI/NotI) and ligated (KpnI/NotI) into pWW029. | This work |
| pKM113 | Vector encoding TetR-UVR8(12–381) under control of PSV40 (PSV40-TetR-UVR8(12–381)-pA). TetR was amplified from pSAM200 using oligos oKM100 (5′-gttcaggaattcgccaccATGTCTAGATTAGATAAAAGTAAAGTGATTAAC-3′) and oKM107 (5′-gttcagggtaccGCTGTACGCGGACCCAC-3′) and digested (EcoRI/KpnI), whereas UVR8(12–381) was amplified from cDNA (GI: 334188609) using oligos oKM102 (5′-gttcagggtaccggcgccggcgccGCTCCTCCTCGTAAGGTTC-3′) and oKM114 (5′-gttcagggatccttaTCCATCGACGCTGAGTGCC-3′) and digested (KpnI/BamHI). pSAM200 was digested (EcoRI/BamHI) and the three fragment were ligated. | This work |
| pKM115 | Vector encoding COP1(WD40)-VP16 under control of PSV40 (PSV40-COP1(WD40)-VP16-pA). COP1(WD40) was amplified from cDNA (GI: 145360562) using oligos oKM116 (5′-gttcaggcggccgcgccaccatgTATAGCAACGGCCTTGCAGATTTTC-3′) and oKM117 (5′-gttcaggcgcgccggcgccCGCAGCGAGTACCAGAACTTTG-3′), digested (NotI/BssHII) and ligated (NotI/BssHII) into pMK233 (Supplementary Table S1). | This work |
| pKM168 | Vector encoding E-UVR8(12–381) under control of PSV40 [PSV40-E-UVR8(12–381)-pA]. E was amplified from pWW043 using oKM190 (5′-GAGCTATTCCAGAAGTAGTGAGG-3′) and oKM191 (5′-atgataagaaccttacgaggaggagcggcgccggcgccggtaccGCTGTACGCGGACGCATG-3′) and introduced into pKM113 (EcoRI/KpnI) by One-Step ISO Assembly (32) (Supplementary Table S1). | This work |
| pKM172 | Vector encoding Ang1 under control of a modified PETR (etr8-PCMVmin-Ang1-pA). Ang1 was amplified from pWB105 using oligos oKM202 (5′-gcatgcatcaattggagctcgcccggggatcctctagagtcagcttctgcgccaccATGTCTGCACTTCTGATCCTAGC-3′) and oKM203 (5′-tgactagcggccgcTCAAAAATCTAAAGGTCGAATCATCATAGTTG-3′), digested (MfeI/NotI) and ligated (EcoRI/NotI) into pKM081. | This work |
| pKM181 | Vector encoding VEGF165 under control of Gal4-UAS5-TATA (Gal4-UAS5-TATA-VEGF165-pA). VEGF165 was amplified from pWB094 using oligos oKM200 (5′-caagtcggtaccgaattcaccgCCATGAACTTTCTGCTGTCTTG-3′) and oKM201 (5′-tgactaccataggatgggcggccgcttaCCGCCTCGGCTTGTCAC-3′), digested (KpnI/PflMI) and ligated (KpnI/PflMI) into pFR-LUC. | This work |
| pMF111 | Vector encoding a PTet-driven SEAP expression unit (PTet-SEAP-pA) | (33) |
| pMK233 | Vector encoding a PSV40-driven PhyB(1–650)-VP16 expression unit [PSV40-PhyB(1–650)-VP16-pA] | (26) |
| pRSet | PT7-driven bacterial expression vector | Novagen |
| pSAM200 | Constitutive TetR-VP16 expression vector (PSV40-TetR-VP16-pA) | (34) |
| pWB094 | Vector for l-tryptophan-inducible VEGF165 expression (PTRT-VEGF165-pA). | (35) |
| pWB105 | Vector for acetaldehyde-inducible Ang1 expression (PAIR-Ang1-pA) | (35) |
| pWW029 | Vector encoding the erythromycin repressor protein E under control of PEF1α (PEF1α -E-pA) | (11) |
| pWW043 | Vector encoding PSV40 driven expression of E-KRAP (PSV40-E-KRAB-pA) | (11) |
Ang1, angiopoietin 1; COP1, E3 ubiquitin ligase COP1; E, macrolide-responsive repressor protein; etr8, operator sequence binding E; FLuc, firefly luciferase; Gal4(65), amino acids 1–65 of the Gal4 DNA-binding domain; Gal4-UAS, Gal4-binding site; HA, human influenza hemagglutinin-derived epitope tag; IRESPV, polioviral internal ribosome entry site; KRAB, transcriptional repressor domain from human Kox1; NLS, nuclear localization signal from simian virus 40 large T antigen; p65, transactivation domain from nuclear factor of activated B cells (NF-κB); pA, polyadenylation signal; PAIR, acetaldehyde-responsive promoter; PEF1α, human elongation factor 1α promoter; PhCMVmin, minimal human cytomegalovirus immediate early promoter; PhyB, phytochrome B; PhyB(1–650), N-terminus of phytochrome B with amino acids 1–650; PIF6, phytochrome-interacting factor 6; PIF6(1–100), N-terminus of phytochrome-interacting factor 6 with amino acids 1–100; PSV40, simian virus 40 early promoter; PTet, tetracycline-responsive promoter; PTRT, tryptophan-responsive promoter; SEAP, human placental secreted alkaline phosphatase; tetO, operator sequence binding TetR; TetR, tetracycline repressor protein; UVR8, photoreceptor protein UV resistance locus 8; UVR8(12–381), core domain of UVR8 consisting of amino acids 12–381; VEGF165, 165 amino acids splice variant of human vascular endothelial growth factor; VP16, Herpes simplex virus-derived transactivation domain; VVD, vivid; WD40; WD40 domain.
Uppercase in oligos, annealing sequence; underlined sequence, restriction site.
Cell culture and transfection
Chinese hamster ovary cells (CHO-K1, ATCC CCL 61) were cultivated in HTS medium (Cell Culture Technologies) supplemented with 10% fetal bovine serum (PAN, cat. no. P30-3602, batch no. P101003TC) and 2 mM l-glutamine (Sigma). African green monkey kidney cells (COS-7, ATCC CRL-1651), human umbilical vein cells (Ea.hy926, ATCC CRL-2922), human embryonic kidney fibroblasts [HEK-293T (36)], mouse embryonic fibroblasts (MEF, ATCC CRL-2214) and the human astrocytoma cell line SNB-19 (DSMZ ACC 325) were maintained in Dulbecco’s modified Eagle’s medium (PAN, cat. no. P03-0710) supplemented with 10% fetal bovine serum (DMEMcomplete). The mouse embryonic fibroblast cell line NIH/3T3 (ATCC CRL-1658) was cultivated in Dulbecco’s modified Eagle’s medium with 10% newborn calf serum (PAN, cat. no. 0402-P100104, batch no. 100104N). All media were supplemented with 100 U/ml penicillin and 0.1 mg/ml streptomycin (PAN). Cells were transfected, using a polyethylene-imine–based method (PEI, linear, MW: 25 kDa) (Polyscience). In brief, 1 mg/ml PEI solution in H2O was adjusted to pH 7.0 with HCl, sterile filtered and stored at −80°C in aliquots. In all, 70 000 cells were seeded per well of a 24-well plate and cultivated overnight. Aliquots of 0.75 µg of DNA were diluted in 50 µl of OptiMEM (Invitrogen) and mixed with 2.5 µl of PEI solution in 50 µl of OptiMEM under vortexing (amounts scaled to one well). After 15 min incubation at room temperature, the precipitate was added to the cells. For other plate formats, the cell number and amount of reagents were scaled up according to the growth area. The culture medium was replaced 5 h after the transfection. All plasmids were transfected in equal amounts (w:w), and only the red/far-red-responsive split transcription factor on pKM022 was transfected in 2-fold excess (w:w). For control experiments, erythromycin (Sigma) was added to a final concentration of 20 µg/ml from a 10 mg/ml stock solution in ethanol.
Illumination conditions
UVB illumination was performed, using a UVB narrowband lamp (Philips, prod. no. PL-S 9W/01) in combination with 310-nm bandpass filters (Ashai Spectra, prod. no. ZBPA310), to eliminate light of higher wavelengths. To modulate light intensity, the UVB light was attenuated by Plexiglas Alltop layers (Evoniks, prod. no. 29080). If not indicated otherwise, light intensity was adjusted for UVB (311 nm) to 2.7 µmol m−2 s−1, for blue (465 nm) to 3.5 µmol m−2 s−1, for red (660 nm) to 8 µmol m−2 s−1 and for far-red (740 nm) to 80 µmol m−2 s−1. The latter illuminations were performed by custom-made LED arrays. Light intensity was measured, using quantum sensors (LI-COR, prod. no. Q45045 or LTF Labortechnik, prod. no. CX-312). All cell handling involving the blue and red light-inducible expression systems was done under safe 522-nm light.
Reporter gene and cell viability assays
The reporter gene secreted alkaline phosphatase (SEAP) was quantified in the cell culture medium, using a colorimetric assay as described elsewhere (37). For quantification of firefly luciferase (FLuc), cells were lysed by addition of 250 µl (per well of a 24-well plate) lysis buffer (25 mM Tris/HCl, pH 7.8, 1% Triton X-100, 15 mM MgSO4, 4 mM ethylene glycol tetraacetic acid (EGTA) and 1 mM dithiothreitol (DTT)). Eighty microliters of the cell lysate was incubated with a 20 µl FLuc substrate (20 mM Tricine, pH 7.8, 2.67 mM MgSO4, 0.1 mM ethylenediaminetetraacetic acid, 33.3 mM DTT, 0.52 mM adenosine triphosphate, 0.27 mM Acetyl-CoA, 5 mM NaOH, 50 mM MgCO3 and 0.47 mM luciferin). Luciferase activity was monitored with an integration time of 1 s, using a Synergy 4 multi-mode microplate reader (BioTeks Instruments Inc). Production of vascular endothelial growth factor (VEGF) was quantified, using the Human VEGF ELISA Development kit (Peprotech, cat. no. 900-K10), and Ang1 was quantified, by using the DuoSet ELISA Development system (R&D systems, cat. no. DY923). Cell viability was determined, by quantifying the mitochondrial activity, using the Cell Proliferation Reagent WST-1 (Roche, cat. no. 11644807001).
RNA isolation and real-time polymerase chain reaction analysis
Cells were lysed with RLT buffer (Qiagen) and homogenized with QiaShredder columns (Qiagen), followed by total RNA isolation, using the RNeasy mini kit (Qiagen). Total mRNA concentration and integrity were determined using an automated electrophoresis system (Experion, BioRad). First-strand cDNA was synthesized from 500 ng of total RNA, using the RevertAid First-strand cDNA Synthesis Kit (Thermo Scientific). Therefore, both the provided random hexamer and the oligo (dT)18-primers were used. The cDNA concentration was fluorescently detected in a microplate reader (Infinite M-200, Tecan) using the Quant-iT PicoGreen dsDNA Assay Kit (Invitrogen), and the cDNA concentration was adjusted to 50 ng/µl for each polymerase chain reaction (PCR). Quantitative PCR (qPCR) analysis was performed with the CFX96 real-time PCR detection system (BioRad). Quantitative amplification detection was achieved, using the SABiosciences qPCR SYBR Green Master Mix (SABiosciences, Qiagen). For robust internal normalization, the PCR was performed with primers for both the unregulated housekeeping genes ß-actin (ACTB) and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) from Chinese hamster (SABiosciences) as described previously (38). For determination of gene expression of SEAP, the primers oKM219 (5′-GTCAGCTTCCTGCATGCTGC-3′) and oKM220 (5′-TCTTGGACAGAGCCACATATGG-3′) were used. For each PCR reaction, both primers were added at a concentration of 0.4 nM. The standard temperature profile included an initial denaturation step of 10 min at 95°C. Data were collected after every cycle of 40 cycles with denaturation at 95°C for 15 s, followed by annealing and extension at 60°C for 60 s, with an adjusted heating ramp of 1°C s−1. Relative SEAP mRNA expression levels were calculated using methods developed by Pfaffl (39) and Livak et al. (40). Data were normalized to the housekeeping genes GAPDH and ACTB and to the time point t = 0 h.
Vascular permeability assay
Ea.hy926 cells (12.000/well) were seeded into a transwell (BD bioscience, cat. No. 353095) and cultivated for 5–7 days until they formed a confluent monolayer, and the medium was replaced every 3 days. Monolayer-containing transwells were transferred to HEK-293T cells that were either mock transfected or transfected for blue light-inducible VEGF165 and UVB-inducible Ang1 expression. Plates were then either kept in the dark or illuminated from the bottom. The quantification of permeability was performed as described before (35).
RESULTS AND DISCUSSION
Design and implementation of UVB-inducible gene expression in mammalian cells
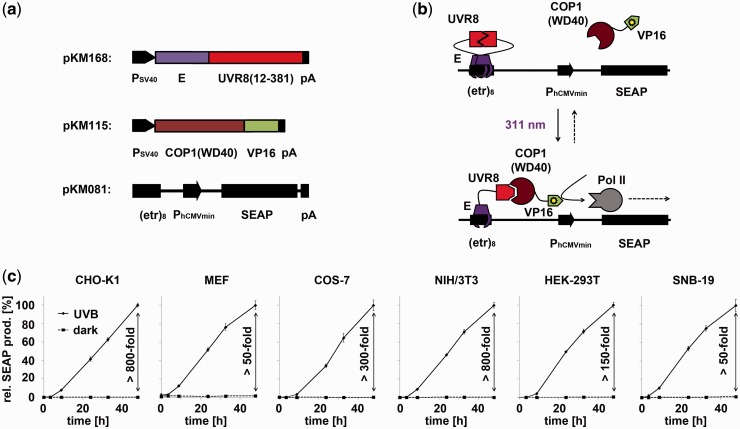
To expand the spectral range of mammalian cell-compatible optogenetic tools, we designed a UVB-inducible mammalian gene control system based on the A. thaliana UVB receptor UVR8 and the WD40 domain of its interacting partner COP1. It is of note that mammals do not possess a UVR8-like UVB receptor (27). By using only the WD40 domain from A. thaliana COP1 that has a sequence consensus <50% compared with its human counterpart, we eliminated the ubiquitin ligase activity of COP1 to further minimize the risk of cross-talk with the host’s signaling pathways. To set-up a UVB-inducible gene expression system, we designed a bipartite UVB-responsive transcription factor, consisting (i) of the UVR8 core domain (amino acids 12–381) fused to the macrolide repressor E (11) (plasmid pKM168) and (ii) the WD40-domain of COP1 (27) fused to the Herpes simplex-derived transactivation domain VP16 (plasmid pKM115, Figure 1a). The response promoter was constructed by fusing an octameric E-responsive operator motif (etr)8 upstream to the minimal cytomegalovirus promoter PhCMVmin (11) and the reporter SEAP (plasmid pKM081, Figure 1a). In the absence of UVB, UVR8 is in the closed configuration and does not interact with COP1(WD40) (Figure 1b) (27). However, absorption of UVB light triggers the transition of UVR8 to the open configuration, the recruitment of COP1(WD40)-VP16 and the subsequent activation of the target promoter, resulting in expression of SEAP. Spontaneous conversion to the closed state of UVR8 in the dark disrupts the interaction between UVR8 and COP1(WD40) and results in a shut-off of gene expression (Figure 1b). For functional validation, the system was transfected into human (HEK-293T, SNB-19), monkey (COS-7), mouse (MEF, NIH/3T3) and hamster (CHO-K1) cell lines that were subsequently cultivated for 48 h in the dark or under 311-nm light (2.7 µmol m−2 s−1). In all configurations, the illuminated cells showed high SEAP production up to 800-fold above the background levels observed in transfected cells that were kept in the dark (Figure 1c), thereby demonstrating the broad applicability of this system.
Figure 1.
Design and implementation of the UVB-inducible gene expression system. (a) Molecular building blocks for UVB-inducible gene expression. The expression vectors (pKM168 and pKM115) for the UVB-inducible split transcription factor were constructed by (i) fusing the gene of the macrolide-responsive repressor E to the coding sequence of the A. thaliana UVR8 core domain and (ii) by linking the gene for the COP1-derived WD40 motif to the DNA sequence encoding the VP16 transactivation domain. The reporter construct (pKM081) was assembled by cloning an octameric E-specific operator site (etr)8 upstream of the minimal human cytomegalovirus promoter PhCMVmin. As reporter gene, human placental secreted alkaline phosphatase (SEAP) was applied. pA, polyadenylation signal; PSV40, simian virus 40 promoter. (b) Mode of function. In the dark, UVR8 is tethered to the operator sequence in the closed configuration but cannot interact with COP1(WD40). On illumination with 311-nm light, the UVR8 transits from the closed to the open state and recruits COP1(WD40)-VP16, to result in the activation of PhCMVmin. On shifting to the dark, UVR8 spontaneously assumes the closed configuration, thereby resulting in a gradual shut down of gene expression. (c) Characterization of UVB-inducible gene expression in mammalian cell lines. The indicated cell lines were transfected with the UVB-inducible expression system (plasmids pKM168, pKM115 and pKM081) and kept in the dark or were illuminated with 311-nm light for 48 h. SEAP production was determined at the indicated points in time and is represented normalized to the values obtained after 48 h under UVB illumination. The corresponding induction factors are indicated. The absolute values for this condition are CHO-K1, 50.2 [U/L]; MEF, 1.6 [U/L]; COS-7, 12.4 [U/L]; NIH/3T3, 17.1 [U/L]; HEK-293T, 687.5 [U/L]; SNB-19, 1.5 [U/L]. Data are means ± standard deviation (SD) (n = 4).
To quantitatively understand the characteristics of the UVB-inducible expression system, as well as to quantify and minimize side effects of UVB radiation on mammalian cells (41,42), we developed an ordinary differential equation (ODE)-based quantitative model and parameterized it by kinetic expression data at the mRNA and protein level.
| (1) |
| (2) |
 |
(3) |
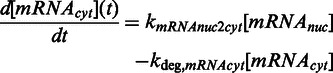 |
(4) |
| (5) |
| (6) |
| (7) |
The model was derived as follows, and details on its derivation are described in Supplementary Information. UVR8 is found either in the closed 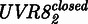 or open
or open 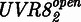 state. The transition rate from
state. The transition rate from 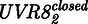 to
to  is proportional to the intensity IUV of the UVB light as previously described for other photoreceptors (43). In the dark,
is proportional to the intensity IUV of the UVB light as previously described for other photoreceptors (43). In the dark, 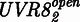 closes with the rate kdeact. COP1(WD40)-VP16, binds to
closes with the rate kdeact. COP1(WD40)-VP16, binds to 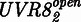 , and triggers expression of the target mRNA, which can be shut-off by the addition of erythromycin [EM = 1, (11)] (Supplementary Figure S1). Basal mRNA production is represented by kbasal,mRNA. The initially nuclear mRNA (mRNAnuc) is subsequently transported to the cytoplasm (mRNAcyt) for translation and secretion of the SEAP protein to the medium. SEAP protein in the medium is thus proportional to the number of cells N, which was assumed to follow exponential growth (growth rate kgrowth). UVB-induced cytotoxicity was described by the first-order degradation rate kdeg,cells, and it was assumed to be proportional to IUV. For modeling plasmid loss in transiently transfected cells, a dilution rate for the gene dose GD per cell was introduced.
, and triggers expression of the target mRNA, which can be shut-off by the addition of erythromycin [EM = 1, (11)] (Supplementary Figure S1). Basal mRNA production is represented by kbasal,mRNA. The initially nuclear mRNA (mRNAnuc) is subsequently transported to the cytoplasm (mRNAcyt) for translation and secretion of the SEAP protein to the medium. SEAP protein in the medium is thus proportional to the number of cells N, which was assumed to follow exponential growth (growth rate kgrowth). UVB-induced cytotoxicity was described by the first-order degradation rate kdeg,cells, and it was assumed to be proportional to IUV. For modeling plasmid loss in transiently transfected cells, a dilution rate for the gene dose GD per cell was introduced.
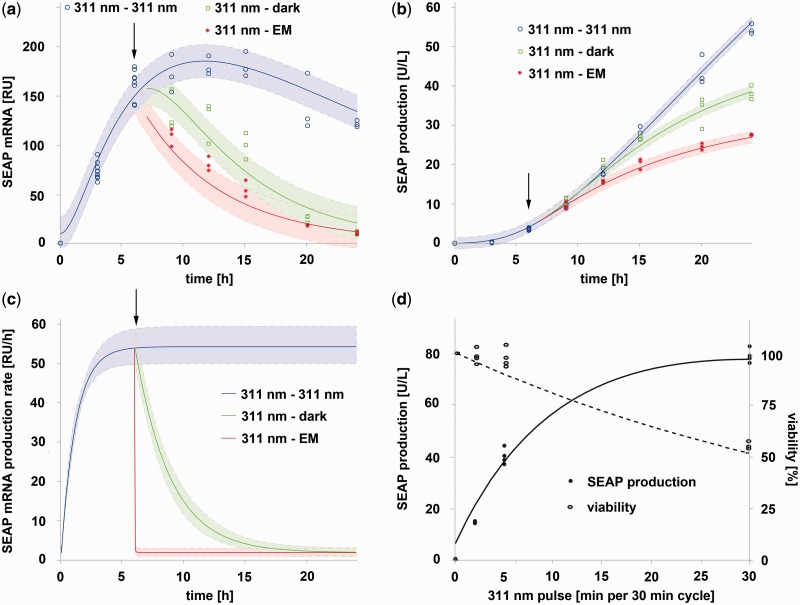
For the parameterization of the model, the ON and OFF kinetics of the UVB system were analyzed. To this end, CHO-K1 cells engineered for UVB-inducible SEAP production were illuminated with 311-nm light (2.7 µmol m−2 s−1) for 6 h to induce gene expression. For the subsequent 18 h, (i) illumination was either continued to quantify sustained expression induction; or (ii) the cells were cultivated in the dark to analyze spontaneous shut down of gene expression by transition of UVR8 to the closed state (31,44); or (iii) the cells were supplemented with erythromycin to immediately shut down promoter activity by preventing the binding of E-UVR8(12–381) to its etr operator (11,26) (Supplementary Figure S1). In all three configurations, the kinetics of SEAP mRNA (Figure 2a) and protein (Figure 2b) levels were determined and applied to parameterize the model.
Figure 2.
Model-based quantitative characterization of UVB-inducible gene expression. (a and b) Light-inducible expression kinetics. CHO-K1 cells were engineered for UVB-inducible SEAP expression (plasmids pKM168, pKM115 and pKM081). After 24 h, the medium was exchanged, and the cells were illuminated for 6 h at 311 nm and were then either kept under 311 nm, moved to darkness or were supplemented with erythromycin (arrow). The SEAP mRNA (a) and protein (b) levels were determined at the indicated points in time. The curves represent the model fit to the data, and the shaded error bands are estimated by a simple error model with a constant Gaussian error. (c) Model-based analysis of expression kinetics. The SEAP mRNA production rate per promoter is shown for cells subjected to the stimuli described in Figure 2a. The shaded bands indicate the 95% prediction confidence interval. (d) Model prediction for SEAP production and cell viability under pulsed light. For this prediction, it was assumed that CHO-K1 cells were transfected with plasmids pKM168, pKM115 and pKM081, cultivated for 24 h in the dark, followed by an 24 h illumination period under pulsed UVB light (indicated in min light per 30 min cycle). The predictions for SEAP production and cell viability are represented by the lines, whereas experimental validation data are shown as circles.
UVB is a potent mutagen, as it is directly absorbed by the aromatic heterocyclic bases of DNA, inducing the formation of cyclobutane-pyrimidine dimers and pyrimidine-(6-4)-pyrimidone photoproducts (45). Furthermore, aromatic amino acids, such as tryptophan and tyrosine, absorb UVB, generating amino acid derivatives that may alter protein function and affect cellular signaling (46). In addition, the activity of stress-related signaling pathways is modulated in response to UVB (47). To quantitatively take into account such cytotoxic side effects of UVB light, CHO-K1 cells transgenic for the UVB-inducible expression system were cultivated for 24 h under 311-nm illumination, ranging from 0 to 4.7 µmol m−2 s−1 (Supplementary Figure S2). At low intensities, SEAP production increased linearly with light dose until reaching a plateau at 2.5 µmol m−2 s−1. In parallel, cell viability decreased with increasing light doses.
Based on these quantitative data, the ODE system (1–7) parameters were estimated by performing a multi-experiment fit based on the maximum likelihood. To obtain uncertainties of the estimated parameters, we calculated the profile likelihood for each parameter (48). It was possible to identify all parameters and calculate finite 95%-confidence intervals for each parameter (Supplementary Information and Supplementary Figure S3 for details on parameter estimation and the estimated parameter values). With the parameterized model, we quantitatively analyzed the ON and OFF kinetics by calculating the activity of the target promoter on UVB illumination and subsequent transfer to the dark (Figure 2c). To quantify the quality of the analysis, we calculated the 95%-prediction confidence interval by propagating the uncertainties of the estimated parameters (49). This analysis revealed fast UVB-responsive inducibility, which can be reverted by two mechanisms, either by the addition of erythromycin (resulting in an immediate shut-off) or by transfer to the dark, that shows a delayed return to the OFF state (characterized by kdeact = 0.350 h−1 corresponding to a half-life time of t1/2 = 1.98 h).
This quantitative model was applied to identify illumination conditions enabling high gene induction while minimizing cytotoxic side effects. Based on the delayed dark reversion (Figure 2c), pulsed light might be used to sustain gene expression while minimizing the overall light dose and the thereto-associated side effects. A quantitative model analysis showed that 5 min light pulses (2.7 µmol m−2 s−1) followed by 25 min cultivation in the dark correlated with cell viabilities >90%, while still reaching an 100-fold expression induction above background (Figure 2d), which compares favorably with established optogenetic gene control systems (24). This model prediction was experimentally verified by cultivating CHO-K1 cells transgenic for UVB-inducible SEAP production for 24 h under pulsed light (2.7 µmol m−2 s−1) of different cycles (2 min light/28 min dark; 5 min light/25 min dark, continuous light). SEAP production and cell viability correlated well with the model prediction (Figure 2d). This observation indicates that this system can be used to achieve high gene induction with minimal cytotoxicity, thereby validating this UVB-inducible system as highly valuable optogenetic tool for mammalian synthetic biology and cell technology.
Multi-chromatic multi-gene control
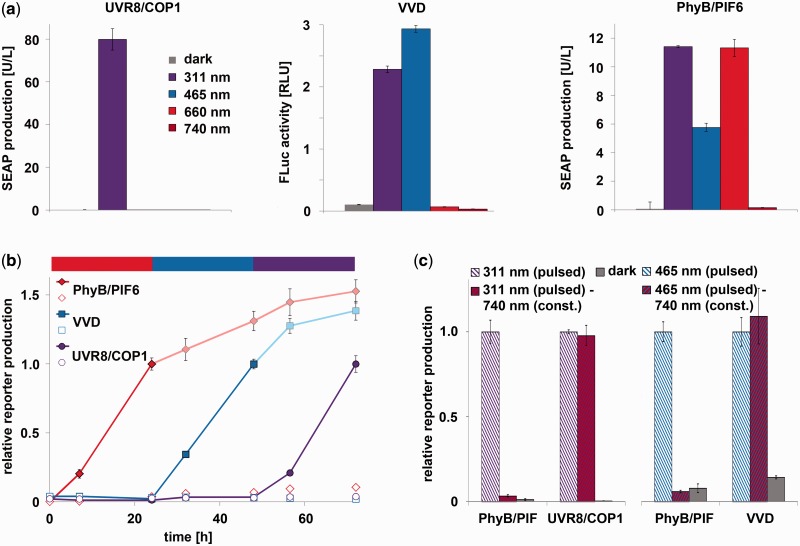
The emergence of mammalian synthetic biology and the design of synthetic mammalian gene networks capitalize on tools and technologies, allowing an external control of transgene activity (14,15,50). Although previous synthetic mammalian gene networks were tuned by a combination of externally applied chemical stimuli (1,2,4,6), the recent emergence of mammalian optogenetic tools (19–21) has paved the way for light-controlled synthetic mammalian systems, allowing an unmatched spatiotemporal resolution in tuning systems performance. To pioneer such mammalian multi-chromatically controlled systems, we combined UVB-inducible transgene control with the previously described blue light-responsive, Neurospora crassa vivid-based expression tool (24) and our recent red/far-red light-responsive gene expression technology, based on A. thaliana phytochrome B (26). To gain insight into the spectral responsiveness of these light switches, CHO-K1 cells were transiently transfected with the respective system’s components, and exposed to UVB (311 nm), blue (465 nm), red (660 nm) and far-red (740 nm) light or incubated in the dark for 24 h. Although the UVB-inducible system was only activated at 311 nm, the blue light system was switched on at 465 and 311 nm, and the red/far-red switch was activated at 660, 465 and 311 nm (Figure 3a). This finding is in agreement with the spectral properties of the involved chromophores, showing absorbance at the respective wavelengths (29,51,52). As a first step toward multi-chromatic multi-gene control inside a single cell, the UVB-, blue- and red/far-red light-responsive gene expression systems were simultaneously transfected into CHO-K1 cells to control the expression of angiopoietin 1 (Ang1, UVB), firefly luciferase (FLuc, blue) and SEAP (red). Twenty-four hours after transfection and reconstitution of functional PhyB by the addition of the chromophore phycocyanobilin (26), cells were illuminated for 24 h with 660-nm light, another 24 h with 465-nm light and finally with 311-nm light for another day. The expression of the reporter proteins was followed over the entire time course, and the induction of reporter expression correlated exactly with the applied respective wavelengths, underlining the functionality of multi-chromatic multi-gene control (Figure 3b).
Figure 3.
Multi-chromatic multi-gene expression control. (a) Spectral responses of the UVB, blue and red light-inducible gene expression systems. CHO-K1 cells were transfected for UVB-inducible SEAP production (left), blue light-inducible firefly luciferase (FLuc) expression (middle) or red/far-red light-switchable gene expression (right). Twenty-four hours post-transfection, the cell culture medium was replaced with fresh medium that was supplemented with 15 µM phycocyanobilin (PCB) for the red/far-red light-switchable gene expression system. After incubation in the dark for 1 h, the cells were illuminated with light of the indicated wavelengths for 24 h before the quantification of the reporter proteins. (b) Multi-chromatic multi-gene control. CHO-K1 cells were transfected for red light-inducible SEAP production (plasmids pKM022 and pKM006), blue light-inducible FLuc expression (plasmids pKM085 and pFR-LUC) and UVB-responsive angiopoietin 1 (Ang1) synthesis (plasmids pKM168, pKM115 and pKM172). Twenty-four hours post-transfection, the medium was exchanged to DMEMcomplete that was supplemented with 15 µM PCB, and after incubation for 1 h in the dark, the cells were illuminated with 660-nm light. After 24 h, illumination was changed to 465 nm, and another 24 h later, the cells were exposed to 311-nm light for another day. Control cells (open symbols) were kept in the dark for the entire experiment. At the indicated points in time SEAP, FLuc and Ang1 production was determined. (c) Avoiding cross-talk between red light and blue/UVB light-inducible gene expression. CHO-K1 cells were transfected for red light-inducible SEAP expression, blue light-responsive FLuc production or UVB-inducible SEAP expression. After 24 h, the medium was replaced, and the cells were either illuminated with pulses of UVB light (2.7 µmol m−2 s−1, 5 min every 30 min; left) or with pulses of blue light (3.5 µmol m−2 s−1, 5 min every 30 min, right) in the absence or presence of constant 740-nm light. Twenty-four hours after illumination start, the reporter production was quantified. The reporter activities are normalized to the samples that received only UVB or blue light, respectively. Data are means ± SD (n = 4).
Given the overlapping action spectra of the photosensory proteins (29,51,52), a cross-activation of the different systems is inherent when using continuous illumination (Figure 3a). However, the rapid toggle switch-like characteristics of the red light-responsive gene expression system combined with the much slower, ODE-model-determined half-life time of the activated state of the UVB-inducible gene expression system (2 h) and of the blue light-responsive system [2 h (24)] suggest that pulsed illumination with UVB or blue light in combination with constant 740-nm light should result in repression of the red light-inducible gene expression system, while triggering expression from the UVB or blue light-responsive systems. To test this concept, we illuminated CHO-K1 cells that had been transfected for red light-inducible SEAP expression or UVB light-responsive SEAP expression with pulses (5 min ON every 30 min) of 311-nm light. At the same time, the cells were constantly illuminated with 740 nm, whereas control cells were either kept in the dark or received pulsed light in the absence of 740-nm illumination. Pulses of UVB light resulted in reporter expression controlled by the red light-inducible gene expression system (Figure 3c). However, reporter expression by the red light-responsive system could be repressed by constant illumination with 740-nm light, whereas this did not influence expression from the UVB-responsive system (Figure 3c). This observation suggests that a pulsed illumination strategy can successfully be applied to independently regulate the expression of UVB and red light-responsive genes. This approach was subsequently expanded for selectively activating blue and red light-responsive genes. To this aim, cells were engineered for blue and red light-inducible gene expression and illuminated under pulsed (5 min ON every 30 min) blue light and continuous 740-nm light. In this configuration, high activation of the blue light-responsive gene was observed while the red light-inducible system remained in the OFF-state (Figure 3c). These findings underline the possibility of selectively activating individual genes by simply adjusting the color and pulsing of the light inducer for achieving multi-chromatic multi-gene control.
Multi-chromatic control of mammalian cell signaling for tissue engineering
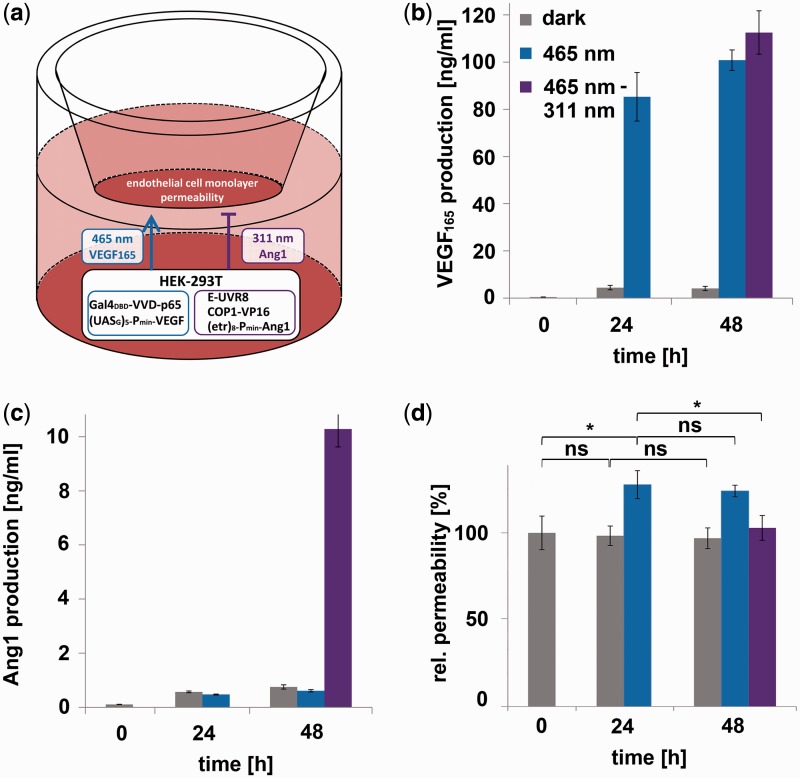
We applied the sequential multi-chromatic multi-gene control in a tissue engineering approach, aiming at reconstructing the signaling processes occurring in neo-vascularization and angiogenesis (35,53). In these processes, the formation of new blood vessels is first induced by the 165 amino acid splice variant of the vascular endothelial growth factor (VEGF165), whereas the maturation of the initially leaky vessels to tight ones requires subsequent angiopoietin 1 (Ang1) signaling (54). First, we demonstrated that blue or UVB illumination does not interfere with endothelial cell monolayer permeability (Supplementary Figure S4). Next, we reconstructed the angiogenic processes in a model system by subjecting the expression of both signaling proteins to multi-chromatic external control. To this end, we engineered human embryonic kidney cells (HEK-293T) for blue light-inducible VEGF165 expression and for UVB-inducible production of Ang1. These cells were subsequently applied to control angiogenic signaling in a transwell-based monolayer of endothelial cells emulating the blood vessel wall (Figure 4a) (35,54). This configuration was first illuminated with blue light for 24 h to induce VEGF165 signaling. Next, the cells were exposed to 311 nm for another 24 h to induce Ang1 signaling, whereas control cells were either kept under blue light or in the dark for the entire experiment. The quantification of the signaling molecules after 24 and 48 h revealed that VEGF165 was induced by blue light (Figure 4b), whereas Ang1 production could only be observed under UVB illumination (Figure 4c). The impact of multi-chromatically controlled sequential VEGF165 and Ang1 expression on endothelial cell function was assessed by measuring the permeability of the cell monolayer using fluorescently labeled dextran. These experiments disclosed an increased permeability after 24 h, correlating with the blue light-mediated signaling of VEGF165, whereas at 48 h, the maturation-inducing effect of UVB-induced Ang1 prevailed by triggering a subsequent decrease in monolayer permeability (Figure 4d). VEGF is the most prominent angiogenic growth factor and induces new blood vessels from pre-existing vasculature (55). Consequently, therapeutic angiogenesis by targeted delivery of VEGF has achieved promising results in chronic myocardial and retinal ischemia. However, complications, such as vascular leakage, may arise from tissue exposure to high VEGF concentrations. Therefore, strategies have been developed for co-administration of growth factor agents, such as Ang1 (56). This approach has been shown to potentially result in the growth of new and non-leaky vasculature (57,58). Because of its unprecedented spatiotemporal resolution and simple fine-tuning by adjusting the illumination intensities and duration, the optogenetic approach presented here for controlled delivery of VEGF and Ang1 holds the potential to further boost the development of therapeutic approaches in angiogenesis. In summary, our experimental observations indicate that multi-chromatically controlled signaling can be applied to re-engineer natural signaling processes, thereby opening a wealth of novel insights and applications in biomedical fundamental and translational research.
Figure 4.
Multi-chromatically controlled angiogenic signaling. (a) Schematic experimental set-up. HEK-293T cells, transgenic for blue light-inducible VEGF165 production (plasmids pKM085 and pKM181) and UVB-responsive Ang1 expression (plasmids pKM085, pKM115 and pKM172) were co-cultured with a monolayer of endothelial Ea.hy926 cells in a transwell system. Blue light-induced production of VEGF165 triggers enhanced permeability of the endothelial cell monolayer, whereas UVB-induced Ang1 signaling induces the formation of tight cell junctions. (b and c) Multi-chromatically controlled VEGF165 and Ang1 production profiles. The angiogenic signaling set-up (a) was illuminated for 24 h with blue (465 nm) light, and subsequently for 24 h with UVB (311 nm) light. Control cells were continuously kept under 465-nm light or in the dark. The concentrations of the signaling molecules VEGF165 (b) and Ang1 (c) were quantified at the indicated points in time. (d) Effect of multi-chromatically controlled angiogenic signaling on endothelial cells. The effect of VEGF165 and Ang1 signaling on the permeability of the endothelial cell layer was determined by quantifying the trans-layer permeation of fluorescently labeled dextran. The permeability is normalized to the control value obtained from monolayers of endothelial cells that were co-cultivated with mock-transfected HEK-293T cells. Data are means ± SD (n = 4). Statistics were performed by the two-tailed t-test. *P < 0.01; ns, not significant.
CONCLUSION
In this study, we pioneer multi-chromatic gene expression control in mammalian cells and apply it to control angiogenic signaling processes. To this end, we developed a novel UVB-inducible expression system based on A. thaliana UVR8. A model-based quantitative characterization of the system kinetics enabled the rational adjustment of control parameters to achieve high gene induction while minimizing adverse side effects. We further demonstrate that the UVB-controlled system can be used in combination with previously described blue and red light-inducible promoters for achieving multi-gene expression kinetics with desired temporal resolution.
Given the previous success of chemically triggered mammalian gene switches in designing synthetic gene networks and developing biomedical applications, we are convinced that the multi-chromatic control of multiple genes and the associated unmatched spatiotemporal precision will ‘enlighten’ the future of mammalian synthetic biology in fundamental and translational research.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1 and 2, Supplementary Figures 1–4, Supplementary Information and Supplementary References [59,60].
FUNDING
Initiating and Networking Fund (IVF) of the Helmholtz Association within the Helmholtz Initiative on Synthetic Biology [SO-078]; European Research Council under the European Community’s Seventh Framework Programme [FP7/2007-2013]/ERC [259043]-CompBioMat; excellence initiative of the German Federal and State Governments [EXC 294, BIOSS]. Funding for open access charge: Initiating and Networking Fund (IVF) of the Helmholtz Association within the Helmholtz Initiative on Synthetic Biology [SO-078].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Martin Fussenegger (ETH Zürich) for providing plasmids pWB094, pWB105 and Ea.hy926 cells.
REFERENCES
- 1.Deans TL, Cantor CR, Collins JJ. A tunable genetic switch based on RNAi and repressor proteins for regulating gene expression in mammalian cells. Cell. 2007;130:363–372. doi: 10.1016/j.cell.2007.05.045. [DOI] [PubMed] [Google Scholar]
- 2.Kramer BP, Viretta AU, Daoud-El-Baba M, Aubel D, Weber W, Fussenegger M. An engineered epigenetic transgene switch in mammalian cells. Nat. Biotechnol. 2004;22:867–870. doi: 10.1038/nbt980. [DOI] [PubMed] [Google Scholar]
- 3.Kramer BP, Fussenegger M. Hysteresis in a synthetic mammalian gene network. Proc. Natl Acad. Sci. USA. 2005;102:9517–9522. doi: 10.1073/pnas.0500345102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weber W, Stelling J, Rimann M, Keller B, Daoud-El Baba M, Weber CC, Aubel D, Fussenegger M. A synthetic time-delay circuit in mammalian cells and mice. Proc. Natl Acad. Sci. USA. 2007;104:2643–2648. doi: 10.1073/pnas.0606398104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tigges M, Marquez-Lago TT, Stelling J, Fussenegger M. A tunable synthetic mammalian oscillator. Nature. 2009;457:309–312. doi: 10.1038/nature07616. [DOI] [PubMed] [Google Scholar]
- 6.Auslander S, Auslander D, Muller M, Wieland M, Fussenegger M. Programmable single-cell mammalian biocomputers. Nature. 2012;487:123–127. doi: 10.1038/nature11149. [DOI] [PubMed] [Google Scholar]
- 7.Weber W, Schoenmakers R, Keller B, Gitzinger M, Grau T, Daoud-El Baba M, Sander P, Fussenegger M. A synthetic mammalian gene circuit reveals antituberculosis compounds. Proc. Natl Acad. Sci. USA. 2008;105:9994–9998. doi: 10.1073/pnas.0800663105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kemmer C, Gitzinger M, Daoud-El Baba M, Djonov V, Stelling J, Fussenegger M. Self-sufficient control of urate homeostasis in mice by a synthetic circuit. Nat. Biotechnol. 2010;28:355–360. doi: 10.1038/nbt.1617. [DOI] [PubMed] [Google Scholar]
- 9.Kemmer C, Fluri DA, Witschi U, Passeraub A, Gutzwiller A, Fussenegger M. A designer network coordinating bovine artificial insemination by ovulation-triggered release of implanted sperms. J. Controlled Release. 2011;150:23–29. doi: 10.1016/j.jconrel.2010.11.016. [DOI] [PubMed] [Google Scholar]
- 10.Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl Acad. Sci. USA. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weber W, Fux C, Daoud-el Baba M, Keller B, Weber CC, Kramer BP, Heinzen C, Aubel D, Bailey JE, Fussenegger M. Macrolide-based transgene control in mammalian cells and mice. Nat. Biotechnol. 2002;20:901–907. doi: 10.1038/nbt731. [DOI] [PubMed] [Google Scholar]
- 12.Fussenegger M, Morris RP, Fux C, Rimann M, von Stockar B, Thompson CJ, Bailey JE. Streptogramin-based gene regulation systems for mammalian cells. Nat. Biotechnol. 2000;18:1203–1208. doi: 10.1038/81208. [DOI] [PubMed] [Google Scholar]
- 13.Horner M, Weber W. Molecular switches in animal cells. FEBS Lett. 2012;586:2084–2096. doi: 10.1016/j.febslet.2012.02.032. [DOI] [PubMed] [Google Scholar]
- 14.Weber W, Fussenegger M. Emerging biomedical applications of synthetic biology. Nat. Rev. Genet. 2012;13:21–35. doi: 10.1038/nrg3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jakobus K, Wend S, Weber W. Synthetic mammalian gene networks as a blueprint for the design of interactive biohybrid materials. Chem. Soc. Rev. 2012;41:1000–1018. doi: 10.1039/c1cs15176b. [DOI] [PubMed] [Google Scholar]
- 16.Shimizu-Sato S, Huq E, Tepperman JM, Quail PH. A light-switchable gene promoter system. Nat. Biotechnol. 2002;20:1041–1044. doi: 10.1038/nbt734. [DOI] [PubMed] [Google Scholar]
- 17.Levskaya A, Chevalier AA, Tabor JJ, Simpson ZB, Lavery LA, Levy M, Davidson EA, Scouras A, Ellington AD, Marcotte EM, et al. Synthetic biology: engineering Escherichia coli to see light. Nature. 2005;438:441–442. doi: 10.1038/nature04405. [DOI] [PubMed] [Google Scholar]
- 18.Ohlendorf R, Vidavski RR, Eldar A, Moffat K, Moglich A. From dusk till dawn: one-plasmid systems for light-regulated gene expression. J. Mol. Biol. 2012;416:534–542. doi: 10.1016/j.jmb.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 19.Levskaya A, Weiner OD, Lim WA, Voigt CA. Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature. 2009;461:997–1001. doi: 10.1038/nature08446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muller K, Weber W. Optogenetic tools for mammalian systems. Mol. Biosyst. 2013;9:596–608. doi: 10.1039/c3mb25590e. [DOI] [PubMed] [Google Scholar]
- 21.Tabor JJ, Levskaya A, Voigt CA. Multichromatic control of gene expression in Escherichia coli. J. Mol. Biol. 2011;405:315–324. doi: 10.1016/j.jmb.2010.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kennedy MJ, Hughes RM, Peteya LA, Schwartz JW, Ehlers MD, Tucker CL. Rapid blue-light-mediated induction of protein interactions in living cells. Nat. Methods. 2010;7:973–975. doi: 10.1038/nmeth.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yazawa M, Sadaghiani AM, Hsueh B, Dolmetsch RE. Induction of protein-protein interactions in live cells using light. Nat. Biotechnol. 2009;27:941–945. doi: 10.1038/nbt.1569. [DOI] [PubMed] [Google Scholar]
- 24.Wang X, Chen X, Yang Y. Spatiotemporal control of gene expression by a light-switchable transgene system. Nat. Methods. 2012;9:266–269. doi: 10.1038/nmeth.1892. [DOI] [PubMed] [Google Scholar]
- 25.Ye HF, Daoud-El Baba M, Peng RW, Fussenegger M. A synthetic optogenetic transcription device enhances blood-glucose homeostasis in mice. Science. 2011;332:1565–1568. doi: 10.1126/science.1203535. [DOI] [PubMed] [Google Scholar]
- 26.Muller K, Engesser R, Metzger S, Schulz S, Kampf MM, Busacker M, Steinberg T, Tomakidi P, Ehrbar M, Nagy F, et al. A red/far-red light-responsive bi-stable toggle switch to control gene expression in mammalian cells. Nucleic Acids Res. 2013;41:e77. doi: 10.1093/nar/gkt002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rizzini L, Favory JJ, Cloix C, Faggionato D, O'Hara A, Kaiserli E, Baumeister R, Schafer E, Nagy F, Jenkins GI, et al. Perception of UV-B by the Arabidopsis UVR8 protein. Science. 2011;332:103–106. doi: 10.1126/science.1200660. [DOI] [PubMed] [Google Scholar]
- 28.Wu D, Hu Q, Yan Z, Chen W, Yan C, Huang X, Zhang J, Yang P, Deng H, Wang J, et al. Structural basis of ultraviolet-B perception by UVR8. Nature. 2012;484:214–219. doi: 10.1038/nature10931. [DOI] [PubMed] [Google Scholar]
- 29.Christie JM, Arvai AS, Baxter KJ, Heilmann M, Pratt AJ, O'Hara A, Kelly SM, Hothorn M, Smith BO, Hitomi K, et al. Plant UVR8 photoreceptor senses UV-B by tryptophan-mediated disruption of cross-dimer salt bridges. Science. 2012;335:1492–1496. doi: 10.1126/science.1218091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Favory JJ, Stec A, Gruber H, Rizzini L, Oravecz A, Funk M, Albert A, Cloix C, Jenkins GI, Oakeley EJ, et al. Interaction of COP1 and UVR8 regulates UV-B-induced photomorphogenesis and stress acclimation in Arabidopsis. EMBO J. 2009;28:591–601. doi: 10.1038/emboj.2009.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heijde M, Ulm R. Reversion of the Arabidopsis UV-B photoreceptor UVR8 to the homodimeric ground state. Proc. Natl Acad. Sci. USA. 2013;110:1113–1118. doi: 10.1073/pnas.1214237110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gibson DG. Enzymatic assembly of overlapping DNA fragments. Methods Enzymol. 2011;498:349–361. doi: 10.1016/B978-0-12-385120-8.00015-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fussenegger M, Mazur X, Bailey JE. A novel cytostatic process enhances the productivity of Chinese hamster ovary cells. Biotechnol. Bioeng. 1997;55:927–939. doi: 10.1002/(SICI)1097-0290(19970920)55:6<927::AID-BIT10>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 34.Fussenegger M, Moser S, Mazur X, Bailey JE. Autoregulated multicistronic expression vectors provide one-step cloning of regulated product gene expression in mammalian cells. Biotechnol. Prog. 1997;13:733–740. doi: 10.1021/bp970108r. [DOI] [PubMed] [Google Scholar]
- 35.Bacchus W, Lang M, El-Baba MD, Weber W, Stelling J, Fussenegger M. Synthetic two-way communication between mammalian cells. Nat. Biotechnol. 2012;30:991–996. doi: 10.1038/nbt.2351. [DOI] [PubMed] [Google Scholar]
- 36.Mitta B, Rimann M, Ehrengruber MU, Ehrbar M, Djonov V, Kelm J, Fussenegger M. Advanced modular self-inactivating lentiviral expression vectors for multigene interventions in mammalian cells and in vivo transduction. Nucleic Acids Res. 2002;30:e113. doi: 10.1093/nar/gnf112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schlatter S, Rimann M, Kelm J, Fussenegger M. SAMY, a novel mammalian reporter gene derived from Bacillus stearothermophilus alpha-amylase. Gene. 2002;282:19–31. doi: 10.1016/s0378-1119(01)00824-1. [DOI] [PubMed] [Google Scholar]
- 38.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-7-research0034. RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 41.Matsumura Y, Ananthaswamy HN. Toxic effects of ultraviolet radiation on the skin. Toxicol. Appl. Pharmacol. 2004;195:298–308. doi: 10.1016/j.taap.2003.08.019. [DOI] [PubMed] [Google Scholar]
- 42.Svobodova AR, Galandakova A, Sianska J, Dolezal D, Lichnovska R, Ulrichova J, Vostalova J. DNA damage after acute exposure of mice skin to physiological doses of UVB and UVA light. Arch. Dermatol. Res. 2012;304:407–412. doi: 10.1007/s00403-012-1212-x. [DOI] [PubMed] [Google Scholar]
- 43.Rausenberger J, Tscheuschler A, Nordmeier W, Wust F, Timmer J, Schafer E, Fleck C, Hiltbrunner A. Photoconversion and nuclear trafficking cycles determine phytochrome A's response profile to far-red light. Cell. 2011;146:813–825. doi: 10.1016/j.cell.2011.07.023. [DOI] [PubMed] [Google Scholar]
- 44.Heilmann M, Jenkins GI. Rapid reversion from monomer to dimer regenerates the Ultraviolet-B photoreceptor UV RESISTANCE LOCUS8 in intact Arabidopsis plants. Plant Physiol. 2013;161:547–555. doi: 10.1104/pp.112.206805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cadet J, Mouret S, Ravanat JL, Douki T. Photoinduced damage to cellular DNA: direct and photosensitized reactions. Photochem. Photobiol. 2012;88:1048–1065. doi: 10.1111/j.1751-1097.2012.01200.x. [DOI] [PubMed] [Google Scholar]
- 46.Pattison DI, Rahmanto AS, Davies MJ. Photo-oxidation of proteins. Photochem. Photobiol. Sci. 2012;11:38–53. doi: 10.1039/c1pp05164d. [DOI] [PubMed] [Google Scholar]
- 47.Schwarz T. UV light affects cell membrane and cytoplasmic targets. J. Photochem. Photobiol. B. 1998;44:91–96. doi: 10.1016/S1011-1344(98)00126-2. [DOI] [PubMed] [Google Scholar]
- 48.Raue A, Kreutz C, Maiwald T, Bachmann J, Schilling M, Klingmuller U, Timmer J. Structural and practical identifiability analysis of partially observed dynamical models by exploiting the profile likelihood. Bioinformatics. 2009;25:1923–1929. doi: 10.1093/bioinformatics/btp358. [DOI] [PubMed] [Google Scholar]
- 49.Raue A, Becker V, Klingmuller U, Timmer J. Identifiability and observability analysis for experimental design in nonlinear dynamical models. Chaos. 2010;20:045105. doi: 10.1063/1.3528102. [DOI] [PubMed] [Google Scholar]
- 50.Weber W, Fussenegger M. Molecular diversity—the toolbox for synthetic gene switches and networks. Curr. Opin. Chem. Biol. 2011;15:414–420. doi: 10.1016/j.cbpa.2011.03.003. [DOI] [PubMed] [Google Scholar]
- 51.Zoltowski BD, Schwerdtfeger C, Widom J, Loros JJ, Bilwes AM, Dunlap JC, Crane BR. Conformational switching in the fungal light sensor Vivid. Science. 2007;316:1054–1057. doi: 10.1126/science.1137128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rockwell NC, Su YS, Lagarias JC. Phytochrome structure and signaling mechanisms. Annu. Rev. Plant. Biol. 2006;57:837–858. doi: 10.1146/annurev.arplant.56.032604.144208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weber CC, Cai H, Ehrbar M, Kubota H, Martiny-Baron G, Weber W, Djonov V, Weber E, Mallik AS, Fussenegger M, et al. Effects of protein and gene transfer of the angiopoietin-1 fibrinogen-like receptor-binding domain on endothelial and vessel organization. J. Biol. Chem. 2005;280:22445–22453. doi: 10.1074/jbc.M410367200. [DOI] [PubMed] [Google Scholar]
- 54.Thurston G. Complementary actions of VEGF and angiopoietin-1 on blood vessel growth and leakage. J. Anat. 2002;200:575–580. doi: 10.1046/j.1469-7580.2002.00061.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thomas KA. Vascular endothelial growth factor, a potent and selective angiogenic agent. J. Biol. Chem. 1996;271:603–606. doi: 10.1074/jbc.271.2.603. [DOI] [PubMed] [Google Scholar]
- 56.Arsic N, Zentilin L, Zacchigna S, Santoro D, Stanta G, Salvi A, Sinagra G, Giacca M. Induction of functional neovascularization by combined VEGF and angiopoietin-1 gene transfer using AAV vectors. Mol. Ther. 2003;7:450–459. doi: 10.1016/s1525-0016(03)00034-0. [DOI] [PubMed] [Google Scholar]
- 57.Shyu KG, Chang H, Isner JM. Synergistic effect of angiopoietin-1 and vascular endothelial growth factor on neoangiogenesis in hypercholesterolemic rabbit model with acute hindlimb ischemia. Life Sci. 2003;73:563–579. doi: 10.1016/s0024-3205(03)00318-7. [DOI] [PubMed] [Google Scholar]
- 58.Yamauchi A, Ito Y, Morikawa M, Kobune M, Huang J, Sasaki K, Takahashi K, Nakamura K, Dehari H, Niitsu Y, et al. Pre-administration of angiopoietin-1 followed by VEGF induces functional and mature vascular formation in a rabbit ischemic model. J. Gene Med. 2003;5:994–1004. doi: 10.1002/jgm.439. [DOI] [PubMed] [Google Scholar]
- 59.Hindmarsh AC, Brown PN, Grant KE, Lee SL, Serban R, Shumaker DE, Woodward CS. SUNDIALS: suite of nonlinear and differential/algebraic equation solvers. ACM Trans. Math. Softw. 2005;31:363–396. [Google Scholar]
- 60.Coleman TF, Li YY. An interior trust region approach for nonlinear minimization subject to bounds. SIAM J. Optim. 1996;6:418–445. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.