Abstract
The universally conserved Kae1/Qri7/YgjD and Sua5/YrdC protein families have been implicated in growth, telomere homeostasis, transcription and the N6-threonylcarbamoylation (t6A) of tRNA, an essential modification required for translational fidelity by the ribosome. In bacteria, YgjD orthologues operate in concert with the bacterial-specific proteins YeaZ and YjeE, whereas in archaeal and eukaryotic systems, Kae1 operates as part of a larger macromolecular assembly called KEOPS with Bud32, Cgi121, Gon7 and Pcc1 subunits. Qri7 orthologues function in the mitochondria and may represent the most primitive member of the Kae1/Qri7/YgjD protein family. In accordance with previous findings, we confirm that Qri7 complements Kae1 function and uncover that Qri7 complements the function of all KEOPS subunits in growth, t6A biosynthesis and, to a partial degree, telomere maintenance. These observations suggest that Kae1 provides a core essential function that other subunits within KEOPS have evolved to support. Consistent with this inference, Qri7 alone is sufficient for t6A biosynthesis with Sua5 in vitro. In addition, the 2.9 Å crystal structure of Qri7 reveals a simple homodimer arrangement that is supplanted by the heterodimerization of YgjD with YeaZ in bacteria and heterodimerization of Kae1 with Pcc1 in KEOPS. The partial complementation of telomere maintenance by Qri7 hints that KEOPS has evolved novel functions in higher organisms.
INTRODUCTION
N6-threonylcarbamoyladenosine (t6A) is one of the 15 universally conserved tRNA modifications found in all three domains of life (1). With the sole exception of bacterial initiator tRNAfMet, t6A is present at position 37 of the anti-codon stem loop of all tRNAs with ANN recognizing anticodons (N being any nucleotide) (1). This modification serves to strengthen the codon–anticodon interaction and promote translational fidelity by the ribosome (2–4). In several subspecies of bacteria, fungi and plants, the t6A modification is processed further by dehydration into cyclic-t6A (5). Cells that are deficient for t6A modification exhibit an increased frequency of frameshift events and are prone to errors in AUG start codon selection, both of which can severely impair normal cellular physiology (6,7).
Comparative genomic analyses have implicated the Kae1/Qri7/YgjD and Sua5/YrdC protein families in the t6A biosynthesis pathway (8–10). In archaea and eukaryotes, Kae1 is part of an ancient macromolecular assembly known as KEOPS (kinase, putative endopeptidase and other proteins of small size). In yeast, KEOPS consists of five subunits, namely the Kae1, Bud32, Cgi121, Pcc1 and Gon7 proteins (11,12). Bud32 is one of the three ancient atypical protein kinases that are conserved in all archaeal and eukaryotic species, while Cgi121 and Pcc1 (a dimerization module) possess novel protein folds with no inferable functions (13). Gon7, to date only recognizable in fungi, remains the sole KEOPS subunit for which no structural information is available. In contrast to Kae1, YgjD orthologues function in a complex with two proteins, YeaZ and YjeE, both of which display no similarity in sequence to subunits within KEOPS.
Mutations within KEOPS in budding yeast gives rise to a set of diverse phenotypes including extreme slow growth, shortened telomeres and defective transcription coactivation by the SAGA and mediator complexes (11,12). The pleiotropic phenotypes of KEOPS have led to its initial classification as a telomere uncapping and elongation factor as well as a recruitment factor for transcriptional coactivators. Interestingly, Sua5/YrdC mutants are also defective in telomere length homeostasis in the budding yeast (14,15). As the posttranscriptional modification of tRNAs can influence seemingly unrelated cellular processes such as the DNA damage response and apoptosis (16–18) by modulating tRNA structure and function (19–21), it is possible that the pleiotropic effects of KEOPS and Sua5 mutations may be due entirely to defects in t6A metabolism, although this hypothesis has yet to be proven formally.
In vitro reconstitution studies have shed light on the mechanics of t6A formation. Reconstitution of t6A modification of tRNA was first achieved using TsaC/YrdC (the bacterial Sua5 orthologue), TsaD/YgjD (the bacterial Kae1 orthologue) and two bacterial-specific proteins TsaB/YeaZ and TsaE/YjeE (herein referred to as YrdC, YgjD, YeaZ and YjeE, respectively) (22) and most recently using archaeal/eukaryotic Sua5 and their corresponding KEOPS complexes (23). YrdC as an isolated protein was shown to generate threonylcarbamoyl-adenylate (TCA), a key early intermediate in the t6A reaction pathway (24). The specific direct or indirect roles that Kae1/YgjD and their known interacting partners play in the t6A synthetic process remains an enigma. To distinguish between direct and indirect contributions to t6A biosynthesis and to provide a framework to explore the accessory/regulatory role that an indirectly functioning subunit may play in t6A formation, we set out to identify and characterize a minimal eukaryotic t6A biosynthesis system. Toward this end, we show that yeast Sua5 and Qri7 (the mitochondrial paralogue of Kae1) are sufficient for t6A biosynthesis in vitro. Structural and functional characterization demonstrates that Qri7 forms dimers in solution, and that the integrity of the dimerization contact surface but not dimerization itself, is an essential component of its t6A biosynthetic function in vitro and biological function in vivo. Interestingly, the mode of dimerization is highly reminiscent of the Kae1–Pcc1 interaction in KEOPS (13) and the YgjD–YeaZ heterodimer (25) in the corresponding complex in bacteria. Together, this work provides new insight into how the universally conserved Kae1/Qri7/YgjD and Sua5/YrdC protein families function at a molecular level and provides a framework for probing how these evolutionarily conserved protein families impinge on multiple aspects of biology.
MATERIALS AND METHODS
Yeast growth rate analyses
The full collection of yeast strains used in this study is found in Supplementary Table 1. Yeast growth was monitored for 24 h by measuring OD595 every 15 min as described (26). Briefly, an overnight culture was diluted to an OD600 of 0.0625 with fresh medium (the kae1Δ, bud32Δ, gon7Δ and cgi121Δ strains were grown in SD-Leu; the pcc1-4 strain was grown in SD-Leu-His) supplemented with 2% galactose. Growth assays were performed in a clear flat-bottom 96-well microplate (Greiner) sealed with adhesive plate seals (Cat. No. AB-0580, ABgene) using a custom-developed platform incorporating microplate reader TECAN GENios (Tecan-US, Durham NC, USA) and ACCESS software (26,27).
Bulk tRNA isolation
Yeast strains were grown in 50 ml cultures to OD600 of 1.5 in their respective media as indicated above. Yeast strains were pelleted by centrifugation and resuspended in 500 μl of 10 mM Bis-Tris-HCl, pH 7.2, 10 mM EDTA and 0.5% sodium dodecyl sulphate. Total bulk RNA was extracted by adding one volume of 65°C phenol. Samples were incubated at 65°C for 30 min and shaken at 700 rpm. Total bulk RNA was isolated from the aqueous phase using a conventional phenol-chloroform extraction protocol. Isolation of bulk tRNA from total cellular RNA was performed using the Nucleobond® RNA/DNA 400 kit (Clontech Laboratories Inc.) according to the manufacturer’s protocol.
Telomere length determination
Telomere length assays were performed as previously described (11).
High performance liquid-chromatography analysis of tRNA composition
Isolated bulk tRNA or in vitro transcribed tRNA was digested and dephosphorylated according to previously described protocols (28). Ribonucleosides were analyzed on a Discovery C18 (15 cm × 4.6 mm, 5 μM) reverse-phase column (Supelco Analytical) equipped with an Ultrasphere ODS guard column (Beckman). Ribonucleosides were separated over a 35-min linear gradient of 98:2 (Buffers A and B, respectively) to 87.5:12.5 at a flow rate of 1.5 ml/min. The compositions of buffers A and B were 250 mM ammonium acetate, pH 6.5, and 40% acetonitrile, respectively. All High performance liquid-chromatography (HPLC) experiments were performed on a Dionex Ultimate 3000 HPLC Unit (Thermo Scientific) and data analysis was performed using the Chromeleon HPLC software.
Recombinant Sua5 and Qri7 production
Yeast Sua5 (full length) and Qri7 (amino acids 30–407) were expressed in Escherichia coli BL21 (DE3) CodonPlus cells harboring an N-terminal hexa-histidine or glutathione-S-transferase tag. Proteins were purified using conventional affinity chromatography (Ni-NTA or glutathione sepharose) coupled to size exclusion chromatography in 25 mM HEPES, pH 7.5, 200 mM NaCl and 2 mM Dithiothreitol (DTT).
In vitro tRNA transcription and purification
Overlap extension PCR was used to generate yeast tRNAIle and tRNAAla hepatitis delta virus ribozyme DNA fragments. The tRNA-ribozyme fragment were ligated into pUC19 and linearized with BamHI before runoff transcription reactions by T7 RNA Polymerase. Transcription reaction conditions were as follows: 100 mM Tris-HCl, pH 8.0, 4 mM ATP, 4 mM GTP, 4 mM CTP, 4 mM UTP, 10 mM DTT, 1 mM spermidine, 0.1% Triton X-100, 25 mM MgCl2, 30 μg/ml linearized DNA template, 0.2 mg/ml T7 RNA Polymerase, 10 U/ml thermostable inorganic phosphate (New England Biolabs) and 200 U/ml RiboLock™ RNase Inhibitor (Thermo Scientific). Transcription reactions were incubated for 4 h at 37°C and terminated by the addition of EDTA to a final concentration of 50 mM. RNA was isolated using a conventional phenol-chloroform extraction protocol. RNA pellets were solubilized in 8 M urea and refolded by rapid dilution into nine volumes of 25 mM Bis-Tris-HCl, pH 6.5, 1 mM MgCl2 and 0.2 mM EDTA, followed by overnight incubation at ambient temperature. The refolded tRNA was purified on a Source15Q column (GE Healthcare) using the following chromatography conditions: (i) four column volumes (CV) at 20% B and (ii) linear gradient over 25 CV from 20 to 30% B. Buffers A and B are 50 mM Bis-Tris-HCl, pH 6.5, 2 mM MgCl2 and 50 mM Bis-Tris-HCl, pH 6.5, 2 mM MgCl2, 2 M NaCl, respectively. Fractions containing tRNA were pooled and further purified using size exclusion chromatography in buffer 20 mM Bis-Tris-HCl, pH 6.5, and 10 mM MgCl2.
In vitro t6A reconstitution assay
t6A biosynthesis assays were performed in 50 mM Tris-HCl, pH 8.0, 200 mM NaCl, 1 mM DTT, 1 mM threonine, 1 mM NaHCO3, 2 mM ATP, 5 mM MnCl2, 2 μM of Sua5/Qri7 and 20 μM of tRNA. Reactions were incubated at ambient temperature for 60 min and stopped by the addition of EDTA to a final concentration of 25 mM. The tRNA was enzymatically digested and analyzed by HPLC as described above.
Liquid chromatography–mass spectrometry analysis of small molecules
Instrumentation consisted of a U-HPLC system, HTS PAL autosampler and a 6538 Q-TOF mass spectrometer with an electrospray ionization source (Agilent Technologies). The reverse-phase ion pairing liquid chromatography method consisted of an Agilent RRHD Extend-C18 column (2.1 × 100 mm, 1.8 μm particle size, Agilent, Santa Clara, CA) and an aqueous/organic gradient run as described below. Mobile phase A consisted of 97:3 water:methanol with 10 mM tributylamine and 15 mM acetic acid. Mobile phase B consisted of methanol or acetonitrile as described below. For the analysis of t6A labeling (Supplementary Figure S3), the flow rate equaled 0.4 ml/min and the gradient using methanol as mobile phase B, extended from 0% B at 0.0 min, to 100% B at 5.5 min, to 0% B at 6.5 min, to 0% B at 7.5 min. For the analysis of TCA, the flow rate equaled 0.5 ml/min with methanol as mobile phase B for 0–23 min and acetonitrile supplied from a second pump as mobile phase B from 23–33 min. The gradient for the analysis of TCA was 1% B at 0 min, 1% B at 4 min, 20% B at 6.67 min, 35% B at 14.67 min, 35% B at 17.33 min, 60% B at 18.67 min, 60% B at 20 min, 90% B at 20.5 min, 99% B at 21.3 min, 99% B at 23.1 min, 1% B at 31 min, 1% B at 32 min. The column temperature was maintained at 37°C, and the autosampler at 4°C. The injection volume was 15 μl in a 10 μl loop. A lock mass mixture with ions at 112.9855 and 1033.9881 was continually supplied at 2 μl/min through a second nebulizer pressurized at 7 psig, and data were continuously recalibrated during the run to these masses. The quadrupole time of flight mass spectrometer was run in negative ionization mode with a data storage threshold of 50. Data were collected in high resolution mode at 2 Hz. Instrument run parameters were drying gas temperature 300°C, drying gas flow 10 L/min, sample nebulizer pressure 50 psig, ESI capillary voltage 3500 V, fragmentor voltage 115 V, skimmer 47 V, octopole 1 RF 750 V. The scan range was 25–1100 m/z. For MS/MS analysis of TCA, ions of m/z 491.1 and 495.1 were targeted using a collision energy of 15 (arbitrary units) using “medium” isolation of 4 m/z as specified by the manufacturer.
Qri7 crystallization, structure determination and refinement
Qri7 (amino acids 30–407) intended for crystallography was purified as a 6×-HIS tagged protein. The 6×-HIS tag was cleaved using TEV protease and subsequently removed using a subtractive nickel purification. Qri7 was then purified in 20 mM HEPES, pH 7.5, 100 mM NaCl and 2 mM DTT using size exclusion chromatography with a Superdex 200 10/300 GL column (GE Healthcare). Crystals were grown at a protein concentration of 10 mg/ml in 0.1 M Tris-HCl, pH 7.5, 8% PEG8000 and 0.2 M MgCl2 by microseeding at 4°C. A seleno-methionine crystal data set was collected at the Advanced Photon Source NE-CAT beamline 24-ID-E using a constant wavelength of 0.97933 Angstroms at 93 Kelvin. The structure of Qri7 was solved by molecular replacement using PDB:2IVP as search model in PHASER (29), and refinement was carried out using BUSTER (30). The Rfree value was calculated using 10% of the data set. The final refined structure showed Ramachandran statistics of 92.84, 5.73 and 1.43% in the preferred, allowed and outlier regions, respectively. The following residues were only modeled to Cβ due to a lack of side chain electron density: N60, L62, L65, K66, R96, N102, I107, I141, I152, D203, R207, K212, K222, Q226, T262, L264, Q277, R282, E310 and F348. The Qri7 coordinates were deposited to the PDB under the accession number 4K25.
Analytical ultracentrifugation
All analytical ultracentrifugation experiments were performed on a XL-I unit (Beckman) at 20°C, and data analysis was performed using SEDFIT and SEDPHAT softwares (31). Sedimentation equilibrium experiments were performed at 0.25, 0.5 and 1 mg/ml protein concentrations at 10 000, 12 000, 15 000 rpm centrifuge speeds. The data were fitted against a monomer–dimer model for dissociation constant calculation. Sedimentation velocity experiments were performed at 0.3125, 0.0625, 0.125, 0.250, 0.5 or 1 mg/ml protein concentrations at speeds of 42 000 rpm at 8 h. Root mean square deviations (RMSD) for all sedimentation velocity data were <0.005.
Refolding of his-Sua5
Purified recombinant 6xHIS-Sua5 was unfolded by dilution into 50 volumes of 50 mM HEPES, pH 7.5, 500 mM NaCl, 50 mM imidazole, 6 M guanidine-hydrochloride and further purified by nickel affinity chromatography. For refolding, the protein was concentrated and then rapidly diluted into 50 volumes of 25 mM HEPES, pH 7.5, 200 mM NaCl, 2 mM DTT and 50 mM arginine. The sample was then subjected to size exclusion chromatography for final purification.
Small molecule docking of TCA into Qri7
Glide software (Schrödinger, Inc.) was used for the small molecule docking in this study. We first validated the performance of Glide against flexible molecules such as ATP and TCA [IUPAC name (2S,3R)-2-[[[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-dihydroxyoxolan-2-yl]methoxy-hydroxyphosphoryl]oxycarbonylamino]-3-hydroxybutanoic acid)] that contain 10 and 12 freely rotatable bonds, respectively. To do so, we extracted the ligand (ATP) from the Kae1 crystal structure (PDB:2IVP) and docked it back into the apo-form of the protein. Before docking, geometries of the ligands were converted from 2D to 3D with LigPrep (Schrödinger, Inc.) using the OPLS2005 force field, and then the structures were ionized to pH 7.0 with Epik and energy minimized with default LigPrep settings. The receptor protein GRID was calculated for a large box that was defined in the presence of ATP at the binding site. A standard box size of 10 Å beyond each side of the ligand was used. Two H-bond constraints were defined that recognize the adenosine portion: the side-chain oxygen atom of Glu176 and the side-chain NH of Asn257, both of which are interactions observed in the 2IVP crystal structure. The hydroxyls of Thr12, Ser129 and Tyr280 located in the vicinity of the ligand were designated as rotatable groups. We used the default scaling of van der Waals radii of 0.8 for atoms with partial atomic changes <0.15. We generated 5000 poses during the docking run, and then selected the 400 best poses for postdocking minimization and saved the top five poses for each ligand. The best docking pose was close to that observed in the 2IVP structure (0.65 Å RMSD based on the all-atom superposition) and preserved all eight H-bonds between the protein and the ligand as well as coordination of the Fe2+ ion by the phosphate group of ATP.
This positive validation result encouraged us to proceed further and dock TCA into the refined structure of Qri7 following the identical procedure. The side chain of residue R218 was converted to an alternative rotamer to prevent steric hindrance to TCA. The resulting structure of TCA in Qri7 overlays closely with the adenosine, sugar and the first phosphate moieties of ATP in the 2IVP structure with Kae1.
Small molecule docking of tobramycin into Qri7
The structure of TobZ bound to tobramycin (PDB:3VET) was superimposed onto the Qri7 structure in Coot by secondary structure matching (SSM superpose), which served to position tobramycin in the Qri7 catalytic cleft. Residues in Qri7 that made contact with the modeled TCA were determined using cCONTACT.
Subcellular fractionation
The isolation of yeast mitochondria using subcellular fractionation was performed as previously described (32).
RESULTS
Bud32, Pcc1, Cgi121 and Gon7 play a supporting role to the essential function of Kae1/Qri7 in growth
In budding yeast, mutational analyses have shown that all five KEOPS subunits contribute to telomere homeostasis and t6A formation (8,9,11). Of the five KEOPS subunits, Kae1 is the sole subunit universally conserved across all three domains of life. Hence, we reasoned that Kae1 contributed an essential core function within KEOPS and that Bud32, Cgi121, Pcc1 and Gon7 evolved to augment or regulate Kae1. Interestingly, eukaryotic cells harbor a Kae1 paralog, Qri7, which is targeted to mitochondria (33). Consistent with the predicted paralogous relationship, redirection of Qri7 from the mitochondria to the cytoplasm via deletion of the mitochondrial targeting sequence enabled rescue of the slow growth phenotype of a kae1Δ yeast strain (34). Intriguingly, GFP-tagged Bud32, Gon7, Cgi121 and Pcc1 proteins do not localize to the mitochondria (33) and the yeast genome does not appear to encode mitochondria-targeted paralogues of Bud32, Gon7, Cgi121 and Pcc1. Taken together, these results raised the possibility that Qri7 and Kae1 share a common essential function, but that Qri7 operates in mitochondria in the absence of additional regulatory KEOPS subunits.
If Bud32, Gon7, Cgi121 and Pcc1 play supporting roles only to Kae1’s essential core function, we reasoned that ectopic expression of Qri7 in the cytoplasm/nucleus should rescue not only the slow growth phenotype of a kae1Δ strain, but also the phenotypes of other KEOPS subunit deletions. We first confirmed that Qri7ΔMTS can rescue the kae1Δ slow growth phenotype and that this was due to a redirection of Qri7 protein from the mitochondria to the cytoplasm as predicted (Figure 1A and Supplementary Figure S1A). Expression of Qri7ΔMTS in a bud32Δ background rescued its associated slow growth phenotype (Figure 1B). Furthermore, the resulting growth rate was nearly identical to a bud32Δ strain harboring a Bud32 expression plasmid. Expression of Qri7ΔMTS in gon7Δ and cgi121Δ strains also resulted in growth rates identical to those observed for strains expressing Gon7 and Cgi121, respectively (Figure 1C and D). The temperature sensitive allele pcc1-4 did not suffer a growth defect at the permissive temperature of 30°C (Figure 1E) but displayed a slow growth phenotype at the nonpermissive temperature of 37°C (12). At 37°C, expression of Qri7ΔMTS resulted in a partial rescue of the slow growth phenotype (Figure 1F). Consistent with Sua5 and Kae1/Qri7 proteins playing non-overlapping roles in the t6A biosynthetic process, Qri7ΔMTS did not rescue the slow growth phenotype associated with a sua5Δ strain (Supplementary Figure S1B). Together, these results indicate that Kae1/Qri7 provide the essential core function(s) of KEOPS that supports growth and are consistent with a model whereby Bud32, Cgi121, Gon7 and Pcc1 play supporting role(s) to the central essential function of Kae1.
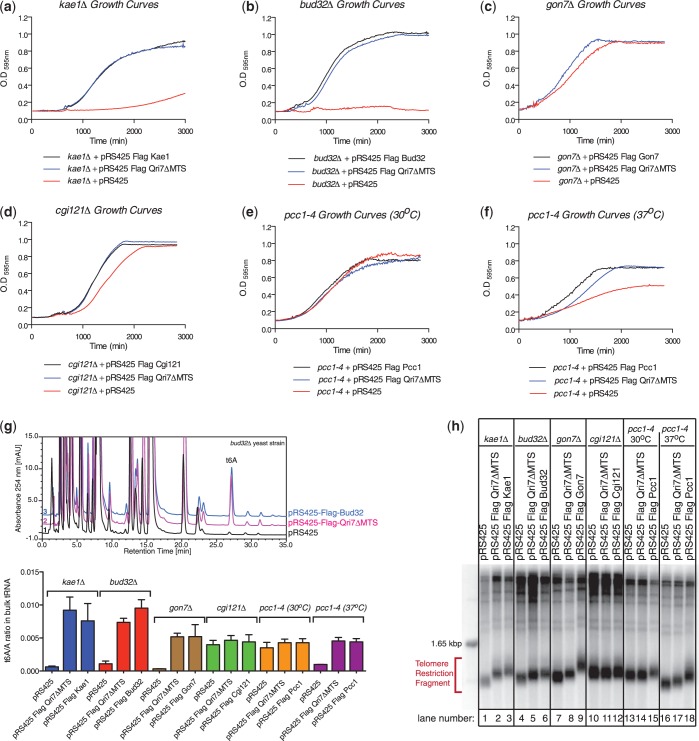
Figure 1.
Qri7 functionally complements the slow growth, diminished t6A level and shortened telomere phenotypes of the KEOPS complex. A plasmid encoding Qri7 lacking its mitochondrial-targeting sequence (Qri7ΔMTS) was expressed in (a) kae1Δ, (b) bud32Δ, (c) gon7Δ, (d) cgi121Δ, (e) pcc1-4 at the permissive temperature of 30°C and (f) pcc1-4 at the nonpermissive temperature of 37°C. Expression of Qri7ΔMTS resulted in a full rescue of the slow growth phenotype in kae1Δ, bud32Δ, gon7Δ and cgi121Δ yeast strains and a partial rescue in the pcc1-4 strain at its nonpermissive temperature. Plotted values represent mean values (n ≥ 4). (g) Representation of bulk tRNA nucleoside composition analysis by HPLC in bud32Δ yeast strains and the corresponding rescue experiments (top panel). Quantification of bulk tRNA t6A nucleoside level measurements normalized to the adenosine nucleoside levels in all null or mutated KEOPS yeast strains (bottom panel). Error bars represent standard deviation (s.d.) (n = 3). (h) Relative to the rescue observed for plasmid expression of the deleted or mutated gene, Qri7ΔMTS partially rescues the shortened telomere phenotype of kae1Δ̣(lanes 1–3), bud32Δ̣(lanes 4–6), gon7Δ̣(lanes 7–9), and pcc1-4 at its 37°C nonpermissive temperature (lanes 16–18). cgi121Δ (lanes 10–12) does not display a significant telomere phenotype (compare lanes 10 and 12). The pcc1-4 strain at its 30°C permissive temperature (lanes 12–15), displays a mild residual telomere phenotype. Within the resolution of the assay, this mild phenotype was not rescued by expression of Qri7ΔMTS.
Bud32, Pcc1, Cgi121 and Gon7 play a supporting role to the essential function of Kae1/Qri7 in t6A biosynthesis and telomere length homeostasis
KEOPS mutant yeast strains additionally suffer from depletion of t6A modification and shortened telomeres (9,11). To determine if these phenotypes can be complemented by the expression of Qri7ΔMTS, we analyzed the t6A nucleoside levels in bulk tRNA extracted out of kae1Δ, bud32Δ, gon7Δ, cgi121Δ and pcc1-4 yeast strains and those harboring the Qri7ΔMTS expression plasmid or the corresponding wild-type protein rescue plasmids (Figure 1G). We note that the t6A nucleoside we detect in our HPLC analyses represents a hydrolytic product of the expected natural cyclic t6A product that is generated during the tRNA hydrolysis reaction (5).
As expected, we observed low levels of t6A nucleoside in the bulk tRNA of kae1Δ, bud32Δ, gon7Δ and pcc1-4 cells, with the latter grown at its nonpermissive temperature. We suspect that the detectable residual levels of t6A nucleosides may originate from mitochondrial tRNAs, which are not expected to be affected by KEOPS mutations. We did not detect a diminished level of t6A nucleoside in our cgi121Δ strain and this parallels the weaker growth and telomere phenotypes observed for this strain (13). In each instance where the levels of t6A is diminished, the ectopic expression of Qri7ΔMTS or the wild-type protein corresponding to the deleted (bud32Δ, gon7Δ) or mutated (pcc1-4) genes resulted in a rescue of normal t6A nucleoside levels (Figure 1G).
We next analyzed telomere length in each of our KEOPS mutant strains and their counterpart rescue strains (Figure 1H). The kae1Δ strain displayed a short telomere phenotype, which could be fully rescued by ectopic expression of Kae1 and partially rescued by ectopic expression of Qri7ΔMTS. We observed a similar trend in our analyses of the bud32Δ and gon7Δ strains. At the permissive temperature for pcc1-4, we observed a minor decrease in telomere length, likely due to residual effects of the temperature sensitive allele. Expression of wild-type Pcc1 at the permissive temperature resulted in a modest increase in telomere length. At the nonpermissive temperature, pcc1-4 undergoes severe telomere shortening, which was rescued by ectopic expression of Pcc1 and partially rescued by ectopic expression of Qri7ΔMTS. The fact that Qri7 cannot fully reestablish telomere homeostasis hints that Kae1 and the larger KEOPS complex may have evolved novel functions that contribute to telomere maintenance in higher organisms.
Together, these results indicate that Qri7 provides an essential function(s) that supports t6A biosynthesis in vivo and, to a partial degree, telomere homeostasis. These results are consistent with a model whereby Bud32, Cgi121, Gon7 and Pcc1 play supporting role(s) to the central essential function of Kae1.
Qri7 and Sua5 are both necessary and sufficient for t6A biosynthesis
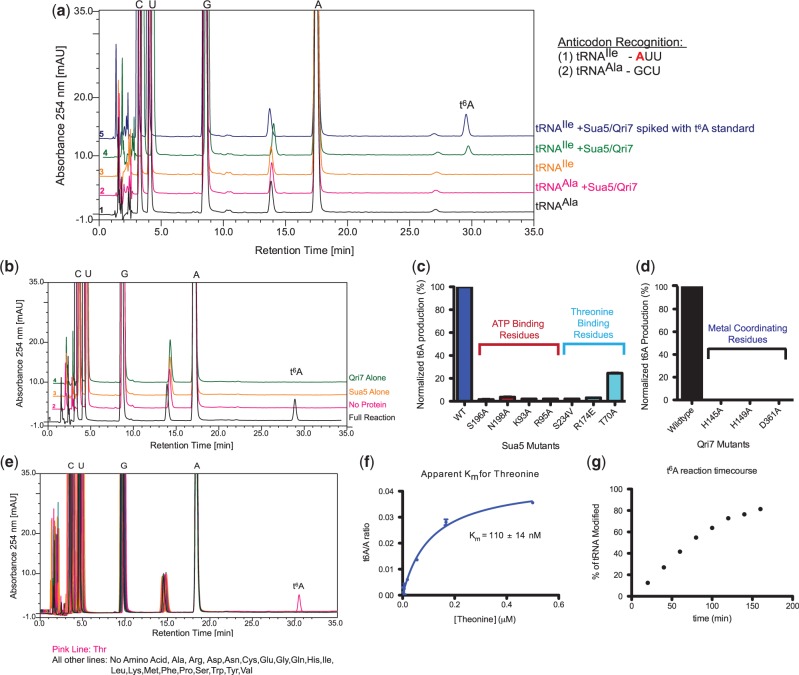
Based on the ability of Qri7 to complement the essential core function(s) of KEOPS in growth, t6A biosynthesis and telomere maintenance, we reasoned that Qri7 may lend itself to the reconstitution of eukaryotic t6A biosynthesis in the most simplistic manner. We thus tested yeast Sua5 and Qri7 for threonylcarbamoylation activity toward two in vitro transcribed yeast tRNAs. tRNAIle recognizes an ANN (specifically AUU) codon and hence is an expected target for t6A modification, whereas tRNAAla does not and hence should not be modified. We carried out in vitro reactions using Sua5, Qri7 and tRNA supplemented with threonine, bicarbonate and ATP, the known substrates for t6A formation (35,36). HPLC nucleoside composition analysis revealed a novel nucleoside species at the 30-min elution mark specifically for the tRNAIle reaction but not that containing tRNAAla (Figure 2A). Addition of a t6A internal standard resulted in an increase in peak area for the 30-min species, suggesting that the newly formed product was t6A. The identity of the novel peak was confirmed using mass spectrometry and by incorporation of heavy atom labeled 13C-threonine and 13C-bicarbonate (Supplementary Figure S3). In addition, the formation of t6A on tRNAIle required both Sua5 and Qri7 (Figure 2B). Notably, mutation of the conserved ATP and threonine-binding pockets of Sua5 [inferred from the structure of Sulfolobus todokaii Sua5; PDB 3AJE (37)] (Figure 2C and Supplementary Figure S2A) and the conserved active site metal-binding residues of Qri7 [inferred from the structure of Pyrococcus abysii Kae1; PDB 2IVP (38)] severely impaired t6A formation (Figure 2D and Supplementary Figure S2B). We conclude that the combination of Qri7 and Sua5 is necessary and sufficient for t6A biosynthesis.
Figure 2.
Sua5 and Qri7 are both necessary and sufficient for in vitro t6A biosynthesis. (a) HPLC nucleoside composition analysis of in vitro transcribed yeast tRNAIle (orange line) and yeast tRNAAla (black line). Incubation of yeast tRNAIle with Sua5/Qri7 (green line) in the presence of threonine, bicarbonate and ATP results in the formation of a novel nucleoside species while incubation of yeast tRNAAla with Sua5/Qri7 (pink line) does not result in the formation of any novel nucleoside species. Spiking the tRNAIle ribonucleoside sample with an internal t6A standard resulted in an increase in peak area for the novel nucleoside (blue line). The identities of the cytidine (C), guanosine (G), adenosine (A), uridine (U) and t6A peaks were confirmed by the addition of internal standards. (b) The formation of t6A in vitro requires both Sua5 and Qri7. HPLC nucleoside composition analysis of tRNAIle alone (pink line), tRNAIle incubated with both Sua5/Qri7 (black line), tRNAIle incubated with Qri7 (green line) and tRNAIle with Sua5 (orange line). (c) Mutation of Sua5 active site residues involved in ATP and threonine binding disrupts t6A biosynthesis. Error bars represent s.d. (n = 2) (d) Mutation of Qri7 active site residues involved in metal coordination completely abolishes t6A biosynthesis. (e) Sua5 and Qri7 specifically conjugate threonine onto tRNAIle (pink line) and not any of the other natural amino acids. (f) The dependency of t6A product formation versus threonine concentration for deriving an apparent Michaelis-Menten constant for threonine. Error bars represent s.d. (n = 2). (g) Time-course analysis of t6A modification by Sua5 and Qri7.
In E. coli tRNA, only trace levels of amino acid cross-reactivity has been detected, namely the replacement of threonine with glycine to generate N6-glycylcarbamoyladenosine (39). This reflects the exquisite specificity of the t6A biosynthetic enzymes for threonine over all other amino acids. Consistent with the observed behavior in vivo, we can detect the formation of novel nucleoside products using Sua5 and Qri7 only upon the addition of threonine (Figure 2E) with an apparent Michaelis–Menten constant for threonine (KmThr) of 110 ± 14 nM (Figure 2F). In our t6A reconstitution reactions, we typically observe >80% modification of input tRNA (Figure 2G).
In sum, we have reconstituted a threonine-specific eukaryotic t6A biosynthetic reaction using only Sua5 and Qri7 proteins. Our results indicate that the Sua5/YrdC and Kae1/Qri7/YgjD protein families form the essential core constituents of the t6A biosynthetic pathway and thus, the additional KEOPS subunits in archaea/eukaryotes, namely Bud32, Pcc1, Gon7, Cgi121 and additional protein factors in bacteria, namely YeaZ and YjeE, likely play indirect/regulatory roles in the t6A biosynthetic process.
Reaction pathway analysis of the eukaryotic Sua5/Qri7 t6A biosynthesis system
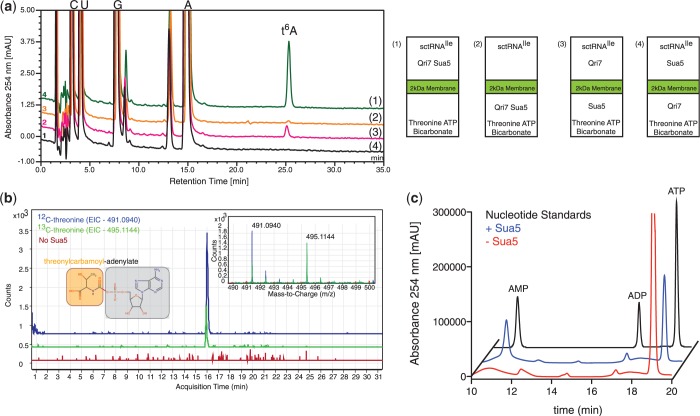
To elucidate the reaction pathway of the eukaryotic Sua5/Qri7 t6A biosynthesis system, we used an experimental setup in which the t6A biosynthesis reaction was separated into adjacent reservoirs divided by a 2000 molecular weight cutoff membrane. We separated threonine, bicarbonate and ATP from Qri7, Sua5 and tRNAIle and were able to sustain t6A formation on tRNAIle (Figure 3A), which is consistent with the ability of threonine, bicarbonate and ATP to diffuse across the barrier membrane. Not unexpectedly, when both Sua5 and Qri7 enzymes were separated from tRNAIle, the ultimate acceptor of t6A modification, t6A formation was abolished. When Sua5 was placed with threonine, bicarbonate and ATP in a reservoir separated from Qri7 and the tRNA, the production of t6A was still supported (Figure 3A). However, when Qri7 was placed with threonine, bicarbonate and ATP in one reservoir separated from Sua5 and tRNA, the production of t6A was abolished (Figure 3A). These experiments support the notion that Sua5 generates a diffusible reaction intermediate, and Qri7 in turn engages this product to carry forth the final conjugation step in t6A biosynthesis.
Figure 3.
Sua5 functions upstream of Qri7 in the t6A biosynthetic pathway. (a) HPLC nucleoside composition analysis of tRNAIle that was incubated with Sua5/Qri7 in a chamber separated from threonine, bicarbonate and ATP by a 2 kDa membrane barrier (green line), which shows the formation of t6A. Separation of both Sua5/Qri7 from tRNAIle (orange line) does not give rise to t6A formation. Separation of Sua5 from Qri7 and tRNAIle (pink line) supports t6A formation while separation of Qri7 from Sua5 and tRNAIle does not support t6A formation (black line) (left panel). Cartoon representation of the experimental setup (right panel). (b) Incubation of Sua5 with threonine, bicarbonate and ATP results in the formation of TCA, which has a m/z value of 491.0940 (mass spectra inset—blue line). Reactions performed in the presence of 13C-labeled threonine resulted in a +4 shift in the m/z value to 495.1144 (mass spectra inset—green line). The extracted ion chromatograms (EIC) for both 12C and 13C-TCA are displayed by blue and green chromatograms respectively. Reactions preformed in the absence of Sua5 did not result in the formation and detection of TCA (mass spectra inlet – red line) and an extracted EIC at 491.0940 m/z is shown by a red chromatogram. (c) Ion-pair HPLC analysis of Sua5 ATP hydrolysis activity. An ATP, ADP and AMP standard mixture is shown (black line). In the presence of threonine and bicarbonate, Sua5 hydrolyzes ATP into AMP (blue line). In the absence of Sua5, ATP does not get hydrolyzed into AMP (red line).
To identify the diffusible reaction intermediate shuttling from Sua5 to Qri7, we incubated Sua5 with threonine, bicarbonate and ATP and analyzed the reaction sample by mass spectrometry. We identified TCA (m/z ratio = 491.0940) (Figure 3B) as the likely intermediate, which is consistent with the earlier finding that bacterial YrdC generates TCA (24). Incubation with 13C-labeled threonine into the reaction conditions caused a +4 m/z shift for this species (Figure 3B), confirming the product was derived from threonine. The product could also be fragmented into an AMP adduct (data not shown) confirming ATP as a precursor. Furthermore, the production of TCA by Sua5 was coupled to the hydrolysis of ATP into AMP, as detected by ion-pair chromatography (Figure 3C). The use of a hexahistidine-tagged Sua5 construct that was purified under denaturating conditions coupled to refolding (Supplementary Figure S4A) showed identical specific activity in t6A biosynthesis and TCA production (Supplementary Figure S4B and C), confirming that Sua5 (and not a bacterial protein contaminant) is both necessary and sufficient for the initial step of t6A synthesis.
The proposed reaction mechanism predicts that Qri7 binds directly to TCA and tRNA as substrates. We speculated that the Qri7 infrastructure previously attributed to ATP binding (38) actually engages TCA. Indeed, we were able to dock threonylcarbomoyl-adenylate into the catalytic cleft using the nucleotide binding mode in the Pyrococcus abyssi Kae1-ATP complex (38), with only minor side-chain reorientations (see methods, Figure 4F).
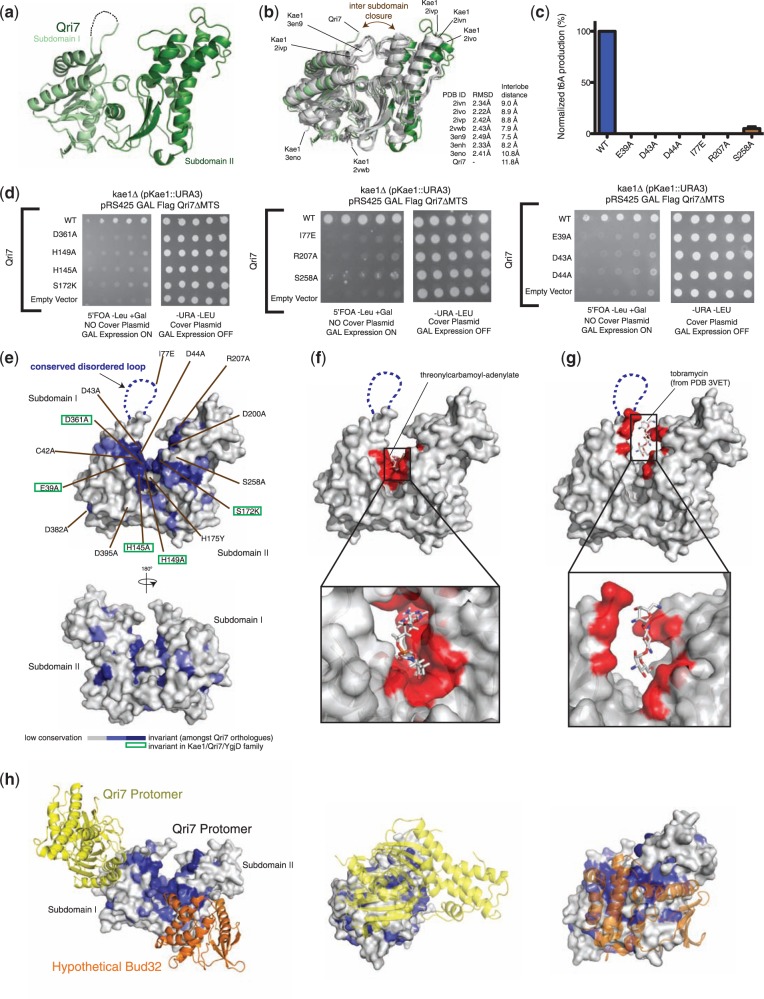
Figure 4.
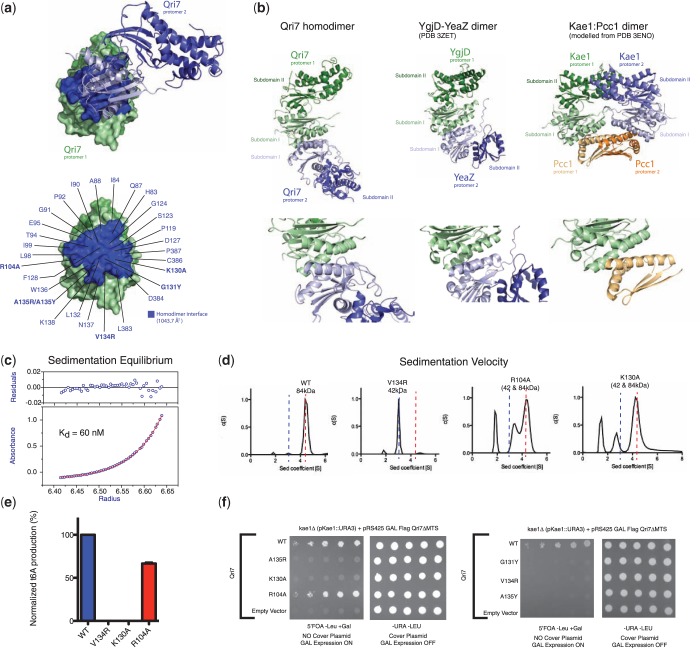
Structure–function analysis of Qri7. (a) Structure of Saccharomyces cerevisiae Qri7 (amino acids 30–407) shown in ribbons representation. Subdomains I and II are shown in light green and dark green, respectively. (b) Structural alignment of scQri7 (green) with all structures of Kae1 solved to date (grey), shown as ribbon representations. RMSDs of Qri7 relative to each Kae1 structure and the intersubdomain distances representing degree of lobe closure are listed. Intersubdomain distances were measured by the distance between Cα atoms of two conserved glycine residues (G117 and G174 in Qri7) on either side of the catalytic cleft. (c) Mutation of catalytic cleft residues on Qri7 abolishes t6A biosynthesis. Error bars represent s.d. (n = 2). (d) Mutation of catalytic cleft residues on Qri7 abolishes its ability to rescue the kae1Δ slow growth phenotype. (e) Projection of conservation onto a surface representation of Qri7. Residues in the catalytic cleft region are labeled and invariant residues across the Kae1/Qri7/YgjD family are boxed in green. (f) Docking of TCA into the catalytic cleft of Qri7. Qri7 catalytic cleft residues in close contact (<5 Å separation) with the docked TCA are shown in red. (g) Modeled tobramycin in the catalytic cleft of Qri7. Residues in close contact (<5 Å separation) with the modeled tobramycin molecule are shown in red. (h) Surface conservation of the Qri7 dimerization interface and hypothetical Bud32 binding site. The second Qri7 protomer in the dimer is shown in yellow ribbon and Bud32 is shown in orange ribbon (left panel). Head-on view of the Qri7 homodimer interface (middle panel) and head-on view of the hypothetical Bud32 binding site on Qri7 (right panel).
In summary, the eukaryotic t6A biosynthesis system functions first through Sua5, which generates TCA using its adenylation function and then secondly through Qri7, which binds to TCA to conjugate the threonyl-carbamoyl moiety onto directly bound tRNA substrates. The activities of Sua5 and Qri7 are not necessarily physically coupled, and the Kae1/Qri7/YgjD protein family contains the required infrastructure for interacting with both TCA and tRNA.
Crystal structure of Qri7
It is intriguing that Qri7 can support t6A biosynthesis as an isolated protein, while Kae1 and YgjD orthologues operate within larger macromolecular assemblies. To shed light onto the structural basis for the unique functional properties of Qri7, we solved the structure of yeast Qri7 (Table 1 and Figure 4A). At first glance, the structure of Qri7 is highly similar to previously reported structures of archaeal Kae1 (13,38,40) (PDB 2IVN, 2IVO, 2IVP, 2VWB, 3EN9, 3ENH and 3ENO) (2.34 Å, 2.22 Å, 2.42 Å, 2.43 Å, 2.49 Å, 2.33 Å and 2.41 Å Cα RMSDs, respectively) (Figure 4B). Qri7 exhibits the prototypical bilobal structure with its catalytic region sandwiched between subdomains I and II. Crystal structures of Kae1 orthologues (like ASKHA fold proteins in general) display variability in interlobe closure, and the Qri7 structure is notable in adopting the most open configuration reported to date for a Kae1 orthologue (Figure 4B).
Table 1.
Data collection and refinement statistics
| scQri7 | |
|---|---|
| Data collection | |
| Space group | P21212 |
| Cell dimensions | |
| a, b, c (Å) | 64.7, 131.2, 48.0 |
| Resolution (Å) | 20.6–2.90 (3.24–2.90) |
| Rsym | 0.059 (0.27) |
| I/σI | 23.4 (3.30) |
| Completeness (%) | 99.9 (99.6) |
| Redundancy | 1.82 (1.85) |
| Refinement | |
| Resolution (Å) | 20.6–2.90 |
| No. reflections | 9434 (2357) |
| Rwork/Rfree | 0.24/0.29 |
| No. atoms | |
| Protein | 2698 |
| Ligand/ion | 74 |
| B-factors | 77.9 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.008 |
| Bond angles (°) | 1.10 |
Structure was determined from a single crystal.
Values in parentheses are for highest-resolution shell.
Rsym = ΣhΣi | Ih,i − Ih| / ΣhΣi Ih,i, where Ih is the mean intensity of the i observations of symmetry related reflections of h. R = ∑ | Fobs − Fcalc | / ∑Fobs, where Fobs = FP and Fcalc is the calculated protein structure factor from the atomic model.
Projection of conservation onto the Qri7 structure identified a large contiguous surface emanating from the catalytic cleft (Figure 4E). Because Qri7, unlike Kae1 and YgjD, functions in the absence of intermolecular regulatory subunits, we reasoned that this surface conservation corresponds to the core catalytic infrastructure required for t6A biosynthesis by the Kae1/Qri7/YgjD family. A portion of the conserved surface was accounted for by the expected binding sites of TCA and phosphate coordinating metal ions (Figure 4F). Additional conservation was apparent on the lateral surface of Qri7 and at the tip of subdomains I and II, the former of which includes a highly conserved disordered loop (Figure 4E and Supplementary Figure S2B). The binding of acceptor substrate (tobramycin) in the evolutionarily related TobZ crystal structure (41) identifies these unaccounted for conserved surfaces at the tips of subdomains I and II as the possible tRNA substrate-binding surface (Figure 4G). Consistent with an essential role for these regions, the site-directed mutants I77E, R207A, S258A and D43A were not able to complement the slow growth phenotype of the kae1Δ strain and were also devoid of t6A biosynthesis activity in vitro (Figures 2D, 4C and D), comparable to mutants with impaired TCA or metal ion binding functions.
Interestingly, modeling of a hypothetical Bud32 [based on the Kae1–Bud32 complex PDB 3ENH (13)] interaction with Qri7 showed that Bud32 overlaps with a large portion of the conserved lateral surface of Qri7. As the lateral conserved surface is functionally required for t6A biosynthesis, this overlap may imply an inhibitory function for Bud32 on the core t6A biosynthetic activity of Kae1 (Figure 4H).
Qri7 forms homodimers in solution
Application of crystallographic symmetry to the Qri7 monomer structure revealed a dimer configuration with a large (2087.5 Å2) hydrophobic buried surface (Figure 5A and Supplementary Figure S2B), which is partially conserved across Qri7 orthologues (Figure 4H). Remarkably, the Qri7 dimerization surface is the same surface used by archaeal and eukaryotic Kae1 proteins to bind Pcc1, a dimerization module within KEOPS (13) (Figure 5B) and the same surface used by the bacterial orthologue YgjD to bind YeaZ (25) (Figure 5B). Interestingly, although the Pcc1 subunit in the Pcc1–Kae1 complex shares no recognizable sequence similarity to the Kae1, Qri7, YgjD or YeaZ proteins, its architecture resembles subdomain I (C-terminal lobe) of these proteins, and engages Kae1 through a four-helix bundle that is surprisingly similar to the Qri7 and YgjD-YeaZ modes of dimerization. This raises the intriguing question of whether Pcc1 evolved independently or from Kae1.
Figure 5.
Qri7 forms homodimers in solution. (a) Two protomers of Qri7 in a homodimer configuration shown in surface and ribbons representation. On the surface represented protomer, the homodimerzation surface is highlighted in blue (top panel). Each residue at the dimerization interface is labeled and residues mutated for further characterization are shown in bold (bottom panel). (b) Ribbons representation and comparison of the homo and heterodimer configurations of Qri7:Qri7, YeaZ:YgjD and Kae1:Pcc1 complexes. Bottom panels display close up views of the dimer interfaces shown on top. (c) Sedimentation equilibrium analysis reveals a dimerization dissociation constant of 60 nM for Qri7. The red line denotes a fitted curve to a self-association model. The residuals for the fit are shown in the top panel. (d) Sedimentation velocity analysis of wild-type Qri7 (residues 30–407) and V134R, K130A and R104A mutants. Wild-type Qri7 behaves as a dimer in solution whereas mutant V134R behaves as a monomer. The K130A and R104A mutants exist in monomer-dimer equilibrium. Peaks with sedimentation coefficients <2 represent artifacts arising from small differences in buffer conditions between test and blank samples. (e) Dimerization mutants V134R and K130A are defective for t6A biosynthesis activity, whereas the R104A mutant retains partial activity. Error bars represent s.d. (n = 2). (f) Mutation of dimer interface residues abolishes the ability of Qri7 to rescue the slow growth phenotype of a kae1Δ yeast strain, with the exception of the R104A mutant.
We confirmed that Qri7 forms homodimers in solution with a dissociation constant of 60 nM by equilibrium analytical ultracentrifugation (Figure 5C). To test whether Qri7 dimerization in solution is mediated by the same surface revealed by the crystal structure, we tested three individual interface mutants (R104A, K130A and V134R) for oligomerization status using velocity analytical ultracentrifugation. While wild-type Qri7 and five additional Qri7 mutant proteins harboring catalytic cleft mutations (D43A, D44A, I77E, R207A and S258A) existed exclusively as dimers (Supplementary Figure S5A), the V134R mutant existed exclusively as a monomer and the R104A and K130A mutants existed in monomer–dimer equilibrium (Figure 5D). The R104A and K130A mutants displayed a dimerization dissociation constant of 3.0 ± 0.3 μM and 2.5 ± 0.2 μM, respectively (Supplementary Figure S5B). These results confirm that Qri7 forms homodimers in solution through the same binding mode revealed in the crystal structure.
To determine whether dimerization is required for Qri7 function, we first tested the R104A, K130A and V134R mutants for t6A biosynthesis activity in vitro. The K130A and V134R mutants were devoid of t6A formation activity in vitro, whereas the R104A mutant displayed only a partial loss of enzymatic function (was 65% active) (Figure 5E). To determine if dimerization contributes to the ability of Qri7 to function in vivo, we tested the three above mutants as well as three additional dimer interface mutants for their ability to rescue the slow growth defect of the kae1Δ strain. Five out of the six mutations (K130A, G131Y, V134R, A135Y and A135R) abolished Qri7’s ability to rescue the slow growth phenotype of a kae1Δ yeast strain (Figure 5F). In contrast to the case for the K130A mutant, the finding that the R104A mutant was still able to rescue the growth of the kae1Δ yeast strain and partially sustain the t6A modification of tRNA, despite being partially compromised for dimerization function, suggests that the R104A mutation may impart a bypass for the necessity of dimerization. Together these studies highlight the importance of the dimerization contact surface but not dimerization per se for Qri7 function in vitro and in vivo.
The similarities in mode of dimerization observed for Qri7 and those observed for Kae1/YgjD orthologues that operate in larger complexes suggest that the potential to participate in protein–protein interactions using a common surface is a general requirement for t6A biosynthesis by the Kae1/Qri7/YgjD family. The ability for Qri7 to homodimerize using this surface provides a rationale for how it may be able to function in the absence of a Pcc1 dimerization module or a YeaZ protein–protein interaction and also how Qri7 can complement the essential function(s) of the entire KEOPS complex.
DISCUSSION
A universally conserved t6A biosynthetic pathway
Our reconstitution and functional studies demonstrate that Sua5 and Qri7 are both necessary and sufficient for the t6A modification of tRNAs. Whereas t6A formation by YgjD requires the additional presence of YeaZ and YjeE proteins and Kae1 requires the additional presence of the KEOPS complex (minimally in vivo), Qri7 appears to function in the mitochondria in the absence of a higher order assembly. The separation of Sua5 and Qri7 in their respective cytoplasmic and mitochondrial compartments suggests that t6A formation occurs via minimally two separable enzymatic steps and that Sua5 and Qri7 enzymes do not require physical coupling, a result supported by our barrier membrane studies. However, we cannot rule out that a small fraction of Sua5 protein resides within the mitochondria, which would eliminate the requirement for TCA, a highly unstable intermediate, to traverse the mitochondrial membrane.
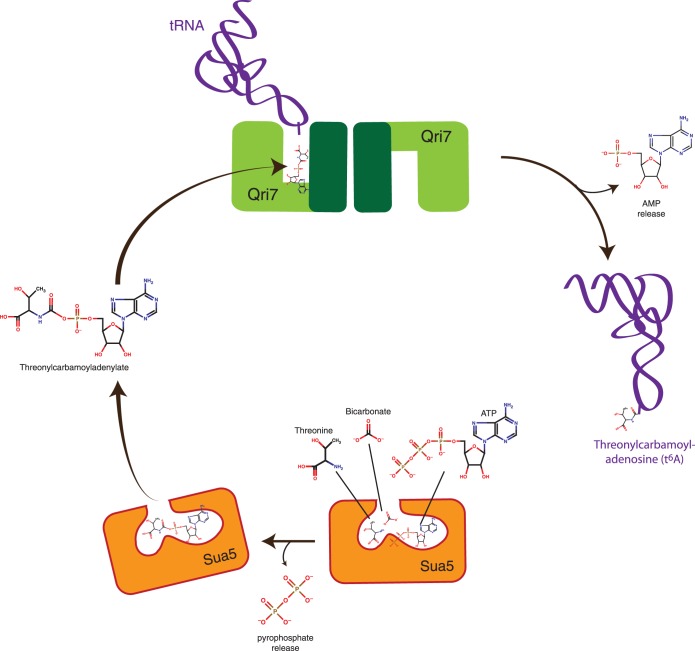
Our data supports the notion that Sua5 acts upstream in the t6A biosynthetic pathway whereby threonine, bicarbonate and ATP are converted to TCA (24) (Figure 6). TCA is released by Sua5 and then bound by Qri7, which can sample a homodimer configuration. Productive binding of the tRNA substrate positions adenosine 37 of the tRNA anticodon loop toward the active site of Qri7 to serve as an acceptor of threonylcarbamoylation (Figure 6). We reason that Qri7 binds to TCA using the nucleotide binding infrastructure of the ASKHA fold and does not require binding of additional ATP molecules to facilitate t6A biosynthesis. This would indicate that the Kae1/Qri7/YgjD family, representing one of the most ancient branches of the ASKHA family, is not an ATP hydrolyzing enzyme, which to our knowledge, is the only current example of a non-ATPase function for an ASKHA-fold protein.
Figure 6.
Minimal eukaryotic threonylcarbamoyladenosine (t6A) biosynthesis pathway using Sua5 and Qri7. Sua5 binds threonine, bicarbonate and ATP to generate TCA using its adenylation function. TCA is then released by Sua5 and bound to a Qri7 homodimer (modeled with one tRNA bound to its active site), which conjugates a threonylcarbamoyl-moiety onto bound tRNA substrates.
Recent studies aimed at analyzing the bacterial t6A biosynthesis system have also discovered that YrdC functions upstream of the YgjD-YeaZ-YjeE complex (24). YrdC, the bacterial orthologue of Sua5, also generates TCA that feeds into the YgjD-YeaZ-YjeE complex. Together with our eukaryotic Sua5/Qri7 t6A biosynthesis system, this collection of findings suggests that t6A biosynthesis occurs through a universally conserved core pathway consisting of the YrdC/Sua5 and Kae1/Qri7/YgjD protein families. As the Kae1/Qri7/YgjD protein family diverged over the course of evolution, bacterial factors YeaZ and YjeE and archaeal/eukaryotic factors Bud32, Cgi121, Pcc1 and Gon7 may have evolved to play indirect roles in the t6A biosynthesis pathway, possibly to regulate the core enzymatic function of YgjD and Kae1, respectively.
Functional relevance of the Kae1/Qri7/YgjD high-order assemblies
While the direct dimerization of Qri7 and the indirect dimerization of Kae1 by Pcc1 (13,42) appear important for protein function, the underlying basis for this requirement remains unclear. A possible function for dimerization may be to allosterically regulate intrinsic catalytic activity. This type of enzyme regulation is exemplified by the eIF2α, RAF and EGFR families of eukaryotic protein kinases. The kinase domains of all three families transition from monomers to specific dimer configurations (43–47), which are required for activation of protein kinase activity. Interestingly, a specific V600E mutation in B-RAF allows it to bypass dimerization as a prerequisite for function (48), a behavior reminiscent of the Qri7 R104A dimer interface mutant.
In the case of RAF and EGFR kinases, specific family members (KSR1 and KSR2 for RAF and Her3/ERBB3 for EGF) have evolved to solely serve as regulators by dispensing with catalytic function while maintaining their ability to dimerize. This phenomenon may account for the functional interplay within YgjD–YeaZ complexes where YeaZ (a structural orthologue of YgjD) has dispensed with catalytic function but still retains the ability to dimerize and hence regulate YgjD function. Interestingly, bacteria with reduced genomes such as Mycobacterium genitalium and Candidatus Sulcia muelleri str. GWSS do not encode YeaZ and YjeE proteins. It is tempting to speculate that YgjD in these minimal organisms have retained an ability to homodimerize and function in isolation, thus alleviating a necessity for YeaZ and YjeE-like proteins. If dimerization truly underpins the regulation of Qri7 and YgjD catalytic function, we reason that the binding of Pcc1 to the corresponding surface of Kae1 might serve a similar regulatory function. The atomic structure of Qri7 and the ability of Qri7 to participate in in vitro t6A biosynthesis as presented in this study provide a conceptual framework for future endeavors to uncover the precise molecular function of KEOPS regulatory subunits and their roles in t6A biosynthesis.
The connection between tRNA biology and telomere maintenance
While Qri7 was able to fully rescue the growth defects and diminished t6A biosynthesis of KEOPS, it was only able to partially rescue the shortened telomere defect. This may reflect the fact that KEOPS has evolved new functions required for telomere maintenance that are not supplied by Qri7. One potential function may be the addition of new layers of regulation, which would allow for more precise temporal and spatial modulation of Kae1 enzymatic output. A second potential function could be the ability to recognize and modify novel RNA substrates, which presumably are not efficiently recognized by Qri7. Whatever the true basis for the incomplete ability of Qri7 to rescue the function of KEOPS in telomere maintenance, our findings provide a beachhead for future studies to decipher the precise role of KEOPS and the missing links between telomere and tRNA biology.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1 and Supplementary Figures 1–5.
ACKNOWLEDGEMENTS
We are grateful to A.C. Gingras for providing us access to a HPLC unit and M. Sanchez, M. Talukdar and M. Canny for assisting with in vitro RNA transcription reactions. We thank E. Styles and C. Boone for providing us with the BY4743 sua5::KANMX/SUA5 yeast strain.
FUNDING
National Center for Research Resources [5P41RR015301-10] and the National Institutes of Health [8 P41 GM103403-10] (to Research conducted at the Advanced Photon Source on the Northeastern Collaborative Access Team beamlines); US DOE [DE-AC02-06CH11357] (to use of the Advanced Photon Source, an Office of Science User Facility operated for the U.S. Department of Energy (DOE) Office of Science by Argonne National Laboratory); Ontario Ministry of Economic Development and Innovation (to Ontario Institute for Cancer Research); The Vanier Canada Graduate Scholarship (to L.W); Canadian Cancer Society Research Institute [20504 to F.S.]. Funding for open access charge: Canadian Cancer Society Research Institute.
Conflict of interest statement. None declared.
REFERENCES
- 1.Cantara WA, Crain PF, Rozenski J, McCloskey JA, Harris KA, Zhang X, Vendeix FA, Fabris D, Agris PF. The RNA modification database, RNAMDB: 2011 update. Nucleic Acids Res. 2011;39:D195–D201. doi: 10.1093/nar/gkq1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Murphy FV, IV, Ramakrishnan V, Malkiewicz A, Agris PF. The role of modifications in codon discrimination by tRNA(Lys)UUU. Nat. Struct. Mol. Biol. 2004;11:1186–1191. doi: 10.1038/nsmb861. [DOI] [PubMed] [Google Scholar]
- 3.Weissenbach J, Grosjean H. Effect of threonylcarbamoyl modification (t6A) in yeast tRNA Arg III on codon-anticodon and anticodon-anticodon interactions. A thermodynamic and kinetic evaluation. Eur. J. Biochem. 1981;116:207–213. doi: 10.1111/j.1432-1033.1981.tb05320.x. [DOI] [PubMed] [Google Scholar]
- 4.Lescrinier E, Nauwelaerts K, Zanier K, Poesen K, Sattler M, Herdewijn P. The naturally occurring N6-threonyl adenine in anticodon loop of Schizosaccharomyces pombe tRNAi causes formation of a unique U-turn motif. Nucleic Acids Res. 2006;34:2878–2886. doi: 10.1093/nar/gkl081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miyauchi K, Kimura S, Suzuki T. A cyclic form of N6-threonylcarbamoyladenosine as a widely distributed tRNA hypermodification. Nat. Chem. Biol. 2013;9:105–111. doi: 10.1038/nchembio.1137. [DOI] [PubMed] [Google Scholar]
- 6.Lin CA, Ellis SR, True HL. The Sua5 protein is essential for normal translational regulation in yeast. Mol. Cell. Biol. 2010;30:354–363. doi: 10.1128/MCB.00754-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hampsey M, Na JG, Pinto I, Ware DE, Berroteran RW. Extragenic suppressors of a translation initiation defect in the cyc1 gene of Saccharomyces cerevisiae. Biochimie. 1991;73:1445–1455. doi: 10.1016/0300-9084(91)90177-3. [DOI] [PubMed] [Google Scholar]
- 8.El Yacoubi B, Hatin I, Deutsch C, Kahveci T, Rousset JP, Iwata-Reuyl D, Murzin AG, de Crecy-Lagard V. A role for the universal Kae1/Qri7/YgjD (COG0533) family in tRNA modification. EMBO J. 2011;30:882–893. doi: 10.1038/emboj.2010.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Srinivasan M, Mehta P, Yu Y, Prugar E, Koonin EV, Karzai AW, Sternglanz R. The highly conserved KEOPS/EKC complex is essential for a universal tRNA modification, t6A. EMBO J. 2011;30:873–881. doi: 10.1038/emboj.2010.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.El Yacoubi B, Lyons B, Cruz Y, Reddy R, Nordin B, Agnelli F, Williamson JR, Schimmel P, Swairjo MA, de Crecy-Lagard V. The universal YrdC/Sua5 family is required for the formation of threonylcarbamoyladenosine in tRNA. Nucleic Acids Res. 2009;37:2894–2909. doi: 10.1093/nar/gkp152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Downey M, Houlsworth R, Maringele L, Rollie A, Brehme M, Galicia S, Guillard S, Partington M, Zubko MK, Krogan NJ, et al. A genome-wide screen identifies the evolutionarily conserved KEOPS complex as a telomere regulator. Cell. 2006;124:1155–1168. doi: 10.1016/j.cell.2005.12.044. [DOI] [PubMed] [Google Scholar]
- 12.Kisseleva-Romanova E, Lopreiato R, Baudin-Baillieu A, Rousselle JC, Ilan L, Hofmann K, Namane A, Mann C, Libri D. Yeast homolog of a cancer-testis antigen defines a new transcription complex. EMBO J. 2006;25:3576–3585. doi: 10.1038/sj.emboj.7601235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mao DY, Neculai D, Downey M, Orlicky S, Haffani YZ, Ceccarelli DF, Ho JS, Szilard RK, Zhang W, Ho CS, et al. Atomic structure of the KEOPS complex: an ancient protein kinase-containing molecular machine. Mol. Cell. 2008;32:259–275. doi: 10.1016/j.molcel.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meng FL, Chen XF, Hu Y, Tang HB, Dang W, Zhou JQ. Sua5p is required for telomere recombination in Saccharomyces cerevisiae. Cell Res. 2010;20:495–498. doi: 10.1038/cr.2010.34. [DOI] [PubMed] [Google Scholar]
- 15.Meng FL, Hu Y, Shen N, Tong XJ, Wang J, Ding J, Zhou JQ. Sua5p a single-stranded telomeric DNA-binding protein facilitates telomere replication. EMBO J. 2009;28:1466–1478. doi: 10.1038/emboj.2009.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghavidel A, Kislinger T, Pogoutse O, Sopko R, Jurisica I, Emili A. Impaired tRNA nuclear export links DNA damage and cell-cycle checkpoint. Cell. 2007;131:915–926. doi: 10.1016/j.cell.2007.09.042. [DOI] [PubMed] [Google Scholar]
- 17.Fu D, Brophy JA, Chan CT, Atmore KA, Begley U, Paules RS, Dedon PC, Begley TJ, Samson LD. Human AlkB homolog ABH8 Is a tRNA methyltransferase required for wobble uridine modification and DNA damage survival. Mol. Cell. Biol. 2010;30:2449–2459. doi: 10.1128/MCB.01604-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mei Y, Yong J, Liu H, Shi Y, Meinkoth J, Dreyfuss G, Yang X. tRNA binds to cytochrome c and inhibits caspase activation. Mol. Cell. 2010;37:668–678. doi: 10.1016/j.molcel.2010.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bjork GR, Wikstrom PM, Bystrom AS. Prevention of translational frameshifting by the modified nucleoside 1-methylguanosine. Science. 1989;244:986–989. doi: 10.1126/science.2471265. [DOI] [PubMed] [Google Scholar]
- 20.Gustilo EM, Vendeix FA, Agris PF. tRNA's modifications bring order to gene expression. Curr. Opin. Microbiol. 2008;11:134–140. doi: 10.1016/j.mib.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Urbonavicius J, Qian Q, Durand JM, Hagervall TG, Bjork GR. Improvement of reading frame maintenance is a common function for several tRNA modifications. EMBO J. 2001;20:4863–4873. doi: 10.1093/emboj/20.17.4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deutsch C, El Yacoubi B, de Crecy-Lagard V, Iwata-Reuyl D. Biosynthesis of threonylcarbamoyl adenosine (t6A), a universal tRNA nucleoside. J. Biol. Chem. 2012;287:13666–13673. doi: 10.1074/jbc.M112.344028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perrochia L, Crozat E, Hecker A, Zhang W, Bareille J, Collinet B, van Tilbeurgh H, Forterre P, Basta T. In vitro biosynthesis of a universal t6A tRNA modification in Archaea and Eukarya. Nucleic Acids Res. 2013;141:1953–1964. doi: 10.1093/nar/gks1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lauhon CT. Mechanism of N6-Threonylcarbamoyladenonsine (t(6)A) Biosynthesis: Isolation and Characterization of the Intermediate Threonylcarbamoyl-AMP. Biochemistry. 2012;51:8950–8963. doi: 10.1021/bi301233d. [DOI] [PubMed] [Google Scholar]
- 25.Nichols CE, Lamb HK, Thompson P, Omari KE, Lockyer M, Charles I, Hawkins AR, Stammers DK. Crystal structure of the dimer of two essential Salmonella typhimurium proteins, YgjD & YeaZ and calorimetric evidence for the formation of a ternary YgjD-YeaZ–YjeE complex. Protein Sci. 2013;22:628–640. doi: 10.1002/pro.2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Giaever G, Flaherty P, Kumm J, Proctor M, Nislow C, Jaramillo DF, Chu AM, Jordan MI, Arkin AP, Davis RW. Chemogenomic profiling: identifying the functional interactions of small molecules in yeast. Proc. Natl Acad. Sci. USA. 2004;101:793–798. doi: 10.1073/pnas.0307490100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Proctor M, Urbanus ML, Fung EL, Jaramillo DF, Davis RW, Nislow C, Giaever G. The automated cell: compound and environment screening system (ACCESS) for chemogenomic screening. Methods Mol. Biol. 2011;759:239–269. doi: 10.1007/978-1-61779-173-4_15. [DOI] [PubMed] [Google Scholar]
- 28.Gehrke CW, Kuo KC. Ribonucleoside analysis by reversed-phase high-performance liquid chromatography. J. Chromatogr. 1989;471:3–36. doi: 10.1016/s0021-9673(00)94152-9. [DOI] [PubMed] [Google Scholar]
- 29.Storoni LC, McCoy AJ, Read RJ. Likelihood-enhanced fast rotation functions. Acta Crystallogr. D Biol. Crystallogr. 2004;60:432–438. doi: 10.1107/S0907444903028956. [DOI] [PubMed] [Google Scholar]
- 30.Smart OS, Womack TO, Flensburg C, Keller P, Paciorek W, Sharff A, Vonrhein C, Bricogne G. Exploiting structure similarity in refinement: automated NCS and target-structure restraints in BUSTER. Acta Crystallogr. D Biol. Crystallogr. 2012;68:368–380. doi: 10.1107/S0907444911056058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys. J. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meisinger C, Pfanner N, Truscott KN. Isolation of yeast mitochondria. Methods Mol. Biol. 2006;313:33–39. doi: 10.1385/1-59259-958-3:033. [DOI] [PubMed] [Google Scholar]
- 33.Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 34.Oberto J, Breuil N, Hecker A, Farina F, Brochier-Armanet C, Culetto E, Forterre P. Qri7/OSGEPL, the mitochondrial version of the universal Kae1/YgjD protein, is essential for mitochondrial genome maintenance. Nucleic Acids Res. 2009;37:5343–5352. doi: 10.1093/nar/gkp557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Powers DM, Peterkofsky A. Biosynthesis and specific labeling of N-(purin-6-ylcarbamoyl)threonine of Escherichia coli transfer RNA. Biochem. Biophys. Res. Commun. 1972;46:831–838. doi: 10.1016/s0006-291x(72)80216-x. [DOI] [PubMed] [Google Scholar]
- 36.Chheda GB, Hong CI, Piskorz CF, Harmon GA. Biosynthesis of N-(purin-6-ylcarbamoyl)-L-threonine riboside. Incorporation of L-threonine in vivo into modified nucleoside of transfer ribonucleic acid. Biochem. J. 1972;127:515–519. doi: 10.1042/bj1270515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuratani M, Kasai T, Akasaka R, Higashijima K, Terada T, Kigawa T, Shinkai A, Bessho Y, Yokoyama S. Crystal structure of Sulfolobus tokodaii Sua5 complexed with L-threonine and AMPPNP. Proteins. 2011;79:2065–2075. doi: 10.1002/prot.23026. [DOI] [PubMed] [Google Scholar]
- 38.Hecker A, Leulliot N, Gadelle D, Graille M, Justome A, Dorlet P, Brochier C, Quevillon-Cheruel S, Le Cam E, van Tilbeurgh H, et al. An archaeal orthologue of the universal protein Kae1 is an iron metalloprotein which exhibits atypical DNA-binding properties and apurinic-endonuclease activity in vitro. Nucleic Acids Res. 2007;35:6042–6051. doi: 10.1093/nar/gkm554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schweizer MP, McGrath K, Baczynskyj L. The isolation and characterization of N-[9-(beta-D-ribofuranosyl)-purin-6-ylcarbamoyl]glycine from yeast transfer RNA. Biochem. Biophys. Res. Commun. 1970;40:1046–1052. doi: 10.1016/0006-291x(70)90899-5. [DOI] [PubMed] [Google Scholar]
- 40.Hecker A, Lopreiato R, Graille M, Collinet B, Forterre P, Libri D, van Tilbeurgh H. Structure of the archaeal Kae1/Bud32 fusion protein MJ1130: a model for the eukaryotic EKC/KEOPS subcomplex. EMBO J. 2008;27:2340–2351. doi: 10.1038/emboj.2008.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parthier C, Gorlich S, Jaenecke F, Breithaupt C, Brauer U, Fandrich U, Clausnitzer D, Wehmeier UF, Bottcher C, Scheel D, et al. The O-carbamoyltransferase TobZ catalyzes an ancient enzymatic reaction. Angew Chem. Int. Ed. Engl. 2012;51:4046–4052. doi: 10.1002/anie.201108896. [DOI] [PubMed] [Google Scholar]
- 42.Costessi A, Mahrour N, Sharma V, Stunnenberg R, Stoel MA, Tijchon E, Conaway JW, Conaway RC, Stunnenberg HG. The human EKC/KEOPS complex is recruited to Cullin2 ubiquitin ligases by the human tumour antigen PRAME. PLoS One. 2012;7:e42822. doi: 10.1371/journal.pone.0042822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dey M, Cao C, Dar AC, Tamura T, Ozato K, Sicheri F, Dever TE. Mechanistic link between PKR dimerization, autophosphorylation, and eIF2alpha substrate recognition. Cell. 2005;122:901–913. doi: 10.1016/j.cell.2005.06.041. [DOI] [PubMed] [Google Scholar]
- 44.Dar AC, Dever TE, Sicheri F. Higher-order substrate recognition of eIF2alpha by the RNA-dependent protein kinase PKR. Cell. 2005;122:887–900. doi: 10.1016/j.cell.2005.06.044. [DOI] [PubMed] [Google Scholar]
- 45.Dey M, Cao C, Sicheri F, Dever TE. Conserved intermolecular salt bridge required for activation of protein kinases PKR, GCN2, and PERK. J. Biol. Chem. 2007;282:6653–6660. doi: 10.1074/jbc.M607897200. [DOI] [PubMed] [Google Scholar]
- 46.Rajakulendran T, Sahmi M, Lefrancois M, Sicheri F, Therrien M. A dimerization-dependent mechanism drives RAF catalytic activation. Nature. 2009;461:542–545. doi: 10.1038/nature08314. [DOI] [PubMed] [Google Scholar]
- 47.Zhang X, Gureasko J, Shen K, Cole PA, Kuriyan J. An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell. 2006;125:1137–1149. doi: 10.1016/j.cell.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 48.Poulikakos PI, Persaud Y, Janakiraman M, Kong X, Ng C, Moriceau G, Shi H, Atefi M, Titz B, Gabay MT, et al. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E) Nature. 2011;480:387–390. doi: 10.1038/nature10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.