Abstract
Glucocorticoid (GC) signaling synchronizes the circadian rhythm of individual peripheral cells and induces the expression of circadian genes, including Period1 (Per1) and Period2 (Per2). However, no GC response element (GRE) has been reported in the Per2 promoter region. Here we report the molecular mechanisms of Per2 induction by GC signaling and its relevance to the regulation of circadian timing. We found that GC prominently induced Per2 expression and delayed the circadian phase. The overlapping GRE and E-box (GE2) region in the proximal Per2 promoter was responsible for GC-mediated Per2 induction. The GRE in the Per2 promoter was unique in that brain and muscle ARNT-like protein-1 (BMAL1) was essential for GC-induced Per2 expression, whereas other GRE-containing promoters, such as Per1 and mouse mammary tumor virus, responded to dexamethasone in the absence of BMAL1. This specialized regulatory mechanism was mediated by BMAL1-dependent binding of the GC receptor to GRE in Per2 promoter. When Per2 induction was abrogated by the mutation of the GRE or E-box, the circadian oscillation phase failed to be delayed compared with that of the wild-type. Therefore, the current study demonstrates that the rapid Per2 induction mediated by GC is crucial for delaying the circadian rhythm.
INTRODUCTION
The circadian clock is composed of an endogenous rhythm that provides approximate 24-h timing cues to various biological activities, including metabolism, physiological processes and behavior. In mammals, the master pacemaker resides in the suprachiasmatic nucleus (SCN) of the hypothalamus. The SCN has self-sustainable oscillators and synchronizes the circadian timing of peripheral tissues by transmitting neuronal and humoral signals. Peripheral tissues also have endogenous clock machinery and are thus able to maintain the circadian rhythm without any external cues (1,2).
The endogenous circadian timing system consists of molecular feedback mechanisms, including the core and auxiliary loop. In the core loop, two basic helix-loop-helix (bHLH)/Per-Arnt-Sim (PAS)-containing transcription factors, circadian locomotor output cycles kaput (CLOCK) and brain and muscle ARNT-like protein-1 (BMAL1), bind to the E-box of clock-controlled genes, such as Period (Per) 1/2, Cryptochrome (Cry) 1/2, Rev-erb α and ROR α. The translated PERs and CRYs translocate to the nucleus and repress CLOCK:BMAL1 activity to return to the starting point. The auxiliary loop reinforces the circadian rhythm by regulating the rhythmic expression of BMAL1 through competitive binding of ROR α and REV-ERB α to RRE (Rev-erb/ROR binding element) on the Bmal1 promoter (3,4).
The endogenous clock does not have an exact 24-h period and has the flexibility to adjust to the phases of the environmental cycle, especially the light/dark photocycle. Light exposure at the time of early/late subjective night results in a delay/advance of the next activity cycle, which is represented as a phase response curve. It has been widely accepted that rapid expression of Per1 and Per2 plays a crucial role in the light-dependent resetting process. In particular, Per1 and Per2 are thought to mainly play a role in phase advance and phase delay, respectively; however, their actual roles only in the resetting process remain to be elucidated (5–9). These molecular events also occur in peripheral tissues and immortalized cell lines through synchronizing signals (10,11).
Glucocorticoid (GC) is a multifunctional hormone that regulates glucose and lipid metabolism, immune activity, the stress response and learning and memory (12–14). The level of GC displays a robust circadian rhythm, and the administration can reset the rhythmic phase of peripheral tissues and immortalized cells (11). Furthermore, the expression of GC receptors (GRs) in most peripheral cells, not in the SCN, enables the entrainment of peripheral clocks without any interference of the master clock. Therefore, GC is considered to be the best candidate for the synchronizing signal between the SCN and peripheral tissues.
During the synchronization process, Per1 and Per2 are rapidly induced and oscillate in a circadian fashion. Whereas GC regulates Per1 through the GC response element (GRE) in its promoter region, the molecular mechanisms of GC-mediated Per2 expression have not been clearly elucidated (15). Chromatin immunoprecipitation (ChIP)-sequencing analysis revealed that three GR-binding sites exist near Per2 gene (16). Several studies have shown that Per2 promoter region, in which the canonical GRE has not been found, is enough for GC responsiveness to Per2 (17,18). On the other hand, So et al. reported that the intronic GR-binding sequence (GBS) can confer GC responsiveness to Per2 (19).
Per2-knockout mice show a significantly shorter circadian rhythm or arrhythmicity of locomotor activities and have defects in the anticipation of feeding (7–9,20,21). Per2 is not only a component of the circadian oscillator but also functions as a mediator for the timed regulation of many types of metabolism. Direct interactions between PER2 and various nuclear receptors, including HNF4α, REV-ERBα and PPARα, enable the circadian oscillation of glucose and lipid metabolism (22). Besides, Per2-knockout mice, which lack 9th intron containing GC-responsive region, exhibited altered GC-induced glucose intolerance and insulin resistance, partly owing to increased leptin levels (19). It is also related to the timing of sleep. Per2-knockout mice wake earlier than wild-type (WT) mice, and the human PER2 S662G mutation prevents the phosphorylation of PER2 by CKIε, resulting in rapid degradation and nuclear export, which is observed in patients with familial advanced sleep phase syndrome (23–25). Therefore, the exact timing of Per2 expression may be critical for maintaining or restoring a physiology that is properly attuned to the environmental light–dark cycle.
In the present study, we investigated the molecular mechanisms underlying GC-mediated Per2 induction and its functional relevance to the regulation of the circadian rhythm. We provide evidence that BMAL1-dependent binding of GR to the overlapping GRE/E-box in the 5′ upstream region of Per2 gene induces the expression of Per2. Furthermore, we demonstrate that GC-mediated Per2 induction by this BMAL1-dependent GR mechanism is responsible for the phase delay.
MATERIALS AND METHODS
Cell culture
WT, Per2::luc knock-in, Per2−/− and Bmal1−/− mouse embryonic fibroblasts (MEFs) were spontaneously immortalized as previously described (26–29). Primary or immortalized cell lines were maintained in Dulbecco’s modified Eagle’s medium (Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin at 37°C in a humidified atmosphere containing 5% carbon dioxide (CO2).
Constructs
Per2 promoter region from −1671 to +26 from the transcription start site (TSS) was cloned into the GL3-basic vector (Promega, Madison, WI, USA), in which the PEST sequence was inserted into the C-terminal of the luciferase gene. The final construct was called Per2 (−1671)::dsluc. A series of 5′ deletion mutants were prepared from Per2 (−1671)::dsluc. E1, E2, GRE and palindromic GRE mutants were generated from Per2 (−271)::dsluc by the site-directed mutagenesis using the following primers: E1 mutant up: 5′-CGGGCTCAGCGCGCGCGGTGCTAGTTTCCACTATGTGACAGCGG-3′, E1 mutant dn: 5′-CCGCTGTCACATAGTGGAAACTAGCACCGCGCGCGCTGAGCCCG-3′; E2 mutant up: 5′-CGGCGAACATGGAGTTCCATAGACGTCTTATGTAAAG-3′, E2 mutant dn: 5′-CTTTACATAAGACGTCTATGGAACTCCATGTTCGCCG-3′; GRE mutant up: 5′-GAGGAACCCGGGCGGCTAGTATGGATATCCATGTGCGTCTTATG-3′, GRE mutant dn: 5′-CATAATACGCACATGGATATCCATACTAGCCGCCCGGGTTCCTC-3′; palindromic GRE up: 5′-GGAACCCGGGCGGAGAACATGGTGTTCTATGTGCGTCTTATG-3′, palindromic GRE dn: 5′-CATAAGACGCACATAGAACACCATGTTCTCCGCCCGGGTTCC-3′.
Luciferase assay
WT and Bmal1−/− MEFs were transfected using Lipofectamine PLUS reagents (Invitrogen). Cells were harvested after treatment with 0.1% ethanol or 1 μM dexamethasone, a synthetic GC (DEX; Sigma-Aldrich, St. Louis, MO, USA), for 10 h, which elicited the maximal induction. Luciferase activities were analyzed by the dual luciferase reporter assay system (Promega). Fold induction was calculated by dividing the luciferase activities in the DEX-treated group by those in the ethanol-treated group.
Recording of real-time luminescence
Per2::luc knock-in MEFs were cultured the day before the monitoring of luminescence. After treatment with the various compounds (0.1% ethanol, 1 μM DEX, 0.1% DMSO, 10 μM forskolin, 50% horse serum, 1 mM dibutyryl cyclic AMP [dbcAMP] and 1 μM ionomycin) for 2 h, and media were changed to normal culture media with 100 μM luciferin (Promega). Per2 (−271)::dsluc and its mutants were transfected into WT MEFs for 24 h. Bioluminescence was measured for 1 min for each dish at 10-min intervals with a real-time luminescence monitoring device (Kronos-Dio; ATTO Corporation, Tokyo, Japan) at 36°C in a humidified atmosphere containing 5% CO2. Data were normalized by the average of the initial minimum value.
Real-time reverse transcriptase-polymerase chain reaction
MEFs were seeded in six-well plates and harvested at the indicated times after treatment with 0.1% ethanol or 1 μM DEX (with or without 5 μM RU486; Sigma-Aldrich). Total RNA was isolated by the single-step acid guanidinium thiocyanate–phenol–chloroform method. Next, 2 μg of RNA was reverse-transcribed using Moloney murine leukemia virus reverse transcriptase (Promega). Real-time polymerase chain reaction was carried out in the presence of SYBR Green I. Gene expression levels were normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) levels. The primers used for real-time reverse transcriptase-polymerase chain reaction were as follows: Per1 up, 5′-GTGTCGTGATTAAATTAGTCAG-3′, Per1 dn, 5′-ACCACTCATGTCTGGGCC-3′; Per2 up, 5′-GCGGATGCTCGTGGAATCTT-3′, Per2 dn, 5′-GCTCCTTCAGGGTCCTTATC-3′; GAPDH up, 5′-CATGGCCTTCCGTGTTCCTA-3′, GAPDH dn, 5′-CCTGCTTCACCACCTTCTTGA-3′.
Chromatin immunoprecipitation
WT and Bmal1−/− MEFs were treated with 0.1% ethanol or 1 μM DEX for 1 h and exposed to 1% formaldehyde for 10 min. Cells were collected and were made to swell with hypotonic buffer (5 mM PIPES, pH 8.0, 85 mM KCl, 0.5% Triton X-100, 1 mM Phenylmethanesulfonyl fluoride (PMSF), 1 mM Na3VO4, 1 mM NaF and protease inhibitor cocktail). After centrifugation, the nuclear pellet was lysed in nuclear lysis buffer (1% Triton X-100, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate, 1 mM PMSF, 1 mM Na3VO4, 1 mM NaF and protease inhibitor cocktail). The chromatin was sheared off by sonication to <500 bp. Precleared samples were immunoprecipitated with normal rabbit serum and anti-GR (Santa Cruz Biotechnology, Santa Cruz, CA, USA). The immunoprecipitated DNA was purified with phenol/chloroform. For PCR, the primers were as follows: Per1 GRE up, 5′-AAGGCTGTGTGCATGTCCT-3′, Per1 GRE dn, 5′-AGAGGGAGGTGACGTCAAAG-3′; Per2 GRE up, 5′-GTGCCAGGTGAATGGAAGTC-3′, Per2 GRE dn, 5′-AGCTACGCTCGTCAATTGGT-3′.
Adenoviral transduction
Per2 recovery constructs were designed to express PER2-LUCIFERASE fusion protein under WT or mutant Per2 promoter (−271 to +26 from the TSS). Adenoviral constructs were generated according to the manufacturer’s instructions (Invitrogen). To determine the effects of the WT or mutant viruses, Per2−/− MEFs were seeded in 35-mm culture dishes, and the adenoviruses were added after 24 h. To analyze the circadian patterns of PER2, we recorded luminescence at 36°C with 5% CO2 using a real-time luminescence monitoring device after a 2-h DEX treatment.
Statistical analysis
Data were analyzed by 1-way analysis of variance with Tukey post hoc tests using GraphPad Prism software (GraphPad Prism Software, Inc., La Jolla, CA, USA). A p-value of <0.05 was considered to be significant.
RESULTS
GC signaling induces a prominent Per2 expression and the delayed circadian phase
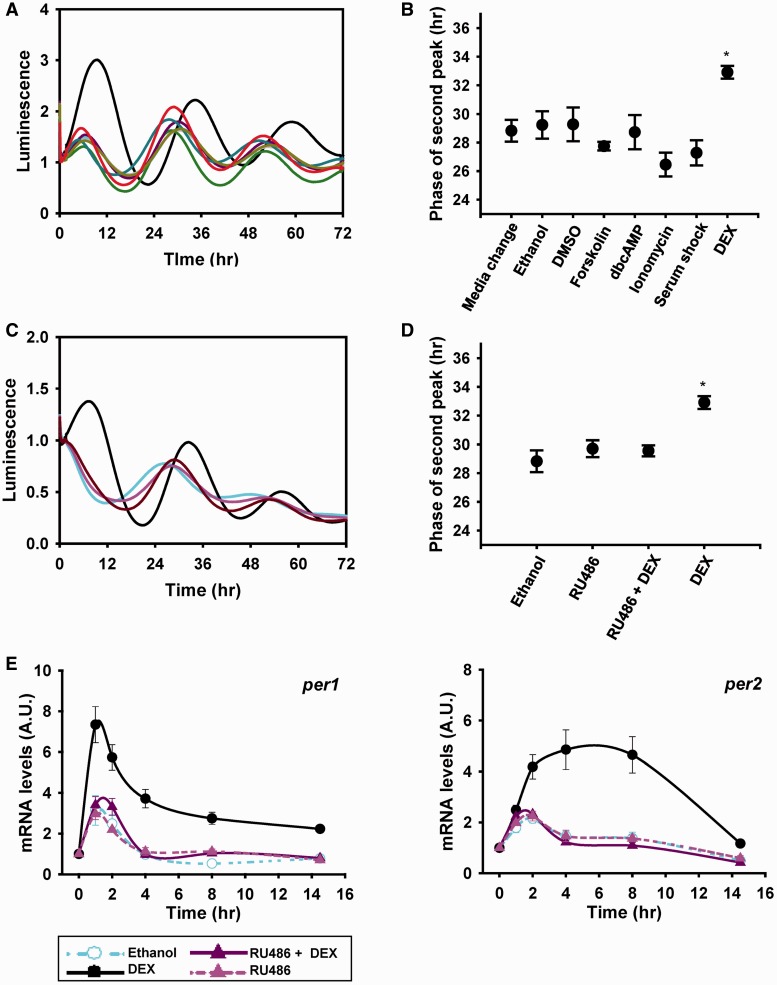
Many signaling pathways regulate clock gene expression (18,30). To examine the effects of GC on Per2 expression, we treated Per2::luc knock-in MEFs with DEX, forskolin, dbcAMP, ionomycin or horse serum and compared the oscillation profiles by recording real-time bioluminescence. DEX significantly increased PER2 protein levels, with a peak at approximately 10 h after treatment, whereas the other stimuli did not elicit gene expression (Figure 1A). Moreover, the phase of PER2 oscillation was significantly delayed in the DEX-treated group (Figure 1B). To verify the functional role of GR on DEX-mediated Per2 expression, Per2::luc knock-in MEFs were co-treated with RU486 (a GR antagonist). Both DEX-induced PER2 expression and the delay in phase were completely blocked by RU486 (Figure 1C and D). These results indicated that the intact GR activity was required for the regulation of Per2 expression and delay in the circadian phase. Consistent with this, the induction of Per1 and Per2 mRNA expression was also blocked by RU486 treatment, indicating that those genes were regulated by GC at the transcriptional level (Figure 1E). Interestingly, we also found that the induction profile of Per1 expression by DEX was more rapid than that of Per2. Moreover, Per1 expression was elicited by other signals that did not increase Per2 levels (Supplementary Figure S1) (15). Hence, although Per1 and Per2 are immediate early genes and have a certain redundancy of function in relation to circadian rhythm, they may produce different outputs depending on the combination of different signal transduction pathways activated. These results suggested that GC-induced Per2 expression was responsible for the phase delay. Therefore, we investigated the molecular mechanism of GC-mediated Per2 induction and its relevance to the regulation of circadian rhythms.
Figure 1.
DEX-induced prominent Per2 induction and the delayed phases of circadian rhythm. (A) Per2::luc knock-in MEFs were treated with the indicated synchronizing signals for 2 h and bioluminescence was measured. Ethanol (0.1%; sky line), DEX (1 μM; black line), DMSO (0.1%; gray line), forskolin (10 μM; green line), dbcAMP (1 mM; dark cyan line), ionomycin (1 μM; purple line), medium change (mustard-colored line) and serum shock (medium containing 50% horse serum; red line). (B) The phase of the second peak was measured for all stimuli in (A) (*P < 0.05; n = 3). (C) PER2::luc knock-in MEFs were treated with RU486 (5 μM) in combination with DEX for 2 h. Ethanol (sky line), DEX (black line), RU486 (pink line) and RU486 + DEX (dark red line). (D) The phase of the second peak was measured for all stimuli in (B) (*P < 0.05; n = 4). (E) WT MEFs were treated with ethanol or DEX for 2 h with or without RU486. Cells were harvested at the indicated times, and Per1 and Per2 mRNA levels were analyzed using real-time PCR. Each value was normalized to the GAPDH expression level. Values are the mean ± standard error of the mean (SEM) of three or four independent experiments performed in triplicates.
The GC-responsive region in the Per2 promoter
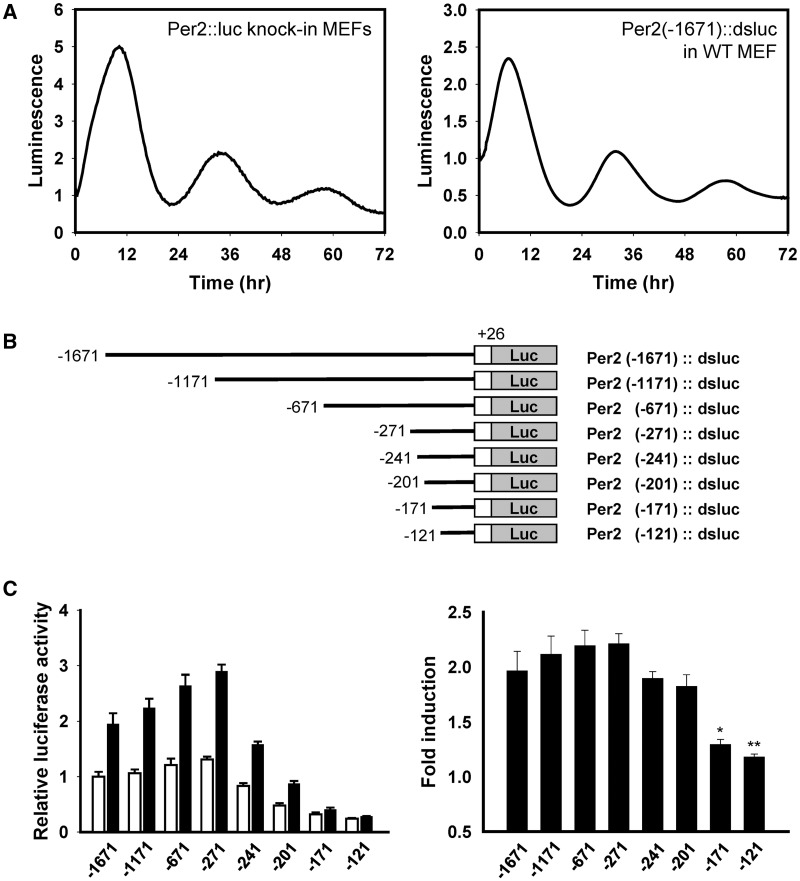
To determine whether GC-induced Per2 expression was regulated by the promoter region, we compared DEX-stimulated oscillation profiles of Per2::luc knock-in and Per2 promoter-driven luciferase (Figure 2A). Although the overall profile of Per2::luc knock-in was delayed, compared with that of Per2 promoter activity, the inductive and circadian oscillatory patterns exhibited similar profiles. Therefore, this result indicated that the 5′ upstream region of Per2 gene was sufficient for DEX-mediated Per2 induction and circadian oscillation. To narrow down the precise region required for Per2 induction, we generated serially deleted promoter constructs (Figure 2B). Although the basal promoter activities of Per2 (−241)::dsluc and Per2 (−201)::dsluc were decreased, the fold induction by DEX treatment was maintained up to Per2 (−201)::dsluc. However, when the region from −201 to −171 was deleted, the fold induction was completely blocked (Figure 2C). These effects were also demonstrated by the recording of real-time bioluminescence. The induction and oscillation profiles of Per2 (−271)::dsluc were almost the same as those of the full-length promoter, i.e. Per2 (−1671)::dsluc, but Per2 (−201)::dsluc and Per2 (−171)::dsluc were not induced (Supplementary Figure S2). Interestingly, these deletion mutants maintained their circadian oscillations, but without Per2 induction, implying that distinct mechanisms regulate the induction event and subsequent oscillations.
Figure 2.
Serial deletion analysis of the mouse Per2 promoter. (A) Comparison of the bioluminescence profiles after DEX treatment between the Per2::luc knock-in MEFs (left) and WT MEFs, which were transfected with Per2 promoter driven-luciferase of approximately 1.7 kb (right). (B) Schematic diagram of the mouse Per2 promoter serial deletion constructs. Nucleotides are numbered from the transcription start site. (C) DEX-responsiveness was analyzed in Per2 serial deletion mutants. WT MEFs were transfected with the serial deletion mutants and treated with ethanol (0.1%; white bar) or DEX (1 μM; black bar) for 10 h, which elicited maximal Per2 induction (left). Fold induction was calculated by dividing the luciferase activities in the DEX-treated group with those in the ethanol-treated group (right). Values are the mean ± SEM of three independent experiments performed in triplicates (*P < 0.05; **P < 0.01).
The conserved region containing the overlapping GRE and E-box is responsible for Per2 induction
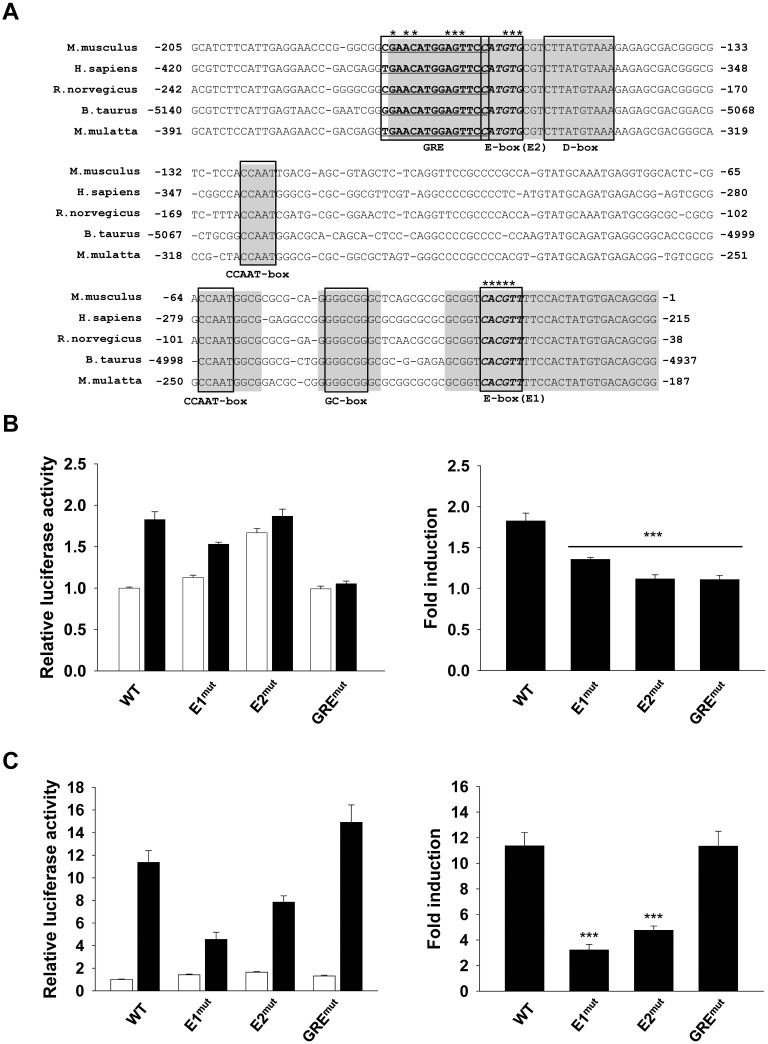
Circadian clock genes are highly conserved in mammals (31–33). To gain insight into the putative role of the GC-responsive region, we compared the 5′ upstream region of Per2 gene in several mammalian species. The proximal region of Per2 promoter was conserved in mice, humans, rats, cows and monkeys (Figure 3A). Sequence analysis revealed that the conserved regions contain a D-box, CCAAT-box, GC-box and a putative GRE, which differed slightly from the consensus sequence (5′-GGTACANNNTGT(T/C)CT-3′). We also found it to be overlapped with one of E-boxes (E2) by1-bp. To investigate the possibility of interaction between the two elements, we generated mutant constructs of Per2 (−271)::dsluc, the shortest construct exhibiting the same circadian oscillation and DEX responsiveness as the full-length promoter (Supplementary Figure S2). The basal levels of the GRE mutant (GREmut) and E1 mutant (E1mut) were similar to the WT; however, the E2 mutant (E2mut) exhibited increased basal activity. As expected, GREmut completely blocked responsiveness to DEX. Interestingly, DEX responsiveness was also blocked in E2mut, whereas E1mut only partially decreased the fold induction (Figure 3B). Therefore, these data suggested that the overlapping GRE/E2 (GE2) was crucial for DEX-mediated Per2 induction and E1 had only a moderate effect on this induction.
Figure 3.
Both GRE and E-box were required for Per2 induction. (A) Sequence alignment of the proximal Per2 promoter region of some mammalian species, including mouse (Mus musculus), human (Homo sapiens), rat (Rattus norvegicus), cow (Bos taurus) and monkey (Macaca mulatta). The conserved regions are shaded gray. Several putative cis-elements are indicated, and two E-boxes are named as E1 and E2. The asterisk indicates the mutated base. (B) DEX-responsiveness of GRE and E-box mutants. WT MEFs were transfected with WT or mutant constructs and treated with ethanol (0.1%; white bar) or DEX (1 μM; black bar) for 10 h (left). Fold induction was calculated by dividing the luciferase activities in the DEX-treated group with those in the ethanol-treated group (right). (C) CLOCK:BMAL1-mediated transcriptional activation of GRE and E-box mutants. Fold induction was calculated by dividing the luciferase activities in CLOCK:BMAL1-transfected group (black bar) with those in pcDNA3-transfected group (white bar). Values are the mean ± SEM of three independent experiments performed in quadruplicates (***P < 0.001).
To elucidate whether the functional interaction between the GRE and E-box was generally found in GC signaling, we tested the effects of the GRE-E-box interaction on the mechanism of Per1 induction. Whereas mutations in GREs completely blocked Per1 induction, mutations in E-boxes did not affect this event (Supplementary Figure S3). These results indicated that the functional interaction between GRE and E-box, as shown in the Per2 promoter, was not a general mechanism of GC signaling.
To confirm that E-boxes on the Per2 promoter were responsible for the binding of the circadian clock machinery, we examined CLOCK:BMAL1-mediated transcriptional activities of these mutants. E1mut and E2mut partially impaired CLOCK:BMAL1 activities, whereas GREmut did not, despite its proximity to E2 (Figure 3C). From these results, we found that whereas both E-boxes had functional roles in CLOCK:BMAL1-mediated transcriptional activity, GRE was not involved in mediating this effect. These results implied that E-box-mediated transcriptional activity was closely related to GC-induced Per2 expression.
BMAL1 is essential for GC-mediated Per2 induction
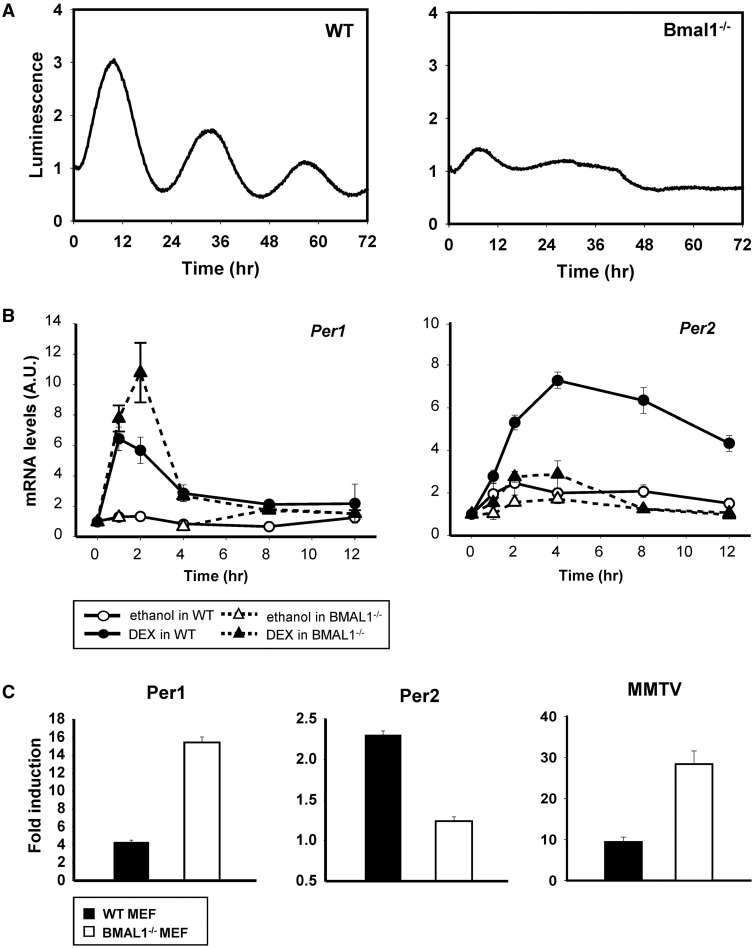
The insights obtained from our determination of the functional interaction between the GRE and E-box suggested that BMAL1 was involved in GC-mediated Per2 induction. To test this hypothesis, we generated Per2::luc knock-in MEFs of two genotypes (WT and Bmal1−/−) and compared the circadian profiles of these MEFs after DEX treatment. As shown in Figure 4A, Bmal1−/− MEFs did not show circadian oscillation or responsiveness to DEX. Consistent with this result, Per2 mRNA was not induced by DEX treatment in Bmal1−/− MEFs, whereas Per1 mRNA was increased (Figure 4B). To determine whether the abrogation of Per2 induction in Bmal1−/− MEFs may result from Per2 promoter activities, we performed reporter assays in WT and Bmal1−/− MEFs (Figure 4C). Whereas Per1 promoter-driven luciferase activities were still increased by DEX treatment in the absence of Bmal1, the induction of Per2 promoter driven-luciferase activity was abolished in Bmal1−/− MEFs. To further examine these properties in relation to GRE-dependent mechanisms, we also tested mouse mammary tumor virus (MMTV) promoter activities in WT and Bmal1−/− MEFs. Similar to the result in the case of Per1, MMTV promoter activities were increased by DEX treatment in Bmal1−/− MEFs. These data indicated that BMAL1 was critical for DEX-induced Per2 expression, and this was distinct from the general GRE mechanism of action.
Figure 4.
BMAL1 was necessary for Per2 induction. (A) Per2::luc knock-in MEFs with a WT (left) or Bmal1−/− (right) genetic background. Bioluminescence was recorded after DEX (1 μM) treatment. (B) WT and Bmal1−/− MEFs were treated with ethanol (0.1%) or DEX (1 μM) for 2 h. Cells were harvested at the indicated times and Per1 and Per2 mRNA levels were analyzed by real-time PCR. Each value was normalized to the GAPDH expression level. Values are the mean±SEM of three or four independent experiments. (C) Per1-luc, Per2-luc and MMTV-luc were transfected into WT (black bar) or Bmal1−/− MEFs (white bar), and cells were treated with ethanol (0.1%) or DEX (1 μM) for 10 h. Luciferase activities were normalized to the renilla luciferase activities, and the fold induction was calculated by dividing the luciferase activities in DEX-treated group with those in ethanol-treated group. Values are the mean ± SEM of three or four independent experiments performed in triplicates.
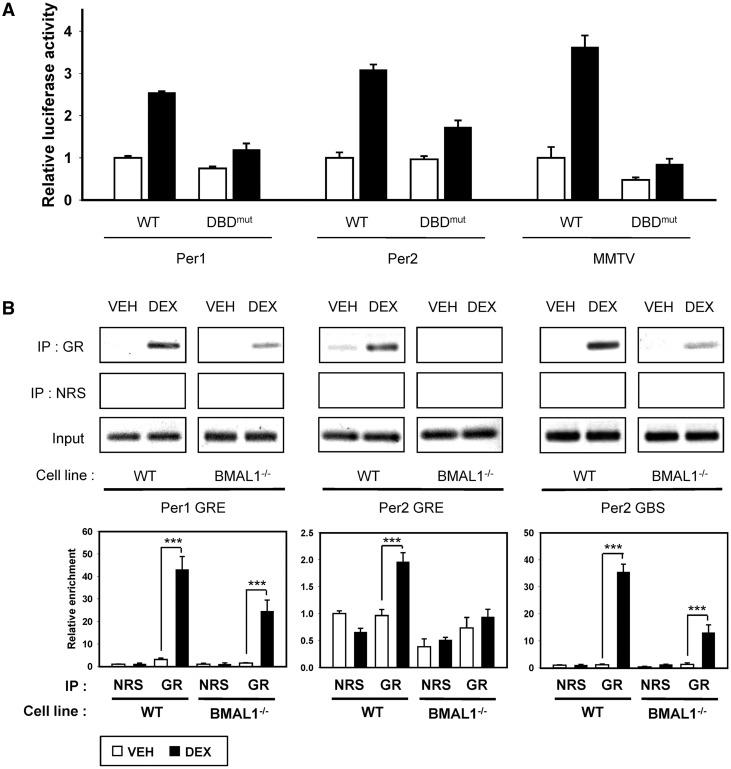
BMAL1 regulates GR occupancy in the GRE of Per2 promoter
To further elucidate the effects of GR on Per2 promoter, we performed a reporter assay with the DNA binding mutant of GR (DBDmut) (12,34). DBDmut decreased DEX responsiveness of Per2, MMTV and Per1 promoters (Figure 5A). To investigate the direct binding of GR to Per2 promoter region and the role of BMAL1 in Per2 induction, we performed ChIP assays in WT and Bmal1−/− MEFs. The recruitment of GR to the GRE in Per2 promoter was increased by DEX treatment in WT MEFs but was completely absent in Bmal1−/− MEFs (Figure 5B). However, GR occupancy of Per1 promoter was increased not only in WT MEFs but also in Bmal1−/− MEFs. We also analyzed GR occupancy of the GBS in the Per2 intron region (19). Similar to the effect observed in Per1 gene, GR occupancy in the intronic GBS of Per2 gene was increased by DEX treatment in WT and Bmal1−/− MEFs. Although the binding of GR to both Per1 GRE and Per2 GBS was increased by DEX treatment in both MEFs, the amount of immunoprecipitated DNA in Bmal1−/− MEFs was decreased than that in WT MEFs, possibly owing to low levels of GR in Bmal1−/− MEFs (Supplementary Figure S5). Consistent with the results of the ChIP assay, GBS-mediated luciferase activities were increased by DEX treatment in WT and Bmal1−/− MEFs (Supplementary Figure S4). These data suggested that BMAL1 was required for the binding of GR to the GRE in Per2 promoter but was not critical for the binding of GR to the GBS in Per2 intronic region. Considering the complete absence of Per2 induction in Bmal1−/− MEFs, GE2 in Per2 promoter was epistatic to the intronic GBS in GC-mediated Per2 induction.
Figure 5.
BMAL1-dependent binding of GR to Per2 promoter. (A) WT GR or DBDmut with MMTV-luc, Per1-luc and Per2-luc were transfected into WT MEFs. Cells were treated with ethanol (0.1%; white bar) or DEX (1 μM; black bar) for 10 h. Luciferase activities were normalized to renilla luciferase activities. Values are the mean ± SEM of three independent experiments performed in triplicates. (B) WT and Bmal1−/− MEFs were treated with ethanol (0.1%; VEH) or DEX (1 μM; DEX) for 1 h. Chromatin was extracted from the harvested cells, and chromatin immunoprecipitation assays were performed with normal rabbit serum (NRS) or anti-GR. Immunoprecipitated DNA was analyzed using the primer sets for Per1 GRE, Per2 GRE and Per2 GBS. Enrichment of GR binding was measured by the agarose gel electrophoresis (upper panel) and quantified by real-time PCR (lower panel). Values are the mean ± SEM of three independent experiments (***P < 0.001).
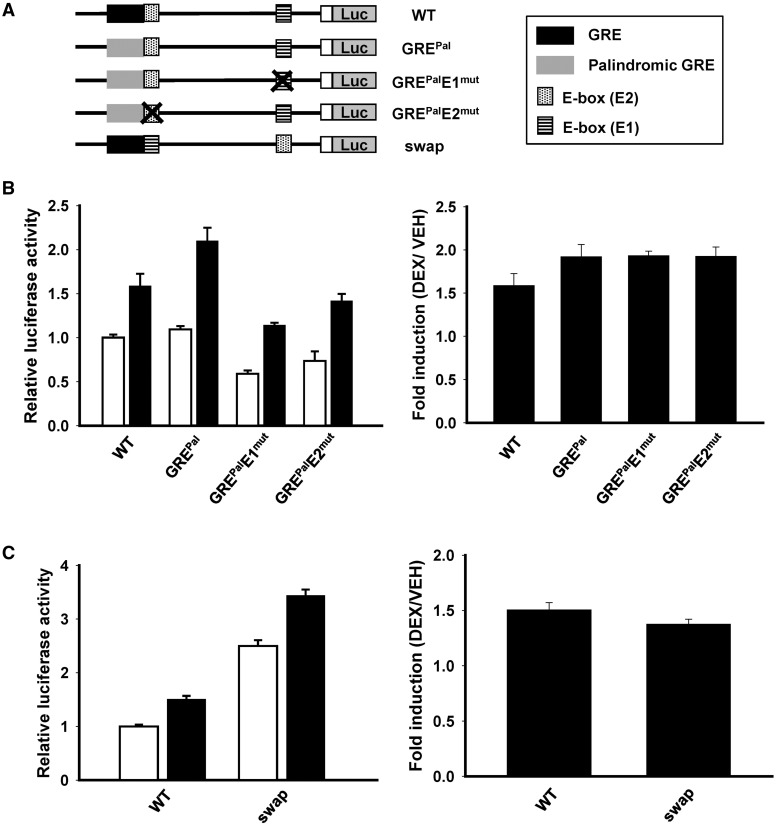
An imperfect palindromic GRE confers the reliance on the overlapping E-box
The GRE in Per2 promoter differs from the palindromic GRE sequence by 4 bp (5′-AGAACANNNTGTTCT-3′). It has been reported that GR has a higher binding affinity for the palindromic GRE than for the imperfect palindromic sequence and that the palindromic GRE decreases the need for the activities of accessory factors (35). To test the possibility that the imperfect palindromic sequence of Per2 GRE leads to dependency on the E-box, we generated palindromic GRE mutants (GREPal) with E1 or E2 mutations (Figure 6A). When the Per2 GRE sequence was replaced with the palindromic sequence, DEX responsiveness was still maintained. Interestingly, additional mutations of E-boxes to GREPal (GREPalE1mut and GREPalE2mut) did not decrease the GC responsiveness of Per2 promoter, although their basal promoter activities were reduced (Figure 6B). This is different from the original Per2 promoter. Therefore, these results suggested that the imperfect GRE sequence increased the reliance on the transcription factor BMAL1.
Figure 6.
The effects of the palindromic GRE and swapped E-boxes on GC-mediated Per2 induction. (A) Schematic diagram of Per2 promoter mutants. (B and C) DEX-responsiveness of Per2 promoter mutants indicated in (A). WT MEFs that were transfected with the reporters were treated with ethanol (0.1%) or DEX (1 μM) for 10 h. Fold induction was calculated by dividing the luciferase activities in the DEX-treated group with those in the ethanol-treated group. Data are represented as the mean ± SEM. Three independent experiments were done in triplicates for each condition.
In addition, we also swapped two E-boxes to analyze the role of the E-box in the function of the GRE (Figure 6A). The swapped construct showed increased basal activities but maintained the fold induction (Figure 6C). Hence, the sequence of E-box only controlled the basal promoter activity and was not informative for GRE action. Rather, it is likely that the distance between GRE and E-box is an important factor for GE2 elements.
Impaired Per2 induction cannot delay the circadian phase
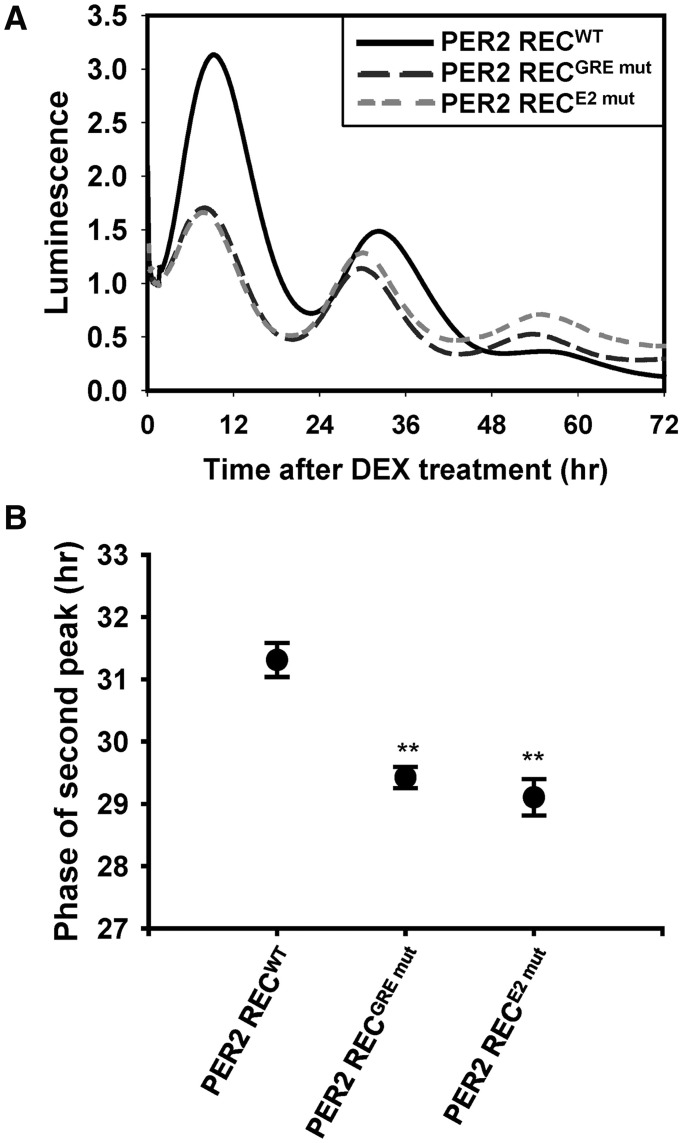
To evaluate the physiological relevance of Per2 induction, we conducted a rescue experiment. First, we examined the oscillation patterns of Per2 promoter constructs. After stimulation with DEX, all the reporter constructs displayed circadian oscillation patterns; however, this induction was not observed in GREmut or E2mut (Supplementary Figure S6A). The mutant reporters exhibited a slightly advanced phase compared with the WT reporter, as previously described (Supplementary Figure S6B) (36,37). Using these constructs, we generated adenoviruses expressing the PER2::LUC fusion protein (PER2 REC) driven by WT or mutant promoters (GREmut and E2mut). Per2−/− MEFs were recovered by these viruses, and the bioluminescence was recorded after DEX treatment. The PER2 RECWT responded to DEX treatment and exhibited similar profiles to PER2::luc knock-in MEFs. However, PER2 RECGRE mut or PER2 RECE2 mut did not display inductive profiles and failed to exhibit a delay in the circadian phase compared with the WT (Figure 7A and B). Although mutant reporter activities were slightly advanced as compared with WT reporter activity, the functional recovery constructs displayed a more pronounced difference between WT and mutant reporters, indicating that the induction of the PER2 regulated the circadian phase (Supplementary Figure S6A and B). Therefore, these data suggested that DEX-mediated Per2 induction was a crucial step in determining the phase of the circadian rhythm.
Figure 7.
Per2 induction mutants cannot delay the circadian rhythm (A) Per2−/− MEFs were recovered with Per2 adenoviruses, in which the expression of Per2 was regulated by its own promoter. WT (Per2 RECWT; solid line), GRE mutant (Per2 RECGRE mut; long dashed line) and E2 mutant (Per2 RECE2 mut; short dashed line). Bioluminescence was recorded after 2 h of DEX treatment. (B) The phase of the second peak was measured (**P < 0.01). Data are represented as the mean ± SEM from three independent experiments.
DISCUSSION
The present study investigated the molecular mechanisms of Per2 induction by GC signaling and its regulatory effects on circadian rhythms. The induction of Per2 expression by DEX was mainly mediated by the overlapping GRE and E-box (GE2). From our understanding of this molecular mechanism, we demonstrated that Per2 induction was crucial for delaying the circadian phase.
Among the clock proteins, the fluctuation of PER expression is thought to be crucial for generating circadian oscillation (38–41). In this regard, it is expected that the resetting process requires an alteration in PER expression to generate new rhythms at the initial stage. In fact, various resetting stimuli increase Per1 and Per2 levels, whereas other resetting signals, such as glucose and exercise, lower their expression, respectively, both in vitro and in vivo (42–44). Several studies have endeavored to substantiate the importance of the induction of Per1 and Per2 in the light-induced phase resetting using Per1- or Per2-knockout mice, but the exact roles of Per1 and Per2 in the resetting process remain controversial. This inconsistency may arise from the fact that the mutant mice used by the research groups were different, and global Per-knockout mice have additional defects besides resetting function defects (5–8). Thus, to understand the functional importance of the rapid response of Per1 and Per2, only the specific site responsible for the resetting signal should be mutated. In this respect, the present study attempted to clarify the molecular mechanisms of Per2 induction and thereby investigate the precise roles of the initial response of Per2 in the regulation of subsequent circadian rhythms.
GC-induced Per2 expression was unique in that it required the additional transcription factor BMAL1 for the binding of the GR to the GE2 element. Previous studies revealed that GRE activity was interrupted by other overlapping transcription factor binding sites. For instance, the GRE in the osteocalcin gene promoter, which overlaps with all the sequences of the TATA boxes, blocks the binding of the general transcription factor IID and represses transcription (45). In a case similar to that of Per2, the cAMP-responsive element (CRE) and E-box (CRE/E-box) of the cyclooxygenase-2 (COX-2) gene overlap by 2 bp. Endotoxin-induced COX-2 gene expression accompanies the activity of the CRE/E-box, in which each element induces a higher level of gene expression than the overlapped sequence (46). In contrast, neither GRE nor E2 of Per2 responded to DEX alone, and only their interaction induced Per2 expression (Figure 3B). To the best of our knowledge, this is the first report of a positive regulatory mechanism, in which the GRE overlaps with other transcription factor binding elements. From a structural viewpoint, this mechanism suggests the cooperative binding of overlapping elements. A previous study showed that GR binds to CLOCK in a ligand-dependent manner, and we also observed direct binding of BMAL1 and GR (data not shown), indicating that the physical interaction between GR and CLOCK/BMAL1 heterodimer can occur (47). However, these data cannot answer the question of how this mechanism applied to GE2 on Per2 promoter. Owing to the immediate vicinity of GRE and E2 elements, it is likely that BMAL1 and GR do not bind to GE2 at the same time, but instead do sequentially. Nevertheless, there still exists the possibility that the inherent GRE sequence in Per2 promoter enables the transcription factors to bind at the same time (48). To clarify this structural issue, further studies are needed.
Moreover, this mechanism differs from the general GRE action. A previous report suggested that the canonical GRE activity is repressed by CLOCK:BMAL1 through the histone acetyl transferase activities of CLOCK, and consistent with this, we found that the MMTV or Per1 promoter can respond to DEX, regardless of the activity of BMAL1 (Figure 4A) (47). In contrast, Per2 cannot be induced in the absence of BMAL1, implying that this mechanism is clearly distinct from the canonical GRE mechanism. It is possible that the imperfect palindromic GRE sequence in Per2 promoter increases the need for the involvement of other transcription factors (Figure 6B). In fact, many genes have GRE and E-box in tandem (49). This suggests that these genes are regulated in a gene-specific manner according to the fidelity of the sequences and the distance between the GRE and E-box.
We previously suggested that the activation of CLOCK:BMAL1 is involved in serum shock-induced Per1 expression (50,51). Moreover, the present study revealed that BMAL1 was necessary in GC-mediated Per2 induction. Therefore, the studies reported by our group suggest that the CLOCK:BMAL1 heterodimer regulates several pathways involved in the resetting process, although it is mainly shown to be a positive regulator of circadian clock genes. Furthermore, each of the resetting signals is likely to generate diverse phases of the circadian rhythm by modulating the expression levels of Per1 and Per2 according to the gene-specific functional interaction between the CLOCK:BMAL1 heterodimer and the specific mediator of the resetting signals, such as GR and CRE-binding protein (52).
Previous studies have reported that the regulation of circadian timing is achieved by a fine-tuning of circadian clock components at the transcriptional level. At least three cis-elements, including the morning-time element (E-box), day-time element (D-box) and night-time elements (RRE), are thought to control this timing. For instance, the peak expression of Cry1, which is adjusted by the combinatorial regulation of D-box and RRE in addition to E-box, exhibits a certain delay relative to that of Per2, which is mainly controlled by E-box (53–55). Furthermore, we propose that GRE is another regulatory element involved in the modulation of circadian timing. It is thought that when the resetting stimuli, i.e. GC, is given, the cooperative interaction between GRE and E-box determines the phase of Per2. Indeed, the phase-delaying role of E2 has been previously suggested in several studies. Akashi et al. (36) reported that the proximal Per2 promoter region consists of a phase-delaying region and an oscillation-driving region. Using a serial deletion analysis of the Per2 promoter, they found that the phase-delaying region comprises from −386 to −106 from the TSS and that the oscillation-driving region comprises a region from −105 to +1. The oscillation-driving region contains a non-canonical E-box (E1), which is an essential and sufficient element for the generation of rhythm, whereas the phase-delaying region contains GE2 (56). Yamajuku et al. (37) demonstrated similar results in that the region from −161 to −143, which contains E2, was shown to be responsible for the phase delay. Although they used different stimuli (i.e. serum shock and dbcAMP), the E2-containing region was thought to be responsible for the phase delay. Consistent with this, we also found that E2mut advances the circadian rhythm of Per2 in DEX-treated cells. Therefore, on the basis of our data and previous studies, DEX-dependent induction of Per2 expression likely accompanies the long-lasting activation of GE2.
Although the mutated recovery constructs failed to delay the circadian rhythm, the phase difference between the WT and mutant constructs was not as large as that of Per2::luc knock-in cells, which were treated with DEX and other signaling molecules (Figures 1A and 7A). There are a number of plausible reasons for this. First, clock genes other than Per2 can affect the resetting process. Previous studies reported that GC induces Per1 expression and downregulates Rev-erb α (15,57). Although we found that Per1 expression, which was stimulated by different signaling molecules, did not exhibit remarkable differences between treatments, there was a slight time lag between the stimuli, which may support the phase delaying effect (Supplementary Figure S1) (30). GC-induced downregulation of Rev-erb α can also directly or indirectly affect phase regulation. Second, it is likely that the excluded region in our recovery constructs was involved in this mechanism. A previous report showed that Per2 induction does not occur in Per2Brdm1 cells, which lack a genomic region of approximately 2 kb containing a GBS (19); however, our constructs did not contain a GBS. Therefore, it is feasible that Per2 induction may be collectively regulated by the interaction between the two elements, GE2 and GBS. To accurately understand how GE2 and GBS regulate GC-mediated Per2 induction, further studies need to be conducted using modified bacterial artificial chromosomes or the whole genome.
The proximal Per2 promoter region is highly conserved in mammals and zebrafish (58). This region includes a non-canonical E-box (E1) that is sufficient for self-sustained circadian rhythm generation and a D-box that is implicated in higher amplitude generation (37,56). In addition, we found that GE2 in this region is also conserved in mammals, but only 5 bp (CATGG) in the middle of the GRE sequence is conserved in zebrafish (58). Although zebrafish has a hypothalamic–pituitary–inter-renal axis that regulates cortisol release in fish, its exact role in the circadian rhythm remains largely unknown. Considering that its peripheral cells can respond to the light directly and that the zebrafish Per2 rhythm depends on the light-dark cycle, it is conceivable that the GC-regulated Per2 induction mechanism evolved because peripheral tissues do not receive direct photic input (59).
Many people suffer from sleep disturbances as well as metabolic and cardiovascular disorders in relation to chronobiological problems that arise under various circumstances, including jet lag and shift work. These disturbances are related to phase misalignment in the master and/or peripheral clocks (60–62). GC is generally accepted as a strong synchronizer of the SCN and peripheral tissues, and an altered GC rhythm is closely related to a variety of circadian disorders. People suffering from Cushing syndrome, diabetes, depression, obesity, Alzheimer’s disease and metabolic syndrome exhibit an altered GC rhythm and abnormal circadian physiology (14,63–66). This might be due to a dysregulation of GC-regulated clock genes, including Per1 and Per2 (67).
In conclusion, this study provides evidence that Per2 induction is responsible for circadian phase delay through a novel regulatory mechanism. It is expected that these findings will help in the effort to achieve a better understanding of the physiological changes that occur in circadian rhythm disorders.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–6.
FUNDING
Korea Ministry of Education, Science, and Technology (MEST) through the Brain Research Center of the 21st Century Frontier Research Program [2012K001134]. Brain Korea 21 Research Fellowships from the MEST (to S.C. and N.P.). Funding for open access charge: Seoul National University.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Dr Joseph S. Takahashi (UT Southwestern Medical Center, Dallas, TX, USA) for kindly providing Per2::luc knock-in mice and Dr Masataka Kinjo (Hokkaido University, Hokkaido, Japan) for kindly providing GR constructs.
REFERENCES
- 1.Welsh DK, Takahashi JS, Kay SA. Suprachiasmatic nucleus: cell autonomy and network properties. Annu. Rev. Physiol. 2010;72:551–577. doi: 10.1146/annurev-physiol-021909-135919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418:935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- 3.Dibner C, Schibler U, Albrecht U. The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annu. Rev. Physiol. 2010;72:517–549. doi: 10.1146/annurev-physiol-021909-135821. [DOI] [PubMed] [Google Scholar]
- 4.Mohawk J, Green C, Takahashi J. Central and peripheral circadian clocks in mammals. Annu. Rev. Neurosci. 2012;35:445–462. doi: 10.1146/annurev-neuro-060909-153128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bae K, Weaver DR. Light-induced phase shifts in mice lacking mPER1 or mPER2. J. Biol. Rhythms. 2003;18:123–133. doi: 10.1177/0748730403252248. [DOI] [PubMed] [Google Scholar]
- 6.Cermakian N, Monaco L, Pando MP, Dierich A, Sassone-Corsi P. Altered behavioral rhythms and clock gene expression in mice with a targeted mutation in the Period1 gene. EMBO J. 2001;20:3967–3974. doi: 10.1093/emboj/20.15.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Albrecht U, Zheng B, Larkin D, Sun ZS, Lee CC. mPer1 and mPer2 are essential for normal resetting of the circadian clock. J. Biol. Rhythms. 2001;16:100–104. doi: 10.1177/074873001129001791. [DOI] [PubMed] [Google Scholar]
- 8.Wakamatsu H, Takahashi S, Moriya T, Inouye ST, Okamura H, Akiyama M, Shibata S. Additive effect of mPer1 and mPer2 antisense oligonucleotides on light-induced phase shift. Neuroreport. 2001;12:127–131. doi: 10.1097/00001756-200101220-00033. [DOI] [PubMed] [Google Scholar]
- 9.Pendergast JS, Friday RC, Yamazaki S. Photic entrainment of period mutant mice is predicted from their phase response curves. J. Neurosci. 2010;30:12179–12184. doi: 10.1523/JNEUROSCI.2607-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Balsalobre A, Damiola F, Schibler U. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell. 1998;93:929–937. doi: 10.1016/s0092-8674(00)81199-x. [DOI] [PubMed] [Google Scholar]
- 11.Balsalobre A, Brown SA, Marcacci L, Tronche F, Kellendonk C, Reichardt HM, Schütz G, Schibler U. Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science. 2000;289:2344–2347. doi: 10.1126/science.289.5488.2344. [DOI] [PubMed] [Google Scholar]
- 12.Kassel O, Herrlich P. Crosstalk between the glucocorticoid receptor and other transcription factors: molecular aspects. Mol. Cell. Endocrinol. 2007;275:13–29. doi: 10.1016/j.mce.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 13.Popoli M, Yan Z, McEwen BS, Sanacora G. The stressed synapse: the impact of stress and glucocorticoids on glutamate transmission. Nat. Rev. Neurosci. 2011;13:22–37. doi: 10.1038/nrn3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chung S, Son GH, Kim K. Circadian rhythm of adrenal glucocorticoid: its regulation and clinical implications. Biochim. Biophys. Acta. 2011;1812:581–591. doi: 10.1016/j.bbadis.2011.02.003. [DOI] [PubMed] [Google Scholar]
- 15.Yamamoto T, Nakahata Y, Tanaka M, Yoshida M, Soma H, Shinohara K, Yasuda A, Mamine T, Takumi T. Acute physical stress elevates mouse period1 mRNA expression in mouse peripheral tissues via a glucocorticoid-responsive element. J. Biol. Chem. 2005;280:42036–42043. doi: 10.1074/jbc.M509600200. [DOI] [PubMed] [Google Scholar]
- 16.Reddy TE, Pauli F, Sprouse RO, Neff NF, Newberry KM, Garabedian MJ, Myers RM. Genomic determination of the glucocorticoid response reveals unexpected mechanisms of gene regulation. Genome Res. 2009;19:2163–2171. doi: 10.1101/gr.097022.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hattori M-A. Hormonal Regulation of Circadian Pacemaker in Ovary and Uterus. In: Aimaretti G, editor. Update on Mechanisms of Hormone Action - Focus on Metabolism, Growth and Reproduction. InTech; 2011. pp. 217–232. [Google Scholar]
- 18.Izumo M, Sato TR, Straume M, Johnson CH. Quantitative analyses of circadian gene expression in mammalian cell cultures. PLoS Comput. Biol. 2006;2:e136. doi: 10.1371/journal.pcbi.0020136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.So AY, Bernal TU, Pillsbury ML, Yamamoto KR, Feldman BJ. Glucocorticoid regulation of the circadian clock modulates glucose homeostasis. Proc. Natl Acad. Sci. USA. 2009;106:17582–17587. doi: 10.1073/pnas.0909733106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zheng B, Larkin DW, Albrecht U, Sun ZS, Sage M, Eichele G, Lee CC, Bradley A. The mPer2 gene encodes a functional component of the mammalian circadian clock. Nature. 1999;400:169–173. doi: 10.1038/22118. [DOI] [PubMed] [Google Scholar]
- 21.Feillet CA, Ripperger JA, Magnone MC, Dulloo A, Albrecht U, Challet E. Lack of food anticipation in Per2 mutant mice. Curr. Biol. 2006;16:2016–2022. doi: 10.1016/j.cub.2006.08.053. [DOI] [PubMed] [Google Scholar]
- 22.Schmutz I, Ripperger JA, Baeriswyl-Aebischer S, Albrecht U. The mammalian clock component PERIOD2 coordinates circadian output by interaction with nuclear receptors. Genes Dev. 2010;24:345–357. doi: 10.1101/gad.564110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Toh KL, Jones CR, He Y, Eide EJ, Hinz WA, Virshup DM, Ptácek LJ, Fu YH. An hPer2 phosphorylation site mutation in familial advanced sleep phase syndrome. Science. 2001;291:1040–1043. doi: 10.1126/science.1057499. [DOI] [PubMed] [Google Scholar]
- 24.Vanselow K, Vanselow JT, Westermark PO, Reischl S, Maier B, Korte T, Herrmann A, Herzel H, Schlosser A, Kramer A. Differential effects of PER2 phosphorylation: molecular basis for the human familial advanced sleep phase syndrome (FASPS) Genes Dev. 2006;20:2660–2672. doi: 10.1101/gad.397006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kopp C, Albrecht U, Zheng B, Tobler I. Homeostatic sleep regulation is preserved in mPer1 and mPer2 mutant mice. Eur. J. Neurosci. 2002;16:1099–1106. doi: 10.1046/j.1460-9568.2002.02156.x. [DOI] [PubMed] [Google Scholar]
- 26.Bae K, Jin X, Maywood ES, Hastings MH, Reppert SM, Weaver DR. Differential functions of mPer1, mPer2, and mPer3 in the SCN circadian clock. Neuron. 2001;30:525–536. doi: 10.1016/s0896-6273(01)00302-6. [DOI] [PubMed] [Google Scholar]
- 27.Yoo SH, Yamazaki S, Lowrey PL, Shimomura K, Ko CH, Buhr ED, Siepka SM, Hong HK, Oh WJ, Yoo OJ. PERIOD2: LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc. Natl Acad. Sci. USA. 2004;101:5339–5346. doi: 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bunger MK, Wilsbacher LD, Moran SM, Clendenin C, Radcliffe LA, Hogenesch JB, Simon MC, Takahashi JS, Bradfield CA. Mop3 Is an essential component of the Master circadian pacemaker in mammals. Cell. 2000;103:1009–1017. doi: 10.1016/s0092-8674(00)00205-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Todaro GJ, Green H. Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J. Cell Biol. 1963;17:299–313. doi: 10.1083/jcb.17.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Balsalobre A, Marcacci L, Schibler U. Multiple signaling pathways elicit circadian gene expression in cultured Rat-1 fibroblasts. Curr. Biol. 2000;10:1291–1294. doi: 10.1016/s0960-9822(00)00758-2. [DOI] [PubMed] [Google Scholar]
- 31.Huang N, Chelliah Y, Shan Y, Taylor CA, Yoo SH, Partch C, Green CB, Zhang H, Takahashi JS. Crystal structure of the heterodimeric CLOCK: BMAL1 transcriptional activator complex. Science. 2012;337:189–194. doi: 10.1126/science.1222804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zylka MJ, Shearman LP, Weaver DR, Reppert SM. Three period homologs in mammals: differential light responses in the suprachiasmatic circadian clock and oscillating transcripts outside of Brain. Neuron. 1998;20:1103–1110. doi: 10.1016/s0896-6273(00)80492-4. [DOI] [PubMed] [Google Scholar]
- 33.Emery P, So WV, Kaneko M, Hall JC, Rosbash M. CRY, a Drosophila clock and light-regulated cryptochrome, is a major contributor to circadian rhythm resetting and photosensitivity. Cell. 1998;95:669–679. doi: 10.1016/s0092-8674(00)81637-2. [DOI] [PubMed] [Google Scholar]
- 34.Mikuni S, Tamura M, Kinjo M. Analysis of intranuclear binding process of glucocorticoid receptor using fluorescence correlation spectroscopy. FEBS lett. 2007;581:389–393. doi: 10.1016/j.febslet.2006.12.038. [DOI] [PubMed] [Google Scholar]
- 35.Scott DK, Strömstedt PE, Wang JC, Granner DK. Further characterization of the glucocorticoid response unit in the phosphoenolpyruvate carboxykinase gene. The role of the glucocorticoid receptor-binding sites. Mol. Endocrinol. 1998;12:482–491. doi: 10.1210/mend.12.4.0090. [DOI] [PubMed] [Google Scholar]
- 36.Akashi M, Ichise T, Mamine T, Takumi T. Molecular mechanism of cell-autonomous circadian gene expression of Period2, a crucial regulator of the mammalian circadian clock. Mol. Biol. Cell. 2006;17:555–565. doi: 10.1091/mbc.E05-05-0396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamajuku D, Shibata Y, Kitazawa M, Katakura T, Urata H, Kojima T, Nakata O, Hashimoto S. Identification of functional clock-controlled elements involved in differential timing of Per1 and Per2 transcription. Nucleic Acids Res. 2010;38:7964–7973. doi: 10.1093/nar/gkq678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamamoto Y, Yagita K, Okamura H. Role of cyclic mPer2 expression in the mammalian cellular clock. Mol. Cell. Biol. 2005;25:1912–1921. doi: 10.1128/MCB.25.5.1912-1921.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen R, Schirmer A, Lee Y, Lee H, Kumar V, Yoo SH, Takahashi JS, Lee C. Rhythmic PER abundance defines a critical nodal point for negative feedback within the circadian clock mechanism. Mol. Cell. 2009;36:417–430. doi: 10.1016/j.molcel.2009.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Numano R, Yamazaki S, Umeda N, Samura T, Sujino M, Takahashi R, Ueda M, Mori A, Yamada K, Sakaki Y. Constitutive expression of the Period1 gene impairs behavioral and molecular circadian rhythms. Proc. Natl Acad. Sci. USA. 2006;103:3716–3721. doi: 10.1073/pnas.0600060103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee Y, Chen R, Lee H, Lee C. Stoichiometric relationship among clock proteins determines robustness of circadian rhythms. J. Biol. Chem. 2011;286:7033–7042. doi: 10.1074/jbc.M110.207217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hirota T, Okano T, Kokame K, Shirotani-Ikejima H, Miyata T, Fukada Y. Glucose down–regulates Per1 and Per2mRNA levels and induces circadian gene expression in cultured rat-1 fibroblasts. J. Biol. Chem. 2002;277:44244–44251. doi: 10.1074/jbc.M206233200. [DOI] [PubMed] [Google Scholar]
- 43.Maywood E, Mrosovsky N, Field M, Hastings M. Rapid down-regulation of mammalian period genes during behavioral resetting of the circadian clock. Proc. Natl Acad. Sci. USA. 1999;96:15211–15216. doi: 10.1073/pnas.96.26.15211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yannielli P, McKinley Brewer J, Harrington M. Is novel wheel inhibition of Per1 and Per2 expression linked to phase shift occurrence? Neuroscience. 2002;112:677–685. doi: 10.1016/s0306-4522(02)00100-8. [DOI] [PubMed] [Google Scholar]
- 45.Strömstedt P, Poellinger L, Gustafsson J, Carlstedt-Duke J. The glucocorticoid receptor binds to a sequence overlapping the TATA box of the human osteocalcin promoter: a potential mechanism for negative regulation. Mol. Cell. Biol. 1991;11:3379–3383. doi: 10.1128/mcb.11.6.3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Calomme C, Dekoninck A, Nizet S, Adam E, Nguyên TLA, Van Den Broeke A, Willems L, Kettmann R, Burny A, Van Lint C. Overlapping CRE and E box motifs in the enhancer sequences of the bovine leukemia virus 5′ long terminal repeat are critical for basal and acetylation-dependent transcriptional activity of the viral promoter: implications for viral latency. J. Virol. 2004;78:13848–13864. doi: 10.1128/JVI.78.24.13848-13864.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nader N, Chrousos GP, Kino T. Circadian rhythm transcription factor CLOCK regulates the transcriptional activity of the glucocorticoid receptor by acetylating its hinge region lysine cluster: potential physiological implications. FASEB J. 2009;23:1572–1583. doi: 10.1096/fj.08-117697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meijsing SH, Pufall MA, So AY, Bates DL, Chen L, Yamamoto KR. DNA binding site sequence directs glucocorticoid receptor structure and activity. Science. 2009;324:407–410. doi: 10.1126/science.1164265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reddy AB, Maywood ES, Karp NA, King VM, Inoue Y, Gonzalez FJ, Lilley KS, Kyriacou CP, Hastings MH. Glucocorticoid signaling synchronizes the liver circadian transcriptome. Hepatology. 2007;45:1478–1488. doi: 10.1002/hep.21571. [DOI] [PubMed] [Google Scholar]
- 50.Jung H, Choe Y, Kim H, Park N, Son GH, Khang I, Kim K. Involvement of CLOCK: BMAL1 heterodimer in serum-responsive mPer1 induction. Neuroreport. 2003;14:15–19. doi: 10.1097/00001756-200301200-00003. [DOI] [PubMed] [Google Scholar]
- 51.Shim HS, Kim H, Lee J, Son GH, Cho S, Oh TH, Kang SH, Seen DS, Lee KH, Kim K. Rapid activation of CLOCK by Ca2+-dependent protein kinase C mediates resetting of the mammalian circadian clock. EMBO Rep. 2007;8:366–371. doi: 10.1038/sj.embor.7400920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Travnickova-Bendova Z, Cermakian N, Reppert SM, Sassone-Corsi P. Bimodal regulation of mPeriod promoters by CREB-dependent signaling and CLOCK/BMAL1 activity. Proc. Natl Acad. Sci. USA. 2002;99:7728–7733. doi: 10.1073/pnas.102075599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ueda HR, Hayashi S, Chen W, Sano M, Machida M, Shigeyoshi Y, Iino M, Hashimoto S. System-level identification of transcriptional circuits underlying mammalian circadian clocks. Nat. Genet. 2005;37:187–192. doi: 10.1038/ng1504. [DOI] [PubMed] [Google Scholar]
- 54.Ukai-Tadenuma M, Yamada RG, Xu H, Ripperger JA, Liu AC, Ueda HR. Delay in feedback repression by cryptochrome 1 is required for circadian clock function. Cell. 2011;144:268–281. doi: 10.1016/j.cell.2010.12.019. [DOI] [PubMed] [Google Scholar]
- 55.Ukai-Tadenuma M, Kasukawa T, Ueda HR. Proof-by-synthesis of the transcriptional logic of mammalian circadian clocks. Nat. Cell Biol. 2008;10:1154–1163. doi: 10.1038/ncb1775. [DOI] [PubMed] [Google Scholar]
- 56.Yoo SH, Ko CH, Lowrey PL, Buhr ED, Song E, Chang S, Yoo OJ, Yamazaki S, Lee C, Takahashi JS. A noncanonical E-box enhancer drives mouse Period2 circadian oscillations in vivo. Proc. Natl Acad. Sci. USA. 2005;102:2608–2613. doi: 10.1073/pnas.0409763102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Torra IP, Tsibulsky V, Delaunay F, Saladin R, Laudet V, Fruchart JC, Kosykh V, Staels B. Circadian and glucocorticoid regulation of Rev-erbα expression in liver. Endocrinol. 2000;141:3799–3806. doi: 10.1210/endo.141.10.7708. [DOI] [PubMed] [Google Scholar]
- 58.Vatine G, Vallone D, Appelbaum L, Mracek P, Ben-Moshe Z, Lahiri K, Gothilf Y, Foulkes NS. Light directs zebrafish period2 expression via conserved D and E boxes. PLoS Biol. 2009;7:e1000223. doi: 10.1371/journal.pbio.1000223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tamai T, Carr A, Whitmore D. Zebrafish circadian clocks: cells that see light. Biochem. Soc. Trans. 2005;33:962–966. doi: 10.1042/BST20050962. [DOI] [PubMed] [Google Scholar]
- 60.Kolla BP, Auger RR. Jet lag and shift work sleep disorders: how to help reset the internal clock. Cleve. Clin. J. Med. 2011;78:675–684. doi: 10.3949/ccjm.78a.10083. [DOI] [PubMed] [Google Scholar]
- 61.Harrington M. Location, location, location: important for jet-lagged circadian loops. J. Clin. Invest. 2010;120:2265–2267. doi: 10.1172/JCI43632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Park N, Cheon S, Son GH, Cho S, Kim K. Chronic circadian disturbance by a shortened light-dark cycle increases mortality. Neurobiol. Aging. 2011;33:1122.e1111–1122.e1122. doi: 10.1016/j.neurobiolaging.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 63.Dickmeis T. Glucocorticoids and the circadian clock. J. Endocrinol. 2009;200:3–22. doi: 10.1677/JOE-08-0415. [DOI] [PubMed] [Google Scholar]
- 64.Herichova I, Zeman M, Stebelova K, Ravingerova T. Effect of streptozotocin-induced diabetes on daily expression of per2 and dbp in the heart and liver and melatonin rhythm in the pineal gland of Wistar rat. Mol. Cell. Biochem. 2005;270:223–229. doi: 10.1007/s11010-005-5323-y. [DOI] [PubMed] [Google Scholar]
- 65.Cermakian N, Lamont EW, Boudreau P, Boivin DB. Circadian clock gene expression in brain regions of Alzheimer’s disease patients and control subjects. J. Biol. Rhythms. 2011;26:160–170. doi: 10.1177/0748730410395732. [DOI] [PubMed] [Google Scholar]
- 66.Tahira K, Ueno T, Fukuda N, Aoyama T, Tsunemi A, Matsumoto S, Nagura C, Matsumoto T, Soma M, Shimba S, et al. Obesity alters the expression profile of clock genes in peripheral blood mononuclear cells. Arch. Med. Sci. 2011;7:933–940. doi: 10.5114/aoms.2011.26603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Barclay JL, Husse J, Bode B, Naujokat N, Meyer-Kovac J, Schmid SM, Lehnert H, Oster H. Circadian desynchrony promotes metabolic disruption in a mouse model of shiftwork. PLoS One. 2012;7:e37150. doi: 10.1371/journal.pone.0037150. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.