Abstract
Both ppGpp and pppGpp are thought to function collectively as second messengers for many complex cellular responses to nutritional stress throughout biology. There are few indications that their regulatory effects might be different; however, this question has been largely unexplored for lack of an ability to experimentally manipulate the relative abundance of ppGpp and pppGpp. Here, we achieve preferential accumulation of either ppGpp or pppGpp with Escherichia coli strains through induction of different Streptococcal (p)ppGpp synthetase fragments. In addition, expression of E. coli GppA, a pppGpp 5′-gamma phosphate hydrolase that converts pppGpp to ppGpp, is manipulated to fine tune differential accumulation of ppGpp and pppGpp. In vivo and in vitro experiments show that pppGpp is less potent than ppGpp with respect to regulation of growth rate, RNA/DNA ratios, ribosomal RNA P1 promoter transcription inhibition, threonine operon promoter activation and RpoS induction. To provide further insights into regulation by (p)ppGpp, we have also determined crystal structures of E. coli RNA polymerase-σ70 holoenzyme with ppGpp and pppGpp. We find that both nucleotides bind to a site at the interface between β′ and ω subunits.
INTRODUCTION
During stress, bacteria accumulate two unusual nucleotides: ppGpp and pppGpp, collectively abbreviated as (p)ppGpp (1). They are synthesized from adenosine triphosphate (ATP) and either guanosine diphosphate (GDP) to make ppGpp or guanosine triphosphate (GTP) to make pppGpp. Their synthesis in Escherichia coli is facilitated by two proteins, RelA and SpoT. RelA is activated during amino acid starvation, whereas SpoT synthesizes (p)ppGpp during starvation for sources of carbon, nitrogen, phosphorus, iron or lipids as well as when stressed by heat or oxidation (2). SpoT is also responsible for degradation of ppGpp and pppGpp to GDP and GTP, respectively. There is yet a third enzyme important in this metabolic cycle—the GppA phosphatase, which converts pppGpp into ppGpp.
Certain mutations or starvation conditions drastically affect the ratios of accumulated ppGpp and pppGpp. For example, when amino acid is starved, some strains accumulate only ppGpp, whereas wild-type strains accumulate both ppGpp and pppGpp (3). This mutant phenotype was originally called ‘spotless’ and mapped on the E. coli chromosome by Laffler and Gallant (4) to a locus thereafter called ‘spoT’. Starvation of wild-type spoT+ strains for a carbon source results in accumulation of only ppGpp, a spoT1 phenocopy (5,6). Early studies repeatedly confirmed that the rate of stable RNA synthesis was inversely correlated with levels of (p)ppGpp whether studying wild-type or spoT mutants and whether starvation was for amino acids or for glucose (5,7). Experimental conditions were not found that gave exclusive accumulation of pppGpp as was the case for ppGpp. Nevertheless, the prevailing view emerged that ppGpp and pppGpp were equally potent nucleotide regulators (2,7).
Despite these correlations, a several decade-long controversy followed as to whether the (p)ppGpp nucleotides together were the only factors responsible for RNA control during both the stringent response and growth rate control in E. coli [see (2,8–10)]. This controversy may now be close to resolution (2,11,12). However, it remains unclear whether ppGpp and pppGpp show equal or preferential regulatory potency for stringent or growth rate control, for positive and negative regulation at the level of expression of other genes, or for putative metabolic regulation that operates through direct enzyme binding (13).
There are only a few reports that attempted to measure differential regulatory effects of ppGpp and pppGpp in vitro. This is in part due to the current lack of commercially available pppGpp. Assays of Bacillus subtilis DNA primase suggest that pppGpp is a more potent inhibitor of this enzyme than ppGpp (14). Regulation of DNA primase may be different in E. coli, where there are two studies available. Indeed, at equimolar ratios of analog:GTP, both found ppGpp to be a more potent inhibitor of primase activity than pppGpp (15,16).
Here, an approach is made to estimate the relative regulatory activities of ppGpp versus pppGpp in E. coli in vitro and in vivo by devising a method to manipulate expression of two ectopic (p)ppGpp synthetases, as well as GppA, to obtain preferential accumulation of one or the other regulator. We also report in parallel a biochemical approach that uses the same enzymes to synthesize pure biochemical amounts of these nucleotides; this simplifies their availability as compared with earlier methods (17). Comparing regulatory potency of ppGpp and pppGpp can be argued to have immediate interest because it can provide clues to details of regulatory mechanisms that are still elusive. In the long term, these studies appear to provide a ‘proof of principle’ of the value of similar approaches to uncover differential regulatory effects of nucleotide regulators in the (p)ppGpp family of structurally related effectors.
Because ratios of ppGpp and pppGpp change with starvation conditions (see earlier in the text), it is necessary to make this assessment with unstarved growing bacterial cells and to induce ppGpp or pppGpp with minimal stress. Any stress that does accompany induction must be equivalent for each nucleotide induced. The regulatory parameters influenced by (p)ppGpp we have chosen to study in vivo and in vitro are inhibition of growth rate, effects on RNA/DNA ratio variation with growth rate, inhibition of ribosomal RNA transcription initiation, stimulation of threonine operon promoter transcription initiation and stabilizing effects on RpoS protein turnover. The results are taken to support a somewhat surprising conclusion that ppGpp is a more potent regulator than pppGpp for all five regulatory phenomena examined. This approach provides tools to ask whether other regulatory features of ppGpp and pppGpp will behave similarly and a way in the future to assess specific regulatory capabilities of other members of the (p)ppGpp-like family of related nucleotides. These efforts are helped by the discovery presented here of a (p)ppGpp binding site on the interface of the β′ and ω subunits of E. coli RNA polymerase σ70 holoenzyme.
MATERIALS AND METHODS
Bacterial strains and growth conditions
Strains (Table 1) used for preferential induction of ppGpp or pppGpp are derivatives of E. coli strain MG1655 deleted for relA, and subsequent chromosomal rearrangements were made by P1vir transduction. RecA mutant derivatives were constructed as hosts to enhance the genetic stability of plasmids expressing growth-toxic products. Liquid cultures were grown at 32°C in MOPS 0.4% glycerol media enriched with 20 amino acids (40 µg/ml) and nucleosides (Ar, Gr, Ur, Cr, Tdr; 20 µg/ml), which contained 2 mM phosphate [see (19)]. In less-extensive studies, equivalent growth effects could also be obtained when the nucleoside mixture was absent. Arabinose sensitivity of various strains was checked on agar plates containing M9 minimal medium, 0.4% glycerol and 0.4% Casaminoacids with or without 0.1% arabinose. In liquid cultures, arabinose was added at indicated concentrations to induce ppGpp or pppGpp. When overnight cultures were used, 0.04% glucose was included in addition to glycerol to catabolite repress low levels of enzyme induction before arabinose addition to minimize accumulation of fast growing suppressor mutants. Antibiotics: ampicillin 100 μg/ml; erythromycin 200 μg/ml, kanamycin 30 μg/ml and tetracycline 20 μg/ml.
Table 1.
Strains and plasmids
| Genotype/plasmid/construct | Reference | |
|---|---|---|
| E. coli | ||
| CF1648 | MG1655 wild-type | Stock collection |
| CF1652 | wt Δrel | Stock collection |
| CF3967 | wt Δrel ΔgppA | (18) |
| CF5766 | BL21(λDE3)/pHM504 (pT7 GppA) Ap | this work |
| CF7955 | BL21(λDE3)/pHM1381 (pUM99) (pT7 RelSeq1-385H) Ap | (19) |
| CF9993 | Wild-type but Δrel rpoS::lac protein fusion | (20) |
| CF10112 | Wild-type but ΔrelΔgppA rpoS::lac protein fusion | (20) |
| CF16755 | CF1652/pBAD18 Ap | (19) |
| CF16756 | CF3967/pBAD18 Ap | This work |
| CF16757 | CF1652/pUM9 Ap | (19) |
| CF16758 | CF1652/pUM76 Ery | This work |
| CF16760 | CF3967/pUM66 Ap | This work |
| CF16761 | CF1652/pUM96 Ap | This work |
| CF16762 | CF1652/pUM9 pUM76 Ap Ery | This work |
| CF16835 | CF1652/pUM96 pUM76 Ap Ery | This work |
| CF16836 | CF3967/pUM95 Ap | This work |
| CF16837 | CF1652/pBAD18 pUM76 Ap Ery | This work |
| CF16838 | CF9993/pUM9 pUM76 Ap Ery | This work |
| CF16839 | CF10112/pUM66 Ap | This work |
| Plasmid sources | ||
| CF6945 | CF1648/pVA8912 (pSC101 pUM76 vector) | (21) |
| CF16781a | pUM76 (pGppA) | this work |
| CF16770a | pBAD-18 (vector parent of RelSeq proteins) | (22) |
| CF16771a | pUM9 (pRelSeq79-385H) H−S+ | (19) |
| CF16772a | pUM66 (pRelSeq1-385H) H+S+ | (19) |
| CF16773a | pUM95 (pUM66 ::H77A,D78A,Y308S) H−S− | this work |
| CF16774a | pUM96 (pUM9::H332R) H−S− | this work |
aIn CF2200 recA56 host.
Manipulation of GppA abundance
The expression of the GppA protein has been altered by either deletion or by modest overexpression in growing cells. The ΔgppA::kan allele in CF3376 has been described (18). Plasmid pUM76, used for low level constitutive expression of GppA, was constructed as follows. An EcoRI-SphI gppA fragment from pHM504 was ligated into a low copy, ColEI-compatible vector pVA8912. The promoter responsible for low level transcription in pVA8912 has not been identified. Purification of the GppA protein lacking an affinity tag was achieved after induction in a BL21(λDE3) transformant with pHM504. This plasmid is a T7 promoter expression vector derivative of pSP72 (Promega) with an optimized ribosomal binding site (23). The primers for inserting the gppA orf were (5′GCATGCCATGGGTTCCACCTCGTCGCTGTATGCAGC-3′) and (5′GCGAAGCTTTGCTAACTAGTCATTAATGCACTTCCAGCGGCCAGTGGAC-3′).
Manipulation of preferential ppGpp and pppGpp accumulation
Accumulation in vivo of mostly ppGpp was mediated with the RelSeq79-385 N-terminal fragment expressed from pUM9 under control of the pBAD, whereas pppGpp was the predominant nucleotide when RelSeq1-385 was expressed from pUM66 (19). The pUM95 plasmid, whose catalytically active isogenic parent is pUM66, has three mutations constructed as follows: the SacI/XhoI containing fragment from pUM29 (RelSeq1-385 Y308S synthetase mutant) was combined with the similarly digested pUM4 (RelSeq1-385 H77A, D78A hydrolase mutant). The pUM95 product is an arabinose-inducible catalytically inactive triple mutant control plasmid. The pUM96 plasmid is a H322R synthetase mutant whose parent is already hydrolase deficient, pUM9 RelSeq79-385 (see Tables 1 and 2).
Table 2.
Effects of inducing mutant (p)ppGpp synthetases on growth rates
| Accumulation specificity | WT/mutant constructs | Doubling time |
|
|---|---|---|---|
| − ara | + ara (Δ μ) | ||
| ↑ ppGpp (WT) | RelSeq79-385 (pUM9) ↑GppA (pUM76) | 79 min | 188 min (2.4×) |
| -ppGpp (mutant) | RelSeq79-385(H322R) (pUM96) ↑GppA (pUM76) | 75 min | 86 min (1.2×) |
| ↑ pppGpp (WT) | RelSeq1-385 (pUM66) ↓GppA ΔgppA | 67 min | 105 min (2×) |
| -ppGpp (mutant) | RelSeq1-385(H77A D78A Y308S) ↓GppA ΔgppA (pUM95) | 65 min | 75 min (1.2×) |
(p)ppGpp measurements
Cellular (p)ppGpp accumulation was measured in MOPS media with 0.2 mM phosphate (19) but adapted for growth and processing in microtiter plates. Overnight flask cultures were diluted 1:100 into 150 μl of fresh media containing incremental amounts of arabinose (between 0 and 0.13%), and growth was monitored. At OD600 of ∼0.05, carrier-free 32P was added (final specific activity 150 μCi/ml), and samples were grown to OD600 ∼0.3; samples (50 μl) were mixed with an equal volume of 13 M formic acid. After three freeze-thaw cycles, samples in microtiter plates were centrifuged at 4°C to pellet the cell debris, and aliquots of the supernatants were applied to polyethyleneimine (PEI) cellulose thin layer chromatography plates, resolved with 1.5 M KH2PO4 (pH 3.4) and quantitated by densitometry as described (19). Estimates of ppGpp and pppGpp abundance relative to the total (pppGpp + ppGpp + GTP) were calculated by first dividing the counts in GTP, ppGpp and pppGpp by 3, 4 and 5, respectively, to obtain the molar equivalents.
Estimation of RNA and DNA
Cultures were grown with or without 0.04% arabinose in enriched MOPS media but labeled at a lower specific activity (∼10 μCi/ml). Triplicate 150 μl aliquots were distributed into microtiter dish wells for further incubation. At a OD600 ∼0.3, two samples of 60 μl were taken from each well and processed differently. One sample (DNA + RNA) was TCA precipitated (by adding an equal volume of 13.4% TCA), placed on ice for 90 min, then filtered (glass GF/F Whatman filters), washed once (5% TCA + 0.5 mM phosphate), washed three times (5% TCA), washed with 95% ethanol and dried, and radioactivity in RNA and DNA was estimated from Cerenkov counts. The second sample (DNA) was treated with 60 μl of 0.66 N NaOH and incubated for 2 h at room temperature to hydrolyze RNA, then TCA precipitated by adding 120 μl of 16% TCA. Further processing was as for the first sample.
RNA was calculated by subtracting counts in the second sample from the first. RNA/DNA ratios were obtained by dividing the RNA counts by the DNA counts in the second sample.
Purification of overexpressed RelSeq1-385
RelSeq1-385 with a C-terminal His-tag was overexpressed from pHM1381 (pUM99) present in strain CF7955 as described previously (19,24). CF7955 was grown at 32°C in 1 l of Luria Bertani Broth (LB) Isopropyl to an OD600 of ∼0.5 and then induced with 0.5 mM isopropyl-thiogalactopyranoside (IPTG) for 3 h. The pelleted cells were resuspended in binding buffer [50 mM Tris–Cl (pH 8.0), 0.5 M NaCl, 5 mM imidazole], pelleted again and frozen. Four grams of frozen cells were then resuspended in 35 ml of binding buffer containing 0.1 mM 4-(2-aminoethyl)-benzenesulfonyl fluoride hydrochloride, 10 mM ethylenediaminetetraacetic acid (EDTA) and 100 µg/ml lysozyme, incubated at 30°C for 30 min and placed on ice. Lysis was completed by sonication using three cycles of 2.5 min each, with 10 s pulses and intervening cooling on ice. Cell debris was removed by low speed centrifugation (12 000 rpm × 30 min, Sorvall SS34 rotor), and the RelSeq1-385 protein was precipitated from the supernatant with 35% ammonium sulfate. The precipitate was resuspended in 5 ml of binding buffer and dialyzed against two changes of 500 ml of the same buffer in the cold. The dialyzed protein was again spun at low speeds, then bound to Ni-NTA agarose (Qiagen), washed with binding buffer containing 60 mM imidazole and eluted with the same buffer but containing 1 M imidazole. The eluted protein was dialyzed against three changes of 50 mM bis-Tris propane (pH 9.0), 300 mM NaCl and 50% glycerol. The yield was ∼30–40 mg of purified RelSeq1-385 with a specific pppGpp synthetase activity of 1.4 μmoles/min/mg.
Purification of GppA
Strain CF5766 carrying pHM504 encoding GppA lacking an affinity tag was grown in 2 l of LB and induced with 0.5 mM IPTG as described for RelSeq1-385. After pelleting, cells were resuspended in 100 ml of cold lysis buffer [50 mM Tris (pH 8.0), 2 mM EDTA], again pelleted and frozen. The pellet was resuspended in 150 ml of lysis buffer containing 0.1 mg/ml lysozyme and was then sonicated using three cycles of 2.5 min each, with 10 s pulses and intervening cooling on ice. Cell debris was pelleted (12 000g for 15 min), and the supernatant was subjected to ammonium sulfate fractionation. The first precipitation was with 35% (w/v) (NH4)2SO4, and the ensuing pellet was discarded. Additional (NH4)2SO4 was added to 45%, and the pellet was resuspended in phosphocellulose-binding buffer [50 mM Tris (pH 7.0), 10 mM KCl, 0.1 mM dithiothreitol (DTT), 5% glycerol], 10 ml of total volume. After dialysis against three changes of the binding buffer, the entire sample was loaded on a 15 ml of phosphocellulose column (Whatman P11, prewashed according to the manufacturer) and washed with 100 ml of binding buffer. GppA was eluted with a 200 ml of linear gradient from 10 to 500 mM KCl in the same buffer. After assaying for pppGpp-ase activity (by monitoring conversion of pppGpp to ppGpp), fractions with GppA were pooled (32 ml), and protein was precipitated by adding an equal volume of saturated (NH4)2SO4. The pellet was resuspended in a minimal volume of storage buffer [10 mM Tris (pH 8.0), 100 mM NaCl, 0.1 mM EDTA, 50% glycerol] and dialyzed against three changes of the same buffer, yielding ∼10 mg of purified GppA as a 1 mg/ml solution with a pppGpp-ase specific activity of 24.5 μmoles/min/mg protein.
Synthesis of pppGpp and ppGpp in vitro
For pppGpp preparation, a 3-ml reaction mixture containing 25 mM bis-Tris propane (pH 9.0), 100 mM NaCl, 15 mM MgCl2, 8 mM ATP and 6 mM GTP was initiated by adding 300 µg of purified RelSeq1-385 and incubated for 1 h at 37°C. When ppGpp was the desired end product, the same pppGpp synthesis reaction was used, and GppA was used for its conversion to ppGpp. Because GppA is sensitive to NaCl, this salt was omitted in the first reaction step; in addition, 0.5 mM dithiothreitol was included. After 1 h, when the GTP was completely consumed (Supplementary Figure S1A), 35 µg/ml GppA was added plus (NH4)2SO4 to a final concentration of 75 mM. This mixture was incubated for an additional 15 min (Supplementary Figure S1B).
Alternatively, it is possible to synthesize ppGpp without using GppA protein but by simply substituting GDP for GTP in the synthesis reaction with purified RelSeq1-385 (Supplementary Figure S4). We favor first synthesizing pppGpp and then converting it to ppGpp with purified GppA because this route seems to result in preparations of ppGpp that are devoid of pppGpp that arises from the presence of contaminating GTP in commercial preparations of GDP.
Reactions yielding pppGpp or ppGpp were stopped by phenol extraction, and the nucleotide products were precipitated by adjusting the LiCl concentration to 1 M, followed by the addition of four volumes of ethanol. The suspension was mixed, incubated on ice for more than 15 min and pelleted at 6000g for 10 min. The pellets were washed twice with absolute ethanol to remove residual LiCl, dried and stored at −20°C before further purification.
The progress of the reaction was monitored by periodic removal of 5 μl of aliquots, which were added to an equal volume of 6 M Na formate (pH 3.7). These were analyzed on PEI cellulose thin layer chromatography plates and run 10 cm above the origin in 1.5 M KH2PO4 (pH 3.4). Resolved nucleotides were visualized by ultraviolet (UV) absorption and/or autoradiography (Supplementary Figure S1). Enzyme specific activity estimates were made with labeled substrates ([α-32P]-GTP or [γ-32P]-ATP) and autoradiographic quantitation.
Purification of pppGpp or ppGpp
Precipitates of the lithium salt from preparative reaction mixtures were resuspended in the binding buffer [0.1 mM LiCl, 0.5 mM EDTA, 25 mM Tris–Cl (pH 7.5)], then loaded onto either a 5-ml Econo-Pac High Q cartridge (Bio-Rad) or a 1.5 × 30 cm column containing 50 ml QAE Sephadex A-25, prewashed and equilibrated in the binding buffer. Nucleotides were resolved with a linear gradient from 0.15 M to 0.5 M LiCl in the binding buffer, and elution was monitored as UV absorbance at 253 nm (BioRad Biologic LP) and verified by TLC mobilities relative to GTP. An elution profile of a pppGpp synthesis reaction is shown (Supplementary Figure S2). If necessary, pooled fractions of ppGpp or pppGpp were batch concentrated by diluting them 1:10 in the binding buffer without salt and reabsorbed to a QAE-sephadex column (1 × 4 cm), then batch eluted with 1 M LiCl and again EtOH precipitated. An example of a chromatogram of purified pppGpp is shown in Supplementary Figure S3. Pure nucleotides were pooled, and concentrations were determined by UV [13.7 A253 /ml/μmole at pH 7 (25)]. This allows precise amounts to be ethanol precipitated as lithium salts, washed twice with ethanol and air dried. Such preparations have been stable for years when stored at −20°C.
In vitro transcription
The rrnB P1 promoter activity was measured as described (26). Briefly, 20 µl of reactions with 30 nM RNA polymerase (Epicentre Technologies) and 10 nM linear DNA template (−180 to +109 bp of the rrnB P1 promoter region) were performed at 30°C in the following buffer: 50 mM Tris-acetate (pH 8.0), 10 mM MgAc, 10 mM β-mercapthoethanol, 10 µg/ml bovine serum albumin, 90 mM potassium glutamate. RNA polymerase was pre-incubated with the indicated concentrations of ppGpp, pppGpp or GDP for 7 min at room temperature. This was followed by the addition of salt, DNA template, NTPs (100 µM ATP, CTP, GTP and 10 µM UTP) and 2 µCi/reaction [α-32P UTP, Perkin Elmer] with amounts of DksA indicated. Reactions were carried out for 8 min and terminated by addition of an equal volume of the stop solution (95% formamide, 20 mM EDTA, 0.05% bromphenol blue and 0.05% xylene cyanol). Samples were analyzed on 7 M urea, 6% polyacrylamide sequencing gels and quantified by phosphorimaging on a GE Healthcare imaging system.
In vitro transcription of the pthr promoter was done in a similar manner, except it was carried out at 37°C in a buffer containing 50 mM Tris–Cl (pH 8.0), 10 mM MgCl2, 10 mM β-mercapthoethanol, 10 µg/ml bovine serum albumin, 60 mM KCl. The linear DNA template spanned −142 to +192 bp of the thrA promoter region.
β-galactosidase assays
Strain CF9993 with a rpoS-lacZ fusion [Δlac, Δ relA255::cat, trp::put::kanR-rpoS-lac (20)] was transformed with pUM9 (pBAD-RelSeq79-385) and pUM76 to yield an arabinose inducible ppGpp overproducing strain, CF16838. CF 10112 (CF9993 deleted for gppA) was transformed with pUM66 (pBAD-RelSeq1-385) and used as arabinose inducible pppGpp producer, CF16839. Cells were grown in MOPS medium containing 0.8 mM phosphate and 0.4% glycerol. Aliquotes were taken during growth between OD600 ∼ 0.1 and 1.1, and processed according to Miller (1972) (27).
X-ray crystal structure determinations of the E. coli RNA polymerase σ70 holoenzyme and (p)ppGpp complexes
The method for preparation of E. coli RNA polymerase σ70 holoenzyme was described (28). RNA polymerase crystals were obtained by using hanging drop vapor diffusion by mixing equal volume of protein solution (∼20 mg/ml) and crystallization solution [0.1 M Hepes–HCl (pH 7.0), 0.2 M CaAcetate, ∼15 % PEG400, 10 mM Tri(2-carboxyethyl)phosphine] and incubating at 22°C over the same crystallization solution. For preparing the RNA polymerase-(p)ppGpp complex crystals, crystals were soaked in crystallization solution containing 30% PEG400 and 1 mM of either ppGpp or pppGpp overnight at 22°C and then flash frozen by liquid nitrogen.
The crystallographic data sets were collected at the Macromolecular Diffraction at Cornell High Energy Synchrotron Source (MacCHESS) A1 beamline (Cornell University, Ithaca, NY), and the data were processed by HKL2000 (29). The E. coli RNA polymerase structure (28) was used as an initial model for the rigid body and positional refinement with non-crystallographic symmetry and secondary structure restraints by using the programs Phenix (30). The resulting map allowed (p)ppGpp that were not present in the search model to be built by Coot (31). Final coordinate and structure factor were submitted to the PDB with ID listed in Supplementary Table S1.
RESULTS
Accumulation of (p)ppGpp involves balancing activities of synthetases, hydrolases and their substrate specificities
Cellular accumulation of (p)ppGpp is determined by a complex balance of total synthetase and hydrolase activities with additional contributions from substrate specificities of both enzymes.
Here, we took advantage of different RelSeq protein fragments from Streptococcus equisimilis to manipulate ppGpp and pppGpp synthesis, as well as hydrolysis. This strategy requires that the E. coli RelA synthetase is deleted in all strains used. The N-terminal half (amino acids 1–385) of the full-length RelSeq protein from S. equisimilis encodes both catalytic domains, active (p)ppGpp hydrolase and active (p)ppGpp synthetase. The RelSeq1-385 synthetase has an in vitro preference for GTP over GDP as pyrophosphate acceptor in vitro and allows for preferential pppGpp accumulation in vivo (19). In contrast, induction of the RelSeq79-385 synthetase that lacks hydrolase leads to accumulation of ppGpp accompanied by little pppGpp (19).
In these experiments, therefore, two sources of (p)ppGpp hydrolases are present: the RelSeq1-385 fragment and the host E. coli SpoT protein. The in vitro specific activity of the SpoT hydrolase is unknown. The RelSeq1-385 hydrolase has a low specific activity: 0.7μm/min/mg (19). The SpoT and RelSeq hydrolases each do not preferentially hydrolyze either ppGpp or pppGpp when both nucleotides are present in equimolar mixtures (21).
Changes in ppGpp or pppGpp levels by arabinose induced expression of RelSeq protein variants
Arabinose induction of RelSeq79-385 leads to preferential accumulation of ppGpp with a trace of pppGpp (Figure 1, lane 1). The residual pppGpp can be further reduced by the presence of the plasmid pUM76, which constitutively overexpresses GppA at low levels. This increases ppGpp from 9 to 13% of the total guanine nucleotide pool and decreases pppGpp from 1 to 0.2% (Figure 1, lane 2).
Figure 1.
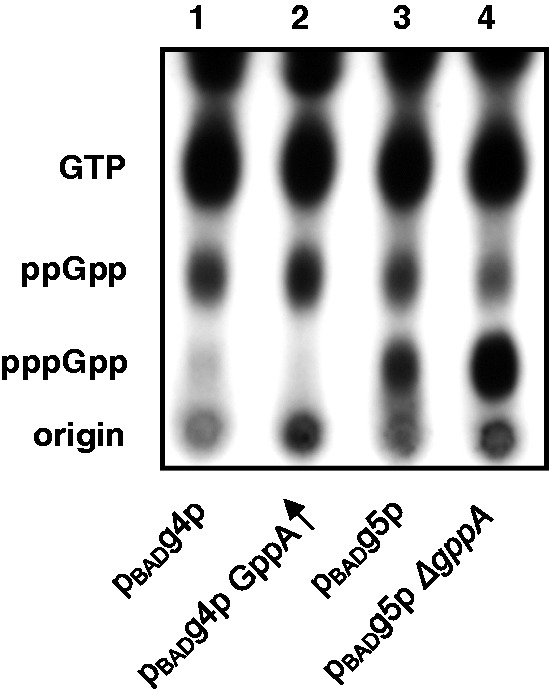
Arabinose induction of cellular ppGpp or pppGpp. An autogradiogram is shown of cell extracts of 32P-labeled GTP, ppGpp and pppGpp resolved by TLC after 0.05% arabinose induction (see ‘Materials and Methods’ section). Lane abbreviations: Lane 1. pBADg4p = (ppGpp synthetase): pBADRelSeq79-385 (pUM9); Lane 2. GppA↑ = (ppGpp synthetase + overexpressed gppA): pBADg4p + PgppA (pUM76); Lane 3. pBADg5p = (pppGpp synthetase): pBADRelSeq1-385 (pUM66); Lane 4. pBADg5p ΔgppA = (pppGpp synthetase + ΔgppA).
When RelSeq1-385 is induced by arabinose, the pppGpp/ppGpp ratio is shifted to favor of pppGpp (Figure 1, lane 3). When gppA is also deleted, conversion of pppGpp to ppGpp is limited, which results in an increase of pppGpp from 16 to 27%, and a decrease of ppGpp from 10 to 5% (Figure 1, lane 4).
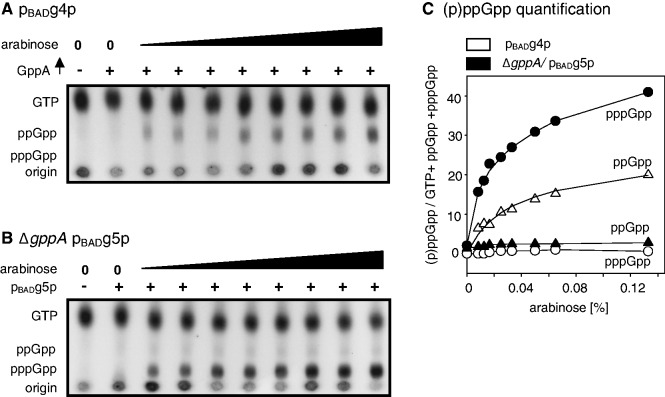
The effects of incremental addition of arabinose
When arabinose is added in concentrations ranging between 0 and 0.133% to cultures that preferentially accumulate ppGpp while GppA is constitutively overproduced, a systematic increase in this nucleotide is observed (Figure 2, panel A and C). Measurements of the relative ppGpp content as a fraction of total guanine nucleotide pool increased from background levels of near zero to 20% (Figure 2, panel C). At the same time, the minority nucleotide, pppGpp, rises to <1%. As controls, the presence of constitutive GppA overproduction or gppA deletion alone had no effect on uninduced cells (Figure 2, panels A and B, lanes 1 and 2).
Figure 2.
ppGpp versus pppGpp: effects of incremental arabinose concentrations. (A) An autoradiogram showing ppGpp accumulation in cells with: lane 1. an uninduced pBAD vector; lane 2. an uninduced pBADg4p + PgppA; lanes 3–10. pBADg4p + PgppA-induced experimental samples containing increasing arabinose concentrations 0.008, 0.012, 0.01, 0.025, 0.033, 0.05, 0.065 and 0.133%. (B) An autoradiogram showing pppGpp accumulation in cells with: lane 1. an uninduced pBAD vector ΔgppA control; lane 2. an uninduced pBADg5p ΔgppA control; lanes 3–10. induced experimental samples with arabinose concentrations as in Panel A. (C) The relation between inducing arabinose concentrations and the fractional content of ppGpp and pppGpp (percentage total of ppGpp/GTP + ppGpp + pppGpp). Data from Panel (A) were used to calculate preferential accumulation of ppGpp (open triangles) and of minority pppGpp (filled circles). Analogous values are taken from data in panel (B) for preferential accumulation of pppGpp (filled circles) and minority ppGpp (filled triangles). The ordinate numerical values for the fractional content of ppGpp or pppGpp are given in percentage.
Parallel measurements of the effects of arabinose addition on pppGpp accumulation were performed when gppA is deleted and RelSeq1-385 is similarly induced; these revealed an accelerated accumulation of pppGpp that reached a higher level than ppGpp, 40% of the total. Here, the minority nucleotide (ppGpp) accumulates to <3% (Figure 2, panels B and C).
Comparing the potency of ppGpp and pppGpp for growth inhibition
To document the extent of growth inhibition, we have monitored the doubling times of cells that accumulate predominately ppGpp or pppGpp as a function of arabinose concentration (0–0.065%). This reveals that ppGpp is more growth inhibitory than pppGpp (Figure 3A). For example, when ppGpp accumulates, the doubling time increases from 80 min to ∼160 min at 0.02% arabinose, whereas at the same arabinose concentration, pppGpp increases doubling times from ∼70 to 110 min.
Figure 3.
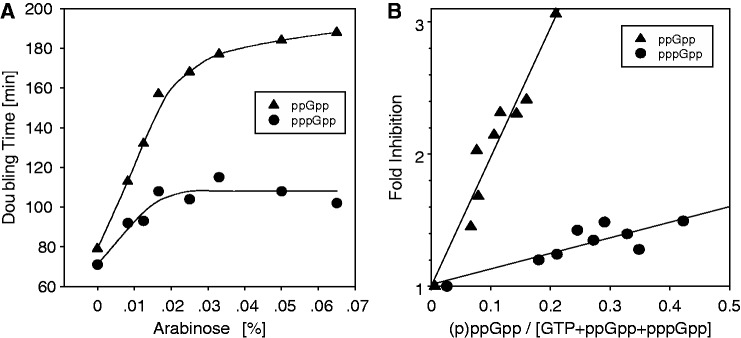
ppGpp versus pppGpp: effects of induction on growth rates and specific activities for growth inhibition. (A) The relation between the observed growth rate (doubling times expressed in minutes) of cells subjected to increasing concentrations of arabinose is shown when ppGpp (filled circles) preferentially accumulates as in Figure 2A as well as when pppGpp (filled triangles) preferentially accumulates as in Figure 2B. (B) The inhibitory activities of increasing the fractional content of ppGpp or pppGpp. The fractional growth rates are calculated from the growth rates expressed as μ (doublings/hr from Figure 3A), which are normalized to μ values for uninduced cultures and expressed as fold-inhibition on the ordinate. These values are plotted against the ppGpp or pppGpp fractional content (from Figure 2C). The slopes of these relations are taken as a measure of the relative specific activity for growth inhibition for ppGpp or pppGpp. Filled circles = pppGpp. These values are corrected for contribution to growth inhibition of the 5% content of ppGpp present in each sample seen in Panel (B) of Figure 2. Filled triangles = ppGpp [not corrected for ∼0.5% content of pppGpp in Panel (A) of Figure 2].
It is evident from both Figures 1 and 2 that under these conditions, accumulation of pppGpp greatly exceeds that of ppGpp. By expressing the growth rates shown in Figure 3A as doublings per hour (µ), we calculated the fold inhibition of growth after normalizing the growth rates of each strain during induction to growth rates observed without arabinose induction. A plot of growth inhibition as a function of the fractional content of ppGpp or pppGpp in the total guanine nucleotide pool is shown in Figure 3B. The slope of each line can be taken as a measure of the specific activity of each nucleotide to inhibit growth. This measure is relative because we do not know the absolute nucleotide concentrations. For ppGpp, the slope is 2.0/0.2 or 10, whereas the slope for pppGpp is 0.4/0.4 or 1.0. Therefore, ppGpp is ∼10-fold more potent an inhibitor of growth than pppGpp. In Figure 3B, the slope for pppGpp has been corrected for the contribution to inhibition owing to the residual ppGpp present, using the relation for nearly 100% ppGpp as the correction coefficient. It is notable that here GTP levels remain higher than ppGpp or pppGpp under even maximal induction conditions.
Growth inhibitory effects of the (p)ppGpp producing proteins are largely due to their catalytic activities
It has been argued previously from correlations (10) and from studies where (p)ppGpp had been completely eliminated (12) that (p)ppGpp is necessary and sufficient for growth rate control. Here, growth perturbations are linked to protein overexpression. This raises the possibility that growth inhibition is due to differential toxic effects of the two RelSeq protein fragments, rather than due to their catalytic activities. Therefore, catalytically defective controls were tested using missense mutants of the two proteins. Plasmid pUM96 expressed RelSeq79-385 H322R, which is a mutant protein defective in ppGpp synthesis. Induction of this altered protein with 0.1% arabinose increased the doubling time of its host from 75 to 86 min, whereas induction of the wild-type gene increased doubling time from 79 to 188 min (Table 2).
The corresponding catalytically defective RelSeq1-385 mutant in pUM95 with inactivated synthetase (Y308S) and hydrolase (H77A, D78A) also failed to slow growth significantly when induced (doubling time of 65 min without arabinose, 75 min with 0.1% arabinose). In contrast, the induction of the wild-type increased the doubling time from 67 to 105 min (Table 2). These data are taken to mean that the growth inhibition observed with induced wild-type proteins can be largly attributed to their catalytic products and not to differential protein toxicity.
As judged from Coomassie stained protein gels, mutant and wild-type proteins show similar extents of expression when induced with arabinose (data not shown). Collectively, these experiments support the notion that the observed growth inhibition is not due to induction artifacts.
Differential inhibitory effects of ppGpp and pppGpp on RNA synthesis
RNA/DNA ratios have been reported to vary inversely with levels of (p)ppGpp (1,10,32). Table 3 compares RNA/DNA ratios of the strains accumulating preferentially ppGpp or pppGpp; these ratios are determined with cells uniformly labeled with 32P-orthophosphate. A differential effect was seen for ppGpp and pppGpp dependent on induction with 0.04% arabinose. The accumulation of ppGpp under these labeling conditions slowed the growth rate from 70 to 154 min (2.2-fold), which was accompanied by RNA/DNA ratios dropping from 5.5 to 2.3 (2.4-fold). The induction of pppGpp was again less effective in slowing growth: from 59 to 91 min (1.5-fold). Induction of pppGpp also resulted in a more modest drop in RNA/DNA ratios than ppGpp, from 5.5 to 3.6 (1.5-fold). The growth rates shown in Table 2 are somewhat faster in this medium than shown in Figure 3A, but the differential inhibition of growth is similar: 2.4-fold for ppGpp and 1.6-fold for pppGpp. The correlation of growth rate and RNA/DNA ratios is internally consistent in each case.
Table 3.
Effects of preferential accumulation of ppGpp or pppGpp on RNA/DNA ratios
| Accumulation specificity | Constructs | Arabinose addition | Doubling time | RNA/ DNA |
|---|---|---|---|---|
| - ppGpp | vector (pBAD18) ↑GppA (pUM76) | + | 78 min | 5.3 |
| - ppGpp | RelSeq79-385 (pUM9) ↑GppA (pUM76) | − | 70 min | 5.5 |
| ↑ ppGpp | RelSeq79-385 (pUM9) ↑GppA (pUM76) | + | 154 min | 2.3 |
| - pppGpp | vector (pBAD18) ↓GppA ΔgppA | + | 64 min | 5.1 |
| - pppGpp | RelSeq1-385 (pUM66) ↓GppA ΔgppA | − | 59 min | 5.5 |
| ↑ pppGpp | RelSeq1-385 (pUM66) ↓GppA ΔgppA | + | 91 min | 3.6 |
Differential inhibition of rrnB P1 activity in vitro
Growth rate control is defined by effects on RNA/DNA ratios, which are thought to be determined largely by (p)ppGpp effects on transcription initiation of ribosomal promoters (1). We are now able to ask with purified ppGpp and pppGpp (see ‘Materials and Methods’ section), if the two nucleotides differentially regulate these promoters in vitro.
The rrnB P1 promoter is a well-studied promoter whose negative regulation by ppGpp requires the additional presence of DksA (33). Here, transcription was performed with increasing amounts of ppGpp or pppGpp, in the presence (60 and 300 nM) or absence of DksA (Figure 4A). GDP was used as control, as it is done routinely, as it can be imagined that the 5′ pyrophosphate residues of ppGpp might impair transcriptional elongation. A control for pppGpp might be GTP, but as the reaction mixture already contains this nucleotide, a standard reaction with no additional nucleotides was used as control. No differences were noted between reactions carried out with or without GDP, and thus only control with GDP is shown.
Figure 4.
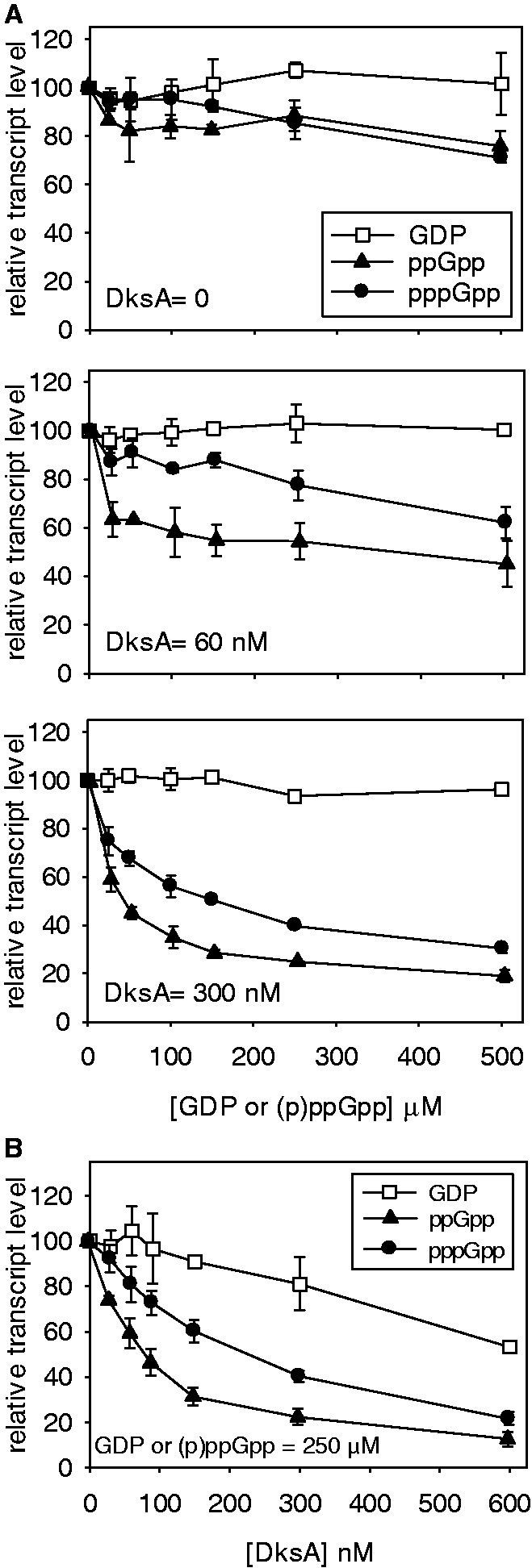
ppGpp versus pppGpp: inhibition of rrnB P1 promoter in vitro. Multi-round in vitro transcription was carried out with (A) increasing (0–500 µM) GDP, ppGpp or pppGpp, and constant DksA concentrations indicated (0, 60 and 300 nM); and (B) increasing DksA (0–600 nM) and constant (250 µM) GDP, ppGpp or pppGpp. Open squares = GDP, filled triangles = ppGpp, filled circles = pppGpp.
In the absence of DksA, ppGpp was a slightly better inhibitor than pppGpp at low (p)ppGpp concentrations (Figure 4A). However, this marginal difference in inhibition is dramatically enhanced in the presence of DksA (Figures 4A and B), and the degree of inhibition seems to depend on the level of DksA present, with greater transcriptional inhibition at 300 nM than 60 nM DksA. This is reinforced by results obtained when high levels of ppGpp or pppGpp were held constant (250 µM) and DksA was titrated (Figure 4B). Low DksA levels enhance the inhibitory effects of ppGpp more than pppGpp, but this differential inhibition was slightly diminished at high DksA levels (Figure 4B). Still, in all instances, ppGpp is a more potent inhibitor than pppGpp. These data are interpreted to qualitatively correlate with the differential inhibitory effects of the two nucleotides on RNA/DNA ratios (Table 3) and growth rate (Figures 3 and 4).
Differential activation of the thr operon promoter by (p)ppGpp
A similar set of experiments as those just mentioned was performed for the threonine operon promoter (pthr) that is positively regulated by ppGpp (34). Again, DksA is thought to participate in this regulatory mechanism (33). In this case, ppGpp was again a far better activator of transcription initiation than pppGpp in the presence of DksA, than in its absence. There was a consistent increase in transcriptional activation with increasing DksA level (Figure 5A and B); at all concentrations, ppGpp was ∼2-fold more efficient than pppGpp judging from the slopes of Figure 5B. In contrast to inhibition of rrnB P1 by (p)ppGpp, the relative difference of activation of pthr activity by the two nucleotides remained roughly constant with increasing nucleotide concentration (Figure 5A).
Figure 5.
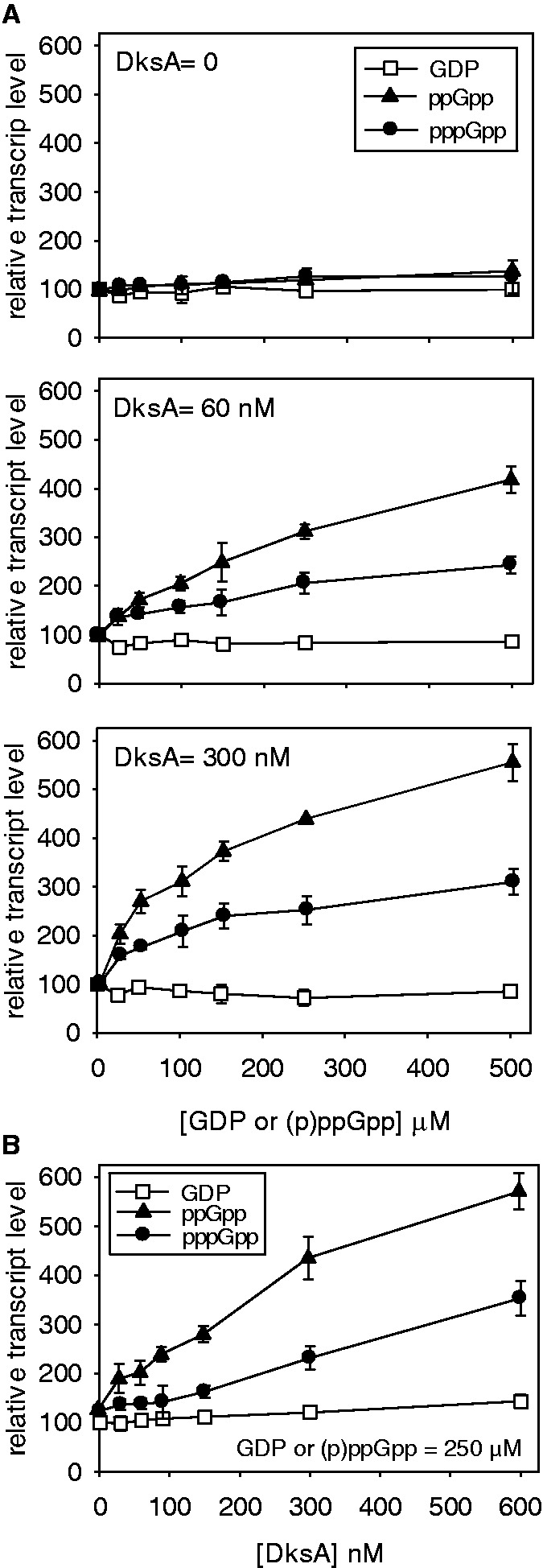
ppGpp versus pppGpp: activation of pthr promoter in vitro. Multi-round in vitro transcription was carried out with (A) increasing (0–500 µM) GDP, ppGpp or pppGpp, and constant DksA concentrations indicated (0, 60 and 300 nM); and (B) increasing DksA (0–600 nM) and constant (250 µM) GDP, ppGpp or pppGpp. Open squares = GDP, filled triangles = ppGpp, filled circles = pppGpp.
These results can be viewed in two ways: either DksA is a better enhancer for ppGpp regulation than for pppGpp or ppGpp is a better enhancer of DksA action than pppGpp.
Stimulation of RpoS-lacZ protein fusion activity in vivo: ppGpp versus pppGpp
The abundance of RpoS, the stationary phase-specific sigma factor σS, is increased whenever ppGpp accumulates (23), except when DksA is deleted (20). The σS protein is regulated by many factors, some of which function at the level of susceptibility to ClpX-mediated proteolysis, which involves adaptor and anti-adaptor proteins, which can require ppGpp (35,36). One measure of RpoS regulation is the activity of an RpoS-lacZ fusion, which in our laboratory has proven sensitive to ppGpp and DksA, although the detailed explanations for this behavior are arguable (20). Here, we compare the abilities of ppGpp and pppGpp to stimulate β-galactosidase activity of this fusion.
Accumulation of ppGpp or pppGpp to varying extents was induced in media containing 0, 0.005, 0.01 or 0.04% arabinose, which resulted in doubling times that ranged from 63 to 190 min for the ppGpp accumulating strain and ranged from 65 to 139 min for the pppGpp strain. In Figure 6, panel A corresponds to the ppGpp producer and panel B to the pppGpp producer strain. The plots of β-galactosidase activity versus cell density for uninduced and induced cultures reveal that for all concentrations of added arabinose, ppGpp is more effective than pppGpp as a positive regulator of fusion activity than pppGpp.
Figure 6.
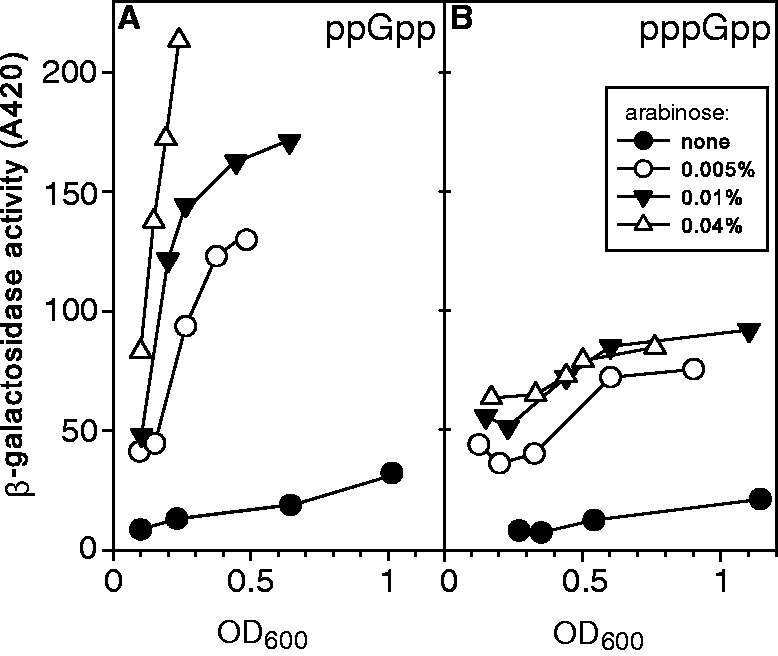
ppGpp versus pppGpp: Stimulation of RpoS-lac protein fusion β-galactosidase activity. Cells containing a single chromosomal copy of a ppGpp-activated RpoS-lac protein fusion were transformed with plasmids to preferentially induce ppGpp (strain CF16838) or pppGpp (CF16839) as in Figure 1. Each culture at indicated cell densities was assayed for β-galactosidase activity as described in ‘Materials and Methods’ section. (A) After adding arabinose (0, 0.005, 0.01 or 0.04%) to preferentially induce ppGpp, cultures were grown with doubling times of 63, 97, 157 and 190 min, respectively. (B) Analogous but preferential induction of pppGpp using the same arabinose concentrations, which resulted in doubling times of 65, 84, 99 and 139 min, respectively.
Finding the ppGpp and pppGpp binding sites of E. coli RNA polymerase
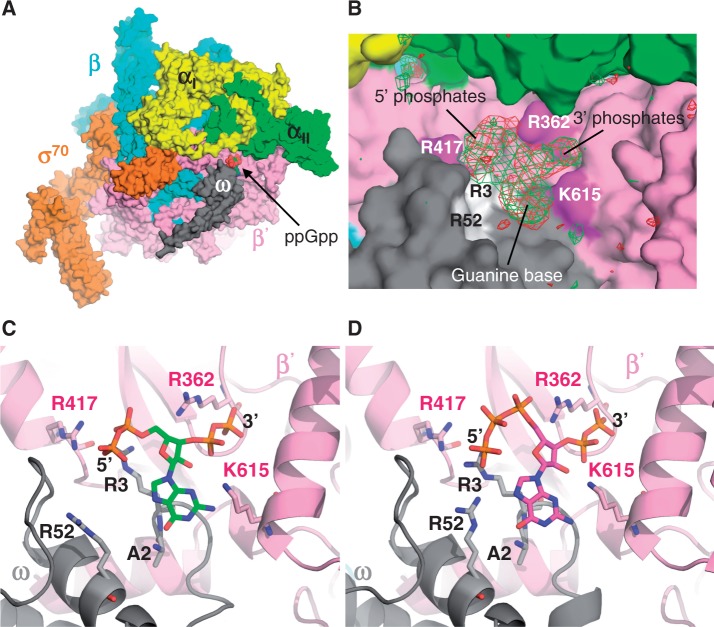
In vitro transcription experiments established that the pppGpp is less potent than ppGpp for rRNA transcription inhibition (Figure 4) and for threonine operon promoter activation (Figure 5). One possible explanation is that these compounds interact differentially with RNA polymerase. To directly identify the ppGpp- and pppGpp-binding sites on RNA polymerase, crystals of E. coli RNA polymerase σ70 holoenzyme complexed with either ppGpp or pppGpp were prepared by soaking 1 mM ppGpp or pppGpp into the preformed RNA polymerase crystals (28). X-ray crystal structures of these complexes were determined at 3.9 and 4.2 Å resolutions, respectively (Supplementary Table S1).
From the RNA polymerase and ppGpp complexes, clear unbiased Fo−Fc electron density for ppGpp was found at the interface between β′ and ω subunits, which is ∼30 Å away from the active site (Figure 7A and B). Clear Fo−Fc map from the RNA polymerase and pppGpp complex was also observed at the same site as from the RNA polymerase and ppGpp complex (Figure 7B), indicating that both ppGpp and pppGpp associate with the RNA polymerase at the same location. We did not observe any Fo−Fc maps for (p)ppGpp around the RNA polymerase active site indicating that (p)ppGpp does not bind the active site under our experimental condition. Our data suggest that no large conformational changes in RNA polymerase structure are induced by binding of (p)ppGpp. However, we cannot rule out that this may be due to the fact that the RNA polymerase-(p)ppGpp complex crystals were prepared by soaking (p)ppGpp into preformed RNA polymerase crystals.
Figure 7.
RNA polymerase – (p)ppGpp complex structures. (A) Overall structure of the E. coli RNA polymerase–ppGpp complex. RNA polymerase is depicted as a molecular surface model (αI: yellow, αII: green, β: cyan, β’: pink, ω: gray, σ70: orange). The ppGpp is depicted as a sphere model and its binding site is indicated. (B) Electron density maps showing ppGpp (green) and pppGpp (red) found in the complexes with RNA polymerase. Basic residues of β′ and ω subunits are indicated and regions of density maps corresponding to the 5′ and 3′ phosphates and the guanosine base are indicated. (C) Binding sites of the ppGpp and (D) pppGpp. Amino acid residues of β′ (pink stick) and ω subunits (gray stick) involved in (p)ppGpp binding are depicted on the cartoon models of β′ (pink) and ω subunits (gray). (p)ppGpp are shown as stick models.
The Fo−Fc maps showed three separated regions that correspond to 5′-di or triphosphates, guanosine (guanine plus ribose rings) and 3′-diphosphates (Figure 7B). Shapes and volumes of the Fo−Fc maps of ppGpp and pppGpp were identical except for the map around the β′ R417. We therefore positioned the 5′-phosphates of (p)ppGpp models at the Fo−Fc map around the β′ R417 and guanosine and 3′-diphosphates on the other two regions (Figure 7). The basic residues of both β′ subunit (R362, R417 and K615) and ω subunit (R3 and R52) around the (p)ppGpp binding pocket may participate in the binding of phosphate groups of (p)ppGpp, but their detailed interactions could not be established with current resolutions of structures.
The N-terminus of the ω subunit accommodates the guanine base of (p)ppGpp. The electron density map of the ω subunit N-terminus Met residue was not observed in the structures of RNA polymerase-(p)ppGpp complexes and RNA polymerase itself (28). The second residue of the ω subunit is Ala, which is known to be one of the preferred residues neighboring N-terminal Met for a protein to become a target for methionine aminopeptidase (37). Therefore, the ω subunit N-terminus Met is likely to be removed post-translationally by a methionine aminopeptidase, which was confirmed by Mass spec analysis (data not shown).
DISCUSSION
If there were no permeability barriers, the most direct way to estimate the potency of ppGpp and pppGpp in principle would be to add each nucleotide to growing cells and measure their regulatory effects. However, E. coli is impermeable to (p)ppGpp (H.A. Raue and M. Cashel unpublished). Instead, gratuitous induction of ppGpp or pppGpp is needed to differentially elevate intracellular concentrations. Ideally the induction system should not have physiological consequences itself other than the preferential accumulation of either pppGpp or ppGpp.
To achieve this, we have eliminated relA, the major wild-type source of E. coli (p)ppGpp synthetase and used a heterologous Relseq source of (p)ppGpp synthetase that does not interact with E. coli ribosomes (21,38). We then manipulated ppGpp and pppGpp abundance using Relseq mutant synthetase variants that, when induced preferentially accumulate one or the other nucleotide (19). We have estimated activity differences under induction conditions that minimize the chance of unintentionally inducing sources of stress other than those that accompany the presence of (p)ppGpp itself. Manipulation of GppA abundance provided fine tuning of ppGpp or pppGpp accumulation. To verify conclusions from in vivo experiments with purified components in vitro, we optimized methods for preparative synthesis of ppGpp and pppGpp using purified RelSeq1-385 and GppA proteins. We are aware that the arabinose induction of gene expression that we use here is autocatalytic (39–41). This means that progressive concentrations of arabinose fully induce a progressively larger portion of cells in a population, rather than an incremental induction of all cells; this is because arabinose induces its own uptake. However, when arabinose concentrations are varied, this approach does provide a test of reproducibility and the equivalence of replicate induction effects at increasing cell densities.
We did encounter problems in these experiments with the spontaneous appearance of fast growing variants. These probably arose either from insertion of the plasmid into the chromosome thereby lowering the copy number and/or from inactivation of (p)ppGpp synthetase on the plasmid. These explanations are inferred by finding that the use of recA mutant host strains confers enhanced stability. Although the recA hosts are stable sources for plasmids, these strains also grow slowly, which complicates estimates of (p)ppGpp effects on growth. Glucose was added to overnight grown cultures to catabolite repress residual inducing activity in the absence of arabinose to largely abolish selection for suppressor mutations. However, glucose must be completely exhausted before arabinose addition so as not to interfere with induction.
The substrate specificities of enzymes that remove the 3′-pyrophosphoryl group of ppGpp or pppGpp to yield GDP or GTP, respectively, could contribute to preferential accumulation of synthetic products. All of the strains used in this study retain an E. coli wild-type chromosomal spoT gene as one source of endogenous hydrolase activity. The induced RelSeq1-385, but not the 79–385, fragment encodes another (p)ppGpp hydrolase. Neither of these purified proteins shows a substrate preference for ppGpp or pppGpp (31). As equimolar mixtures of ppGpp and pppGpp do mutually compete as substrates, the predominant of the two substrates should reduce the rate of hydrolysis of the minority nucleotide. It is consistent that a large excess of pppGpp over ppGpp is seen to be accompanied by persistent levels of ppGpp with relatively minor effects of deleting gppA in addition (Figure 1, lanes 3 and 4; Figure 2, panel B). In contrast, preferential induction of ppGpp as the major substrate is not accompanied by analogous high levels of pppGpp (Figure 1, lanes 1 and 2; Figure 2A). We suggest that this disparity might be due to overexpression of plasmid borne gppA, which is specific for pppGpp conversion to ppGpp and thereby counteracts the otherwise stabilizing effects of excess pppGpp competing with ppGpp for hydrolysis. Somerville and Ahmed (42) noticed that in the absence of gppA, the elevation of pppGpp to levels less than observed here (nearly equal to ppGpp) gave measurable stabilizing effects on ppGpp decay.
This raises the question of the functional roles of GppA in E. coli. It could now be argued that one role could be to fine tune the more potent regulatory activity of ppGpp. GppA is not found in all bacteria, and that even encountered homologs may have different catalytic properties (43).
The ppx gene product, polyphosphate phosphatase, has also been noted to degrade pppGpp to ppGpp by removal of the 5′-γ-phosphate residue, like GppA (44). Conversely, guanosine pentaphosphate phosphohydrolase (GppA) of E.coli is a long-chain exopolyphosphatase (44). Unlike gppA, however, the presence or absence of the chromosomal ppx does not alter ppGpp/pppGpp ratios when (p)ppGpp is elevated during a stringent response provoked by amino acid starvation (Cashel, M, unpublished). The presence of wild-type chromosomal ppx in our strains therefore probably does not influence these experiments.
The early work on (p)ppGpp regulation revealed its accumulation during severe starvation protocols that themselves limited cell growth; it then was a surprise to find that gratuitous IPTG-mediated induction of E. coli relA expression, which led to (p)ppGpp accumulation even in the absence of starvation also severely limited growth, and thereby facilitated adaptive stress responses (45). Here, we encounter another surprise, which is that pppGpp may not be the regulatory equivalent, or the ‘regulatory sister’ of ppGpp, which seemed to be the prevailing view (see ‘Introduction’ section). We found instead that cells with nearly as much pppGpp as GTP have a doubling time of about two hrs (Figures 2B and 3, Table 2 and form small colonies on plates incubated overnight at 37°C. These are high levels of pppGpp, similar to ppGpp accumulation in wild-type cells during a full blown stringent response to amino acid starvation (2). Moreover, sustained balanced growth occurs, despite high pppGpp, apparently without deleterious effects on cells (data not shown). In contrast, colonies do not form after 2 days when ppGpp is induced in recA hosts under these conditions leading to maximum levels of only about one-fifth of GTP (as in Figures 2A and 3 and Tables 2 and 3). Quantitative estimates of relative potency for growth rate inhibition reveal that ppGpp is ∼10 times more potent an inhibitor than pppGpp.
The greater effects of ppGpp versus pppGpp on growth rate regulation in our experiments are not an isolated finding. Instead, we find that ppGpp appears to also function in E. coli as a more effective regulator than pppGpp in all four of the remaining regulatory phenomena tested besides inhibition of growth rate. These include the following: (i) RNA/DNA ratios, thought by most to define the growth rate control phenomenon (Table 3, Figure 3B); (ii) inhibition of rrnB P1 transcription initiation in vitro (Figure 4); (iii) activation of initiation of pthr promoter transcription (Figure 5); and (iv) activation of RpoS protein stabilization as judged by RpoS-LacZ-protein fusions (Figure 6). The differential regulatory effects found are quantatitively different in the various assays, and there are quantitative disparities between regulatory effects that might be expected to be related (such as effects on growth rate and on rrnBP1 transcription initiation). Nevertheless, it is evident that ppGpp is a more potent regulator than pppGpp when tested in vitro and in vivo.
With respect to transcription, (p)ppGpp regulation at the level of gene expression in E. coli is now known to entail major contributions from the DksA protein, and our assays accordingly compare the in vitro potency of ppGpp versus pppGpp on RNA polymerase transcription including DksA as a variable. We find it interesting that differences in levels of inhibition (rrnBP1 activity) diminish with increasing both ppGpp and pppGpp concentrations when DksA is present at high levels (Figure 4), whereas differences in activation of the threonine operon promoter appear to persist systematically (Figure 5). This behavior could be explained by early saturation of inhibition of rrnBP1, i.e. a point is reached when the inhibition cannot be made more severe. In contrast, activation of pthr activity does not appear to be saturated in our system even at the highest DksA concentrations studied.
The ppGpp-binding site in the Thermus RNA polymerase crystal structure found previously was near the catalytic site [ref. (46)] but subsequent mutant studies failed to verify its relevance to E. coli transcription regulation by ppGpp and DksA (47,48). Here, a binding site for pppGpp and ppGpp was found at the interface between β′ and ω subunits (Figure 7, Supplementary Movie 1) by soaking each into existing crystals of E. coli holoenzyme. The conformations of the N-termini of the ω subunits of E. coli and Thermus thermophilus are different (28). This probably accounts for the absence of a ppGpp-binding site at the interface between β′ and ω subunits in Thermus RNA polymerase structure. In E. coli, ω is also known to facilitate the assembly and maturation of the β′ subunit of RNA polymerase (49).
The (p)ppGpp binding site shown in the figure is ∼30 Å away from the catalytic pocket for RNA polymerization. Is it possible that binding to this site can nevertheless influence the RNA polymerase catalysis? Such a scenario seems probable because the (p)ppGpp binding site is located on the surface of a double-psi β-barrel (DPBB) domain of the β’ subunit (see Supplementary Movie 1), which forms the active site of RNA polymerase coordinating the catalytic Mg2+ by the—DFDGD—motif, found in all cellular RNA polymerases (50). Binding of (p)ppGpp on the surface of the DPBB domain might induce a signal, which is transmitted to the other side of the DPBB domain to regulate transcription. This possibility is further enhanced by the notion that during Mycobacterium tuberculosis pathogenesis, rifampicin exposures result in the appearance of more virulent strains comprised of antibiotic resistant mutants that grow slowly. Increased virulence is due to compensatory (suppressor) mutations, for which the DPBB domain was found to be a hot spot (51). Although the source of the signal in this example is different, it can be imagined that both (p)ppGpp and the compensatory mutations use a similar allosteric signal to modify the transcription activity of RNA polymerase.
The study presented here raises two fundamental questions. The first is whether the (p)ppGpp-binding site found here is functional for (p)ppGpp-mediated regulation of transcription. The second question is whether the binding of ppGpp and pppGpp can explain how it is that pppGpp is a less potent regulator than ppGpp, despite the similarities of their binding. A limited literature and a large number of experimental approaches are available that bear on these questions.
There are three reports supporting the notion that the (p)ppGpp-binding site found in this study is functional for (p)ppGpp-mediated transcription regulation. These studies showed that the presence of ω is required for ppGpp-dependent transcription inhibition in vitro (52,53) and ppGpp-dependent transcription activation in vivo (54). However, there is also a report that the complete absence of ω did not alter (p)ppGpp regulation of RNA accumulation during the stringent response in vivo (55). We therefore plan future studies to evaluate the physiological role of the (p)ppGpp-binding site found here from RNA polymerase crystal structures.
The structures of ppGpp and pppGpp contacting RNA polymerase residues are similar, but not identical due to the presence of an extra 5’ phosphate group in pppGpp (Figure 7). A goal of this study is to provide the means to explore the relation between regulator structure and function. Future mutant studies are planned to address the key question of whether the (p)ppGpp binding site identified here is functional. Comparison of the E. coli RNA polymerase crystal structures of the (p)ppGpp-free and the ppGpp and pppGpp bound complexes formed by soaking into pre-existing protein crystals may not be sufficient to provoke structural changes of RNA polymerase, as mentioned earlier in the text. It is also possible that static X-ray crystal structures may be insufficient to understand the mechanism of action of (p)ppGpp. Additional experimental approaches such as Raman crystallography and molecular dynamic simulation might also be necessary to investigate active site dynamics on binding of (p)ppGpp. Raman crystallographic approaches have been used to characterize subtle chemical changes of macromolecule occurring in single crystals on binding of ligands (56) and to monitor the transcription initiation process of a single-unit RNA polymerase (57). The molecular dynamic simulations can also explain how a point mutation remote from the active site is able to influence the transcription fidelity of viral RNA-dependent RNA polymerase (58).
Another class of explanations for differences in regulatory potency is that perhaps affinities of ppGpp and pppGpp for RNA polymerase are different or that ppGpp and pppGpp interact differently with RNA polymerase in the presence of low physiological concentrations of DksA than in its absence or presence in high concentrations. Future studies of (p)ppGpp affinities for RNA polymerase are planned. Still more possibilities are raised by the fact that there is evidence of binding of ppGpp to multiple RNA polymerase sites but yet without functional verification (46,48,59,60). In addition, suppressor alleles that reverse ppGpp0 phenotypes are found at multiple locations in RNA polymerase subunits encoded by rpoB, rpoC and rpoD for reasons not yet clear (2). These placements could hint that multiple binding sites or multiple RNA polymerase conformations might mimic (p)ppGpp binding, again suggestive of multiple mechanisms of action. The relatively simple means to prepare pure (p)ppGpp reported here should facilitate in vitro experiments to explore differential effects of ppGpp and pppGpp on RNA polymerase binding and on individual steps in regulatory behavior.
A basic question for future work is whether the regulatory effects of ppGpp and pppGpp on gene expression are qualitative or quantitative. Clues to regulatory differences between ppGpp and pppGpp could in principle, come from transcriptional profiling during strongly preferential accumulation of ppGpp or pppGpp, or even effects of the absence of ω. These might well reveal effects on gene expression that are different in different organisms, particularly E. coli and B. subtilis. Such studies could better define the spectrum of regulatory events governed by each nucleotide. So far, our data suggest that pppGpp may simply be a less effective regulator with the same range of targets as ppGpp.
Whatever the general comparisons of regulatory potency for ppGpp versus pppGpp turn out to be in E. coli, this relationship is unlikely to hold for all bacteria, given the abundant bacterial diversity with respect to enzymes in (p)ppGpp metabolism (61), regulation (62) and the different essential roles played by ppGpp for a variety of bacterial pathogens (63).
More generally, it now appears feasible to employ analogous approaches as a tool, possibly combined with protein engineering to alter synthetase or hydrolase substrate specificities, to explore the regulatory effects on fundamental processes in a single model host. There are already several additional putative members of the ppGpp family of nucleotides as candidates for study. These include isomers of GTP (ppGp and pGpp), NUDIX degradation products of ppGpp, (pGp) and nucleotide chimeras (A5′-ppp5′-Gpp) (64–68).
ACCESSION NUMBERS
4JK1, 4JK2 (PDB).
SUPPLEMENTARY DATA
Supplementary Data are available on NAR Online: Supplementary Table 1, Supplementary Figures 1–4 and Supplementary Movie 1.
FUNDING
Intramural Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development of the National Institutes of Health, and by National Institutes of Health grant [GM087350-A1 (to K.S.M.)]. Funding for open access charge: Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health, Bethesda, Maryland 20892 (USA). Intramural Program. Contact M. Cashel.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Martin Blum for his help in media preparation. They thank Tomoko Bowser for preparing crystals, the staff at the MacCHESS and the BCSB for support crystallographic data collection. PyMOL (http://www.pymol.org/) was used for preparing figures and the movie.
REFERENCES
- 1.Potrykus K, Cashel M. (p)ppGpp: still magical? Annu. Rev. Microbiol. 2008;62:35–51. doi: 10.1146/annurev.micro.62.081307.162903. [DOI] [PubMed] [Google Scholar]
- 2.Cashel M, Genrty DR, Hernandez VJ, Vinella D. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. 2nd edn. Neidhardt FC, et al., editors. Washington, DC: ASM Press; 1996. pp. 1458–1496. [Google Scholar]
- 3.Cashel M. The control of ribonucleic acid synthesis in Escherichia coli. IV. Relevance of unusual phosphorylated compounds from amino acid-starved stringent strains. J. Biol. Chem. 1969;244:3133–3141. [PubMed] [Google Scholar]
- 4.Laffler T, Gallant JA. Stringent control of protein synthesis in E. coli. Cell. 1974;3:47–49. doi: 10.1016/0092-8674(74)90036-1. [DOI] [PubMed] [Google Scholar]
- 5.Lazzarini RA, Cashel M, Gallant J. On the regulation of guanosine tetraphosphate levels in stringent and relaxed strains of Escherichia coli. J. Biol. Chem. 1971;246:4381–4385. [PubMed] [Google Scholar]
- 6.Winslow RM. A consequence of the rel gene during a glucose to lactate downshift in Escherichia coli. The rates of ribonucleic acid synthesis. J. Biol. Chem. 1971;246:4872–4877. [PubMed] [Google Scholar]
- 7.Gallant JA. Stringent control in E. coli. Annu Rev Genet. 1979;13:393–415. doi: 10.1146/annurev.ge.13.120179.002141. [DOI] [PubMed] [Google Scholar]
- 8.Baracchini E, Bremer H. Stringent and growth control of rRNA synthesis in Escherichia coli are both mediated by ppGpp. J. Biol. Chem. 1988;263:2597–2602. [PubMed] [Google Scholar]
- 9.Gaal T, Gourse RL. Guanosine 3′-diphosphate 5′-diphosphate is not required for growth rate-dependent control of rRNA synthesis in Escherichia coli. Proc. Natl Acad. Sci. USA. 1990;87:5533–5537. doi: 10.1073/pnas.87.14.5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ryals J, Little R, Bremer H. Control of rRNA and tRNA syntheses in Escherichia coli by guanosine tetraphosphate. J. Bacteriol. 1982;151:1261–1268. doi: 10.1128/jb.151.3.1261-1268.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ehrenberg MN, Bremer H, Dennis PP. Medium-dependent control of the bacterial growth rate. Biochimie. 2013;95:643–658. doi: 10.1016/j.biochi.2012.11.012. [DOI] [PubMed] [Google Scholar]
- 12.Potrykus K, Murphy H, Philippe N, Cashel M. ppGpp is the major source of growth rate control in E. coli. Environ. Microbiol. 2011;13:563–575. doi: 10.1111/j.1462-2920.2010.02357.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kanjee U, Ogata K, Houry WA. Direct binding targets of the stringent response alarmone (p)ppGpp. Mol. Microbiol. 2012;85:1029–1043. doi: 10.1111/j.1365-2958.2012.08177.x. [DOI] [PubMed] [Google Scholar]
- 14.Wang JD, Sanders GM, Grossman AD. Nutritional control of elongation of DNA replication by (p)ppGpp. Cell. 2007;128:865–875. doi: 10.1016/j.cell.2006.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rymer RU, Solorio FA, Tehranchi AK, Chu C, Corn JE, Keck JL, Wang JD, Berger JM. Binding mechanism of metal-NTP substrates and stringent-response alarmones to bacterial DnaG-type primases. Structure. 2012;20:1478–1489. doi: 10.1016/j.str.2012.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maciag M, Kochanowska M, Lyzen R, Wegrzyn G, Szalewska-Palasz A. ppGpp inhibits the activity of Escherichia coli DnaG primase. Plasmid. 2010;63:61–67. doi: 10.1016/j.plasmid.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 17.Cashel M. Preparation of guanosine tetraphosphate (ppGpp) and guanosine pentaphosphate (pppGpp) from Escherichia coli ribosomes. Anal. Biochem. 1974;57:100–107. doi: 10.1016/0003-2697(74)90056-6. [DOI] [PubMed] [Google Scholar]
- 18.Kuroda A, Murphy H, Cashel M, Kornberg A. Guanosine tetra- and pentaphosphate promote accumulation of inorganic polyphosphate in Escherichia coli. J. Biol. Chem. 1997;272:21240–21243. doi: 10.1074/jbc.272.34.21240. [DOI] [PubMed] [Google Scholar]
- 19.Mechold U, Murphy H, Brown L, Cashel M. Intramolecular regulation of the opposing (p)ppGpp catalytic activities of Rel(Seq), the Rel/Spo enzyme from Streptococcus equisimilis. J. Bacteriol. 2002;184:2878–2888. doi: 10.1128/JB.184.11.2878-2888.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brown L, Gentry D, Elliott T, Cashel M. DksA affects ppGpp induction of RpoS at a translational level. J. Bacteriol. 2002;184:4455–4465. doi: 10.1128/JB.184.16.4455-4465.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mechold U, Cashel M, Steiner K, Gentry D, Malke H. Functional analysis of a relA/spoT gene homolog from Streptococcus equisimilis. J. Bacteriol. 1996;178:1401–1411. doi: 10.1128/jb.178.5.1401-1411.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gentry DR, Hernandez VJ, Nguyen LH, Jensen DB, Cashel M. Synthesis of the stationary-phase sigma factor sigma s is positively regulated by ppGpp. J. Bacteriol. 1993;175:7982–7989. doi: 10.1128/jb.175.24.7982-7989.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hogg T, Mechold U, Malke H, Cashel M, Hilgenfeld R. Conformational antagonism between opposing active sites in a bifunctional RelA/SpoT homolog modulates (p)ppGpp metabolism during the stringent response [corrected] Cell. 2004;117:57–68. doi: 10.1016/s0092-8674(04)00260-0. [DOI] [PubMed] [Google Scholar]
- 25.Cashel M, Kalbacher B. The control of ribonucleic acid synthesis in Escherichia coli. V. Characterization of a nucleotide associated with the stringent response. J. Biol. Chem. 1970;245:2309–2318. [PubMed] [Google Scholar]
- 26.Potrykus K, Vinella D, Murphy H, Szalewska-Palasz A, D'Ari R, Cashel M. Antagonistic regulation of Escherichia coli ribosomal RNA rrnB P1 promoter activity by GreA and DksA. J. Biol. Chem. 2006;281:15238–15248. doi: 10.1074/jbc.M601531200. [DOI] [PubMed] [Google Scholar]
- 27.Miller JH. Experiments in molecular genetics. New York: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 28.Murakami KS. The X-ray Crystal Structure of Escherichia Coli RNA Polymerase σ70 Holoenzyme. J. Biol. Chem. 2013;288:9126–9134. doi: 10.1074/jbc.M112.430900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 30.Afonine PV, Mustyakimov M, Grosse-Kunstleve RW, Moriarty NW, Langan P, Adams PD. Joint X-ray and neutron refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 2010;66:1153–1163. doi: 10.1107/S0907444910026582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 32.Bremer H, Dennis PP. In: Escherichia coli and Salmonella: cellular and molecular biology. 2nd edn. Neidhardt FC, et al., editors. Vol. 1. Washington, DC: ASM Press; 1996. pp. 1553–1569. [Google Scholar]
- 33.Paul BJ, Barker MM, Ross W, Schneider DA, Webb C, Foster JW, Gourse RL. DksA: a critical component of the transcription initiation machinery that potentiates the regulation of rRNA promoters by ppGpp and the initiating NTP. Cell. 2004;118:311–322. doi: 10.1016/j.cell.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 34.Barker MM, Gaal T, Josaitis CA, Gourse RL. Mechanism of regulation of transcription initiation by ppGpp. I. Effects of ppGpp on transcription initiation in vivo and in vitro. J. Mol. Biol. 2001;305:673–688. doi: 10.1006/jmbi.2000.4327. [DOI] [PubMed] [Google Scholar]
- 35.Bougdour A, Gottesman S. ppGpp regulation of RpoS degradation via anti-adaptor protein IraP. Proc. Natl Acad. Sci. USA. 2007;104:12896–12901. doi: 10.1073/pnas.0705561104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Battesti A, Majdalani N, Gottesman S. The RpoS-mediated general stress response in Escherichia coli. Annu. Rev. Microbiol. 2011;65:189–213. doi: 10.1146/annurev-micro-090110-102946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Flinta C, Persson B, Jornvall H, Heijne G. Sequence determinants of cytosolic N-terminal protein processing. Eur. J. Biochem. 1986;154:193–196. doi: 10.1111/j.1432-1033.1986.tb09378.x. [DOI] [PubMed] [Google Scholar]
- 38.Mechold U, Malke H. Characterization of the stringent and relaxed responses of Streptococcus equisimilis. J Bacteriol. 1997;179:2658–2667. doi: 10.1128/jb.179.8.2658-2667.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Novick A, Weiner M. Enzyme Induction as an All-or-None Phenomenon. Proc. Natl Acad. Sci. USA. 1957;43:553–566. doi: 10.1073/pnas.43.7.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Siegele DA, Hu JC. Gene expression from plasmids containing the araBAD promoter at subsaturating inducer concentrations represents mixed populations. Proc. Natl Acad. Sci. USA. 1997;94:8168–8172. doi: 10.1073/pnas.94.15.8168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morgan-Kiss RM, Wadler C, Cronan JE., Jr Long-term and homogeneous regulation of the Escherichia coli araBAD promoter by use of a lactose transporter of relaxed specificity. Proc. Natl Acad. Sci. USA. 2002;99:7373–7377. doi: 10.1073/pnas.122227599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Somerville CR, Ahmed A. Mutants of Escherichia coli defective in the degradation of guanosine 5′-triphosphate, 3′-diphosphate (pppGpp) Mol. Gen. Genet. 1979;169:315–323. doi: 10.1007/BF00382277. [DOI] [PubMed] [Google Scholar]
- 43.Choi MY, Wang Y, Wong LL, Lu BT, Chen WY, Huang JD, Tanner JA, Watt RM. The two PPX-GppA homologues from Mycobacterium tuberculosis have distinct biochemical activities. PLoS One. 7:e42561. doi: 10.1371/journal.pone.0042561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Keasling JD, Bertsch L, Kornberg A. Guanosine pentaphosphate phosphohydrolase of Escherichia coli is a long-chain exopolyphosphatase. Proc. Natl Acad. Sci. USA. 1993;90:7029–7033. doi: 10.1073/pnas.90.15.7029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schreiber G, Metzger S, Aizenman E, Roza S, Cashel M, Glaser G. Overexpression of the relA gene in Escherichia coli. J. Biol. Chem. 1991;266:3760–3767. [PubMed] [Google Scholar]
- 46.Artsimovitch I, Patlan V, Sekine S, Vassylyeva MN, Hosaka T, Ochi K, Yokoyama S, Vassylyev DG. Structural basis for transcription regulation by alarmone ppGpp. Cell. 2004;117:299–310. doi: 10.1016/s0092-8674(04)00401-5. [DOI] [PubMed] [Google Scholar]
- 47.Kasai K, Nishizawa T, Takahashi K, Hosaka T, Aoki H, Ochi K. Physiological analysis of the stringent response elicited in an extreme thermophilic bacterium, Thermus thermophilus. J. Bacteriol. 2006;188:7111–7122. doi: 10.1128/JB.00574-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vrentas CE, Gaal T, Berkmen MB, Rutherford ST, Haugen SP, Vassylyev DG, Ross W, Gourse RL. Still looking for the magic spot: the crystallographically defined binding site for ppGpp on RNA polymerase is unlikely to be responsible for rRNA transcription regulation. J. Mol. Biol. 2008;377:551–564. doi: 10.1016/j.jmb.2008.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mathew R, Chatterji D. The evolving story of the omega subunit of bacterial RNA polymerase. Trends Microbiol. 2006;14:450–455. doi: 10.1016/j.tim.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 50.Iyer LM, Koonin EV, Aravind L. Evolutionary connection between the catalytic subunits of DNA-dependent RNA polymerases and eukaryotic RNA-dependent RNA polymerases and the origin of RNA polymerases. BMC Struct. Biol. 2003;3:1. doi: 10.1186/1472-6807-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Comas I, Borrell S, Roetzer A, Rose G, Malla B, Kato-Maeda M, Galagan J, Niemann S, Gagneux S. Whole-genome sequencing of rifampicin-resistant Mycobacterium tuberculosis strains identifies compensatory mutations in RNA polymerase genes. Nat. Genet. 2012;44:106–110. doi: 10.1038/ng.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Igarashi K, Fujita N, Ishihama A. Promoter selectivity of Escherichia coli RNA polymerase: omega factor is responsible for the ppGpp sensitivity. Nucleic Acids Res. 1989;17:8755–8765. doi: 10.1093/nar/17.21.8755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vrentas CE, Gaal T, Ross W, Ebright RH, Gourse RL. Response of RNA polymerase to ppGpp: requirement for the omega subunit and relief of this requirement by DksA. Genes Dev. 2005;19:2378–2387. doi: 10.1101/gad.1340305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Costanzo A, Nicoloff H, Barchinger SE, Banta AB, Gourse RL, Ades SE. ppGpp and DksA likely regulate the activity of the extracytoplasmic stress factor sigmaE in Escherichia coli by both direct and indirect mechanisms. Mol. Microbiol. 2008;67:619–632. doi: 10.1111/j.1365-2958.2007.06072.x. [DOI] [PubMed] [Google Scholar]
- 55.Gentry D, Xiao H, Burgess R, Cashel M. The omega subunit of Escherichia coli K-12 RNA polymerase is not required for stringent RNA control in vivo. J. Bacteriol. 1991;173:3901–3903. doi: 10.1128/jb.173.12.3901-3903.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Carey PR, Chen Y, Gong B, Kalp M. Kinetic crystallography by Raman microscopy. Biochim. Biophys. Acta. 2011;1814:742–749. doi: 10.1016/j.bbapap.2010.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen Y, Basu R, Gleghorn ML, Murakami KS, Carey PR. Time-resolved events in the reaction pathway of transcript initiation by a single-subunit RNA polymerase: Raman crystallographic evidence. J. Am. Chem. Soc. 2011;133:12544–12555. doi: 10.1021/ja201557w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Moustafa IM, Shen H, Morton B, Colina CM, Cameron CE. Molecular dynamics simulations of viral RNA polymerases link conserved and correlated motions of functional elements to fidelity. J. Mol. Biol. 2011;410:159–181. doi: 10.1016/j.jmb.2011.04.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chatterji D, Fujita N, Ishihama A. The mediator for stringent control, ppGpp, binds to the beta-subunit of Escherichia coli RNA polymerase. Genes Cells. 1998;3:279–287. doi: 10.1046/j.1365-2443.1998.00190.x. [DOI] [PubMed] [Google Scholar]
- 60.Toulokhonov II, Shulgina I, Hernandez VJ. Binding of the transcription effector ppGpp to Escherichia coli RNA polymerase is allosteric, modular, and occurs near the N terminus of the beta'-subunit. J. Biol. Chem. 2001;276:1220–1225. doi: 10.1074/jbc.M007184200. [DOI] [PubMed] [Google Scholar]
- 61.Atkinson GC, Tenson T, Hauryliuk V. The RelA/SpoT homolog (RSH) superfamily: distribution and functional evolution of ppGpp synthetases and hydrolases across the tree of life. PLoS One. 2011;6:e23479. doi: 10.1371/journal.pone.0023479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dalebroux ZD, Swanson MS. ppGpp: magic beyond RNA polymerase. Nat. Rev. Microbiol. 2012;10:203–212. doi: 10.1038/nrmicro2720. [DOI] [PubMed] [Google Scholar]
- 63.Dalebroux ZD, Yagi BF, Sahr T, Buchrieser C, Swanson MS. Distinct roles of ppGpp and DksA in Legionella pneumophila differentiation. Mol. Microbiol. 2010;76:200–219. doi: 10.1111/j.1365-2958.2010.07094.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ito D, Kato T, Maruta T, Tamoi M, Yoshimura K, Shigeoka S. Enzymatic and molecular characterization of arabidopsis ppGpp pyrophosphohydrolase, AtNUDX26. Biosci. Biotechnol. Biochem. 2012;76:2236–2241. doi: 10.1271/bbb.120523. [DOI] [PubMed] [Google Scholar]
- 65.Kramer GF, Baker JC, Ames BN. Near-UV stress in Salmonella typhimurium: 4-thiouridine in tRNA, ppGpp, and ApppGpp as components of an adaptive response. J. Bacteriol. 1988;170:2344–2351. doi: 10.1128/jb.170.5.2344-2351.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ooga T, Ohashi Y, Kuramitsu S, Koyama Y, Tomita M, Soga T, Masui R. Degradation of ppGpp by nudix pyrophosphatase modulates the transition of growth phase in the bacterium Thermus thermophilus. J. Biol. Chem. 2009;284:15549–15556. doi: 10.1074/jbc.M900582200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pao CC, Gallant J. A new nucleotide involved in the stringent response in Escherichia coli. Guanosine 5′-diphosphate-3′-monophosphate. J. Biol. Chem. 1979;254:688–692. [PubMed] [Google Scholar]
- 68.Sajish M, Kalayil S, Verma SK, Nandicoori VK, Prakash B. The significance of EXDD and RXKD motif conservation in Rel proteins. J. Biol. Chem. 2009;284:9115–9123. doi: 10.1074/jbc.M807187200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.