Abstract
The adaptive immunity of bacteria against foreign nucleic acids, mediated by CRISPR (clustered regularly interspaced short palindromic repeats), relies on the specific incorporation of short pieces of the invading foreign DNA into a special genomic locus, termed CRISPR array. The stored sequences (spacers) are subsequently used in the form of small RNAs (crRNAs) to interfere with the target nucleic acid. We explored the DNA-binding mechanism of the immunization protein Csn2 from the human pathogen Streptococcus agalactiae using different biochemical techniques, atomic force microscopic imaging and molecular dynamics simulations. The results demonstrate that the ring-shaped Csn2 tetramer binds DNA ends through its central hole and slides inward, likely by a screw motion along the helical path of the enclosed DNA. The presented data indicate an accessory function of Csn2 during integration of exogenous DNA by end-joining.
INTRODUCTION
Clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated (Cas) proteins constitute a prokaryotic adaptive immunity system to prevent viral infections or the invasion by mobile DNA elements (1–3). The immunization is achieved by the specific storage of foreign nucleic acid sequences as ‘portraits’ of the targets, which are later used in the form of small RNAs for the specific recognition and inactivation of the invading DNA. Ten CRISPR-Cas systems have been described, which differ in the organization of the CRISPR operon, constituted by a set of diverse CRISPR-associated genes (cas) and the CRISPR arrays (4). The cas genes encode the protein components of the system, which exhibit various biochemical activities (5).
The mechanism of the CRISPR-mediated immunity is divided into three stages (2,6). The first stage describes the immunization of the host cells by the integration of foreign DNA-derived spacer sequences into the CRISPR array (adaptation stage). The second stage comprises the transcription of the CRISPR array to the precursor CRISPR RNA (pre-crRNA), the expression of Cas proteins and the processing of the pre-crRNA to small crRNAs (expression/processing stage). In the third stage, the crRNA-loaded Cas protein complexes screen the foreign DNA for spacer-matching sequences and initiate the nucleolytic hydrolysis of the target DNA (interference stage). According to the recent classification of the different CRISPR-Cas systems, three major types (type I, II and III) have been defined (4), which exhibit mechanistic variability in the maturation of the crRNAs and the inactivation of the target nucleic acids (7–15).
The type II CRISPR-Cas systems, mainly found in pathogenic bacteria (16), consist of the CRISPR array, the cas genes encoding for the proteins Cas9, Cas1, Cas2 and Csn2 (in type II-A) or Cas4 (in type II-B) and a gene for the trans-encoded crRNA (tracrRNA). The tracrRNA contains sequence elements complementary to the repeat of the pre-crRNA (11). The RNA duplex formation of the tracrRNA and the pre-crRNA mediates the maturation of the crRNAs by RNaseIII and the Cas9 protein (11,17). Loaded with the tracrRNA:crRNA duplex, the Cas9 protein introduces double-strand DNA breaks at the spacer-matching protospacer region (12). Several studies have shown that the Cas9 protein along with designed crRNA:tracrRNA is active in eukaryotic cells and can be used for multiplex genome editing in bacteria and eukaryotes (18–23). Moreover, engineered Cas9 protein with inactivated nuclease domains has been used for sequence-specific control of transcription in Escherichia coli and eukaryotic cells (24).
The activities of the proteins Cas1, Cas2 and Csn2 are dispensable for maturation of the crRNAs and the cleavage of the target DNA, and thus are likely required in the immunization stage (10,11). The nucleases Cas1 and Cas2 are involved in incorporation of new spacer sequences into the CRISPR array (25–27). Little is known about the function of the Csn2 protein; although early studies showed that a knockout of the csn2 gene prevents the development of phage-insensitive mutants by an uptake of new spacer sequences (7,28), the role of the Csn2 protein during immunization is unknown. The structures and DNA-binding activities of Csn2 proteins from Enterococcus faecalis, S. agalactiae and Streptococcus pyogenes have been analyzed independently (29–31). These Csn2 proteins are ring-shaped homotetramers, and the structure of a protomer consists of a ‘head’ and a ‘leg’ domain. Whereas the leg domain consists of a pair of long stretched helices that form the interface for the tetrameric assembly, the function of the head domain is still unclear. More recently, based on the crystal structure of the Cas protein Stu0660 from Streptococcus thermophilus, a second Csn2 protein family has been proposed, which shares no sequence similarity to Csn2 proteins; however, its members have a similar tetrameric ring-shaped structure (32). All Csn2 proteins exhibit Ca2+-dependent double-stranded DNA (dsDNA)-binding activities without any notable sequence dependence. The tetrameric structure contains a large positively charged central hole of ∼4 nm in diameter, which is stabilized by Ca2+-ions bound to the helices in the leg domain. This hole is large enough to encircle the dsDNA. An alternative DNA binding mode has been suggested by molecular dynamics simulations, which revealed that the Csn2 tetramers can adopt a conformation with a groove outside the ring that could interact with the dsDNA (30).
To elucidate the role of the Csn2 protein in the CRISPR immunization stage, we have analyzed the DNA-binding mechanism of Csn2 from the human pathogen S. agalactiae in detail, using different biochemical techniques, atomic force microscopy (AFM) and molecular dynamics (MD) simulations. We provide evidence that Csn2 binds to the ends of linear dsDNA and moves inward through rotation-coupled translocation. The encircling of the dsDNA by Csn2 tetramers was probed by capping the DNA ends of Csn2–DNA complexes with streptavidin, keeping the Csn2 rings arrested on the DNA. The MD simulations give mechanistic insights at the molecular level how a Csn2 ring may rotate along the helical path of the DNA in a Ca2+-dependent manner. The data suggest that Csn2 is a DNA end-loading toroidal protein, whose function is directly related to DNA end metabolism. Csn2 thus resembles proteins involved in the repair of dsDNA breaks and recombination.
MATERIALS AND METHODS
Purification of Csn2
Streptococcus agalactiae Csn2 protein was expressed and purified as previously described (30).
Preparation of relaxed plasmid DNA for competition experiments
Relaxation of supercoiled plasmid DNA was performed with E. coli Topoisomerase I (New England Biolabs, NEB). Supercoiled pUC18-1 plasmid DNA (33) was extracted and purified from E. coli cells grown to stationary phase in YT medium. Ten micrograms of supercoiled plasmid DNA were incubated in 1 × NEBuffer 4 (50 mM potassium acetate, 20 mM Tris acetate, pH 7.9, 10 mM magnesium acetate, 1 mM DTT) and 1 µg ml−1 bovine serum albumin in a total volume of 30 µl. Five units E. coli Topoismerase I were added and the reaction mixture was incubated for 1 h at 37°C to allow complete relaxation. After extraction with phenol/chloroform and ethanol precipitation, the DNA was resuspended in deionized water (Millipore). The relaxation was verified on a 1% agarose gel. DNA bands were stained with ethidium bromide and visualized under UV light.
Analyses of protein–DNA interaction by electrophoretic mobility shift assays
The 155 bp DNA fragment was obtained by EcoRI-DraI digestion of the plasmid pUC18-1. DNA fragments were purified by agarose gel electrophoresis and end labeled by Klenow (Promega) incorporation of [α-32P]-dATP. Binding reaction mixtures contained 2 nM DNA, 10 mM CaCl2, 20 ng µl−1 heparin and indicated concentrations of Csn2 or unlabeled plasmid DNA (supercoiled, relaxed or linearized) as given in the individual experiments. After incubation for 15 min at room temperature, the complexes were separated from free DNA on native 5% (w/v) Tris/glycine polyacrylamide gels. DNA bands were visualized by autoradiography.
Preparation of DNA for AFM
The pCR001 plasmid (5125 bp) used in the AFM analyses was constructed by ligation of cas1-cas2 genes into the NdeI-PacI cleaved pACYCDuet-1 vector (Novagen). The insert with cas1-cas2 genes were obtained by PCR from genomic DNA of E. coli MG1655 using the following primers: forward 5′-GAATGCCATATGACCTGGCTTCCCCTTAAT-3′ and reverse 5′-CCGACCTTAATTAACATTTCCTTATTATTAAAGATCAGCT-3′.
The plasmid pCR001 was extracted and purified from E. coli cells grown to stationary phase in YT medium with the Qiagen Plasmid Midi Kit. To obtain relaxed DNA, 10 µg of the supercoiled plasmid was treated with E. coli Topoisomerase I as described above. Linear plasmid DNA used in the AFM analyses was produced by cleavage with the restriction enzyme Ecl136II. After relaxation or linearization reactions, the DNA was extracted with phenol/chloroform, purified with PCR purification Kit (Qiagen) and precipitated with ethanol. The relaxed and linearized DNA probes were resuspended in deionized water (Millipore).
Atomic force microscopic (AFM) analyses of Csn2–DNA complexes
Linearized or relaxed pCR001 was incubated with the Csn2 protein in the presence of 2 mM CaCl2 in a total volume of 5 µl. The DNA and protein concentrations are given in the individual experiments. The samples were incubated 10 min at room temperature to allow complex formation and diluted with 95 µl of adsorption buffer (5 mM CaCl2, 2 mM Tris–HCl, pH 8.0, 10 mM NaCl). Forty-microliter aliquots of the samples were immediately transferred to freshly cleaved mica. After 1 min incubation the mica surface was washed three times with 100 µl deionized water (Millipore). Excess water was dried with compressed N2. Imaging of Csn2–DNA complexes was performed with Nanowizard II (JPK Instruments AG, Berlin) in intermittent contact in air, using standard silicon cantilevers (OLYMPUS OMCL-AC160).
Biotinylation of dsDNA fragment and Csn2 binding studies
The 256 bp DNA fragment was obtained by EcoRI-BamHI digestion of the plasmid pUC18-1. After purification of the DNA fragments by agarose gel electrophoresis, the biotinylation of the ends was achieved by Klenow incorporation of Biotin-11-dUTP. Ten microgram of the DNA fragment was incubated with 5 U Klenow (Promega), each 0.2 mM dCTP, dGTP, dATP and Biotin-11-dUTP (GeneOn) in 1 × Klenow buffer (Promega, 50 mM Tris–HCl, pH 7.2, 10 mM MgSO4 and 100 µM DTT) in a total volume of 50 µl for 1 h at room temperature. After extraction with phenol/chloroform and ethanol precipitation, the DNA was dissolved in 10 µl TE buffer.
The binding studies with the biotinylated DNA were performed by sequential incubation of 168 ng of the biotinylated DNA in the presence of 7.2 mM CaCl2 with 4.7 µg Csn2, 14 mM ethylene glycol tetraacetic acid (EGTA) and/or 2 µg streptavidin (Sigma-Aldrich) in a total volume of 14.4 µl. The volumes of the binding reaction without EGTA or streptavidin were adjusted by addition of deionized water (Millipore). The incubation of the samples occurred in three steps, each for 15 min at room temperature. The complexes were separated from free DNA on a native 2% Tris/Acetate agarose gel. The bands were visualized by ethidium bromide staining.
MD simulations
MD simulations were performed with the AMBER 11 suite of programs (34) together with the force field as described by Cornell et al. (35) using modifications suggested by Simmerling et al. (36). To generate a starting structure of Csn2 binding to dsDNA, canonical B-DNA of 36 bp length (sequence: 5′-GTTTTAGAGCTGTGCTGTTTCGAATGGTTCCAAAAC-3′) was inserted into the central hole of tetrameric Csn2 such that the DNA is perpendicular to a least-squares fit plane through the protein atoms and that Csn2 is displaced by ∼6 Å from the center of the DNA along the double strand. The tetrameric Csn2 was taken from Ellinger et al. (30) (PDB code: 3QHQ), including the Ca2+-ions. In addition, a starting structure of Csn2 binding to dsDNA without Ca2+-ions was generated. The starting structures were placed into octahedral periodic boxes of TIP3P water molecules (37). The distance between the edges of the water box and the closest atom of the solutes was at least 11 Å, resulting in system sizes of ∼186 000 atoms. The systems were minimized by 50 steps of steepest descent minimization followed by 450 steps of conjugate gradient minimization. The particle mesh Ewald method (38) was used to treat long-range electrostatic interactions, and bond lengths involving bonds to hydrogen atoms were constrained using SHAKE (39). The time-step for all MD simulations was 2 fs, with a direct-space, non-bonded cutoff of 8 Å. Applying harmonic restraints with force constants of 5 kcal mol−1 Å−2 to all solute atoms, canonical ensemble (NVT)-MD was carried out for 50 ps, during which the system was heated from 100 to 300 K. Subsequent isothermal isobaric ensemble (NPT)-MD was used for 150 ps to adjust the solvent density. Finally, the force constants of the harmonic restraints on solute atom positions were gradually reduced to zero during 100 ps of NVT-MD. The following NVT-MD at 300 K with a time constant of 10 ps for heat-bath coupling was used for analysis, with conformations extracted every 20 ps. The simulation lengths amount to 100 ns of which the first 30 ns were discarded. Each simulation was repeated once with varied starting velocities, resulting in four independent simulations in total.
For analyzing the trajectories, conformations were superimposed with respect to the phosphorous atoms of the dsDNA or the Cα atoms of Csn2. The conformational variability of Csn2 binding to dsDNA agrees with that observed in MD simulations of Csn2 alone (30). For determining the translocation along the dsDNA, the distance between the center of the dsDNA and the average coordinates of the Cα atoms of all four helices H5 was computed. For determining the rotation of Csn2 relative to the dsDNA, the angle defined by the point triple (average coordinates of the phosphorous atoms of the five most central nucleotides on one strand; average coordinates of the phosphorous atoms of the five most central nucleotides on the other strand; average coordinates of the Cα atoms of one pair of helices H5 located in one of the legs of the tetrameric structure) was computed. For determining the kink angle of the dsDNA, the angle defined by the point triple (average coordinates of the phosphorous atoms of the five outermost base pairs at one end of the DNA; average coordinates of the phosphorous atoms of the five most central base pairs of the DNA; average coordinates of the phosphorous atoms of the five outermost base pairs at the other end of the DNA) was computed.
RESULTS
Binding of Csn2 requires free DNA ends
The Csn2 tetramers from S. agalactiae are stable in the range of pH 7.0–9.0, a wide range of ionic strengths as well as in the presence of Ca2+-chelating EGTA (30). Owing to the high stability of the ring-shaped Csn2 tetramers, it is unlikely that the quaternary structure can be opened wide enough to bind and enclose dsDNA as it is known to occur in several DNA-enclosing proteins (40,41). Therefore, if the Csn2 tetramers encircle the dsDNA the tetramers should bind DNA by threading onto the DNA ends. In contrast, a binding of the DNA outside the central hole will be independent on the presence of free DNA ends. To distinguish between these two alternative DNA-binding modes, we performed electrophoretic mobility shift assays (EMSAs) and tested whether the binding of Csn2 requires free DNA ends.
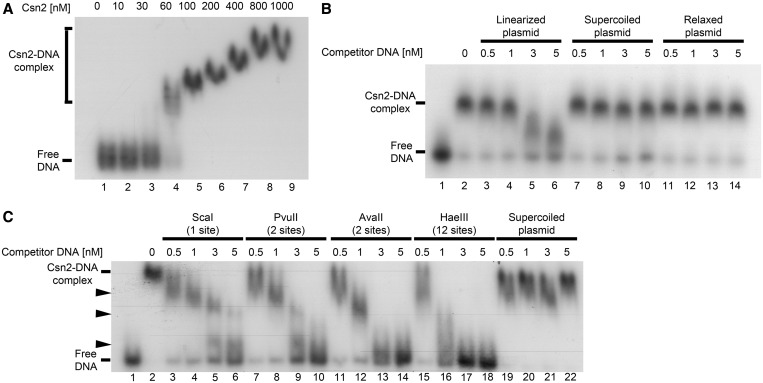
First, we incubated a 155 bp 32P-radiolabeled DNA fragment with increasing amounts of Csn2 and separated the protein–DNA complexes on a 5% native Tris/glycine polyacrylamide gel. As can be seen in Figure 1A, Csn2 formed one complex band with an estimated apparent K0.5 value between 30 and 60 nM (Figure 1A, lanes 3 and 4). The declining mobility of the complex band with increasing protein concentration indicates the binding of multiple Csn2 molecules to the DNA fragment. To test a putative requirement for a free DNA end for the interaction, we performed competition experiments with unlabeled plasmid DNA, either in closed circular or linearized form. As shown in Figure 1B (lanes 3–6), the complex formation of Csn2 with the linear radiolabeled DNA was challenged with increasing concentrations of the linearized pUC18-1 plasmid. However, the same plasmid in supercoiled or relaxed form almost did not compete for binding of Csn2 (Figure 1B, lanes 7–14; Figure 1C, lanes 19–22). This indicates that free DNA ends are necessary for Csn2-binding and supports the threading of dsDNA into the central channel of the Csn2 tetramers.
Figure 1.
Electrophoretic mobility shift assays of a radiolabeled 155 bp DNA fragment with Csn2 either in the absence (A) or in the presence of competitor DNA (B and C) are presented. In each reaction 2 nM 32P-labeled DNA, 20 ng/µl heparin, and 10 mM CaCl2 were employed. (A) Titration of Csn2 in the range of 0 to 1 µM is shown. (B) Csn2 binding to the radiolabeled DNA fragment was competed with indicated amounts of 2915 bp unlabeled plasmid DNA either in ScaI-linearized (lanes 3–6), supercoiled (lanes 7–10) or in relaxed (lanes 11–14) form. The concentration of Csn2 was constant at 60 nM in lanes 2–14. Lanes 1 and 2 show the control reactions, performed either in the absence of Csn2 (lane 1) or in the absence of competitor DNA (lane 2). (C) The same competition experiment as in (B) but with PvuII-, AvaII-, or HaeIII-cleaved competitor plasmid. The numbers of cleavage sites of the different endonucleases are given in the brackets. The black arrowheads indicate intermediate Csn2-DNA complexes, resulting from decomposition of the fully occupied complexes.
To further evaluate the dependence of the Csn2–DNA interaction on free dsDNA ends, we repeated the competition experiments with the plasmid DNA cleaved by restriction enzymes at two sites, with the aim to double the concentration of the DNA ends. The competition efficiency was higher with DNA cleaved with double cutters PvuII or AvaII compared with the ScaI-linearized DNA at the same final competitor concentration (Figure 1C, lanes 7–14). Moreover, plasmid DNA cleaved with HaeIII at 12 sites reduced the binding to the radiolabeled DNA already at the lowest competitor concentration tested (Figure 1C, lanes 15–18). Thus, the challenging efficiency depended on the concentration of free DNA ends at equal total competitor DNA concentration. The appearance of three intermediate complexes during the decay of the Csn2–DNA complexes is consistent with a binding of multiple Csn2 tetramers to the 155 bp dsDNA, and suggests a sliding of the bound proteins along the DNA after loading the DNA from its ends.
Cleavage of the pUC18-1 plasmid with the restriction enzymes ScaI, PvuII and HaeIII produces blunt ends, while cleavage with AvaII gives 5′-overhangs of three bases. The comparable challenging extent of the PvuII- and AvaII-cleaved DNA demonstrated that the DNA end-binding activity of Csn2 is not restricted to blunt ends but short 5′-protruding DNA ends are also bound by Csn2 (Figure 1C, lanes 7–14). The ability of Csn2 to bind dsDNA with 3′- and/or 5′-overhangs was further examined with different synthetic DNA substrates (Supplementary Figure S1). Csn2 did not form complexes with single-stranded DNA (ssDNA) (Supplementary Figure S1A and B, lanes 1–3). However, the hybridization of complementary oligonucleotides to ssDNA restored the Csn2–DNA interaction (Supplementary Figure S1A and B, lanes 4–6). Moreover, Csn2 formed faint complex bands with a DNA substrate that contains a 36-bp dsDNA region flanked by 29 or 34 nt ssDNA tails (Supplementary Figure S1B, lanes 7–9). These results indicate that Csn2 is in principle able to enter dsDNA regions by threading ssDNA tails in its central hole.
Dissociation of Csn2 occurs at the DNA ends: evidence for encircling the DNA by Csn2 rings
The crystal structure of the Csn2 from S. agalactiae revealed three Ca2+-ions bound to the leg domain of each protomer (30). These ions have also been found in the crystal structures of Csn2 proteins from E. faecalis and S. pyogenes (29,31). The loss of one of the Ca2+-ions within one protomer of Csn2 resulted in a reorientation of α-helices in the leg domain, which form the inner rim of the central hole (30) (Supplementary Figure S2). Thus, the Ca2+-ions are considered to be important for the stabilization of the tetramer conformation that is able to bind the dsDNA (29–31).
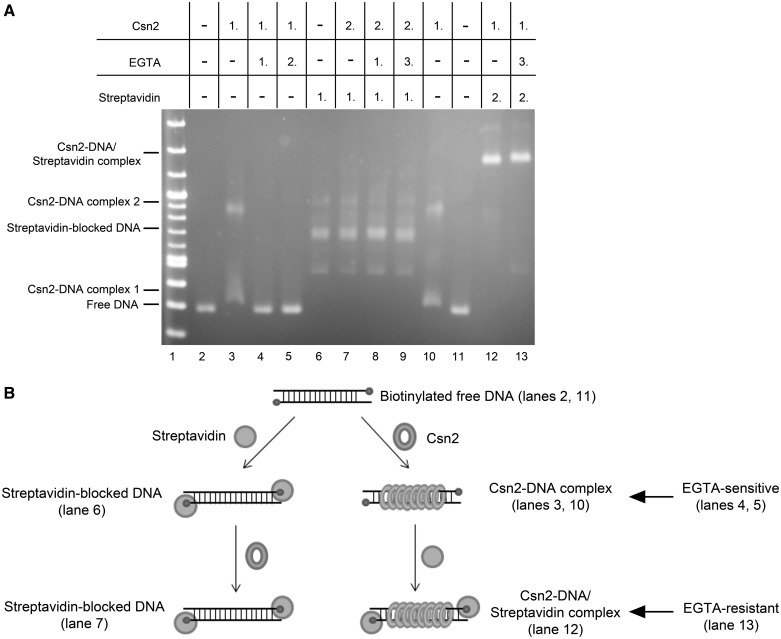
Because capturing the Ca2+-ions by EGTA did not disassemble the tetramers but abolished their DNA-binding activity (30,31), we suggested that in the presence of EGTA the conformation of the central hole becomes disordered, resulting in weakened interactions of the tetramers with the DNA backbone and leading to the dissociation of the sliding rings from the DNA ends. If this were the case, the obstruction of the DNA ends of Csn2–DNA complexes should result in arrested Csn2 rings on the DNA, and the complexes should not dissociate in the presence of EGTA. To test this, we attached biotin to both ends of a dsDNA fragment, allowing the blocking of both DNA-termini with streptavidin.
Incubation of Csn2 with biotinylated dsDNA led to the formation of two complex bands, demonstrating that biotin itself did not interfere with Csn2 binding (Figure 2A, lanes 2, 10). As shown previously (29–31), in the presence of EGTA the complex formation was completely inhibited (Figure 2, lane 4). To test whether EGTA causes the dissociation of DNA-bound Csn2 molecules, we incubated the DNA with Csn2 in the presence of Ca2+ for 15 min and then added EGTA to the binding reaction followed by incubation for additional 15 min. Indeed, the lack of Csn2–DNA complexes indicated the decomposition of the complexes in the presence of EGTA (Figure 2A, lane 5).
Figure 2.
(A) Binding analyses of Csn2 in the presence and absence of EGTA and free DNA ends on 2% Tris-acetate agarose gel. In each lane 168 ng linear DNA and 7.2 mM CaCl2 were employed. The numbers above the lanes indicate the order of addition of streptavidin (2 µg), Csn2 (4.7 µg), or EGTA (14 mM) in a total volume of 14.4 µl. Lanes 2–5: Influence of EGTA on Csn2-DNA interaction is shown. Lanes 6–9: 168 ng of the end-biotinylated DNA fragment were incubated first with streptavidin to block the DNA ends. Lanes 10 and 11: Streptavidin was added after binding of Csn2. After separation of the complexes the agarose gel was stained with ethidium bromide. (B) Schematic presentation of the binding analysis, shown in (A).
As expected, the tethering of streptavidin to the DNA ends inhibited the binding of Csn2 (Figure 2A, lanes 6 and 7), consistent with the requirement of free DNA ends for Csn2 binding. In contrast, adding streptavidin after the binding of Csn2 resulted in supershifted Csn2-DNA/streptavidin complexes (Figure 2A, lane 12). Moreover, the Csn2 proteins in these complexes were captured on the DNA because adding EGTA did not lead to a decay of the complexes (Figure 2A, lane 13). Thus, both the association as well as the dissociation of Csn2 occurs at the DNA ends, indicating a sliding activity of the Csn2 rings (Figure 2B). In the absence of Ca2+-ions, the tetramers slip down from the DNA ends, likely due to weakened electrostatic interactions between the distorted α-helices of the ring channel and the phosphate backbone of the DNA helix.
AFM of Csn2–DNA complexes
To confirm the DNA end requirement and to probe a binding of multiple Csn2 molecules, which would confirm a sliding activity of the Csn2 rings, we imaged the Csn2–DNA complexes by AFM in intermittent contact mode in air.
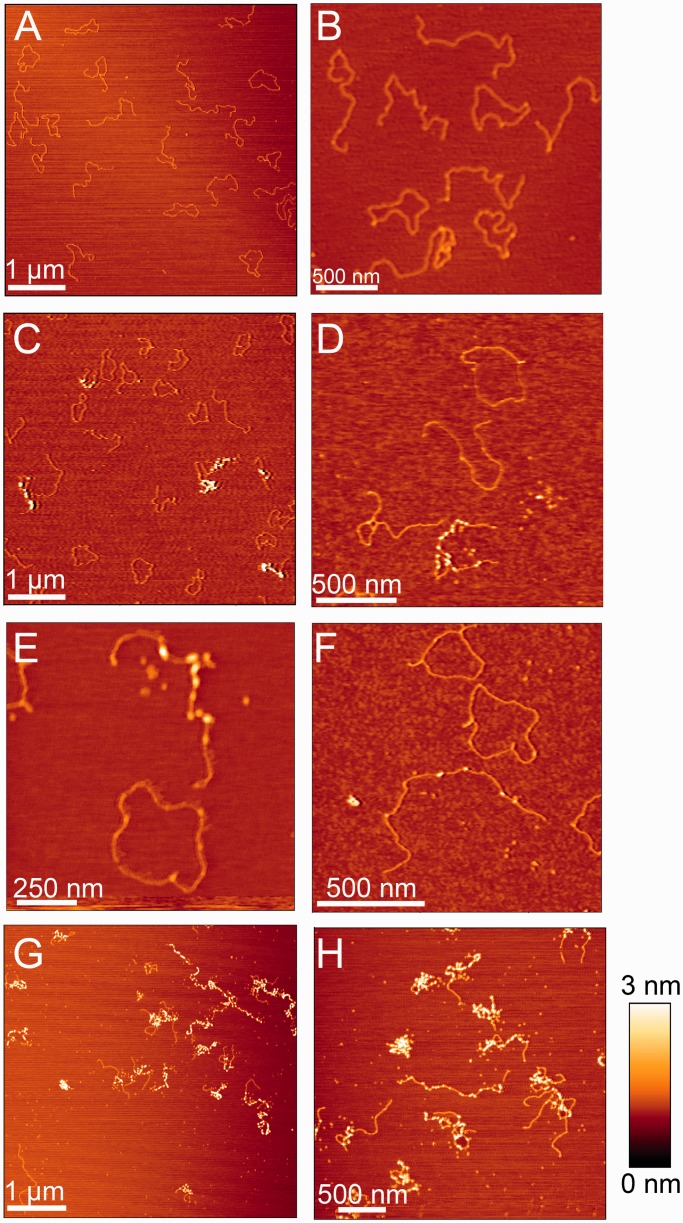
The AFM analyses were performed with the 5125 bp plasmid pCR001 as a substrate, either in linearized or relaxed circular form. AFM images of equal amounts of both DNA forms (each 1.3 nM) in the absence of Csn2 are shown Figure 3A and B. Some of the molecules of both DNA forms contained nodes, likely due to a crossover of DNA helices or a distortion/kinking of the DNA during the drying process of the probe on the mica surface (Figure 3B). AFM images of the same DNA mixture in the presence of 176 nM Csn2 showed the formation of protein–DNA complexes (Figure 3C–F). In average the ratio of the heights and widths of bound Csn2 and free DNA (Csn2-DNA/DNA) were 4.2 and 1.3, respectively. In contrast, the ratio of heights and widths of the DNA nodes relative to straight DNA were in average 1.9 and 1.5, respectively. Thus, these DNA nodes, which were also observed in the protein–free sample (Figure 3B), were not considered as protein–DNA complexes.
Figure 3.
Representative AFM images of 5125 bp plasmid DNA in the absence or presence of Csn2 are shown. (A) and (B) show images of 1.3 nM relaxed and 1.3 nM linear plasmid DNA in the absence of Csn2; (C–F) images of equal amounts of relaxed and linear plasmid DNA (each 1.3 nM) incubated with 176 nM Csn2; (G) and (H) images of 2.6 nM linear plasmid DNA incubated with 800 nM Csn2. The relative color scale range is 0–3 nm in all images.
Csn2 was exclusively associated with linear DNA (Figure 3C–F), consistent with its DNA end-dependent–binding activity as observed in the mobility shift assays. The binding was not restricted to the DNA ends but internal sites along the entire DNA fragment were also occupied by several Csn2 proteins (Figure 3D–F), indicating a sliding activity of the tetramers after initial binding at the DNA ends. Moreover, the binding of Csn2 seems to be highly cooperative, as some linear DNA molecules were free of any protein, whereas others were bound by several Csn2 proteins in the same sample (Figure 3D–F). Such an apparent cooperative binding activity is also known for the ‘recombination associated protein’ RdgC from E. coli, a ring-shaped DNA-binding protein with preference for DNA ends (42). As control, incubation of Csn2 with the relaxed plasmid in the absence of linear DNA confirmed that Csn2 does not bind to circular DNA (Supplementary Figure S3).
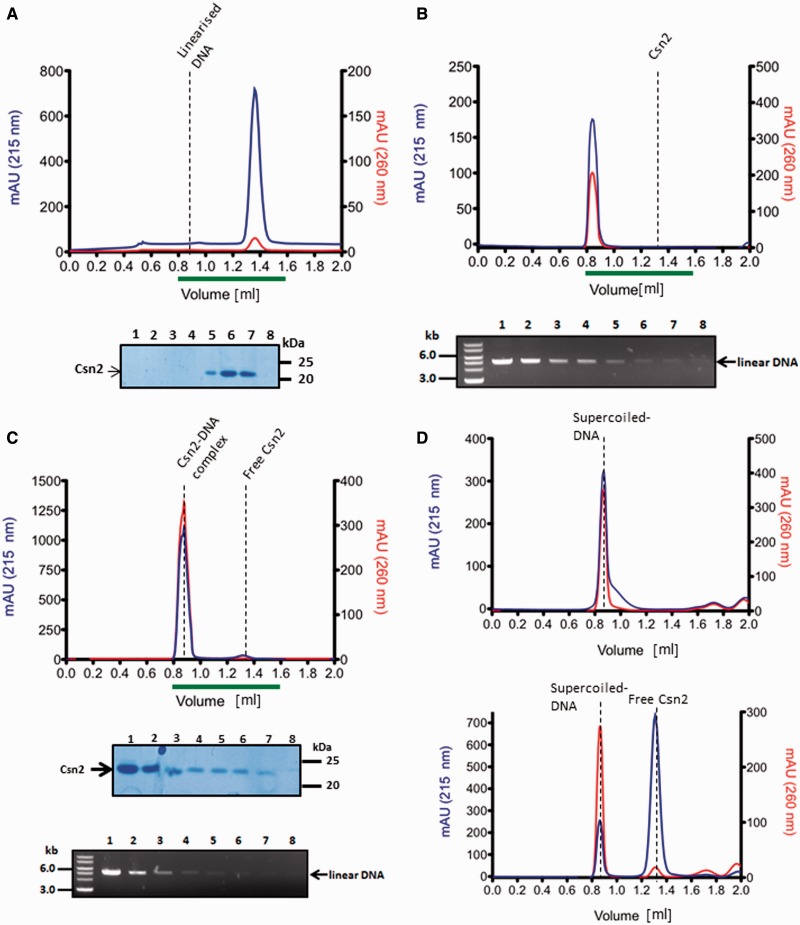
At higher Csn2 concentration, highly condensed nucleoprotein complexes were observed, likely caused by clustering of multiple Csn2 proteins and DNA-dependent self-association of DNA-bound Csn2 (Figure 3G and H). The formation of these higher-order nucleoprotein complexes should not be caused by an unspecific aggregation of the protein, as previous study has shown that Csn2 exists as stable tetramer in solution (30). To further evaluate the DNA end-dependence and to exclude an unspecific aggregation of Csn2 in the presence of DNA, we analyzed the elution profile of Csn2 in the presence of linear or closed circular plasmid DNA by size exclusion chromatography (Figure 4). Incubation of Csn2 with the linear DNA caused a co-elution of the entire Csn2 tetramers in the void fraction (Figure 4C). In contrast, the same DNA in supercoiled form did not change the elution profile of the tetrameric Csn2 (Figure 4D). The results are consistent with a selective binding of the Csn2 tetramers to the linear dsDNA and exclude an unspecific protein aggregation in the presence of DNA.
Figure 4.
Analytical gel filtration analysis of Csn2 and Csn2-DNA complexes performed with a Superdex 200 PC 3.2/30 column is shown. Elution profiles of 20 µM Csn2 (A), 30 nM linearized pCR001 plasmid (B) and Csn2-DNA complexes (C) are shown. In (D) the elution profiles of 30 nM supercoiled pCR001 alone (upper part) or in the presence of 20 µM Csn2 (lower part) are shown. 100 µl fractions were collected starting at an elution volume of 0.8 ml. Aliquots of the fractions 1 to 8, indicated by the green lines below the elution profiles, were analyzed on 10% SDS gels (A, C) and on 1% agarose gels (B, C).
MD simulations reveal Ca2+-dependent coupled translocation-rotation motions of Csn2 and kinking of the DNA
To further study Csn2 binding to DNA and the sliding activity of the tetramers, we performed two independent all-atom MD simulations of 100 ns length each of the tetrameric Csn2 with dsDNA of 36 bp length bound within the central hole. The sequence of the dsDNA corresponds to the CRISPR repeat sequence of the type II-A CRISPR system of S. agalactiae. Although we do not have an indication for a sequence-specific binding of Csn2, a bioinformatics analysis revealed a correlation between the presence of the csn2 gene and a particular CRISPR repeat sequence (43).
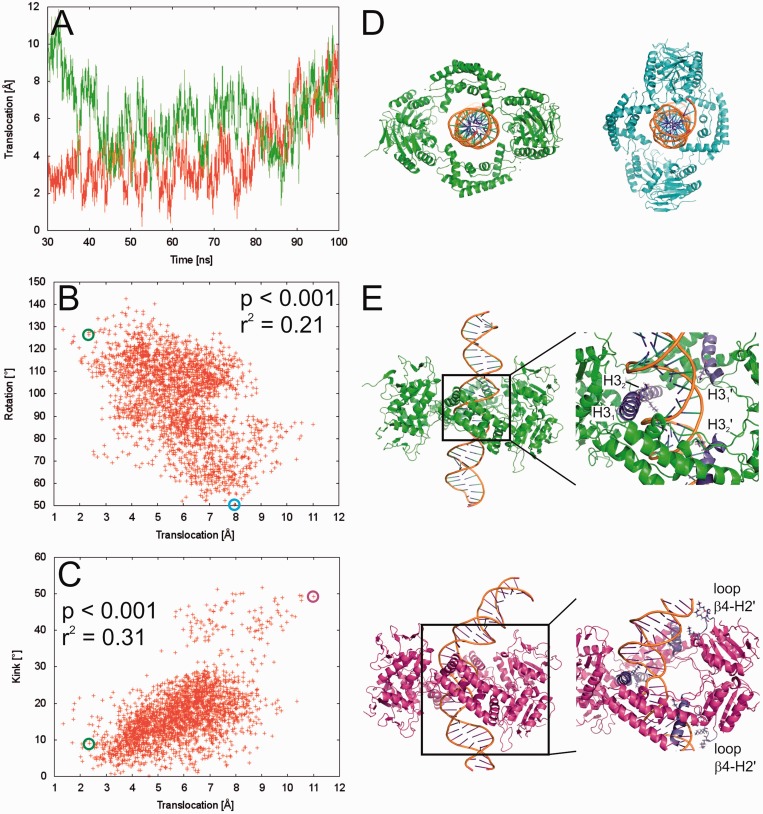
The simulations reveal sliding motions of Csn2 along the DNA of a magnitude of >10 Å (Figure 5A), which is equivalent to about one-third of the B-DNA pitch. The motion pattern shows both inward and outward movements accompanied by repeated changes in the direction of motion, as expected for a process driven by thermal energy. Yet, during the last 70 ns of the simulations, the Csn2 molecules prefer to be more closely located to the center of the DNA: although distances up to 12 Å away from the center are observed during the simulations (Figure 5A), the combined likelihood from both trajectories of Csn2 being within 6 Å of the center is 66%. Thus, with the present length of the simulations, no preference for Csn2 to move toward one of the DNA ends could be detected.
Figure 5.
(A) Movement of Csn2 along the DNA determined from two independent MD simulations (red and green lines, respectively) over a time course of 70 ns. A translocation value of zero refers to Csn2 being centered on the DNA; translocation values >0 indicate a shift of Csn2 toward the termini of the DNA. (B) Coupled motions of translocation and rotation of Csn2 when moving along the DNA over the last 50 ns of the MD simulation. To determine the rotational motion, all conformations of the trajectories were aligned with respect to the phosphorous atoms of the DNA. (C) Coupling between DNA bending as determined by a kink angle and the translocation of Csn2 along the DNA over the last 50 ns of the MD simulation. (D) View along the DNA axis on Csn2-DNA conformations extracted from the MD trajectory at 50.3 ns (green) and 97.3 ns (cyan). The complexes were aligned with respect to the phosphorous atoms of the DNA and display a rotation of ∼75° of Csn2 during a translocation of ∼5.7 Å. In panel B, the respective data points are marked by circles. (E) Side view on Csn2-DNA complexes extracted from the MD trajectory at 50.3 ns (green) and 98.5 ns (magenta). The complexes display a kinking of the DNA by ∼40° during a translocation of Csn2 of ∼8.7 Å. In the close-up figures, helices H3 and loops β4-H2 are colored in blue, and Lys residues in these structural elements are depicted as sticks; unprimed labels mark helices and loops that belong to one dimer, primed labels mark objects that belong to the other dimer. In panel C, the respective data points are marked by circles.
The sliding motions of Csn2 are weakly (r2 = 0.21) but significantly (P < 0.001) correlated to a rotation of the protein around the DNA (Figure 5B, D), as found for both independent MD simulations (data for the second simulation is shown in Supplementary Figure S4). The slope of the correlation line is ∼13°/Å, which is only slightly larger than the turn angle per 1 Å rise of ∼11° for B-DNA (44). The N-termini of helices H3 are likely responsible for this screw motion as the two helices within one dimer of Csn2, first, are almost perfectly collinearly oriented, second, are tilted by ∼30° with respect to the axis of the dsDNA binding within the central hole and, third, possess a Lys (K78) at the N-terminus (Figure 5D). Taken together, this allows the pair of helices H3 of one dimer of Csn2 to fit with their N-termini in between the phosphate backbones of the major groove, while the helix pair of the other dimer is closer to the phosphate backbone of the minor groove. Therefore, on sliding, Csn2 must rotate around the dsDNA for helices H3 need to follow the course of the phosphate backbones, ultimately leading to a screw motion.
To probe the effect of removing Ca2+-ions on Csn2 binding to the DNA, the above MD simulations were repeated without Ca2+. No gross structural distortions of the tetrameric state of Csn2 were observed (Supplementary Figure S5), in agreement with the observed high stability of Csn2 even in the presence of EGTA (30). No release of Csn2 from the DNA was observed either, which is not surprising given the limited size of the water box enclosing the Csn2–DNA complex in the simulation that prevents a slipping down of Csn2. However, in contrast to the simulations with Ca2+, no correlation between a sliding motion of Csn2 and its rotation could be detected in either one of the simulations (r2 < 0.07). Thus, the absence of Ca2+ apparently disturbs the screw motion of Csn2, pointing to weakened interactions between Csn2 and DNA.
Both MD simulations furthermore reveal weak (r2 = 0.31) or very weak (r2 equals; 0.06) but significant (P < 0.001) correlations between the translocation of Csn2 from the DNA center and DNA bending in that a large kink angle in the DNA is preferentially observed if Csn2 moves toward a DNA end (Figure 5C and E). The DNA bending is fostered by the insertion of the N-terminus of at least one helix H3 of one of the Csn2 dimers into the major groove, and interactions between the phosphate backbones of the minor groove at two positions with Lys-rich loops between β4 and H2 of the other dimer, respectively (Figure 5E). As no correlation could be detected when plotting the kink angle against the distance of Csn2 from one end of the DNA, no indication was found either that the bending occurs preferentially at one end (and thus at a specific sequence) of the DNA.
DISCUSSION
In this study, we have characterized the DNA end-binding activity of the Cas protein Csn2 and investigated the structure of the Csn2–DNA complexes by atomic force microscopic imaging and MD simulations. Our results assign Csn2 as a new member of the class of multimeric toroidal proteins involved in DNA end-metabolism and indicate a pivotal role of Csn2 as an accessory protein during the integration of spacer sequences into the CRISPR array.
Csn2 tetramers bind at dsDNA ends and slide inward along the DNA
The toroidal structure of the DNA-binding protein Csn2 in different studies led to the suggestion that it binds to DNA through its positively charged central hole and encloses the DNA, although alternative binding mechanisms were also considered (29–31). Furthermore, the crystal structure of the Cas protein Stu0660 of the S. thermophilus has revealed the presence of a second group of the Csn2 protein family (Stu0660-like Csn2), the members of which share no sequence similarity but have a highly similar tetrameric structure (32). In contrast to the canonical Csn2 proteins, the Csn2-like Stu0660 protein has an extended C-terminal domain and binds to dsDNA without the need of Ca2+-ions. Moreover, due to the lack of a mobility shift with a circular DNA, the authors suggested that Stu0660 ‘does not or barely’ binds to circular DNA but selectively to linear dsDNA (32). Our study is consistent with this suggestion and clearly demonstrates that the Csn2 tetramer binds DNA through its positively charged central hole from the DNA ends and then slides inward along the DNA.
The sliding activity of Csn2 tetramers is not energy-dependent and thus likely not a directed translocation but rather a thermal energy-driven random walk fostered by electrostatic interactions between positively charged amino acids of Csn2 and the negatively charged phosphate backbone of the DNA. MD simulations revealed a rotation of Csn2 when moving along the helical axis of the DNA, similar to a rotation-coupled sliding of the sliding clamp PCNA (45), endonuclease EcoRV (46) or human oxoguanine DNA glycosylase 1 (47). One-dimensional diffusion of DNA-binding proteins is considered to facilitate finding of the specific binding sites (48). Although we have no evidence for a sequence-specific interaction in vitro, the reported correlation of the presence of the csn2 gene and a particular CRISPR repeat sequence (43) could point to a specific role of Csn2 at the repeat sequence. The MD simulations were performed with the CRISPR repeat sequence, and one of the simulations indicated a bending of the DNA preferentially when Csn2 was located at an end. AFM is in principle suitable to study protein-induced DNA bending (49); however, the predicted Csn2-induced kinking is located at the ends of the DNA, and thus difficult to detect with AFM. Therefore, the structural deformation of the DNA ends on Csn2-binding, as proposed by the MD simulations, needs further experimental validation.
The cooperative binding of Csn2, observed in the EMSA and AFM analyses, indicates that multiple Csn2 tetramers are needed at the DNA ends to form stable Csn2–DNA complexes. We suggest that the binding of the first Csn2 tetramer at one DNA end is stabilized by loading of additional tetramers onto the same DNA end, which could impede a slipping down of the initially bound tetramers from the DNA end. This is consistent with the observation that the dissociation of Csn2–DNA complexes occurs through slipping down of the sliding tetramers at the DNA termini (Figure 2). In addition, at high concentrations, Csn2 tends to cluster along the DNA, leading to highly condensed nucleoprotein complexes. This apparent DNA-dependent self-association of the tetramers could point to a physical contact of the tetramers, contributing to the cooperative DNA end-loading of Csn2, as known for the Ku protein (50). The heterodimeric Ku protein forms a ring-like structure, binds preferentially at DNA ends, encircles the DNA (51), can slide along the DNA and promotes looped DNA structures (52). Moreover, it tends to cluster along the DNA as visualized by AFM (53). The DNA-binding properties of Ku are thus similar to the features of Csn2 observed in this study. The function of Ku in vivo is to mediate the formation of DNA end-synapsis and recruitment of recombination proteins to allow the repair of double-strand DNA breaks (54).
DNA end-metabolism and spacer integration
A series of multimeric ring-shaped proteins is known, which meet different functions in DNA metabolism, such as replication, transcription, recombination or DNA repair (40,41). In principle, there are three general ways how the DNA can get encircled by a ring-shaped protein: the quaternary structure can be opened wide enough to bind and enclose the DNA [e.g. RdgC (55)], the ring-shaped structure gets directly assembled around the DNA [e.g. PCNA (56)] or the DNA end has to pass through the central hole of the protein [e.g. Ku protein (51)]. In the latter case, the function of the protein is directly linked to DNA ends. Thus, the results presented here strongly suggest that the biological function of the CRISPR adaptation protein Csn2 is related to free DNA ends, which are likely formed during spacer acquisition.
The study of Yosef et al. (27) characterized a minimal requirement for spacer integration in type I-E CRISPR systems, and demonstrated the involvement of leader DNA and the nucleases Cas1 and Cas2 in the immunization process. The uptake of new spacers at the leader proximal end is also observed for other CRISPR subtypes, and together with the universality of Cas1 and Cas2, it seems that the main principle mechanism for spacer acquisition is similar in the different CRISPR subtypes. However, some of the CRISPR-Cas subtypes require in addition to Cas1 and Cas2 the proteins Csn2 or Cas4 for the acquisition of new spacer sequences. The Cas4 protein is proposed to be a subunit of the multiprotein complex Cascis (CRISPR-associated complex for the integration of spacers), mediating the CRISPR-mediated immunization (57). A recent study of the Cas4 protein from Sulfolobus solfataricus demonstrated that it contains a 5′–3′ DNA exonuclease activity, implying the requirement of DNA end modification/resection during integration of new spacer sequences into the CRISPR array (58). Moreover, the crystal structure of the Cas4 protein from S. solfataricus has been deposited in PDB database, showing that the Cas4 protein forms, like Csn2, a ring-shaped structure (PDB ID: 4IC1). Thus, the structural similarity of Cas4 to AddB, its RecB-like exonuclease activity (58) and the DNA end-binding of Csn2 presented in this study are in agreement with DNA end-processing steps during the spacer integration. Another indication for the participation of DNA end-processing proteins in the adaptation stage is the genetic association of nurA and herA genes with type III-A CRISPR operons in Thermoproteales (59), encoding for an exonuclease–helicase complex involved in DNA end-resection (60).
The lack of enzymatic activity of Csn2 suggests that Cas4 and Csn2 are likely not functional homologs. However, the conserved co-localization within the type II systems and the DNA end-related activities of both proteins indicate that they are involved in DNA end-resection processes during integration of spacers. The reported protein–protein interaction of Cas1 with RecB in E. coli (61) and the AddB-like structure and activity of the Cas4 protein (58) could support an involvement of DNA repair systems during the uptake of CRISPR spacer sequences. The study of Chayot et al. (62) has demonstrated that the insertion of non-related exogenous sequences in E. coli depends on the RecBCD complex. The role of RecBCD is to process the DNA ends exonucleolytically on the 3′-end to produce microhomology with the incoming DNA, a mechanism termed alternative end-joining (A-EJ) (62). A similar mechanism could be catalyzed by Cas4 in type II-B or AddAB proteins in type II-A systems lacking Cas4.
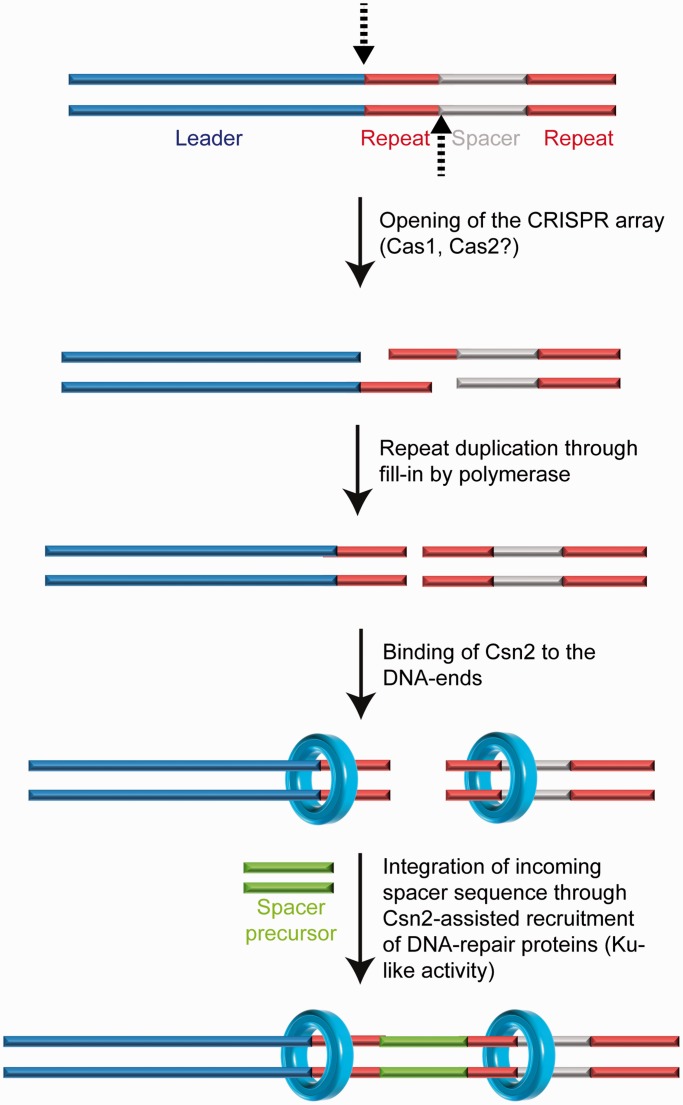
The DNA end-binding activity of Csn2 suggests that it has a function as accessory protein on DNA ends. Csn2 could fulfill a task analogous to Ku during acquisition of exogenous DNA by non-homologous end-joining, by binding to the double-strand breaks to hold the ends together and/or by recruiting recombination proteins (63) (Figure 6). Alternatively, the DNA end binding activity of Csn2 could be related to the nuclease activity of Cas9, introducing specific double-strand DNA breaks into the target DNA (12,17). It is possible that after recognition and cleavage of the foreign DNA by Cas9-tracrRNA:crRNA ribonuleoprotein complex, Csn2 proteins are loaded at the DNA ends, e.g. to mark the foreign DNA as a substrate for new spacer sequences and/or to recruit the nucleases for specific generation of new spacer precursors. Indeed, such a coupling of the CRISPR interference with the immunization stage has been described to occur in the type I-E systems, known as ‘primed acquisition’ of spacer sequences (25). Although our analyses do not directly address the mechanisms of CRISPR spacer integration, the DNA end-binding of the adaptation protein Csn2 and the structure of the Csn2–DNA complexes resemble known recombination proteins acting on DNA end-metabolism. Therefore, we propose that the integration of new spacer DNA may occur by a recombination mechanism similar to non-homologous end-joining.
Figure 6.
A model for the proposed function of Csn2 DNA end-binding in spacer integration is shown. The leader proximal repeat sequence serves as template for duplication of the repeat sequence (25,26,64), suggesting a cleavage at the leader-repeat and repeat-spacer borders (indicated by the dashed arrows). Complementary strand synthesis and binding of Csn2 to the DNA ends could assist in the integration of new spacer DNA fragments through holding the DNA ends together, while simultaneously recruiting DNA-repair proteins and protecting the double-stranded DNA breaks from exonucleolytic degradation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–5.
FUNDING
Deutsche Forschungsgemeinschaft (DFG) [PU 435/1-1]; Strategischer Forschungsfonds at the Heinrich Heine University (to Ü.P.); initiative ‘Fit for Excellence’ at the Heinrich Heine University (to H.G.). Funding for open access charge: Institutional funds and Strategischer Forschungsfonds of the Heinrich Heine University.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Britta Ries and Andre Abts for assistance during the experiments. S.S. acknowledges the continuing support from the Heinrich Heine University. H.G. is grateful to the ‘Zentrum für Informations- und Medientechnologie’ (ZIM) at the Heinrich Heine University for computational support. We gratefully thank the members of the DFG Research unit FOR 1680 for helpful discussions.
REFERENCES
- 1.Bhaya D, Davison M, Barrangou R. CRISPR-Cas systems in bacteria and archaea: versatile small RNAs for adaptive defense and regulation. Annu. Rev. Genet. 2011;45:273–297. doi: 10.1146/annurev-genet-110410-132430. [DOI] [PubMed] [Google Scholar]
- 2.Al-Attar S, Westra ER, van der Oost J, Brouns SJ. Clustered regularly interspaced short palindromic repeats (CRISPRs): the hallmark of an ingenious antiviral defense mechanism in prokaryotes. Biol. Chem. 2011;392:277–289. doi: 10.1515/BC.2011.042. [DOI] [PubMed] [Google Scholar]
- 3.Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482:331–338. doi: 10.1038/nature10886. [DOI] [PubMed] [Google Scholar]
- 4.Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, Moineau S, Mojica FJ, Wolf YI, Yakunin AF, et al. Evolution and classification of the CRISPR-Cas systems. Nat. Rev. Microbiol. 2011;9:467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol. Direct. 2006;1:7. doi: 10.1186/1745-6150-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van der Oost J, Jore MM, Westra ER, Lundgren M, Brouns SJ. CRISPR-based adaptive and heritable immunity in prokaryotes. Trends Biochem. Sci. 2009;34:401–407. doi: 10.1016/j.tibs.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 7.Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 8.Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, van der Oost J. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Westra ER, van Erp PB, Kunne T, Wong SP, Staals RH, Seegers CL, Bollen S, Jore MM, Semenova E, Severinov K, et al. CRISPR immunity relies on the consecutive binding and degradation of negatively supercoiled invader DNA by Cascade and Cas3. Mol. Cell. 2012;46:595–605. doi: 10.1016/j.molcel.2012.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sapranauskas R, Gasiunas G, Fremaux C, Barrangou R, Horvath P, Siksnys V. The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli. Nucleic Acids Res. 2011;39:9275–9282. doi: 10.1093/nar/gkr606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hale CR, Majumdar S, Elmore J, Pfister N, Compton M, Olson S, Resch AM, Glover CV, III, Graveley BR, Terns RM, et al. Essential features and rational design of CRISPR RNAs that function with the Cas RAMP module complex to cleave RNAs. Mol. Cell. 2012;45:292–302. doi: 10.1016/j.molcel.2011.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hale CR, Zhao P, Olson S, Duff MO, Graveley BR, Wells L, Terns RM, Terns MP. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell. 2009;139:945–956. doi: 10.1016/j.cell.2009.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jore MM, Lundgren M, van Duijn E, Bultema JB, Westra ER, Waghmare SP, Wiedenheft B, Pul U, Wurm R, Wagner R, et al. Structural basis for CRISPR RNA-guided DNA recognition by Cascade. Nat. Struct. Mol. Biol. 2011;18:529–536. doi: 10.1038/nsmb.2019. [DOI] [PubMed] [Google Scholar]
- 16.Haft DH, Selengut J, Mongodin EF, Nelson KE. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput. Biol. 2005;1:e60. doi: 10.1371/journal.pcbi.0010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl Acad. Sci. USA. 2012;109:E2579–E2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J. RNA-programmed genome editing in human cells. Elife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat. Biotechnol. 2013;31:230–232. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- 20.Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat. Biotechnol. 2013;31:233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, Peterson RT, Yeh JR, Joung JK. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol. 2013;31:227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Datsenko KA, Pougach K, Tikhonov A, Wanner BL, Severinov K, Semenova E. Molecular memory of prior infections activates the CRISPR/Cas adaptive bacterial immunity system. Nat. Commun. 2012;3:945. doi: 10.1038/ncomms1937. [DOI] [PubMed] [Google Scholar]
- 26.Swarts DC, Mosterd C, van Passel MW, Brouns SJ. CRISPR interference directs strand specific spacer acquisition. PLoS One. 2012;7:e35888. doi: 10.1371/journal.pone.0035888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yosef I, Goren MG, Qimron U. Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic Acids Res. 2012;40:5569–5576. doi: 10.1093/nar/gks216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deveau H, Barrangou R, Garneau JE, Labonte J, Fremaux C, Boyaval P, Romero DA, Horvath P, Moineau S. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J. Bacteriol. 2008;190:1390–1400. doi: 10.1128/JB.01412-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nam KH, Kurinov I, Ke A. Crystal structure of clustered regularly interspaced short palindromic repeats (CRISPR)-associated Csn2 protein revealed Ca2+-dependent double-stranded DNA binding activity. J. Biol. Chem. 2011;286:30759–30768. doi: 10.1074/jbc.M111.256263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ellinger P, Arslan Z, Wurm R, Tschapek B, MacKenzie C, Pfeffer K, Panjikar S, Wagner R, Schmitt L, Gohlke H, et al. The crystal structure of the CRISPR-associated protein Csn2 from Streptococcus agalactiae. J. Struct. Biol. 2012;178:350–362. doi: 10.1016/j.jsb.2012.04.006. [DOI] [PubMed] [Google Scholar]
- 31.Koo Y, Jung DK, Bae E. Crystal structure of Streptococcus pyogenes Csn2 reveals calcium-dependent conformational changes in its tertiary and quaternary structure. PLoS One. 2012;7:e33401. doi: 10.1371/journal.pone.0033401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee KH, Lee SG, Eun Lee K, Jeon H, Robinson H, Oh BH. Identification, structural, and biochemical characterization of a group of large Csn2 proteins involved in CRISPR-mediated bacterial immunity. Proteins. 2012;80:2573–2582. doi: 10.1002/prot.24138. [DOI] [PubMed] [Google Scholar]
- 33.Pul U, Wurm R, Wagner R. The role of LRP and H-NS in transcription regulation: involvement of synergism, allostery and macromolecular crowding. J. Mol. Biol. 2007;366:900–915. doi: 10.1016/j.jmb.2006.11.067. [DOI] [PubMed] [Google Scholar]
- 34.Case DA, Cheatham TE, III, Darden T, Gohlke H, Luo R, Merz KM, Jr, Onufriev A, Simmerling C, Wang B, Woods RJ. The Amber biomolecular simulation programs. J. Comput. Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cornell WD, Cieplak CI, Bayly IR, Gould IR, Merz KM, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J. Am. Chem. Soc. 1995;117:5179–5197. [Google Scholar]
- 36.Simmerling C, Strockbine B, Roitberg AE. All-atom structure prediction and folding simulations of a stable protein. J. Am. Chem. Soc. 2002;124:11258–11259. doi: 10.1021/ja0273851. [DOI] [PubMed] [Google Scholar]
- 37.Jorgensen WL, Chandrasekhar J, Madura J, Klein ML. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 38.Darden T, York D, Pederson L. Particle Mesh Ewald—a Nlog(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 39.Ryckaert JP, Ciccotti G, Berendsen HJC. Numerical-Integration of Cartesian Equations of Motion of a System with Constraints: Molecular Dynamics of n-Alkanes. J. Comput. Phys. 1977;23:327–341. [Google Scholar]
- 40.Hingorani MM, O'Donnell M. Toroidal proteins: running rings around DNA. Curr. Biol. 1998;8:R83–R86. doi: 10.1016/s0960-9822(98)70052-1. [DOI] [PubMed] [Google Scholar]
- 41.Hingorani MM, O'Donnell M. A tale of toroids in DNA metabolism. Nat. Rev. Mol. Cell. Biol. 2000;1:22–30. doi: 10.1038/35036044. [DOI] [PubMed] [Google Scholar]
- 42.Tessmer I, Moore T, Lloyd RG, Wilson A, Erie DA, Allen S, Tendler SJ. AFM studies on the role of the protein RdgC in bacterial DNA recombination. J. Mol. Biol. 2005;350:254–262. doi: 10.1016/j.jmb.2005.04.043. [DOI] [PubMed] [Google Scholar]
- 43.Kunin V, Sorek R, Hugenholtz P. Evolutionary conservation of sequence and secondary structures in CRISPR repeats. Genome Biol. 2007;8:R61. doi: 10.1186/gb-2007-8-4-r61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saenger W. Principles of Nucleic Acid Structure. New York: Springer-Verlag; 1984. [Google Scholar]
- 45.Kochaniak AB, Habuchi S, Loparo JJ, Chang DJ, Cimprich KA, Walter JC, van Oijen AM. Proliferating cell nuclear antigen uses two distinct modes to move along DNA. J. Biol. Chem. 2009;284:17700–17710. doi: 10.1074/jbc.M109.008706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dikic J, Menges C, Clarke S, Kokkinidis M, Pingoud A, Wende W, Desbiolles P. The rotation-coupled sliding of EcoRV. Nucleic Acids Res. 2012;40:4064–4070. doi: 10.1093/nar/gkr1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blainey PC, Luo G, Kou SC, Mangel WF, Verdine GL, Bagchi B, Xie XS. Nonspecifically bound proteins spin while diffusing along DNA. Nat. Struct. Mol. Biol. 2009;16:1224–1229. doi: 10.1038/nsmb.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.von Hippel PH, Berg OG. Facilitated target location in biological systems. J. Biol. Chem. 1989;264:675–678. [PubMed] [Google Scholar]
- 49.Dame RT, van Mameren J, Luijsterburg MS, Mysiak ME, Janicijevic A, Pazdzior G, van der Vliet PC, Wyman C, Wuite GJ. Analysis of scanning force microscopy images of protein-induced DNA bending using simulations. Nucleic Acids Res. 2005;33:e68. doi: 10.1093/nar/gni073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ma Y, Lieber MR. DNA length-dependent cooperative interactions in the binding of Ku to DNA. Biochemistry. 2001;40:9638–9646. doi: 10.1021/bi010932v. [DOI] [PubMed] [Google Scholar]
- 51.Walker JR, Corpina RA, Goldberg J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature. 2001;412:607–614. doi: 10.1038/35088000. [DOI] [PubMed] [Google Scholar]
- 52.Dynan WS, Yoo S. Interaction of Ku protein and DNA-dependent protein kinase catalytic subunit with nucleic acids. Nucleic Acids Res. 1998;26:1551–1559. doi: 10.1093/nar/26.7.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cary RB, Peterson SR, Wang J, Bear DG, Bradbury EM, Chen DJ. DNA looping by Ku and the DNA-dependent protein kinase. Proc. Natl Acad. Sci. USA. 1997;94:4267–4272. doi: 10.1073/pnas.94.9.4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Feldmann E, Schmiemann V, Goedecke W, Reichenberger S, Pfeiffer P. DNA double-strand break repair in cell-free extracts from Ku80-deficient cells: implications for Ku serving as an alignment factor in non-homologous DNA end joining. Nucleic Acids Res. 2000;28:2585–2596. doi: 10.1093/nar/28.13.2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Briggs GS, Yu J, Mahdi AA, Lloyd RG. The RdgC protein employs a novel mechanism involving a finger domain to bind to circular DNA. Nucleic Acids Res. 2010;38:6433–6446. doi: 10.1093/nar/gkq509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kong XP, Onrust R, O'Donnell M, Kuriyan J. Three-dimensional structure of the beta subunit of E. coli DNA polymerase III holoenzyme: a sliding DNA clamp. Cell. 1992;69:425–437. doi: 10.1016/0092-8674(92)90445-i. [DOI] [PubMed] [Google Scholar]
- 57.Plagens A, Tjaden B, Hagemann A, Randau L, Hensel R. Characterization of the CRISPR/Cas subtype I-A system of the hyperthermophilic crenarchaeon Thermoproteus tenax. J. Bacteriol. 2012;194:2491–2500. doi: 10.1128/JB.00206-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang J, Kasciukovic T, White MF. The CRISPR associated protein cas4 Is a 5′ to 3′ DNA exonuclease with an iron-sulfur cluster. PLoS One. 2012;7:e47232. doi: 10.1371/journal.pone.0047232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bernick DL, Cox CL, Dennis PP, Lowe TM. Comparative genomic and transcriptional analyses of CRISPR systems across the genus Pyrobaculum. Front. Microbiol. 2012;3:251. doi: 10.3389/fmicb.2012.00251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Blackwood JK, Rzechorzek NJ, Abrams AS, Maman JD, Pellegrini L, Robinson NP. Structural and functional insights into DNA-end processing by the archaeal HerA helicase-NurA nuclease complex. Nucleic Acids Res. 2012;40:3183–3196. doi: 10.1093/nar/gkr1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Babu M, Beloglazova N, Flick R, Graham C, Skarina T, Nocek B, Gagarinova A, Pogoutse O, Brown G, Binkowski A, et al. A dual function of the CRISPR-Cas system in bacterial antivirus immunity and DNA repair. Mol. Microbiol. 2011;79:484–502. doi: 10.1111/j.1365-2958.2010.07465.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chayot R, Montagne B, Mazel D, Ricchetti M. An end-joining repair mechanism in Escherichia coli. Proc. Natl Acad. Sci. USA. 2010;107:2141–2146. doi: 10.1073/pnas.0906355107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Doherty AJ, Jackson SP. DNA repair: how Ku makes ends meet. Curr. Biol. 2001;11:R920–R924. doi: 10.1016/s0960-9822(01)00555-3. [DOI] [PubMed] [Google Scholar]
- 64.Goren MG, Yosef I, Auster O, Qimron U. Experimental definition of a clustered regularly interspaced short palindromic duplicon in Escherichia coli. J. Mol. Biol. 2012;423:14–16. doi: 10.1016/j.jmb.2012.06.037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.