Abstract
We report the development of simple fluorogenic probes that report on the activity of both bacterial and mammalian uracil–DNA glycosylase (UDG) enzymes. The probes are built from short, modified single-stranded oligonucleotides containing natural and unnatural bases. The combination of multiple fluorescent pyrene and/or quinacridone nucleobases yields fluorescence at 480 and 540 nm (excitation 340 nm), with large Stokes shifts of 140–200 nm, considerably greater than previous probes. They are strongly quenched by uracil bases incorporated into the sequence, and they yield light-up signals of up to 40-fold, or ratiometric signals with ratio changes of 82-fold, on enzymatic removal of these quenching uracils. We find that the probes are efficient reporters of bacterial UDG, human UNG2, and human SMUG1 enzymes in vitro, yielding complete signals in minutes. Further experiments establish that a probe can be used to image UDG activity by laser confocal microscopy in bacterial cells and in a human cell line, and that signals from a probe signalling UDG activity in human cells can be quantified by flow cytometry. Such probes may prove generally useful both in basic studies of these enzymes and in biomedical applications as well.
INTRODUCTION
Uracil–DNA glycosylase (UDG) enzymes serve important roles in two major cellular functions: the repair of hydrolytic damage to nucleobases and the addition of genetic diversity in the immune system (1–3) In the former role, the enzymes remove uracil that results from spontaneous hydrolytic deamination of cytosine. This is a critical function that prevents dangerous mutagenesis arising from U:G mispairs in genomic DNA. This deamination is a large potential source of errors that can lead to diseases, such as cancer; in this regard, several hundred such deamination events occur in DNA per day in each cell (4). In addition, two processes, deamination of the deoxynucleoside triphosphate dCTP and phosphorylation of dUDP, result in incorporation by polymerases of uracil (via dUTP intermediate) into DNA, forming U–A pairs (3,5). In a second major role, UDG activities act to increase sequence diversity in IgG-encoding DNAs. In this biological pathway, the enzyme AID acts to actively deaminate cytosines, resulting in mutagenic U–G base pairs. The resulting mutations add to the genetic diversity of the variable regions of immunoglobulins (6).
Although eubacteria, such as Escherichia coli, express two UDG enzymes, (7) mammalian cells possess several enzymes that remove uracil from DNA, some with overlapping activity (3) Although other known UDG enzymes exist [including TDG (8), MBD4 (9) and the mitochondrial UNG1 (10)], it is generally believed that the major uracil DNA glycosylase enzymes involved in nuclear damage repair are UNG2 (11,12) and SMUG1 (10,13) Both can cleave uracil from single-stranded or duplex DNA (12,13) Overall, because there are several UDG enzymes and they have overlapping activity, the specific cellular roles of the different enzymes remain under investigation (3).
Detection of UDG activity is of interest for multiple reasons. First, it is a useful tool in the basic scientific study of UNG activities in the overlapping enzymes. Second, it is a possible diagnostic and aids in biomedical study: defects in UNG activity have been linked to lymphoma and likelihood of contracting infections (3,14). Finally, as UNG2 is known to be packaged in the HIV-1 viral particle and affects its replication, the enzyme has been proposed as a therapeutic target for this and other viruses (15,16). UDG enzymes have been studied for some time, and their activities are generally measured by gel-based assays (17). In contrast to such classical methods, fluorogenic methods confer the advantage of simplicity and offer the possibility of real-time reporting. In this light, more recent detection methods have been designed around synthetic DNAs containing uracil bases (18–21). However, these approaches have been limited by the fact that they give low to moderate signal over background; in addition, most yield indirect fluorescence reporting and have little or no cellular use. One report of a beacon-type 39-nt probe design required removal of up to eight uracils followed by strand cleavage for signal; signals for UNG activity were reported in fixed cells (18). However, in such a design, nucleases can yield high-background signal by cleavage of the natural DNA structure.
Our new approach to detection of uracil excision uses small single-stranded modified DNAs containing fluorescent DNA base replacements as base excision repair enzyme sensors (22). To induce a change in fluorescence, we made use of the fact that thymine, and by extension, uracil, can efficiently quench certain fluorophores by a photoinduced charge-transfer mechanism (23,33). This results in a dark initial state for the unreacted probe, which then yields a direct and robust light-up signal when uracil is released by enzymatic glycosylase activity.
Early studies in our laboratory showed that a short DNA probe containing a single unnatural pyrene nucleotide could report on bacterial UDG enzymatic activity in vitro and in bacterial cells (22). However, the pyrene emission maximum is 381 nm, and as a result, the signal is limited to short wavelengths in the long ultraviolet (UV) and far-violet part of the visible spectrum, rendering it difficult to image by microscopy or detect by fluorescence in a cellular setting where autofluorescence can interfere. Furthermore, the probe design was not tested with eukaryotic enzymes, which may or may not tolerate a large unnatural nucleotide in or near the active site. Finally, it was not known whether such synthetic DNA compounds would be sensitive enough and sufficiently stable to report on intracellular UNG activity in mammalian cells.
Here, we describe new experiments aimed at shifting the fluorogenic signal in such probes to longer, more practically useful wavelengths, and we evaluate their activity with eukaryotic UDG enzymes in vitro and in cell culture. We find that the use of multiple fluorophores incorporated into such sensor designs can result in robust signals shifted to longer wavelengths in the visible region of the spectrum, rendering the probes easily visualized and imaged. We document the use of two forms of energy transfer in the new multichromophore probe designs, yielding both light-up probes and colour-change (ratiometric) responses. Further, we show that probes can be used readily to detect activity in mammalian UNG2 and SMUG1 enzymes in vitro. Finally, we also find that these new probes can be used to detect UDG activity directly in bacterial and mammalian cells in real time as repair occurs.
MATERIALS AND METHODS
Monomer synthesis
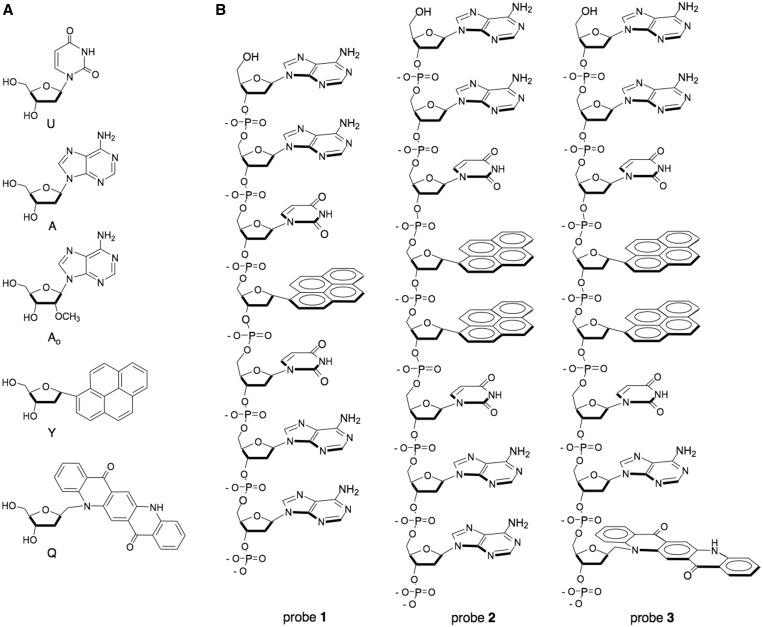
Synthesis of the fluorescent pyrene deoxyriboside Y (Figure 1) was carried out as previously reported (24). Synthesis of the quinacridone monomer Q is described in the Supplementary Data. The nucleosides were converted to the 5′-dimethoxytrityl-3′-phosphoramidite derivatives as described. The other natural and unnatural phosphoramidites [A (deoxyadenosine), Ao (2′-O-methyladenosine), U (deoxyuridine) and S (a tetrahydrofuran abasic spacer)] were purchased from Glen Research.
Figure 1.
(a) Structures of natural and unnatural nucleosides incorporated in probes; (b) structures of probes 1–3. Probes 1 and 2 are fluorogenic designs, whereas probe 3 yields a ratiometric signal.
Probe synthesis and purification
Probes 1–7 and reference 8 and 9 were synthesized on an Applied Biosystems 394 DNA/RNA synthesizer, using 3′-phosphate CPG column (Glen Research) on 1 mmol scale with DMT-off method using standard 3′–5′ cyanoethyl phosphoramidite chemistry. Coupling of each monomer was done with extended coupling time (999 s). Oligomers were de-protected by saturated NH4OH for 17 h at 55°C. Purification was carried out on a Shimadzu Series high-performance liquid chromatography (HPLC) with an Alltech C5 column with acetonitrile and TEAA buffer (100 mM, pH 7.4) as eluents. The identities were confirmed by MALDI-TOF mass spectrometry (Table S1).
Probe optical characterization
Absorption spectra were obtained on a Cary 100 Bio UV–Vis spectrometer (Figure S1). The concentrations of probes and references were determined by pyrene UV absorbance at 342 nm (e = 47000 cm−1 M−1). Fluorescence emission spectra were obtained on a Jobin Yvon-Spex Fluorolog 3 spectrometer with 340-nm excitation equipped with a thermostat accurate to 0.1°C. Quinine sulphate was used as a fluorescence quantum yield standard. Quantum yields were calculated according to the equation
where Φ is the quantum yield for photoemission, A is the magnitude of the absorbance at the excitation wavelength, E is the integrated emission intensity, I is the intensity of the excitation wavelength and n is the refractive index of the solvent; the subscripts R and S indicate values for the reference and sample, respectively. Quenching efficiency was determined by dividing the fluorescence intensity of each probe by the fluorescence intensity of the attributed reference compound, multiplying the result by 100 and then subtracting the result from 100.
In vitro fluorescence enzyme assays
The probes were dissolved in water to prepare a stock solution of 10 mM. The probes (concentration: 400 nM) were incubated at 37°C for UDG assay in 1 × UDG reaction buffer (20 mM Tris–HCl, 1 mM ethylenediaminetetraacetic acid and 1 mM dithiothreitol). The probes (concentration: 4 nM) were incubated at 37°C for human UNG2 and human SMUG1 assays in 1 × New England Biolabs (NEB) reaction buffer 1 (10 mM Bis–Tris–propane–HCl, 1 mM MgCl2 and 1 mM dithiothreitol). When the fluorescence intensity was stable, the UDG (New England Biolabs), human UNG2 (Origene) or human SMUG1 (Origene) was added to the solution. After mixing rapidly, the fluorescence spectra or the time courses of the fluorescence intensity of solutions were recorded.
Serum stability assay
The probes were incubated at 37.0°C with a mixture of human serum (Sigma Aldrich) diluted in 1 × phosphate-buffered saline (PBS) to 10%, and the fluorescence spectra of solutions were recorded at various time points.
Fluorescence confocal microscopy
A-253 cells (ATCC) were grown in McCoy’s 5a (modified) medium supplemented with 10% fetal bovine serum, in a humidified atmosphere at 37°C with 5% CO2. Cells were plated in a chambered cover glass and allowed to reach 60% confluency. The growth medium was removed, and 1 × PBS containing 7 μM probe and Lipofectamine 2000 (Invitrogen) (ratio of probe and Lipofectamine 2000 is 1 mg and 1.2 ml), was added to the cells and incubated for 2 h at 37°C with 5% CO2. Cells were rinsed twice with PBS and visualized directly in a chambered cover glass using a Leica SP5 multiphoton/confocal laser scanning microscope viewed through a ×63 objective. A 710-nm laser was used to excite the probes. Optical sections through the z-dimension were acquired in step sizes of ∼0.335 µm. Image acquisition was performed at the Cell Sciences Imaging Facility of Beckman Center for Molecular and Genetic Medicine, Stanford University School of Medicine.
Bacterial experiments
An E. coli strain lacking the UDG enzyme (BW310DE3) was transformed with pET28a-afuung plasmid using standard procedures (25). Bacteria and plasmid were kindly provided by S. S. David (University of California-Davis). The starter culture was diluted 1:100 with fresh LB–kan medium and was grown at 37°C until OD600 of the culture reached 0.6. Isopropyl-1-thio-β-galactopyranoside (IPTG) was then added to a final concentration of 1 mM, and the culture was incubated for an additional 5 h at 30°C. Aliquots (10–20 ml) were centrifuged, and the supernatant was decanted. Pellets were washed with water and resuspended in 1 × ThermoPol II (Mg-free) reaction buffer [20 mM Tris–HCl, 10 mM (NH4)2SO4, 10 mM KCl and 0.1% Triton X-100] (+IPTG bacteria). The cells without addition of IPTG were also harvested and resuspended in 1 × ThermoPol II (Mg-free) reaction buffer (−IPTG bacteria). The bacterial solutions were used as a stock solution. Probes were added [final concentration: (probe) = 2 μM, (bacteria, OD600) = 0.31], and the samples were incubated at 65°C without shaking and protected from light. Aliquots of incubated bacteria were spotted on cover slides without washing or fixation. Samples were imaged under a Nikon Eclipse 80i epifluorescence microscope equipped with × 100 objective. Images were captured with a QIclick digital CCD camera and QCapture Pro 7 Imaging software with excitation 330–380 nm and emission >420 nm.
Flow cytometry
HeLa cells were grown in Dulbecco’s Modified Eagle’s Medium supplemented with 10% fetal bovine serum, 100 U/ml of penicillin and 100 μg/ml of streptomycin in a humidified incubator at 37°C with 5% CO2. Cells were seeded in a six-well plate at a density of ∼0.5 × 106 cells/well at least 24 h before transfection and grown to 80% confluency. In all, 1.2 ml of Lipofectamine 2000 Reagent per 1 mg of probe in OptiMEM was used. Lipofectamine was mixed with half of the volume of transfection media and incubated at room temperature for 15 min. Probes were mixed with the other half of the transfection media to a final concentration of 2 μM probe. The Lipofectamine and probe mixtures were combined and incubated 15 min at room temperature. The cells were washed three times with PBS to remove antibiotics and serum, and the Lipofectamine-probe mixture was added. The cells were incubated 6 h at 37°C. After washing once with PBS, the cells were collected by trypsinization at room temperature. The cells were resuspended in PBS and analysed by flow cytometry at the Stanford Shared FACS Facility on a BD LSR II flow cytometer. Fluorescence was measured with laser excitation at 355 nm, using a 525/50 bandpass emission filter. Ten thousand events/samples were recorded.
RESULTS
Testing new multichromophore designs with bacterial UDG
Our previous experiments in quenched probe design resulted in the development of probe 1 (Figure 1), which was shown to yield positive signals with emission maximum at 381 nm in the presence of bacterial UDG (24). Although the light-up response of this probe was excellent (Figure 2), its short wavelength made it difficult to image by microscopy. Our initial goal for the current experiments was to design new probes that would yield fluorescence clearly in the visible range. To address this, we tested two new probe designs, probes 2 and 3 (Figure 1), starting with the initial architecture of probe 1, which has uracil residues arranged on either side of a pyrene monomer for efficient quenching, and which retains non-quenching adenines on both ends to enhance enzymatic recognition. To shift emission into the visible range, we adopted two strategies: first, we added an additional pyrene residue adjacent to the first one; such an arrangement is expected to yield efficient pyrene excimer fluorescence, which typically has an emission maximum at ∼490 nm, and which has a usefully large Stokes shift of ∼150 nm (26). This yielded the design of probe 2. Our second strategy was to replace a terminal adenine with a yellow–orange quinacridone monomer (Q) (probe 3); our hope was that this would yield energy transfer between pyrene and quinacridone chromophores, possibly resulting in yet further bathochromic shifts in emission. Probe 4, containing two 2′-O-methyladenosine residues at the end phosphodiester linkages, was a variant of pyrene excimer probe 2, prepared as a strategy for enhancing nuclease resistance (see later in the text). As controls for the aforementioned, we prepared probes 5–7, which are identical to 1–3 but with thymines replacing uracil nucleobases. As thymine is not recognized by UNG enzymes (1–3), such probes are expected to remain dark in the presence of UNG activity.
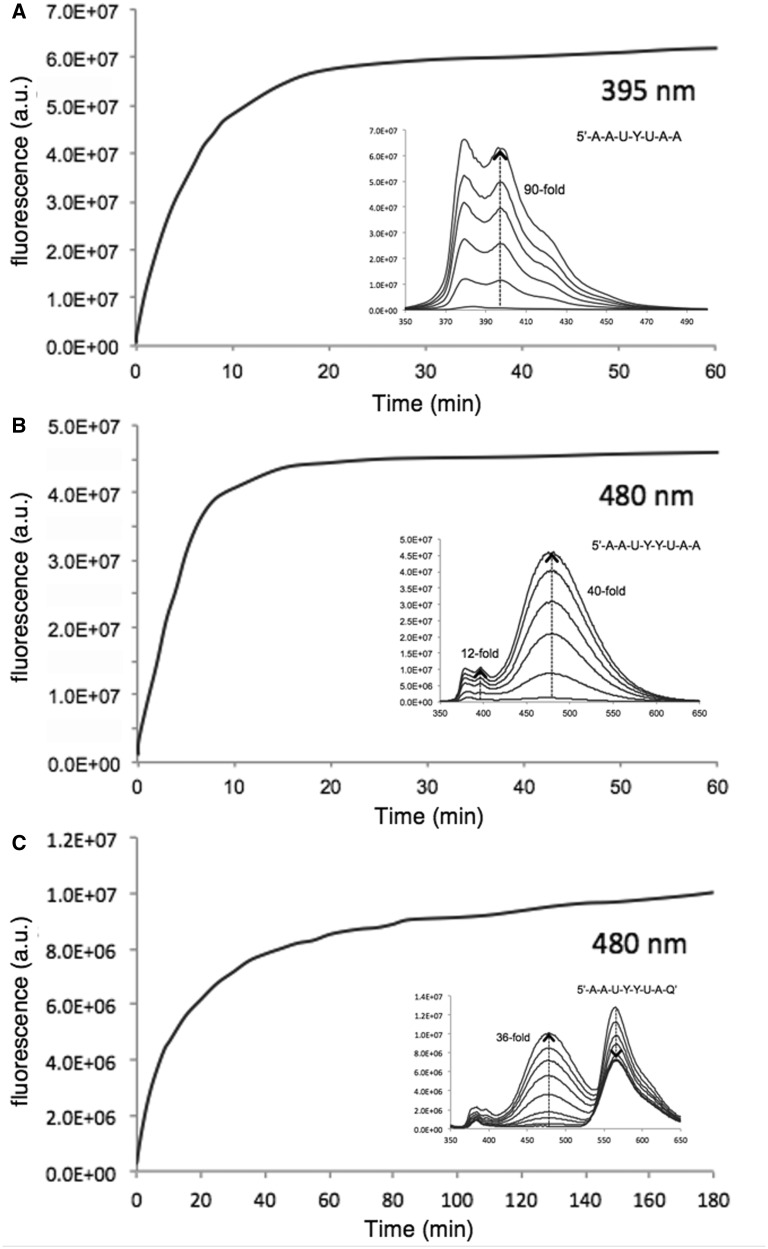
Figure 2.
In vitro enzymatic responses of probes 1 (A), 2 (B), and 3 (C) with E. coli UDG. Time courses of fluorescence response are shown at the indicated wavelengths; insets show full spectral changes over the same time course. Conditions: 400 nM probe, UDG 1 U/ml (0.4 nM), 37°C, excitation 340 nm.
Probes were prepared on an automated DNA synthesizer, purified by HPLC and characterized by mass spectrometry, by absorption spectra (Supplementary Data) and by residual emission properties (Table 1). The new unreacted probes showed excellent quenching with very low-quantum yields, approaching the values seen previously for probe 1.
Table 1.
Sequences and optical data for quenched probes
| Compound | Probe sequence | Quantum yielda | Quenching efficiencya (%) |
|---|---|---|---|
| Probe 1 | 5′-AAUYUAA | 0.004 | 98.7 |
| Probe 2 | 5′-AAUYYUAA | 0.006 | 97.7 |
| Probe 3 | 5′-AAUYYUAQ | 0.035 | 86.0 |
| Probe 4 | 5′-AoAUYYUAoAb | 0.006 | 97.6 |
| Probe 5 | 5′-AATYTAA | 0.006 | 98.0 |
| Probe 6 | 5′-AATYYTAA | 0.011 | 95.6 |
| Probe 7 | 5′-AoATYYTAoA | 0.003 | 98.8 |
| Reference 8 | 5′-YSc | 0.310 | – |
| Reference 9 | 5′-YYS | 0.250 | – |
aValues for the unreacted probes.
bMonomer ‘Ao’ is 2′-O-methyladenosine, incorporated to increase nuclease resistance.
cMonomer ‘S’ is a tetrahydrofuran (abasic) spacer, added to enhance solubility.
As an initial test of enzyme substrate capability, fluorescence response and emission wavelength, we first tested probes 1–3 with E. coli UDG enzyme in vitro (Figure 2 and Supplementary Figure S2). We found that all three probes were good enzyme substrates. Interestingly, probe 2 (despite its two unnatural pyrene monomers) was even more efficient as a substrate for the bacterial enzyme than the previous probe 1, yielding 90% reaction in only ∼10 min as compared with 20 min for 1. Probe 3, with its three unnatural fluorophore monomers, was found to be slowed in its reaction somewhat (90% reaction in ∼80 min) albeit not drastically. Spectral changes were followed over these time courses (Figure 2, see insets), and they revealed that the new probe 2 yielded almost exclusively a long-wavelength band characteristic of excimer fluorescence, with a robust 40-fold light-up signal. Interestingly, probe 3 showed two clear bands of emission (480 and 565 nm); initially, the 565-nm band was dominant, but over the time course of reaction, this band decreased in intensity and the 480-nm band strongly increased. Ratios of peak intensity (565:480) shifted markedly from 59 to 0.72, yielding an 82-fold change over the 3-h time course. Images of probes in cuvettes before and after reaction (Supplementary Figure S2) confirmed that the two new probes were indeed readily imaged at visible wavelengths; probe 2 yielded a strong cyan colour after UDG reaction, whereas probe 3 shifted from yellow–orange to whitish in hue, consistent with its initial single strong band shifting to dual 480/565-nm bands at the end of the reaction. Overall, the results confirmed that the new multifluorophore probes are able to yield signals well into the visible spectrum, with large Stokes shifts of 140–220 nm.
Activity with human UNG2 and SMUG1
Our second major aim was to investigate whether the new fluorogenic probes could act as substrates for mammalian UDG enzymes. To our knowledge, no fluorescent probes have been studied with either UNG2 or SMUG1 enzymes. To test this, we reacted probes 2 and 4 (along with thymine control 6) with human SMUG1 and UNG2 enzymes in vitro, respectively. Time courses and spectral changes of the fluorescence responses are shown in Figures 3 and 4. We found that the commercial enzyme preparation of SMUG1 yielded reaction that proceeded relatively slowly (reaching completion after ∼10 h), whereas UNG2 generated signals rapidly, yielding the full fluorescence signal in only ∼30 min. Probe 2 displayed the excimer band at 480 nm for both enzymes, but interestingly, it also showed significant amounts of pyrene monomer emission (380 nm) as well. Importantly, the thymine-containing control (probe 6) yielded essentially no signal for either enzyme. Probe 4, with 2′-O-methyladenosine residues, yielded slower reactions than the probe with unmodified sugars, and in both cases, the fluorescence appeared to plateau somewhat below the intensity reached with probe 2, suggesting that the modified sugars in probe 4 may inhibit complete removal of the quenching uracil residues. Nevertheless, responses were relatively strong and far above the control in these latter cases. Spectral changes for the two enzymes with probe 4 are given in Supplementary Figure S3.
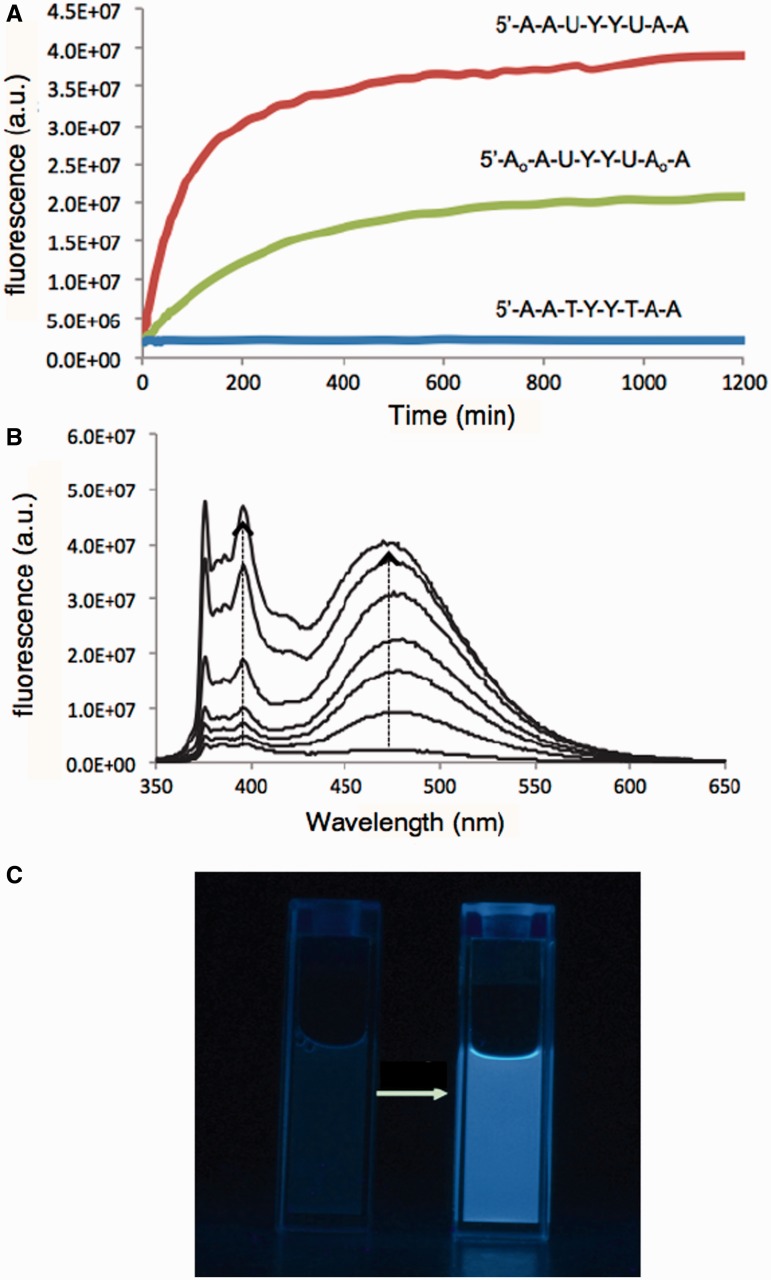
Figure 3.
In vitro responses of probes 2 and 4 to hSMUG1, along with thymine-containing control probe 6. (A) Time courses of response at 480 nm. (B) Spectral changes in response of probe 2. Conditions: [hSMUG1] = 500 U/ml (400 nM), [probe] = 4 μM, 37°C, excitation 340 nm. (C) Photograph showing visible change (illumination by 365 nm UV lamp). For photograph, [hSMUG1] = 500 U/ml (400 nM), [probe 2] = 4 μM.
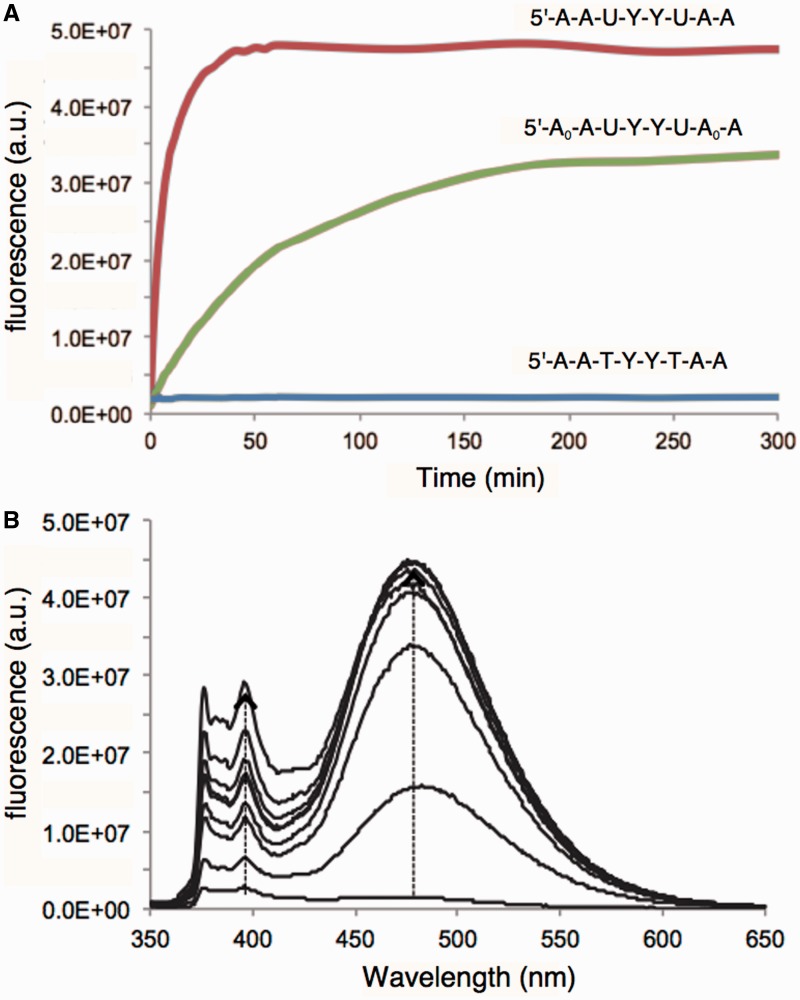
Figure 4.
In vitro responses of probes 2, 4 and control 6 to human UNG2 enzyme. (A) Time courses of response of probes at 480 nm. (B) Spectral changes in response of probe 2 over the same time course. Excitation = 340 nm. Condition: [hUNG2] = 9.7 mg/ml (90 nM); [probe] = 4 μM; 37°C, excitation 340 nm.
To further investigate the mechanism of signal generation by probes of this class, we evaluated the products of probe 2 and nuclease-resistant probe 4, both containing two uracils and two pyrenes, in reactions with the enzyme SMUG1, by MALDI-mass spectrometry. After extended incubation to ensure final products were achieved, we found for probe 2 peaks corresponding to loss of one or both uracils, as designed. For probe 4, we found mass peaks corresponding both to loss of a uracil and resulting chain cleavage (Supplementary Figure S8). Approximately equal intensities were found for both sites of cleavage, suggesting no strong preference for removal of one or other of the two uracils. The analysis is consistent with the observation that for this probe, signal reached approximately half the intensity of that from probe 2 (Figure 3).
To further characterize the enzyme substrate properties of the most efficient long-wavelength probe (probe 2) with the bacterial and human enzymes, we carried out studies at varied substrate concentrations and time points to obtain Michaelis–Menten parameters for this fluorogenic substrate (Table 2 and Supplementary Figure S7). Note that the kinetic behaviour is potentially complicated by the fact that two uracils must be removed before the main signals are generated. Nevertheless, reasonable kinetic fits were achieved, providing estimates for kinetic parameters. The results showed that despite the presence of two large and strongly stacking pyrene ‘bases’, probe 2 has an apparent Km with the bacterial enzyme that is essentially identical to that of the previously studied probe 1, and close to the literature value with natural single-stranded DNA (21). The Km(app) values of probe 2 with SMUG1 and UNG2 are 0.82 and 4.67 mM−1, respectively, which are also in good agreement with the literature values with natural single-stranded DNA: the published Km of SMUG1 with DNA ranges from 1.7 to 2.3 mM, and for UNG2 from 0.21 to 13.8 mM−1 (5). It should be noted, however, that buffer conditions used here were not identical to those used previously.
Table 2.
In vitro enzyme substrate parameters of selected probes with bacterial and human uracil–DNA glycosylases
| Enzyme | Substrate | Km(app) (μM) | kcat(app) (min−1) | kcat/Km (min−1 μM−1) |
|---|---|---|---|---|
| E. coli UDG | 5′-AAUYUAA | 0.21 | 2040 | 9710 |
| E. coli UDG | 5′-AAUYYUAA | 0.30 | 882 | 2940 |
| hSMUG1 | 5′-AAUYYUAA | 0.82 | 35 | 43 |
| hUNG2 | 5′-AAUYYUAA | 4.67 | 4.6 | 0.99 |
One possible application of fluorogenic enzyme substrates in general is in the study and screening of enzyme inhibitors. To test this in a preliminary way, we carried out enzyme reactions of probe 2 with UNG2 enzyme in the presence of varying concentrations of 5-fluorouracil (5FU), a known weak inhibitor of UNG glycosylases (27). We found that increasing concentrations of 5FU in the low millimolar range resulted in a dose-dependent drop in probe response (Supplementary Figure S4), with half diminishment in signal occurring at ∼2 mM 5FU. This is consistent with the published value for this inhibitor with UDG (19,27).
Nuclease susceptibility experiments
One of our goals for this work was the use of fluorogenic probes for measuring and imaging UDG activity directly in cells. As cells contain multiple nuclease enzymes, and our probes are built from DNA, it seemed possible that they might be degraded to some extent in the cellular context. Although they contain 2–3 unnatural nucleobases, which might inhibit such nuclease activity, they also contain natural nucleotide monomers as well, and if the phosphodiester bond between uracil and pyrene nucleotides were cleaved, a fluorogenic signal would likely result. Thus, we tested this issue explicitly with two sources of nuclease: human serum and T4 DNA polymerase, which contain strong 3′ and 5′ exonuclease activities (28). We investigated the nuclease susceptibility of probe 2 and of the further-modified probe 4, which was designed to block exonuclease activity by virtue of its 2′-O-methyl groups situated at the end phosphodiester linkages. Spectral changes of the fluorescence responses against 10% human serum are shown in Supplementary Figure S5 and against T4 DNA polymerase are shown in Supplementary Figure S6. Little or no fluorescence changes were observed for probe 2 and probe 4 > 6 h in 10% human serum, indicating that the probes are relatively stable even without end protection. In the stronger nuclease environment of a purified T4 DNA polymerase preparation, probe 2 showed substantial signal increase >3 h, indicating partial degradation, whereas probe 4 showed a much smaller increase, indicating that the 2′-O-methyl groups at the terminal phosphate linkages are sufficient to largely (although not completely) block exonuclease degradation.
As a further test of the effect of the 2′-O-methyl modifications on the stabilization of probes (e.g. 4 and 7) against nucleases, we tested thymine-containing probe 6 and its 2′-O-methyl-modified version (probe 7) directly in cell lysate, measuring fluorescence increases over time by spectrofluorimeter as pyrenes are separated from thymines by backbone cleavage. The data show clearly (Supplementary Figure S9) that the 2′-O-methyl groups markedly slow nuclease degradation, reducing the 460-nm excimer signal increase from 5.3-fold for the unprotected probe to 1.6-fold for the protected one after 2 h.
Cellular imaging studies
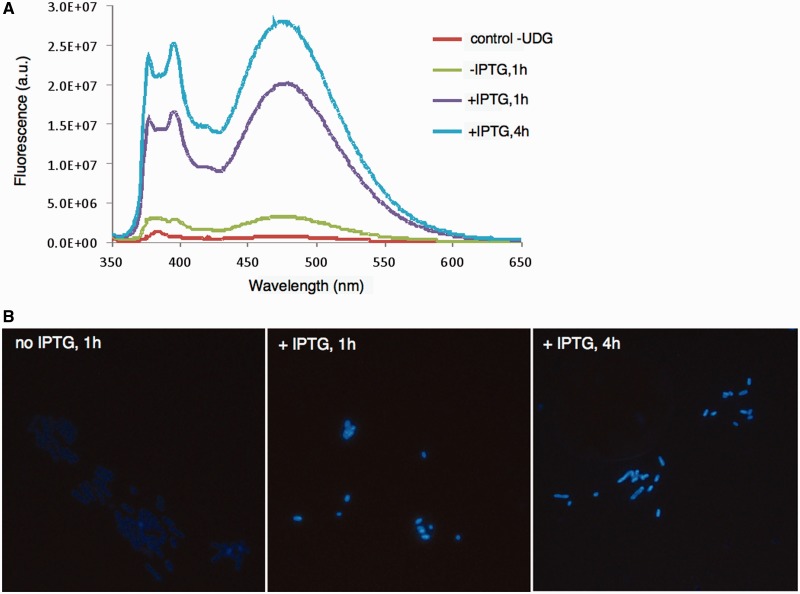
The fact that the new probes emit fluorescence at visible wavelengths and with large Stokes shifts suggested the possibility of their use in detection of UDG activities directly associated with cells. We first tested this possibility with E. coli cells expressing bacterial UDG enzyme from an inducible plasmid; the enzyme was that from Archaeoglobus fulgidus, which is stable at 65°C (29). The E. coli strain used was a knockout strain lacking endogenous UDG. First, we measured the fluorescence of bacterial cell suspensions in the presence of probe 2 (Figure 5A). Results showed that the bacteria produced little signal without the expression plasmid; however, with this plasmid, the 480-nm band characteristic of probe 2 was seen, and with IPTG induction this signal increased substantially.
Figure 5.
Fluorescence detection and imaging of UDG activity associated with bacterial cells using probe 2. A UDG knockout strain of E. coli [BW(310)DE] was used as control; it was transfected with the AfuUDG gene, and fluorescence was observed for bacterial suspensions in solution (A) and by epifluorescence microscopy (excitation 340–380 nm, emission >400 nm) (B). Enhanced signals are seen with induction of gene expression by IPTG and at longer incubation times with the probe.
We then imaged the bacteria directly by epifluorescence microscopy (Figure 5B). The bacteria lacking UDG enzymes were too dark to be imaged (data not shown); however, bacteria expressing AfuUDG were clearly visible with cyan colour, and further incubation with IPTG induction of gene expression yielded robust cyan staining of the bacteria.
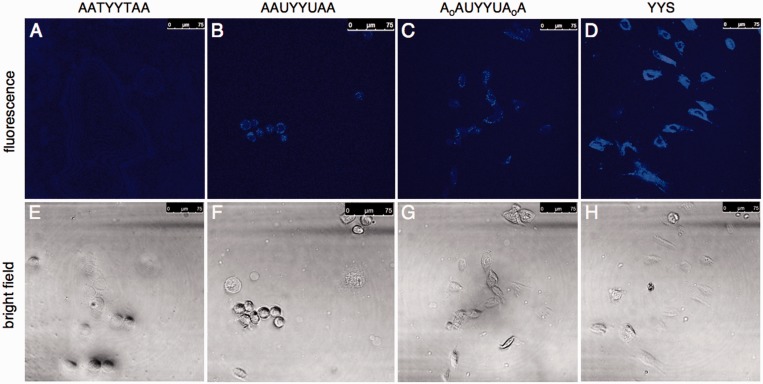
Next, we proceeded to investigate the possibility of using probes 2 and/or 4 in human cells. We used a commercial cationic lipid transfection reagent to deliver probes into A253 cells by incubating for 2 h at 2 mM probe concentration (37°C), and then imaged the cells by confocal microscopy with two-photon excitation at 710 nm. The results are shown in Figure 6. The control thymine-containing probe (6) yielded little signal, whereas after 2-h incubation, both probes 2 and 4 yielded visible cyan signals. The signals were well above the fluorescence of the control, although they were less intense than the fluorescence of a positive control dye (YYS, a trimer oligonucleotide containing two pyrenes), which suggests either incomplete reaction of probes 2 and 4 under these conditions, or less efficient intracellular delivery of the longer 2 and 4 probes as compared with the short positive control. Signals were mainly observed outside the nucleus; it is reported that SMUG1 is localized both to the cytoplasm and nucleus, UNG1 is primarily mitochondrial and UNG2 primarily nuclear (5,10,30). Thus, we hypothesize that the observed signals arise mainly from SMUG1/UNG1 activity, and lack of nuclear signals likely reflect the localization of the probe delivered by this cationic lipid. Intracellular localization of short DNA–polyfluorophore oligomers similar to those here has been observed mainly in the cytoplasm (31). In the current cellular images, there was no obvious difference between probes 2 and 4, which suggests that the benefit of added nuclease resistance in 4 (if there is any in this context) may be offset by its poorer enzyme substrate capabilities. Overall, the results confirm that such probes can be used to image UDG enzymatic activity directly associated with a human cell line.
Figure 6.
Imaging UDG activity in A253 cells by confocal fluorescence microscopy. (B and C) Images of cells treated with probes 2 and 4 (sequences shown); (A) results with thymine-containing control probe 6, and (D) positive control (fully fluorescent probe). Bright field images (E–H) of the same cells are shown below each corresponding fluorescence image. Cells were treated with 2 μM probe and with Lipofectamine as a transfection reagent for 2 h before imaging. Two-photon excitation at 710 nm.
Quantifying activity by flow cytometry
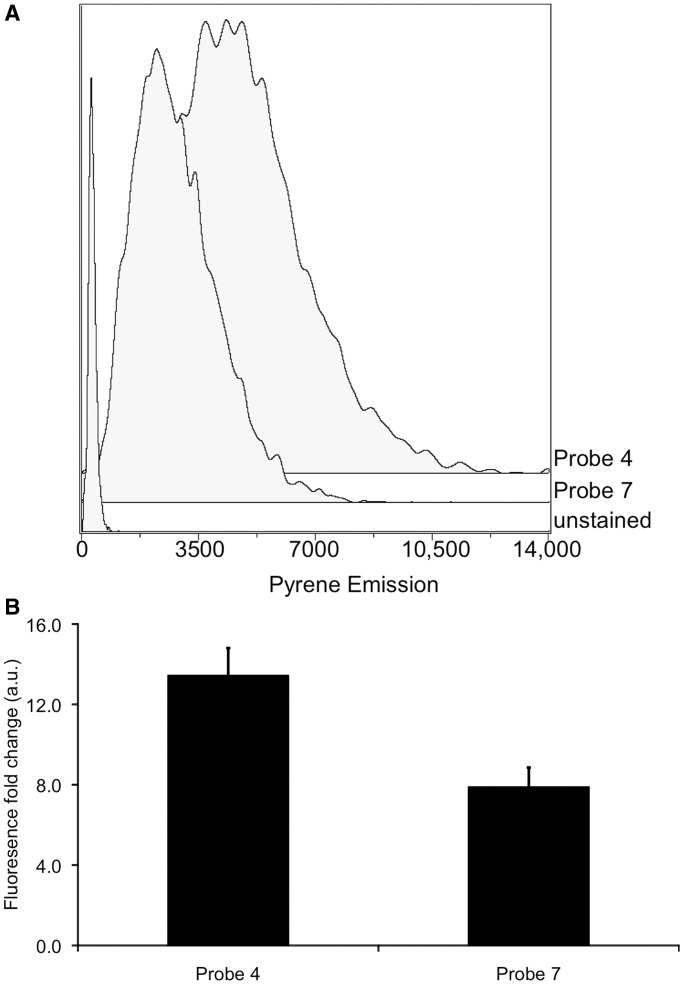
Given our microscopic observation of signals from fluorogenic probes at visible wavelengths, we proceeded to attempt to quantify the signals by flow cytometry. The 2′-O-methyl-protected uracil-containing probe 4 and a thymine-containing 2′-O-methyl-protected control (probe 7) were transfected into HeLa cells with a commercial cationic lipid reagent. After 6 h of incubation post-transfection, we monitored signals by laser excitation at 355 nm, following emission with a 525/50 bandpass filter. The data revealed a 13-fold increase in pyrene excimer emission over untreated cells with probe 4 and an 8-fold increase with probe 7, thus indicating a 64% increase in signal because of UNG repair activity (Figure 7). The increase in signal from the control probe indicates that intracellular nucleases still target the probe despite the 2′-O-methyl modifications, leading to high background. However, probe 4 still yields uracil repair activity signals above the background.
Figure 7.
Analysis by flow cytometry of signals from cells transfected with probes 4 and 7, measured 6 h post-transfection. (A) Representative histograms of pyrene emission intensity of cells transfected with no probe, probe 4 and control probe 7. (B) Averaged fold changes in fluorescence in cells transfected with probes 4 and 7 relative to cells without a probe. Error bars show standard deviations (n=3).
DISCUSSION
Our experiments show that the new probes successfully report on UNG repair activity in vitro, and that they function well with bacterial and human enzymes. Quantitative function with all the enzymes is excellent (Table 2), with Km values that are similar to that of native DNA despite the presence of multiple large unnatural nucleobases in the probes. This high activity could make such probes useful in assays of enzyme activity for basic studies of enzyme function, and for screens of activity in enzyme mutants. Moreover, we have shown that such a probe can report on small-molecule inhibition of UNG2; thus, the probes could be useful in future screens for new inhibitors of UNG enzymes.
Compared with previous fluorescent probes of UDG activity, the current probes are smaller and simpler, and they yield considerably higher signal over background. Previous reports have described quencher-conjugated double-stranded DNA constructs 28–39 nt in size, yielding a relatively small 4- to 8-fold enhancement in signal with enzymatic repair (18–20). Their signalling was indirect, as the signal enhancement required further degradation and unwinding of the DNA to separate the quencher. By contrast, the current probes directly report on repair activity by making the removal of the damaged base the mechanism for signal generation. Another earlier approach used smaller single-stranded constructs containing the fluorescent base 2-aminopurine (2AP) (21); those probes showed relatively small 3- to 8-fold enhancements with bacterial UDG repair. Moreover, 2AP emits fluorescence in DNA with low efficiency and has a short emission wavelength in the UV region (10). In marked contrast to this, the current probes show 40- to 82-fold changes in signal. Relative to our previous single-fluorophore probes, the new molecules yield signals well into the visible region, at 480–565 nm. This red-shifted emission, which results from energy transfer between the two or three dyes in the probes, can allow the experimenter to avoid much of the background from cellular autofluorescence, and the large 140–220 nm Stokes shifts separate the signal far from interference by the excitation light. Probe 3, in particular, may have special use in quantifying signals, as it produces a ratiometric output. Future studies measuring signals at both 480/565 nm wavelengths will be needed to explore this possibility.
To our knowledge, these are the first fluorogenic probes to have been validated specifically with human UNG2 or SMUG1 repair enzymes in vitro. Moreover, we show positive signals over background in a human cell line both by confocal microscopy and by flow cytometry. One earlier report describes testing double-stranded probes in mammalian cell extracts (19), but no controls with identical thymine-containing DNAs were performed to rule out simple degradation by cellular nucleases, or unwinding by helicases and other DNA-binding proteins, as the source of signal. Similarly, a beacon-type 39mer probe was reported to yield signals in fixed cells (18), but background signal from cleavage of the probes by nucleases or unwinding by DNA-binding proteins is a concern. Our imaging and flow cytometry experiments show positive signal over background compared with thymine-containing controls, and the presence of unnatural bases and 2′-O-methyl groups inhibit nuclease activity. In addition, as the probes are single-stranded, no interference from helix unwinding can occur.
Our preliminary cellular experiments suggest the possible use of such probes in the study of cellular UNG activities in varied cell types and tissues. Moreover, one can envision the use of this molecular strategy (quenching of fluorescence by a damaged or altered base) in the detection of other damage repair pathways as well. In the present case, more studies will be needed to investigate which cellular UNG enzyme or enzymes are responsible for generating the signal; in addition, more tests of new nuclease-protected variants may be helpful in lowering the background signal further. Finally, as with most DNA constructs, exploring various methods of intracellular delivery is an issue worthy of further investigation; delivery of greater amounts of probes into cells may enable the possibility of obtaining higher signal over background, and in addition, control of intracellular location would be valuable in investigating the biological roles of the different human UNG enzymes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1, Supplementary Figures 1–9, Supplementary Scheme 1, Supplementary Methods, Supplementary Data and Supplementary Reference [32].
FUNDING
U.S. National Institutes of Health [GM067201 to E.T.K.]; Japan Society for the Promotion of Science (to T.O.); National Science Foundation (to S.K.E.). Funding for open access charge: Stanford University.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Lisa Engstrom and Prof. S. S. David (UC-Davis) for providing bacteria expressing the Afu-UDG enzyme.
REFERENCES
- 1.Pearl LH. Structure and function in the uracil-DNA glycosylase superfamily. Mutat. Res. 2000;460:165–181. doi: 10.1016/s0921-8777(00)00025-2. [DOI] [PubMed] [Google Scholar]
- 2.Krokan HE, Standal R, Slupphaug G. DNA glycosylases in the base excision repair of DNA. Biochem. J. 1999;325:1–16. doi: 10.1042/bj3250001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sousa MM, Krokan HE, Slupphaug G. DNA-uracil and human pathology. Mol. Aspects Med. 2007;28:276–306. doi: 10.1016/j.mam.2007.04.006. [DOI] [PubMed] [Google Scholar]
- 4.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 5.Kavli B, Sundheim O, Akbari M, Otterlei M, Nilsen H, Skorpen F, Aas PA, Hagen L, Krokan HE, Slupphaug G. hUNG2 is the major repair enzyme for removal of uracil from U:A matches, U:G mismatches, and U in single-stranded DNA, with hSMUG1 as a broad specificity backup. J. Biol. Chem. 2002;277:39926–39936. doi: 10.1074/jbc.M207107200. [DOI] [PubMed] [Google Scholar]
- 6.Muramatsu M, Sankaranand VS, Anant S, Sugai M, Kinoshita K, Davidson NO, Honjo T. Specific expression of activation-induced cytidine deaminase (AID), a novel member of the RNA-editing deaminase family in germinal center B cells. J. Biol. Chem. 1999;274:18470–18476. doi: 10.1074/jbc.274.26.18470. [DOI] [PubMed] [Google Scholar]
- 7.Gallinari P, Jiricny J. A new class of uracil-DNA glycosylases related to human thymine-DNA glycosylase. Nature. 1996;383:735–738. doi: 10.1038/383735a0. [DOI] [PubMed] [Google Scholar]
- 8.Neddermann P, Jiricny J. Efficient removal of uracil from G.U mispairs by the mismatch-specific thymine DNA glycosylase from hela cells. Proc. Natl Acad. Sci. USA. 1994;91:1642–1646. doi: 10.1073/pnas.91.5.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hendrich B, Hardeland U, Ng HH, Jiricny J, Bird A. The thymine glycosylase MBD4 can bind to the product of deamination at methylated cpg sites. Nature. 1999;401:301–304. doi: 10.1038/45843. [DOI] [PubMed] [Google Scholar]
- 10.Caradonna S, Ladner R, Hansbury M, Kosciuk M, Lynch F, Muller S. Affinity purification and comparative analysis of two distinct human uracil-dna glycosylases. Exp. Cell Res. 1996;222:345–359. doi: 10.1006/excr.1996.0044. [DOI] [PubMed] [Google Scholar]
- 11.Haushalter KA, Todd Stukenberg P, Kirschner MW, Verdine GL. Identification of a new uracil-dna glycosylase family by expression cloning using synthetic inhibitors. Curr. Biol. 1999;9:174–185. doi: 10.1016/s0960-9822(99)80087-6. [DOI] [PubMed] [Google Scholar]
- 12.Nilsen H, Haushalter KA, Robins P, Barnes DE, Verdine GL, Lindahl T. Excision of deaminated cytosine from the vertebrate genome: Role of the SMUG1 uracil-dna glycosylase. EMBO J. 2001;20:4278–4286. doi: 10.1093/emboj/20.15.4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krokan HE, Otterlei M, Nilsen H, Kavli B, Skorpen F, Andersen S, Skjelbred C, Akbari M, Aas PA, Slupphaug G. Properties and functions of human uracil-dna glycosylase from the UNG gene. Prog. Nucleic Acid Res. Mol. Biol. 2001;68:365–386. doi: 10.1016/s0079-6603(01)68112-1. [DOI] [PubMed] [Google Scholar]
- 14.Imai K, Slupphaug G, Lee WI, Revy P, Nonoyama S, Catalan N, Yel L, Forveille M, Kavli B, Krokan HE, et al. Human uracil-dna glycosylase deficiency associated with profoundly impaired immunoglobulin class-switch recombination. Nat. Immunol. 2003;4:1023–1028. doi: 10.1038/ni974. [DOI] [PubMed] [Google Scholar]
- 15.Willetts KE, Rey F, Agostini I, Navarro JM, Baudat Y, Vigne R, Sire J. DNA repair enzyme uracil DNA glycosylase is specifically incorporated into human immunodeficiency virus type 1 viral particles through a vpr-independent mechanism. J. Virol. 1999;73:1682–1688. doi: 10.1128/jvi.73.2.1682-1688.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anders CK, Winer EP, Ford JM, Dent R, Silver DP, Sledge GW, Carey LA. Poly(adp-ribose) polymerase inhibition: “Targeted” therapy for triple-negative breast cancer. Clin. Cancer Res. 2010;16:4702–4710. doi: 10.1158/1078-0432.CCR-10-0939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krokan H, Wittwer CU. Uracil dna-glycosylase from HeLa cells: General properties, substrate specificity and effect of uracil analogs. Nucleic Acids Res. 1981;9:2599–2613. doi: 10.1093/nar/9.11.2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maksimenko A, Ishchenko AA, Sanz G, Laval J, Elder RH, Saparbaev MK. A molecular beacon assay for measuring base excision repair activities. Biochem. Biophys. Res. Commun. 2004;319:240–246. doi: 10.1016/j.bbrc.2004.04.179. [DOI] [PubMed] [Google Scholar]
- 19.Liu B, Yang X, Wang K, Tan W, Li H, Tang H. Real-time monitoring of uracil removal by uracil-dna glycosylase using fluorescent resonance energy transfer probes. Anal. Biochem. 2007;366:237–243. doi: 10.1016/j.ab.2007.04.049. [DOI] [PubMed] [Google Scholar]
- 20.Kim Y, Hong IS. Detection of BER enzymes based on fluorescence resonance energy transfer. Bull. Korean Chem. Soc. 2009;30:2149–2151. [Google Scholar]
- 21.Drohat AC, Jagadeesh J, Ferguson E, Stivers JT. Role of electrophilic and general base catalysis in the mechanism of Escherichia coli uracil DNA glycosylase. Biochemistry. 1999;38:11866–11875. doi: 10.1021/bi9910878. [DOI] [PubMed] [Google Scholar]
- 22.Ono T, Wang S, Koo CK, Engstrom L, David SS, Kool ET. Direct fluorescence monitoring of DNA base excision repair. Angew. Chem. Int. Ed. 2012;51:1689–1692. doi: 10.1002/anie.201108135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Telser J, Cruickshank KA, Morrison LE, Netzel TL, Chan CK. DNA duplexes covalently labeled at two sites: Synthesis and characterization by steady-state and time-resolved optical spectroscopies. J. Am. Chem. Soc. 1989;111:7226–7232. [Google Scholar]
- 24.Moran S, Ren RX, Sheils CJ, Rumney S, Kool ET. Non-hydrogen bonding ‘terminator’ nucleosides increase the 3′-end homogeneity of enzymatic RNA and DNA synthesis. Nucleic Acids Res. 1996;24:2044–2052. doi: 10.1093/nar/24.11.2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sambrook J, Maniatis T. Molecular Cloning, A Laboratory Manual. 2nd edn. Cold Spring Harbor: Cold Spring Harbor Press; 1989. [Google Scholar]
- 26.Paris PL, Langenhan JM, Kool ET. Probing DNA sequences in solution with a monomer-excimer fluorescence color change. Nucleic Acids Res. 1998;26:3789–3793. doi: 10.1093/nar/26.16.3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shroyer MJ, Bennett SE, Putnam CD, Tainer JA, Mosbaugh DW. Mutation of an active site residue in Escherichia coli uracil-DNA glycosylase: Effect on DNA binding, uracil inhibition and catalysis. Biochemistry. 1999;38:4834–4845. doi: 10.1021/bi982986j. [DOI] [PubMed] [Google Scholar]
- 28.Huang WM, Lehman IR. On the exonuclease activity of phage T4 deoxyribonucleic acid polymerase. J. Biol. Chem. 1972;247:3139–3146. [PubMed] [Google Scholar]
- 29.Sandigursky M, Franklin WA. Uracil-DNA glycosylase in the extreme thermophile Archaeoglobus fulgidus. J. Biol. Chem. 2000;275:19146–19149. doi: 10.1074/jbc.M001995200. [DOI] [PubMed] [Google Scholar]
- 30.Anderson CT, Friedberg EC. The presence of nuclear and mitochondrial uracil-DNA glycosylase in extracts of human KB cells. Nucleic Acids Res. 1980;8:875–888. [PMC free article] [PubMed] [Google Scholar]
- 31. Teo,Y.N. (2010) Biomolecular detection with combinatorial oligodeoxyfluorosides. Ph.D. Thesis, Stanford University.
- 32.Shigdel UK, He C. A new 1′-methylenedisulfide deoxyribose that forms an efficient cross-link to DNA cytosine-5 methyltransferase (DNMT) J. Am. Chem. Soc. 2008;130:17634–17635. doi: 10.1021/ja8064304. [DOI] [PubMed] [Google Scholar]
- 33.Ward DC, Reich E, Stryer L. Fluorescence studies of nucleotides and polynucleotides. J. Biol. Chem. 1969;244:1228–1237. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.